Abstract
Litter decomposition is a key ecosystem process, relevant for the release and storage of nutrients and carbon in soil. Soil fungi are one of the dominant drivers of organic matter decomposition, but fungal taxa differ substantially in their functional ability to decompose plant litter. Knowledge is mostly based on observational data and subsequent molecular analyses and in vitro studies have been limited to forest ecosystems. In order to better understand functional traits of saprotrophic soil fungi in grassland ecosystems, we isolated 31 fungi from a natural grassland and performed several in vitro studies testing for i) leaf and wood litter decomposition, ii) the ability to use carbon sources of differing complexity, iii) the enzyme repertoire. Decomposition strongly varied among phyla and isolates, with Ascomycota decomposing the most and Mucoromycota decomposing the least. The phylogeny of the fungi and their ability to use complex carbon were the most important predictors for decomposition. Our findings show that it is crucial to understand the role of individual members and functional groups within the microbial community. This is an important way forward to understand the role of microbial community composition for the prediction of litter decomposition and subsequent potential carbon storage in grassland soils.
Keywords: litter decomposition, saprotrophic fungi, grassland, fungal traits, fungal phylogeny, carbon use
The ability to use complex carbon, fungal phylogeny and the fungal enzyme repertoire can be used as predictors for litter decomposition in grassland soils.
Introduction
The decomposition of litter in soil is a key ecosystem process, relevant for the supply of nutrients to plants and for soil carbon storage. Grasslands are of crucial importance for soil carbon sequestration as they store about two thirds of terrestrial carbon (Bai and Cotrufo 2022). One of the main drivers of organic matter decomposition are soil saprobic fungi (Treseder and Lennon 2015). However, our knowledge on fungal litter decomposition is mainly based on observational field data, studies on fungi living aboveground, e.g. saprotrophs growing on senescent leaf litter, and sequencing approaches (Hudson 1968, Frankland 1998, Bradford et al. 2014, Johnston et al. 2018, Osono 2020, Lodato et al. 2021), thus lacking information on the functional capabilities of individual, soil inhabiting taxa. There are a number of studies with a focus on fungal litter decomposition by individual fungal isolates, but these studies are mostly limited to forest ecosystems (e.g. Hiscox et al. 2017, Lustenhouwer et al. 2020, Osono 2020). Another important parameter to assess the role of fungi in organic matter turnover is their ability to use different single carbon sources. However, the research regarding this question is also limited to fungi originating from forests or aquatic ecosystems (Sati and Bisht 2006, Hanson et al. 2008, Algora Gallardo et al. 2021). Therefore, we currently lack detailed information on functional traits of individual, soil inhabiting fungal taxa of grassland ecosystems and the links between these traits.
Many of the early studies on litter decomposition have revealed how fungi tackle the degradation of organic matter based on successional community dynamics. Also, fungi were grouped according to their abilities to break down simple or complex organic matter, i.e. Mucoromycota were identified as ‘sugar fungi’ with a clear preference for readily available substrates (Hudson 1968). Cellulose degraders were found in both Ascomycota and Basidiomycota, with Ascomycota more frequently being found to break down cellulose (Hudson 1968), but Agaricomycetes (Basidiomycota) have an especially pronounced ability in this context (Treseder and Lennon 2015). Basidiomycetes are traditionally seen as wood decomposers (Boddy and Watkinson 1995), while the actual ability to break down the complex compound lignin is most frequently found in the class of the Agaricomycetes (Treseder and Lennon 2015), but can also be found throughout Asco- and Basidiomycota.
The use of different carbon sources is linked to the ability to produce extracellular enzymes (hereafter referred as enzymes). Although a lot is known about the relationship between enzyme activities and litter decomposition in situ (e.g. Sinsabaugh 1994, Sinsabaugh et al. 1994, 2016, Allison and Vitousek 2004, Kang and Freeman 2009), we only have limited data on this relationship for individual fungal isolates (e.g. Lustenhouwer et al. 2020). Additionally, most studies use either single or combined substrates and fungal species and sometimes measure only a few individual enzymes. However, litter decomposition is a complex process driven by a suite of different enzymes (Sinsabaugh et al. 1994, Moore et al. 2020, Zheng et al. 2020).
The functional ability to decompose litter further depends on the litter chemistry, such as the litter quality. Litter quality is usually subclassified in high-quality litter, which is easily degradable, and low-quality litter, which is difficult to degrade and more persistent (Castellano et al. 2015). Another important aspect of litter chemistry is the amount of water-soluble carbon in the litter (Allison and Vitousek 2004) and the identity and availability of particular carbon sources, which can influence the relative abundance of individual species and thus fungal community composition (Hanson et al. 2008).
Litter decomposition further depends on the identity of the plant, the plant tissue (e.g. leaves or twigs), the number and the diversity of litter types present and also the nutrient status of litter and soil (Gartner and Cardon 2004, Yin and Koide 2019, Grossman et al. 2020, Osono 2020, Porre et al. 2020). For example, Yin and Koide (2019) describe that the microbial community requires specific nutrients for the decomposition of more complex litter. Fungal decomposers have different strategies to deal with the challenges of varying nutrient and carbon (C) supply (Frankland 1998). Such strategies include changing growth and respiration (C allocation), metabolism (e.g. enzyme excretion) and cell stoichiometry (Camenzind et al. 2021, Manzoni et al. 2021).
There is large variability in the ecosystem process of litter decomposition. The functional ability of individual fungal taxa can differ substantially and thus potentially contributes to this variability (Allison 2012, Crowther et al. 2014). Trait-based approaches have shown important functional differences among fungal lineages and have linked them to traits such as competitiveness, enzymatic capability, stress tolerance or growth rates (Lustenhouwer et al. 2020). Due to these functional differences among fungal taxa, the composition of microbial communities will be important to assess and forecast soil organic matter dynamics. Currently, only microbial biomass or fungal: bacterial ratios are incorporated into carbon cycling models. However, for the development of microbial-explicit models in grasslands it will be important to understand the role of individual members and functional groups within fungal communities in this ecosystem as well, in order to adequately predict litter decomposition and subsequent carbon release and storage (Hicks et al. 2022, Wan and Crowther 2022).
Thus, with this study, we aimed to improve our understanding of functional differences among saprotrophic soil fungi originating from a grassland ecosystem, and their ability to decompose substrates differing in complexity. Our set of fungi includes 31 isolates of the phyla Ascomycota and Basidiomycota—which are generally well known to be able to process various organic compounds, including complex substrates—and Mucoromycota. For this last group there has been some debate in the past, and a large study using fungi from forest ecosystems found that the ability of Mucoromycota to process complex litter is low compared with Asco- and Basidiomycota (Osono 2020). However, a recent study could show that there are some families within the Mucoromycota that also can process complex organic matter (Pawłowska et al. 2019).
