ABSTRACT
Marine oxygen-deficient zones (ODZs) are portions of the ocean where intense nitrogen loss occurs primarily via denitrification and anammox. Despite many decades of study, the identity of the microbes that catalyze nitrogen loss in ODZs is still being elucidated. Intriguingly, high transcription of genes in the same family as the nitric oxide dismutase (nod) gene from Methylomirabilota has been reported in the anoxic core of ODZs. Here, we show that the most abundantly transcribed nod genes in the Eastern Tropical North Pacific ODZ belong to a new order (UBA11136) of Alphaproteobacteria, rather than Methylomirabilota as previously assumed. Gammaproteobacteria and Planctomycetia also transcribe nod, but at lower relative abundance than UBA11136 in the upper ODZ. The nod-transcribing Alphaproteobacteria likely use formaldehyde and formate as a source of electrons for aerobic respiration, with additional electrons possibly from sulfide oxidation. They also transcribe multiheme cytochrome (here named ptd) genes for a putative porin-cytochrome protein complex of unknown function, potentially involved in extracellular electron transfer. Molecular oxygen for aerobic respiration may originate from nitric oxide dismutation via cryptic oxygen cycling. Our results implicate Alphaproteobacteria order UBA11136 as a significant player in marine nitrogen loss and highlight their potential in one-carbon, nitrogen, and sulfur metabolism in ODZs.
IMPORTANCE
In marine oxygen-deficient zones (ODZs), microbes transform bioavailable nitrogen to gaseous nitrogen, with nitric oxide as a key intermediate. The Eastern Tropical North Pacific contains the world’s largest ODZ, but the identity of the microbes transforming nitric oxide remains unknown. Here, we show that highly transcribed nitric oxide dismutase (nod) genes belong to Alphaproteobacteria of the novel order UBA11136, which lacks cultivated isolates. These Alphaproteobacteria show evidence for aerobic respiration, using oxygen potentially sourced from nitric oxide dismutase, and possess a novel porin-cytochrome protein complex with unknown function. Gammaproteobacteria and Planctomycetia transcribe nod at lower levels. Our results pinpoint the microbes mediating a key step in marine nitrogen loss and reveal an unexpected predicted metabolism for marine Alphaproteobacteria.
KEYWORDS: nitric oxide, Alphaproteobacteria, marine, oxygen-deficient zone, nitrogen, oxygen, denitrification
INTRODUCTION
Marine oxygen-deficient zones (ODZs) contribute up to half of the ocean’s nitrogen loss (1) and are a major source of marine emissions of the potent greenhouse gas nitrous oxide (N2O) (2). The primary source of the N2O at the oxic–anoxic interface and in anoxic waters in ODZs is denitrification (3, 4). The microbial enzyme responsible for N2O production during denitrification is nitric oxide reductase (Nor), which uses electrons from cytochrome c (cNor) or quinol (qNor), to reduce nitric oxide (NO) to N2O (5–7). In the qNor family, there are bona fide qNor enzymes and NO dismutase (Nod). Nod proteins lack the quinol-binding site, seemingly preventing the enzyme from taking up external electrons; instead, Nod is theorized to disproportionate NO into dinitrogen and O2 in methane-oxidizing Methylomirabilota bacteria (8, 9) and in the alkane-oxidizing Gammaproteobacterium HdN1 (10).
The Eastern Tropical North and South Pacific (ETNP and ETSP) ODZs are the world’s largest and second largest ODZs, and the subjects of extensive microbial ecology studies. Abundant NO reductase-like genes and transcripts in the ETNP and ETSP ODZ cluster in the same enzyme subfamily as Nod (11–14). Due to the similarity of ODZ Nod proteins to those of Methylomirabilota (NC10), it was initially presumed that ODZ bacteria also used Nod proteins to disproportionate NO into N2 and O2 for use in intra-aerobic methane oxidation (11, 13, 15). However, Fuchsman et al. (12) found that the peak of nod gene abundance in the ETNP ODZ correlates with a peak of modeled N2O production (4) and does not correlate with abundance of methane monooxygenase genes, suggesting that Nod proteins in the ETNP ODZ are potentially an important source of N2O and are unlikely to be involved in methane oxidation. The plausibility that Nod proteins can reduce NO to N2O is supported by a study of a novel eukaryotic denitrification pathway in foraminifera (Globobulimina spp.) that produce N2O while expressing Nod (16). Yet, the phylogenetic identity and metabolic context of marine Nod proteins, which are a key biological source of either N2O or O2+N2 in marine ODZs, remain unresolved.
In this study, we sought to determine the identity, predicted metabolism, and environmental niche of the ODZ organism responsible for the highly transcribed nod genes first discovered by Padilla et al. (11). We found that the most abundantly transcribed nod genes in the ETNP ODZ belong to Alphaproteobacteria in the novel order UBA11136. Significant transcription of nod genes was limited to waters with <1 µM O2. These nod-transcribing Alphaproteobacteria also transcribe genes involved in aerobic respiration, which was unexpected given that they inhabit anoxic waters, as well as genes involved in oxidation of formaldehyde, likely indicating methylotrophy. Genes encoding multiheme cytochrome proteins potentially implicated in nitrogen or iron cycling were also transcribed.
