Abstract
In this brief report, we provide an analysis of the influence of a novel CYP2C haplotype (CYP2C:TG) on proton pump inhibitor (PPI) pharmacokinetics (PK) in children. The CYP2C:TG haplotype has been proposed to be associated with increased CYP2C19 activity. We sought to determine if this CYP2C:TG haplotype resulted in similar alterations in metabolism for proton pump inhibitors, which are primarily metabolized by CYP2C19. In a cohort of 41 children aged 6–21 participating in a PPI pharmacokinetic study, effects of the CYP2C:TG allele were assessed by fitting two linear regression models for each of the six PK outcomes assessed, the second of which accounted for the presence of the CYP2C:TG allele. The difference in R 2 values between the two models was computed to quantify the variability in the outcome that could be accounted for by the CYP2C:TG allele after adjustment for the CYP2C19 genotype. We found the CYP2C:TG haplotype to have no measurable additive impact on CYP2C19‐mediated metabolism of PPIs in vivo in older children and adolescents. The findings of this study do not support the clinical utility of routine testing for the CYP2C:TG haplotype to guide PPI dose adjustments in children.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
A prior study reported a novel CYP2C haplotype (CYP2C:TG) that was functionally similar to that of the increased function CYP2C19*17 allele. The CYP2C:TG haplotype was described to confer increased activity toward CYP2C19 substrates (e.g., escitalopram) similar to that of CYP2C19*17.
WHAT QUESTION DID THIS STUDY ADDRESS?
Does the CYP2C:TG haplotype influence the metabolism of frequently prescribed proton pump inhibitors which are also metabolized by CYP2C19?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
The novel CYP2C:TG haplotype does not appear to impact the CYP2C19‐mediated metabolism of proton pump inhibitors in children.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
Current findings, including those of this study, do not support routine pharmacogenetic testing of CYP2C:TG to individualize prescribing for PPIs. Additional studies are warranted, however, to elucidate substrate‐specific impacts of the CYP2C:TG allele on other CYP2C19 substrates and to further explore its clinical utility.
INTRODUCTION
Proton pump inhibitors (PPIs) are frequently prescribed to children, with the prevalence of PPI prescriptions in American children more than doubling in recent decades. 1 As PPI prescribing increases in children, continued expansion of knowledge surrounding pharmacogenetic influences on PPI pharmacokinetics (PK) is imperative to optimize drug safety and efficacy in individual patients. Developmental expression (ontogeny) of CYP2C19, the primary enzyme metabolizing PPIs, genetic variation in the CYP2C19 gene, and body habitus have been found to alter PPI clearance and exposure. 2 , 3 , 4 , 5 For example, the CYP2C19*17/*17 diplotype is associated with an ultrarapid metabolizer (UM) phenotype for PPIs compared to individuals with a CYP2C19*1/*1 normal function (NM) diplotype. 3 Known CYP2C19 allelic variation and its impacts on PPI PK and metabolic phenotype form the basis for the Clinical Pharmacogenetic Implementation Consortium (CPIC) CYP2C19‐based PPI dosing recommendations. 6 However, the source of some PPI PK variability in adults and children remains unclear.
A novel haplotype referred to as CYP2C:TG (defined by rs2860840 T and rs11188059 G, and found only on CYP2C19*1 alleles) has been associated with ultrarapid metabolism for the CYP2C19 substrate escitalopram, with increased activity similar to the CYP2C19*17/*17 diplotype. 7 A clinically relevant increase of activity in adults with the CYP2C:TG haplotype was, however, not observed by a subsequent study using PK data for several CYP2C19 substrates. 8 Further studies are needed to clarify whether the CYP2C:TG haplotype meaningfully impacts the CYP2C19 function of other CYP2C19 substrates including PPIs. Based on prior work from Bråten et al., 7 we aimed to determine if the CYP2C:TG haplotype is associated with an increased likelihood of increased CYP2C19 function for PPIs, compared to individuals without CYP2C:TG. A secondary aim was to investigate whether individuals with the CYP2C:TG allele have activity comparable to CYP2C19*17.
