Abstract
Hepatitis C virus (HCV) infection frequently leads to chronic hepatitis and cirrhosis of the liver and has been linked to development of hepatocellular carcinoma. We previously identified a small yeast RNA (IRNA) capable of specifically inhibiting poliovirus (PV) internal ribosome entry site (IRES)-mediated translation. Here we report that IRNA specifically inhibits HCV IRES-mediated translation both in vivo and in vitro. A number of human hepatoma (Huh-7) cell lines expressing IRNA were prepared and characterized. Constitutive expression of IRNA was not detrimental to cell growth. HCV IRES-mediated cap-independent translation was markedly inhibited in cells constitutively expressing IRNA compared to control hepatoma cells. However, cap-dependent translation was not significantly affected in these cell lines. Additionally, Huh-7 cells constitutively expressing IRNA became refractory to infection by a PV-HCV chimera in which the PV IRES is replaced by the HCV IRES. In contrast, replication of a PV-encephalomyocarditis virus (EMCV) chimera containing the EMCV IRES element was not affected significantly in the IRNA-producing cell line. Finally, the binding of the La autoantigen to the HCV IRES element was specifically and efficiently competed by IRNA. These results provide a basis for development of novel drugs effective against HCV infection.
Hepatitis C virus (HCV) is the primary causative agent of parenterally transmitted non-A, non-B hepatitis and affects a significant part of the worldwide population. HCV infection frequently leads to chronic hepatitis, cirrhosis of the liver, and hepatocellular carcinoma (8, 17, 33). There is currently no effective therapy or vaccine available for HCV other than alpha interferon. HCV has been a difficult virus to study due to the lack of an appropriate tissue culture system and an adequate, simple, and low-cost animal model. The RNA genome of HCV has been cloned and characterized and shown to be infectious when injected into the livers of chimpanzees (17, 20, 22, 41). The single-stranded, plus-polarity RNA genome of HCV, a member of the Flaviviridae, is approximately 9,500 nucleotides (nt) long. The 5′ untranslated region (UTR) of HCV RNA is approximately 340 nt long, is highly structured, and contains multiple AUG codons (5–7, 20, 28, 37, 38). The 5′ UTR is highly conserved among different strains of HCV (6, 15). It is followed by a single large open reading frame that encodes a polyprotein which is proteolytically processed to produce the mature structural and nonstructural proteins of HCV. Nucleotides 40 to 370 of the 5′ UTR of HCV have been shown to contain an internal ribosome entry site (IRES) (13, 21, 32, 38, 39). Although the initiation codon for HCV polyprotein synthesis is located at nt 342, an additional 28 nt from the coding sequence is required for efficient synthesis of HCV proteins (21, 31).
IRES-mediated translation was first discovered in picornaviruses (18, 29). Recent studies on picornavirus-IRES-mediated translation have demonstrated that cellular trans-acting proteins distinct from canonical translation initiation factors play an important role in IRES-mediated translation. These proteins bind to the IRES and presumably help to facilitate the binding of ribosomes to the IRES. Some of the trans-acting proteins have been identified as the La autoantigen, polypyrimidine tract-binding (PTB) protein, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and poly(rC)-binding protein (PCBP) (3, 14, 16, 18, 26, 28a, 29, 34, 40). The La polypeptide binds to both poliovirus (PV) and HCV IRES elements and stimulates IRES-mediated translation (2, 26, 35). Similarly, the PTB protein interacts with HCV and other picornavirus IRES sequences and stimulates viral 5′ UTR-mediated translation (1, 19, 40). Three other polypeptides of unknown function, p25, p87, and p120, have been shown to interact specifically with the HCV IRES (13, 42).
Previous results from our laboratory have shown that PV IRES-mediated translation is restricted in the yeast Saccharomyces cerevisiae, in part due to a trans-acting factor capable of inhibiting PV IRES-mediated translation in HeLa cell extracts (9). The inhibitor was purified and subsequently shown to be a small (60-nt) RNA which specifically inhibited cap-independent, IRES-mediated translation but had little or no effect on cap-dependent translation of cellular mRNAs (10). The yeast RNA (called IRNA) was found to bind strongly several cellular polypeptides which interact with the PV IRES element, including the La autoantigen (11). It appears, therefore, that IRNA inhibits PV IRES-mediated translation by competing for critical cellular polypeptides that are required for viral IRES-mediated translation.
Because HCV and PV IRES elements bind similar polypeptides, we reasoned that IRNA might also interfere with HCV IRES-mediated translation. Using transient transfection of hepatoma cells and a hepatoma cell line constitutively expressing IRNA, we demonstrate specific inhibition of HCV IRES-mediated translation by IRNA. Additionally, hepatoma cells constitutively expressing IRNA became refractory to infection by both PV and PV-HCV chimera in which the PV IRES is replaced by the HCV IRES element. Finally, the binding of the La autoantigen to the HCV IRES element was specifically and efficiently competed by IRNA.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells were grown in spinner culture in minimum essential medium supplemented with 1 g of glucose per liter and 6% newborn calf serum. HeLa monolayer cells were maintained in tissue culture flask or plates in minimum essential medium (GIBCO/BRL) supplemented with 10% fetal bovine serum. The hepatocellular carcinoma cells (Huh-7) were grown in RPMI medium supplemented with 10% fetal bovine serum. PV1 (type 1 Mahoney) and its chimeric derivatives HCV-PV and encephalomyocarditis virus (EMCV)-PV (generous gift from Eckard Wimmer) were amplified in HeLa cells, and the virus titer was calculated by plaque assay as described previously (23).
Plasmid construction.
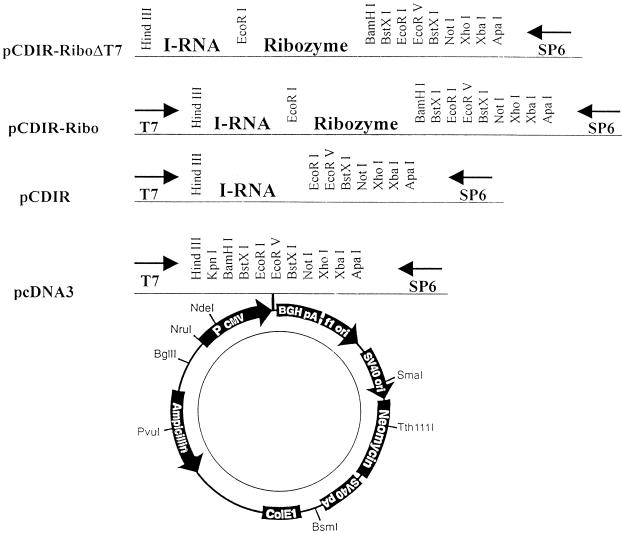
IRNA-encoding sequences were cloned into pCDNA3 (Invitrogen) vector under the cytomegalovirus promoter, yielding pCDIR. To generate the correct 3′ end of the IRNA transcript in vivo, the ribozyme of hepatitis delta virus (pSA1 [30]) was cloned at the 3′ end of the IRNA gene (pCDIR.Ribo). The existing T7 promoter of the parental vector (pCDNA3) was further deleted to generate the correct 5′ end of the IRNA (pCDIR.Ribo.ΔT7) for experiments involving in vivo expression (see Fig. 3). A control plasmid, pCDRibo.ΔT7, was also constructed by cloning the hepatitis delta ribozyme sequence into the XhoI-HindIII sites of the same pCDNA3 vector (lacking the T7 promoter). Plasmid pCD HCV-luc was constructed by cloning the HCV-luciferase fragment into the KpnI-XhoI sites of pCDNA3. pCD-luc lacks the HCV UTR sequences.