RESULTS
Transcribed nod sequences in the ETNP ODZ belong to Alphaproteobacteria, Gammaproteobacteria, and Planctomycetia
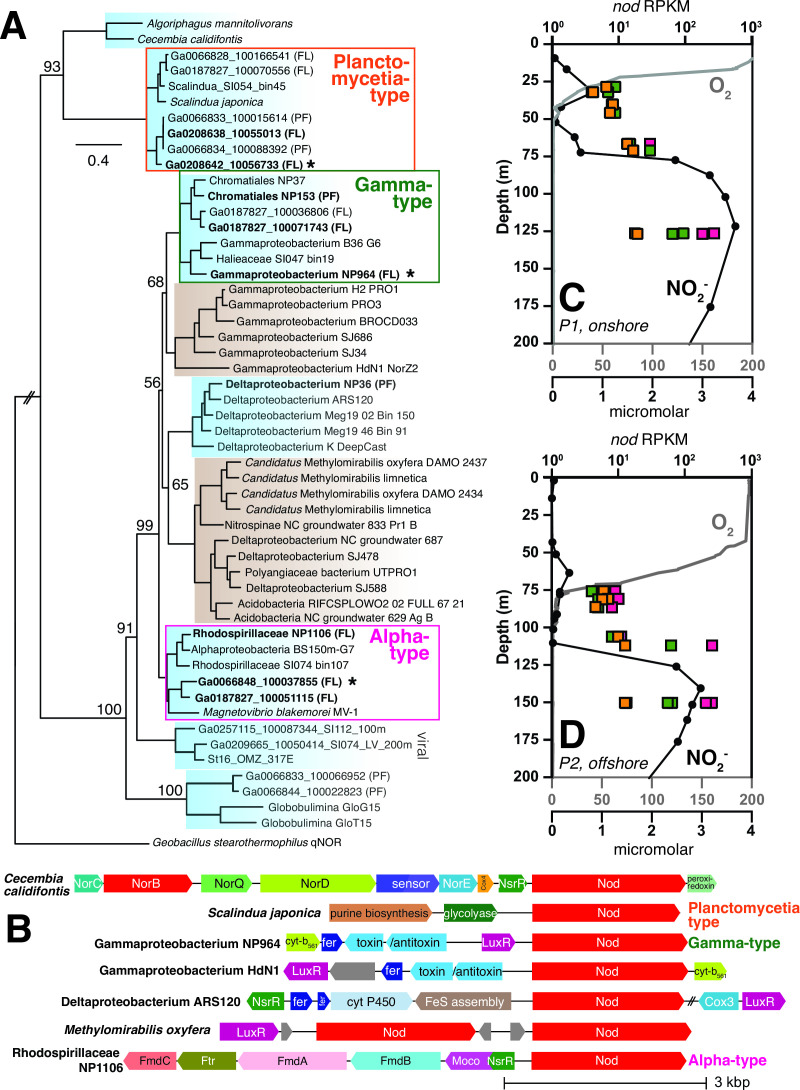
Our reanalysis of highly transcribed nod genes in the ETNP ODZ (11) shows that these genes belong to Alphaproteobacteria rather than a member of Methylomirabilota as previously assumed. Querying the Nod amino acid sequences from Padilla et al. (11) against ETNP ODZ metagenomes in the JGI IMG/MER database returned multiple 100% identity matches, including a nod gene (Ga0066848_100037855) on a scaffold with hypothetical genes with 100% identity to Alphaproteobacteria metagenome-assembled genomes (MAGs) from the ETNP ODZ (17) (Table S1). We binned previously sequenced ETNP ODZ metagenomes Ga0066848 (ETNP201310SV72) and Ga0066829 (ETNP201306SV43) (18) into MAGs. Contigs with the most highly transcribed nod genes were present in two Alphaproteobacteria MAGs (GTDB taxonomy: UBA11136 sp002686135; species representative: Rhodospirillaceae bacterium isolate ARS27) with 97% average nucleotide identity. Querying the Nod amino acid sequences from Padilla et al. (11) against NCBI’s nonredundant protein database returned matches to other MAGs assigned to Alphaproteobacteria order UBA11136 from low-oxygen marine settings (ETNP, Saanich Inlet, and the Black Sea; 78%–80% identity), the marine magnetotactic alphaproteobacterium Magnetovibrio blakemorei MV-1 (75% identity), Gammaproteobacterium HdN1 (66% identity), and Methylomirabilis spp. (66% identity; Table S2).
To glean additional insights into evolutionary relationships, we updated a previous Nod phylogeny (19) with additional amino acid sequences from marine MAGs (20–22) and ETNP ODZ metagenomes (18), subdivided into cells that are free-living (FL; 0.2–1.6 μm) and from the particle fraction (PF; >1.6 μm; Fig. 1A; Table S3). The Nod topology was generally consistent with a previous phylogeny from Fuchsman et al. (12), with additional taxonomic data from MAGs in the Tara Oceans data set further constraining Nod placement (22). As expected based on the binning and BLAST results, the Nod sequence from Padilla et al. (11) (Ga0066848_100037855) clustered phylogenetically with marine Alphaproteobacteria (OTU III in Fuchsman et al. [12], hereafter “Alpha-type Nod”); this clade contained three unique sequences, all of which were present in multiple metagenomes and all from the free-living fraction, and one of which was identical to that of Rhodospirillaceae NP1106 (GenBank: MBV28360). Four unique ODZ Nod sequences clustered with marine Gammaproteobacteria (OTU II in Fuchsman et al. [12], hereafter “Gamma-type Nod”); these sequences were monophyletic with a cluster of Gammaproteobacteria Nod cluster sequences from sewage sludge, including Gammaproteobacterium HdN1 (23) and other wastewater Gammaproteobacteria. Multiple ETNP ODZ metagenomes contained Gamma-type Nod sequences identical to those of Gammaproteobacteria NP964 (GenBank: MBP20251). Gamma-type Nod had ~70% identity to Alpha-type Nod. Several ODZ Nod sequences, all from the particle fraction, clustered with marine Deltaproteobacteria in a clade of monophyletic nod genes from groundwater Methylomirabilota, Deltaproteobacteria, and Acidobacteria MAGs (~65% identity to Alpha-type Nod). Six unique Nod ODZ protein sequences (two of which were present in multiple metagenomes) clustered with Planctomycetia (OTU I in Fuchsman et al. [12], hereafter “Planctomycetia-type Nod”), were primarily found in free-living cells, and had ~40% identity to Alpha-type Nod. Intriguingly, two ODZ sequences clustered in the eukaryotic Globobulimina clade (~50% identity to Alpha-type Nod). Viral Nod sequences from Saanich Inlet (~55% identity to Alpha-type Nod) clustered with the viral Nod sequence previously reported by Gazitúa et al. (24) from the ETSP ODZ (St16 OMZ 317E-viral).
Fig 1.
Marine Nod clades, gene neighborhoods, and depth profiles of transcription. (A) A maximum likelihood phylogeny of nitric oxide dismutase (Nod) amino acid sequences in marine (blue) and select terrestrial (brown) taxa, primarily from marine MAGs (20–22) and ETNP ODZ metagenomes (18). Branch support was evaluated using 1,000 rapid bootstrap replicates, with bootstrap values shown for deep branches. The tree is drawn to scale, with branch lengths in number of substitutions per site. Bold sequences represent those present in multiple ETNP ODZ metagenomes (see Table S3 for duplicate accession numbers). “PF” indicates genes from the particle fraction (>1.6 μm fraction) of filters. “FL” indicates genes from the free-living fraction (0.2–1.6 μm) collected on Sterivex filters. The most highly transcribed ETNP ODZ sequence is indicated with an asterisk. The qNor sequence Geobacillus stearothermophilus was used as the outgroup. (B) Gene neighborhoods surrounding nod genes in select taxa. GenBank contigs: Cecembia calidifontis SGXG01000001, Scalindua japonica BAOS01000045, Gammaproteobacteria NP964 PBRC01000062, Gammaproteobacterium HdN1 FP929140, Deltaproteobacteria NZCL01000067, Candidatus Methylomirabilis oxyfera FP565575, and Rhodospirillaceae NP1106 PCBZ01000014. Unlabeled gray genes are hypothetical. (C, D) Oxygen concentrations (gray lines), nitrite concentrations (black circles), and nod transcripts (squares, as reads per kilobase per million mapped reads [RPKM]) with depth in ETNP ODZ P1 (onshore) and P2 (offshore) sites (25).
