Summary
Lipids and lipid metabolites have essential roles in plant–pathogen interactions. GDSL‐type lipases are involved in lipid metabolism modulating lipid homeostasis. Some plant GDSLs modulate lipid metabolism altering hormone signal transduction to regulate host‐defence immunity. Here, we functionally characterized a rice lipase, OsGELP77, promoting both immunity and yield. OsGELP77 expression was induced by pathogen infection and jasmonic acid (JA) treatment. Overexpression of OsGELP77 enhanced rice resistance to both bacterial and fungal pathogens, while loss‐of‐function of osgelp77 showed susceptibility. OsGELP77 localizes to endoplasmic reticulum and is a functional lipase hydrolysing universal lipid substrates. Lipidomics analyses demonstrate that OsGELP77 is crucial for lipid metabolism and lipid‐derived JA homeostasis. Genetic analyses confirm that OsGELP77‐modulated resistance depends on JA signal transduction. Moreover, population genetic analyses indicate that OsGELP77 expression level is positively correlated with rice resistance against pathogens. Three haplotypes were classified based on nucleotide polymorphisms in the OsGELP77 promoter where OsGELP77 Hap3 is an elite haplotype. Three OsGELP77 haplotypes are differentially distributed in wild and cultivated rice, while OsGELP77 Hap3 has been broadly pyramided for hybrid rice development. Furthermore, quantitative trait locus (QTL) mapping and resistance evaluation of the constructed near‐isogenic line validated OsGELP77, a QTL for broad‐spectrum disease resistance. In addition, OsGELP77‐modulated lipid metabolism promotes JA accumulation facilitating grain yield. Notably, the hub defence regulator OsWRKY45 acts upstream of OsGELP77 by initiating the JA‐dependent signalling to trigger immunity. Together, OsGELP77, a QTL contributing to immunity and yield, is a candidate for breeding broad‐spectrum resistant and high‐yielding rice.
Keywords: disease resistance, lipase, OsGELP77, OsWRKY45, QTL, rice
Introduction
GDSL‐type lipases/esterases represent a variety of lipolytic enzymes that can hydrolyse diverse lipidic substrates, featured with a conserved GDSL motif GxSxxxxG at the N‐terminus (Akoh et al., 2004; Brick et al., 1995). A subgroup GDSLs is further classified as SGNH hydrolase due to four invariant catalytic residues Ser‐Gly‐Asn‐His in the corresponding four conserved blocks. The four catalytic residues are flexible active sites that can change conformation after the binding of different substrates, which contribute to the lipid hydrolase activity and influence the substrate specificity for GDSLs (Akoh et al., 2004).
GDSLs are widely distributed in all living species from prokaryotes to eukaryotes. In plants, GDSL gene family consists of a wide range of members, such as 105 members in Arabidopsis (Arabidopsis thaliana) and 115 members in rice (Oryza sativa L.) (Chepyshko et al., 2012; Lai et al., 2017; Zhang et al., 2020). Accumulating evidence has verified the importance of GDSLs in multiple processes from plant growth and development to biotic and abiotic stress responses (Shen et al., 2022). However, only a few plant GDSLs have been referenced with that their substrates are decoded and their broad biological functions are characterized. Some plant GDSLs play important roles in plant immunity through altering lipid metabolism and hormone signalling (Shah, 2005; Shen et al., 2022). Arabidopsis GDSL LIPASE1 (GLIP1) is a secreted lipase. GLIP1 regulates pathogen resistance in association with ethylene signalling (Kim et al., 2013; Kwon et al., 2009). Enhancement of GLIP1 increased resistance to necrotrophic fungus Alternaria brassicicola, necrotrophic bacteria Erwinia carotovora, and hemibiotrophic bacteria Pseudomonas syringae (Kwon et al., 2009), while loss function of GLIP1 led to susceptible to necrotrophic fungus Alternaria brassicicola (Oh et al., 2005). Arabidopsis GLIP2 possesses lipase activity as GLIP1, however, GLIP2 and GLIP1 play distinct roles in response to pathogen attacks that GLIP2 regulates resistance in association with auxin signalling (Lee et al., 2009). Arabidopsis AtGDSL1 has lipase activity, playing positive roles in against infection of necrotrophic fungus Sclerotinia sclerotiorum (Ding et al., 2020). Ectopic expression of BnGDSL1, the functional homologue of AtGDSL1 of Brassica napus, in Nicotiana benthamiana displayed a significantly reduced disease severity (Ding et al., 2020). Rice OsGLIP1 and OsGLIP2 are identified as negative regulators of plant immune responses. Simultaneous down‐regulation of both of them enhanced rice resistance to bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo) and fungal pathogen Magnaporthe oryzae (M. oryzae), while overexpression of these two genes showed compromised defence (Gao et al., 2017). However, whether other rice GDSL genes play roles in defence response and even play positive roles in against pathogen attacks are unknown.
GDSLs are lipid‐degrading enzymes that have a wide range of substrates due to their flexible conformation (Neves Petersen et al., 2001). Several plant GDSLs including Arabidopsis GLIP2, rice OsGLIP1 and OsGLIP2 have lipolytic activity toward universal substrates such as p‐nitrophenyl acetate or p‐nitrophenyl butyrate by in vitro lipase activity assays (Gao et al., 2017; Lee et al., 2009). However, the identification of native substrates for plant GDSLs in planta is a great challenge. The biochemical functions of plant GDSLs are largely concluded by in vivo lipidomics analysis that based on which kinds of lipid species have altered contents when GDSL genes are deleted or overexpressed (Gao et al., 2017; Lee et al., 2009; Shen et al., 2022).
Lipid metabolism supplies precursors for jasmonic acid (JA) biosynthesis (Lavell and Benning, 2019). Plastid‐targeted rice lipase EG1 participates in JA biosynthesis. The loss‐of‐function of eg1 displayed defects in spikelet development and had decreased free JA in spikelets, while exogenous MeJA treatment rescued the abnormal spikelet (Cai et al., 2014). As the best‐studied lipid metabolite, JA has vital functions as a signal molecule in plant response to pathogen attacks. Rice allene oxide synthase genes OsAOS1 and OsAOS2 are two key JA biosynthesis genes in rice. Overexpression of OsAOS1 and OsAOS2 in rice increased the endogenous JA level and enhanced rice resistance to Xoo and M. oryzae (Hou et al., 2019; Mei et al., 2006). In plants, some minor perturbation in lipid homeostasis has direct consequences on JA signalling (Lavell and Benning, 2019). Both Arabidopsis AtPAD4 and rice OsPAD4 are lipases, while they adopt different mechanisms against pathogen infection. AtPAD4 plays a negative role in response to pathogen infection relying on JA signalling pathway (Brodersen et al., 2006), while OsPAD4 plays a positive role in the fight against pathogen by enhancing JA accumulation (Ke et al., 2014). The deciphered mechanisms support that both JA accumulation and JA signal transduction play critical roles in mediating defence responses against different pathogens (Ghorbel et al., 2021; Wan and Xin, 2022). Moreover, JA plays vital roles in determining grain yield by regulating spikelet formation and development (Deveshwar et al., 2020).
Rice serves as the major food crop, its production is restrained by various diseases, such as fungal blast caused by M. oryzae, bacterial blight caused by Xoo, and bacterial leaf streak caused by Xanthomonas oryzae pv. oryzicola (Xoc), leading to yield loss up to 30% (Deng et al., 2020; Kou et al., 2010; Zhao et al., 2022). Accumulating evidence validates that the most effective and sustainable strategy to control rice diseases is the genetic improvement, including pyramiding R genes and resistance quantitative trait locus (QTL). Compared with R gene‐mediated pathogen race‐specific resistance which can be overcome by pathogens in a short time, rice resistance QTL can confer pathogen species‐non‐specific resistance, that is, durable and broad‐spectrum resistance (Deng et al., 2020). Although a large number of QTL have been mapped in different rice accessions, the causal QTL genes have finitely been identified and the underlying mechanisms of them against pathogen attack are seldomly characterized (Deng et al., 2020; Kou et al., 2010; Zhao et al., 2022).
Here, we functionally characterized OsGELP77, a QTL contributing to broad‐spectrum resistance in rice, encodes a GDSL‐type lipase. Both in vitro biochemical assays and in vivo lipidomic profiling assays demonstrated that OsGELP77 modulates lipid metabolism. Overexpression of OsGELP77 or pyramiding of a natural elite haplotype of OsGELP77, OsGELP77 Hap3 which has a high expression level, could significantly increase lipase activity and JA accumulation, resulting in increased resistance to various pathogens and improved yield. In addition, OsWRKY45 directly binds to the OsGELP77 promoter and activates OsGELP77 expression, initiating the JA‐dependent signalling pathway to trigger immunity. Therefore, our results suggest that OsGELP77 could be applied for genetic improvement of rice.
Results
Induction of OsGELP77 expression against pathogen challenge in rice
OsGELP77 expression was induced upon bacterial pathogen Xoo infection from our previous transcriptome data (Hong et al., 2015). Here, we detailedly analysed OsGELP77 transcript level in a time course upon Xoo infection along with water inoculation as mock control using RT‐qPCR assay. In line with transcriptome data, Xoo infection induced OsGELP77 expression, reaching the peak at 12 h post infection (hpi) (Figure S1a), while mock inoculation could not reprogram its transcription. Simultaneously, we assessed OsGELP77 expression patterns after bacterial pathogen Xoc or fungal pathogen M. oryzae infection. Similarly, OsGELP77 expressions were markedly induced when rice leaves were challenged with Xoc or M. oryzae but not the corresponding mock control (Figure S1b,c), suggesting OsGELP77 may act as a pathogenic responsive gene. The expression of many pathogenic responsive genes is usually regulated by defence signal molecules such as salicylic acid (SA) or JA (Yang et al., 2013). In order to determine whether OsGELP77 was regulated by these inducers of defence responses, rice leaves were treated with SA and JA along with water as mock control, and then OsGELP77 expression patterns were analysed. Results showed that OsGELP77 expressions were elevated upon JA treatment, while were not altered after SA or mock treatment (Figure S1d). Together, these results demonstrate that OsGELP77 might have roles in rice immune responses.
OsGELP77 positively regulates rice resistance to various pathogens
Because of the activation of OsGELP77 after bacterial or fungal pathogens infection, it would be reasonable to expect that OsGELP77 is involved in plant defence against various pathogens. To analyse OsGELP77 roles in disease resistance, we generated OsGELP77 overexpressing transgenic plants (OsGELP77‐OE) in ZH11 where the maize ubiquitin promoter was adopted to drive OsGELP77 expression. Eight independent OsGELP77‐OE T0 plants where OsGELP77 expression levels were significantly increased were selected for further study (Figure S2). Parallelly, loss‐of‐function mutants of OsGELP77 were generated by CRISPR/Cas9‐mediated mutagenesis in ZH11 background where two sgRNA matching the first exon of OsGELP77 were designed for Cas9 cleavage (Figure S3a). Two loss‐of‐function allelic mutants osgelp77‐1 and osgelp77‐2 which contained 99‐bp and 100‐bp deletions, respectively, in the OsGELP77 genomic DNA, were selected for the following study (Figure S3b,c). To remove the potential off‐targets mutant, the osgelp77‐1 and osgelp77‐2 mutants were backcrossed with wild type ZH11 and generated Cas9‐free mutant and wild type pairs for OsGELP77 gene for subsequent analysis.
We first evaluated disease responses of the OsGELP77‐OE plant and osgelp77 mutant to Xoo. All the OsGELP77‐OE T0 plants had shorter lesion lengths after Xoo PXO99 inoculation than wild type at the booting stage (Figure S2). Furthermore, we inoculated the representative OsGELP77‐OE plant and osgelp77 mutant with a set of Xoo strains (PXO71, PXO61, PXO341, PXO347). The OsGELP77‐OE plants displayed enhanced resistance to different Xoo strains, as evidenced by 3.2‐ to 4.2‐cm shorter lesion lengths than wild type (Figure 1a,b). In agreement, Xoo growth in the OsGELP77‐OE leaves was decreased, with about 58‐fold lower Xoo population at 12 dpi than wild type (Figure 1c). Reversely, the osgelp77 mutants displayed decreased resistance to these Xoo strains, as evidenced by 2.5‐ to 5.1‐cm longer lesion lengths and 26‐fold higher in planta Xoo population at 12 dpi than wild type (Figure 1a–c).
Figure 1.
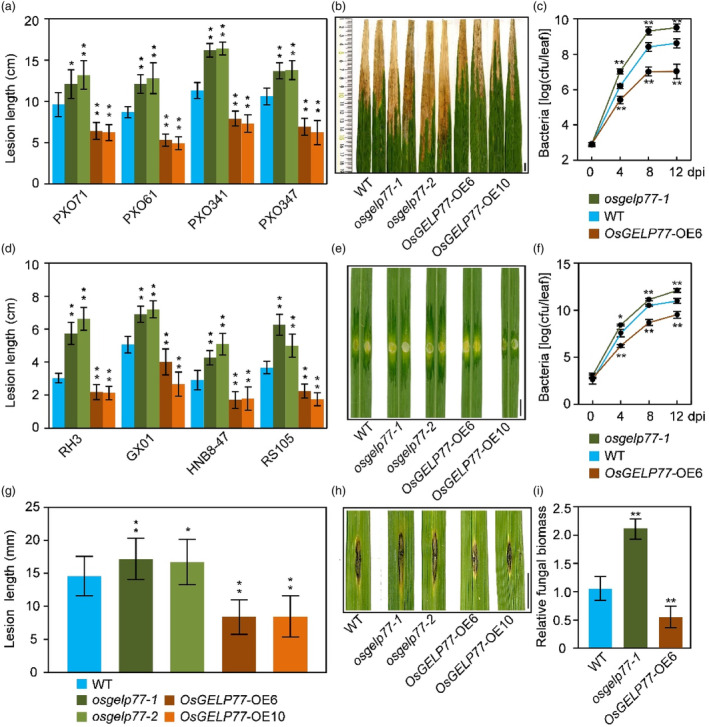
OsGELP77 positively confers rice resistance to various pathogens. (a) Response of the OsGELP77‐OE plants and osgelp77 mutants to different Xoo strains. Plants were inoculated with Xoo at the booting stage. (b) Phenotype of the OsGELP77‐OE plants and osgelp77 mutants after Xoo PXO341 infection. Scale bar: 1 cm. (c) Growth of Xoo PXO341 in leaves of the OsGELP77‐OE6 plant and osgelp77‐1 mutant. (d) Response of the OsGELP77‐OE plants and osgelp77 mutants to different Xoc strains. Plants were inoculated with Xoc at the tillering stage. (e) Phenotype of the OsGELP77‐OE plants and osgelp77 mutants after Xoc RS105 infection. Scale bar: 1 cm. (f) Growth of Xoc RS105 in leaves of the OsGELP77‐OE6 plant and osgelp77‐1 mutant. (g) Response of the OsGELP77‐OE plants and osgelp77 mutants to M. oryzae isolate 99–20‐2. Plants were inoculated with M. oryzae at the tillering stage. (h) Phenotype of the OsGELP77‐OE plants and osgelp77 mutants after M. oryzae infection. Scale bar: 1 cm. (i) Relative fungal biomass of M. oryzae in leaves of the OsGELP77‐OE6 plant and osgelp77‐1 mutant. dpi, days post infection. Data represent means ± SD. n = 30 (a, d, g), n = 6 (c, f, i). Asterisks in (a, c, d, f, g, i) indicate significant differences between wild type (WT) and the OsGELP77‐OE plants or osgelp77 mutants determined by two‐tailed Student's t‐test at **P < 0.01 or *P < 0.05.