FIG. 3.
Schematic diagram of the constructs used for in vivo expression of IRNA. The diagram (not to scale) shows the cloning of IRNA encoding sequences into eukaryotic expression vector pCDNA3 (Invitrogen). The polylinker site following the cytomegalovirus (CMV) promoter sequence is illustrated above the parental plasmid. Additional construction of different IRNA-encoding plasmids are shown, and their names are given at the left. BGH, bovine growth hormone; pA, poly(A) site; SV40, simian virus 40; ori, origin.
Cloning of hepatoma cell lines expressing IRNA.
Plasmids pCDIR, PCDIR.Ribo, and pCDIR.Ribo.ΔT7 were electroporated into Huh-7 cells and selected for neomycin resistance with 400 μg of G418 (Invitrogen) per ml for 4 to 6 weeks. The antibiotic-resistant cell clones were harvested and further selected by dilutional cloning. A control cell line was also prepared in a similar method using plasmid pCD.Ribo.ΔT7.
Detection of IRNA in the cell lines.
IRNA expression in the cell lines was measured by isolating total RNA from these cells and quantitating the IRNA level by reverse transcriptase (RT)-mediated PCR (RT-PCR) using IRNA-specific oligonucleotide primers. One to 5 ng of in vitro-transcribed, purified IRNA, 1 to 2 μg of total RNA isolated from the IRNA-expressing pCDIR.Ribo.ΔT7 cell line, and 2 μg of total RNA from Huh-7 control cells were reverse transcribed by murine leukemia virus RT using 2.5 μM random hexamer primers in a 20-μl reaction according to the Perkin-Elmer Cetus RNA PCR kit protocol. Eight hundred nanograms of each primer (corresponding to 5′ nt 1 to 20 and 3′ nt 1 to 20 of the IRNA sequence) was used to amplify the 60-nt fragment in a 100-μl PCR. The cycling parameters were as follows: denaturation, 95°C for 1 min; annealing, 65°C for 1 min; extension, 72°C for 1 min; total of 50 cycles. Twenty microliters of each reaction product was loaded onto an 8% native acrylamide gel and visualized by ethidium bromide staining.
DNA transfection.
For each transfection assay, 106 Huh-7 cells in 30-mm-diameter plates were transfected with 15 μl of Lipofectin (GIBCO/BRL) and 2 to 5 μg of plasmid DNA. At 16 h posttransfection, cell lysates were prepared according to the Promega protocol and assayed for both β-galactosidase (β-Gal) and luciferase expression.
Detection of various mRNA levels by RT-PCR.
The luciferase, GAPDH, and β-actin mRNA levels in the total RNA isolated from control Huh-7 and IRNA-expressing cells were quantitated by RT-PCR. Three micrograms of total RNA isolated from both the IRNA-expressing cell line, pCDIR.Ribo.ΔT7, and control Huh-7 cells transfected with plasmids encoding the luciferase and β-Gal genes were reverse transcribed by Moloney murine leukemia virus (M-MLV) RT, using 250 pmol of random hexamer primers. Prior to cDNA synthesis, the RNA and primers were denatured at 95°C for 5 min and cooled slowly to room temperature over a 15-min period to allow the primers to anneal. Two hundred units of M-MLV RT was added, and the 20-μl reaction mixture was incubated at 42°C for 1 h. Prior to PCR amplification, the first-strand reactions were heated at 95°C for 5 min to inactivate the M-MLV RT, and 2 μl of each reaction was amplified by Taq DNA polymerase (Perkin-Elmer Cetus) in a standard 50-μl PCR. The following specific oligonucleotide primers were used in the PCRs: luciferase primers (5′ nt 962 to 981 and 3′ nt 1397 to 1416) to generate a 400-bp fragment; GAPDH primers (5′ nt 212 to 236 and 3′ 787 to 811) to generate a 600-bp fragment; and β-actin primers (5′ nt 1038 to 1067 and 3′ nt 1876 to 1905) to generate a 661-bp fragment. A total of 50 cycles were performed (each cycle consisting of denaturation [94°C for 1 min], annealing [55°C for 45 s], and extension [72°C for 1 min] for luciferase detection and denaturation [94°C for 45 s], annealing [60°C for 45 s], and extension [72°C for 1.5 min] for both GAPDH and β-actin detection). Ten-microliter aliquots of the RT-PCR mixtures were loaded on a 1× Tris-borate-EDTA–1.2% agarose gel and visualized by ethidium bromide staining.
Plaque assay.
Plaque assays were performed as described below (unless stated otherwise). Huh-7 cells (106 cells) were infected with either PV or HCV-PV chimera, and after 72 h, cell extracts were prepared. Two hundred fifty microliters of cell extract was used to further infect HeLa monolayer cells (2 × 106 cells in 60-mm-diameter plates). After 3 days of incubation at 37°C, the plaques were developed by staining with 1% crystal violet.
In vitro transcription.
RNA transcripts were synthesized in vitro with T7 or SP6 RNA polymerase from linearized plasmid DNA which was gel purified after digestion with the appropriate restriction enzyme. The pSDIR clone (10) was linearized with HindIII and transcribed with T7 RNA polymerase to generate IRNA. The HCV IRES-containing bicistronic construct pT7DC1-341 (a generous gift from A. Siddiqui) was linearized with HpaI, and the runoff capped transcript was generated with T7 RNA polymerase. The HCV 5′ UTR was cloned into pSP-luc+ vector (Promega) between the BglII and HindIII sites. To generate HCV 5′ UTR RNA, the construct was linearized with HindIII, gel purified, and transcribed with SP6 RNA polymerase. The nonspecific RNA was synthesized from pSP-luc+ vector (Promega), linearized with HindIII, and transcribed by SP6 RNA polymerase.
In vitro translation.
The T7DC1-341 RNA was translated in HeLa cell extract as described previously (9). Approximately 80 μg of HeLa cell extract was used to translate 2 μg of capped bicistronic mRNA in 25 μl of reaction mixture, in the presence of 25 μCi of [35S]methionine (800 Ci/mmol; Amersham) and 40 U of RNasin (Promega). The reaction mixture was incubated for 1 h at 37°C, and the products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography.
In vivo labeling of cellular proteins.
Monolayers of hepatoma cells (2 × 106 cells/60-mm-diameter plate) were preincubated in methionine-free medium (GIBCO) for 45 min at 37°C. Then 100 μCi of trans-labeled methionine (specific activity, >1,000 Ci/mmol) was added to each plate, and incubation was continued for another hour. [35S]methionine-labeled cell extract was prepared as described previously (10, 11).
In vivo labeling and immunoprecipitation of the viral proteins.
Monolayer hepatoma cells (5 × 105 cells/30-mm-diameter plate) were infected with 150 μl of either PV-HCV or PV-EMCV chimera (titer, 2.5 × 104 PFU/ml), and the infection was continued for 24 h. Cellular and viral proteins were labeled as described above. In vivo-labeled viral proteins in infected cells were detected by immunoprecipitation with anti-PV capsid antibody as described previously (10, 11).