METHODS
Study design and patient population
Children aged 6–21 years were recruited to participate in a PK and pharmacogenetic (PGx) study of PPIs (oral lansoprazole, oral pantoprazole, and intravenous (IV) pantoprazole) at a single quaternary pediatric institution. Participating children were generally healthy and included those with and without obesity; children with impaired liver or kidney function based on relevant lab values or diagnostic codes were excluded. The study was approved for human subjects research by the Institutional Review Board (IRB). Participants were not receiving any of the study drugs as part of their routine medical care and were asked to abstain from taking inhibitors/inducers of CYP2C19 for ≥3 days prior to PK visit (e.g., citalopram, escitalopram, fluoxetine, fluvoxamine, ketoconazole, ticlopidine, felbamate, trazodone, valproic acid, topiramate, phenobarbital, carbamazepine, phenytoin). Study drugs were administered in a fasted state (≥4 h) at one or more visits during the study period of July 2018–December 2022. The collection of serum samples for PGx testing was obtained at the first study visit. A dense sampling strategy was employed for the collection of PK samples (i.e., goal of 12–14 samples collected between the administration and 10 h post‐dose). One study drug was administered at each visit, with some participants returning at least 1 week later for subsequent visits and receiving more than one study drug.
Genotyping
Study participants were genotyped on a QuantStudio 12K flex Real‐Time PCR system for CYP2C19*2, *3, *4, *5, *8, *9, *12, *17, and *35, and two variants in the CYP2C18 gene, rs2860840 (c.*31C>T) and rs11188059 (c.819+2182G>A) using commercially available TaqMan assays (system and assays from Thermo Fisher Scientific, Waltham, MS). All reactions were performed in the 96‐well format as recommended by the manufacturer. For a subset of participants, the CYP2C19 genotype was obtained using ADMEseq, a next‐generation sequencing panel as described previously. 9
Pharmacokinetic analysis
Serum levels of each study drug were measured using validated assays developed in our clinical pharmacology laboratory (Table S1). Lansoprazole and its relevant metabolite levels were measured using LC/MS/MS and Xevo TQ‐XS triple quadrupole mass spectrometer (Waters, Manchester UK). Pantoprazole and its relevant metabolites were measured using reverse‐phase HPLC based on the method published by Xie et al. 10
PPI plasma concentration versus time data were curve fit using a peeling algorithm to generate initial poly‐exponential parameter estimates. Final estimates of the apparent terminal elimination rate constant (k el) were determined from an iterative, linear least squares regression algorithm. A model‐independent approach was used to estimate parameters of interest. Peak plasma concentration (C max) and the time to achieve C max (T max) were obtained by direct examination of the pharmacokinetic profile. Area under the plasma concentration versus time curve during the sampling period (AUClast) was calculated using the mixed log‐linear method where the last refers to the final sampling time with quantifiable PPI plasma concentrations. Extrapolation of the AUC to infinity (AUCtotal) was achieved by the summation of AUClast + C n/k el where C n is the last observable plasma concentration calculated from the curve fit. Clearance and apparent oral clearance were calculated according to dose/AUCtotal and normalized for weight‐adjusted dose where noted. All analyses were conducted using Kinetica version 5.0 (Thermo Electron, Philadelphia, PA, USA).
Statistical analysis
Effects of the CYP2C:TG allele were assessed by fitting two linear regression models for each PK outcome. The first model included a count of CYP2C19*1 alleles and the count of CYP2C19*17 alleles as explanatory variables; these variables capture CYP2C19 genotype because the other alleles present in the sample (*2 or *35) are nonfunctional. The second model also included a count of CYP2C:TG alleles. The difference in R 2 values between the two models was computed to quantify the variability in the outcome that could be accounted for by the CYP2C:TG allele after adjustment for the CYP2C19 genotype. Non‐parametric bootstrapping was used to obtain 95% confidence intervals for the regression coefficients for CYP2C:TG allele count and for the change in model R 2 resulting from the addition of CYP2C:TG allele count to the model. Analyses were carried out in R, with bootstrapping implemented using the boot package. PK outcomes were log‐transformed as needed to reduce skewness.