We know from previous studies with our set of saprotrophic fungi that they have varying enzymatic capabilities, e.g. the 7 isolates of Mucoromycota are unable to produce laccase (Zheng et al. 2020). The 31 isolates also have varying aggregate formation capabilities, a key ecosystem process that is linked to many other processes, including stabilization and storage of organic matter in soil (Lehmann et al. 2020). Moreover, these fungi have flexible stoichiometric reactions to differing nutrient supply, suggesting they can adapt to changing resource supply (Camenzind et al. 2021).
With this set of fungi, we performed three laboratory experiments to analyze i) the ability of fungal isolates to decompose leaf and wood litter, ii) their usage of different carbon substrates varying in complexity and iii) enzymatic capabilities. We additionally relate our experimental data to a dataset of existing chemical and biotic traits of the same set of fungi. We hypothesized that (i) there will be important differences in litter decomposition among isolates, even within phyla that are known for being typical cellulose degraders, i.e. Ascomycota and Agaricomycetes, or wood decomposers, i.e. Basidiomycota. (ii) Mucoromycota will show low decomposition rates of woody litter, since they are known as ‘sugar fungi’, also reflected by a preference for simple C substrates. And (iii) we expected to see correlations of enzymatic traits and the ability to use complex C sources with fungal litter decomposition rates.
Material and methods
Fungal isolates
We used 31 fungal isolates previously characterized in detail (Andrade-Linares et al. 2016, Camenzind et al. 2022). Saprotrophic fungi were isolated from a grassland soil (Mallnow Lebus, Brandenburg, Germany, 52°27.778′N, 14°29.349′E) and cover the phyla Mucoromycota, Basidiomycota and Ascomycota (see Supporting Information, Table S1). Briefly, soil was washed and diluted in order to reduce spore abundance and to increase the quantity of living fungal hyphae attached to soil particles (Gams and Domsch 1967, Thorn et al. 1996). Subsequently, soil suspensions were spread on different media with the addition of antibiotics to prevent bacterial growth. Isolates were grown on potato-dextrose-agar (PDA) at room temperature thereafter (22°C).
Experiment I: In vitro experiment on litter decomposition
We performed an in vitro study with soil and 31 saprotrophic fungal isolates decomposing two different types of litter (N=10 for each isolate), with 20 control plates (no fungus) per litter type, resulting in 660 experimental units. The stock of fungal cultures was grown on PDA and kept at 4°C. For the experiment, we used Ø 6 cm petri dishes with water agar, on top of which we added an inoculum plug (0.5 cm diameter) of the stock culture, and placed a mesh bag (2×2 cm, 38 µm pore size (Sefar NITEX, Switzerland)) containing litter next to the inoculum. For this experiment we chose two litter types of plants that are common at the field site: a low-quality litter: linden wood (Tilia cordata) and a high-quality litter: leaves of a grass (Arrhenatherum elatius). For leaf litter we collected whole plants at the field site in summer when leaves were green. After collection, plants were dried at room temperature in order to mimic senescence, and finally, leaves were cut to 1 cm pieces. Wood stems were purchased as uniform stems 4×3×100 mm (Meyer & Weigand GmbH, Germany) and cut to small pieces of 5–10 mm. 35 mg of dried litter were added to the mesh bag, the bags closed with a stainless steel clip and autoclaved twice within 48 h. After the addition of the mesh bag to the plate, we finally added 10 grams of sterilized soil (autoclaved twice at 121°C for 20 min with a time delay of 24 h between events; see Fig. S2 for a picture of the stepwise filling of the plates). The soil originated from the same field where the fungi were isolated. Soil properties were 0.11% N, 1.45% C, 19 mg kg−1 available P (Mehlich III extraction) and pH 6.79. The soil was wetted to 60% field capacity. The plates were incubated in the dark for 10 weeks at room temperature. We know from other experiments with these fungi that respiration, and thus microbial activity, starts to decline after 4 weeks under similar experimental conditions (data in preparation for publication). Considering that our wood litter pieces are smaller (4×3×5–10 mm) compared to other studies’ litter sizes (usually 10–20×5 mm) and comparing experiment durations (e.g. Lustenhouwer et al. 2020, Osono 2020), we estimated that 10 weeks were sufficient for the revelation of differences in wood degradation. Upon harvest the soil and litter were dried and the litter mass loss was determined gravimetrically. We refer to litter decomposition as effect size calculated as
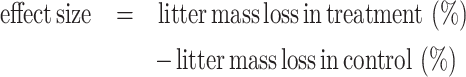 |
Hyphae were extracted from 4.0 g of soil (Jakobsen et al. 1992) and hyphal length (m g−1 soil) of all present hyphae in soil was measured according to Rillig et al. (1999). We corrected the results for background concentration of hyphae from the control plates without fungi.
Experiment II: In vitro experiment on carbon substrate use
In a second experiment we tested the ability of the same set of fungi to utilize different carbon sources for growth (N=3 for each isolate; 30 fungal isolates (RLCS10 failed to grow on the medium type that was used); 6 different C sources incl. control without C source; yielding 540 experimental units). For this experiment, fungal isolates were grown on phytagel based media in Ø 6 cm petri dishes, overlain with a 1 µm mesh to be able to extract fungal biomass at the end of experiments (see Supporting Information S4 for methodological details). Growth media were designed following principles according to Liebig's Law of the Minimum (Camenzind et al. 2020). All elements were provided in sufficient amounts to ensure C to be the only limiting element in order to observe effects exerted by C substrate manipulation. Element supply rates were determined based on the direct analysis of respective element contents in fungal tissues, avoiding osmotic stress by high salt additions (for details see Table S3). We prepared six different media using phytagel as gelling agent (2 g L−1) and final amounts of carbon compounds as follows: control (no C source), glucose 5 g L−1, cellobiose 4.75 g L−1, xylan (a hemicellulose) 4.4 g L−1, cellulose 4.5 g L−1, and litter 4.42 g L−1. Litter was assembled from a mixture of plant litter collected at the site of fungal isolation that was dried at 40 °C and ground to a powder. C additions were standardized by molar amounts, resulting in comparable concentrations of C for each treatment.
We used the biomass gain of fungal isolates on each C substrate minus the biomass gain on control treatments (no C source added) as a response variable, and standardized these values based on maximum biomass growth in order to make these values comparable among isolates. The use of a carbon source j for a fungal isolate i was calculated as follows, based on average values of all three replicates:
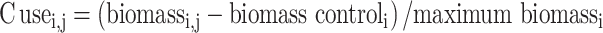 |
where maximum biomass is the largest biomass the isolate had on any of the five C sources.
For this experiment we also calculated an indicator for the ability of fungal isolates to grow on more complex C compounds, hereafter referred to as “complex carbon use ability” (CCUA), which we defined as the weighted average of values according to the complexity of C compounds used:
 |
This indicator serves as an index and has no quantitative meaning, since the numbers indicating complexity are chosen arbitrarily.