Then, we investigated OsGELP77 roles in resistance to Xoc by inoculating the OsGELP77‐OE plant, osgelp77 mutant, and wild type with different Xoc strains (RH3, GX01, HNB8‐47, RS105). The OsGELP77‐OE plants showed enhanced resistance to various Xoc strains, with nearly 60%–73% decreased lesion length (Figure 1d,e) and 15‐ to 40‐fold lower in planta Xoc population than wild type (Figure 1f). However, the osgelp77 mutants showed reverse phenotype relative to the OsGELP77‐OE plants against Xoc infection, as evidenced by longer lesion length and more Xoc growth than wild type (Figure 1d–f).
In parallel, we punch‐inoculated the transgenic plants with M. oryzae at the tillering stage using wild type as control. The OsGELP77‐OE plants showed increased resistance to M. oryzae with approximately 41% decreased lesion length, whereas the osgelp77 mutants showed decreased resistance to M. oryzae with about 18% increased lesion length than wild type (Figure 1g,h). Consistently, the OsGELP77‐OE plants had obviously reduced fungal biomass while the osgelp77 mutants had increased fungal biomass than wild type (Figure 1i). Taken together, these results indicate that OsGELP77 positively regulates rice resistance to multiple pathogens.
OsGELP77 has lipase activity
To investigate the spatial expression profile of OsGELP77, we examined its expression pattern in different tissues including nodes, sheaths, roots, leaves, and panicles. The RT‐qPCR results presented that OsGELP77 was ubiquitously expressed, with the relatively highest transcription in nodes and the lowest expression in roots (Figure 2a). Moreover, we designed an OsGELP77pro:GUS construct where the OsGELP77 promoter was used to drive the reporter GUS gene expression and then transformed the construct into ZH11 to generate transgenic plants. GUS staining of transgenic rice plants expressing OsGELP77pro:GUS revealed that the GUS signals were ubiquitously detected in various rice tissues (Figure 2b). The GUS staining results were completely consistent with the RT‐qPCR results, confirming the widely expressed pattern of OsGELP77.
Figure 2.
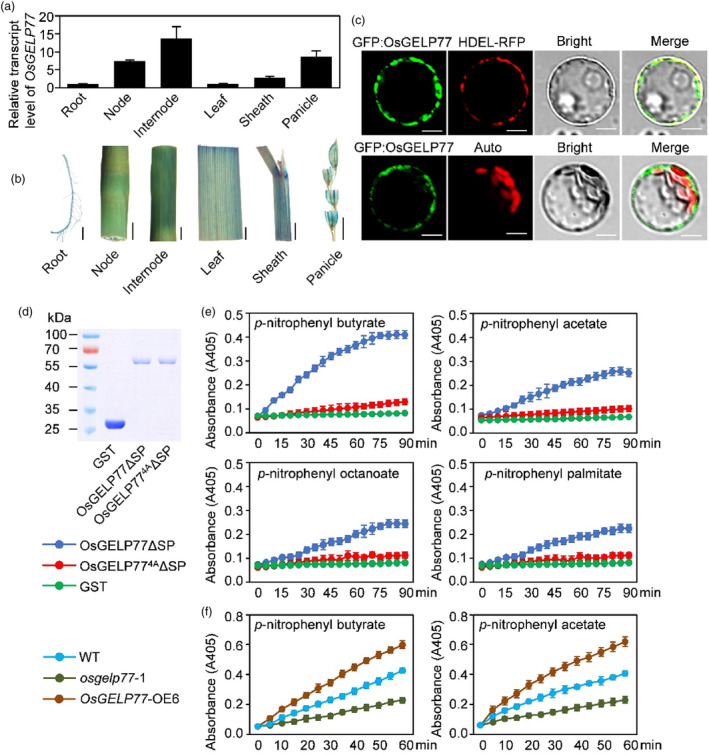
Expression patterns of OsGELP77 and lipase activity of OsGELP77. (a) OsGELP77 expression in different tissues. Data represent means ± SD (n = 3). Gene expression analysis was performed by RT‐qPCR and normalized to Actin. (b) Histochemical staining of different tissues of OsGELP77pro:Gus transgenic lines. Scale bars: 1 cm. (c) Subcellular localization of OsGLEP77 in rice protoplasts. HDEL protein fused to RFP was used as an endoplasmic reticulum marker. Auto, chlorophyll autofluorescence. Scale bars: 10 μm. (d) Expression and purification of recombinant OsGELP77ΔSP‐GST and OsGELP774AΔSP‐GST proteins in E. coli. (e) Recombinant OsGELP77ΔSP‐GST and OsGELP774AΔSP‐GST proteins were incubated with p‐nitrophenyl butyrate, p‐nitrophenyl acetate, p‐nitrophenyl octanoate, or p‐nitrophenyl palmitate. The absorbance readings were collected every 5 min in a time course of 90 min. (f) Total proteins from the leaves of transgenic plants and wild type (WT) were incubated with p‐nitrophenyl butyrate and p‐nitrophenyl acetate. The absorbance readings were collected every 5 min in a time course of 60 min. Data represent means ± SD. n = 6 (e, f).
To analyse the subcellular localization of OsGELP77, we designed a GFP:OsGELP77 construct where the OsGELP77 cDNA was downstream fused with the green fluorescent protein (GFP) gene and transiently transfected the construct into rice protoplasts. GFP:OsGELP77 exclusively colocalized with HDEL which was upstream fused to the red fluorescent protein (RFP) as an endoplasmic reticulum (ER)‐localized marker protein, while GFP:OsGELP77 did not colocalized with chloroplast (Figure 2c), confirming OsGELP77 is an ER protein. Moreover, pathogen‐associated molecular pattern flg22 treatment could not alter ER‐localized OsGELP77 sublocalization (Figure S4a).
Comparison of OsGELP77 protein sequence with those of functionally characterized paralogues or orthologues showed a high degree of similarity (Figure S4b). Typically, a conserved GDSL motif in the N‐terminal and four invariant key catalytic residues (Ser, Gly, Asn, and His) in the functional blocks were invariably presented among these proteins. It hints that OsGELP77 might have lipase activity copying its paralogues or orthologues. To assess the potential lipase activity of OsGELP77, we expressed OsGELP77 in E. coli and purified it for activity assay. Because of the signal peptide in the N‐terminal, we expressed GST‐tagged OsGELP77ΔSP recombinant protein without the signal peptide (Figure 2d). After purification, the recombinant proteins were incubated with four lipid substrates p‐nitrophenyl butyrate, p‐nitrophenyl acetate, p‐nitrophenyl octanoate, and p‐nitrophenyl palmitate which are generally used for lipase activity assay (Gao et al., 2017). We found OsGELP77ΔSP could hydrolyse these four lipid substrates with high efficiency compared to GST control, and OsGELP77 preferred to hydrolyse p‐nitrophenyl butyrate in these four substrates (Figure 2e), suggesting that OsGELP77 processes a lipase activity. Simultaneously, we expressed GST‐tagged OsGELP774AΔSP recombinant protein where the four invariant key catalytic residues in the functional blocks were mutated to alanine. Unsurprisingly, the purified OsGELP774AΔSP protein completely could not hydrolyse these four lipid substrates (Figure 2e), demonstrating the key role of the four conserved catalytic residues. In addition, we measured the lipase activity of total proteins in the leaves of OsGELP77 transgenic plants. Lipase activity of total proteins from the OsGELP77‐OE plants was higher, while that from the osgelp77 mutants was lower than wild type (Figure 2f). Together, these results suggest that ER‐localized OsGELP77 is a functional lipase.
OsGELP77 modulates lipid metabolism
To uncover the role of OsGELP77 in lipid metabolism, we conducted lipidomic profiling on leaves of the OsGELP77 transgenic plants. A total of 12 classes of lipids including phosphatidic acid (PA), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylinositol (PI), phosphatidylserine (PS), free fatty acids (FFA), diacylglycerol (DAG), triacylglycerol (TAG), digalactosyldiacylglycerol (DGDG), monogalactosyldiacylglycerol (MGDG), and sulfoquinovosyldiacylglycerol (SQDG) were identified and quantified by LC‐ESI‐MS/MS. Total contents of PA, PS, FFA, and TAG were significantly higher in the OsGELP77‐OE plants, while lower in the osgelp77 mutants than wild type. Reversely, total levels of PG, PI, and DAG were obviously decreased in the OsGELP77‐OE plants, while increased in the osgelp77 mutants than wild type (Figure 3a). Moreover, total contents of PC, PE, DGDG, MGDG, and SQDG had no significant difference between in these transgenic plants and wild type.
Figure 3.
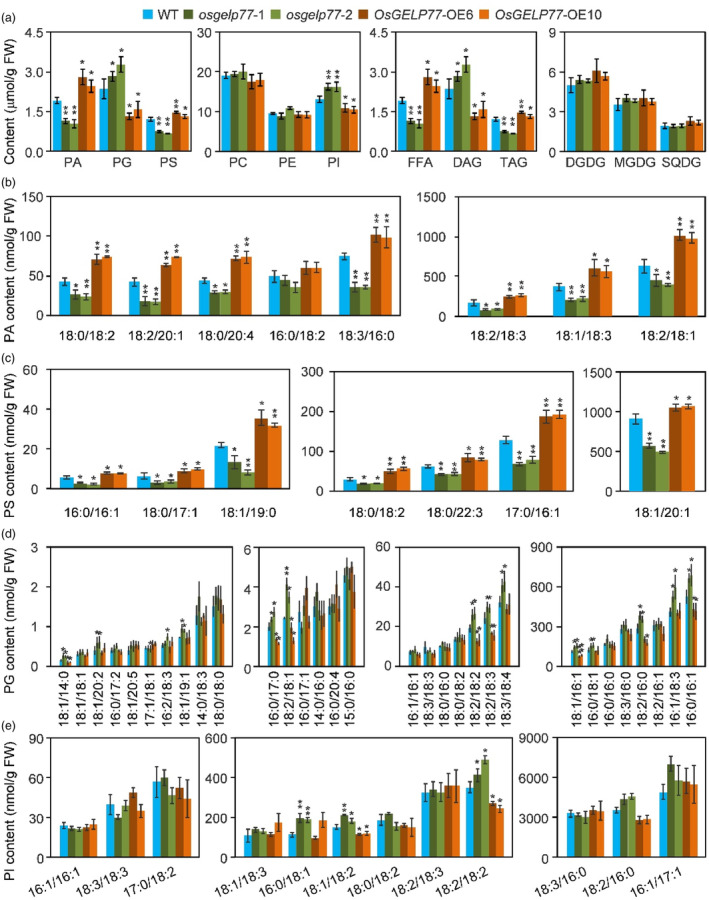
Lipidomic profiling of the OsGELP77‐OE plants and osgelp77 mutants. (a) Total lipid composition in the leaves of transgenic plants and wild type (WT). (b) Contents of eight PA species. (c) Contents of seven PS species. (d) Contents of 31 PG species. (e) Contents of 12 PI species. The individual lipid species is presented as the XX:Y nomenclature where XX is the number of carbon atoms and Y is the number of double bonds in the fatty acyl groups. Data represent means ± SD (n = 3). Asterisks in (a–e) indicate significant differences between WT and the OsGELP77‐OE plants or osgelp77 mutants determined by two‐tailed Student's t‐test at **P < 0.01 or *P < 0.05.
We further quantified the contents of 379 individual lipid molecular species in the OsGELP77‐OE plant and osgelp77 mutant. Seven of eight PA species were accumulated in the OsGELP77‐OE plants and decreased in the osgelp77 mutants than wild type, except for 16:0/18:2 PA which had comparable levels in the transgenic plants and wild type (Figure 3b). Seven PS species all had enhanced accumulation in the OsGELP77‐OE plants and declined levels in the osgelp77 mutants relative to wild type (Figure 3c). For PG, we quantified and analysed 31 PG species, eight of them had lower contents in the OsGELP77‐OE plants, while 13 of them had higher levels in the osgelp77 mutants than wild type (Figure 3d). For 12 PI species, only two of them (18:1/18:2, 18:2/18:2) had decreased contents in the OsGELP77‐OE plants, and increased levels in the osgelp77 mutants than wild type (Figure 3e). The quantified 29 PC and 20 PE species had similar contents between in transgenic plants and wild type (Figure S5a,b). Moreover, we quantified 28 FFA and found that the OsGELP77‐OE plants had a significant accumulation of 18:1 and 18:2, whereas the osgelp77 mutants had an obvious decline of 18:1 and 18:2 (Figure S6). We further quantified and analysed 26 DGDG, 37 MGDG, and 25 SQDG in the transgenic plant leaves. Although there were similar total contents of DGDG, MGDG, or SQDG between in the OsGELP77‐OE plants or the osgelp77 mutants and wild type, five DGDG species (Figure S7), six MGDG species (Figure S8), and four SQDG species (Figure S9) had altered contents. In addition, we analysed 49 DAG and 107 TAG contents, with the results that at least three DAG had lower levels in the OsGELP77‐OE plants and higher levels in the osgelp77 mutants (Figure S10), whereas one‐third of TAG species had enhanced contents in the OsGELP77‐OE plants and decreased contents in the osgelp77 mutants (Figure S11), relative to those in wild type.
