Abstract
Neutrophils are the most abundant white blood cell, and they differentiate in homeostasis in the bone marrow from hematopoietic stem cells (HSCs) via multiple intermediate progenitor cells into mature cells that enter the circulation. Recent findings support a continuous model of differentiation in the bone marrow of heterogeneous HSCs and progenitor populations. Cell fate decisions at the levels of proliferation and differentiation are enforced through expression of lineage-determining transcription factors and their interactions, which are influenced by intrinsic (intracellular) and extrinsic (extracellular) mechanisms. Neutrophil homeostasis is subjected to positive-feedback loops, stemming from the gut microbiome, as well as negative-feedback loops resulting from the clearance of apoptotic neutrophils by mature macrophages. Finally, the cellular kinetics regarding the replenishing of the mature neutrophil pool is discussed in light of recent contradictory data.
Introduction
Neutrophils play a critical role in our immune system, yet the details regarding neutrophil lineage determination and regulation of neutropoiesis remain to be elucidated.1 The field generally distinguishes 2 types of neutropoiesis: steady-state neutropoiesis and emergency neutropoiesis. The latter is described as accelerated neutropoiesis under conditions of high demand, such as during systemic inflammation and infection,2 and will be discussed in the section “Emergency neutropoiesis.”
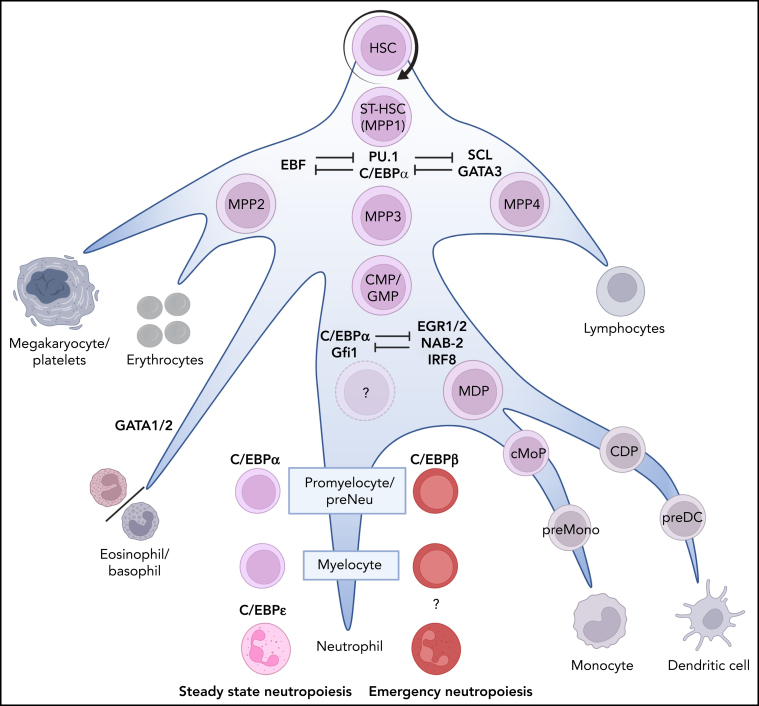
Previously, the description of hematopoiesis, including neutropoiesis, made use of hierarchical models.3 In such primarily murine models, hematopoietic stem cells (HSCs) give rise to a common myeloid progenitor (CMP) or a common lymphoid progenitor. The common lymphoid progenitor precursors become T cells, B cells, or natural killer (NK) cells, whereas the CMPs differentiate into granulocyte-monocyte progenitors (GMPs) or megakaryocyte-erythroid progenitors. However, as more details of this complicated puzzle were identified, scientists struggled to fit their data into these simplified models, such as the existence of a population of lympho-myeloid multipotent progenitors that give rise to lymphoid and myeloid cell lines.4 Recently, a novel dynamic model for hematopoiesis in the mouse was described in detail by Cheng and colleagues.5 In this dynamic model, the majority of HSCs remain dormant in the bone marrow (BM), and only a small population differentiate via short-term HSCs (ST-HSCs) into multipotent progenitors (MPPs). Based on immune phenotyping, several MPP subsets have been identified with different lineage biases: MPP2s and MPP3s have increased granulocyte/macrophage potential compared with MPP4s, with MPP3s showing a predominantly granulocytic output. MPP2s show extensive megakaryocyte lineage potential, and MPP4s primarily show lymphoid output (Figure 1).6 The advent of single-cell RNA sequencing (scRNAseq) has allowed analysis of hematopoietic stem and progenitor cell states in unprecedented detail and revealed a structured hierarchy within the MPP compartment (for additional information, see the excellent review by Watcham et al7). High-dimensional clustering analysis, as well as inferred transcriptional trajectories, revealed that the earliest split in fate separates erythroid-megakaryocyte progenitors from leukocyte progenitors, which separate further into lymphoid, dendritic cell (DC), and monocytic/granulocytic progenitors.8 Interestingly, initial scRNAseq data from mice8 and human9, 10, 11 BM indicate that eosinophils and basophils, which are traditionally thought to be similar to neutrophils, have progenitors that originate from erythroid-megakaryocyte progenitors instead of from myeloid progenitors. However, later reports suggested that basophils can originate from an erythroid-megakaryocyte-basophil progenitor, as well as a basophil-neutrophil-monocyte progenitor.12 Taken together, this indicates that the hematopoietic tree might be more complex than originally thought, with cell types originating from multiple lineages. However, it should be noted that scRNAseq only provides transcriptome data, with changes in a cells' transcriptome reflecting a fate decision that may have taken place earlier.
Figure 1.
Neutrophil production in the BM. The recognized stages of stem cell differentiation that contribute to neutropoiesis are indicated by the names of the neutrophil progenitors in color. Bifurcations into nonneutrophil-producing lineages are indicated in gray. Importantly, the differentiation occurs in a gradual manner (priming) rather than as discrete steps associated with division. The dominant lineage-determining transcription factors are indicated in blue. The first bias toward neutropoiesis starts with a slow and gradual commitment of MPP3s toward the myeloid lineage. MPP3s can proliferate or differentiate into GMPs, which can proliferate and/or physically cluster together into loose patchecs (p) of GMP.89 This clustering facilitates differentiation into compact clusters of (c)GMP that, in turn, differentiate into promyelocytes and myelocytes, thereby forming clusters of these cells in the BM.89 During these last differentiation steps, the progenitors lose their propensity to proliferate (mediated by the expression of C/EBPε)2 and continue to mature toward mature neutrophils via metamyelocytes and banded cells (see Figure 2). CDP, common dendritic cell progenitor; cMoP, common monocyte progenitor; EGR1/2, early growth response ½; GATA1/2, GATA binding receptor ½; IRF8, interferon regulatory factor-8; MDP, monocyte/macrophage/DC; NAB-2, NGFI-A binding protein 2; preMono, preMonocyte; SCL, stem cell leukemia.