Experiment III: In vitro experiment on extracellular enzyme activity
We determined the activity of a number of hydrolytic enzymes using the laboratory kit API ZYMTM (BioMerieux) for semi-quantitative analyses of selected enzymes produced by microorganisms. We used this assay, because it covers a large range of fungal enzymes and the targeted enzyme activities provide insights into functional traits that are not only related to carbon use and fungal growth, but also to fungal competition. Among the enzymes included in this set are acid phosphatase, alkaline phosphatase and phosphohydrolase, which can be related to P mineralization, and leucine arylamidase, valine arylamidase, trypsin, α-chymotrypsin, and N-acetyl-β-glucosaminidase, which are important for fungal N turnover in soil. In addition, the carbon-targeting enzymes esterase, lipase, α-galactosidase, α-glucosidase, α-mannosidase, β-galactosidase, β-glucoronidase and β-glucosidase were included in the assay. Fungi were grown on PDA and fungal material was collected from the margin of an actively growing colony, the agar was removed and the fungal biomass homogenized with 2 ml of NaCl solution (0.85%). Following the manufacturer's instructions, 65 µl of the resulting suspension were added to each cupule of the kit and incubated at 37°C for 4.5 h. After the incubation one drop of ZYM A (25 g Tris hydroxymethyl-aminomethane, 11 ml 37% HCl, 10 g sodium lauryl sulfate, 100 ml H2O) and ZYM B (0.12 g Fast Blue BB, 50 ml methanol, 50 ml dimethyl sulfoxide) reagent were dispensed into each of the cupules. The resulting color reactions were read after 5 min and the color intensity compared to the color code provided by the manufacturer. Results were documented in steps as numbers from 0–5 as the amount of substrate hydrolyzed: 0 (0 nmol), 1 (5 nmol), 2 (10 nmol), 3 (20 nmol), 4 (30 nmol), and 5 (≥40 nmol). The intensity of enzyme activity (nmol) was taken as response variable. This response variable is semi-quantitative, as the addition of fungal material was standardized by size only and the quantitative assessment of fungal biomass in the cupules was difficult. We therefore show relative enzyme activities in order to visualize the enzymatic capabilities and calculated an index for the joint effect of enzymes. This calculated index was used for correlations with other traits (see ‘Statistical analyses’ for more details). For the controls, we repeated the above described procedure, but used plates with PDA medium only, free of fungal material.
Trait collection
To complete our trait database, we selected further traits from previously published data using the same set of 31 fungi (Table 1; Lehmann et al. 2020, Zheng et al. 2020). We selected traits that likely influence litter decomposition or can be linked with decomposition, either through biotic properties of the fungus (e.g. colony extension rate) or through chemical properties (e.g. enzyme activities).
Table 1.
Traits used from the trait database
| Trait # | Fungal trait | Description | Relevance |
|---|---|---|---|
| 1 | Aggregate formation | Ability to form new soil aggregates (Lehmann et al. 2020) | Contribution to the stabilization of OM in the soil (Lehmann et al. 2020) |
| 2 | Hydrophobicity | Hydrophobicity of fungal surface (Alcohol droplets test) (Lehmann et al. 2020) | Potential contribution to the stabilization of soil aggregates (Lehmann et al. 2020) |
| 3 | Biomass density | Biomass density on potato dextrose agar (µg mm−2) (Lehmann et al. 2020) | Fungal abundance has been used as proxy for OM turnover (McGuire and Treseder 2010) |
| 4 | Colony extension rate | Colony growth rate on potato dextrose agar (µm h−1) (Lehmann et al. 2020) | Fast growing fungi have been shown to be better decomposers (Lustenhouwer et al. 2020) |
| 5 | Competitive ability | Sum of wins against competitors in pairwise interactions (Soliveres et al. 2018) | Dominant fungi have been shown to be better decomposers (Lustenhouwer et al. 2020) |
| 6–10 | Cellobiohydrolase, laccase, leucine aminopeptidase, acid phosphatase | Enzyme activity measured on fungal material growing on potato dextrose agar (U mg−1 biomass) (Zheng et al. 2020) | Enzyme activity is often related with the degradation of litter (Sinsabaugh et al. 1994, Allison and Vitousek 2004, Kang and Freeman 2009) |
Statistical analyses
We used the software R for all statistical analyses (version 4.3.1 R Core Team 2023). As a first step, we visualized the raw mass loss data using swarm plots and an estimation method that generates unpaired mean differences (treatment minus control) with bootstrapping (5000 iterations, package ‘dabestr’; Ho et al. 2019, Ho 2020). This plot allowed us to see the magnitude and precision of the fungal taxa effect on litter decomposition compared to the control. The bootstrap estimates are by default bias- and skewness-corrected and produce 95% confidence intervals (CI), which, if situated above the zero line (line of no effect), can show that the treatment induced a detectable response.
As a second step, we visualized raw decomposition data (treatment—control) for individual isolates, and subsequently analyzed mean decomposition of each isolate (mean of 10 replicates) for correlations using Spearman's rank correlations (Spearman's ρ). We analyzed correlations of decomposition with carbon use (data from experiment II), enzyme activities of experiment III and traits of our trait collection. The enzyme activities of experiment III were analyzed as i) joint effect of all enzymes using the first principal component of a principal component analysis (PCA) applied to the 16 enzyme activities and ii) as joint effect of those enzymes targeting C degradation using the first principal component of a PCA applied to the enzyme activities of esterase, lipase, α-galactosidase, β-galactosidase, β-glucoronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase and α-mannosidase. The enzymes included in the trait collection were also analyzed as joint effect using the first principal component of a PCA applied to the four enzyme activities of that dataset. To summarize this large dataset we created a correlation matrix of all variables using Spearman's correlation type in the packages ‘Hmisc’ and ‘corrplot’ (Harrell 2014, Wei et al. 2021).
We further tested correlations of average decomposition rates of each isolate for phylogenetic signals using phylogenetic generalized linear models (pgls function in the package ‘caper’; Orme 2018). Pgls models are similar to linear regression, where we investigate the relation between a predictor variable and a response variable, with the addition of controlling for phylogenetic signals in the response, i.e. we test for the non-independence of the residuals under the influence of the potential phylogenetic relatedness of the taxa involved (Mundry 2014). We mainly used three output parameters for the interpretation of pgls models: i) λ: If λ = 0 then residuals are independent, i.e. there is no phylogenetic signal in the variation of the data; if λ ≥ 1 the data has more covariation than expected under Brownian motion (Orme 2018, Cooper 2022). We decided to use λ based on Münkemüller et al. (2012) and because it is used in numerous other publications and therefore comparable to those results. ii) Adjusted R-squared and P-values that show whether the model detects a significant relationship between predictor and response after phylogenetic correction. The phylogenetic tree used for this analysis was constructed using the unweighted pair group method with arithmetic mean (upgma(), package ‘phangorn’; Schliep 2010), based on long sequence reads. Taxon names are based on taxonomic classification of ITS1, 5.8S, ITS2 and partial LSU marker according to the UNITE database (Nilsson et al. 2019) and the RDP LSU dataset (Cole et al. 2014). Phylum names are given as in Spatafora et al. (2017); if synonymous names occurred the ‘current name’ according to Species Fungorum was used (https://www.speciesfungorum.org/; see also Camenzind et al. 2022).