We investigated gene neighborhoods surrounding ODZ nod genes in the three main phylogenetic clusters of ODZ sequences: Planctomycetia-type Nod, Gamma-type Nod, and Alpha-type Nod. Although “unknown Nor-related” marine Bacteroidota sequences were located on an operon with other nor genes, there was no consistent gene neighborhood for nod sequences (Fig. 1B). Planctomycetia-type nod genes were not located in the vicinity of any genes with recognizable related function. Gamma-type nod gene neighborhoods contained ferredoxins and cytochrome b561 genes for electron transport. Upstream of the Alpha-type nod in Rhodospirillaceae NP1106 is a cluster of formylmethanofuran dehydrogenase genes (fmd/fwd) used in C1 metabolism via tetrahydromethanopterin/methanofuran-linked reactions. Immediately upstream or downstream of nod genes, helix–turn–helix transcriptional regulators were common (Fig. 1B). Neighboring Gamma-type and Methylomirabilis nod genes, LuxR-type regulators were common; these regulators have diverse functions and their potential connection to Nod remains unclear. Neighboring Alpha-type and Bacteroidota (e.g., Cecembia calidifontis) nod genes, Rrf2-type regulators were present. The protein NsrR in the Rrf2 family regulates global cellular response to NO toxification by directly sensing NO with an iron-sulfur cluster (26, 27). The presence of this NsrR-like regulator suggests that Nod in marine Alphaproteobacteria and Bacteroidota may be involved in nitrosative stress response and NO detoxification.
Alphaproteobacterial nod is highly transcribed in anoxic waters
We assessed transcription of Alpha-, Gamma-, and Planctomycetia-type nod genes from the oxycline to upper ODZ (secondary nitrite maximum) using ETNP ODZ metatranscriptomes from an onshore station with a shallower oxycline (P1; Fig. 1C) and an offshore station with a deeper oxycline (P2; Fig. 1D) (25). In both oxyclines, transcription was low (4–10 reads per kilobase per million mapped reads [RPKM], n = 8) for all three nod types (Fig. 1C and D). Below the oxyclines, nod transcripts began to rise and were highest at the secondary nitrite maxima, with Alpha-type (184–274 RPKM, n = 4) > Gamma-type (55–95 RPKM, n = 4) > Planctomycetia-type (13–19 RPKM, n = 4; Table S4).
MAGs with highly transcribed nod gene represent a new order of Alphaproteobacteria
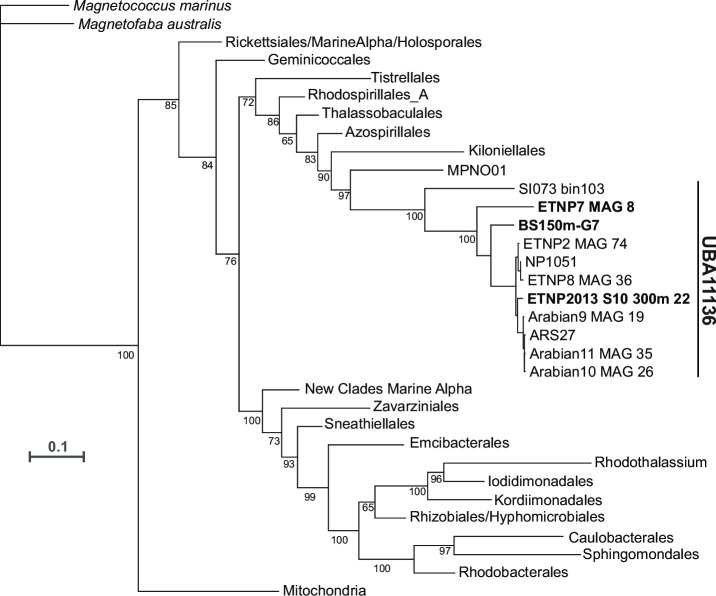
In order to assess the phylogeny of the nod-containing Alphaproteobacteria MAGs, we constructed an alphaproteobacterial phylogeny using the conserved protein NADH ubiquinone oxidoreductase subunit L (NuoL) as in Cevallos and Degli Esposti (28), with additional representation of order UBA11136 including our MAG ETNP2013_S10_300m_22 (Fig. 2). MAG ETNP2013_S06_300m_15 was not included in the phylogeny because its nuoL gene was truncated. The phylogeny confirmed that nod-containing Alphaproteobacteria belong to the order UBA11136 and showed that UBA11136 is situated near other Alphaproteobacteria orders found in ODZs.
Fig 2.
Alphaproteobacteria phylogeny with order UBA11136 expanded and nod-containing MAGs bolded. The phylogeny was constructed using the alphaproteobacterial phylogenetic marker NADH ubiquinone oxidoreductase subunit L as in Cevallos and Degli Esposti (28). Taxonomic names are from Cevallos and Degli Esposti (28) and GTDB Release 08-RS214. The scale bar represents amino acid substitutions per site. The full phylogeny is shown in Fig. S1.