Most models of hematopoiesis are based on the hypothesis that (stochastic) fluctuations in cross-antagonistic pairs of lineage-determining transcription factors (LDTFs) are responsible for the determination of cell fate.13, 14 However, elegant murine studies using live imaging, long-term quantification, and single-cell analysis show that the expression patterns of LDTF pairs are not compatible with discrete (stochastic) switches.14 Rather, they suggest that cell fate is decided earlier by unknown processes and is merely reinforced/executed through complex LDTF circuits.5
Because the MPP pool has a large potential for self-renewal (ie, keeping a pool of cells with a similar cell fate despite multiple divisions) (Figure 1), it is important for the compensation for the loss of cells at the end of differentiation.15, 16 These MPPs do not pass through a discrete stepwise tree-like hierarchy, described in previous models of differentiation; instead, they slowly differentiate in response to multiple intrinsic and extrinsic cues. The detailed description of the control of the early HSC–ST-HSC–MPP axis is beyond the scope of this review and can be found elsewhere.17, 18
In addition to being able to undergo self-renewing proliferation, monocyte/neutrophil-biased MPPs can differentiate into CMPs, which, in turn, can differentiate into GMPs.6 CMPs and GMPs were initially thought to be oligopotent, but recent single-cell analyses, such as cellular barcoding in mice,19 as well as scRNAseq and single-cell assay for transposase-accessible chromatin using sequencing (ATACseq) experiments in mice20 and humans,10, 21 have shown that most of these cells can be primed toward certain lineages. The exact structure of the hematopoietic tree at this level is not clear, with murine monocytes being shown to originate from monocyte-neutrophil or monocyte-DC primed progenitors.22, 23
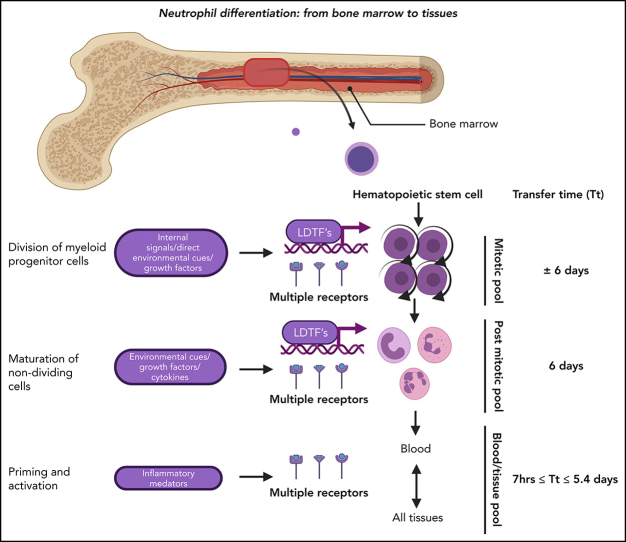
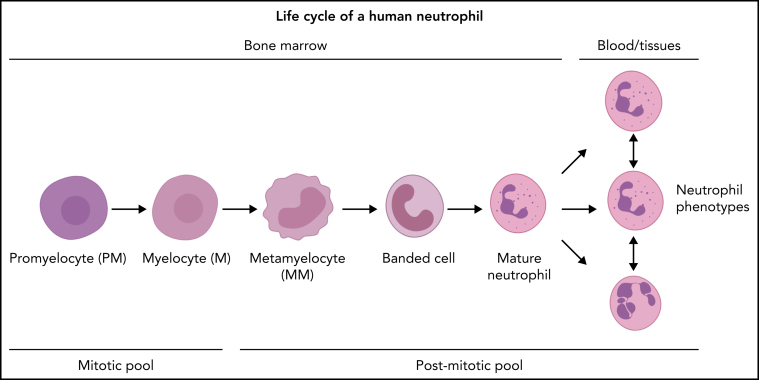
As stated before, hematopoietic stem and progenitor cells can undergo self-renewing proliferation or differentiation. Control of the “choice” between self-renewing proliferation and differentiation has yet to be determined, but humoral and spatial factors (niches) are likely to be involved. GMPs are highly proliferative cells,16, 24, 25 yet it remains unclear how many divisions take place in the GMP pool. The very poor repopulation of neutrophils after transplantation of GMPs into sublethally irradiated mice implies that the GMP pool is very limited in self-renewing proliferation.16 However, because the first time point was measured 5 days posttransplantation, which is much longer than the postmitotic transfer time of neutrophils in the mouse,26, 27 transient amplification divisions resulting in 1 wave of mature neutrophils might have been missed.16 GMPs, in turn, can mature into neutrophil-specific promyelocytes/preneutrophils.24 During the maturation step of GMPs into neutrophil progenitors, the proliferation capacity of these cells remains under debate, because reported mitotic and/or proliferation indices of promyelocytes and myelocytes vary markedly between studies.15, 28, 29, 30 In marked contrast to the GMP compartment, which contains few nonproliferative cells,25 it has been postulated that the (pro)myelocyte pool contains 30% to 50% nonproliferating cells; this can be explained by a relatively long G1 phase or the presence of resting cells.28 Although not confirmed by other studies, such a “lazy pool” could be an attractive solution to quickly mobilize extra neutrophils upon high demand. The myelocyte is the last stage in neutropoiesis that has the capacity to divide. Next, the neutrophil progenitors lose the propensity to divide and only mature into metamyelocytes, banded neutrophils, mature neutrophils, and hypersegmented neutrophils.27, Figure 2 shows the differentiation of fully committed neutrophil progenitors, and Figure 1 summarizes a simplified hierarchical visualization of continuous neutropoiesis. Every step in neutropoiesis is controlled by complex mechanisms that are now being identified with novel genetic methodology and will be discussed below.
Figure 2.