UV-induced cross-linking.
HeLa and hepatoma translation lysates (S10) were prepared as previously described (9); 40 fmol of 32P-labeled RNA probe (8 × 104 cpm) was incubated with 30 to 60 μg of S10 extract from HeLa cells as described earlier (10). Following incubation, samples were irradiated with UV light from a UV lamp (multiband UV, 254/366-nm model UGL; 25 UVP, Inc.) at a distance of 3 to 4 cm for 10 min at room temperature. Unbound RNAs were digested with a mixture of 20 μg each of RNase A and RNase T1 at 37°C for 30 min, and protein-nucleotidyl complexes were analyzed by SDS-PAGE on a 14% polyacrylamide gel.
RESULTS
Inhibition of HCV IRES-mediated translation by IRNA in vivo and in vitro.
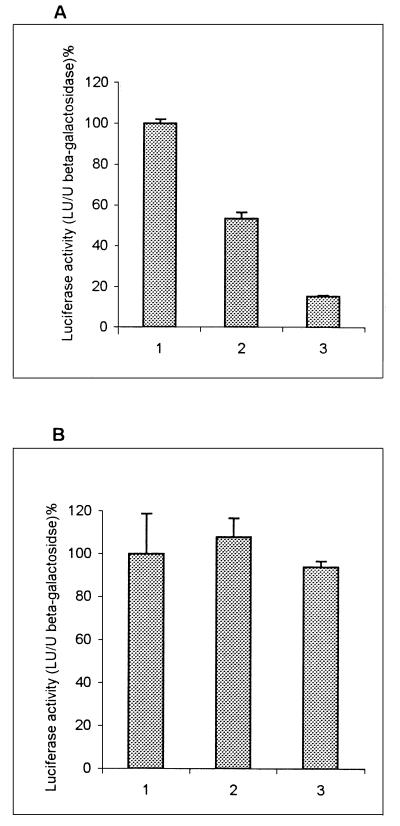
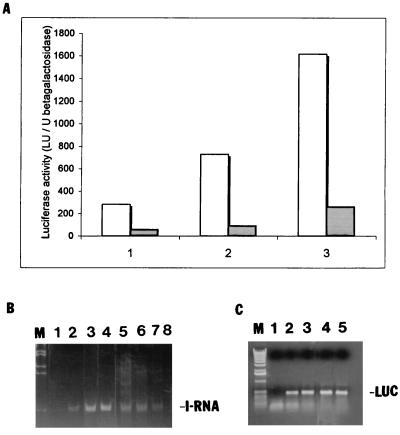
To test the possibility that IRNA interferes with HCV IRES-mediated translation, human hepatocellular carcinoma cells (Huh-7) were transiently cotransfected with 3 plasmids: a reporter gene expressing luciferase programmed by the HCV IRES element (pCD HCV-luc), pSV40/β-gal to measure transfection efficiency, and the plasmid expressing IRNA (pCDIR.Ribo.ΔT7). All transfections were done in triplicate and contained equal amounts of the luciferase reporter and β-Gal plasmids. Increasing concentrations of plasmid pCDIR.Ribo.ΔT7 were used in various reactions, and the total amount of DNA in each reaction was kept constant by addition of an appropriate amount of a nonspecific DNA (pCDNA3). Following transfection, luciferase activity was measured in cell extracts. At the lowest concentration of the IRNA plasmid, inhibition of luciferase activity from plasmid pCD HCV-luc was approximately 50% compared to the control (Fig. 1A). However, at the highest concentration, 90% of luciferase activity was inhibited. Translation of luciferase from a control plasmid (pCDNA3-luc) without the HCV IRES was not significantly inhibited by IRNA (Fig. 1B). Expression of either ribozyme alone or a nonspecific RNA similar in length to IRNA did not interfere with luciferase expression (data not shown). These results suggested that HCV IRES-mediated translation was specifically inhibited by IRNA in hepatoma cells, whereas cap-dependent translation of luciferase from the control plasmid lacking the HCV IRES element was not significantly affected by IRNA.
FIG. 1.
Effect of IRNA on HCV IRES-mediated translation in Huh-7 cells. Monolayer cells (106) were transfected with three different plasmid DNAs: pCDIR.Ribo.ΔT7 expressing IRNA, pCD HCV-luc reporter plasmid in which luciferase translation is programmed by the HCV IRES, and a β-Gal reporter gene to measure transfection efficiency. After 24 h of transfection, extracts were made and luciferase and β-Gal activities were measured. Luciferase activity (light units) is expressed as percentage of the control after normalizing for β-Gal activity and protein content for each transformation. (A) Vertical bars 1, 2, and 3 show the dose-response effect of pCDIR.Ribo.ΔT7 on HCV IRES-mediated translation of luciferase gene at 0, 1.25, and 1.88 μg of pCDIR.Ribo.ΔT7, respectively. Total DNA concentration was made up to 2.5 μg by adding 2.5, 1.25, and 0.62 μg of pCDNA3 (bars 1, 2, and 3, respectively). (B) A similar experiment was performed in which plasmid pCD HCV-luc was replaced by pCDNA3-luc conferring cap-dependent translation of luciferase. All transfection reactions contained 1 μg each of β-Gal and luciferase reporter plasmids.
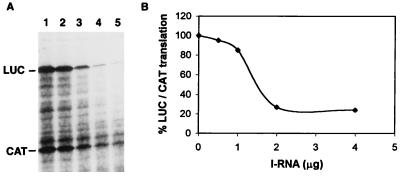
To confirm the results obtained in vivo, the effect of IRNA on HCV IRES-mediated translation was determined in vitro. A bicistronic construct consisting of the HCV IRES flanked by chloramphenicol acetyltransferase (CAT) and luciferase genes was used in this experiment. While synthesis of CAT is mediated by cap-dependent translation, the downstream luciferase synthesis occurs by HCV IRES-mediated translation. Translation was measured by quantitating radioactivity incorporated into luciferase and CAT polypeptides at 0, 0.5, 1, 2, and 4 μg of IRNA (Fig. 2A). The specific inhibition of luciferase synthesis was normalized by determining the ratio of luciferase to CAT at each IRNA concentration (Fig. 2B). Translation conditions were chosen so that approximately equal amounts of radioactivity were incorporated into luciferase and CAT proteins in the absence of IRNA (Fig. 2A, lane 1). In addition to full-length luciferase polypeptide, some lower-molecular-weight products were also observed. This is presumably due to premature termination during luciferase synthesis. At 1 μg of IRNA, luciferase synthesis was inhibited to 20% of the control, whereas at 2 μg of IRNA, specific inhibition of HCV IRES-mediated luciferase synthesis was 76% compared to the control. Although both luciferase and CAT syntheses were inhibited by IRNA in vitro, luciferase synthesis was affected much more than CAT synthesis at higher concentrations of IRNA. The inhibition of cap-dependent translation by IRNA could be due to its interaction with general RNA-binding proteins which have been implicated in facilitating cap-dependent translation (36). In addition, IRNA’s interaction with La may affect AUG start site selection during translation initiation, as suggested by a recent study (25).
FIG. 2.