To further analyse the lipidomic data, we performed KEGG enrichment analysis to annotate the potential implication of different lipids. The major enriched metabolic pathways both in the OsGELP77‐OE plant and the osgelp77 mutant were glycerolipid metabolism, glycerophospholipid metabolism, and sphingolipid metabolism (Figure S12a,b), suggesting that the altered metabolic pathways were associated with the biochemical function of OsGELP77.
OsGELP77‐modulated lipid metabolism alters JA homeostasis
To decipher the underlying mechanism of OsGELP77‐modulated lipid metabolism participating in immune response, we analysed the transcriptome profile of the osgelp77 mutant and wild type leaves. Totally, 106 up‐regulated differentially expressed genes (DEGs) and 100 down‐regulated DEGs were identified in the osgelp77 mutant compared with wild type (Tables S1 and S2). The GO term analysis showed that enriched GO terms for biological process included JA‐mediated signalling pathway. KEGG pathway enrichment analysis presented that the mainly enriched pathway contained plant hormone signal transduction. Both the GO and KEGG enrichment analysis for DEGs of OsGELP77 were related with JA (Figure S13), which echoed with the infer that OsGELP77 modulates lipid metabolism as JA is lipid‐derived phytohormone.
Among the DEGs, OsJAZ8, acting as a repressor of JA signalling and negatively regulating the JA‐induced resistance to Xoo (Sun et al., 2022; Yamada et al., 2012), had enhanced level in the osgelp77 mutant, while OsMYC2 encoding a global regulator of JA signalling and positively regulating rice resistance against Xoo (Uji et al., 2016), had compromised expression in the osgelp77 mutant. RT‐qPCR assays confirmed higher expression of OsJAZ8 and lower expression of OsMYC2 in the osgelp77 mutant, and reversely, lower expression of OsJAZ8 and higher expression of OsMYC2 in the OsGELP77‐OE plant (Figure S14). Since OsJAZ8 and OsMYC2 are induced by JA and pathogens, participating in JA‐dependent signal transduction (Uji et al., 2016; Yamada et al., 2012). The expressions of OsJAZ8 and OsMYC2 were induced in the leaves of OsGELP77‐OE plants, osgelp77 mutants, and wild type after Xoo inoculation, however, the induced tendency was obviously higher for OsMYC2 in the OsGELP77‐OE plant and OsJAZ8 in the osgelp77 mutant than those in wild type (Figure S14).
The above results promoted us to evaluate JA content in the transgenic plants. Consistent with altered lipid homeostasis and changed expression patterns of JA‐dependent signalling pathway genes, JA contents were 2.1‐ to 2.3‐fold higher in the OsGELP77‐OE plants and 1.9‐ to 2.7‐fold lower in the osgelp77 mutants than wild type (Figure 4a). In agreement, expression levels of OsLOX2 and OsAOS2, which two are JA biosynthesis genes (Ghorbel et al., 2021), were significantly higher in the OsGELP77‐OE plants and lower in the osgelp77 mutants relative to wild type (Figure 4b). Accordingly, JA‐responsive pathogen‐related PR genes such as OsPR1a, OsPR1b, and OsPR5 had significantly enhanced expression levels in the OsGELP77‐OE plants and attenuated expressions in the osgelp77 mutants than wild type (Figure 4c). Moreover, SA content and transcript levels of two representative SA‐responsive genes were comparable between in the OsGELP77‐OE plants or osgelp77 mutants and wild type (Figure 4d,e). These results further support that OsGELP77‐modulated lipid metabolism alters JA content.
Figure 4.
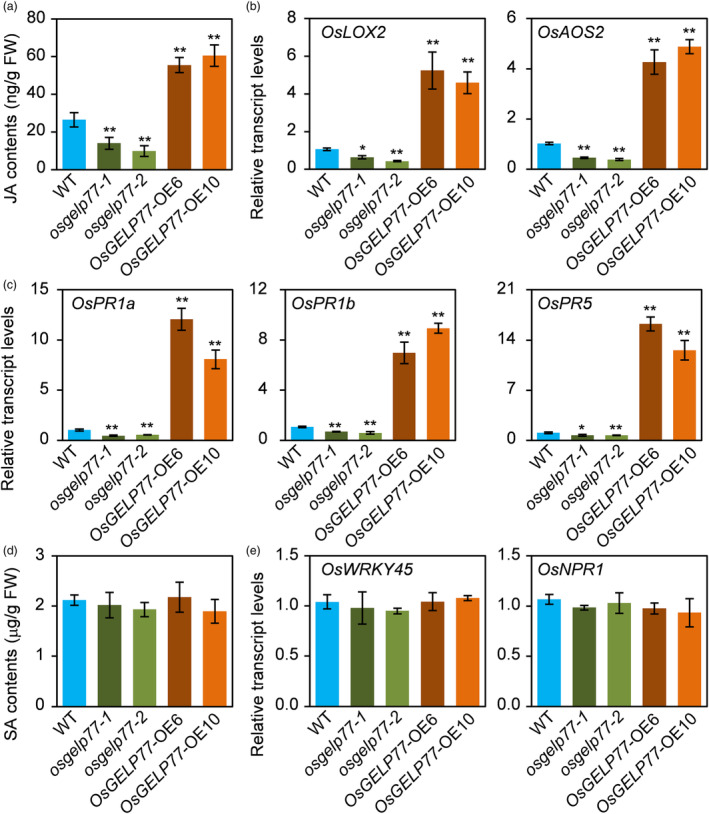
OsGELP77 Alters JA homeostasis. (a) JA contents in the leaves of transgenic plants and wild type (WT). (b) Relative transcript levels of OsLOX2 and OsAOS2 in the leaves of transgenic plants and WT. (c) Relative transcript levels of JA‐responsive PR genes in the leaves of transgenic plants and WT. (d) SA contents in the leaves of transgenic plants and WT. (e) Relative transcript levels of OsWRKY45 and OsNPR1 in the leaves of transgenic plants and WT. Data represent means ± SD (n = 3). Gene expression analysis was performed by RT‐qPCR and normalized to Actin. Asterisks in (a–c) indicate significant differences between WT and the OsGELP77‐OE plants or osgelp77 mutants determined by two‐tailed Student's t‐test at **P < 0.01 or *P < 0.05.
Since JA inhibits seedling growth in plants and this property has been widely used in JA synthesis‐ and signalling‐related mutant selection (Deng et al., 2012). A seed germination assay was carried out to analyse the effects of methyl jasmonate (MeJA) on the growth of OsGELP77 transgenic plants. The results showed that root and shoot elongation of the OsGELP77‐OE plants were more sensitive to MeJA treatment, while root and shoot elongation of the osgelp77 mutants were less sensitive to MeJA application relative to wild type (Figure S15a,b). Taken together, these results suggest that OsGELP77‐modulated lipid metabolism positively regulates JA homeostasis.
OsGELP77‐modulated resistance depends on JA signalling
Since both enhanced JA levels and activated JA signalling mediate plant resistance to pathogen infections (Campos et al., 2014; Zhang et al., 2017b). To further confirm whether OsGELP77‐mediated resistance depends on JA levels or JA signalling, we crossed the OsGELP77‐OE plant with coi1‐13 which is a JA‐insensitive rice line in Nipponbare (Nip) background. In coi1‐13, JA signalling receptor genes OsCOI1A and OsCOI1B were knocked down and JA signal transduction was disrupted (Yang et al., 2012). We found overexpression of OsGELP77 could significantly accumulate JA in coi1‐13 as that in Nip (Figure 5a). In agreement, overexpression of OsGELP77 could activate the transcription of JA‐responsive gene OsVSP which functions downstream of OsCOI1 in JA signal transduction (Yang et al., 2012), in Nip. However, overexpression of OsGELP77 could not induce OsVSP expression in coi1‐13 although there was abundance of JA content (Figure 5b). Accordingly, overexpression of OsGELP77 increased rice resistance to Xoo in Nip, while could not enhance resistance in coi1‐13 (Figure 5c), suggesting that OsGELP77‐mediated resistance largely depends on JA signal transduction.
Figure 5.
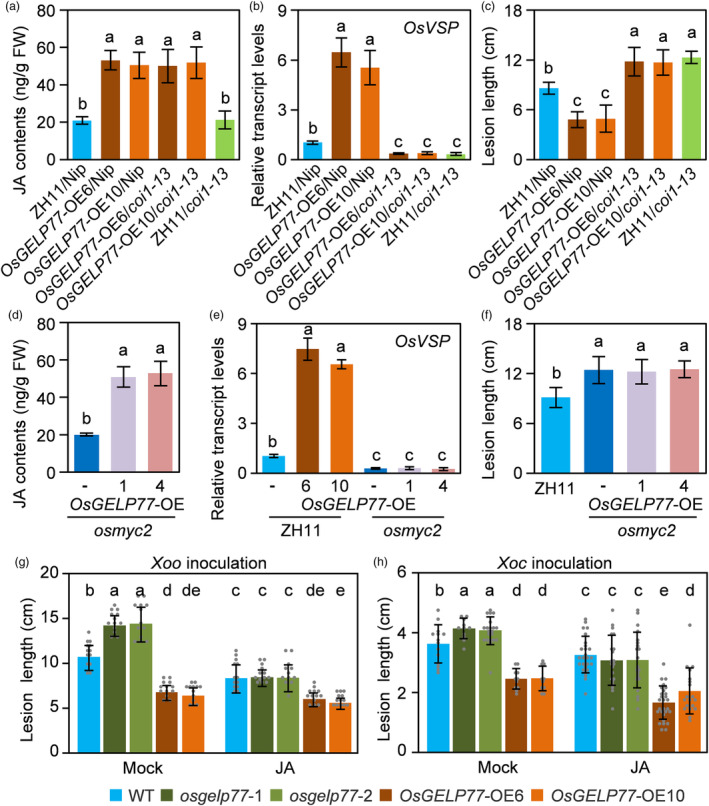
OsGELP77‐mediated resistance is JA signalling dependent. (a) JA contents in the leaves of transgenic plants and the corresponding background. (b) Relative transcript level of OsVSP in the leaves of transgenic plants and the corresponding background. (c) Responses of the transgenic plants and the corresponding background to Xoo strain PXO99. (d) JA contents in the leaves of transgenic plants. (e) Relative transcript level of OsVSP in the leaves of transgenic plants. (f) Responses of the transgenic plants to Xoo strain PXO99. (g) Response of the transgenic plants and wild type (WT) to Xoo strain PXO99. (h) Response of the transgenic plants and WT to Xoc strain GX01. Plants were pretreated with 250 mM JA and two days later were inoculated with Xoo or Xoc. Water inoculation was used as mock control. Data represent means ± SD. n = 3 (a, b, d, e). n = 30 (c, f, g, h). The different letters above each bar in (a, b, c, d, e, f, g, h) indicate statistically significant differences, as determined by one‐way ANOVA analysis followed by Tukey's multiple test (P < 0.05).
Parallelly, we overexpressed OsGELP77 in the osmyc2 mutant (Qiu et al., 2022), to evaluate the relationship between JA signalling disruption and OsGELP77‐mediated resistance (Figure S16). Overexpression of OsGELP77 in osmyc2 mutant could also enhance JA accumulation (Figure 5d). However, OsGELP77 overexpressing could not heighten OsVSP expression in osmyc2 (Figure 5e), and could not reverse the susceptibility of osmyc2 mutant to Xoo (Figure 5f). These data together suggest that JA signalling disruption can attenuate OsGELP77‐mediated rice resistance.
In addition, to experimentally confirm whether OsGELP77‐mediated resistance was JA signalling dependent, we analysed the response of the OsGELP77‐OE plant or osgelp77 mutant to exogenous application of JA. Previous study has validated that exogenous JA treatment reduces the susceptibility of rice to Xoo (Ke et al., 2014). Consistent with previous results, exogenous JA treatment enhanced rice resistance to Xoo in the osgelp77 mutant and wild type, as evidenced by the shorter lesion length compared with mock‐treated plants (Figure 5g). JA application could complement the susceptibility of the osgelp77 mutant, that the mutant even showed shorter lesion length than mock‐treated wild type. Knockout of OsGELP77 compromised the enhanced resistance to Xoo conferred by exogenous JA application, however, the osgelp77 mutant had fewer reduced lesion length compared with JA‐treated wild type. A similar pattern was observed that exogenous JA treatment could clearly enhance resistance to Xoc in the osgelp77 mutant (Figure 5h). Due to high abundance of JA, the enhanced resistance to Xoo and Xoc was not obviously calculated in the OsGELP77‐OE plant after exogenous application of JA. Collectively, these results support the inference that OsGELP77‐modulated lipid metabolism increases JA content which subsequently heightens JA signal transduction triggering plant immunity.
OsGELP77‐modulated lipid metabolism modifies agronomic traits
To further investigate the effect of OsGELP77‐modulated lipid metabolism on rice growth and development, we planted the OsGELP77‐OE plants and osgelp77 mutants in the paddy field along with wild type and examined a set of agronomic traits of them. The OsGELP77‐OE plants had similar agronomic traits including plant height, flag leaf length and width, tiller number, productive panicles per plant, grain width as wild type, while increased panicle length (+4.0% ~ 4.4%), grain length (+2.7% ~ 2.9%), seed setting rate (+4.5% ~ 4.7%), 1000‐grain weight (+2.4% ~ 2.8%), and yield per plant (+3.5% ~ 4.8%). Reversely, the osgelp77 mutants showed decreased tiller number (−19.7% ~ 26.8%), panicle length (−6.1% ~ 9.9%), productive panicles per plant (−14.2% ~ 22.1%), grain length (−1.4% ~ 2.8%), seed setting rate (−9.5% ~ 13.3%), 1000‐grain weight (−2.4% ~ 3.9%), and yield per plant (−7.1% ~ 8.1%) compared with wild type (Figure S17), suggesting that OsGELP77 has positive effects on agronomic traits.