RESULTS
Subject characteristics, PK outcomes, and baseline laboratory values are summarized in Appendix S1. One highly influential outlier, the sole poor metabolizer with a CYP2C19*2/*2 genotype, was excluded from analyses for six of the PK outcome variables. Among the 45 individuals, 14 had at least one CYP2C:TG allele. As shown in Table 1, the CYP2C:TG allele was present in 41% of subjects genotyped as CYP2C19*1/*1, 36% of *1/*17, and 29% of *1/*2; none of the six samples with other genotypes (including those without any CYP2C19*1 alleles) had CYP2C:TG, which is consistent with previous observations. 8
TABLE 1.
CYP2C19 genotype by CYP2C:TG alleles.
| CYP2C19 genotype | Number of subjects | Predicted phenotype | Subjects with zero CYP2C:TG alleles | Subjects with one CYP2C:TG allele | Subjects with two CYP2C:TG alleles |
|---|---|---|---|---|---|
| *1/*1 | 22 | NM | 13 (59%) | 6 (27%) | 3 (14%) |
| *1/*2 | 7 | IM | 5 (71%) | 2 (29%) | 0 |
| *1/*17 | 11 | RM | 7 (64%) | 4 (36%) | 0 |
| *1/*35 | 2 | IM | 2 (100%) | 0 | 0 |
| *2/*2 | 1 | PM | 1 (100%) | 0 | 0 |
| *2/*17 | 2 | IM | 2 (100%) | 0 | 0 |
| *17/*17 | 1 | UM | 1 (100%) | 0 | 0 |
As shown in Figure 1, the impact of the CYP2C:TG haplotype on the PK of PO pantoprazole, PO lansoprazole, or IV pantoprazole was negligible. Although the CYP2C:TG allele count variable increased R 2 by 0.02–0.03, this was only observed for four PK outcomes (unadjusted C max for oral pantoprazole, unadjusted and adjusted CL for IV pantoprazole, and adjusted C max for lansoprazole) and was not consistent across the investigated PPIs. The magnitudes of the scaled CYP2C:TG coefficient for these outcomes were in the 0.24–0.35 range, but none was estimated with enough precision to rule out an effect of comparable or larger magnitude in the opposite direction as that suggested by the coefficient. Results of the regression models are available in Table S2.
FIGURE 1.
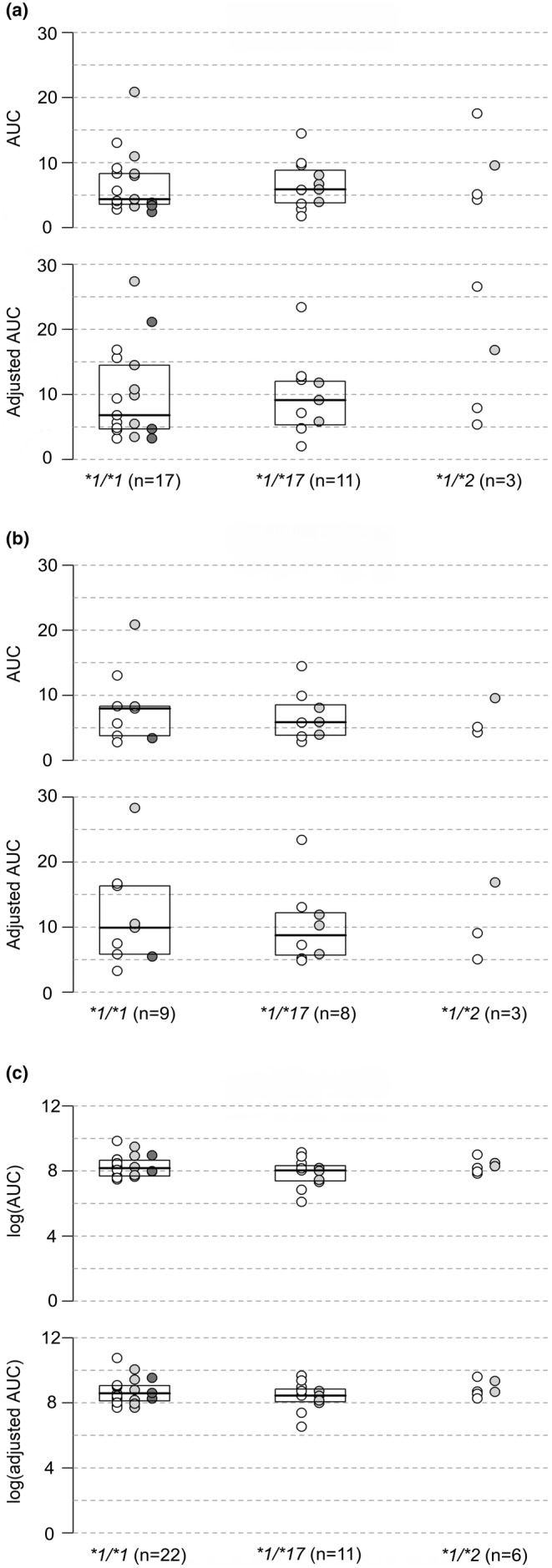
Visual representation of PPI AUC by CYP2C19 genotype. This figure shows a visual representation of PPI Area under the curve (AUC; μg/mL h) and weight‐base dose‐adjusted AUC (μg/mL h per mg/kg) by CYP2C19 genotypes: *1/*1, *1/*17, and *1/*2. White circles represent individuals with 0 CYP2C:TG alleles, light gray circles represent individuals with 1 CYP2C:TG allele, and dark gray circles represent individuals with two CYP2C:TG alleles. Panel (a) shows AUC results for oral pantoprazole, Panel (b) shows results for intravenous pantoprazole, and Panel (c) shows results for oral lansoprazole. There was no significant difference in AUC or adjusted AUC based on the number of CYP2C:TG alleles for any of the included PPIs.
DISCUSSION