Thirdly, we performed a PCA to learn about links of decomposition with other variables of the whole trait data set available for this set of fungi. In order to increase readability of the PCA we reduced the number of traits included in the analysis and thus selected those traits for the PCA that showed a significant relationship with decomposition in the Spearman or the pgls analysis. The selected variables were scaled to unit variance in the prcomp function of the package ‘stats’ (R Core Team 2023).
Results
Litter decomposition
Across all isolates, mass loss was 56.8% for leaf litter and 15.6% for wood litter, while in the control mass loss was 35.5% and 4.9%, respectively (See Fig. 1A and B). Mass loss across Ascomycota was on average 61.7% for leaf litter and 18.9% for wood litter; Basidiomycota caused an average mass loss of 64.5% in leaf and 17.5% in wood litter, and Mucoromycota had an average mass loss of 38.7% for leaf and 5.1% for wood litter. Both Asco- and Basidiomycota had CIs above the zero line for leaf and wood litter, i.e. they actively decomposed both litter types, whereas Mucoromycota had CIs overlapping with the zero line, showing a neutral response.
Figure 1.
Unpaired mean difference plot for mass loss of leaf and wood litter in control, Ascomycota, Basidiomycota and Mucoromycota. The swarm plot represents the raw data, the unpaired mean difference represents means (black dots), standard errors (black lines) and confidence intervals (grey areas around the mean).
Decomposition (treatment—control) varied substantially among isolates (see Fig. 2A and B). For leaf litter there was also variation among the replicates of one isolate, for example for RLCS01 (Mucor fragilis), RLCS02 (Mortierella elongata strain 2) and RLCS04 (Mortierella exigua) there were some replicates that decomposed litter and some not at all. These strains are all within the Mucoromycota, which decomposed on average only slightly more than the control (plus 3.2%). One strain (RLCS09, Trametes versicolor) had considerable variation in decomposition of both leaf and wood litter. The leaf and wood litter in the control plates also had mass loss, which can be explained by degradation through fragmentation caused by wetting the experimental units and through handling the litter at harvest when the litter was very brittle. Therefore, a few replicates showed negative results for decomposition.
Figure 2.
Violin plot with raw data points for decomposition (treatment—control) per isolate, means and standard errors (black points and bars) of leaf litter (upper panel) and wood litter (lower panel). Information about phylum affiliation is color coded (yellow: Ascomycota, red: Basidiomycota, dark blue: Mucoromycota). The phylogenetic tree (top of Figure) was constructed using the unweighted pair group method with arithmetic mean (upgma(), package ‘phangorn’; Schliep 2010) based on long sequence reads (ITS1, 5.8S, ITS2, and partial LSU marker according to the UNITE database and the RDP LSU data set. For more details see Methods section.
Carbon substrate use
The carbon sources were preferred in the order glucose > cellobiose > xylan > cellulose > ground litter (see Fig. 3A). Almost all fungi were able to grow on glucose media very well, while pure cellulose and ground litter based media were poor growth substrates for the majority of isolates. However, we also found differences between isolates, e.g. RLCS29 (Macrolepiota excoriata, Basidiomycota), RLCS12 (Didymellaceae, Ascomycota) and RLCS06 (Chaetomium angustispirale, Ascomycota) grew comparatively well on more complex substrates, while not growing as well on glucose as the other isolates.
Figure 3.
Enzymatic capacities of the different fungal isolates determined either by the use of different C sources (A) or enzymatic activity profiles (b). (a) Heat map of the use of the carbon sources glucose (glu), cellobiose (cb), xylan (xyl), cellulose (cel) and ground litter (lit) and the index for the complex carbon use ability (CCUA); values depict relative biomass production. Values for each isolate are added to the heatmap, where the brown tone illustrates the complexity of C sources (from left to right: glucose with light brown, to litter with dark brown), and the color intensity of the respective brown tone represents the values of individual isolates in each column. Differences in CCUA values are also visualized by respective color intensity. (B), Relative activity of different extracellular enzymes (% of total activity) determined by semi-quantitative analyses using the API ZYMTM kit.
Abbreviations for enzymes are: Ak_pho = alkaline phosphatase, Ac_phos = acid phosphatase, PhoHyd = phosphohydrolase, Leu_Aryl = leucine arylamidase, Val_Aryl = valine arylamidase, Try = trypsin, α_Chy = α-chymotrypsin, β_GluAm = N-acetyl-β-glucosaminidase, α_Glu = α-glucosidase, β_Glu = β-glucosidase, α_Gal = α-galactosidase, β_Gal = β-galactosidase, β-Glucoro = β-glucoronidase, α_Man = α-mannosidase, Est = esterase, Lip = lipase
Extracellular enzyme activities
A number of enzymes were produced by >70% of isolates, although in contrasting intensities. Among them were alkaline phosphatase, acid phosphatase, esterase, leucine arylamidase, valine arylamidase, phosphohydrolase, β-glucosidase and N-acetyl-β-glucosaminidase (Fig. 3B). Activity of lipase, trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase and β-glucuronidase was detected in very few species (≤10 isolates).
The Mucoromycota showed a more uniform enzyme pattern compared to the other phyla. Their enzyme repertoire mainly consisted of acid and alkaline phosphatase, phosphohydrolase and leucine and valine arylamidase. Basidiomycota and Ascomycota had more diverse enzyme repertoires and large variations among their isolates.
In Basidiomycota, for all isolates of our set, activities of the following enzymes were detected: alkaline phosphatase, leucine arylamidase, valine arylamidase, acid phosphatase, phosphohydrolase, β-glucosidase and N-acetyl-β-glucosaminidase. In Ascomycota, for most isolates of our set, activities of the following enzymes were detected: alkaline phosphatase, esterase, leucine arylamidase, acid phosphatase, phosphohydrolase, N-acetyl-β-glucosaminidase and β-glucosidase.
When we correlated litter decomposition rates with other functional traits, the growth on hemicellulose (xylan), the index for the ability to use complex carbon (CCUA) and the joint effect of the enzymes of our trait collection (PC1_EnzTraitCol) had the strongest relationship with decomposition rates of both litter types (Fig. 4). Leaf decomposition also had strong links with the joint effect of the C targeting enzymes of experiment III (PC1_CEnz_exp3) and the ability to form new soil aggregates (Aggr_formation). A summary of correlations of all variables with each other is shown in Fig. S6 as a correlation matrix.
Figure 4.
