Abstract
The UDP-glucuronosyltransferase (UGT) family of enzymes play a central role in the metabolism and detoxification of a wide range of endogenous and exogenous compounds. UGTs exhibit a high degree of structural similarity and display overlapping substrate specificity, often making estimations of potential drug-drug interactions difficult to fully elucidate. One such interaction yet to be examined may be occurring between UGTs and cannabinoids, as the legalization of recreational and medicinal cannabis and subsequent co-usage of cannabis and therapeutic drugs increases in the United States and internationally. In the present study, the inhibition potential of the major cannabinoids Δ9-tetrahydrocannabinol (THC), cannabidiol (CBD), and cannabinol (CBN), as well as their major metabolites, was determined in microsomes isolated from HEK293 cells overexpressing individual recombinant UGTs and in microsomes from human liver and kidney specimens. The highest inhibition was seen by CBD against the glucuronidation activity of UGTs 1A9, 2B4, 1A6, and 2B7, with binding-corrected IC50 values of 0.12 ± 0.020 µM, 0.22 ± 0.045 µM, 0.40 ± 0.10 µM, and 0.82 ± 0.15 µM, respectively. Strong inhibition of UGT1A9 was also demonstrated by THC and CBN, with binding-corrected IC50 values of 0.45 ± 0.12 μM and 0.51 ± 0.063 μM, respectively. Strong inhibition of UGT2B7 was also observed for THC and CBN; no or weak inhibition was observed with cannabinoid metabolites. This inhibition of UGT activity suggests that in addition to playing an important role in drug-drug interactions, cannabinoid exposure may have important implications in patients with impaired hepatic or kidney function.
SIGNIFICANCE STATEMENT
Major cannabinoids found in the plasma of cannabis users inhibit several UDP-glucuronosyltransferase (UGT) enzymes, including UGT1A6, UGT1A9, UGT2B4, and UGT2B7. This study is the first to show the potential of cannabinoids and their metabolites to inhibit all the major kidney UGTs as well as the two most abundant UGTs present in liver. This study suggests that as all three major kidney UGTs are inhibited by cannabinoids, greater drug-drug interaction effects might be observed from co-use of cannabinods and therapeutics that are cleared renally.
Introduction
UDP-glucuronosyltransferases (UGTs) are an important family of phase II metabolizing enzymes that facilitate the detoxification of a wide variety of endogenous and exogenous compounds, including steroid hormones, drugs, and environmental carcinogens (Meech et al., 2019). Mammalian UGTs are classified based on structural and amino acid sequence homology into two main families, the UGT1 and UGT2 families, which are further divided into three subfamilies UGT1A, UGT2A, and UGT2B, and catalyze the transfer of glucuronic acid from UDP glucuronic acid (UDPGA) to an electrophilic moiety of a given substrate, resulting in a more polar conjugate that is more easily excreted from the body in the urine or bile (Bushey and Lazarus, 2012). An additional subfamily, the UGT3A subfamily, contains two members, UGT3A1 and UGT3A2, which use the alternative sugar donors UDP-N-acetylglucosamine, UDP-glucose, and UDP-xylose as co-substrates (MacKenzie et al., 2011). Mammalian UGTs are membrane-bound enzymes localized in the endoplasmic reticulum and expressed with a high degree of tissue specificity (Meech et al., 2019). Although many UGTs are highly expressed in the liver, some are also expressed in extrahepatic tissues, including kidney and tissues of the aerodigestive tract (Meech et al., 2019; Vergara et al., 2020). The UGTs that exhibit the highest level of hepatic expression are UGTs 2B7 (17% of total hepatic UGT expression), 2B4 (16.1%), 2B15 (11.2%), and 1A1 (11%) (Kasteel et al., 2020). A number of UGTs are also expressed in the kidney, including UGT1A9 (45% of total renal UGT expression), UGT2B7 (41%), and UGT1A6 (7%) (Rowland et al., 2013).
UGTs account for the metabolism of 15% of pharmaceuticals, and one-seventh of the drugs prescribed in the United States in 2002 are cleared by the UGTs (Williams et al., 2004). Although studies of drug-drug interactions (DDIs) are a major emphasis of research for phase I metabolizing enzymes including the cytochrome P450 enzyme family, UGT enzymes have historically received less scrutiny for their DDI potential, even though drug interactions via the inhibition of glucuronidation have been increasingly identified. Impaired glucuronidation activity can cause undesired effects resulting from the slow elimination of endogenous substances such as bilirubin (Sun et al., 2017) as well as the buildup of toxic drug metabolites, as has been documented in studies correlating individuals with UGT1A1-deficient phenotypes and irinotecan toxicity (Iyer et al., 1998; Tallman et al., 2007). DDIs between therapeutics and UGT inhibitors have also been observed in the case of UGT2B7 inhibition by both valproic acid and probenecid (Cimoch et al., 1998; Rowland et al., 2006).