IRNA selectively inhibits HCV IRES-mediated translation in vitro. The bicistronic construct containing the HCV IRES flanked by CAT and luciferase genes was transcribed by T7 RNA polymerase, and the bicistronic RNA was translated in vitro in 25 μl of HeLa cell lysate in the absence (A, lane 1) or presence of increasing concentrations of IRNA (lanes 2 to 5; 0.5, 1, 2, and 4 μg, respectively). (B) The band intensities of luciferase (LUC) and CAT were quantitated by densitometry, and the ratios of luciferase to CAT were calculated. The percentage of luciferase/CAT translation was plotted against the concentration of IRNA.
Construction of hepatoma cell lines expressing IRNA constitutively.
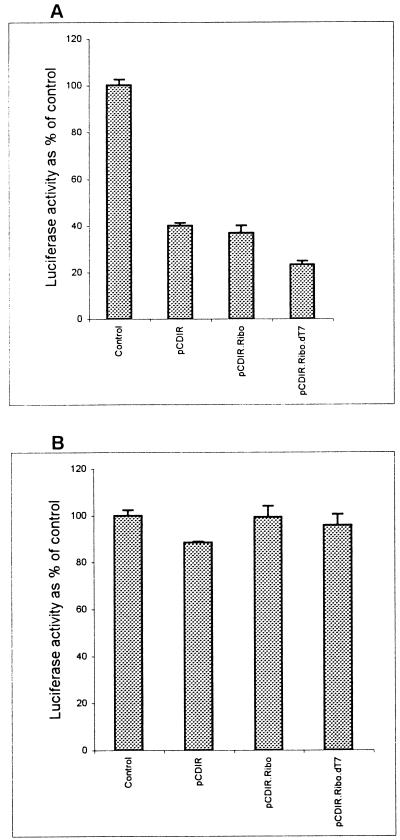
To determine the long-term effect of expression of IRNA in Huh-7 cells, cell lines constitutively expressing IRNA were generated by using a pCDNA-based vector as described in Materials and Methods. The cell line made initially contained the T7 sequences at the 5′ end of the IRNA gene (pCDIR [Fig. 3]). We made an additional cell line in which the hepatitis delta ribozyme sequence was added to the 3′ end of IRNA sequence for generation of the exact 3′ end (pCDIR.Ribo [Fig. 3]). Another cell line was prepared by using the pCDIR.Ribo construct which lacked the T7 promoter sequences (pCDIR.Ribo.ΔT7 [Fig. 3]). The control cells (Huh-7) and cell lines expressing IRNA were cotransfected with pCD HCV-luc and pSV40/β-gal. Cell extracts were used to measure both luciferase and β-Gal activities. The results were plotted as percent of control after normalizing for β-Gal activity and protein concentration. Both pCDIR and pCDIR. Ribo cells showed approximately 60% inhibition of luciferase activity compared to the control (Fig. 4A). Maximum inhibition (∼80%) of luciferase expression was observed in the pCDIR.Ribo.ΔT7 cell line compared to the control (Fig. 4A). Titration of the reporter construct (pCD HCV-luc) in the cell line pCDIR.Ribo.ΔT7 consistently showed 80 to 85% inhibition of HCV IRES-mediated translation of luciferase (Fig. 5A). No significant inhibition of cap-dependent translation from the pCDNA-luc construct was observed in cell lines expressing IRNA (Fig. 4B). These results suggest that IRNA interferes with luciferase expression programmed by HCV IRES in cells expressing IRNA. Because RT-PCR analysis showed no significant difference in the HCV IRES-luc reporter mRNA levels between control cells and the cells expressing IRNA (Fig. 5C), we concluded that IRNA was capable of inhibiting HCV IRES-mediated translation in vivo. The level of IRNA was determined in the pCDIR.Ribo.ΔT7 cells by RT-PCR and was found to be approximately 0.05% of the total cellular RNA (Fig. 5B).
FIG. 4.
HCV IRES-mediated translation is inhibited in Huh-7 cells constitutively expressing IRNA. Plasmid pCD HCV-luc was cotransfected with a β-Gal reporter gene into either control Huh-7 cells or IRNA-expressing cell lines (pCDIR, pCDIR.Ribo, and pCDIR.Ribo.ΔT7), and luciferase activity was plotted as percentage of control (A). Similarly, plasmid pCDNA3-luc conferring cap-dependent translation of the luciferase gene was transfected into either the control Huh-7 or hepatoma-IRNA cell line (B). β-Gal activity was measured to normalize transfection efficiencies in both experiments.
FIG. 5.
HCV-luciferase reporter dose response and quantitation of IRNA and luciferase mRNA in the cell line. (A) Increasing concentrations (1, 2, and 3 μg) of the pCD HCV-luc reporter plasmid were transfected into either Huh-7 hepatoma cells (dotted bars) or the IRNA-expressing hepatoma cell line (pCDIR.Ribo.ΔT7) (white bars). A control β-Gal plasmid was also cotransfected to normalize transfection efficiencies. Luciferase activity (103 light units [LU]) was plotted against increasing concentrations of the test plasmids. (B) Detection of IRNA in hepatoma cell line by RT-PCR. IRNA expression level was detected by RT-PCR using IRNA-specific oligonucleotide primers. Lane 1, no-RNA control; lanes 2 to 4, 1, 2.5, and 5 ng, respectively, of in vitro-transcribed, purified IRNA; lanes 5 to 7, 2, 1.5, and 1 μg, respectively, of total RNA from an IRNA-expressing hepatoma cell, pCDIR.Ribo.ΔT7. Lane 8 contained 2 μg of total RNA from the control Huh-7 cells. Lane M represents marker DNA. (C) Detection of luciferase (LUC) mRNA by RT-PCR. Total RNA isolated from control (Huh-7) and pCDIR.Ribo.ΔT7 cells transfected with the luciferase reporter plasmid was used to detect luciferase mRNA levels by using luciferase-specific oligonucleotide primers. Lane 1, no-RNA control; lanes 2 and 3, 1 and 10 ng of luciferase mRNA standard; lanes 4 and 5, 1 μg of total RNA isolated from Huh-7 cells and cells expressing IRNA, respectively, after transfection with pCDHCV-luc.
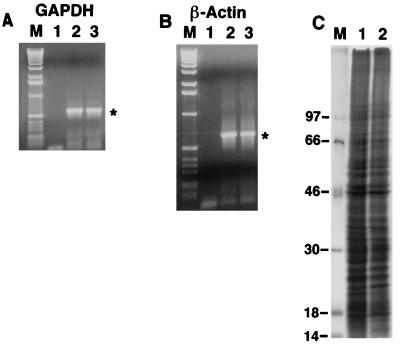
To determine whether constitutive expression of IRNA interfered with cellular transcription, levels of two cellular mRNAs, GAPDH and β-actin, were determined by RT-PCR using appropriate oligonucleotide primers. As can be seen in Fig. 6A and B, no significant differences were detected in the overall expression of these mRNAs between Huh-7 (control) and IRNA- expressing pCDIR.Ribo.ΔT7 cells. We also determined the effect of IRNA expression on the level of overall cellular protein synthesis by labeling cells with [35S]methionine. Global protein synthesis was largely unaffected in the pCDIR.Ribo.ΔT7 cells compared to the control Huh-7 cells (Fig. 6C). However, the intensity of a couple of polypeptides was reduced in the cell line compared to the control (Fig. 6C, lanes 1 and 2, indicated by dots). These results are consistent with the finding that translation of a capped mRNA was not significantly affected in the cells expressing IRNA (Fig. 1 and 4).