Bars represent Spearman's rank correlation coefficients (ρ) for the average decomposition of leaf litter (A) and wood litter (B) with other traits. Corresponding P-values of the Spearman correlation are listed on the right. Coefficients span a range from positive values (right side of y-axis), indicating positive relationships, to negative values (left side of the y-axis), indicating negative relationships. Abbreviations: Aggr_formation = ability to form new soil aggregates, Biom_dens = fungal biomass density, Cb_use = fungal growth on cellobiose based medium, CCUA = complex carbon use ability, Cel_use = fungal growth on cellulose based medium, Col_ext_rate = colony extension rate, Compet_ability = competitiveness, Glu_use = fungal growth on glucose based medium, Hydrophob = hydrophobicity of fungal biomass, Hyphal_length = hyphal length in soil, Lit_use = fungal growth on litter medium, PC1_EnzTraitCol = principal component 1 of PCA of the four enzymes of trait collection, PC1_AllEnz_exp3 = principal component 1 of PCA of the 16 enzymes of experiment III, PC1_Cenz_exp3 = principal component 1 of PCA of nine C targeting enzymes of experiment III (esterase, lipase, alpha_galactosidase, beta_galactosidase, beta_glucoronidase, alpha_glucosidase, beta_glucosidase, beta_glucosamine, alpha_mannosidase), Xy_use = fungal growth on xylan based medium.
When we considered phylogenetic dependencies of the data in the pgls models, we saw a different picture for leaf and wood decomposition (see Table S5). The ability to form new aggregates was a strong predictor for leaf decomposition (R2 = 0.31, P-value = 0.0008), independent of the phylogeny. When taking phylogenetic dependencies into account, the analysis revealed a positive relationship of competitiveness and a negative relationship of hyphal length with leaf decomposition (R2 = 0.22, P-value = 0.0055; R2 = 0.30, P-value = 0.0008, respectively). Relationships of leaf decomposition with growth on xylan based media, CCUA and joint effects of enzymes (PC1_EnzTraitCol and PC1_CEnz_exp3) were significant in the spearman analysis, but not in the pgls model, suggesting a strong influence of the phylogeny of the data in these cases. For wood decomposition, the growth on xylan based media (R2 = 0.42, P-value = 0.0001) and the ability to use complex carbon (CCUA) (R2 = 0.28, P-value = 0.0015) had the strongest explanatory power, independent of the phylogeny. The consideration of phylogenetic dependencies also revealed a negative correlation of wood decomposition with the joint effect of all enzymes of experiment III (PC1_AllEnz_exp3; R2 = 0.13, P-value = 0.027) and a negative relationship with growth on glucose based media (R2 = 0.21, P-value = 0.006).
In a PCA of all (relevant) traits, the PC axis 1 explained 33% of the variance, while axis 2 explained 19% of the variance (Fig. 5). The primary PC axis 1 shows that the largest trait variability in this dataset was reflected by the ability to use complex C sources, with negative loadings of isolates with a clear preference for glucose, and positive loadings of the ability to use xylan and complex C sources. Likewise, litter decomposition ability positively loaded on the 1st axis, as well as some of the joint enzyme activities (Fig. 5). Likewise, the correlation matrix (Fig. S6) showed that leaf and wood decomposition correlated well with the use of xylan and the more general ability to use more complex carbon (CCUA), but also with specific joint effects of enzymes: the joint effect of C degrading enzymes of experiment III (PC1_CEnz_exp3) correlated with leaf litter decomposition, and the joint effect of the enzymes of our trait collection, which included the oxidative enzymes laccase and cellobiohydrolase (PC1_EnzTraitCol), correlated with wood decomposition. Fungal growth on the glucose based media negatively aligned with wood litter decomposition and hyphal length in leaf litter samples (Hy_le_leaf) negatively aligned with leaf decomposition. The PCA results further illustrated the clear separation of trait spaces among phyla. Especially the Mucoromycota were separate, characterized by negative values on PC1.
Figure 5.
PCA showing leaf and wood litter decomposition for the three phyla (yellow, red and dark blue) with isolates depicted as dots. Abbreviations: Aggr_form = ability to from new soil aggregates, CCUA = complex carbon use ability, Compet = competitive ability expressed as number of wins in pairwise interactions, Decomp_leaf = decomposition of leaf litter, Decomp_wood = decomposition of wood litter, Glucose_use = fungal growth on glucose based medium, Hy_le_leaf = hyphal length in soil samples with leaf litter, Hy_le_wood = hyphal length in soil samples with wood litter, PC1_EnzTraitCol = principal component 1 of PCA of the four enzymes of trait collection, PC1_AllEnz_exp3 = principal component 1 of PCA of the 16 enzymes of experiment III, PC1_CEnz_exp3 = principal component 1 of PCA of nine C targeting enzymes of experiment III (esterase, lipase, alpha_galactosidase, beta_galactosidase, beta_glucoronidase, alpha_glucosidase, beta_glucosidase, beta_glucosamine, alpha_mannosidase), Xyl_use = fungal growth on xylan based medium.
Discussion
Grassland soils harbor Ascomycota and Basidiomycota fungi broadly involved in leaf litter decomposition, but less in wood litter decomposition
The functional ability to decompose litter varied substantially among isolates, with differences conserved at the phylum level. While within the Ascomycota and Basidiomycota most isolates showed the ability to decompose leaf litter (increase in 26% and 29%, respectively, in relation to the controls), only 10 out of 24 isolates within these groups were also able to decompose wood. Mucoromycota had much lower ability to decompose leaf litter (increase of 3.2% compared to control) and showed no ability to decompose wood.
These results confirmed a clear preference of the grassland fungal isolates used here for leaf litter. Since fungal isolates were isolated from a natural grassland soil, an adaptation to this kind of high-quality litter from grassland plants is likely. Additionally, growth in their “home” soil may have facilitated the decomposition of this litter type. However, grassland soils also harbor very complex organic matter, partly resulting from woody litter such as woody roots or woody stem pieces of certain shrubs. Therefore, lower-quality litters such as wood litter must also represent a valuable C source for these soil fungi. It has been shown before that certain substrates, for example cellulose, can be preferred by a specific community where species are organized into functional guilds of decomposers (Bhatnagar et al. 2018). The 10 isolates that were able to decompose a noticeable amount of wood (>5%) are distributed among the orders Sordariales, Pleosporales, Hypocreales, Helotiales (Ascomycota) and Polyporales (Basidiomycota). These fungi seem to form a functional group in our set of fungi.
Interestingly, there is a connection between leaf decomposition and the ability to form new soil aggregates. As we showed in a previous study by Lehmann et al. (2020), the ability to form new aggregates is strongly influenced by fungal phylogeny, with Ascomycota being the best and Mucoromycota the worst aggregators. Based on their analyses of fungal trait contributions to soil aggregation, the authors concluded that a critical local colony biomass density and the phylogeny, rather than overall soil hyphal length, contribute to the formation of new soil aggregates. Similarly, we could not find a positive influence of hyphal length on decomposition. In other, similar experiments by our lab, we saw these fungi switching from growth to reproduction, i.e. the production of spores at the time of resource depletion (data in preparation for publication). Hence, the biomass and hyphal length measured at the end of the experiment do not necessarily reflect the fungal abundance throughout the duration of the experiment. For leaf decomposition the relationship with hyphal length was negative after phylogenetic correction. This might be indicative of a trade-off between decomposition of high-quality litters and hyphal growth, similar to the trade-off found for enzymatic activity and fungal growth in Zheng et al. (2020).