This detailed PK analysis of 45 children who received one or more of the three study PPIs (oral pantoprazole, IV pantoprazole, oral lansoprazole) at different study visits found the CYP2C:TG haplotype to have no measurable impact on CYP2C19‐mediated metabolism of PPIs in vivo. Our findings contrast Bråten et al., who reported that the CYP2C:TG haplotype was associated with increased CYP2C19 activity using escitalopram or sertraline serum concentrations as a measurement of CYP2C19 activity 7 , 11 and concluded that activity of CYP2C19*1 alleles with the “TG” haplotype have activity levels comparable to those seen for the increased function CYP2C19*17 allele. The authors thus proposed that testing for the “TG” haplotype will improve CYP2C19 phenotype prediction. Also in contrast to our results is an observational study that aimed to tie CYP2C:TG haplotype (especially homozygotes) to PPI treatment failure in adults with gastroesophageal reflux disease (GERD). 12 Their data are difficult to interpret though as a higher prevalence of the CYP2C:TG haplotype, but not CYP2C19*17, was found in patients with treatment failure, and differences were only found in a subset of confirmed GERD cases and not in their general population of cases.
Other recent studies have investigated associations between the CYP2C:TG haplotype and CYP2C19 substrates PK. Our results are consistent with those reported by Zubiaur et al. in adults who did not find any differences among CYP2C19*1 alleles with and without the CYP2C:TG haplotype using PK data for pantoprazole (n = 60), rabeprazole (n = 35) and omeprazole (n = 31). 8 Zubiaur also investigated the effect of the CYP2C:TG haplotype on various other CYP2C19 substrates, including citalopram, sertraline, and voriconazole. No appreciable impact of CYP2C:TG alleles on CYP2C19 PK or metabolic phenotype were found for any of these drugs either. 8
Our findings may differ from prior work from Bråten et al. for several reasons. Differences may be due to other enzymes contributing to PPIs metabolism, such as CYP3A4, which do not contribute to escitalopram metabolism. Additionally, our PK analysis and outcomes were based on a dense sampling scheme, as opposed to therapeutic drug monitoring (TDM) trough levels. The rich sampling strategy permits more accurate characterization of PK parameters thereby providing a more complete picture of the relationship between PPI exposure and the CYP2C:TG haplotype. The use of a dense sampling strategy allows for the capture of precise peak and trough drug levels and, making estimations of inter‐individual variability and PK parameter determination more accurate than those based on trough levels alone as in TDM. Finally, CYP2C19 ontogeny may play a role in PPI PK in infants, however, our study included primarily adolescents who have been shown to express CYP2C19 at adult levels. 13 It is possible also that the currently unknown ontogeny of the CYP2C:TG haplotype could impact PK in children/adolescents.