Alphaproteobacteria transcribe genes for formate metabolism, aerobic respiration, and a multiheme cytochrome complex
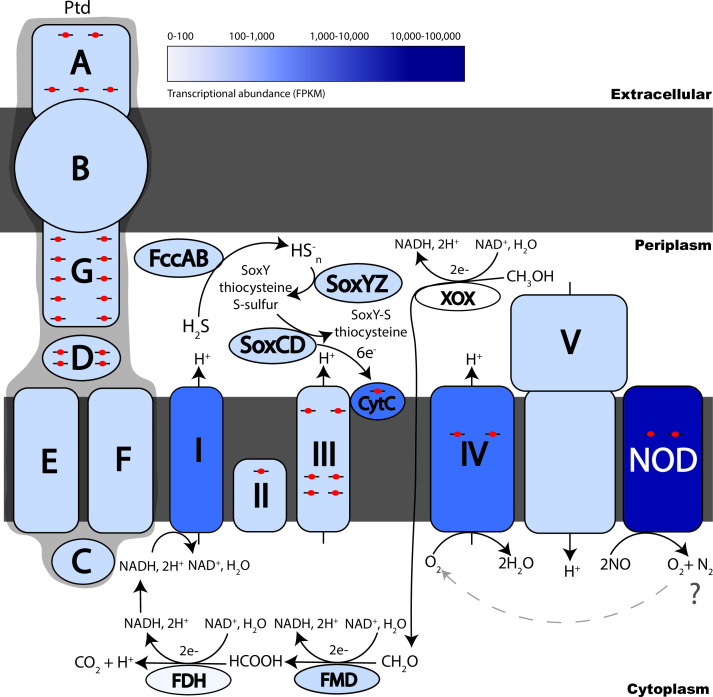
To glean insight into potential roles for Nod in a cellular context, we sought to reconstruct the electron transport chain of the Alphaproteobacteria with the most highly transcribed nod genes (Alphaproteobacterium MAG ETNP2013_S10_300m_22 and Alphaproteobacterium MAG ETNP2013_S06_300m_15, 73% and 69% estimated completeness, respectively) at the secondary nitrite maximum. Of total metagenomic reads, 0.38% map to ETNP2013_S10_300m_22 and 0.39% map to ETNP2013_S06_300m_15. In both MAGs, nod was in the top three most transcribed genes in the ETNP ODZ (~44,000 FPKM; Table S5), after a bacterial nucleoid DNA-binding protein and a potassium-gated channel protein. In addition to nod, we found that genes for formaldehyde oxidation via tetrahydromethanopterin/methanofuran-linked reactions, including formylmethanofuran dehydrogenase (fwd/fmd) and formylmethanofuran–tetrahydromethanopterin N-formyltransferase (ftr), were transcribed in both MAGs (Table S5). Both MAGs also transcribed NAD-dependent formate dehydrogenase (Table S5). Thus, the alphaproteobacterium appears to be capable of conversion of formaldehyde to formate and use of formate as a source of electrons for NADH:ubiquinone oxidoreductase (Complex I; Fig. 3). The source of formaldehyde is likely methanol oxidation, as pyrroloquinoline quinone (PQQ)-dependent ethanol/methanol dehydrogenases were found in Alphaproteobacteria MAGs from low-oxygen marine settings (Table S6). Methane monooxygenase genes were not found in the partial Alphaproteobacteria MAGs, precluding our ability to rule out the possibility of these genes in the missing portions of the genomes. The Alphaproteobacteria PQQ-dependent dehydrogenase genes contained the motif DYDG (Table S6), which is characteristic of the lanthanide-containing form of the enzymes rather than the calcium form (29).
Fig 3.
Schematic of the electron transport chain in nod-containing Alphaproteobacteria. Increasing transcriptional activity is indicated from lighter to darker blue (Table S5). Red circles with black lines indicate hemes. Hypothetical Ptd proteins are labeled A, B, C, D, E, F, and G (Table S8). Proposed electron transfer from formate to Complex I is shown. Highly transcribed Nod protein and predicted O2 generation is shown as feeding into A1-type CCO Complex IV. Additional electrons for CytC and the electron transport chain are proposed to come from sulfur oxidation carried out by the flavocytochrome c sulfide dehydrogenase (FccAB, FCC), and sulfane-sulfur dehydrogenase (SoxCD) with the multi-enzyme carrier complex (SoxYZ).
A full aerobic electron transport chain (Complex I, II, III, and IV) and F0F1-type ATP synthase were transcribed in both bins (Fig. 3; Table S5). Complex IV (cytochrome c oxidase) was type A1 according to the Sousa et al. (30) classification, and the cox operon in the GTDB species representative Rhodospirallaceae ARS27 was subtype b (COX2-COX1-CtaB-CtaG_Cox11-COX3-DUF983-SURF1-CtaA1-M32-Tsy-M16B) according to the Geiger et al. (31) classification. Sulfur oxidation genes, including flavocytochrome c sulfide dehydrogenase (FccAB), sulfane hydrogenase (SoxCD), and carrier protein SoxYZ, were also transcribed, as were numerous transposes (Fig. 3; Table S5).
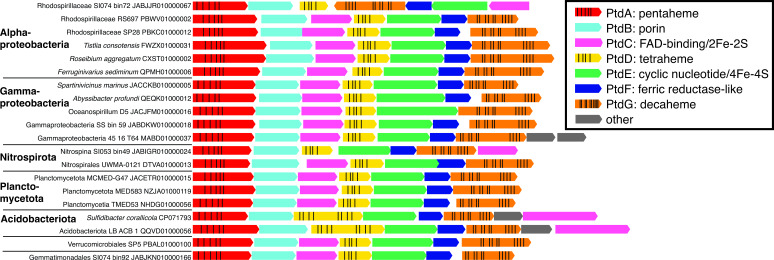
Genes for a multiheme cytochrome complex were transcribed in both bins. To our knowledge, this putative operon has not been previously described. Hereafter, we designate it the ptdABCDEFG operon for its sequence of penta/tetra/deca-heme proteins, interspersed with other conserved proteins. ptdAB genes are highly transcribed in our Alphaproteobacteria MAGs, but it is unclear if the rest of the operon is also highly transcribed, because it was truncated in our MAGs’ scaffolds. The ptd gene cluster consists of a penta-heme protein with a C-terminal beta-sandwich (PtdA), a porin (PtdB), a FAD/NAD(P)-binding oxidoreductase (PtdC), a periplasmic tetra-heme protein (PtdD), a cyclic nucleotide-binding domain protein with two 4Fe–4S clusters (PtdE), a cytoplasmic transmembrane ferric reductase-like protein (PtdF), and a periplasmic deca-heme protein (PtdG; Fig. 3; Table S7 and S8). The function of this complex is unknown, but the presence of genes encoding a porin and multiple multiheme proteins resembles porin-cytochrome protein complexes involved in extracellular reduction electron transfer during Fe(III) and Mn(IV) reduction (32, 33). PtdA has a homolog to a penta-heme cytochrome c552 protein of unknown function in a thermophilic purple sulfur gammaproteobacterium (34) and is in the same COG family (COG3303) as formate-dependent nitrite reductase, NrfA. ptdABCDEFG genes were prevalent in Alphaproteobacteria, Gammaproteobacteria, Nitrospirales, and Planctomycetes MAGs from marine or high salinity environments (Fig. 4; Table S7).
Fig 4.
Gene neighborhoods of pentaheme–tetraheme–decaheme genes from select organisms. Depicted heme spacing is approximate. All organisms are from saline environments (seawater, marine sediment, or saline spring).
DISCUSSION
This study predicts the previously ambiguous identity of the microorganisms that make the dominant nitric oxide-transforming protein (Nod) in the world’s largest ODZ, the Eastern Tropical North Pacific. Extensive horizontal gene transfer of nod genes between microbial genomes is evident from the lack of conservation of gene neighborhood and patchy phylogeny (12), which may be mediated by viral infection (24). We found that the most transcriptionally active nod genes in the ETNP upper ODZ belong to the novel Alphaproteobacteria order UBA11136. Alpha-type nod transcript abundances (~200 RPKM) are similar to those of dissimilatory nitrate reductase (narG) in the ODZ (35). The nod-transcribing Alphaproteobacteria are also transcribing genes for formaldehyde oxidation, likely as a source of electrons to the respiratory chain via NAD reduction by formate dehydrogenase. Sulfide may be used as a supplemental electron donor and/or may be concomitantly oxidized for detoxification (36, 37).