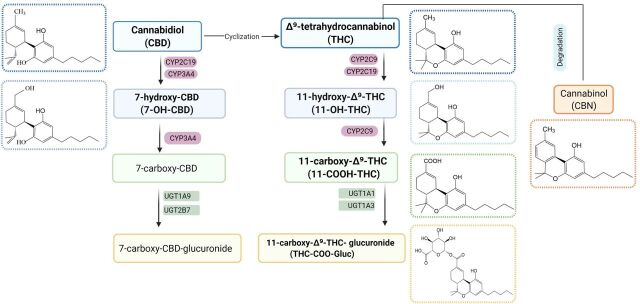
The recent legalization of cannabis has caused a dramatic increase in the use of cannabis-derived products in both recreational and medicinal situations, where cannabis is frequently used or targeted for more chronic diseases like cancer, arthritis, and depression and often concurrently used with important groups of conventional medications including anticancer agents, antidepressants, and pain medications (Bridgeman and Abazia, 2017). Situations in which polypharmacy is occurring within a patient could result in deleterious DDIs between cannabinoids and any number of therapeutic agents. Δ9-tetrahydrocannabinol (THC) is the best described psychoactive constituent of cannabis, and plasma concentrations of THC and its active metabolite, 11-hydroxy (OH)-THC, quickly peak after usage and decrease rapidly over a short duration, dependent on the specific mode of consumption (Fig. 1) (Sharma et al., 2012). In contrast, the inactive metabolites, 11-nor-9-carboxy-Δ9-tetrahydrocannabinol (THC-COOH) and 11-COO-Δ9-tetrahydrocannabinol-glucuronide (THC-COO-Gluc), peak much more slowly, to a lower level than the active cannabinoids, and remain present in plasma over a much longer duration of time (Huestis, 2007). Actual plasma levels of active and inactive cannabinoids are highly variable (in the micromolar to submicromolar range) and will vary widely depending on the user, dose, and method of ingestion. Cannabinol (CBN) appears to be a degradation product of THC within the Cannabis plant (Russo and Marcu, 2017) and has been shown to be only weakly psychoactive. Cannabidiol (CBD) is often termed as medical marijuana and interacts with the CB1 and CB2 receptors in the brain with a much lower affinity as compared with THC and 11-OH-THC, resulting in extremely low psychoactive effects (Pertwee, 2008). However, CBD usage is rapidly expanding among many patient populations due in part to its good safety profile (Larsen and Shahinas, 2020). Recent clinical and preclinical trials have shown that CBD has a broad range of potential applications, displaying anti-inflammatory properties, antipsychotic, and antiepileptic effects, as well as modulation of the immune system and the central nervous system (Esposito et al., 2013; Boychuk et al., 2015; Campos et al., 2016; Devinsky et al., 2016). Similarly, CBD and its metabolites, 7-hydroxy-cannabidiol (7-OH-CBD) and 7-carboxy-cannabidiol (CBD-COOH), are present in the plasma after cannabis inhalation, with unchanged CBD and glucuronidated CBD (CBD-COO-Gluc), as the main excretion products in urine (Harvey and Mechoulam, 1990; Huestis, 2007). All cannabinoids are highly lipophilic and concentrate in tissues with slow release back into the bloodstream (Huestis, 2007). This leads to varying plasma concentrations of active and inactive cannabinoids that persist in the bloodstream, potentially incurring deleterious DDIs over a much wider time frame than that of the initial cannabis consumption.
Fig. 1.
Metabolic pathways and structures of major cannabinoids and their metabolites.
Previous studies have shown that THC, CBD, and CBN can strongly inhibit several major hepatic cytochrome P450s (P450s) (Yamaori et al., 2010; Yamaori et al., 2011a; Yamaori et al., 2011b; Yamaori et al., 2011c; Jiang et al., 2013; Cox et al., 2019; Nasrin et al., 2021). In addition, the major active metabolite of THC, 11-OH-THC, and two major inactive metabolites, THC-COOH and THC-COO-Gluc, also exhibited strong inhibition of a number of hepatic P450 enzymes (Nasrin et al., 2021). In the present study, the inhibition potential of major cannabinoids and their metabolites against major hepatic and renal human UGT enzymes were evaluated.
Material and Methods
Chemicals and Reagents.
THC, 11-OH-THC, THC-COOH, THC-COO-Gluc, CBD, 7-OH-CBD, and CBN were purchased from Cayman Chemicals (Ann Arbor, MI) or Sigma-Aldrich (St. Louis, MO). Pooled human liver microsomes (HLMs) [n = 50, mixed gender (21 female and 29 male), race (42 Caucasian, 4 Hispanic, 2 African American, and 2 Asian), and age (5–77 years)] and pooled human kidney microsomes (HKMs) [n = 8, mixed gender (50% each), race (3 African American, 3 Caucasian, and 2 Hispanic), and age (42–70 years)] were obtained from Sekisui Xenotech, LLC (Lenexa, KS). β-estradiol, chenodeoxycholic acid, trifluoperazine, serotonin, propofol, codeine, zidovudine (AZT), nicotine, oxazepam, dihydroexemestane, ketoconazole, diclofenac, acetaminophen, and furosemide were all purchased from Sigma-Aldrich. Optima grade methanol, acetonitrile, and formic acid were obtained from Fisher Scientific (Waltham, MA). Ultra-low-binding microcentrifuge tubes, Dulbecco’s modified Eagle’s medium, Dulbecco’s phosphate-buffered saline, UDPGA, alamethacin, MgCl2, and geneticin (G418) were purchased from VWR (Radnor, PA). BCA protein assays were purchased from Pierce (Rockford, IL), premium grade FBS was purchased from Seradigm (Radnor, PA), and ChromatoPur bovine serum albumin (BSA) was purchased from MB Biomedicals (Santa Ana, CA).
Inhibition Assays.