Neutrophil production in the BM under homeostatic conditions. The neutrophil committed compartment in BM consists of dividing progenitors (mitotic pool: promyelocytes and myelocytes) and maturing nondividing progenitors (postmitotic pool: metamyelocytes, banded cells, mature cells, and hypersegmented neutrophils). The postmitotic transfer time is 5 to 6 days.51, 52, 53 The origin of functional neutrophil phenotypes is uncertain, but they might be evoked by tissue instructions81 or parallel differentiation74 or transdifferentiation.79, 80
The role of LDTFs in controlling cell fate decisions in steady-state neutropoiesis
As mentioned above, LDTFs play an important role in the reinforcement of cell fate decisions. Figure 1 includes expression patterns of the dominant LDTFs at each differentiation step driving, among others, the differentiation of the neutrophil lineage.31 This hierarchical route is compliant with intrinsic signals of unknown origin, as well as with epigenetic and external regulation in response to environmental cues, such as cytokines, cell-cell interactions, and regulation of the expression of microRNAs.17, 32, 33 LDTFs play an essential role in cell fate decisions of MPPs, and genetic targeting of these LDTFs disrupts differentiation. A key LDTF in all myeloid lineages is PU.1, which is encoded by the SPI1 gene.34, 35 High levels of PU.1 in MMPs enforce myelo-lymphoid cell commitment, primarily through an antagonistic interaction with GATA binding factor 1 (GATA-1),34, 36 a key player in the enforcement toward the megakaryocyte/erythroid cell (MegE) fate. Thus, the interplay between PU.1 and GATA-1 shapes hematopoietic lineage restriction toward the myelo-lymphoid or MegE lineage. PU.1 and GATA-1 actively inhibit each other's molecular activities via several direct and indirect mechanisms that are described in detail by Chou and colleagues.37 The outcome of such interactions are dose dependent, and shifting the balance of these transcription factors (TFs) favors reprograming of committed myeloid cells38 and differentiation into the neutrophil-biased GMP (Figure 1). It is important to emphasize that the human neutrophil CD34+ GMP has not been identified. On the other hand, early progenitors designated as preneutrophils have been described in murine BM.24
Differentiation toward a neutrophilic over a monocyte/macrophage/DC (MDP) lineage ensures the activation of neutrophil-specific genes while blocking monocyte/DC-specific genes, and vice versa. An important factor in the development of the MDP lineage is interferon regulatory factor-8 (IRF8), especially at the MDP stage and in common monocyte progenitors.23, 39 Data obtained from IRF8-deficiency and IRF8-knockout studies in mice show that a lack of IRF8 results in a significant increase in neutrophil counts, at the expense of macrophages. IRF8 is able to block neutrophil differentiation through a direct interaction with C/EBPα, which blocks the ability of C/EBPα to stimulate transcription of neutrophil-specific genes.39
The distinction between MDP and neutrophil cell fate is further enforced through a regulatory subcircuit made up of downstream counteracting repressors and feed-forward loops.40 In this circuit, high PU.1 expression levels favor a monocyte/DC fate over a neutrophil cell fate41 by inducing expression of the TF early growth response 1/2 (EGR1/2) and the corepressor NGFI-A binding protein 2 (NAB2). EGR1/2 interacts with NAB2, and these complexes (EGR/NAB) regulate MDP differentiation and functioning.36 A key factor for ensuring a neutrophil fate over a MDP cell fate is C/EBPα and its downstream repressor Gfi-1.36, 41 Gfi-1 ensures the activation of neutrophil-specific genes and represses MDP cell fate through the inhibition of EGR/NAB complexes.42 The EGR/NAB complex, in urn, is able to repress Gfi-1 and, thus, a neutrophilic cell fate.42 Taken together, EGR/NAB and Gfi-1 form a regulatory subcircuit to ensure lineage determination and restriction. It is highly likely that other cell lineages are dependent on similar subcircuits, such as already described in the erythrocyte/megakaryocyte lineage.43
Gfi-1 does not only play a key role in this subcircuit-mediated restriction of the monocytic lineage. In maturing eosinophils, its expression is downregulated to ensure eosinophilic differentiation associated with expression of eosinophil major basic protein, a hallmark of the eosinophil lineage.44 Furthermore, in murine studies, Gfi-1 was found to ensure neutrophil development through repression of the cytokine CSF1, as well as microRNAs miR-21 and miR-196b, which are involved in the repression of neutrophil-specific genes (although the mechanisms for this repression are incompletely understood).45 Finally, Gfi-1 has been shown to act as the main downstream regulator of C/EBPα-induced expression of C/EBPε.46 C/EBPε is a critical regulator of terminal neutropoiesis and is able to indirectly upregulate the expression of neutrophil-specific genes, such as ELANE (encoding neutrophil elastase) and GSF3R (encoding the granulocyte colony-stimulating factor [G-CSF] receptor).47
Disruption of C/EBPε expression in mice results in the abnormal differentiation of neutrophils beyond the promyelocyte stage.48 Mutations in C/EBPε have been linked to cases of neutrophil-specific granule deficiency, a rare disorder in which developing neutrophils exhibit an absence of specific and/or secondary granules.49 C/EBPε is able to indirectly upregulate the expression of neutrophil-specific genes, such as ELANE (encoding neutrophil elastase) and the GSF3R (encoding the G-CSF receptor) through its interaction with the DNA-binding domain of the c-Myb repressor.50 Finally, C/EBPε is involved in the regulation of cell cycle protein expression, because it induces the expression of cyclin-dependent kinase inhibitor 1B (p27kip1), while inhibiting cyclin-dependent kinase 4 (CDK4), CDK6, cyclin A, and cyclin D.3 Through these interactions, C/EBPε irreversibly halts cell proliferation during the metamyelocyte stage of neutropoiesis in favor of definitive neutropoiesis.
Emergency neutropoiesis
Under conditions of high demand, the neutrophil compartment can increase the output of terminally differentiated neutrophils by a mechanism generally referred to as emergency neutropoiesis (EN) (Figure 2).2 An essential mechanism underlying EN is activation of the LDTF C/EBPβ that interferes with C/EBPα-mediated steady-state limited neutropoiesis, thereby favoring proliferation and accelerated maturation. This is mediated by direct mechanisms initiated by direct sensing of damage/microbe-associated molecular patterns or by indirect mechanisms mediated by the production of cytokines/growth factors produced by bystander cells. Despite the consensus regarding the importance of EN for the increased production of neutrophils under conditions of infection and/or inflammation, it needs to be emphasized that the maturation phase in humans (postmitotic pool) is inherently slow (postmitotic transfer time of mature neutrophils ∼5 days),51, 52, 53 and no data on disease-induced shortening of this period have been accurately described. This implicates that the early blood neutrophilia found under conditions of acute infection/inflammation is caused by recruitment of cells from an already differentiated pool in the body. The most likely source of these cells is the BM, because it contains 5 to 10 times more neutrophils than does the peripheral blood.53 A description of the precise mechanisms underlying EN is beyond the scope of this article but was reported elsewhere.2
Intrinsic vs extrinsic signals involved neutrophil fate decisions
Increasing evidence implies that an intrinsic epigenetic myeloid bias of ST-HSCs and MPPs results in a baseline neutropoiesis in a highly adaptable system. The underlying mechanisms can be studied by genome-wide location of TF binding sites (cis-regulatory elements) and/or histone modifications of TFs that are referred to as the “TF cistrome.” The application of these TF cistromes leads to the identification of the cell- and tissue-specific patterns of regulatory TFs between MPP subsets and downstream mature immune cells.46 This is illustrated by the PU.1 cistrome, which shows great differences between neutrophils and B cells; in B cells, the majority of PU.1 binds to the enhancers EBF and E2A, whereas in neutrophil progenitors these enhancers are primarily localized in binding sites for C/EBP and Gfi-1.46 Epigenetic mapping of >22 000 promoters in MPPs, MegEs, and T cells revealed unique epigenetic maps that correlate with the developmental plasticity of ST-HSCs, early progenitors, and mature hematopoietic cells.54 Furthermore, HSC-specific homeobox genes, a family of TFs that is associated with differentiation,55 show increased DNA methylation upon differentiation (resulting in decreased expression), indicating that the accumulation of DNA methylation prevents aberrant activation at later stages of differentiation.56 Taken together, these data strengthen the hypothesis that cell fate decisions in homeostasis are significantly mediated by these heterogenic cistromes.