Our discovery of a putative porin–cytochrome complex (ptd operon) in marine bacteria was unexpected. Porin–cytochrome complexes have been best studied for their role in extracellular electron transport, particularly for respiratory metal reduction and oxidation (32, 33). It is conceivable that the Ptd complex is involved in iron reduction in ODZs; there is iron reduction at the secondary nitrite maximum and it is hypothesized to be bacterially mediated, but the microbes involved have yet to be determined (38, 39). Alternatively, the presence of ptdABCDEFG genes in numerous nitrite-oxidizing bacteria (Nitrospirales) could imply the involvement of these genes in nitrogen cycling; PtdA was in the same COG family as formate-dependent nitrite reductase (40), and PtdC is similar to a flavohemoprotein with predicted nitric oxide dioxygenase activity, also annotated as hydroxylamine oxidoreductase-linked cytochrome. The function of PtdABCDEFG remains completely unknown and requires future biochemical characterization.
On the other end of the electron transport chain, high transcription of a heme/copper terminal oxidase suggests that O2 is being used as the terminal electron acceptor in nod-transcribing Alphaproteobacteria MAGs. The transcribed heme/copper oxidase is A1-type (low O2 affinity), also present in mitochondria, and adapted for high O2 concentrations. Low O2 affinity A1-type heme/copper oxidases are transcribed in other anoxic environments (41). Because ODZs have extremely low concentrations of O2 below the oxycline, O2 for aerobic respiration may be generated in situ and rapidly consumed. Given that the function of Nod is proposed to be dismutation of two NO molecules into N2 and O2 (8), it is possible that the O2 source for aerobic respiration in the UBA11136 MAGs is NO dismutation, although other sources of O2 (e.g., in situ photosynthesis, mixing) in anoxic waters are also conceivable (42). The physiological uses of Gamma-type and Planctomycetia-type Nod may be different from Alpha-type Nod, although this remains to be investigated.
The source of NO, the presumed substrate for Nod, may be generated in the same organism using Nod or generated by a different organism (or chemical pathway). Nitric oxide was positively correlated with nitrite in the ETSP ODZ and was only detectable when O2 was <1–2 µM (43). In the ETNP ODZ, NO concentration and turnover rates were elevated at O2 < 100 µM (44). Both studies suggest that the NO in ODZs likely originates from nitrification or nitrifier denitrification, while genomic analyses indicate that the copper-containing nitrite reductase (nirK) in SAR11 bacteria (presumably performing denitrification) may be a key source of NO (12). Because most ODZ denitrifiers specialize in only one of the three steps (NO2− reduction, NO reduction, and N2O reduction) (45) and known nitrite reductases were not identified in our MAGs, existing data indicate that the NO that is used as a substrate for alphaproteobacterial Nod is not generated in vivo. (Only 4 out of 32 nod-containing MAGs contained a nitrite reductase gene: two Gammaproteobacteria MAGs contained nirK, one Myxococcota MAG contained nirS, and one Scalindua MAG contained nirS). It is also possible that another uncharacterized enzyme produces NO.
This study suggests that marine Alphaproteobacteria from order UBA11136 are actively reducing NO under anoxia, as implied by their abundant transcription of nod genes. Although there is strong evidence that the substrate for Nod in ODZs is NO based on its abundance, the products of this enzyme (N2O vs N2+O2) remain uncertain. Nod is theorized to disproportionate NO into N2 and O2 in methane-oxidizing Methylomirabilota bacteria (8, 9), but no biochemical characterizations of Nod have been published to date, and foraminifera expressing Nod produce N2O (16). The apparent lack of other denitrification genes in nod-transcribing Alphaproteobacteria is consistent with the observation that denitrification in ODZs is largely divided into distinct microbial taxa (12, 13, 45). For example, although nitrate reductase (narG) genes are widely distributed amongst ODZ microbes (45), SAR11 bacteria appear to dominate in narG transcriptional activity (35). Our finding that the transcription of nod is catalyzed primarily by marine Alphaproteobacteria implies that this taxon contributes significantly to marine nitrogen loss.
MATERIALS AND METHODS
Nod phylogeny and gene neighborhood
Amino acid sequences of highly transcribed nod genes “ETNP 2014 Stn10 150m” and “ETNP 2013 Stn6 300m” were acquired from the authors of Padilla et al. (11) (see Table S2 for sequences). These sequences were used for BLASTP searches of ODZ metagenomes in the JGI IMG/MER database and the NCBI nonredundant protein (nr) database. Sequences (n = 53, 731 gap-free sites) were aligned using the MAFFT online server with the L-INS-i method (46). A phylogeny was generated with 1,000 bootstraps using model LG+I+G4 with W-IQ-Tree (47). The phylogeny was visualized using FigTree v.1.4.4, and the fasta file (Nod_alignment) is available as a supplemental data set. Gene neighborhoods were generated using the EFI Gene Neighborhood Tool (48) with single sequence BLAST of the UniProt database using the amino acid sequence Ga0066848_100037855 (JGI IMG/MER) as the Nod query with an e-value cutoff of 10−5 and with 10 genes upstream and downstream the gene of interest.
Transcription of nod genes in ETNP ODZ depth profiles
Magic Basic Local Alignment Search Tool (49) was used to search ETNP ODZ metatranscriptomes (PRJNA727903; Mattes et al. [25]) using representative nucleotide sequences for Planctomycetia-like (Ga0066826_100064333 [JGI IMG/MER]), Gamma-like (PBRC01000062.1:19833–22205 [NCBI]), and Alpha-like (Ga0066848_100037855 [JGI IMG/MER]) nod genes. Default parameters were used except for the score threshold (18). Read hits were normalized to reads per kilobase million (RPKM).
Metagenomic binning
Binning of metagenome-assembled genomes (MAGs) was performed using the KBase platform (50). ETNP ODZ metagenomes were collected in 2013 and sequenced by Joint Genome Institute (JGI) using an Illumina HiSeq 2500 as described in Ruiz-Perez et al. (18). Assemblies for the ETNP ODZ metagenomes (18) containing Alpha-type nod genes (ETNP201310SV72 [GOLD Analysis Project ID Ga0066848; stn10 300m] and ETNP201306SV43 [GOLD Analysis Project ID Ga0066829; stn6 300m]) were imported from JGI IMG/MER into KBase. Metagenomic assemblies were binned into MAGs using MaxBin2 v2.2.4 (51). The two MAGs containing nod genes (MAG ETNP2013_S10_300m_22 from ETNP201310SV72, and ETNP2013_S06_300m_15 from ETNP201306SV43) were selected for further analysis. Average nucleotide identity was calculated using FastANI (52). MAG taxonomy and genome quality were evaluated by GTDB-Tk v2.3.2 (53). MAGs were annotated with RASTtk v1.073 (54). Metagenomic reads were mapped to MAGs using Bowtie2 (55).