Natural variation in OsGELP77
Given positive roles of OsGELP77 in disease resistance and agronomic traits, we then investigated its variation in rice germplasm. We compared the sequences of OsGELP77 in 2178 rice accessions from the RiceVarMap database (http://ricevarmap.ncpgr.cn/), including 80 tropical japonica, 481 temperate japonica, 823 indica, 686 intermediate varieties, and 108 Aus rice. Polymorphisms were detected exclusively in the 2‐kb promoter but not in the 1942‐bp open reading frame, including 6 single‐nucleotide polymorphisms (SNPs) distributed in the OsGELP77 promoter. Therefore, three distinct haplotypes were classified on the basis of nucleotide polymorphisms in the OsGELP77 promoter (Figure 6a). Of which, OsGELP77 Hap1 and OsGELP77 Hap2 were almost absent in both tropical japonica and temperate japonica, and OsGELP77 Hap1 was absent in Aus rice. An analysis of the geographical distribution of these rice accessions showed that OsGELP77 Hap1 and OsGELP77 Hap2 were mainly distributed in accessions from Asia including China, India, Philippines, Indonesia, and Bangladesh, whereas OsGELP77 Hap3 was distributed in accessions from almost all rice‐producing countries/regions (Figure 6b). We then randomly evaluated disease responses of 109 mini‐core rice accessions which consist of tropical japonica, temperate japonica, indica, intermediate varieties, and Aus rice representing a broad range of rice germplasm (Xie et al., 2015). After Xoo infection, accessions harbouring OsGELP77 Hap3 had shorter lesion lengths than those containing OsGELP77 Hap1 or OsGELP77 Hap2 (Figure 6c). Accordingly, significantly higher expressions of OsGELP77 were observed in leaves of accessions harbouring OsGELP77 Hap3 relative to those accessions containing OsGELP77 Hap1 or OsGELP77 Hap2 (Figure 6d). OsGELP77 expression is positively correlated with the resistance to Xoo, supporting its involvement in the regulation of resistance to Xoo infection.
Figure 6.
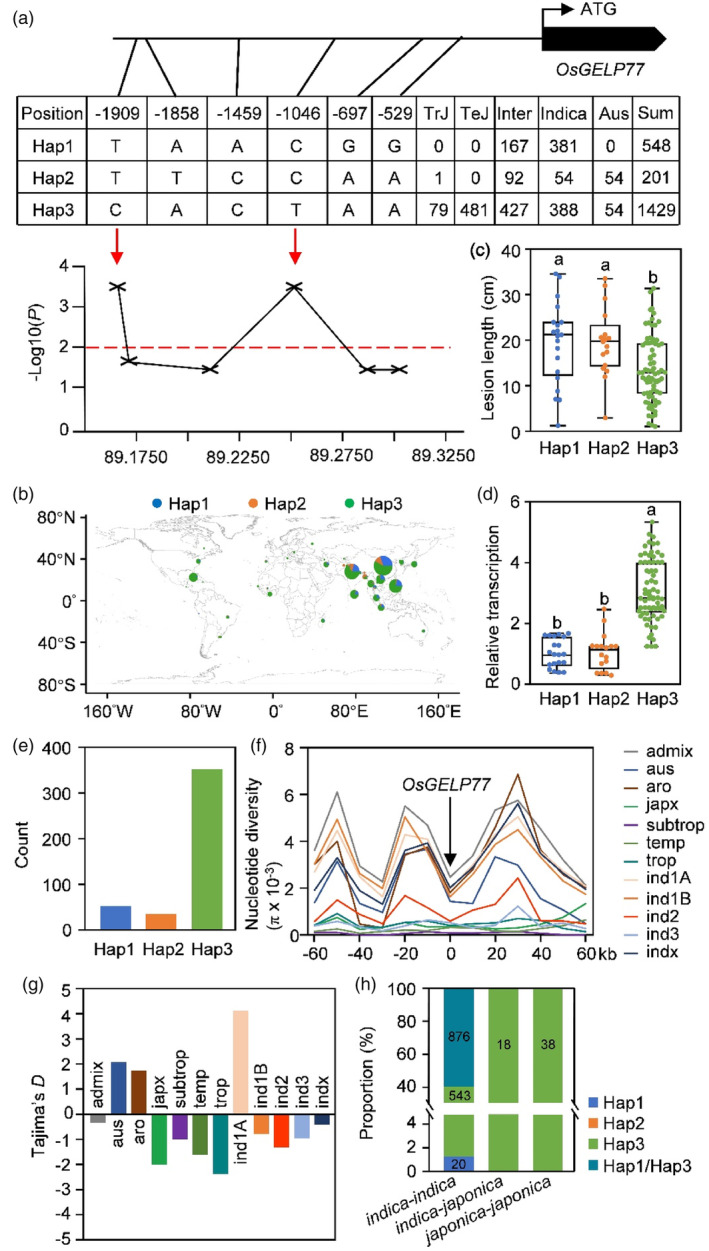
Natural variation in OsGELP77. (a) Natural variation and haplotype analysis of OsGELP77 in 2178 rice accessions. The three haplotypes were based on the six SNPs distributed in the promoter. (b) Geographic distributions of 2178 rice accessions. (c) Disease responses of 109 mini‐core rice accessions harbouring three classes of OsGELP77 haplotype after Xoo PXO99 infection. Plants were inoculated with Xoo at the booting stage. Data represent means ± SD (n = 30). (d) Relative OsGELP77 transcript level of the three classes of haplotypes in leaves. Data represent means ± SD (n = 3). (e) Distributions of three OsGELP77 haplotypes in wild rice. (f) Nucleotide diversity of OsGELP77 and its surrounding 120‐kb region in different rice subgroups. (g) Tajima's D values of OsGELP77 genomic sequences in the subgroups of cultivated rice. (h) Distributions of three OsGELP77 haplotypes in hybrid rice varieties. The different letters above each bar in (c, d) indicate statistically significant differences, as determined by one‐way ANOVA analysis followed by Tukey's multiple test (P < 0.05).
Domestication selection of OsGELP77
Based on association analysis, two functional SNPs for OsGELP77 that SNP (T/C) at −1909 bp and SNP (C/T) at −1046 bp were significantly associated with resistance to Xoo (Figure 6a). To track the evolution origin of two functional SNPs, we compared OsGELP77 sequences from 446 geographically diverse accessions of wild rice species Oryza rufipogon (Huang et al., 2012). The majority of wild rice contains OsGELP77 Hap3 (Figure 6e).
To investigate whether artificial selection has contribution to the domestication of OsGELP77, the genomic sequences of 3 k rice accessions were used to analyse the genetic diversity of this gene and its flanking regions. The genetic diversity (π) for OsGELP77 was relatively lower than its flanking regions (Figure 6f), suggesting a selective sweep at this locus. Among the 12 subgroups of 3 k rice accessions, negative Tajima's D values were observed mainly in the japonica subpopulation and indica subpopulation from South Asia and Southeast Asia but not in the Aus rice and indica subpopulation from East Asia (Figure 6g), implying OsGELP77 underwent differential selection during japonica and South or Southeast Asia indica rice adaptation.
In addition, we investigated the distribution of different OsGELP77 haplotypes in 1495 hybrid rice varieties F1 (Huang et al., 2015). Of the 1439 varieties from indica‐indica hybrid, 20 varieties contain OsGELP77 Hap1 and 543 varieties harbour OsGELP77 Hap3, while the majority (876 varieties) have OsGELP77 Hap1 and OsGELP77 Hap3. The 18 varieties from indica‐japonica hybrid and 38 varieties from japonica‐japonica hybrid all contain OsGELP77 Hap3 (Figure 6h). The results indicate that the elite allele OsGELP77 Hap3 has been broadly pyramided to develop hybrid rice contributing to rice breeding.
OsGELP77 colocalizes with disease‐resistance QTL
Rice accessions containing OsGELP77 Hap3 had high OsGELP77 expression and showed resistance to Xoo compared with those harbouring OsGELP77 Hap1 or OsGELP77 Hap2, which promoted us a tip that whether OsGELP77 is a QTL for disease resistance. To support the hypothesis, we retrieved data on disease‐related QTL from PubMed, Q‐TARO, and RiceQTLPro (Kim et al., 2014; Yonemaru et al., 2010). We found that OsGELP77 loci colocalized with previously reported disease‐related QTL which were derived using different genetic populations (Figure S18). Rice var Minghui 63 shows moderate resistance to M. oryzae isolate V86013 and Xoo strain PXO61, rice var Zhenshan 97 is susceptible to these pathogens. Several QTL for disease resistance against M. oryzae or Xoo were identified in Minghui 63/Zhenshan 97 population (Chen et al., 2003; Kou et al., 2010). Minghui 63 carries OsGELP77 Hap3 and Zhenshan 97 carries OsGELP77 Hap1. Moreover, OsGELP77 expression level was obviously higher in different tissues of Minghui 63 than in Zhenshan 97 (Figure S19). To support that OsGELP77 is a QTL conferring disease resistance, we mapped OsGELP77 in a recombinant inbred line (RIL) segregation population developed from a cross between Minghui 63 and Zhenshan 97. The mapping showed that OsGELP77 colocalized with the curve peak of a resistance QTL against Xoo strain PXO61 and a resistance QTL against M. oryzae isolate V86013 which explained 6.7% and 6.1% of the phenotypic variation of resistance to PXO61 and V86013 in the population, respectively (Figure 7a). The present results suggest that OsGELP77 allele from Minghui 63 is most likely a candidate for the gene underlying the resistance QTL.
Figure 7.
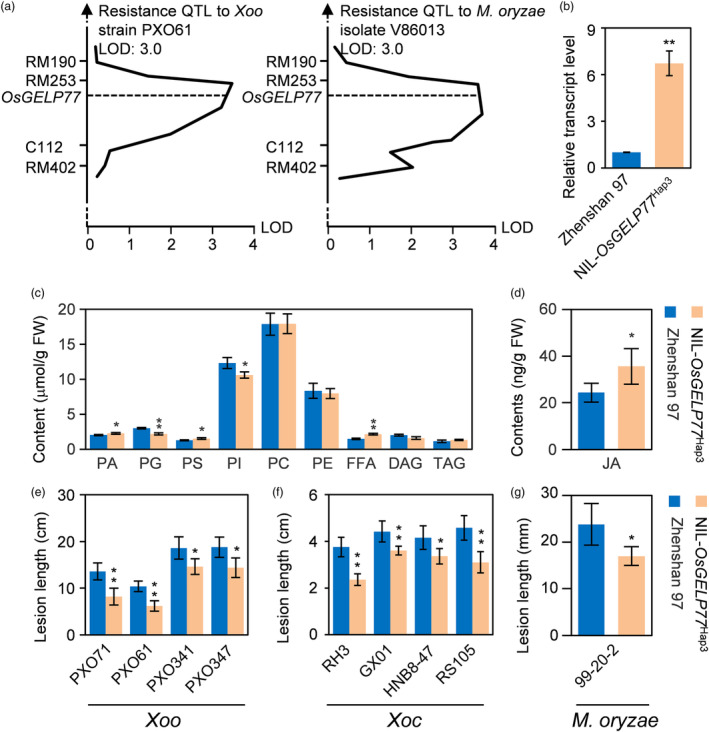
OsGELP77 is a disease‐resistance QTL. (a) Colocalization of OsGELP77 and disease resistance QTL. LOD, Logarithm of odds. (b) Relative transcript level of OsGELP77. (c) Contents of lipid species in the leaves of NIL‐OsGELP77 Hap3 and Zhenshan 97. (d) JA content in the leaves of NIL‐OsGELP77 Hap3 and Zhenshan 97. (e) Response of NIL‐OsGELP77 Hap3 and Zhenshan 97 to different Xoo strains. Plants were inoculated with Xoo at the booting stage. (f) Response of NIL‐OsGELP77 Hap3 and Zhenshan 97 to different Xoc strains. Plants were inoculated with Xoc at the tillering stage. (g) Response of NIL‐OsGELP77 Hap3 and Zhenshan 97 to M. oryzae isolate 99–20‐2. Plants were inoculated with M. oryzae at the tillering stage. Data represent means ± SD. n = 3 (b, c, d), n = 30 (e, f, g). Asterisks in (b, c, d, e, f, g) indicate significant differences between Zhenshan 97 and NIL‐OsGELP77 Hap3 determined by two‐tailed Student's t‐test at **P < 0.01 or *P < 0.05.
To experimentally validate OsGELP77 is a disease‐resistance QTL, we constructed the near‐isogenic line (NIL) NIL‐OsGELP77 Hap3 in the Zhenshan 97 background. Higher OsGELP77 expression was observed in the NIL‐OsGELP77 Hap3 than in Zhenshan 97 (Figure 7b). Accordingly, changes of lipid species levels were observed in the leaves between NIL‐OsGELP77 Hap3 and Zhenshan 97. NIL‐OsGELP77 Hap3 had higher PA and PS contents and lower PG and PI contents than Zhenshan 97 (Figure 7c). NIL‐OsGELP77 Hap3 also had 1.5‐fold higher JA accumulation than Zhenshan 97 (Figure 7d). Furthermore, the responses of NIL‐OsGELP77 Hap3 and Zhenshan 97 to different pathogens were evaluated. After inoculation with both bacterial pathogens Xoo and Xoc and fungal pathogen M. oryzae, the NIL‐OsGELP77 Hap3 exhibited shorter lesion length than its background Zhenshan 97 (Figure 7e–g). These results together support that OsGELP77 is a QTL contributing to broad‐spectrum disease resistance.
Given that OsGELP77 has been reported acting as a trans‐eQTL hot spot controlling seed‐related agronomic traits (Wang et al., 2010). We here confirmed that pyramiding of OsGELP77 Hap3 could enhance seed‐related phenotypes, including increased panicle length, 1000‐grain weight, and yield per plant (Figure S20). Collectively, these data indicate that OsGELP77 Hap3 could improve rice immunity and yield.