In this study, overall enzymatic capacities of our fungal collection had significant relationships with decomposition of both litter types, corroborating numerous findings in the literature showing correlations between enzyme activity and decomposition (e.g. Sinsabaugh et al. 1994). When we joined the enzyme activities as ‘C targeting enzymes’, we found a good correlation with leaf decomposition only, indicating that the specification of C targeting enzymes (analyzed here) can be used as predictor for the decomposition of high-quality litter. The relationship between enzymatic activities and decomposition of both litter types was strongly influenced by the phylogeny of the data, showing that certain taxa within our data set have similar traits that can drive these correlations. The phylogeny also drives the relationship of the competitiveness of the fungi with decomposition. After taking phylogeny into account, leaf decomposition had a strong relationship with competitiveness, indicating that good competitors can decompose high-quality substrates better than more complex ones, such as wood litter. The degradation of litter in our grassland soil will thus be strongly influenced by the microbial community composition.
Complex carbon and xylan utilization by fungi are associated with litter decomposition
The ability to grow on xylan based media and more complex substrates in general (CCUA) were positively correlated with leaf and wood litter decomposition. Within our set of fungi, we observed strong differences between the phyla, but also large variations between isolates for this trait, showing that there are likely specialists among our set of fungi that are able to use one resource more effectively than others. This corroborates previous findings showing that members of the Mucoromycota, Ascomycota, and Basidiomycota can have very different resource utilization capabilities (Osono and Takeda 2002, Hanson et al. 2008, Khosravi et al. 2015, Pawłowska et al. 2019).
We found that the relation of growth on xylan based media and CCUA with wood litter decomposition was conserved when we considered fungal phylogeny, showing that the correlation between wood decomposition and xylan use and CCUA is not only driven by phylogenetic relatedness. Thus, we can use the ability to process complex carbon and the use of hemicelluloses like xylan as predictors for wood decomposition for our set of grassland fungi. For leaf litter decomposition CCUA and xylan use were also important predictors, but in this case, correlations were mainly driven by phylogeny. Hence, decomposition of high-quality litter such as leaf litter in our soil may depend on fungal community composition.
Contrary to growth on xylan, maximum biomass gain on glucose (related to reduced capacities to use complex C sources (Fig. 5)) was negatively related to wood decomposition, showing that fungi preferring a simple sugar as substrate did not degrade the more complex leaf and wood litter very well. In our study, this can be attributed to the Mucoromycota, which had their highest biomass on glucose amended medium, and which were unable to decompose wood litter and only slightly decomposed leaf litter.
Phylogenetic effects on decomposition differ between leaf and wood litter
In our study, we used three different phyla, with the Mucoromycota clearly forming an ecologically separate group. They have different enzyme profiles and different abilities to process carbon and decompose litter, i.e. they were among the worst decomposers. This result agrees with the general notion in the literature of Mucoromycota being ‘sugar fungi’, and having less enzymatic capability for C degradation than Asco- and Basidiomycota (Pawłowska et al. 2019, Moore et al. 2020). However, this very general grouping has recently been questioned by Pawlowska et al. (2019). The authors screened the capability of 52 Mucoralean strains to use different C sources using the Biolog phenotypic microarray system, and found surprisingly diverse metabolic activity. However, similar to enzymatic profiles, Biolog data represent no direct evidence for the decomposition potential of complex litter types. On the other hand, Pawloska et al. (2019) mainly included members of Mucorales and Umbellopsidales, while we also tested several Mortierellales. Indeed, Mucor (RLCS01) and Umbelopsis (RLCS19) showed slightly different traits than Mortierella, but still clearly grouped within the Mucoromycota. Additionally, some species of Mucoromycota are specifically known to be able to degrade hemicelluloses (Dix and Webster 1995). These different outcomes show that our results strongly depended on the species we included in our study. Thus, it may be relevant to test the observed relationships with more fungal isolates, or complement these analyses with the growing availability of genomic traits (Treseder et al. 2021).
The ecological separation of the Mucoromycota in our dataset clearly drives the phylogenetic signal that we find in leaf litter decomposition and the correlations with this variable. Therefore, we assume that the specific isolates among the Mucoromycota in our grassland soil will only have minor effects on dynamics of high-quality litter decomposition.
One Basidiomycete of our set is a member of the family Polyporales (RLCS09), a group known to be wood decomposers (Money 2016). This was the only Basidiomycete that decomposed wood in our experiment. The other three Basidiomycetes of our set are in the group Agaricales (Agaricomycetes) and did not decompose wood, but they decomposed leaf litter. This supports the general assumption that Agaricomycetes have widely diverging abilities to utilize lignin and cellulose (Floudas et al. 2020), and that only a minority of fungi possess the ability to produce lignin degrading peroxidases (Treseder and Lennon 2015).
Ascomycota were the largest group in our data set (20 out of 31 fungi) and were among the best decomposers, especially for leaf litter. This corroborates existing knowledge on these fungi, which are known to be involved in, for example, straw decomposition (Sordariales and Hypocreales; Ma et al. 2013) or wood decomposition (Xylariales; Helotiales; Richter and Glaeser 2015, Cedeño–Sanchez et al. 2020), and represent a major group found in wood and leaf litter (Pleosporales; Zhang et al. 2012), or the organic layer of forest soils (Chaetothyriales, Heliotiales; Baldrian et al. 2012). Thus, we conclude that this phylum is of crucial importance for the prediction of litter decomposition in grassland soils.
Conclusion and further research
The direct assessment of fungal utilization of xylan—and more complex C sources in general—appears to be a strong predictor of litter decomposition, even stronger than the enzyme repertoire of the fungi. Thus, future studies should preferably use more direct assessments of fungal carbon use abilities, in order to predict litter degradation patterns. We found that the phylogeny of fungi in our dataset was very important for the prediction of litter decomposition. There are likely specialists in wood decomposition in our grassland soil that could be indicative for the degradation of complex substrates in grassland soils. In order to fully understand the role of the phylogeny for decomposition in grassland soils, future research should include fungi from different grassland ecosystems, i.e. further species, and more importantly, this research should look at the role of fungal richness and diversity for litter decomposition. Another important factor for follow-up studies is litter diversity, namely the presence of more than one litter type at a time, which can also be decisive for the decomposition rate (Pei et al. 2017).
Supplementary Material
Acknowledgement
We thank Johanna Kückes for help with experimental lab work.
Contributor Information
Eva F Leifheit, Institute of Biology, Freie Universität Berlin, Altensteinstr. 6, 14195 Berlin, Germany; Berlin-Brandenburg Institute of Advanced Biodiversity Research (BBIB), Berlin 14195, Germany.
Tessa Camenzind, Institute of Biology, Freie Universität Berlin, Altensteinstr. 6, 14195 Berlin, Germany; Berlin-Brandenburg Institute of Advanced Biodiversity Research (BBIB), Berlin 14195, Germany.
Anika Lehmann, Institute of Biology, Freie Universität Berlin, Altensteinstr. 6, 14195 Berlin, Germany; Berlin-Brandenburg Institute of Advanced Biodiversity Research (BBIB), Berlin 14195, Germany.