Alphaproteobacterial NuoL phylogeny
Alphaproteobacterial NADH ubiquinone oxidoreductase subunit L (NuoL) and mitochondrial ND5 marker proteins (n = 320) were aligned as in Cevallos and Degli Esposti (28), with additional representation of order UBA11136. A maximum likelihood phylogeny with 1000 bootstraps was constructed in IQ-tree (56) using the LG+F model with ultrafast bootstrap (57). Taxonomic names and clades are from Cevallos and Degli Esposti (28) and GTDB Release 08-RS214. The fasta file (NuoL_alignment) is available as a supplemental data set. Alphaproteobacteria MAGs containing nod genes (Table S2) were classified as belonging to order UBA11136 using GTDB-Tk v2.3.2 (53).
Mapping transcripts to metagenomic bins
Metatranscriptomic mapping to MAGs was performed using the KBase platform (50). RNA-seq data (25) were imported from the depth with the highest nod transcription, the secondary nitrite maximum (126 m, NCBI run SRR14460584), and aligned to MAGs using the Bowtie2 (55) app in KBase. The Cufflinks v.2.2.1 (58) app in KBase was then used to assemble the aligned RNA-seq data into a set of transcripts and to calculate the relative abundances of the transcripts expressed in fragments per kilobase per million fragments mapped (FPKM).
Cellular localization and heme numbers
Cellular locations of Ptd proteins were predicted using PSORTb v.3.0.3 analysis (59). Numbers of heme-binding motifs per protein were identified by counting CXXCH sequences. Ptd gene neighborhoods were generated using the EFI Gene Neighborhood Tool (48) with single sequence BLAST of the UniProt database using the amino acid sequence Ga0066848_100031354 (JGI IMG/MER) as the PtdA query with an e-value cutoff of 10−5 and with 10 genes upstream and downstream the gene of interest.
ACKNOWLEDGMENTS
We thank Laura Bristow for helpful discussions. We thank Mauro Degli Esposti for assistance with the NADH ubiquinone oxidoreductase subunit L phylogeny. We thank Cory Padilla, Anthony Bertagnolli, and Neha Sarode for sharing previous data.
We acknowledge funding from an NSF Graduate Research Fellowship to C.E.E., the Simons Foundation, and NSF Awards 2022991 and 2054927 to F.J.S.
Contributor Information
Jennifer B. Glass, Email: jennifer.glass@eas.gatech.edu.
Jennifer F. Biddle, University of Delaware, Newark, Delaware, USA
DATA AVAILABILITY
The KBase bioinformatic pipeline and MAGs are at https://narrative.kbase.us/narrative/106999. Original metagenomic reads are available at BioProject PRJNA375524 (ETNP201306SV43, SAMN06344130) and BioProject PRJNA375542 (ETNP201310SV72, SAMN06344148). MAG Alphaproteobacteria bacterium ETNP2013_S06_300m_15 was deposited into BioProject PRJNA375524 (BioSample SAMN38229257, WGS Accession JAZDBU000000000) and Alphaproteobacteria bacterium ETNP2013_S10_300m_22 was deposited into BioProject PRJNA375542 (BioSample SAMN38228782, WGS Accession JAZDCE000000000). All ETNP ODZ data sets used in this manuscript are listed in Table S9.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.02099-23.
Figure S1.
NuoL amino acid alignment FASTA file.
Nod amino acid alignment FASTA file.
Table S1-S9.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. DeVries T, Deutsch C, Rafter PA, Primeau F. 2013. Marine denitrification rates determined from a global 3-D inverse model. Biogeosciences 10:2481–2496. doi: 10.5194/bg-10-2481-2013 [DOI] [Google Scholar]
- 2. Yang S, Chang BX, Warner MJ, Weber TS, Bourbonnais AM, Santoro AE, Kock A, Sonnerup RE, Bullister JL, Wilson ST, Bianchi D. 2020. Global reconstruction reduces the uncertainty of oceanic nitrous oxide emissions and reveals a vigorous seasonal cycle. Proc Natl Acad Sci U S A 117:11954–11960. doi: 10.1073/pnas.1921914117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Frey C, Bange HW, Achterberg EP, Jayakumar A, Löscher CR, Arévalo-Martínez DL, León-Palmero E, Sun M, Sun X, Xie RC, Oleynik S, Ward BB. 2020. Regulation of nitrous oxide production in low-oxygen waters off the coast of Peru. Biogeosciences 17:2263–2287. doi: 10.5194/bg-17-2263-2020 [DOI] [Google Scholar]
- 4. Babbin AR, Bianchi D, Jayakumar A, Ward BB. 2015. Rapid nitrous oxide cycling in the suboxic ocean. Science 348:1127–1129. doi: 10.1126/science.aaa8380 [DOI] [PubMed] [Google Scholar]
- 5. Wasser IM, de Vries S, Moënne-Loccoz P, Schröder I, Karlin KD. 2002. Nitric oxide in biological denitrification: Fe/Cu metalloenzyme and metal complex NOx redox chemistry. Chem Rev 102:1201–1234. doi: 10.1021/cr0006627 [DOI] [PubMed] [Google Scholar]
- 6. Kraft B, Strous M, Tegetmeyer HE. 2011. Microbial nitrate respiration–genes, enzymes and environmental distribution. J Biotechnol 155:104–117. doi: 10.1016/j.jbiotec.2010.12.025 [DOI] [PubMed] [Google Scholar]
- 7. Zumft WG. 2005. Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme–copper oxidase type. J Inorg Biochem 99:194–215. doi: 10.1016/j.jinorgbio.2004.09.024 [DOI] [PubMed] [Google Scholar]
- 8. Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJCT, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJM, Janssen-Megens EM, Francoijs K-J, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883 [DOI] [PubMed] [Google Scholar]