It is important to emphasize the high plasticity of the aforementioned cistrome profiles. Upon intra- and extracellular cues, TF cistromes can be rapidly reprogrammed, typically resulting in changes in transcriptional output.57 Thus, TF cistromes allow a rapid demand-driven response in neutrophil homeostasis and EN (Figure 1). However, it remains to be elucidated whether this specific cistrome signature is fixed toward a certain direction during early differentiation or whether this is a stochastic dynamic process that is regulated at later stages. To further complicate the situation, most TFs involved in HSC cell fate determination and differentiation have been found through knockdown and knockout studies and are not usually studied under homeostatic conditions. This asks for studies performed in homeostatic models.31
In summary, recent literature indicates that lineage choice is a gradual process that is controlled through the expression levels of multiple LDTFs, initiated by primarily unknown mechanisms.
Control and kinetics of the neutrophil compartment in homeostasis
At steady-state conditions, the total neutrophil count in the body is a balance of neutropoiesis, neutrophil release from the BM into the blood, redistribution in the tissues, and the clearance of mature neutrophils from circulation and target tissues.3 It is generally believed that mature neutrophils progress to the peripheral tissues or go back to the BM,58 where they undergo apoptosis and are phagocytosed by activated macrophages in a process that is generally referred to as “efferocytosis.”59 Efferocytosis results in anti-inflammatory signaling and the downregulation of proinflammatory interleukin-23 (IL-23) production by activated macrophages. IL-23 is a potent inducer of IL-17 production by T cells, NK cells, and NK T cells. In turn, IL-17 increases G-CSF and G-CSF–dependent neutropoiesis.60 Because efferocytosis results in the downregulation of this IL-23/IL-17/G-CSF axis, it forms a negative-feedback loop. This has also been called the neutrophil rheostat or “neutrostat”61 and is believed to be an important regulator of neutrophil numbers in homeostasis.
Interestingly, commensal bacteria in the gut exert a crucial positive-feedback loop that is nicely illustrated by the relative neutropenia in germ-free mice, which showed only 10% of normal neutrophil blood levels and low levels of plasma G-CSF.62 Toll-like receptor 4 (TLR-4) signaling in endothelial cells in the BM63 plays a crucial role in this process, and the activation of TLR-4 by bacterial-derived lipopolysaccharides has been shown to stimulate G-CSF–dependent neutropoiesis.64 Because germ-free mice and G-CSF gene–targeted mice remain capable of (low levels of) neutropoiesis, it has been hypothesized that trace amounts of lipopolysaccharides or intrinsic TLR agonists are sufficient to maintain a positive-feedback signal.62
In addition to positive- and negative-feedback loops that are operational in the early phase of differentiation, a multitude of regulatory (epi)genetic mechanisms65 are involved in the control of neutropoiesis in the postmitotic phase (see review see Hidalgo et al1 for additional details). These mechanisms range from the involvement of microRNAs66 and the phosphoinositide inositol-triphosphate 3-kinase,67 to the proinflammatory cytokine IL-32θ that negatively regulates PU.1 expression (inhibiting neutrophil expression).68
It is generally accepted that, under homeostasis in humans, ∼1011 neutrophils are produced and cleared daily, although this is based on a very short half-life of the neutrophils.1, 69 This short half-life has been challenged recently; therefore, this huge daily production of neutrophils is again under debate.26, 52 Currently, our knowledge about the maintenance of the neutrophil compartment in homeostasis is fragmentary. G-CSF is often mentioned as a key regulator of neutrophil production. Although G-CSF is a highly potent inducer of neutropoiesis (and can be administered to shorten chemotherapy-induced neutropenia in the clinic70), G-CSF–deficient mice and G-CSF receptor–deficient mice still show 10% to 30% of the normal neutrophil counts, and the resulting neutrophils are fully functional.71 This indicates that G-CSF plays an important, but not indispensable, role in neutropoiesis.
It has not been determined at which differentiation stage the neutrophil compartment is primarily compensating for the loss of mature neutrophils. Höfer and Rodewald31 hypothesized that 50% of the MPPs differentiate to the downstream stages,31 and these investigators put forward the idea that self-renewing divisions do not take place beyond the MPP stage. This is supported by the finding that only transplantation of ST-HSCs and MPPs leads to an appreciable repopulation of mature neutrophils in sublethally irradiated mice.16 GMPs have been shown to divide extensively at the population level in mice and humans using DNA labeling, mitotic indexes, Ki67, and the Fucci reporter.15, 28, 29, 30 However, all of these methods provide snapshots of division at the population level and do not give insight into the number of amplification divisions that GMPs undergo before differentiating into promyelocytes. Intriguingly, the generation times of these committed progenitors seem very short because ∼50% of the cells already become BrdU/EdU+ within 1 to 2 hours of staining, which opens up the question whether this technique produces artificial results.24, 25
In conclusion, it has not been determined at what phase in the neutrophil compartment neutrophil (progenitor) numbers are increased for compensation of the huge loss of neutrophils at the end of the lifespan of mature neutrophils that equals the daily production of these cells (estimated to be ∼1011 per day).