Another notable finding of our analysis, as shown in Figure 1, was the lack of difference in PK parameters between CYP2C19*1/*1 and *1/*17 individuals, raising concerns regarding the classification of the CYP2C19*1/*17 diplotype as rapid metabolizers (RM), at least for PPIs. This finding is also consistent with those presented by Zubiaur et al. 8 Since there was only one homozygous CYP2C19*17/*17 participant in our study, we were unable to interrogate whether the CYP2C:TG haplotype is associated with increased activity levels comparable to that of the *17/*17 diplotype for PPIs. This necessitates further exploration in future PPI PK studies.
The findings of this study and those reported by Zubiaur et al. 8 do not support the clinical utility of routine testing for the CYP2C:TG haplotypes to guide PPI dose adjustments. Future work evaluating the potential effects of this novel CYP2C:TG haplotype should incorporate in vivo, robust PK sampling strategies of more diverse CYP2C19 substrates.
This study should be viewed in light of some limitations. First, the smaller number of TG:TG homozygotes, as well as *17/*17 homozygotes, limited our ability to compare PK outcomes directly between these groups which have both been associated with increased CYP2C19 metabolism and UM metabolizer phenotypes in other studies. Additionally, prior studies have shown only small differences in metabolism between the CYP2C19*1 and *17 alleles, which may make it difficult to assess meaningful contributions of the CYP2C:TG haplotype on the activity of CYP2C19*1 alleles in our analysis. 6 , 14 Finally, single‐dose studies are not always representative of real‐world experience, as it cannot account for dose accumulation over time. Therefore, further studies are needed regarding the clinical implications of CYP2C:TG to PPI PK and pharmacodynamics. Among the study's strengths are inclusion of multiple PPIs, robust PK sampling scheme focusing on meaningful PK outcomes including AUC, and a relatively large sample size for a pediatric PK study, though a larger sample size would allow for more precise effect size estimates and stronger evidence regarding the magnitude and direction of any effects.
CONCLUSION
The results of this detailed PK and PGx analysis of children do not support previously described impacts of the novel CYP2C:TG haplotype on CYP2C19‐mediated metabolism of PPIs, and thus, do not support routine pharmacogenetic testing for this new variant. However future studies could focus specifically on the most relevant haplotypes for discerning any differences related to CYP2C:TG and CYP2C19*17.
AUTHOR CONTRIBUTIONS
K.E.K. wrote the manuscript. V.S., V.S.S., S.A.‐R., R.E.P., A.G., and J.S.L. designed the research. V.S. performed the research. All authors analyzed the data. P.T. contributed new reagents/analytical tools.
FUNDING INFORMATION
Research reported in this publication was supported by the National Institute of Diabetes and Digestive and Kidney Diseases K23 DK115827 (Shakhnovich), and Eunice Kennedy Shriver National Institute of Child Health and Human Development T32 HD069038 (Dr. Kyler).
CONFLICT OF INTEREST STATEMENT
The authors declared no competing interests for this work.
Supporting information
Table S1
Table S2
Appendix S1
ACKNOWLEDGMENTS
Erin Boone and Karim Pirani for technical assistance with genotype analyses. Jansynn Radford and Chance S. Friesen for assisting with data collection and laboratory technical assistance. Tao Lin and Kim Gibson for assistance with drug assay development and drug level measurements in the clinical pharmacology lab. Veronica Williams for study enrollment, data collection and organization, and management of IRB and regulatory matters.