FIG. 6.
Effect of IRNA expression on cellular transcription and translation. (A and B) Detection of GAPDH (A) and β-actin (B) mRNAs by RT-PCR. Lane 1, no-RNA control; lanes 2 and 3, 3 μg of the total RNA isolated from Huh7 control cells and cell lines expressing IRNA (pCDIR.Ribo.ΔT7), respectively. (C) In vivo labeling of proteins. Monolayer Huh7 hepatoma cells (lane 1) and hepatoma pCDIR.Ribo.ΔT7 cells (lane 2) were labeled with [35S]methionine for 1 h, and in vivo-labeled proteins were analyzed on an SDS–14% polyacrylamide gel. Lane M shows the migration of 14C-labeled protein markers (Gibco/BRL), with approximate molecular masses as indicated to the left in kilodaltons.
Hepatoma cells constitutively expressing IRNA are refractory to infection by a PV-HCV chimera under translational control of HCV IRES.
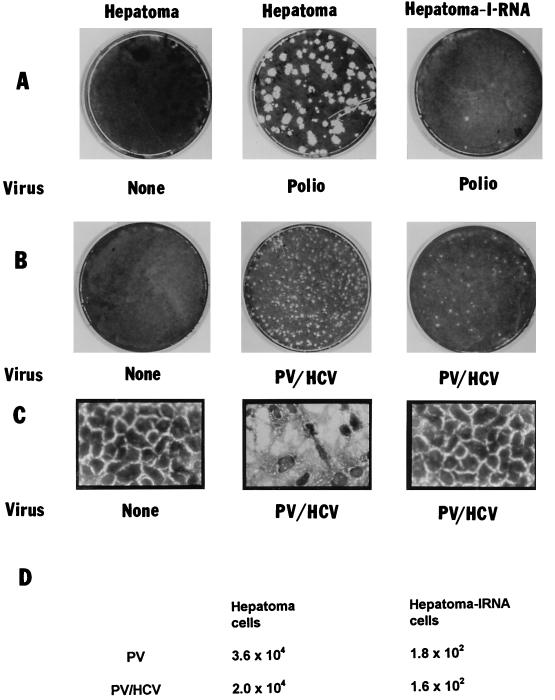
To determine the effect of IRNA on HCV IRES-mediated translation during virus infection, the pCDIR.Ribo.ΔT7 cells were infected with a chimeric PV (PV-HCV 701) in which the PV IRES is replaced by the HCV IRES. PV-HCV 701 contained the 5′ cloverleaf structure of PV, followed by HCV IRES (nt 9 to 332) plus 123 amino acids of HCV core protein followed by the entire poliovirus open reading frame plus the 3′ UTR and poly(A) site (23). Translation of viral proteins in cells infected with the PV-HCV chimera is mediated by the HCV IRES element (23). Huh-7 control cells and the hepatoma-IRNA cells (pCDIR. Ribo.ΔT7) were infected with PV and the PV-HCV chimera. Following infection, cell extracts were prepared from infected and mock-infected cells which were then used to further infect HeLa monolayer cells. Plaques characteristic of wild-type PV and PV-HCV 701 were apparent in HeLa cells infected with cell extract from control hepatoma cells (Fig. 7A and B, middle panel). Evidently viral replication was drastically affected in the cell line expressing IRNA (hepatoma-IRNA) with either virus (Fig. 7A and B, right panel). In a parallel experiment, the virus titers were measured by the serial dilution method, and the results demonstrated more than a 100-fold decrease in virus yield in IRNA-expressing hepatoma cells compared to the control cells (Fig. 7D). While the control hepatoma cells show extensive damage after infection, the cells expressing IRNA are almost totally protected from the cytopathic effect of the chimeric virus (Fig. 7C). Thus, hepatoma cells constitutively expressing IRNA were significantly resistant to both PV and the PV-HCV chimera under the conditions used for infection.
FIG. 7.
Hepatoma cells constitutively expressing IRNA prevent PV and HCV-PV chimera infection. Huh-7 control cells or the IRNA expressing hepatoma cell line (pCDIR.Ribo.ΔT7) (∼106 cells) were infected with 500 PFU of either PV (Polio) (A) or HCV-PV chimera (B and C). After 72 h, either cells were stained for the observation of cytopathic effects (C) or cell extracts were made to further infect HeLa monolayer cells for the plaque assay (A and B). Cells were stained by crystal violet. (D) Average virus titers obtained from three independent plaque assay experiments.
To rule out the possibility that the cloned hepatoma cell line is simply less able to support the viral infectious life cycle, its ability to support replication of another chimeric PV was examined. For this purpose, a chimeric PV [PV1 (ENPO)] containing the EMCV IRES was used (14a). We had previously shown that EMCV IRES-mediated in vitro translation was not inhibited by IRNA (9). Both the PV-HCV and PV-EMCV chimeras were used to infect the Huh-7 control cells, hepatoma-IRNA cells, and hepatoma cells expressing only the ribozyme. As can be seen in Table 1, the PV-HCV chimera titer was reduced 100-fold in hepatoma-IRNA cells compared to Huh-7 control cells. In contrast, the PV-EMCV titer was not significantly reduced in hepatoma-IRNA cells compared to Huh-7 cells. This is consistent with our previous finding that EMCV IRES-mediated translation is not inhibited by IRNA in vitro (9). Also, the hepatoma-ribozyme cell line was as active in supporting PV-HCV (or PV-EMCV) replication as the control Huh-7 cells. These results suggest that the cloned hepatoma cell line expressing IRNA is not simply less able to support virus replication in general.
TABLE 1.
Virus titers
| Chimera | Virus titer (PFU/ml)a
|
||
|---|---|---|---|
| Huh-7 | Hepatoma-IRNA | Hepatoma-ribozyme | |
| PV-HCV | 5.9 × 108 | 1.0 × 106 | 5.2 × 108 |
| PV-EMCV | 1.8 × 108 | 0.8 × 108 | 1.6 × 108 |
3 × 105 cells in 30-mm-diameter plates were infected with 150 μl of PV-HCV or PV-EMCV chimera (2.5 × 104 PFU/ml) for 24 h. Cell extracts were prepared from infected cells, and virus titers were determined by infecting 2 × 106 HeLa cells and counting plaques after 48 h of infection. Each titer is an average of three experiments.
To confirm the results obtained by using the plaque assay, viral proteins were labeled with [35S]methionine during infection of Huh-7 and hepatoma-IRNA cells with the PV-HCV and PV-EMCV chimeras. Labeled capsid proteins were then immunoprecipitated with anti-PV capsid antiserum and analyzed by SDS-PAGE (Fig. 8). Quantitation of the results showed that the inhibition of individual capsid protein synthesis in hepatoma-IRNA cells infected with PV-HCV varied from 60% (VP0), 82% (VP1), 78% (VP2), and 77% (VP3) compared to that in Huh-7 control cells (Fig. 8; compare lanes 1 and 2). Consistent with our plaque assay results, only marginal inhibition of capsid protein synthesis was observed with the PV-EMCV chimera (lanes 3 and 4). In case of the PV-EMCV chimera, VP0, VP1, VP2, and VP3 were inhibited by 6, 35, 33, and 15% in hepatoma-IRNA cells compared to Huh-7 cells.