Human embryonic kidney (HEK) 293 cells individually overexpressing recombinant UGTs 1A1, 1A3, 1A4, 1A6, 1A9, 2B4, 2B7, 2B10, 2B15, and 2B17 were developed and described previously (Dellinger et al., 2006). Microsomal membrane fractions of UGT-overexpressing cell lines were prepared by differential centrifugation as previously described, with total microsomal protein concentrations determined using the BCA assay as per the manufacturer’s recommendations. An initial screen, performed in duplicate, of the inhibition potential of individual cannabinoids and their metabolites against UGTs 1A1, 1A3, 1A4, 1A6, 1A9, 2B4, 2B7, 2B10, 2B15, and 2B17 were determined using microsomes (50–100 µg) from UGT-overexpressing HEK293 cell lines in reactions containing either 10 µM or 100 µM of cannabinoid or metabolite, probe substrate (Supplemental Table 1), 50 mM Tris-HCl buffer (pH 7.4), MgCl2 (5 mM), 2% BSA, and 4 mM UDPGA in a final reaction volume of 25 µl. All substrates were used at concentrations near their respective Michaelis-Menten constant (Km; Supplemental Table 1). As cannabinoids exhibit extensive nonspecific binding (70–90%) to protein and labware (Garrett and Hunt, 1974), microsomal incubation conditions were optimized to prevent underestimation of inhibitory potency (IC50). To reduce nonspecific binding and adsorption to labware, low-binding microcentrifuge tubes were used for all reactions, with BSA added to increase the solubility of cannabinoids as well as to sequester inhibitory long-chain unsaturated fatty acids (Rowland et al., 2008; Patilea-Vrana et al., 2019).
Microsomes were preincubated with alamethicin (50 µg/mg of microsomal protein) on ice for 20 minutes prior to incubation. The reaction was initiated by the addition of UDPGA and incubated for 60–120 minutes (Supplemental Table 1) at 37°C. Reactions were terminated and proteins precipitated by the addition of an equal volume (25 µl) of ice-cold stop solution (acetonitrile:methanol; 1:1). Samples were mixed on a vortex mixer and centrifuged at 17,000g for 15 minutes. The supernatant (∼50 µl) was transferred to an ultra-high-performance liquid chromatograph (UPLC) sample vial, and the probe metabolite was detected using a UPLC (Waters Acquity; Waters Corp, Milford, MA) coupled to a triple-quadrupole mass spectrometer (Waters Xevo TQD; Waters Corp) by multiple reaction monitoring analysis. As a positive control for every inhibition experiment, 10 µM or 100 µM probe inhibitors (ketoconazole/diclofenac) were added instead of the cannabinoid compounds. Reactions containing only vehicle (3% methanol) and without any inhibitor were used as an indicator of 100% activity for each substrate/enzyme combination. All IC50 analyses were performed in triplicate. Incubation conditions were optimized for HLMs, HKMs, and overexpressing cell lines for both microsomal protein and reaction time, with optimal conditions chosen based on the following criteria: 1) metabolite formation was linear with time and enzyme concentration, 2) substrate consumption was no more than 20% of the initial amount, and 3) metabolite formation was reliably and reproducibly detected by the UPLC–tandem mass spectrometry (UPLC-MS/MS) method used.
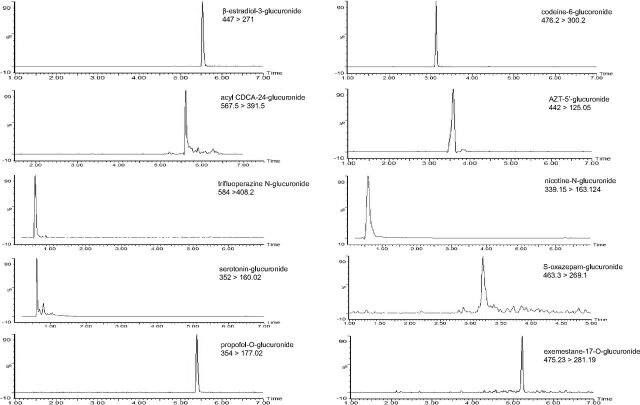
For UPLC-MS/MS, samples (2–5 µl) were injected onto an Acquity UPLC column (BEH C18, 1.7 µM, 2.1 × 100 mm; Waters Corp). A 9-minute gradient elution was used with mobile phases A (0.1% formic acid in water) and B (100% methanol) as follows: 1 minute at 95% A:5% B followed by a linear gradient for 7 minutes to 5% A:95% B, 1 minute at 5% A:95% B, and re-equilibration for 1 minute at 95% A:5% B. The flow rate was 0.4 ml/min, and the column temperature was 40°C. Analytes were detected using a Waters Xevo TQD tandem mass spectrometer equipped with a Zspray electrospray ionization interface operated in the positive ion mode for all the UGT metabolites tested in this study except furosemide glucuronide, which was analyzed in negative ion mode, with the capillary voltage at 0.6 kV. Nitrogen was used as both the cone and desolvation gas at 50 and 800 L/h, respectively. Ultrapure argon was used for collision-induced dissociation. The desolvation temperature was 500°C. For detection of the metabolite peaks, the mass spectrometer was operated in multiple reaction monitoring mode using the ion-related parameters for each transition. The following transitions were used for the detection of each probe metabolite: β-estradiol-3-glucuronide (m/z 447 > 271), acyl chenodeoxycholic acid-24-glucuronide (m/z 567.5 > 391.5), trifluoperazine N-glucuronide (m/z 584 > 408.2), serotonin-glucuronide (m/z 352 > 160.02), propofol-O-glucuronide (m/z 354 > 177.02), codeine-6-glucoronide (m/z 476.2 > 300.2), AZT-5′-glucuronide (m/z 442 > 125.05), nicotine-N-glucuronide (m/z 339.15 > 163.124), S-oxazepam-glucuronide (m/z 463.3 > 269.1), and exemestane-17-O-glucuronide (m/z 475.23 > 281.19).