Kyler KE, Gaedigk A, Abdel‐Rahman S, et al. Influence of novel CYP2C‐haplotype on proton pump inhibitor pharmacokinetics in children. Clin Transl Sci. 2024;17:e13782. doi: 10.1111/cts.13782
REFERENCES
- 1. Hales CM, Kit BK, Gu Q, Ogden CL. Trends in prescription medication use among children and adolescents—United States, 1999‐2014. JAMA. 2018;319(19):2009‐2020. doi: 10.1001/jama.2018.5690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Litalien C, Théorêt Y, Faure C. Pharmacokinetics of proton pump inhibitors in children. Clin Pharmacokinet. 2005;44(5):441‐466. doi: 10.2165/00003088-200544050-00001 [DOI] [PubMed] [Google Scholar]
- 3. Hagymási K, Müllner K, Herszényi L, Tulassay Z. Update on the pharmacogenomics of proton pump inhibitors. Pharmacogenomics. 2011;12(6):873‐888. doi: 10.2217/pgs.11.4 [DOI] [PubMed] [Google Scholar]
- 4. El Rouby N, Lima JJ, Johnson JA. Proton pump inhibitors: from CYP2C19 pharmacogenetics to precision medicine. Expert Opin Drug Metab Toxicol. 2018;14(4):447‐460. doi: 10.1080/17425255.2018.1461835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shakhnovich V, Abdel‐Rahman S, Friesen CA, et al. Lean body weight dosing avoids excessive systemic exposure to proton pump inhibitors for children with obesity. Pediatr Obes. 2019;14(1):e12459. doi: 10.1111/ijpo.12459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lima JJ, Thomas CD, Barbarino J, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2C19 and proton pump inhibitor dosing. Clin Pharmacol Ther. 2021;109(6):1417‐1423. doi: 10.1002/cpt.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bråten LS, Haslemo T, Jukic MM, et al. A novel CYP2C‐haplotype associated with ultrarapid metabolism of escitalopram. Clin Pharmacol Ther. 2021;110(3):786‐793. doi: 10.1002/cpt.2233 [DOI] [PubMed] [Google Scholar]
- 8. Zubiaur P, Soria‐Chacartegui P, Boone EC, et al. Impact of CYP2C:TG haplotype on CYP2C19 substrates clearance in vivo, protein content, and in vitro activity. Clin Pharmacol Ther. 2023;114:1033‐1042. doi: 10.1002/cpt.3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaedigk A, Boone EC, Scherer SE, et al. CYP2C8, CYP2C9, and CYP2C19 characterization using next‐generation sequencing and haplotype analysis: a GeT‐RM collaborative project. J Mol Diagn. 2022;24(4):337‐350. doi: 10.1016/j.jmoldx.2021.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xie Z, Chen X, Jin F, Zhong D. Simultaneous determination of pantoprazole and its two metabolites in dog plasma by HPLC. J Chromatogr Sci. 2005;43(5):271‐275. doi: 10.1093/chromsci/43.5.271 [DOI] [PubMed] [Google Scholar]
- 11. Bråten LS, Ingelman‐Sundberg M, Jukic MM, Molden E, Kringen MK. Impact of the novel CYP2C:TG haplotype and CYP2B6 variants on sertraline exposure in a large patient population. Clin Transl Sci. 2022;15(9):2135‐2145. doi: 10.1111/cts.13347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kee PS, Maggo SDS, Kennedy MA, et al. Omeprazole treatment failure in gastroesophageal reflux disease and genetic variation at the CYP2C locus. Front Genet. 2022;13:869160. doi: 10.3389/fgene.2022.869160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hines RN. Ontogeny of human hepatic cytochromes P450. J Biochem Mol Toxicol. 2007;21(4):169‐175. doi: 10.1002/jbt.20179 [DOI] [PubMed] [Google Scholar]
- 14. Shakhnovich V, Smith PB, Guptill JT, et al. Obese children require lower doses of pantoprazole than nonobese peers to achieve equal systemic drug exposures. J Pediatr. 2018;193:102‐108.e1. doi: 10.1016/j.jpeds.2017.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Appendix S1