Until now, few studies have touched upon the mechanisms that underlie the kinetics of the neutrophil and its progenitors. This is essential for the determination of their numbers and life spans at different tissue locations. Two studies26, 52 on this subject adopted the assumption that every cell at a certain stage has the same chance to proliferate or differentiate. This chance model is generally applied in lymphocyte kinetics.72, 73 However, it is unknown whether subsequent division/differentiation steps of an individual neutrophil (progenitor) are a matter of chance and are best described by ordinary differential equations.26, 52 These uncertainties have led to very different estimations of the life span of the neutrophil, ranging from 7 to 9 hours to 5.4 days.26, 52, 69 No experimental data have unequivocally shown that such a model is adequate for the description of the neutrophil compartment. In fact, an old study69 supports a conveyor belt model based on a first-in first-out principle. This model is now nicely supported by a recent in vitro study showing a clear homogeneity in transit times through the different proliferation and differentiation phases within progenitor families.74
Activity of the microbiome and the functioning of immune cells are influenced by circadian rhythms. A study by Cossío et al suggests that aged neutrophils are primarily cleared from the circulation at night.75 Because this clearance generally occurs through efferocytosis by BM macrophages, this implies that G-CSF–dependent neutropoiesis, mentioned in the neutrostat hypothesis, is also dependent on circadian rhythms. This concept is supported by the demonstration that circadian rhythms influence the synthesis and release of cytokines, chemokines, and cytolytic factors.76 An interesting review series on the increasingly important role of circadian rhythms in immunology was published recently.77
Knowledge gaps in the understanding of neutropoiesis
The role of plasticity of fully differentiated neutrophils
Under defined in vitro conditions, different neutrophil functional phenotypes exist (Figure 2).78 They can even transdifferentiate into cells with characteristics of DCs, including the propensity for antigen presentation,79, 80 cell surface marker expression profiles, and nuclear morphology. This transdifferentiation is poorly understood in terms of the intracellular signals driving differentiation. These neutrophils might be lost from analysis as they change their typical neutrophil characteristics.79, 80 Importantly, a recent study applying pseudo-time analysis on RNA sequencing data of single neutrophils provided evidence that the neutrophil compartment in the mouse differentiates as a single continuum, denying the presence of true neutrophil phenotypes in the mouse that are characterized by unique transcriptional signatures.81 This study implies that heterogeneity in the neutrophil compartment is caused by instruction in the tissues rather than differentiation along separate differentiation paths. This conclusion is at odds with our study in humans that supports parallel differentiation of neutrophil phenotypes.51
Determination of generation times of neutrophil progenitors in the BM in homeostasis
Transplantation of progenitors into sublethally irradiated mice has the disadvantage of differentiation of cells in an “empty” neutrophil compartment, leading to increased production rates. Data regarding this issue is lacking in humans.
Data obtained with BrdU and EdU labeling in mice imply extremely fast cell divisions in MPP and GMP pools
Within 1 to 2 hours of labeling with BrdU or EdU, 24% of MPPs and 46% of GMPs become positive for this marker of DNA synthesis.24, 25 Likewise, 30% of terminally differentiated neutrophils become positive within 24 hours after start of BrdU/EdU labeling. These data suggest extremely short generation times and postmitotic transfer times in mice in vivo that are not recapitulated in vitro.82
How does the system compensate for the clearance of 1011 neutrophils per day?
This large number is based, in part, on the assumption that a neutrophil lives for 7 to 9 hours.69 Extrapolation of data described by Dmytrus and colleagues,83 with the assumption that the complete BM in humans consists of ∼1012 nucleated CD45+ cells, leads to an estimate of 2 × 109 MPPs in the BM of a healthy human; 50% of the cells in this compartment divide each day.31 Following the hypothesis that most of the self-renewal divisions (Figure 2) are in the MMP compartment, 3 or 4 consecutive divisions per day would be necessary to produce sufficient numbers of neutrophil progenitors that originate from differentiation-linked divisions to compensate for the daily loss of 1011 neutrophils. This putative large number of divisions in the MPP pool can be more accurately determined when it is clear how many sequential divisions are present in the GMP pool. It is important to emphasize that, if the hypothesis that the half–life span of neutrophils is much longer than 7 to 9 hours is accepted,26 the requirement for such a high number of divisions in the MPP/GMP pools is reduced significantly.
Where does the majority of neutrophil clearance occur in the body?
It is clear that CXCL12-CXCR4 play an important role in neutrophil retention84 in, and homing85 to, the BM, but the efferocytosis of neutrophils by BM macrophages seems to be a rare event. The role of efferocytosis of neutrophils in peripheral tissues is uncertain, although many tissues harbor fully differentiated neutrophils.86
Most in vivo studies are performed in mice, and the translation of these results to the functioning of human BM remains difficult
There is marked difference between the isolation of human and mouse BM material. In humans, BM is primarily harvested by aspiration of the central BM.87 On the other, mice are euthanized, followed by flushing of their bones to isolate central BM, and endosteal cells are collected from bone fragments.88 Because aspiration collects fluid from the central BM, these samples cannot be used to compare the endosteal niche in mice and humans.
Concluding remarks
In this review, neutropoiesis is described as a continuous model of lineage restriction, incorporating an intricate network in which neutropoiesis in homeostasis is closely regulated by intrinsic signals and environmental cues. Within the limitations of the knowledge gaps, we hypothesize a constant baseline production of neutrophils from HSCs, independent of extrinsic cues, that is based on, among other factors, their TF cistromes profiles, natural fluctuations, and circadian rhythms. This is supported by studies on the role of G-CSF and germ-free mice, which indicate a production of ∼10% to 20% of wild-type levels. This baseline is increased by a constant demand for neutrophil production that is driven by danger-associated molecular pattern signaling and is counterbalanced by the clearance of neutrophils by macrophages, leading to inhibition of the IL-23–IL-17–G-CSF pathway. Future studies must be focused on the gap in knowledge to understand a system that produces and eliminates 1011 cells per day in homeostasis and even more so in times of acute or chronic infection and/or inflammation.
Acknowledgments
The authors thank Nienke Vrisekoop for critically reading the manuscript. The figures were created with BioRender.
Footnotes
Overbeeke et al reviewed our current understanding of the process of neutrophil ontogeny in the bone marrow. The traditional models have changed in the face of better molecular characterization of cell fate decisions, and a broader understanding of the intrinsic and extrinsic signals that regulate neutrophil proliferation, maturation, and clearance.