Determination of IC50 Values.
For those cannabinoids or metabolites that inhibited UGT activity ≥50% at cannabinoid concentrations ≤100 µM, IC50 determinations were performed in HLMs, HKMs, and microsomes from HEK293 UGT-overexpressing cell lines, using multiple concentrations of cannabinoid inhibitor ranging between 0.5 and 120 µM.
Experiments were performed to determine nonspecific binding constants (fu,inc) for the individual cannabinoids in HEK293 microsomes, HLMs, and HKMs as previously described (Nasrin et al., 2021).
Statistical Analysis.
Data were exported and analyzed using an Excel spreadsheet (Microsoft). The amount of metabolite formed at each concentration of inhibitor relative to the control (percent relative activity) was calculated as Peak area of metabolite with inhibitor/Peak area of metabolite without inhibitor × 100%.
IC50 values were calculated by plotting the percent relative activity of UGT enzymes versus the log concentration of the test inhibitors using GraphPad Prism 7.04 software (GraphPad Software Inc., San Diego, CA).
Results
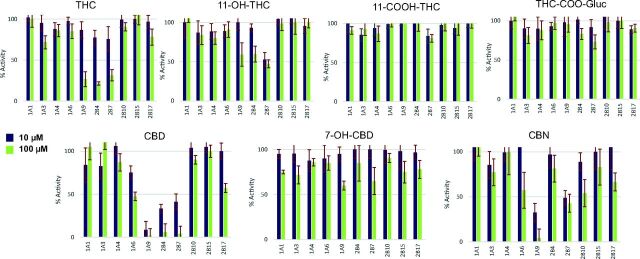
Glucuronide metabolite peaks were detected by liquid chromatography–tandem mass spectrometry in incubations of each probe substrate analyzed in these studies (Fig. 2). Using recombinant UGT (rUGT)–overexpressing cell microsomes and probe UGT substrates, preliminary screening studies demonstrated that 100 μM THC decreased the relative activity of microsomes from rUGT 1A9, 2B4, and 2B7 overexpressing cells by 74%, 79%, and 69%, respectively, as compared with control reactions without added cannabinoid (Fig. 3). A similar pattern was observed for CBD, with 10 µM CBD exhibiting 25%, 91%, 66%, and 58% inhibition and 100 μM CBD exhibiting 54%, 98%, 94%, and 96% inhibition, against microsomes from rUGTs 1A6, 1A9, 2B4, and 2B7 overexpressing cells, respectively, as compared with control reactions without added cannabinoid (Fig. 3). Similar to that observed for THC and CBD, CBN exhibited significant inhibition against rUGT1A9 and rUGT2B7 microsomes. Unlike that observed for THC and CBD, significant inhibition was not observed for rUGT2B4 microsomes with CBN. Although no significant inhibition was observed in rUGT1A1, rUGT1A3, rUGT1A4, and rUGT2B15 microsomes by THC, CBD, and CBN, marginal inhibition was observed for 100 μM CBD and CBN against rUGT2B17 microsomes (43% and 34%, respectively). Marginal inhibition (43% and 47%, respectively) was also observed for 100 μM CBN against rUGT1A6 and rUGT2B10 microsomes.
Fig. 2.
Chromatograms of probe metabolites in microsomes from UGT-overexpressing HEK293 cell lines. Probe substrates (at concentrations close to their known Km; see Supplemental Table 1) were incubated in rUGTs for 60–120 minutes at 37°C, and individual corresponding metabolites were analyzed by UPLC-MS/MS as described in Supplemental Table 1 and in the Materials and Methods.
Fig. 3.
Screening of cannabinoid inhibition of major hepatic UGTs in microsomes from UGT-overexpressing HEK293 cell lines. Probe substrates were β-estradiol for UGT1A1, chenodeoxycholic acid for UGT1A3, trifluoperazine for UGT1A4, serotonin for UGT1A6, propofol for UGT1A9, codeine for UGT2B4, zidovudine for UGT2B7, nicotine for UGT2B10, oxazepam for UGT2B15, and dihydroexemestane for UGT2B17. Incubations were performed using 10 or 100 µM of cannabinoid, with probe substrate concentrations at or close to their known Km for their corresponding enzyme (see Supplemental Table 1). Shown are the mean inhibition of two individual experiments performed for each probe substrate. Data are expressed as a percentage of metabolite formation formed in assays with cannabinoid compared with assays without cannabinoid.
For THC and CBD metabolites, no significant inhibition was observed using up to 100 µM THC-COOH or THC-COO-Gluc against any of the UGT enzymes tested. However, 100 µM 11-OH-THC resulted in marginal inhibition of the activities of rUGT1A9 (41%), rUGT2B4 (40%), and rUGT2B7 (53%) microsomes, whereas 100 µM 7-OH-CBD resulted in marginal decreases in the activities of rUGT1A9 (40%) and rUGT2B7 (45%) microsomes (Fig. 3).