- 9. Ettwig KF, Speth DR, Reimann J, Wu ML, Jetten MSM, Keltjens JT. 2012. Bacterial oxygen production in the dark. Front Microbiol 3:273. doi: 10.3389/fmicb.2012.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zedelius J, Rabus R, Grundmann O, Werner I, Brodkorb D, Schreiber F, Ehrenreich P, Behrends A, Wilkes H, Kube M, Reinhardt R, Widdel F. 2011. Alkane degradation under anoxic conditions by a nitrate‐reducing bacterium with possible involvement of the electron acceptor in substrate activation. Environ Microbiol Rep 3:125–135. doi: 10.1111/j.1758-2229.2010.00198.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Padilla CC, Bristow LA, Sarode N, Garcia-Robledo E, Gómez Ramírez E, Benson CR, Bourbonnais A, Altabet MA, Girguis PR, Thamdrup B, Stewart FJ. 2016. NC10 bacteria in marine oxygen minimum zones. ISME J 10:2067–2071. doi: 10.1038/ismej.2015.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fuchsman CA, Devol AH, Saunders JK, McKay C, Rocap G. 2017. Niche partitioning of the N cycling microbial community of an offshore oxygen deficient zone. Front Microbiol 8:2384. doi: 10.3389/fmicb.2017.02384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dalsgaard T, Stewart FJ, Thamdrup B, De Brabandere L, Revsbech NP, Ulloa O, Canfield DE, DeLong EF. 2014. Oxygen at nanomolar levels reversibly suppresses process rates and gene expression in anammox and denitrification in the oxygen minimum zone off northern Chile. mBio 5:e01966. doi: 10.1128/mBio.01966-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ganesh S, Parris DJ, DeLong EF, Stewart FJ. 2014. Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J 8:187–211. doi: 10.1038/ismej.2013.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thamdrup B, Steinsdóttir HGR, Bertagnolli AD, Padilla CC, Patin NV, Garcia‐Robledo E, Bristow LA, Stewart FJ. 2019. Anaerobic methane oxidation is an important sink for methane in the ocean’s largest oxygen minimum zone. Limn & Ocean 64:2569–2585. doi: 10.1002/lno.11235 [DOI] [Google Scholar]
- 16. Woehle C, Roy A-S, Glock N, Wein T, Weissenbach J, Rosenstiel P, Hiebenthal C, Michels J, Schönfeld J, Dagan T. 2018. A novel eukaryotic denitrification pathway in foraminifera. Curr Biol 28:2536–2543. doi: 10.1016/j.cub.2018.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uzun M, Alekseeva L, Krutkina M, Koziaeva V, Grouzdev D. 2020. Unravelling the diversity of magnetotactic bacteria through analysis of open genomic databases. Sci Data 7:252. doi: 10.1038/s41597-020-00593-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruiz-Perez CA, Bertagnolli AD, Tsementzi D, Woyke T, Stewart FJ, Konstantinidis KT. 2021. Description of Candidatus mesopelagibacter carboxydoxydans and Candidatus anoxipelagibacter denitrificans: nitrate-reducing SAR11 genera that dominate mesopelagic and anoxic marine zones. Syst Appl Microbiol 44:126185. doi: 10.1016/j.syapm.2021.126185 [DOI] [PubMed] [Google Scholar]
- 19. Hu Q-Q, Zhou Z-C, Liu Y-F, Zhou L, Mbadinga SM, Liu J-F, Yang S-Z, Gu J-D, Mu B-Z. 2019. High microbial diversity of the nitric oxide dismutation reaction revealed by PCR amplification and analysis of the nod gene. Inter Biod & Biod 143:104708. doi: 10.1016/j.ibiod.2019.05.025 [DOI] [Google Scholar]
- 20. Lin H, Ascher DB, Myung Y, Lamborg CH, Hallam SJ, Gionfriddo CM, Holt KE, Moreau JW. 2021. Mercury methylation by metabolically versatile and cosmopolitan marine bacteria. ISME J 15:1810–1825. doi: 10.1038/s41396-020-00889-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cabello-Yeves PJ, Callieri C, Picazo A, Mehrshad M, Haro-Moreno JM, Roda-Garcia JJ, Dzhembekova N, Slabakova V, Slabakova N, Moncheva S, Rodriguez-Valera F. 2021. The microbiome of the black sea water column analyzed by shotgun and genome centric metagenomics. Environ Microbiome 16:5. doi: 10.1186/s40793-021-00374-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tully BJ, Graham ED, Heidelberg JF. 2018. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci Data 5:170203. doi: 10.1038/sdata.2017.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ehrenreich P, Behrends A, Harder J, Widdel F. 2000. Anaerobic oxidation of alkanes by newly isolated denitrifying bacteria. Arch Microbiol 173:58–64. doi: 10.1007/s002030050008 [DOI] [PubMed] [Google Scholar]
- 24. Gazitúa MC, Vik DR, Roux S, Gregory AC, Bolduc B, Widner B, Mulholland MR, Hallam SJ, Ulloa O, Sullivan MB. 2021. Potential virus-mediated nitrogen cycling in oxygen-depleted oceanic waters. ISME J 15:981–998. doi: 10.1038/s41396-020-00825-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mattes TE, Burke S, Rocap G, Morris RM. 2022. Two metatranscriptomic profiles through low-dissolved-oxygen waters (DO, 0 to 33 μM) in the eastern tropical North Pacific ocean. Microbiol Resour Announc 11:e0120121. doi: 10.1128/mra.01201-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tucker NP, Le Brun NE, Dixon R, Hutchings MI. 2010. There’s NO stopping NsrR, a global regulator of the bacterial NO stress response. Trends Microbiol 18:149–156. doi: 10.1016/j.tim.2009.12.009 [DOI] [PubMed] [Google Scholar]
- 27. Bodenmiller DM, Spiro S. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J Bacteriol 188:874–881. doi: 10.1128/JB.188.3.874-881.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cevallos MA, Degli Esposti M. 2022. New Alphaproteobacteria thrive in the depths of the ocean with oxygen gradient. Microorganisms 10:455. doi: 10.3390/microorganisms10020455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keltjens JT, Pol A, Reimann J, Op den Camp HJM. 2014. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol 98:6163–6183. doi: 10.1007/s00253-014-5766-8 [DOI] [PubMed] [Google Scholar]
- 30. Sousa FL, Alves RJ, Ribeiro MA, Pereira-Leal JB, Teixeira M, Pereira MM. 2012. The superfamily of heme–copper oxygen reductases: types and evolutionary considerations. Bio et Biophysica Acta (BBA) - Bio 1817:629–637. doi: 10.1016/j.bbabio.2011.09.020 [DOI] [PubMed] [Google Scholar]
- 31. Geiger O, Sanchez-Flores A, Padilla-Gomez J, Degli Esposti M. 2023. Multiple approaches of cellular metabolism define the bacterial ancestry of mitochondria. Sci Adv 9:eadh0066. doi: 10.1126/sciadv.adh0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richardson DJ, Butt JN, Fredrickson JK, Zachara JM, Shi L, Edwards MJ, White G, Baiden N, Gates AJ, Marritt SJ, Clarke TA. 2012. The 'porin-cytochrome' model for microbe-to-mineral electron transfer. Mol Microbiol 85:201–212. doi: 10.1111/j.1365-2958.2012.08088.x [DOI] [PubMed] [Google Scholar]