Diana R Andrade-Linares, Helmholtz Zentrum München, Deutsches Forschungszentrum für Gesundheit und Umwelt (GmbH), Research Unit for Comparative Microbiome Analyses – COMI, 85764 Neuherberg, Germany.
Max Fussan, Institute of Biology, Freie Universität Berlin, Altensteinstr. 6, 14195 Berlin, Germany; Berlin-Brandenburg Institute of Advanced Biodiversity Research (BBIB), Berlin 14195, Germany.
Sophia Westhusen, Institute of Biology, Freie Universität Berlin, Altensteinstr. 6, 14195 Berlin, Germany.
Till M Wineberger, Institute of Biology, Freie Universität Berlin, Altensteinstr. 6, 14195 Berlin, Germany.
Matthias C Rillig, Institute of Biology, Freie Universität Berlin, Altensteinstr. 6, 14195 Berlin, Germany; Berlin-Brandenburg Institute of Advanced Biodiversity Research (BBIB), Berlin 14195, Germany.
Author contributions
Eva F Leifheit, Tessa Camenzind, Anika Lehmann, Diana R Andrade-Linares, Max Fussan, Sophia Westhusen, Till M Wineberger, and Matthias C Rillig
Conflict of interest
The authors do not have any conflict of interest.
Funding
EFL acknowledges funding from the Deutsche Forschungsgemeinschaft (LE 3859/1-1). TC acknowledges funding by the Deutsche Forschungsgemeinschaft (465123751, SPP2322 SoilSystems).
Data availability
The full data sheet has been deposited at https://github.com/Dr-Eva-F-Leifheit/fungal-traits-decomposition.
References
- Algora Gallardo C, Baldrian P, López-Mondéjar R. Litter-inhabiting fungi show high level of specialization towards biopolymers composing plant and fungal biomass. Biol Fertil Soils. 2021;57:77–88. [Google Scholar]
- Allison SD, Vitousek PM. Extracellular enzyme activities and carbon chemistry as drivers of tropical plant litter decomposition. Biotropica. 2004;36:285–96. [Google Scholar]
- Allison SD. A trait-based approach for modelling microbial litter decomposition. Ecol Lett. 2012;15:1058–70. [DOI] [PubMed] [Google Scholar]
- Andrade-Linares DR, Veresoglou SD, Rillig MC. Temperature priming and memory in soil filamentous fungi. Fungal Ecol. 2016;21:10–5. [Google Scholar]
- Bai Y, Cotrufo MF. Grassland soil carbon sequestration: current understanding, challenges, and solutions. Science. 2022;377:603–8. [DOI] [PubMed] [Google Scholar]
- Baldrian P, Kolařík M, Štursová M et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012;6:248–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar JM, Peay KG, Treseder KK. Litter chemistry influences decomposition through activity of specific microbial functional guilds. Ecolog Monogr. 2018;88:429–44. [Google Scholar]
- Boddy L, Watkinson SC. Wood decomposition, higher fungi, and their role in nutrient redistribution. Canad J Botan-Rev. 1995;73:S1377–83. [Google Scholar]
- Bradford MA, Warren Ii RJ, Baldrian P et al. Climate fails to predict wood decomposition at regional scales. Nat Clim Change. 2014;4:625–30. [Google Scholar]
- Camenzind T, Lehmann A, Ahland J et al. Trait-based approaches reveal fungal adaptations to nutrient-limiting conditions. Environ Microbiol. 2020;22:3548–60. [DOI] [PubMed] [Google Scholar]
- Camenzind T, Philipp Grenz K, Lehmann J et al. Soil fungal mycelia have unexpectedly flexible stoichiometric C:N and C:P ratios. Ecol Lett. 2021;24:208–18. [DOI] [PubMed] [Google Scholar]
- Camenzind T, Weimershaus P, Lehmann A et al. Soil fungi invest into asexual sporulation under resource scarcity, but trait spaces of individual isolates are unique. Environ Microbiol. 2022;24:2962–78. [DOI] [PubMed] [Google Scholar]
- Castellano MJ, Mueller KE, Olk DC et al. Integrating plant litter quality, soil organic matter stabilization, and the carbon saturation concept. Global Change Biol. 2015;21:3200–9. [DOI] [PubMed] [Google Scholar]
- Cedeño–Sanchez M, Wendt L, Stadler M et al. Three new species of Hypoxylon and new records of Xylariales from Panama. Mycosphere. 2020;11:1457–76. [Google Scholar]
- Cole JR, Wang Q, Fish JA et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper N. Phylogenetic comparative methods in R using phylogeny as a statistical fix. 2022. https://www.scribd.com/document/591943365/Phylogenetic-Comparative-Methods-in-R-Using-phylogeny-as-a-statistical-fix.
- Crowther TW, Maynard DS, Crowther TR et al. Untangling the fungal niche: the trait-based approach. Front Microbiol. 2014;5:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix NJ, Webster J. Fungal Ecology. London: Chapman & Hall, 1995. [Google Scholar]
- Floudas D, Bentzer J, Ahrén D et al. Uncovering the hidden diversity of litter-decomposition mechanisms in mushroom-forming fungi. ISME J. 2020;14:2046–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankland JC. Fungal succession—unravelling the unpredictable. Mycol Res. 1998;102:1–15. [Google Scholar]
- Gams W, Domsch KH. Beiträge zur Anwendung der Bodenwaschtechnik für die Isolierung von Bodenpilzen. Archiv Für Mikrobiologie. 1967;58:134–44. [Google Scholar]
- Gartner TB, Cardon ZG. Decomposition dynamics in mixed-species leaf litter. Oikos. 2004;104:230–46. [Google Scholar]
- Grossman JJ, Cavender-Bares J, Hobbie SE. Functional diversity of leaf litter mixtures slows decomposition of labile but not recalcitrant carbon over two years. Ecol Monogr. 2020;90:e01407. [Google Scholar]
- Hanson CA, Allison SD, Bradford MA et al. Fungal taxa target different carbon sources in forest soil. Ecosystems. 2008;11:1157–67. [Google Scholar]
- Harrell FE. Hmisc: a package of miscellaneous R functions. 2014. https://hbiostat.org/R/Hmisc/.
- Hicks LC, Frey B, Kjøller R et al. Toward a function-first framework to make soil microbial ecology predictive. Ecology. 2022;103:e03594. [DOI] [PubMed] [Google Scholar]
- Hiscox J, Savoury M, Toledo S et al. Threesomes destabilise certain relationships: multispecies interactions between wood decay fungi in natural resources. FEMS Microbiol Ecol. 2017;93:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J, Tumkaya T, Aryal S et al. Moving beyond P values: data analysis with estimation graphics. Nat Methods. 2019;16:565–6. [DOI] [PubMed] [Google Scholar]
- Ho J. Using dabestr. How to create estimation plots. 2020.