The inhibitory effects of THC, 11-OH-THC, CBD and CBN were extended to establish IC50 values and binding-corrected IC50 values (IC50,u) for each cannabinoid against the UGT enzymes shown to be inhibited by ≥ 50% using 100 μM cannabinoid in the rUGT screening assays (described above). The unbound fraction in the incubation mixture were 0.042 ± 0.003, 0.038 ± 0.002, and 0.085 ± 0.005 in overexpressing HEK cell lines for THC, CBD and CBN respectively. For HLMs, the unbound fraction of THC, CBD and CBN in the incubation mixture were 0.048 ± 0.002, 0.051± 0.008, and 0.092 ± 0.006, respectively, and for HKMs the unbound fractions were 0.052 ± 0.005, 0.062 ± 0.009, and 0.12 ± 0.015, respectively.
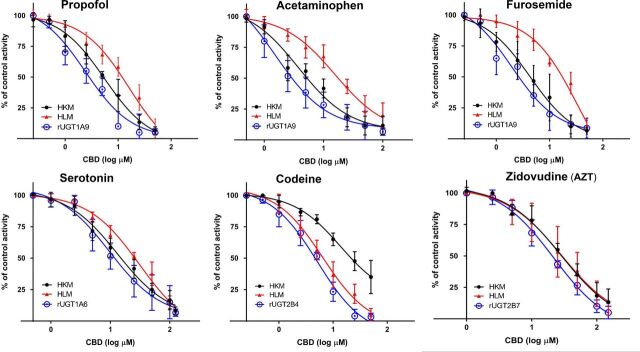
The strongest inhibition was observed by CBD against rUGTs 1A9 and 2B4, with IC50 values of 3.2 ± 0.52 µM and 5.8 ± 1.2 µM, and IC50,u values of 0.12 ± 0.020 µM and 0.22 ± 0.045 µM, using propofol and codeine as UGT1A9 and UGT2B4 probe substrates, respectively (Table 1). CBD also exhibited significant inhibition of the glucuronidation of serotonin (a probe substrate for rUGT1A6) in rUGT1A6 microsomes (IC50 = 10 ± 2.6 µM and IC50,u = 0.40 ± 0.10 µM), and AZT glucuronidation as a probe substrate in rUGT2B7 microsomes (IC50 = 21 ± 3.9 µM and IC50,u = 0.82 ± 0.15 µM). The IC50 values for CBD for propofol glucuronidation were similar in HKMs (IC50 = 5.5 ± 0.56 µM and IC50,u = 0.34 ± 0.035 µM) but higher in HLMs (IC50 = 19 ± 4.6 µM and IC50,u = 1.0 ± 0.24 µM) as compared with that observed for rUGT1A9 microsomes (Table 1), a pattern that was reversed in HKMs (IC50 = 39 ± 5.9 µM and IC50,u = 2.5 ± 0.37 µM) versus HLMs (IC50 = 8.0 ± 1.1 µM and IC50,u = 0.40 ± 0.058 µM) for CBD inhibition of codeine glucuronidation. The decreased level of inhibition of propofol glucuronidation by CBD in HLMs versus HKMs and the similar inhibition pattern of HKMs and rUGT1A9 microsomes is apparent when examining plots of percent glucuronidation activity versus CBD concentrations (Fig. 4). Similar to that observed for rUGT2B7 microsomes, more moderate inhibition was observed for CBD of AZT glucuronidation in HLMs (IC50 = 30 ± 4.1 µM and IC50,u = 1.5 ± 0.21 µM) and HKMs (IC50 = 35 ± 3.5 µM and IC50,u = 2.2 ± 0.22 µM), with IC50 values that were only slightly higher than that observed for rUGT2B7 microsomes (Table 1; Fig. 4). The IC50 values for serotonin glucuronidation of 28 ± 6.5 µM (IC50,u = 1.4 ± 0.33 μM), and 17 ± 3.7 µM (IC50,u = 1.0 ± 0.23 μM) in HLMs and HKMs, respectively, were slightly higher than that observed for rUGT1A9 microsomes (Table 1).
TABLE 1.
IC50 values (µM) of cannabinoids against major hepatic UGT enzymes in microsomes from recombinant UGT–overexpressing cells, HLMs, or HKMs.