Authorship
Contribution: C.O., T.T., and L.K. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
REFERENCES
- 1.Hidalgo A, Chilvers ER, Summers C, Koenderman L. The neutrophil life cycle. Trends Immunol. 2019;40(7):584–597. doi: 10.1016/j.it.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Manz MG, Boettcher S. Emergency granulopoiesis. Nat Rev Immunol. 2014;14(5):302–314. doi: 10.1038/nri3660. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence SM, Corriden R, Nizet V. The ontogeny of a neutrophil: mechanisms of granulopoiesis and homeostasis. Microbiol Mol Biol Rev. 2018;82(1):1–22. doi: 10.1128/MMBR.00057-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11(7):585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 5.Cheng H, Zheng Z, Cheng T. New paradigms on hematopoietic stem cell differentiation. Protein Cell. 2020;11(1):34–44. doi: 10.1007/s13238-019-0633-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietras EM, Reynaud D, Kang YA, et al. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell. 2015;17(1):35–46. doi: 10.1016/j.stem.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watcham S, Kucinski I, Gottgens B. New insights into hematopoietic differentiation landscapes from single-cell RNA sequencing. Blood. 2019;133(13):1415–1426. doi: 10.1182/blood-2018-08-835355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tusi BK, Wolock SL, Weinreb C, et al. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature. 2018;555(7694):54–60. doi: 10.1038/nature25741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hay SB, Ferchen K, Chetal K, Grimes HL, Salomonis N. The Human Cell Atlas bone marrow single-cell interactive web portal. Exp Hematol. 2018;68:51–61. doi: 10.1016/j.exphem.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellin D, Loperfido M, Baricordi C, et al. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat Commun. 2019;10(1):2395. doi: 10.1038/s41467-019-10291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velten L, Haas SF, Raffel S, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol. 2017;19(4):271–281. doi: 10.1038/ncb3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf FA, Hamey FK, Plass M, et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019;20(1):59. doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber R, Pietsch D, Günther J, Welz B, Vogt N, Brand K. Regulation of monocyte differentiation by specific signaling modules and associated transcription factor networks. Cell Mol Life Sci. 2014;71(1):63–92. doi: 10.1007/s00018-013-1322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoppe PS, Schwarzfischer M, Loeffler D, et al. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature. 2016;535(7611):299–302. doi: 10.1038/nature18320. [DOI] [PubMed] [Google Scholar]
- 15.Busch K, Klapproth K, Barile M, et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518(7540):542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- 16.Boyer SW, Rajendiran S, Beaudin AE, et al. Clonal and quantitative in vivo assessment of hematopoietic stem cell differentiation reveals strong erythroid potential of multipotent cells. Stem Cell Reports. 2019;12(4):801–815. doi: 10.1016/j.stemcr.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laurenti E, Göttgens B. From haematopoietic stem cells to complex differentiation landscapes. Nature. 2018;553(7689):418–426. doi: 10.1038/nature25022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adolfsson J, Månsson R, Buza-Vidas N, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121(2):295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Naik SH, Perié L, Swart E, et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496(7444):229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- 20.Paul F, Arkin Y, Giladi A, et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors [published correction appears in Cell. 2016;164(1-2):325] Cell. 2015;163(7):1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 21.Buenrostro JD, Corces MR, Lareau CA, et al. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell. 2018;173(6):1535–1548.e16. doi: 10.1016/j.cell.2018.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menezes S, Melandri D, Anselmi G, et al. The heterogeneity of Ly6Chi monocytes controls their differentiation into iNOS+ macrophages or monocyte-derived dendritic cells. Immunity. 2016;45(6):1205–1218. doi: 10.1016/j.immuni.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yáñez A, Ng MY, Hassanzadeh-Kiabi N, Goodridge HS. IRF8 acts in lineage-committed rather than oligopotent progenitors to control neutrophil vs monocyte production. Blood. 2015;125(9):1452–1459. doi: 10.1182/blood-2014-09-600833. [DOI] [PubMed] [Google Scholar]
- 24.Evrard M, Kwok IWH, Chong SZ, et al. Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking, and effector functions. Immunity. 2018;48(2):364–379.e8. doi: 10.1016/j.immuni.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Passegué E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202(11):1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pillay J, den Braber I, Vrisekoop N, et al. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood. 2010;116(4):625–627. doi: 10.1182/blood-2010-01-259028. [DOI] [PubMed] [Google Scholar]
- 27.Tak T, Tesselaar K, Pillay J, Borghans JAM, Koenderman L. What's your age again? Determination of human neutrophil half-lives revisited. J Leukoc Biol. 2013;94(4):595–601. doi: 10.1189/jlb.1112571. [DOI] [PubMed] [Google Scholar]
- 28.Mary JY. Normal human granulopoiesis revisited. II. Bone marrow data. Biomed Pharmacother. 1985;39(2):66–77. [PubMed] [Google Scholar]
- 29.Dresch C, Troccoli G, Mary JY. Growth fraction of myelocytes in normal human granulopoiesis. Cell Tissue Kinet. 1986;19(1):11–22. doi: 10.1111/j.1365-2184.1986.tb00711.x. [DOI] [PubMed] [Google Scholar]
- 30.Matarraz S, Fernandez C, Albors M, et al. Cell-cycle distribution of different cell compartments in normal versus reactive bone marrow: a frame of reference for the study of dysplastic hematopoiesis. Cytometry B Clin Cytom. 2011;80(6):354–361. doi: 10.1002/cyto.b.20605. [DOI] [PubMed] [Google Scholar]