FIG. 8.
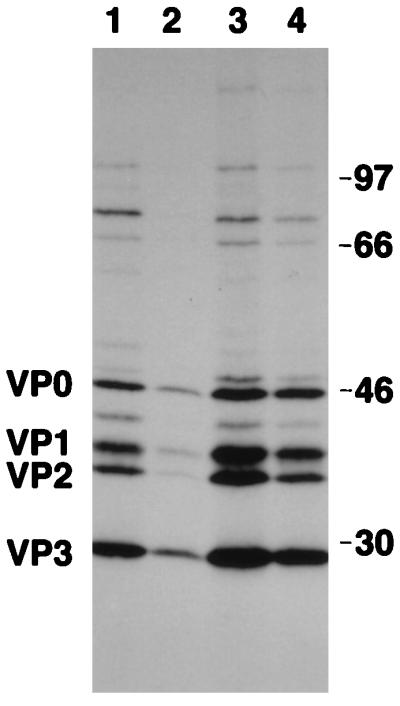
Hepatoma cells expressing IRNA inhibit translation of the PV-HCV chimera. Approximately 3.5 × 105 monolayer hepatoma cells (Huh-7) (lanes 1 and 3) or IRNA-expressing hepatoma cells (lanes 2 and 4) were infected with approximately 3.75 × 103 PFU of either PV-HCV (lanes 1 and 2) or PV-EMCV (lanes 3 and 4) chimera. After 24 h of infection, cells were labeled with [35S]methionine. In vivo-labeled proteins were immunoprecipitated with anti-PV capsid antibody and analyzed on an SDS–14% polyacrylamide gel. The positions of the PV capsid proteins are indicated on the left. Numbers at the right refer to the approximate molecular masses (in kilodaltons) of the 14C-labeled protein markers (Amersham).
IRNA competes with HCV IRES for the La protein.
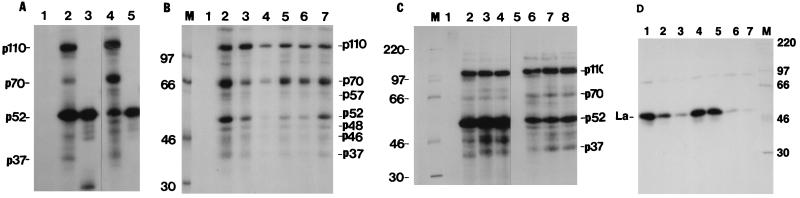
Since the IRNA sequence is not complementary to the 5′ UTR sequences of HCV RNA and is therefore not likely to act as an antisense RNA, we determined whether IRNA was capable of binding cellular proteins believed to be required for HCV IRES-mediated translation. [α-32P]UTP-labeled HCV IRES and IRNA were used to form UV-cross-linked complexes with a HeLa S10 fraction (10, 11). When 32P-labeled HCV IRES was used in the UV-cross-linking experiment, major protein-nucleotidyl complexes were observed at 110, 70, and 52 kDa and minor bands were detected at 100, 57, 55, 48, 46, and 37 kDa (Fig. 9A, lane 4). Similar complexes were also observed when 32P-labeled IRNA was used as the probe (Fig. 9A, lane 2). When purified La was used in the UV-cross-linking experiment, both [32P]IRNA and [32P]HCV IRES (Fig. 9A, lanes 3 and 5) bound the La protein which comigrated with the p52 detected in the S10 fraction. Unlabeled HCV IRES competed with the labeled probe ([32P]HCV IRES) for binding to p110, p70, p57 (a doublet), p52, p48, p46, and p37 (Fig. 9B, lanes 2 to 4). Unlabeled IRNA strongly competed with [32P]HCV IRES for the binding of p52, whereas weak competition was observed with p70, p57, p48, p46, and p37 (Fig. 9B, lanes 5 and 6). A nonspecific RNA was not as effective as HCV IRES or IRNA in the competition assay (Fig. 9B, lane 7). Approximately 80% of La (p52) bound to [32P]HCV-IRES was competed with unlabeled IRNA (Fig. 9B; compare lanes 2 and 6), whereas only 22% competition was observed with a nonspecific RNA (lane 7). For other proteins (p48, p46, p37, p70, and p110), however, specific competition with IRNA was marginal compared to the control. These results suggest that IRNA specifically competes with HCV IRES for La binding, an observation consistent with a recent result that La specifically stimulates HCV IRES-mediated translation in vitro (2).
FIG. 9.
IRNA binds proteins that interact with HCV 5′ UTR. (A) 32P-labeled IRNA (lanes 1 to 3) and HCV 5′ UTR RNA (lanes 4 and 5) were UV cross-linked to cellular polypeptides, using 30 μg of HeLa S10 fraction (lanes 2 and 4) and 0.3 μg of purified La protein (lanes 3 and 5). Numbers to the left correspond to the molecular masses (in kilodaltons) of the polypeptides indicated. Lane 1 contains the IRNA probe but no S10 extract. (B) Competition UV-cross-linking studies were performed with 32P-labeled HCV 5′ UTR RNA and 15 μg of HeLa S10 in the absence (lane 2) and presence of various unlabeled competitor RNAs (lanes 3 to 7). Lanes 3 and 4, 250- and 500-fold molar excess of unlabeled HCV 5′ UTR; lanes 5 and 6, 250- and 500-fold molar excess of unlabeled IRNA; lane 7, 500-fold molar excess of a nonspecific RNA (polylinker region of pSPluc; Promega). Lane 1 contains the probe but no S10 extract; lane M shows the migration (in kilodaltons) of marker proteins. The numbers to the right correspond to the molecular masses (in kilodaltons) of proteins which cross-link to the labeled HCV 5′ UTR probe. (C) 32P-labeled IRNA (lanes 1 to 4) and HCV 5′ UTR RNA (lanes 5 to 8) were UV cross-linked to cellular polypeptides, using 30 μg of S10 extract of either HeLa cells (lanes 2 and 6), Huh7 cells (lanes 3 and 7), or Huh7 cell line pCDIR.Ribo.ΔT7 (lanes 4 and 8). Lanes 1 and 5 contain the probe but no S10 extract. Numbers to the right correspond to the molecular masses (in kilodaltons) of the polypeptides indicated. Lane M shows the migration (in kilodaltons) of marker proteins. (D) Competition UV-cross-linking studies were performed with 32P-labeled HCV 5′ UTR RNA and purified La protein (150 ng) in the absence (lane 1) and presence of 100-fold (lanes 2, 4, and 6) and 200-fold (lanes 3, 5, and 7) molar excesses of unlabeled IRNA (lanes 2 and 3), nonspecific RNA (lanes 4 and 5), and HCV 5′ UTR RNA (lane 6 and 7). Lane M shows the migration (in kilodaltons) of marker proteins. The migration of La protein cross-linked with HCV 5′ UTR is indicated.