| Probe substrate | Microsomes | THC | CBD | CBN | |||
|---|---|---|---|---|---|---|---|
| IC50 | IC50,u | IC50 | IC50,u | IC50 | IC50,u | ||
| µM | µM | µM | µM | µM | µM | ||
| Serotonin | rUGT1A6 | 10 ± 2.6 | 0.40 ± 0.10 | ||||
| HKM | NA | 17 ± 3.7 | 1.0 ± 0.23 | NA | |||
| HLM | 28 ± 6.5 | 1.4 ± 0.33 | |||||
| Codeine | rUGT2B4 | 11 ± 2.7 | 0.47 ± 0.11 | 5.8 ± 1.2 | 0.22 ± 0.045 | ||
| HKM | 55 ± 5.2 | 2.9 ± 0.27 | 39 ± 5.9 | 2.5 ± 0.37 | NA | ||
| HLM | 13 ± 2.6 | 0.61 ± 0.13 | 8.0 ± 1.1 | 0.40 ± 0.058 | |||
| AZT | rUGT2B7 | 33 ± 8.5 | 1.4 ± 0.36 | 21 ± 3.9 | 0.82 ± 0.15 | 49 ± 12 | 4.2 ± 1.1 |
| HKM | 51 ± 12 | 2.6 ± 0.65 | 35 ± 3.5 | 2.2 ± 0.22 | 57 ± 7.5 | 6.9 ± 0.90 | |
| HLM | 59 ± 6.6 | 2.8 ± 0.32 | 30 ± 4.1 | 1.5 ± 0.21 | 59 ± 8.6 | 5.5 ± 0.79 | |
| Propofol | rUGT1A9 | 11 ± 3.0 | 0.45 ± 0.12 | 3.2 ± 0.52 | 0.12 ± 0.020 | 6.0 ± 0.75 | 0.51 ± 0.063 |
| HKM | 12 ± 3.4 | 0.64 ± 0.18 | 5.5 ± 0.56 | 0.34 ± 0.035 | 7.5 ± 1.7 | 0.90 ± 0.20 | |
| HLM | 30 ± 6.4 | 1.4 ± 0.31 | 19 ± 4.6 | 1.0 ± 0.24 | 31 ± 4.1 | 2.9 ± 0.38 | |
| Furosemide | rUGT1A9 | 8.0 ± 0.47 | 0.33 ± 0.020 | 2.4 ± 0.66 | 0.090 ± 0.025 | 9.2 ± 2.1 | 0.78 ± 0.18 |
| HKM | 10 ± 4.1 | 0.54 ± 0.22 | 3.6 ± 0.80 | 0.22 ± 0.049 | 15 ± 0.77 | 1.9 ± 0.092 | |
| HLM | 32 ± 6.3 | 1.5 ± 0.30 | 29 ± 4.0 | 1.5 ± 0.20 | 34 ± 6.3 | 3.1 ± 0.58 | |
| Acetaminophen | rUGT1A9 | 12 ± 3.7 | 0.49 ± 0.15 | 1.9 ± 0.29 | 0.073 ± 0.011 | 6.9 ± 0.54 | 0.59 ± 0.046 |
| HKM | 15 ± 3.0 | 0.79 ± 0.16 | 3.8 ± 0.82 | 0.24 ± 0.05 | 21 ± 3.4 | 2.6 ± 0.41 | |
| HLM | 29 ± 8.9 | 1.4 ± 0.43 | 12 ± 3.2 | 0.64 ± 0.16 | 30 ± 4.5 | 2.8 ± 0.96 | |
IC50 values are presented as means ± S.D. of three independent experiments. IC50,u, binding-corrected IC50; NA, not analyzed.
Fig. 4.
Inhibitory effects of CBD on the glucuronidation of UGT probe substrates in microsomes from UGT-overexpressing HEK293 cell lines (rUGT), HLMs, and HKMs. Shown are representative plots comparing CBD concentration with the percent glucuronidation activity against probe substrates in rUGT microsomes, HLMs, and HKMs. Incubations were performed for 60–120 minutes at 37°C using 80–90 µg of rUGT microsomes or 90–200 µg HLMs or HKMs with the following probe substrates: propofol, acetaminophen, and furosemide for UGT1A9; serotonin for UGT1A6; codeine for UGT2B4; and AZT for UGT2B7 (see Supplemental Table 1 for concentrations). Individual metabolites were analyzed by UPLC-MS/MS as described in the Materials and Methods.
THC exhibited IC50 values that were slightly higher than CBD for propofol, codeine, and AZT glucuronidation in rUGT microsomes, HLMs, and HKMs (Table 1). Similar to that observed for CBD, THC exhibited similar IC50 values for propofol in rUGT1A9 microsomes (IC50 = 11 ± 3.0 μM and IC50,u = 0.45 ± 0.12 μM) and codeine glucuronidation in rUGT2B4 microsomes (IC50 = 11 ± 2.7 μM and IC50,u = 0.47 ± 0.11 μM), with a higher value observed for AZT glucuronidation in rUGT2B7 microsomes (IC50 = 33 ± 8.5 µM and IC50,u = 1.4 ± 0.36 μM). Also similar to that observed for CBD, the IC50 values for THC for propofol glucuronidation was similar in HKMs (IC50 = 12 ± 3.4 µM and IC50,u = 0.64 ± 0.18 μM) but higher in HLMs (IC50 = 30 ± 6.4 µM and IC50,u = 1.4 ± 0.31 μM) as compared with that observed for rUGT1A9 microsomes, a pattern that was reversed for THC inhibition of codeine glucuronidation in HKMs (IC50 = 55 ± 5.2 µM and IC50,u = 2.9 ± 0.27 μM) versus HLMs (IC50 = 13 ± 2.6 µM and IC50,u= 0.61 ± 0.13 μM). Again similar to that observed for CBD, more moderate inhibition was observed for THC inhibition of AZT glucuronidation in HLMs and HKMs, with IC50 values that were only slightly higher than that observed for rUGT2B7 microsomes [IC50 values = 59 ± 6.6 (IC50,u = 2.8 ± 0.32 μM) and 51 ± 12 µM (IC50,u = 2.6 ± 0.65 μM), respectively].