OsWRKY45 activates OsGELP77 transcription
In previous studies, OsWRKY45 has been reported acting as a positive regulator in rice resistance against Xoo, Xoc, and M. oryzae, which was accompanied by increased JA accumulation (Shimono et al., 2012; Tao et al., 2009). The similar mechanism of JA‐dependent immunity between OsWRKY45 and OsGELP77 encouraged us to test whether OsWRKY45 genetically acts upstream of OsGELP77. OsWRKY45 functions as a transcriptional activator binding to the canonical W‐box motif with TTGACC core sequence of its downstream target genes (Tao et al., 2009). Notably, the 2‐kb OsGELP77 promoter contains three W‐box motifs which might be OsWRKY45 binding sites. To validate that OsGELP77 is the downstream target gene of OsWRKY45, we conducted a suite of assays. Firstly, we detected OsGELP77 transcription in leaf of OsWRKY45 transgenic plants. RT‐qPCR results showed that OsGELP77 had significantly increased expression in the OsWRKY45‐OE plants, while decreased expression in the oswrky45 mutant than in wild type (Figure 8a). Then, to investigate the direct interaction between OsWRKY45 and OsGELP77 promoter, we conducted EMSA assays to test the binding of OsWRKY45 to the W‐box motif in vitro. The prokaryote‐expressed and purified recombinant fusion protein consisting of His‐TF and OsWRKY45 efficiently bound to W‐box motif‐containing probes, but not mutated probes. Increasing amounts of unlabelled probes could weaken OsWRKY45 binding on labelled W‐box motif (Figure 8b), suggesting that OsWRKY45 specifically targets to W‐box motif within the OsGELP77 promoter. In addition, we performed ChIP‐qPCR assays to evaluate the binding of OsWRKY45 to OsGELP77 promoter. OsWRKY45 binding was enriched at the three W‐box motifs, suggesting that OsWRKY45 bound to the OsGELP77 promoter in vivo (Figure 8c). Lastly, to determine the effect of OsWRKY45 on the OsGELP77 expression, we performed transient expression assays in rice protoplast cells using a construct that OsWRKY45 driven by the Ubiquitin promoter as an effector and a construct that the firefly luciferase gene driven by the intact or mutated OsGELP77 promoter as a reporter. The P OsGELP77 :LUC construct but not the mP OsGELP77 :LUC construct resulted in significantly enhanced LUC activity in protoplasts, conforming transcriptional activation for OsWRKY45 toward on OsGELP77 (Figure 8d). Collectively, these results indicate that OsWRKY45 can directly bind to the promoter of OsGELP77 to activate its transcription.
Figure 8.
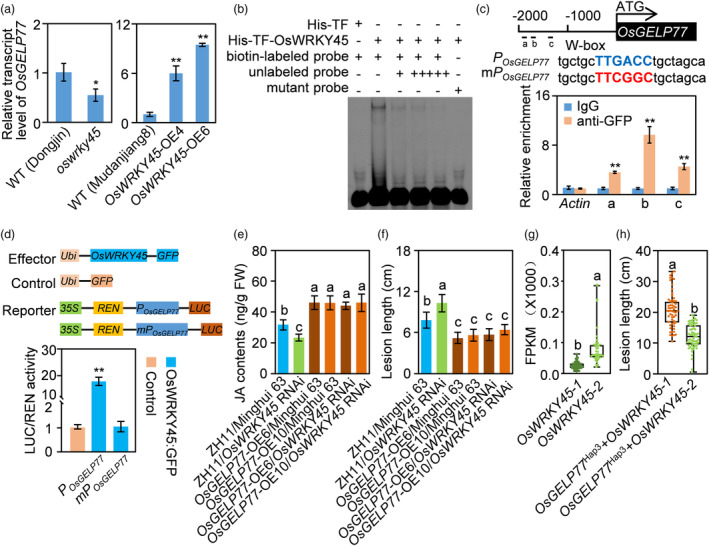
OsWRKY45 genetically acts upstream of OsGELP77. (a) OsGELP77 expression in OsWRKY45‐OE and oswrky45 mutant. (b) DNA binding activity assay of OsWRKY45 by EMSA assay. (c) Binding assay of OsWRKY45 to the promoter of OsGELP77 by ChIP‐qPCR in OsWRKY45:GFP plants using the anti‐GFP antibody. Anti‐GFP antibody was used for immunoprecipitation and IgG acted as a control. The blue capital letters indicate the intact W‐box, the red capital letters represent the mutated W‐box. (d) Activity assay of OsWRKY45 in regulating OsGELP77 expression. (e) JA contents in the leaves of transgenic plants and the corresponding background. (f) Responses of the transgenic plants and the corresponding background to Xoo strain PXO99. Plants were inoculated with Xoo at the booting stage. (g) FPKM values of OsWRKY45‐1 and OsWRKY45‐2 in rice leaf. (h) Responses of rice varieties harbouring OsGELP77 Hap3 and OsWRKY45‐1 or OsWRKY45‐2 to Xoo strain PXO99. Plants were inoculated with Xoo at the booting stage. Data represent means ± SD. n = 3 (a, c, d, e), n = 30 (f). Asterisks in (a, c, d) indicate significant differences between transgenic plants and WT or control determined by two‐tailed Student's t‐test at **P < 0.01 or *P < 0.05. The different letters above each bar in (e, f, g, h) indicate statistically significant differences, as determined by one‐way ANOVA analysis followed by Tukey's multiple test (P < 0.05).
OsWRKY45 genetically acts upstream of OsGELP77
To determine the genetic epistasis between OsWRKY45 and OsGELP77 in the JA‐dependent immunity, we crossed the OsGELP77‐OE plant with OsWRKY45 RNAi line which exhibited susceptible to pathogens in Minghui 63 background. We found overexpression of OsGELP77 could markedly accumulate JA in OsWRKY45 RNAi line as that in Minghui 63, although the OsWRKY45 RNAi line had attenuated JA level relative to the corresponding background (Figure 8e). In agreement, overexpression of OsGELP77 enhanced rice resistance to Xoo in Minghui 63, and could also reverse the susceptibility of OsWRKY45 RNAi line to Xoo (Figure 8f). Collectively, these results support that OsWRKY45 genetically acts upstream of OsGELP77 modulating disease resistance.
Previously, we have demonstrated that japonica rice varieties exclusively carry OsWRKY45‐1, indica rice varieties all carry OsWRKY45‐2. The major difference in DNA sequences between OsWRKY45‐1 and OsWRKY45‐2 is a TE‐siR815b insertion in the first intron of OsWRKY45‐1 (Tao et al., 2009; Zhang et al., 2016). When evaluating the OsWRKY45 expression in leaf transcriptomes of 287 rice varieties (Liu et al., 2022), we found OsWRKY45‐2 expression was higher than OsWRKY45‐1 (Figure 8g). In agreement, rice varieties harbouring OsGELP77 Hap3 and OsWRKY45‐2 had significantly shorter lesion lengths compared with those harbouring OsGELP77 Hap3 and OsWRKY45‐1 after Xoo infection (Figure 8h). These results further confirm the OsWRKY45‐OsGELP77 cascade positively triggers rice immunity.
Discussion
In response to pathogen attacks, plants have evolved a wide range of defence strategies to protect themselves, such as activating plant immune signalling networks, reprogramming transcriptional or translational regulatory systems, modulating phytohormone or secondary metabolite homeostasis (Wang et al., 2022). In this work, we showed that host rice regulates OsWRKY45‐activated OsGELP77 expression to modulate lipid metabolism, resulting in increased JA accumulation and strengthened JA signal transduction to boost immunity.
OsGELP77 modulates lipid metabolism
Of the 115 GDSL genes in rice, nine of them have been functionally characterized, including that OsGELP34/RMS2, OsGELP110, and OsGELP115 control pollen exine development (Zhang et al., 2020; Zhao et al., 2020b), OsGELP64/MHZ11 modulates ethylene signalling in roots (Zhao et al., 2020a), OsGELP62/DARX1 and OsGELP33/DR/BS1/GER1 regulate secondary wall formation and patterning (Riemann et al., 2007; Yu et al., 2020; Zhang et al., 2017a, 2019), OsGLIP1 and OsGELP78/OsGLIP2 negatively modify rice immunity (Gao et al., 2017), and OsGELP112/WDL1 affects cutin formation (Park et al., 2010). Here, we revealed that a new member, OsGELP77, positively regulates rice broad‐spectrum resistance and promotes grain yield. Like rice GDSL lipases reported previously, OsGELP77 exhibits lipase activity toward universal substrates such as p‐nitrophenyl butyrate, p‐nitrophenyl acetate, p‐nitrophenyl octanoate, or p‐nitrophenyl palmitate via the in vitro and in vivo assays (Figure 2e,f). However, the in vivo substrates for OsGELP77 have not yet been identified in this study that is the same as other rice GDSL lipases with substrates unknown. Although lipid profiling has enabled us to compare the abundance of individual lipid species in the OsGELP77‐OE plant and osgelp77 mutant, the precise substrates that OsGELP77 catalysed or the biochemical reactions that OsGELP77 involved are still unclear. We speculate that both the technical limitation of mass spectrometry‐based lipidomics and broad substrates of GDSL lipases might have effects on disclosing the precise substrates for OsGELP77 (Hu and Zhang, 2018).
Based on mass spectrometry‐based lipidomics, 379 individual lipid species belonging to 12 classes of lipids could be identified and quantified in the OsGELP77 transgenic plants. Seven of the 12 classes of lipids had altered accumulations that PA, PS, FFA, and TAG contents were positively correlated with OsGELP77 expression, while PG, PI, and DAG contents were negatively correlated with OsGELP77 expression (Figure 3). The widely altered contents of different classes of lipids in the transgenic plants are possibly related to broad substrates of OsGELP77 because recombinant OsGELP77 could catalyse the hydrolysis of both short‐chain substrates (p‐nitrophenyl butyrate, p‐nitrophenyl acetate, p‐nitrophenyl octanoate) and long‐chain substrate (p‐nitrophenyl palmitate). When evaluating lipid profiling, the OsGELP77 transgenic plants had different kinds of accumulated lipid species compared with such as OsGLIP1, OsGLIP2, or OsGELP64/MHZ11 transgenic plants (Gao et al., 2017; Zhao et al., 2020a), which may the result that these GDSL lipases prefer different substrates in planta. In addition, OsGELP110 and OsGELP115 are localized in the peroxisomes and OsGELP33/DR/BS1/GER1 is localized in the Golgi apparatus, the other seven GDSL lipases are all localized in the ER. The different subcellular localization of GDSL lipases may also have effects on lipid metabolism.
OsGELP77 positively regulates rice immunity and yield
Several GDSL lipases have been reported involved in plant defence responses under different manners. Arabidopsis GLIP1, GLIP2, and AtGDSL1, and Brassica napus BnGDSL1 play positive roles in defence responses (Ding et al., 2020; Kwon et al., 2009; Lee et al., 2009; Oh et al., 2005). In contrast, rice OsGLIP1 and OsGLIP2 play negative roles in against pathogen attacks (Gao et al., 2017). In this study, we proposed that OsGELP77 is a positive regulator of rice immunity that overexpression of OsGELP77 significantly increased rice resistance to bacterial pathogens Xoo or Xoc and fungal pathogen M. oryzae, while OsGELP77 loss‐of‐function mutants decreased disease resistance to various pathogens (Figure 1). Moreover, OsGELP77 acts as a lipase thus modulates lipid metabolism, which ultimately alters JA homeostasis. As JAs are lipid‐derived phytohormones, it is not surprise that overexpression of OsGELP77 enhanced JA accumulation. The OsGELP77 overexpressing plants accumulated more JA and had elevated expressions of plant immune regulators, which supports the widely accepted view that JA accumulation or JA signalling activation indicates constitutive defence activation (Yang et al., 2013). However, because OsGELP77‐modulated lipid metabolism occurred in ER, and JA biosynthesis takes place in chloroplasts and peroxisomes (Wan and Xin, 2022; Yang et al., 2013), the direct relationship between OsGELP77‐modulated lipid metabolism and OsGELP77‐regulated JA accumulation is urgently waiting for exploration in the future.
Similar to OsGLIP1 or OsGLIP2, OsGELP77 possesses lipase activity, however, these three rice GDSL lipases play reverse roles in plant immunity due to their diverse features. Firstly, OsGELP77 expression was induced in response to pathogen infection, while both OsGLIP1 and OsGLIP2 expressions were suppressed against pathogen attacks. Secondly, OsGELP77 expression was elevated upon treatment of defence signalling molecule JA while not SA, however, OsGLIP1 and OsGLIP2 expressions were inhibited after SA treatment. Lastly, OsGELP77 possibly modulated the contents of lipid species including PA, PG, PI, PS, FFA, DAG, and TAG, while OsGLIP1 and OsGLIP2 largely influence the levels of DGDG and MGDG. Therefore, the different expression patterns of these three rice GDSL genes in response to pathogen attacks and defence signalling molecule treatment, and the difference of native lipid substrates for them may coordinately determine their diverse roles in immunity.
Although Arabidopsis GLIP1, GLIP2, AtGDSL1, and rice OsGELP77 positively modulate defence response, they regulate plant immunity through regulation of different phytohormone signalling. GLIP1 modulates immunity through feedback regulation of ethylene signalling (Kim et al., 2013; Kwon et al., 2009). GLIP2 confers pathogen resistance via regulation of auxin signalling (Lee et al., 2009). AtGDSL1‐mediated resistance against pathogens may depend on SA signalling (Ding et al., 2020). However, we here revealed that OsGELP77‐triggered resistance may largely rely on JA signalling. Except for increased expression of JA biosynthetic genes in the OsGELP77‐OE plants, there were also reprogrammed expression of key JA signalling genes, such as increased OsMYC2 and decreased four OsJAZs which have been reported playing critical roles in rice immune responses (Sun et al., 2022; Uji et al., 2016; Yamada et al., 2012). Moreover, the assessment of growth inhibition upon exogenous treatment of JA to seedlings of OsGELP77‐OE plant and osgelp77 mutant revealed that OsGELP77 is involved in JA signal transduction (Figure S15). Clearly, these GDSL genes stimulate host immune responses through the regulation of different phytohormone signalling. OsGELP77 acts as a lipase involved in lipid metabolism, and JA is a class of lipid‐derived phytohormone. Thus, it is logical that OsGELP77 modulates JA accumulation or JA signalling against pathogen attacks. Because OsGELP77 had broad substrates, whether OsGELP77‐catalysed lipid metabolism affects the homeostasis or signalling of other phytohormones that in turn boosts rice immunity should be further investigated.
JA has been reported to increase grain yield through regulating spikelet formation and development in rice (Deveshwar et al., 2020). Both the OsGELP77‐OE plant and NIL‐OsGELP77 Hap3 line had higher JA accumulation. Therefore, overexpression of OsGELP77 and pyramiding an elite haplotype OsGELP77 Hap3 all could promote grain yield. The high proportion of OsGELP77 Hap3 may be attributed to the advocated breeding strategy. In the present study, Minghui 63 presents OsGELP77 Hap3 and Zhenshan 97 presents OsGELP77 Hap1, Shanyou 63, an elite mega rice hybrid in China, is derived from the cross by Minghui 63 and Zhenshan 97. Thus, we could not exclude the contribution of OsGELP77 Hap3 to heterosis. OsGELP77 positively regulates broad‐spectrum disease resistance and promotes yield, thus, it is not surprise that OsGELP77 Hap3 has been widely used to breed hybrid rice.