To confirm that the 52-kDa polypeptide cross-linked to HCV IRES was indeed La, purified recombinant La protein was UV cross-linked to 32P-labeled HCV IRES and the cross-linked protein was visualized by SDS-PAGE (Fig. 9D). Both unlabeled IRNA (lanes 2 and 3) and HCV IRES (lanes 6 and 7) competed well with the labeled probe for binding to La. A nonspecific RNA, however, was not effective in the competition reaction (lanes 4 and 5).
Because most of our experiments were conducted with hepatoma cells, we compared protein binding to IRNA and HCV IRES between HeLa and hepatoma cells (note that the UV-cross-linking studies described above used a HeLa S10 fraction). Translation cell extracts were prepared from HeLa (Fig. 9C, lanes 2 and 6), hepatoma (Huh-7) (lanes 3 and 7), and hepatoma cells expressing IRNA (pCDIR.Ribo.ΔT7) (lanes 4 and 8) and used in UV-cross-linking experiments with either [32P]IRNA (lanes 1 to 4) or [32P]HCV IRES (lanes 5 to 8) as the probe. As is evident from Fig. 9C, the protein binding profile of the S10 fraction derived from hepatoma cells was almost identical to that from HeLa cells. However, the band intensities of four cross-linked polypeptides (p37 and a doublet just above it, and a band migrating above p70) were significantly greater in hepatoma cell extracts than in HeLa cell extracts (compare lanes 3 and 4 with lanes 7 and 8).
DISCUSSION
We have shown here that HCV IRES-mediated translation is blocked in vivo and in vitro by a small yeast RNA which was initially characterized as a specific inhibitor of PV IRES-mediated, cap-independent translation (9–11). Because there is no appropriate tissue culture system with which to study HCV infection, we used a PV-HCV chimera in which the PV IRES is replaced by HCV IRES to evaluate the efficacy of IRNA in inhibiting viral replication. Hepatoma cells constitutively expressing IRNA became relatively resistant to the chimeric virus compared to the control hepatoma cells. In vitro translation from a bicistronic mRNA containing CAT- and luciferase-encoding genes flanked by the HCV IRES showed specific inhibition of HCV-mediated translation by IRNA. Finally we demonstrated that binding of the La autoantigen (p52) by the HCV IRES was inhibited in the presence of IRNA. In fact, both IRNA and HCV IRES bound similar proteins when incubated with HeLa cell extract. UV-cross-linking assays using cell extracts prepared from hepatoma cells showed almost the same profile as seen with HeLa cell extract. These results demonstrate that IRNA is capable of specifically inhibiting HCV IRES-mediated translation both in vivo and in vitro.
Although we have demonstrated that IRNA preferentially inhibits HCV IRES-mediated translation both in vivo and in vitro, we cannot completely rule out the possibility that it also inhibits some other critical steps in PV-HCV chimera replication. For example, the direct inhibition of viral RNA synthesis by IRNA would also result in inhibition of viral protein synthesis. In fact, recent evidence suggests that PCBP may be involved in both PV translation and replication (3, 4, 14, 28a). It is not known at present whether IRNA interacts with PCBP; however, such an interaction might interfere with both translation and replication of the viral RNA genome. Since transfection of cells with viral RNA showed results similar to infection with the intact virus, the difference in virus titer seen in hepatoma versus hepatoma-IRNA cells could not be due to a disruption in the PV receptor function (data not shown). Additionally, a PV-EMCV chimera replicated well in the cell line expressing IRNA, suggesting that the cells contained a functional PV receptor. In addition, Northern analysis showed that the stability of viral RNA was unchanged in the hepatoma-IRNA cells compared to control cells (data not shown). Moreover, translation of the PV-HCV 701 chimera was specifically inhibited by IRNA (Fig. 8). Therefore, the reduction in the number of plaques seen in hepatoma-IRNA cells is most likely due to the reduced translation of viral proteins.
At low multiplicities of infection of the viruses (PV and PV-HCV), cells expressing IRNA showed significant resistance (∼100-fold) to virus infection compared to control cells (Fig. 7; Table 1). This could be due to inhibition of primary translation of the input viral RNA in cells expressing IRNA. Multiple rounds of replication and reinfection of control cells, but not of IRNA-expressing cells, resulted in an amplified effect seen in the plaque assay shown in Fig. 7 and Table 1. At higher multiplicities of infections (1 to 5), the virus titer was approximately 10-fold lower in pCDIR.Ribo.ΔT7 cells compared to control cells. This could be due to one or more of the following reasons: (i) the level of IRNA expression was relatively low in the cell line; (ii) the viral 5′ UTR, which competes with IRNA for binding of relevant protein factors, has significantly higher affinity for these proteins than IRNA; (iii) the amount of IRNA in the cytoplasm is low compared to the total amount of expressed IRNA; and finally (iv) all of the IRNA molecules in the cell line may not have been properly folded to assume the right secondary structure required for its translation-inhibitory activity.
The hepatoma cells expressing IRNA grow as well as normal hepatoma cells in tissue culture. The mRNA levels of two genes, encoding GAPDH and β-actin, were found to be identical in both control Huh-7 and hepatoma cells expressing IRNA. Also the long-term expression of IRNA did not affect significantly the overall translation of cellular mRNAs as measured by [35S]methionine incorporation (Fig. 6C). This was surprising since some cellular mRNAs such as the immunoglobulin heavy-chain-binding protein, mouse androgen receptor, and Drosophila antennapedia mRNAs have been shown to use IRES-mediated translation (24, 27). It is possible that cellular mRNAs having IRES elements are also translated in a cap-dependent manner, as almost all mRNAs synthesized in vivo are capped. The normal function of IRNA in yeast is not known. However, sequences spanning the active site of IRNA (11) have been found to be highly homologous with a yeast chromosome 3 fragment (data not shown).
Although PV and HCV IRES elements have little or no sequence homology, their sequences can be organized into similar higher-order structures (5). Recent results from various laboratories and the data presented here suggest that specific factors (such as La) believed to be required for IRES-mediated translation must be common between the two viruses. Although in UV-cross-linking studies with labeled HCV IRES, La was found to be the major polypeptide competed by IRNA, binding of other polypeptides (p37, p46, p48, p57, p70, and p110) was also affected by unlabeled I-RNA (Fig. 9). Whether these proteins play important roles in HCV IRES-mediated translation is not known at present. Our attempts to deplete a HeLa cell extract by passing it through an IRNA-affinity column to determine if addition of La and other proteins would restore translation in depleted extracts have failed (data not shown). The studies presented here do not rule out the possibility of involvement of one or more of these polypeptides in IRES-mediated translation. We have recently determined the secondary structure of IRNA and have found that both a stem and a loop formed by intramolecular folding of IRNA are important for inhibition of viral IRES-mediated translation (unpublished data). The secondary structure of IRNA appears to be very similar to portions of both PV and HCV IRES elements. Future studies involving protein-IRNA interaction and determining the three-dimensional structure of IRNA (or IRNA-La complex) will help elucidate the mechanism of IRES-mediated translation and provide new strategies to develop novel drugs effective against HCV infection.
ACKNOWLEDGMENTS
This work was supported by grant AI-38056 from the National Institutes of Health to A.D. S.G. was supported by grant DK 46952 and the Irma T. Hirschl Charitable Trust.