The pattern of inhibition observed for CBN for propofol glucuronidation was virtually identical to that observed for both THC and CBD, with similar IC50 values observed for rUGT1A9 microsomes (IC50 = 6.0 ± 0.75 μM and IC50 u = 0.51 ± 0.063 μM) and HKMs (IC50 = 7.5 ± 1.7 μM and IC50 u = 0.90 ± 0.20 μM) and a higher IC50 value observed for HLMs (IC50 = 31 ± 4.1 μM and IC50 u = 2.9 ± 0.38 µM; Table 1). Similar to that observed for both THC and CBD, CBN exhibited more moderate inhibition of AZT glucurondation, with similar IC50 values observed for rUGT2B7 microsomes (IC50 = 49 ± 12 μM and IC50 u = 4.2 ± 1.1 μM), HLMs (IC50 = 59 ± 8.6 μM and IC50 u = 5.5 ± 0.79 μM) and HKMs (IC50 = 57 ± 7.5 μM and IC50 u = 6.9 ± 0.090 μM). Since CBN did not exhibit inhibitory activity against codeine glucuronidation in the screening assays, IC50 values were not determined for CBN against codeine glucuronidation in rUGT2B4 microsomes, HLMs, or HKMs.
The only THC metabolite that exhibited ≥ 50% inhibition for any UGT in the rUGT microsomal screening assays was 11-OH-THC for UGT2B7. This metabolite exhibited weak inhibition of rUGT2B7 microsomal activity (IC50 = 79 ± 11 µM and IC50,u = 4.9 ± .41 μM, calculated using the 11-OH-THC fu,inc value from previous studies (Nasrin et al., 2021). The IC50 values for AZT glucuronidation in HLMs and HKMs were not determined as inhibition did not occur at > 50% at the concentration range tested (up to 100 μM AZT).
To better validate the inhibitory effects of cannabinoids on UGT1A9-mediated glucuronidation, two additional UGT1A9 probe substrates, furosemide and acetaminophen, were examined. As shown in Table 1, the glucuronidation of both agents was strongly inhibited by THC, CBD and CBN at levels similar to those observed for propofol glucuronidation in rUGT1A9 microsomes. The highest level of inhibition was again observed with CBD, with IC50 values in rUGT1A9 microsomes of 2.4 ± 0.66 μM (IC50,u = 0.090 ± 0.025 μM) and 1.9 ± 0.29 μM (IC50,u = 0.073 ± 0.011 μM) for furosemide and acetaminophen glucuronidation, respectively (Table 1). The IC50 values observed in HKMs were very similar to those determined in rUGT1A9 microsomes, with CBD exhibiting the highest level of inhibition at 3.6 ± 0.80 μM (IC50,u = 0.22 ± 0.049 μM) and 3.8 ± 0.82 μM (IC50,u = 0.24 ± 0.05 μM) for furosemide and acetaminophen glucuronidation, respectively, and less inhibition in HLMs, with IC50 values of 29 ± 4.0 μM (IC50,u = 1.5 ± 0.20 μM) and 12 ± 3.2 μM (IC50,u = 0.64 ± 0.16 μM), respectively. The decreased level of inhibition of furosemide and acetaminophen glucuronidation by CBD in HLMs versus HKMs and the similar inhibition pattern of HKMs and rUGT1A9 microsomes is apparent when examining plots of percent glucuronidation activity versus CBD concentrations (Fig. 4). THC and CBN were slighly less potent inhibitors in rUGT1A9 microsomes, with IC50 values of 8.0 ± 0.47 μM (IC50,u = 0.33 ± 0.020 μM) and 9.2 ± 2.1 μM (IC50,u = 0.78 ± 0.18 μM), respectively, against furosemide, and 12 ± 3.7 μM (IC50,u = 0.49± 0.15 μM) and 6.9 ± 0.54 μM (IC50,u = 0.59 ± 0.046 μM), respectively, against acetaminophen (Table 1). However, the same trend observed in the tissue microsomes was also observed for rUGT1A9 microsomes, with similar IC50 values for HKMs against furosemide (IC50 = 10 ± 4.1 μM and IC50,u = 0.54 ± 0.22 μM for THC; IC50 = 15 ± 0.8 μM and IC50,u = 1.9 ± 0.092 μM for CBN) and acetaminophen (IC50 = 15 ± 3.0 μM and IC50,u = 0.79 ± 0.16 μM for THC; IC50 = 21 ± 3.4 μM and IC50,u = 2.6 ± 0.41 μM for CBN), but somewhat higher for HLMs against furosemide (IC50 = 32 ± 6.3 μM and IC50,u = 1.5 ± 0.30 μM for THC; IC50 = 30 ± 4.5 μM and IC50,u = 2.8 ± 0.96 μM for CBN) and acetaminophen (IC50 = 29 ± 8.9 μM and IC50,u = 1.4 ± 0.43 μM for THC; IC50 = 34 ± 6.3 μM and IC50,u = 3.1 ± 0.58 μM for CBN; Table 1). The decreased level of inhibition of furosemide and acetaminophen glucuronidation by THC and CBN in HLMs versus HKMs and the similar inhibition pattern with both THC and CBN of HKMs and rUGT1A9 microsomes is apparent when examining plots of percent glucuronidation activity versus CBD concentrations (Supplemental Fig. 1).