- 33. Shi L, Fredrickson JK, Zachara JM. 2014. Genomic analyses of bacterial porin-cytochrome gene clusters. Front Microbiol 5:657. doi: 10.3389/fmicb.2014.00657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen JH, Yu LJ, Boussac A, Wang-Otomo ZY, Kuang T, Shen JR. 2019. Properties and structure of a low-potential, penta-heme cytochrome C(552) from a thermophilic purple sulfur photosynthetic bacterium Thermochromatium tepidum. Photosynth Res 139:281–293. doi: 10.1007/s11120-018-0507-y [DOI] [PubMed] [Google Scholar]
- 35. Tsementzi D, Wu J, Deutsch S, Nath S, Rodriguez-R LM, Burns AS, Ranjan P, Sarode N, Malmstrom RR, Padilla CC, Stone BK, Bristow LA, Larsen M, Glass JB, Thamdrup B, Woyke T, Konstantinidis KT, Stewart FJ. 2016. SAR11 bacteria linked to ocean anoxia and nitrogen loss. Nature 536:179–183. doi: 10.1038/nature19068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Callbeck CM, Canfield DE, Kuypers MMM, Yilmaz P, Lavik G, Thamdrup B, Schubert CJ, Bristow LA. 2021. Sulfur cycling in oceanic oxygen minimum zones. Limn & Ocean 66:2360–2392. doi: 10.1002/lno.11759 [DOI] [Google Scholar]
- 37. Schmitz RA, Peeters SH, Mohammadi SS, Berben T, van Erven T, Iosif CA, van Alen T, Versantvoort W, Jetten MSM, Op den Camp HJM, Pol A. 2023. Simultaneous sulfide and methane oxidation by an extremophile. Nat Commun 14:2974. doi: 10.1038/s41467-023-38699-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moffett JW, Goepfert TJ, Naqvi SWA. 2007. Reduced iron associated with secondary nitrite maxima in the Arabian sea. Deep Sea Res Part I Oceanogr Res Pap 54:1341–1349. doi: 10.1016/j.dsr.2007.04.004 [DOI] [Google Scholar]
- 39. Glass JB, Kretz CB, Ganesh S, Ranjan P, Seston SL, Buck KN, Landing WM, Morton PL, Moffett JW, Giovannoni SJ, Vergin KL, Stewart FJ. 2015. Meta-omic signatures of microbial metal and nitrogen cycling in marine oxygen minimum zones. Front Microbiol 6:998. doi: 10.3389/fmicb.2015.00998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simon J, Gross R, Einsle O, Kroneck PM, Kröger A, Klimmek O. 2000. A NapC/NirT-type cytochrome c (NrfH) is the mediator between the quinone pool and the cytochrome c nitrite reductase of Wolinella succinogenes. Mol Microbiol 35:686–696. doi: 10.1046/j.1365-2958.2000.01742.x [DOI] [PubMed] [Google Scholar]
- 41. Berg JS, Ahmerkamp S, Pjevac P, Hausmann B, Milucka J, Kuypers MMM. 2022. How low can they go? Aerobic respiration by microorganisms under apparent anoxia. FEMS Microbiol Rev 46:1–14. doi: 10.1093/femsre/fuac006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Garcia-Robledo E, Padilla CC, Aldunate M, Stewart FJ, Ulloa O, Paulmier A, Gregori G, Revsbech NP. 2017. Cryptic oxygen cycling in anoxic marine zones. Proc Natl Acad Sci U S A 114:8319–8324. doi: 10.1073/pnas.1619844114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lutterbeck HE, Arévalo-Martínez DL, Löscher CR, Bange HW. 2018. Nitric oxide (NO) in the oxygen minimum zone off Peru. Deep Sea Res Part II Top Stud Oceanogr 156:148–154. doi: 10.1016/j.dsr2.2017.12.023 [DOI] [Google Scholar]
- 44. Ward BB, Zafiriou OC. 1988. Nitrification and nitric oxide in the oxygen minimum of the eastern tropical North Pacific. Deep Sea Res Part A Oceanogr Res Pap 35:1127–1142. doi: 10.1016/0198-0149(88)90005-2 [DOI] [Google Scholar]
- 45. Zhang IH, Sun X, Jayakumar A, Fortin SG, Ward BB, Babbin AR. 2023. Partitioning of the denitrification pathway and other nitrite metabolisms within global oxygen deficient zones. ISME Commun 3:76. doi: 10.1038/s43705-023-00284-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. doi: 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235. doi: 10.1093/nar/gkw256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zallot R, Oberg N, Gerlt JA. 2019. The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 58:4169–4182. doi: 10.1021/acs.biochem.9b00735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boratyn GM, Thierry-Mieg J, Thierry-Mieg D, Busby B, Madden TL. 2019. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinformatics 20:405. doi: 10.1186/s12859-019-2996-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Arkin AP, Cottingham RW, Henry CS, Harris NL, Stevens RL, Maslov S, Dehal P, Ware D, Perez F, Canon S, et al. 2018. KBase: the United States department of energy systems biology knowledgebase. Nat Biotechnol 36:566–569. doi: 10.1038/nbt.4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu YW, Simmons BA, Singer SW. 2016. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32:605–607. doi: 10.1093/bioinformatics/btv638 [DOI] [PubMed] [Google Scholar]
- 52. Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. 2022. GTDB-Tk V2: memory friendly classification with the genome taxonomy database. Bioinformatics 38:5315–5316. doi: 10.1093/bioinformatics/btac672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA III, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. doi: 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi: 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
NuoL amino acid alignment FASTA file.
Nod amino acid alignment FASTA file.
Table S1-S9.
Data Availability Statement
The KBase bioinformatic pipeline and MAGs are at https://narrative.kbase.us/narrative/106999. Original metagenomic reads are available at BioProject PRJNA375524 (ETNP201306SV43, SAMN06344130) and BioProject PRJNA375542 (ETNP201310SV72, SAMN06344148). MAG Alphaproteobacteria bacterium ETNP2013_S06_300m_15 was deposited into BioProject PRJNA375524 (BioSample SAMN38229257, WGS Accession JAZDBU000000000) and Alphaproteobacteria bacterium ETNP2013_S10_300m_22 was deposited into BioProject PRJNA375542 (BioSample SAMN38228782, WGS Accession JAZDCE000000000). All ETNP ODZ data sets used in this manuscript are listed in Table S9.