- 31.Höfer T, Rodewald HR. Differentiation-based model of hematopoietic stem cell functions and lineage pathways. Blood. 2018;132(11):1106–1113. doi: 10.1182/blood-2018-03-791517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laslo P, Pongubala JMR, Lancki DW, Singh H. Gene regulatory networks directing myeloid and lymphoid cell fates within the immune system. Semin Immunol. 2008;20(4):228–235. doi: 10.1016/j.smim.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Hoogendijk AJ, Pourfarzad F, Aarts CEM, et al. Dynamic transcriptome-proteome correlation networks reveal human myeloid differentiation and neutrophil-specific programming. Cell Rep. 2019;29(8):2505–2519.e4. doi: 10.1016/j.celrep.2019.10.082. [DOI] [PubMed] [Google Scholar]
- 34.Fisher RC, Scott EW. Role of PU.1 in hematopoiesis. Stem Cells. 1998;16(1):25–37. doi: 10.1002/stem.160025. [DOI] [PubMed] [Google Scholar]
- 35.Krumsiek J, Marr C, Schroeder T, Theis FJ. Hierarchical differentiation of myeloid progenitors is encoded in the transcription factor network. PLoS One. 2011;6(8):e22649. doi: 10.1371/journal.pone.0022649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kastner P, Chan S. PU.1: a crucial and versatile player in hematopoiesis and leukemia. Int J Biochem Cell Biol. 2008;40(1):22–27. doi: 10.1016/j.biocel.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 37.Chou ST, Khandros E, Bailey LC, et al. Graded repression of PU.1/Sfpi1 gene transcription by GATA factors regulates hematopoietic cell fate. Blood. 2009;114(5):983–994. doi: 10.1182/blood-2009-03-207944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burda P, Laslo P, Stopka T. The role of PU.1 and GATA-1 transcription factors during normal and leukemogenic hematopoiesis. Leukemia. 2010;24(7):1249–1257. doi: 10.1038/leu.2010.104. [DOI] [PubMed] [Google Scholar]
- 39.Kurotaki D, Yamamoto M, Nishiyama A, et al. IRF8 inhibits C/EBPα activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nat Commun. 2014;5(1):4978. doi: 10.1038/ncomms5978. [DOI] [PubMed] [Google Scholar]
- 40.Mak KS, Funnell APW, Pearson RCM, Crossley M. PU.1 and haematopoietic cell fate: dosage matters. Int J Cell Biol. 2011;2011:808524. doi: 10.1155/2011/808524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Möröy T, Vassen L, Wilkes B, Khandanpour C. From cytopenia to leukemia: the role of Gfi1 and Gfi1b in blood formation. Blood. 2015;126(24):2561–2569. doi: 10.1182/blood-2015-06-655043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laslo P, Spooner CJ, Warmflash A, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126(4):755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 43.Cooper S, Guo H, Friedman AD. The +37 kb Cebpa enhancer is critical for Cebpa myeloid gene expression and contains functional sites that bind SCL, GATA2, C/EBPα, PU.1, and additional Ets factors. PLoS One. 2015;10(5):e0126385. doi: 10.1371/journal.pone.0126385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Q, Dong F. Gfi-1 inhibits the expression of eosinophil major basic protein (MBP) during G-CSF-induced neutrophilic differentiation. Int J Hematol. 2012;95(6):640–647. doi: 10.1007/s12185-012-1078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Hu N, Dong F. Gfi1-mediated repression of c-Fos, Egr-1 and Egr-2, and inhibition of ERK1/2 signaling contribute to the role of Gfi1 in granulopoiesis. Sci Rep. 2019;9(1):1–9. doi: 10.1038/s41598-018-37402-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ostuni R, Natoli G, Cassatella MA, Tamassia N. Epigenetic regulation of neutrophil development and function. Semin Immunol. 2016;28(2):83–93. doi: 10.1016/j.smim.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 47.Verbeek W, Gombart AF, Chumakov AM, Müller C, Friedman AD, Koeffler HP. C/EBPepsilon directly interacts with the DNA binding domain of c-myb and cooperatively activates transcription of myeloid promoters. Blood. 1999;93(10):3327–3337. [PubMed] [Google Scholar]
- 48.Hock H, Orkin SH. Zinc-finger transcription factor Gfi-1: versatile regulator of lymphocytes, neutrophils and hematopoietic stem cells. Curr Opin Hematol. 2006;13(1):1–6. doi: 10.1097/01.moh.0000190111.85284.8f. [DOI] [PubMed] [Google Scholar]
- 49.Gombart AF, Shiohara M, Kwok SH, Agematsu K, Komiyama A, Koeffler HP. Neutrophil-specific granule deficiency: homozygous recessive inheritance of a frameshift mutation in the gene encoding transcription factor CCAAT/enhancer binding protein–epsilon. Blood. 2001;97(9):2561–2567. doi: 10.1182/blood.v97.9.2561. [DOI] [PubMed] [Google Scholar]
- 50.Lekstrom-Himes JA. The role of C/EBP(epsilon) in the terminal stages of granulocyte differentiation. Stem Cells. 2001;19(2):125–133. doi: 10.1634/stemcells.19-2-125. [DOI] [PubMed] [Google Scholar]
- 51.Tak T, Wijten P, Heeres M, et al. Human CD62Ldim neutrophils identified as a separate subset by proteome profiling and in vivo pulse-chase labeling. Blood. 2017;129(26):3476–3485. doi: 10.1182/blood-2016-07-727669. [DOI] [PubMed] [Google Scholar]
- 52.Lahoz-Beneytez J, Elemans M, Zhang Y, et al. Human neutrophil kinetics: modeling of stable isotope labeling data supports short blood neutrophil half-lives. Blood. 2016;127(26):3431–3438. doi: 10.1182/blood-2016-03-700336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dancey JT, Deubelbeiss KA, Harker LA, Finch CA. Neutrophil kinetics in man. J Clin Invest. 1976;58(3):705–715. doi: 10.1172/JCI108517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weishaupt H, Sigvardsson M, Attema JL. Epigenetic chromatin states uniquely define the developmental plasticity of murine hematopoietic stem cells. Blood. 2010;115(2):247–256. doi: 10.1182/blood-2009-07-235176. [DOI] [PubMed] [Google Scholar]
- 55.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26(47):6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 56.Cullen SM, Mayle A, Rossi L, Goodell MA. Hematopoietic stem cell development: an epigenetic journey. Curr Top Dev Biol. 2014;107:39–75. doi: 10.1016/B978-0-12-416022-4.00002-0. [DOI] [PubMed] [Google Scholar]
- 57.Garcia-Bassets I, Wang D. Cistrome plasticity and mechanisms of cistrome reprogramming. Cell Cycle. 2012;11(17):3199–3210. doi: 10.4161/cc.21281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rankin SM. The bone marrow: a site of neutrophil clearance. J Leukoc Biol. 2010;88(2):241–251. doi: 10.1189/jlb.0210112. [DOI] [PubMed] [Google Scholar]
- 59.Doran AC, Yurdagul A, Jr, Tabas I. Efferocytosis in health and disease. Nat Rev Immunol. 2020;20(4):254–267. doi: 10.1038/s41577-019-0240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stark MA, Huo Y, Burcin TL, Morris MA, Olson TS, Ley K. Phagocytosis of apoptotic neutrophils regulates granulopoiesis via IL-23 and IL-17. Immunity. 2005;22(3):285–294. doi: 10.1016/j.immuni.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 61.Wirths S, Bugl S, Kopp HG. Neutrophil homeostasis and its regulation by danger signaling. Blood. 2014;123(23):3563–3566. doi: 10.1182/blood-2013-11-516260. [DOI] [PubMed] [Google Scholar]