We thank E. Wimmer and H. Lu for the PV-HCV 701 chimera and Aleem Siddiqui for the pT7DC1-341 HCV bicistronic construct. We are grateful to Arun Venkatesan and Megan Igo for critically reading the manuscript. We thank Rajeev Banerjee, Raquel Izumi, Richard Kimura, and Kathy Weidmen for help and cooperation. We also thank E. Berlin for excellent secretarial assistance.
REFERENCES
- 1.Ali N, Siddiqui A. Interaction of polypyrimidine tract binding protein with 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J Virol. 1996;69:6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blyn L B, Swiderek K M, Richards O, Stahl D C, Semler B L, Ehrenfeld E. Poly r(C) binding protein 2, binds to stem-loop IV of the poliovirus RNA 5′noncoding region: identification by automated ligand chromatography-tandem mass spectrometry. Proc Natl Acad Sci USA. 1996;93:11115–11120. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blyn L B, Towner J S, Semler B L, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown E A, Zhang H, Ping L, Lemon S. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;19:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukh J, Purcell R H, Miller R H. Sequence analysis of the 5′ non-coding region of hepatitis C virus. Proc Natl Acad Sci USA. 1992;89:4942–4946. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choo Q-L, et al. Genomic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo Q L, Kuo G, Weiner A, Wang K S, Overby L, Bradley D, Houghton M. Identification of the major, parenteral non-A, non-B hepatitis agent (hepatitis C virus) using a recombinant cDNA approach. Semin Liver Dis. 1992;12:279–288. doi: 10.1055/s-2007-1007399. [DOI] [PubMed] [Google Scholar]
- 9.Coward P, Dasgupta A. Yeast cells are incapable of translating RNAs containing the poliovirus 5′-untranslated region: evidence for a translational inhibitor. J Virol. 1992;66:286–295. doi: 10.1128/jvi.66.1.286-295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das S, Coward P, Dasgupta A. A small yeast RNA selectively blocks internal initiation of translation programmed by poliovirus RNA: specific interaction with cellular proteins that bind to viral 5′-untranslated region. J Virol. 1994;68:7200–7211. doi: 10.1128/jvi.68.11.7200-7211.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das S, Kenan D J, Bocskai D, Keene J D, Dasgupta A. Sequences within a small yeast RNA required for inhibition of internal initiation of translation: interaction with La and other cellular proteins influences its inhibitory activity. J Virol. 1996;70:1624–1632. doi: 10.1128/jvi.70.3.1624-1632.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukushi S, Katayama K, Kurihara C, Ishiyama N, Hoshino F B, Ando T, Oya A. Complete 5′noncoding region is necessary for the efficient internal initiation of hepatitis C virus RNA. Biochem Biophys Res Commun. 1994;199:425. doi: 10.1006/bbrc.1994.1246. [DOI] [PubMed] [Google Scholar]
- 13.Fukushi S, Kurihara C, Ishiyama N, Hoshino F B, Oya A, Katayama K. The sequence element of the internal ribosome entry site and a 25-kilodalton cellular protein contribute to efficient internal initiation of translation of hepatitis C virus RNA. J Virol. 1997;71:1662–1666. doi: 10.1128/jvi.71.2.1662-1666.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamarnik A V, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 14a.Gromeier M, Alexander L, Wimmer E. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc Natl Acad Sci USA. 1996;93:2370–2375. doi: 10.1073/pnas.93.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han J H, et al. Characterization of the terminal region of hepatitis C viral RNA: identification of conserved sequences in the 5′-untranslated region and poly(A) tails at the 3′end. Proc Natl Acad Sci USA. 1991;88:1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellen C U T, Witherell G W, Schmid M, Shin S H, Pestova T V, Gil A, Wimmer E. A cytoplasmic 57 kd protein (p57) that is required for translation of picornavirus RNA by internal ribosome entry is identical to the nuclear polypyrimidine tract binding protein. Proc Natl Acad Sci USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houghton M, Weiner A, Han J, Kuo G, Choo Q L. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991;14:381–388. [PubMed] [Google Scholar]
- 18.Jang S K, Krausslich H G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaminski A, Hunt S L, Patton J G, Jackson R J. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- 20.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kettinen H, Grace K, Grunert S, Clarke B, Rowlands D, Jackson R. In: Proceedings of the International Symposium on Viral Hepatitis and Liver Diseases, Tokyo. Nishioka K, Suzuki H, Mishiro S, Oda T, editors. 1994. p. 125. [Google Scholar]
- 22.Lolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 23.Lu H H, Wimmer E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of internal ribosomal entry site of hepatitis C virus. Proc Natl Acad Sci USA. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macejak D G, Sarnow P. Internal initiation of translation mediated by the 5′-leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 25.McBartney S, Sarnow P. Evidence for involvement of trans-acting factors in selection of the AUG start codon during eukaryotic translational initiation. Mol Cell Biol. 1996;16:3523–3534. doi: 10.1128/mcb.16.7.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh S K, Scott M P, Sarnow P. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- 28.Okamoto H, Okada S, Sugiyama Y, Kurai K, Iizuka H, Machida A, Miyakawa Y, Mayumi M J. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent region. Gen Virol. 1991;72:2697–2704. doi: 10.1099/0022-1317-72-11-2697. [DOI] [PubMed] [Google Scholar]
- 28a.Parsley T B, Towner J S, Blyn L B, Ehrenfled E, Semler B L. Poly (rc) binding protein 2 forms a ternary complex with the 5′-terminal sequence of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 29.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 30.Perrotta A T, Been M D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991;350:434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Rowlands D J, Jackson R J. Unique features of internal initiation of translation of hepatitis C virus translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rijnbrand R, Bredenbck P, van der Straten T, Whetter L, Inchanspe G, Lemon S, Spaan W. Almost the entire 5′non-translated region of hepatitis C virus is required for cap independent translation W. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 33.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe T Y, Koi S, Onji M, Ohta Y, Choo Q-L, Houghton M, Kuo G. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schultz D B, Hardin C C, Lemon S M. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5′-nontranslated RNA of hepatitis A virus. J Biol Chem. 1996;271:14134–14142. doi: 10.1074/jbc.271.24.14134. [DOI] [PubMed] [Google Scholar]
- 35.Svitkin Y V, Meerovitch K, Lee H S, Dholakia J N, Kenan D J, Agol V I, Sonenberg N. Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J Virol. 1994;68:1544–1550. doi: 10.1128/jvi.68.3.1544-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svitkin Y V, Ovchinnikov L P, Dreyfuss G, Sonenberg N. General RNA binding proteins render translation cap dependent. EMBO J. 1996;15:7147–7155. [PMC free article] [PubMed] [Google Scholar]
- 37.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsukiyama-Kohara Z, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witherell G W, Gil A, Wimmer E. Interaction of polypyrimidine tract binding protein with the encephalomyocarditis virus mRNA internal ribosomal entry site. Biochemistry. 1993;32:8268–8275. doi: 10.1021/bi00083a030. [DOI] [PubMed] [Google Scholar]
- 41.Yanagi M, Purcell R H, Emerson S U, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yen J H, Chang S C, Hu C R, Chu S C, Lin S S, Hsieh Y S, Chang M F. Cellular proteins specifically bind to the 5′-noncoding region of hepatitis C virus RNA. Virology. 1995;208:723–732. doi: 10.1006/viro.1995.1204. [DOI] [PubMed] [Google Scholar]