OsGELP77 is a disease‐resistance QTL
With the accessibility of huge amounts of genome‐wide SNPs and InDels and large amounts of high‐throughput phenotypic data against different pathogens for a large number of rice accessions, a lot of resistance QTL have been identified based on genome‐wide association studies (GWAS) or linkage QTL mapping in rice (Deng et al., 2020; Kou et al., 2010; Liu et al., 2021). However, a great amount of the causal QTL have not been functionally characterized. Contrary to QTL identification by forward genetics through GWAS and QTL mapping, we here by reverse genetics revealed that OsGELP77 is a disease‐resistance QTL which is supported by the following evidence: (1) OsGELP77 co‐located with several disease‐resistance QTL that were derived using different rice accessions (Figure S18); (2) OsGELP77 colocalized with the curve peak of resistance QTL against both bacterial pathogen and fungal pathogen by QTL mapping using a previously developed RIL population (Figure 7a); and (3) OsGELP77 acts as a disease‐resistance QTL which was experimentally validated by evaluating the defence response of the constructed a NIL‐OsGELP77 Hap3 (Figure 7).
OsWRKY45 activates OsGELP77 triggering immunity
OsWRKY45 positively regulates broad‐spectrum resistance to bacterial and fungal pathogens via SA‐ and JA‐dependent signalling pathways (Shimono et al., 2012; Tao et al., 2009). Transcription factors IPA1 and OsARF8 could activate and repress the expression of OsWRKY45, respectively (Feng et al., 2022; Wang et al., 2018). Moreover, OsWRKY45 could activate its downstream target genes including OsWRKY13, OsWRKY62, and OsNAC4 (Cheng et al., 2015; Nakayama et al., 2013). The deciphered transcriptional cascades mainly explain the SA‐dependent signalling pathway for OsWRKY45‐mediated broad‐spectrum resistance. Here, we uncover the JA‐dependent signalling pathway for OsWRKY45‐mediated rice resistance that OsWRKY45 directly targets and activates OsGELP77 expression to modulate lipid metabolism, resulting in increased JA accumulation and strengthened JA signal transduction. The biochemical assays demonstrate that OsWRKY45 binds to OsGELP77 promoter and activates its transcription, the genetic assays confirm that OsWRKY45 genetically acts upstream of OsGELP77 and the OsWRKY45‐OsGELP77 cascade confers rice immunity via JA‐dependent signalling pathway (Figure 8). Published data and present results jointly demonstrate that OsWRKY45 activates different target genes to modulate SA and JA signalling pathways to trigger rice immunity.
In summary, our findings identify that OsGELP77, a QTL contributing to broad‐spectrum disease resistance and yield, encodes a GDSL‐type lipase which modulates lipid metabolism to promote JA accumulation, promoting both immunity and yield. OsWRKY45 can directly bind to the promoter of OsGELP77 and activate its expression, leading to an enhanced JA signalling pathway which triggers rice immunity. The elite haplotype OsGELP77 Hap3 is considered to be a valuable resource for breeding disease‐resistant and high‐yielding rice.
Materials and methods
Plant materials and growth conditions
Rice (Oryza sativa ssp. Geng) cultivar Zhonghua 11 (ZH11) and the transgenic rice in ZH11 genetic background were used for experiments. The OsGELP77‐overexpressing (OsGELP77‐OE) plants and osgelp77 knockout mutants were generated in ZH11 as described below. The coi1‐13 mutants were generated in Nip background (Yang et al., 2012). The osmyc2 mutants were generated in ZH11 background (Qiu et al., 2022). The OsWRKY45‐OE, OsWRKY45‐RNAi, OsWRKY45:GFP, and oswrky45 transgenic plants were generated previously in different background (Cheng et al., 2015; Tao et al., 2009).
Rice materials were planted in the Experimental Station of Huazhong Agricultural University in normal rice growing seasons (from May to October) under natural conditions (Chu et al., 2022).
Generation of constructs and transgenic plants
The full‐length cDNA of OsGELP77 without stop codon was amplified from cDNA of ZH11 leaves by PCR using the specific primers (Table S3), and then the amplified PCR products were cloned into the pU1301 vector via restriction–ligation method to generate the OsGELP77‐OE construct (Yuan et al., 2010). CRISPR/Cas9‐mediated mutagenesis strategy was used to knock out OsGELP77 gene. Two 20‐bp sgRNA designed using the web tool CRISPR‐P (http://crispr.hzau.edu.cn/CRISPR2/) which specifically target the first exon of OsGELP77, were constructed on the sgRNA expression cassettes of OsU6a and OsU3 by overlapping PCR, and then these two cassettes were subcloned into the pYLCRISPR/Cas9 expression vector via Gibson Assembly cloning method to generate the OsGELP77 knockout construct (Ma et al., 2015). The OsGELP77 promoter was amplified from DNA of ZH11 leaves by PCR using the specific primers (Table S3), and then the amplified PCR products were inserted upstream of the reporter β‐glucuronidase (GUS) gene into the DX2181 vector via over‐lapping PCR method to generate the OsGELP77pro:GUS construct (Yang et al., 2022). The insertion fragments of all the constructs were verified by DNA sequencing using the vector‐specific primers (Table S3). All the constructs were transformed into Agrobacterium tumefaciens strain EHA105, which were then transformed into rice calli from mature embryos of ZH11. Transgenic rice plants were generated by Agrobacterium‐mediated transformation (Chu et al., 2022). The positive lines of OsGELP77‐OE plants were verified by PCR using the gene‐specific primer and vector‐specific primer. The homozygous osgelp77 mutants were confirmed by sequencing the genome DNA of OsGELP77.
Pathogen inoculation
For bacterial pathogen inoculation assay, eight to ten uppermost fully expanded leaves of each plant were inoculated with Xoc at the tillering stage or Xoo at the booting stage. Briefly, Xoc or Xoo stocks were streaked on nutrient agar medium (3 g/L beef extract, 5 g/L polypeptone, 1 g/L yeast extract, 10 g/L sucrose, and 20 g/L agar) and incubated at 28 °C for 2 days (Yang et al., 2022). Bacterial cells were collected from the agar medium, suspended in sterilized water, and adjusted to an optical density of 0.5 at 600 nm. Rice leaves were inoculated with Xoc by the penetration method or Xoo by the leaf‐clipping method (Hui et al., 2019). Xoc or Xoo strains isolated from different rice‐growing regions show virulence on rice variety ZH11 and Zhenshan 97 (Hui et al., 2019). Xoc used in this study included four strains (RH3, HNB8‐47, GX01, and RS105). Xoo used in this study included five strains (PXO61, PXO99, PXO341, PXO71, and PXO347). Disease was assessed by counting colony‐forming units every 4 days after Xoc or Xoo inoculation and measuring the lesion length of rice leaves 14 days post bacterial pathogen inoculation (Yuan et al., 2016).
For the fungal pathogen inoculation assay, rice leaves were inoculated with M. oryzae at the tillering stage. Briefly, M. oryzae isolate 99‐20‐2 was cultured on oatmeal tomato agar medium (15% tomato juice, 40 g/L oatmeal, and 20 g/L agar) at 28 °C for 5 days. Conidia were collected from the medium, suspended in sterilized water containing 0.05% Tween‐20, and adjusted to an optical density of 5.0 × 105 spores per millilitre. Rice leaves were inoculated with M. oryzae by the punch inoculation method (Chu et al., 2022). Disease was scored by measuring the lesion length and quantifying fungal biomass at 7 days after M. oryzae inoculation. The fungal biomass in infected rice leaves was analysed using the threshold cycle value of M. oryzae Pot2 DNA against the cycle value of rice genomic ubiquitin DNA (Chu et al., 2022).
MeJA treatment and hormone quantification
Rice seeds were surface‐sterilized with 75% ethanol and 2.5% sodium hypochlorite and pregerminated on 1/2 Murashige and Skoog medium for 2 days. The identically sprouted seeds were transplanted on Murashige and Skoog medium supplemented with different concentrations of MeJA for 7 days. The lengths of primary roots and shoots were measured and seedlings were photographed.
Rice seedlings growing in a controlled growth greenhouse at the 5‐leaf stage were sprayed with 250 μM MeJA which was dissolved in 0.05% (v/v) methanol and 0.05% (v/v) Tween 20 or mock control solution containing 0.05% (v/v) methanol and 0.05% (v/v) Tween 20 until uniformly wet. The treated rice plants were kept in sealed plastic shade for 2 days, and then inoculated with Xoc or Xoo.
For JA and SA quantification, leaves of rice growing in the paddy field at the tillering stage were sampled and ground in liquid nitrogen. About 100 mg powder of each sample was mixed with the extraction buffer (methanol:water:acetic acid = 40:9:1, v/v/v) that supplemented with stable isotope‐labelled internal standards, vigorously shaken in the dark at 4 °C for 16 h, and then centrifuged at 4 °C for 20 min. The supernatants were filtered through a syringe‐facilitated nylon filter, dried by evaporation under the flow of nitrogen gas, and finally dissolved in methanol. JA and SA were quantified by an ultrafast liquid chromatography coupled to electrospray ionization tandem mass spectrometry (UFLC‐ESI‐MS/MS) in an ABI 4000 Q‐TRAR system (Applied Biosystems) according to a previously reported method (Huang et al., 2018).
Gene expression analysis
Total RNA was extracted from different rice tissues using Trizol reagent (Invitrogen). About 5 μg total RNA was pre‐treated with RNase‐free DNase I (Invitrogen) to eliminate potentially contaminating DNA, and then was reverse‐transcribed into cDNA using oligo(dT)15 primer and M‐MLV reverse transcriptase (Promega) according to the manufacturer's protocol. For gene expression analysis, quantitative real‐time PCR (RT‐qPCR) was performed using MonAmp™ ChemoHS qPCR Mix (Monad, China) in the ABI QuantStudio™ (Applied Biosystems). RT‐qPCR was carried out using gene‐specific primers (Table S4) which were designed using the Primer Express v3.0 software (Applied Biosystems). The rice actin gene was used to standardize relative RNA measures as inner control (Chu et al., 2022). Each RT‐qPCR assay was biologically repeated at least twice with similar results, with each repetition having three technical replicates (Yuan et al., 2016). The Student's t‐test was used for the RT‐qPCR analysis.
Transcriptome analysis
Rice leaves at the booting stage of the osgelp77 mutant and wild type were sampled with three biological replicates for whole transcriptome analysis. The assays including library construction and Illumina HiSeq4000 platform‐based sequencing were carried out by Novogene (Beijing, China). The analysis including read quality evaluation, clean reads mapping, transcript abundance measurement, and differential expression analysis was conducted at the Galaxy platform (https://usegalaxy.org/) according to the workflow. The gene with relative expression log2FC ≥1 or log2FC ≤ −1 and the false discovery rate (FDR) < 0.05 considered as differentially expressed genes (Hong et al., 2015). Goatools was used for the Gene Ontology (GO) analysis and KOBAS was used for the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis with an adjusted P value cutoff of 0.05.
Transient expression assays in rice protoplasts
The OsGELP77 promoter (2 kb upstream of the translational ATG start codon) was amplified by PCR using gene‐specific primers (Table S3), cloned into the pGreen II 0800 vector to generate the P OsGELP77 :LUC construct as reporter. The three W‐box motifs of OsGELP77 promoter were mutated using the GeneTailor Site‐Directed Mutagenesis System (Invitrogen Life Technologies) to generate the mP OsGELP77 :LUC construct. The coding sequence of OsWRKY45 was amplified with gene‐specific primers (Table S3), then cloned into the pU1301 vector to generate the Ubi:OsWRKY45 construct as effector. The corresponding reporter and effector constructs were co‐transfected into rice protoplasts as described previously (Ke et al., 2020). After culturing for 16 h in dark at 25 °C, the transfected protoplasts were gathered by centrifugation at 100 g for 8 min. The luciferase activities were measured using Dual Luciferase Reporter Assay System (Promega) by a Spark Multimode Microplate Reader (Tecan, Switzerland) according to the manufacturer's instructions. The relative reporter gene expression levels were expressed as the ratio of firefly luciferase (LUC) to renilla luciferase (REN).
Subcellular localization assays
The full‐length OsGELP77 cDNA was amplified by PCR using the specific primers (Table S3), and then the amplified PCR products were cloned into the pRTVnGFP vector (He et al., 2018), to generate the GFP:OsGELP77 construct where OsGELP77 was in‐frame and downstream of the green fluorescent protein (GFP) that driven by the maize Ubiquitin promoter. After sequencing using the gene‐specific primer, the GFP:OsGELP77 construct was transiently introduced into rice protoplasts which were isolated from 12‐day‐old green rice leaf sheaths via PEG‐mediated transformation (Yang et al., 2022). The protoplasts were incubated at 28 °C for 12 h, and then were visualized for fluorescent signals by a Leica Microsystem (LAS AF, Germany) according to a previously reported protocol (Yang et al., 2022). Alternatively, after incubation for 12 h, the protoplasts were treated with 100 nM flg22 for 2 h, and then visualized for fluorescent signals.
Chromatin immunoprecipitation (ChIP)‐qPCR
The ChIP assays were performed as described previously (Ke et al., 2020). In brief, chromatin was extracted from the OsWRKY45:GFP seedlings (Cheng et al., 2015), and fragmented to 200–400 bp via ultrasound. The OsWRKY45:GFP protein bonding DNAs were enriched using protein A Dynabeads (Invitrogen) coupling with anti‐GFP antibody or IgG as control at 4 °C overnight. After abundant washing and de‐crosslinking, the input and precipitated DNA samples were amplified by qPCR using gene‐specific primers (Table S3).
Electrophoretic mobility shift assays (EMSA)
EMSA assays were performed using Light Shift Chemiluminescent EMSA Kit (Thermo Scientific), as described previously (Ke et al., 2020). In brief, the synthesized 5‐carboxyfluorescein biotin‐labelled probes were incubated with purified His‐TF‐OsWRKY45 recombinant proteins for 30 min at room temperature. The equal and increasing amounts of unlabelled probes were mixed with labelled probes for competition reaction. The mutant probes were used as negative control. The protein‐DNA complex was separated on a 6% PAGE gel for 3 h at 4 °C in the dark. Gels were photographed using FLA‐5100 (FUJIFILM, Japan).