- 62.Bugl S, Wirths S, Radsak MP, et al. Steady-state neutrophil homeostasis is dependent on TLR4/TRIF signaling. Blood. 2013;121(5):723–733. doi: 10.1182/blood-2012-05-429589. [DOI] [PubMed] [Google Scholar]
- 63.Boettcher S, Gerosa RC, Radpour R, et al. Endothelial cells translate pathogen signals into G-CSF-driven emergency granulopoiesis. Blood. 2014;124(9):1393–1403. doi: 10.1182/blood-2014-04-570762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Märklin M, Bugl S, Wirths S, et al. Oral intake of lipopolysaccharide regulates toll-like receptor 4-dependent granulopoiesis. Exp Biol Med (Maywood). 2020;245(14):1254–1259. doi: 10.1177/1535370220931043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grassi L, Pourfarzad F, Ullrich S, et al. Dynamics of transcription regulation in human bone marrow myeloid differentiation to mature blood neutrophils. Cell Rep. 2018;24(10):2784–2794. doi: 10.1016/j.celrep.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Larsen MT, Mattias H, Glenth⊘j A, et al. miRNA-130a regulates C/EBPε expression during granulopoiesis. Blood. 2014;123(7):1079–1089. doi: 10.1182/blood-2013-08-523233. [DOI] [PubMed] [Google Scholar]
- 67.Elich M, Sauer K. Regulation of hematopoietic cell development and function through phosphoinositides. Front Immunol. 2018;9:931. doi: 10.3389/fimmu.2018.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim MS, Kang JW, Park YS, et al. IL-32θ inhibits monocytic differentiation of leukemia cells by attenuating expression of transcription factor PU.1. Oncotarget. 2015;6(6):4394–4405. doi: 10.18632/oncotarget.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cartwright GE, Athens JW, Wintrobe MM. The kinetics of granulopoiesis in normal man. Blood. 1964;24(6):780–803. [PubMed] [Google Scholar]
- 70.Bugl S, Wirths S, Müller MR, Radsak MP, Kopp HG. Current insights into neutrophil homeostasis. Ann N Y Acad Sci. 2012;1266(1):171–178. doi: 10.1111/j.1749-6632.2012.06607.x. [DOI] [PubMed] [Google Scholar]
- 71.Craig M, Humphries AR, Mackey MC. A mathematical model of granulopoiesis incorporating the negative feedback dynamics and kinetics of G-CSF/neutrophil binding and internalization. Bull Math Biol. 2016;78(12):2304–2357. doi: 10.1007/s11538-016-0179-8. [DOI] [PubMed] [Google Scholar]
- 72.Asquith B, Debacq C, Macallan DC, Willems L, Bangham CRM. Lymphocyte kinetics: the interpretation of labelling data. Trends Immunol. 2002;23(12):596–601. doi: 10.1016/s1471-4906(02)02337-2. [DOI] [PubMed] [Google Scholar]
- 73.Asquith B, Borghans JAM, Ganusov VV, Macallan DC. Lymphocyte kinetics in health and disease. Trends Immunol. 2009;30(4):182–189. doi: 10.1016/j.it.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 74.Tak T, Prevedello G, Simon G, et al. HSPCs display within-family homogeneity in differentiation and proliferation despite population heterogeneity. eLife. 2021;10:e60624. doi: 10.7554/eLife.60624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cossío I, Lucas D, Hidalgo A. Neutrophils as regulators of the hematopoietic niche. Blood. 2019;133(20):2140–2148. doi: 10.1182/blood-2018-10-844571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Labrecque N, Cermakian N. Circadian clocks in the immune system. J Biol Rhythms. 2015;30(4):277–290. doi: 10.1177/0748730415577723. [DOI] [PubMed] [Google Scholar]
- 77.Milling S. It's time to think about circadian rhythms. Immunology. 2020;161(4):259–260. doi: 10.1111/imm.13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hellebrekers P, Vrisekoop N, Koenderman L. Neutrophil phenotypes in health and disease. Eur J Clin Invest. 2018;48:e12943. doi: 10.1111/eci.12943. suppl 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsushima H, Geng S, Lu R, et al. Neutrophil differentiation into a unique hybrid population exhibiting dual phenotype and functionality of neutrophils and dendritic cells. Blood. 2013;121(10):1677–1689. doi: 10.1182/blood-2012-07-445189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mysore V, Cullere X, Mears J, et al. FcγR engagement reprograms neutrophils into antigen cross-presenting cells that elicit acquired anti-tumor immunity. Nat Commun. 2021;12(1):4791. doi: 10.1038/s41467-021-24591-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grieshaber-Bouyer R, Radtke FA, Cunin P, et al. ImmGen Consortium The neutrotime transcriptional signature defines a single continuum of neutrophils across biological compartments. Nat Commun. 2021;12(1):2856. doi: 10.1038/s41467-021-22973-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crossen PE, Durie BG, Trent JM. Generation time (GT) of human bone marrow cells cultured in the CFC-gm assay. Cell Tissue Kinet. 1986;19(5):533–538. doi: 10.1111/j.1365-2184.1986.tb00753.x. [DOI] [PubMed] [Google Scholar]
- 83.Dmytrus J, Matthes-Martin S, Pichler H, et al. Multi-color immune-phenotyping of CD34 subsets reveals unexpected differences between various stem cell sources. Bone Marrow Transplant. 2016;51(8):1093–1100. doi: 10.1038/bmt.2016.88. [DOI] [PubMed] [Google Scholar]
- 84.McDermott DH, Pastrana DV, Calvo KR, et al. Plerixafor for the treatment of WHIM syndrome. N Engl J Med. 2019;380(2):163–170. doi: 10.1056/NEJMoa1808575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Furze RC, Rankin SM. The role of the bone marrow in neutrophil clearance under homeostatic conditions in the mouse. FASEB J. 2008;22(9):3111–3119. doi: 10.1096/fj.08-109876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ballesteros I, Rubio-Ponce A, Genua M, et al. Co-option of neutrophil fates by tissue environments. Cell. 2020;183(5):1282–1297.e18. doi: 10.1016/j.cell.2020.10.003. [DOI] [PubMed] [Google Scholar]
- 87.Bain BJ. Bone marrow aspiration. J Clin Pathol. 2001;54(9):657–663. doi: 10.1136/jcp.54.9.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grassinger J, Haylock DN, Williams B, Olsen GH, Nilsson SK. Phenotypically identical hemopoietic stem cells isolated from different regions of bone marrow have different biologic potential. Blood. 2010;116(17):3185–3196. doi: 10.1182/blood-2009-12-260703. [DOI] [PubMed] [Google Scholar]
- 89.Hérault A, Binnewies M, Leong S, et al. Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature. 2017;544(7648):53–58. doi: 10.1038/nature21693. [DOI] [PMC free article] [PubMed] [Google Scholar]