- Hudson HJ. The ecology of fungi on plant remains above the soil. The New Phytologist. 1968;67:837–74. [Google Scholar]
- Jakobsen I, Abbott LK, Robson AD. External hyphae of vesicular-arbuscular mycorrhizal fungi associated with Trifolium subterraneum L. New Phytol. 1992;120:371–80. [Google Scholar]
- Johnston SR, Hiscox J, Savoury M et al. Highly competitive fungi manipulate bacterial communities in decomposing beech wood (Fagus sylvatica). FEMS Microbiol Ecol. 2018;95:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Freeman C. Soil enzyme analysis for leaf litter decomposition in global wetlands. Commun Soil Sci Plant Anal. 2009;40:3323–34. [Google Scholar]
- Khosravi C, Benocci T, Battaglia E et al. Chapter one—sugar catabolism in Aspergillus and other fungi related to the utilization of plant biomass. In: Sariaslani S, Gadd GM (eds.), Advances in Applied Microbiology. Vol. 90, Cambridge, Massachusetts, United States of America: Academic Press, 2015, 1–28. [DOI] [PubMed] [Google Scholar]
- Lehmann A, Zheng W, Ryo M et al. Fungal traits important for soil aggregation. Front Microbiol. 2020;10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato MB, Boyette JS, Smilo RA et al. Functional importance and diversity of fungi during standing grass litter decomposition. Oecologia. 2021;195:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustenhouwer N, Maynard DS, Bradford MA et al. A trait-based understanding of wood decomposition by fungi. Proc Natl Acad Sci. 2020;117:11551–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A, Zhuang X, Wu J et al. Ascomycota members dominate fungal communities during straw residue decomposition in arable soil. PLoS One. 2013;8:e66146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoni S, Chakrawal A, Spohn M et al. Modeling microbial adaptations to nutrient limitation during litter decomposition. Front Forest Glob Chan. 2021;4:1–23. [Google Scholar]
- McGuire KL, Treseder KK. Microbial communities and their relevance for ecosystem models: decomposition as a case study. Soil Biol Biochem. 2010;42:529–35. [Google Scholar]
- Money NP. Chapter 1–fungal diversity. In: Watkinson SC, Boddy L, Money NP (eds.), The Fungi, 3rd edn, Boston: Academic Press, 2016, 1–36. [Google Scholar]
- Moore D, Robson GD, Trinci APJ. 21st Century Guidebook to Fungi. Cambridge: Cambridge University Press, 2020. [Google Scholar]
- Mundry R. Statistical issues and assumptions of phylogenetic generalized least squares. In: Garamszegi LZ (ed.), Modern Phylogenetic Comparative Methods and Their Application in Evolutionary Biology: concepts and Practice. Berlin, Heidelberg: Springer Berlin Heidelberg, 2014, 131–53. [Google Scholar]
- Münkemüller T, Lavergne S, Bzeznik B et al. How to measure and test phylogenetic signal. Methods Ecol Evol. 2012;3:743–56. [Google Scholar]
- Nilsson RH, Larsson KH, Taylor AFS et al. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019;47:D259–d264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme D. The caper package: comparative analysis of phylogenetics and evolution in R. 2018.
- Osono T, Takeda H. Comparison of litter decomposing ability among diverse fungi in a cool temperate deciduous forest in Japan. Mycologia. 2002;94:421–7. [PubMed] [Google Scholar]
- Osono T. Functional diversity of ligninolytic fungi associated with leaf litter decomposition. Ecol Res. 2020;35:30–43. [Google Scholar]
- Pawłowska J, Okrasińska A, Kisło K et al. Carbon assimilation profiles of mucoralean fungi show their metabolic versatility. Sci Rep. 2019;9:11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z, Leppert KN, Eichenberg D et al. Leaf litter diversity alters microbial activity, microbial abundances, and nutrient cycling in a subtropical forest ecosystem. Biogeochemistry. 2017;134:163–81. [Google Scholar]
- Porre RJ, van der Werf W, De Deyn GB et al. Is litter decomposition enhanced in species mixtures? A meta-analysis. Soil Biol Biochem. 2020;145:107791. [Google Scholar]
- R Core Team . R: a Language and Environment for Statistical Computing. Vol. 4.3.1, Vienna, Austria: R Foundation for Statistical Computing, 2023. [Google Scholar]
- Richter DL, Glaeser JA. Wood decay by Chlorociboria aeruginascens (Nyl.) Kanouse (Helotiales, Leotiaceae) and associated basidiomycete fungi. Int Biodeterior Biodegrad. 2015;105:239–44. [Google Scholar]
- Rillig MC, Field CB, Allen MF. Soil biota responses to long-term atmospheric CO2 enrichment in two California annual grasslands. Oecologia. 1999;119:572–7. [DOI] [PubMed] [Google Scholar]
- Sati SC, Bisht S. Utilization of various carbon sources for the growth of waterborne conidial fungi. Mycologia. 2006;98:678–81. [DOI] [PubMed] [Google Scholar]
- Schliep KP. Phangorn: phylogenetic analysis in R. Bioinformatics. 2010;27:592–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsabaugh RL, Moorhead DL, Linkins AE. The enzymic basis of plant litter decomposition: emergence of an ecological process. Appl Soil Ecol. 1994;1:97–111. [Google Scholar]
- Sinsabaugh RL, Turner BL, Talbot JM et al. Stoichiometry of microbial carbon use efficiency in soils. Ecolog Monogr. 2016;86:172–89. [Google Scholar]
- Sinsabaugh RL. Enzymic analysis of microbial pattern and process. Biol Fertil Soils. 1994;17:69–74. [Google Scholar]
- Spatafora JW, Aime MC, Grigoriev IV et al. The fungal tree of life: from molecular systematics to genome-scale phylogenies. Microbiol Spectr. 2017;5:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn RG, Reddy CA, Harris D et al. Isolation of saprophytic basidiomycetes from soil. Appl Environ Microb. 1996;62:4288–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treseder KK, Alster CJ, Cat LA et al. Nutrient and stress tolerance traits linked to fungal responses to global change: four case studies. Elementa: Sci Anthropocene. 2021;9:1–19. [Google Scholar]
- Treseder KK, Lennon JT. Fungal traits that drive ecosystem dynamics on land. Microbiol Mol Biol Rev. 2015;79:243–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Crowther TW. Uniting the scales of microbial biogeochemistry with trait-based modelling. Funct Ecol. 2022;36:1457–72. [Google Scholar]
- Wei T, Simko V, Levy M. A visualization of a correlation matrix. 2021. https://github.com/taiyun/corrplot.
- Yin N, Koide RT. The role of resource transfer in positive, non-additive litter decomposition. PLoS One. 2019;14:e0225337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Crous PW, Schoch CL et al. Pleosporales. Fungal Divers. 2012;53:1–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng WS, Lehmann A, Ryo M et al. Growth rate trades off with enzymatic investment in soil filamentous fungi. Sci Rep. 2020;10:11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full data sheet has been deposited at https://github.com/Dr-Eva-F-Leifheit/fungal-traits-decomposition.