Histochemical GUS staining
Histochemical GUS staining was performed according to a previously reported method (Yuan et al., 2009). In brief, rice tissues including nodes, roots, leaves, sheaths, and panicles were immersed in GUS staining solution (50 mM of sodium phosphate buffer at pH 7.0, 10 mM of EDTA, 0.1% Triton X‐100, 1 mg/mL of X‐Gluc, 100 mg/mL of chloramphenicol, 1 mM of potassium ferrocyanide, 1 mM of potassium ferricyanide, and 20% methanol), infiltrated under a vacuum for 30 min, and incubated at 37 °C for 24 h. Then, the stained rice tissues were immersed in a fixation solution (50% ethanol, 3.7% formaldehyde, and 5% acetic acid) at 25 °C for 24 h. Various rice tissues were photographed using a stereo microscope (Leica M205 FCA, Germany).
Recombinant protein purification and lipase activity assay
The full‐length OsGELP77 cDNA without the N‐terminal signal peptide was amplified using the specific primers (Table S3), then the amplified PCR products were cloned into the pGEX‐6p‐1 vector (GE Healthcare). Simultaneously, OsGELP774A where the four invariant key catalytic residues were mutated to alanine using overlapping‐PCR method was cloned into the same vector. The constructs were transformed into Escherichia coli BL21 (DE3) cells. The expression of the recombinant OsGELP77ΔSP or OsGELP774AΔSP proteins was induced using 1 mM isopropylthio‐β‐galactoside at 16 °C. After induction for 20 h, cells were collected, suspended in PBS buffer, and sonicated. The recombinant proteins were purified using GST‐tag beads (GE Healthcare) according to the manufacturer's instructions and confirmed by SDS‐PAGE gel. The recombinant His‐TF‐OsWRKY45 protein and the control His‐TF protein were prokaryote‐expressed and purified using Ni Sepharose 6 fast Flow (GE Healthcare), as described previously (Cheng et al., 2015).
Lipase activity was measured according to a previously reported method (Gao et al., 2017). In brief, p‐nitrophenyl butyrate, p‐nitrophenyl acetate, p‐nitrophenyl octanoate, or p‐nitrophenyl palmitate was dissolved in a buffer containing 0.5 M HEPES, reaching a final concentration of 1 mM. Aliquots of the recombinant proteins were incubated with the substrates at 30 °C for 90 min. Absorbance was quantified spectrometrically at 405 nm every 5 min using a Spark™ Multimode Microplate Reader (Tecan, Switzerland).
Lipidomics analysis
The lipidomics analysis of rice leaves was performed according to previously reported methods (Chen et al., 2013; Gao et al., 2017). In brief, rice leaves growing in the paddy field at the tillering stage were sampled and ground in liquid nitrogen. About 50 mg powder of each sample was mixed with 1 mL methanol, 1 mL chloroform, and 0.8 mL ultra‐purified water, and vigorously shaken for 10 min for lipid extraction. Following centrifugation, the organic phase was orderly treated with chloroform and 1 M KCl to purify lipid. The organic solution was filtered, then dried under the flow of nitrogen gas, and finally dissolved in chloroform. Lipid species quantification was conducted using a liquid chromatography‐electrospray ionization triple quadrupole mass spectrometry (LC‐ESI‐MS/MS) system (LC, Shim‐pack UFLC SHIMADZU CBM30A; MS, ABI 6500 QTRAP) by Metware (Wuhan, China). Individual lipid species were analysed based on metabolite structure referring to public mass spectrometry databases including Metware‐made database (Chen et al., 2013), Metlin (http://metlin.scripps.edu/), and MassBank (http://www.massbank.jp/).
Haplotype analysis and multiple sequence alignments
The genomic sequence of OsGELP77 in 2178 rice accessions was acquired from RiceVarMap v2.0 (http://ricevarmap.ncpgr.cn/). Haplotypes of the accessions were analysed from InDels and SNPs in OsGELP77 promoter. Protein sequences of functionally characterized plant GDSL proteins were retrieved from NCBI. Multiple sequence alignments of protein were analysed using Clustal Omega (www.clustal.org/omega).
Co‐mapping OsGELP77 and resistance QTL
A rice recombinant inbred line (RIL) population consisting of 241 lines developed from a cross between resistant Minghui 63 and susceptible Zhenshan 97 was used to analyse co‐mapping OsGELP77 and resistance QTL (Kou et al., 2010). The RIL population has been previously used to identify the quantitative disease resistance for bacterial pathogen Xoo and fungal pathogen M. oryzae (Chen et al., 2003; Kou et al., 2010). A molecular linkage map containing 221 simple‐sequence repeats markers that covering the whole rice genome was previously developed with the RIL population for QTL mapping. OsGELP77 was mapped on the molecular linkage map developed from the RIL population using PCR‐based derived cleaved amplification polymorphism sequence (dCAPS) marker which was designed using dCAPS Finder (http://helix.wustl.edu/dcaps/dcaps.html). Mapmaker/Exp 3.0 was used for linkage analysis and Windows QTL Cartographer 2.5 was used for QTL analysis (Kou et al., 2010).
Agronomic traits analysis
All rice plants were cultivated in three replicates at three different plots under natural field conditions in normal rice growing season. Agronomic traits including plant height, tiller number, flag leaf length, flag leaf width, panicle length, productive panicles per plant, grain length, grain width, grain thickness, primary branch number, and 1000‐grain weight were measured at the ripening stage. Of which, traits such as grain length, grain width, grain thickness, primary branch number, and 1000‐grain weight were measured by the high‐throughput rice phenotyping facility (Yang et al., 2014).
Statistical analysis
Statistical parameters are reported in the figures and figure legends. The differences between samples were analysed for statistical significance using ANOVA or Student's t‐test.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
MY, MZ, and DC designed the experiments, MZ, DC, and JT performed the experiments, JC, KX, and YH assisted the experiments, MZ and DC analysed the data, MY wrote and revised the manuscript.
Supporting information
Figure S1 OsGELP77 expression is induced by different pathogens.
Figure S2 Enhanced resistance to Xoo of OsGELP77‐OE plants.
Figure S3 Generation of OsGELP77 knockout lines.
Figure S4 Alignment of amino acid sequences of functionally characterized plant GDSL proteins.
Figure S5 Contents of individual PC and PE species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S6 Contents of 28 FFA species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S7 Contents of 26 DGDG species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S8 Contents of 37 MGDG species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S9 Contents of 25 SQDG species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S10 Contents of 49 DAG species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S11 Contents of 107 TAG species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S12 KEGG enrichment analysis of differential lipids between the osgelp77 mutant and wild type (a) or between the OsGELP77‐OE plant and wild type (b).
Figure S13 GO enrichment and KEGG pathway analysis of osgelp77 mutant DEGs.
Figure S14 Modulating OsGELP77 expression influenced the expression of JA signalling pathway genes.
Figure S15 The OsGELP77‐OE and osgelp77 mutant plants showed opposite responses to MeJA treatment in primary shoot and root development.
Figure S16 Relative transcription of OsGELP77 in the osmyc2 mutant.
Figure S17 Agronomic traits of OsGELP77 transgenic lines grown in field.
Figure S18 Colocalization of OsGELP77 loci with previously reported disease‐related QTL derived using different genetic populations.
Figure S19 Expression profiles of OsGELP77 in different tissues of rice varieties Minghui 63 and Zhenshan 97.
Figure S20 Agronomic traits of NIL‐OsGELP77 Hap3 and Zhenshan 97 grown in field.
Table S1 Detailed list of Up‐DEGs in the osgelp77 mutant relative to wild type.
Table S2 Detailed list of Down‐DEGs in the osgelp77 mutant relative to wild type.
Table S3 PCR primers used for vectors construction for OsGELP77.
Table S4 PCR primers used for RT‐qPCR assay.
Acknowledgements
We are grateful to Prof. Donglei Yang at Nanjing Agricultural University for providing coi1‐13 mutant and Prof. Yanjun Kou at China National Rice Research Institute for providing osmyc2 mutant. This work was supported by grants from the STI 2030‐Major Projects (2022ZD0400201), the National Natural Science Foundation of China (31821005, 32172421), the Major Project of Hubei Hongshan Laboratory (2022hszd016), the earmarked fund for the China Agriculture Research System (CARS‐01‐01), the Fundamental Research Funds for the Central Universities (2662023PY006) and the open funds of the National Key Laboratory of Crop Genetic Improvement.
Data availability statement
The RNA‐seq data from this study is deposited to NCBI Sequence Read Archive (BioProject ID: PRJNA895969).
References
- Akoh, C.C. , Lee, G.C. , Liaw, Y.C. , Huang, T.H. and Shaw, J.F. (2004) GDSL family of serine esterases/lipases. Prog. Lipid Res. 43, 534–552. [DOI] [PubMed] [Google Scholar]
- Brick, D.J. , Brumlik, M.J. , Buckley, J.T. , Cao, J.X. , Davies, P.C. , Misra, S. , Tranbarger, T.J. et al. (1995) A new family of lipolytic plant enzymes with members in rice, arabidopsis and maize. FEBS Lett. 377, 475–480. [DOI] [PubMed] [Google Scholar]
- Brodersen, P. , Petersen, M. , Bjørn Nielsen, H. , Zhu, S. , Newman, M.A. , Shokat, K.M. , Rietz, S. et al. (2006) Arabidopsis MAP kinase 4 regulates salicylic acid‐ and jasmonic acid/ethylene‐dependent responses via EDS1 and PAD4. Plant J. 47, 532–546. [DOI] [PubMed] [Google Scholar]
- Cai, Q. , Yuan, Z. , Chen, M. , Yin, C. , Luo, Z. , Zhao, X. , Liang, W. et al. (2014) Jasmonic acid regulates spikelet development in rice. Nat. Commun. 5, 3476. [DOI] [PubMed] [Google Scholar]
- Campos, M.L. , Kang, J.H. and Howe, G.A. (2014) Jasmonate‐triggered plant immunity. J. Chem. Ecol. 40, 657–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Wang, S. , Xing, Y. , Xu, C. , Hayes, P.M. and Zhang, Q. (2003) Comparative analyses of genomic locations and race specificities of loci for quantitative resistance to Pyricularia grisea in rice and barley. Proc. Natl. Acad. Sci. U. S. A. 100, 2544–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Gong, L. , Guo, Z. , Wang, W. , Zhang, H. , Liu, X. , Yu, S. et al. (2013) A novel integrated method for large‐scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant, 6, 1769–1780. [DOI] [PubMed] [Google Scholar]
- Cheng, H. , Liu, H. , Deng, Y. , Xiao, J. , Li, X. and Wang, S. (2015) The WRKY45‐2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 167, 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepyshko, H. , Lai, C.P. , Huang, L.M. , Liu, J.H. and Shaw, J.F. (2012) Multifunctionality and diversity of GDSL esterase/lipase gene family in rice (Oryza sativa L. japonica) genome: new insights from bioinformatics analysis. BMC Genomics, 13, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, C. , Huang, R. , Liu, L. , Tang, G. , Xiao, J. , Yoo, H. and Yuan, M. (2022) The rice heavy‐metal transporter OsNRAMP1 regulates disease resistance by modulating ROS homoeostasis. Plant Cell Environ. 45, 1109–1126. [DOI] [PubMed] [Google Scholar]
- Deng, H. , Liu, H. , Li, X. , Xiao, J. and Wang, S. (2012) A CCCH‐type zinc finger nucleic acid‐binding protein quantitatively confers resistance against rice bacterial blight disease. Plant Physiol. 158, 876–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y. , Ning, Y. , Yang, D.L. , Zhai, K. , Wang, G.L. and He, Z. (2020) Molecular basis of disease resistance and perspectives on breeding strategies for resistance improvement in crops. Mol. Plant 13, 1402–1419. [DOI] [PubMed] [Google Scholar]
- Deveshwar, P. , Prusty, A. , Sharma, S. and Tyagi, A.K. (2020) Phytohormone‐mediated molecular mechanisms involving multiple genes and QTL govern grain number in rice. Front. Genet. 11, 586462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L.N. , Li, M. , Guo, X.J. , Tang, M.Q. , Cao, J. , Wang, Z. , Liu, R. et al. (2020) Arabidopsis GDSL1 overexpression enhances rapeseed Sclerotinia sclerotiorum resistance and the functional identification of its homolog in Brassica napus . Plant Biotechnol. J. 18, 1255–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Q. , Wang, H. , Yang, X.M. , Hu, Z.W. , Zhou, X.H. , Xiang, L. , Xiong, X.Y. et al. (2022) Osa‐miR160a confers broad‐spectrum resistance to fungal and bacterial pathogens in rice. New Phytol. 236, 2216–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M. , Yin, X. , Yang, W. , Lam, S.M. , Tong, X. , Liu, J. , Wang, X. et al. (2017) GDSL lipases modulate immunity through lipid homeostasis in rice. PLoS Pathog. 13, e1006724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbel, M. , Brini, F. , Sharma, A. and Landi, M. (2021) Role of jasmonic acid in plants: the molecular point of view. Plant Cell Rep. 40, 1471–1494. [DOI] [PubMed] [Google Scholar]
- He, F. , Zhang, F. , Sun, W. , Ning, Y. and Wang, G.L. (2018) A versatile vector toolkit for functional analysis of rice genes. Rice 11, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, H. , Liu, Y. , Zhang, H. , Xiao, J. , Li, X. and Wang, S. (2015) Small RNAs and gene network in a durable disease resistance gene‐mediated defense responses in rice. PLoS One, 10, e0137360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y. , Wang, Y. , Tang, L. , Tong, X. , Wang, L. , Liu, L. , Huang, S. et al. (2019) SAPK10‐mediated phosphorylation on WRKY72 releases its suppression on jasmonic acid biosynthesis and bacterial blight resistance. iScience 16, 499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, T. and Zhang, J.L. (2018) Mass‐spectrometry‐based lipidomics. J. Sep. Sci. 41, 351–372. [DOI] [PubMed] [Google Scholar]
- Huang, X. , Kurata, N. , Wei, X. , Wang, Z.X. , Wang, A. , Zhao, Q. , Zhao, Y. et al. (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Yang, S. , Gong, J. , Zhao, Y. , Feng, Q. , Gong, H. , Li, W. et al. (2015) Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun. 6, 6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, R. , Li, Y. , Tang, G. , Hui, S. , Yang, Z. , Zhao, J. , Liu, H. et al. (2018) Dynamic phytohormone profiling of rice upon rice black‐streaked dwarf virus invasion. J. Plant Physiol. 228, 92–100. [DOI] [PubMed] [Google Scholar]
- Hui, S. , Shi, Y. , Tian, J. , Wang, L. , Li, Y. , Wang, S. and Yuan, M. (2019) TALE‐carrying bacterial pathogens trap host nuclear import receptors for facilitation of infection of rice. Mol. Plant Pathol. 20, 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, Y. , Liu, H. , Li, X. , Xiao, J. and Wang, S. (2014) Rice OsPAD4 functions differently from Arabidopsis AtPAD4 in host‐pathogen interactions. Plant J. 78, 619–631. [DOI] [PubMed] [Google Scholar]
- Ke, Y. , Yuan, M. , Liu, H. , Hui, S. , Qin, X. , Chen, J. , Zhang, Q. et al. (2020) The versatile functions of OsALDH2B1 provide a genic basis for growth‐defense trade‐offs in rice. Proc. Natl. Acad. Sci. U. S. A. 117, 3867–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.G. , Kwon, S.J. , Jang, Y.J. , Nam, M.H. , Chung, J.H. , Na, Y.C. , Guo, H. et al. (2013) GDSL LIPASE1 modulates plant immunity through feedback regulation of ethylene signaling. Plant Physiol. 163, 1776–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.K. , Seol, Y.J. , Lee, D.J. , Lee, J.H. , Lee, T.H. and Park, D.S. (2014) RiceQTLPro: an integrated database for quantitative trait loci marker mapping in rice plant. Bioinformation, 10, 664–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou, Y. , Li, X. , Xiao, J. and Wang, S. (2010) Identification of genes contributing to quantitative disease resistance in rice. Sci. China Life Sci. 53, 1263–1273. [DOI] [PubMed] [Google Scholar]
- Kwon, S.J. , Jin, H.C. , Lee, S. , Nam, M.H. , Chung, J.H. , Kwon, S.I. , Ryu, C.M. et al. (2009) GDSL lipase‐like 1 regulates systemic resistance associated with ethylene signaling in Arabidopsis . Plant J. 58, 235–245. [DOI] [PubMed] [Google Scholar]
- Lai, C.P. , Huang, L.M. , Chen, L.O. , Chan, M.T. and Shaw, J.F. (2017) Genome‐wide analysis of GDSL‐type esterases/lipases in Arabidopsis . Plant Mol. Biol. 95, 181–197. [DOI] [PubMed] [Google Scholar]
- Lavell, A.A. and Benning, C. (2019) Cellular organization and regulation of plant glycerolipid metabolism. Plant Cell Physiol. 60, 1176–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.S. , Kim, B.K. , Kwon, S.J. , Jin, H.C. and Park, O.K. (2009) Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem. Biophys. Res. Commun. 379, 1038–1042. [DOI] [PubMed] [Google Scholar]
- Liu, Z. , Zhu, Y. , Shi, H. , Qiu, J. , Ding, X. and Kou, Y. (2021) Recent progress in rice broad‐spectrum disease resistance. Int. J. Mol. Sci. 22, 11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. , Zhu, X. , Zhang, J. , Shen, M. , Chen, K. , Fu, X. , Ma, L. et al. (2022) eQTLs play critical roles in regulating gene expression and identifying key regulators in rice. Plant Biotechnol. J. 20, 2357–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Zhang, Q. , Zhu, Q. , Liu, W. , Chen, Y. , Qiu, R. , Wang, B. et al. (2015) A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant, 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Mei, C. , Qi, M. , Sheng, G. and Yang, Y. (2006) Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 19, 1127–1137. [DOI] [PubMed] [Google Scholar]
- Nakayama, A. , Fukushima, S. , Goto, S. , Matsushita, A. , Shimono, M. , Sugano, S. , Jiang, C.J. et al. (2013) Genome‐wide identification of WRKY45‐regulated genes that mediate benzothiadiazole‐induced defense responses in rice. BMC Plant Biol. 13, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves Petersen, M.T. , Fojan, P. and Petersen, S.B. (2001) How do lipases and esterases work: the electrostatic contribution. J. Biotechnol. 85, 115–147. [DOI] [PubMed] [Google Scholar]
- Oh, I.S. , Park, A.R. , Bae, M.S. , Kwon, S.J. , Kim, Y.S. , Lee, J.E. , Kang, N.Y. et al. (2005) Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola . Plant Cell 17, 2832–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.J. , Jin, P. , Yoon, J. , Yang, J.I. , Jeong, H.J. , Ranathunge, K. , Schreiber, L. et al. (2010) Mutation in Wilted Dwarf and Lethal 1 (WDL1) causes abnormal cuticle formation and rapid water loss in rice. Plant Mol. Biol. 74, 91–103. [DOI] [PubMed] [Google Scholar]
- Qiu, J. , Xie, J. , Chen, Y. , Shen, Z. , Shi, H. , Naqvi, N.I. , Qian, Q. et al. (2022) Warm temperature compromises JA‐regulated basal resistance to enhance Magnaporthe oryzae infection in rice. Mol. Plant 15, 723–739. [DOI] [PubMed] [Google Scholar]
- Riemann, M. , Gutjahr, C. , Korte, A. , Riemann, M. , Danger, B. , Muramatsu, T. , Bayer, U. et al. (2007) GER1, a GDSL motif‐encoding gene from rice is a novel early light‐ and jasmonate‐induced gene. Plant Biol. 9, 32–40. [DOI] [PubMed] [Google Scholar]
- Shah, J. (2005) Lipids, lipases, and lipid‐modifying enzymes in plant disease resistance. Annu. Rev. Phytopathol. 43, 229–260. [DOI] [PubMed] [Google Scholar]
- Shen, G. , Sun, W. , Chen, Z. , Shi, L. , Hong, J. and Shi, J. (2022) Plant GDSL esterases/lipases: evolutionary, physiological and molecular functions in plant development. Plan. Theory 11, 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono, M. , Koga, H. , Akagi, A. , Hayashi, N. , Goto, S. , Sawada, M. , Kurihara, T. et al. (2012) Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 13, 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, B. , Shang, L. , Li, Y. , Zhang, Q. , Chu, Z. , He, S. , Yang, W. et al. (2022) Ectopic expression of OsJAZs alters plant defense and development. Int. J. Mol. Sci. 23, 4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Z. , Liu, H. , Qiu, D. , Zhou, Y. , Li, X. , Xu, C. and Wang, S. (2009) A pair of allelic WRKY genes play opposite roles in rice‐bacteria interactions. Plant Physiol. 151, 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uji, Y. , Taniguchi, S. , Tamaoki, D. , Shishido, H. , Akimitsu, K. and Gomi, K. (2016) Overexpression of OsMYC2 results in the up‐regulation of early JA‐responsive genes and bacterial blight resistance in rice. Plant Cell Physiol. 57, 1814–1827. [DOI] [PubMed] [Google Scholar]
- Wan, S. and Xin, X.F. (2022) Regulation and integration of plant jasmonate signaling: a comparative view of monocot and dicot. J. Genet. Genomics, 49, 704–714. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Yu, H. , Xie, W. , Xing, Y. , Yu, S. , Xu, C. , Li, X. et al. (2010) A global analysis of QTLs for expression variations in rice shoots at the early seedling stage. Plant J. 63, 1063–1074. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Zhou, L. , Shi, H. , Chern, M. , Yu, H. , Yi, H. , He, M. et al. (2018) A single transcription factor promotes both yield and immunity in rice. Science 361, 1026–1028. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Pruitt, R.N. , Nürnberger, T. and Wang, Y. (2022) Evasion of plant immunity by microbial pathogens. Nat. Rev. Microbiol. 20, 449–464. [DOI] [PubMed] [Google Scholar]
- Xie, W. , Wang, G. , Yuan, M. , Yao, W. , Lyu, K. , Zhao, H. , Yang, M. et al. (2015) Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection. Proc. Natl. Acad. Sci. U. S. A. 112, E5411–E5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, S. , Kano, A. , Tamaoki, D. , Miyamoto, A. , Shishido, H. , Miyoshi, S. , Taniguchi, S. et al. (2012) Involvement of OsJAZ8 in jasmonate‐induced resistance to bacterial blight in rice. Plant Cell Physiol. 53, 2060–2072. [DOI] [PubMed] [Google Scholar]
- Yang, D.L. , Yao, J. , Mei, C.S. , Tong, X.H. , Zeng, L.J. , Li, Q. , Xiao, L.T. et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. U. S. A. 109, E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D.L. , Yang, Y. and He, Z. (2013) Roles of plant hormones and their interplay in rice immunity. Mol. Plant 6, 675–685. [DOI] [PubMed] [Google Scholar]
- Yang, W. , Guo, Z. , Huang, C. , Duan, L. , Chen, G. , Jiang, N. , Fang, W. et al. (2014) Combining high‐throughput phenotyping and genome‐wide association studies to reveal natural genetic variation in rice. Nat. Commun. 5, 5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. , Hui, S. , Lv, Y. , Zhang, M. , Chen, D. , Tian, J. , Zhang, H. et al. (2022) miR395‐regulated sulfate metabolism exploits pathogen sensitivity to sulfate to boost immunity in rice. Mol. Plant 15, 671–688. [DOI] [PubMed] [Google Scholar]
- Yonemaru, J. , Yamamoto, T. , Fukuoka, S. , Uga, Y. , Hori, K. and Yano, M. (2010) Q‐TARO: QTL annotation rice online database. Rice, 3, 194–203. [Google Scholar]
- Yuan, M. , Chu, Z. , Li, X. , Xu, C. and Wang, S. (2019) Pathogen‐induced expressional loss of function is the key factor in race‐specific bacterial resistance conferred by a recessive R gene xa13 in rice. Plant Cell Physiol, 50, 947–955. [DOI] [PubMed] [Google Scholar]
- Yu, Y. , Woo, M.O. , Rihua, P. and Koh, H.J. (2020) The DROOPING LEAF (DR) gene encoding GDSL esterase is involved in silica deposition in rice (Oryza sativa L.). PLoS One, 15, e0238887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, M. , Chu, Z. , Li, X. , Xu, C. and Wang, S. (2010) The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell, 22, 3164–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, M. , Ke, Y. , Huang, R. , Ma, L. , Yang, Z. , Chu, Z. , Xiao, J. et al. (2016) A host basal transcription factor is a key component for infection of rice by TALE‐carrying bacteria. Elife, 5, e19605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Tao, Z. , Hong, H. , Chen, Z. , Wu, C. , Li, X. , Xiao, J. et al. (2016) Transposon‐derived small RNA is responsible for modified function of WRKY45 locus. Nat. Plants, 2, 16016. [DOI] [PubMed] [Google Scholar]
- Zhang, B. , Zhang, L. , Li, F. , Zhang, D. , Liu, X. , Wang, H. , Xu, Z. et al. (2017a) Control of secondary cell wall patterning involves xylan deacetylation by a GDSL esterase. Nat. Plants, 3, 17017. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Zhang, F. , Melotto, M. , Yao, J. and He, S.Y. (2017b) Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 68, 1371–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Gao, C. , Mentink‐Vigier, F. , Tang, L. , Zhang, D. , Wang, S. , Cao, S. et al. (2019) Arabinosyl deacetylase modulates the arabinoxylan acetylation profile and secondary wall formation. Plant Cell, 31, 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Wang, M. , Li, Y. , Yan, W. , Chang, Z. , Ni, H. , Chen, Z. et al. (2020) GDSL esterase/lipases OsGELP34 and OsGELP110/OsGELP115 are essential for rice pollen development. J. Integr. Plant Biol. 62, 1574–1593. [DOI] [PubMed] [Google Scholar]
- Zhao, H. , Ma, B. , Duan, K.X. , Li, X.K. , Lu, X. , Yin, C.C. , Tao, J.J. et al. (2020a) The GDSL lipase MHZ11 modulates ethylene signaling in rice roots. Plant Cell, 32, 1626–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Long, T. , Wang, Y. , Tong, X. , Tang, J. , Li, J. , Wang, H. et al. (2020b) RMS2 encoding a GDSL lipase mediates lipid homeostasis in anthers to determine rice male fertility. Plant Physiol. 182, 2047–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Zhu, X. , Chen, X. and Zhou, J.M. (2022) From plant immunity to crop disease resistance. J. Genet. Genomics, 49, 693–703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 OsGELP77 expression is induced by different pathogens.
Figure S2 Enhanced resistance to Xoo of OsGELP77‐OE plants.
Figure S3 Generation of OsGELP77 knockout lines.
Figure S4 Alignment of amino acid sequences of functionally characterized plant GDSL proteins.
Figure S5 Contents of individual PC and PE species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S6 Contents of 28 FFA species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S7 Contents of 26 DGDG species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S8 Contents of 37 MGDG species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S9 Contents of 25 SQDG species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S10 Contents of 49 DAG species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S11 Contents of 107 TAG species in the leaves of OsGELP77‐OE plants and osgelp77 mutants.
Figure S12 KEGG enrichment analysis of differential lipids between the osgelp77 mutant and wild type (a) or between the OsGELP77‐OE plant and wild type (b).
Figure S13 GO enrichment and KEGG pathway analysis of osgelp77 mutant DEGs.
Figure S14 Modulating OsGELP77 expression influenced the expression of JA signalling pathway genes.
Figure S15 The OsGELP77‐OE and osgelp77 mutant plants showed opposite responses to MeJA treatment in primary shoot and root development.
Figure S16 Relative transcription of OsGELP77 in the osmyc2 mutant.
Figure S17 Agronomic traits of OsGELP77 transgenic lines grown in field.
Figure S18 Colocalization of OsGELP77 loci with previously reported disease‐related QTL derived using different genetic populations.
Figure S19 Expression profiles of OsGELP77 in different tissues of rice varieties Minghui 63 and Zhenshan 97.
Figure S20 Agronomic traits of NIL‐OsGELP77 Hap3 and Zhenshan 97 grown in field.
Table S1 Detailed list of Up‐DEGs in the osgelp77 mutant relative to wild type.
Table S2 Detailed list of Down‐DEGs in the osgelp77 mutant relative to wild type.
Table S3 PCR primers used for vectors construction for OsGELP77.
Table S4 PCR primers used for RT‐qPCR assay.
Data Availability Statement
The RNA‐seq data from this study is deposited to NCBI Sequence Read Archive (BioProject ID: PRJNA895969).