Discussion
The present study is the first to conduct a comprehensive examination of the inhibitory effects of major cannabinoids (THC, CBD and CBN) on the enzymatic activities of each of the primary hepatic UGT enzymes (UGTs 1A1, 1A3, 1A4, 1A9, 2B4, 2B7, 2B10, 2B15, and 2B17). In addition, the major metabolites of THC and CBD (11-OH-THC, THC-COOH, THC-COO-Gluc, and 7-OH-CBD) were also screened as potential inhibitors. The results from the present study indicate that the parent cannabinoids (THC, CBD and CBN) exhibit strong inhibition of the glucuronidation activities of UGTs 1A6, 1A9, 2B4 and 2B7, and marginal inhibition of a number of additional UGTs including UGT2B17 by both CBD and CBN, and UGT2B10 by CBN. In contrast to that observed previously for major hepatic P450 enzymes (Yamaori et al., 2011a; Yamaori et al., 2011b; Yamaori et al., 2011c; Yamaori et al., 2012; Bansal et al., 2020; Nasrin et al., 2021), major THC metabolites exhibited little inhibition of hepatic UGT enzymes, with only 11-OH-THC exhibiting significant inhibition against a single UGT (UGT2B7). Similar to that observed for THC metabolites, the major CBD metabolite, 7-OH-CBD, exhibited no significant inhibition against the UGTs tested, with only marginal inhibition observed for UGTs 1A9 and 2B7. In a pattern similar to that observed for P450 enzymes, THC-COOH exhibited no significant inhibition against any of the UGT enzymes tested in the present study.
Tissue and plasma concentrations of cannabinoids vary widely by user and are dependent upon a number of factors including dose, cannabis strain, mode of consumption and expertise of the user (Sharma et al., 2012). Average plasma concentrations of THC from a 10 mg dose by inhalation are 110 ug/L (0.35 µM), which are about 3-fold higher than those observed for oral dosing (360 ug/L; 1.1 μM). The average CBD plasma level after a 400 mg oral dose is 181 ug/L (0.76 µM) (Manini et al., 2015). The IC50,u values observed in the present study for THC and CBD against several UGT enzymes are in the micromolar to submicromolar range, suggesting that unwanted DDIs with xenobiotics metabolized by the same UGT enzymes may occur in co-users of cannabis.
Of the cannabinoids tested, the strongest inhibition in rUGT microsomes was observed by CBD against the glucuronidation of propofol (UGT1A9), serotonin (UGT1A6), codeine (UGT2B4), and AZT (UGT2B7), followed by THC, which also exhibited strong inhibition against the same suite of enzymes. CBN was shown to be similarly effective at inhibiting the glucuronidation of propofol and AZT in rUGT1A9 and rUGT2B7 microsomes, respectively; however, unlike that observed for CBD and THC, no inhibition of rUGT2B4 microsomal activity was observed by CBN using codeine as the probe substrate.
Atlhough the liver is considered the most important organ for the metabolism of drugs and other xenobiotics, the kidney also plays an important role, especially when glucuronidation is a primary component of a drug’s metabolism and elimination (Margaillan et al., 2015). UGT protein expression in both the human liver and human kidney has large interindividual variability; however, current literature estimates that 13 UGTs are expressed in significant amounts in liver, whereas only 3 UGTs are appreciably expressed in human kidney, including UGTs 1A9 and 2B7, which are expressed at similar levels, and UGT1A6, which is expressed at a much lower level; UGT2B4 shows negligible expression in human kidney (Margaillan et al., 2015; Basit et al., 2020). Consistent with the relatively high expression pattern of UGT1A9 in human kidney, the IC50 values observed in HKMs for propofol, furosemide, and acetaminophen, all UGT1A9 substrates, were similar to that observed for each agent in rUGT1A9 microsomes for CBD, THC and CBN. This contrasts with HLMs, where the IC50 values were higher (approximately 3-fold) than those observed in rUGT1A9 microsomes in all cases. In addition, the 6-fold lower IC50 exhibited by UGT1A9 as compared with UGT2B7 in rUGT microsomes by THC, CBD, and CBN corresponds with the larger IC50 values observed in HKMs using a UGT2B7 probe substrate (AZT) versus that observed for UGT1A9 probe substrates, reflecting the relative inhibition of the two enzymes by cannabinoids. These data support the possibility that the major cannabinoids in Cannabis, CBD, THC, and CBN, may all act to inhibit the two highly expressed UGT enzymes in human kidney, UGT1A9, and UGT2B7, in vivo.
The relative inhibition of codeine glucuronidation observed in HKMs was approximately 5–7-fold higher for THC and CBD as compared with rUGT2B4 microsomes, suggesting that UGT2B4 is likely not a major glucuronidating enzyme in kidney. This pattern is consistent with the low relative expression of UGT2B4 in this organ (Basit et al., 2020). This activity pattern contrasts to the very similar IC50 values observed in HLMs for codeine glucuronidation as compared with those observed in rUGT2B4 microsomes, a pattern consistent with the high expression of UGT2B4 in human liver. Although codeine glucuronidation is considered a probe substrate of UGT2B4 activity, UGT2B7 is also likely a major contributor to the hepatic glucuronidation of this agent (Court et al., 2003). When comparing codeine glucuronidation activities for both THC and CBD in the present study, the IC50 values are nearly identical in HLMs to those determined for rUGT2B4 microsomes. In addition, the IC50 values were approximately 3.6-fold lower for rUGT2B4 microsomes than for rUGT2B7 microsomes for both cannabinoids. These data indicate that although these two UGT enzymes are highly homologous, they may have unique binding interactions with cannabinoids and suggest that CBD, THC, and CBN strongly inhibit hepatic UGT2B4 activity.
UGT2B7 is arguably the most important UGT enzyme involved in phase II metabolism, as it is expressed at high levels in the liver and is the most commonly listed UGT involved in the biotransformation of the top 200 drugs currently prescribed in the United States (Williams et al., 2004). Inhibition of this enzyme has the potential to impact thousands of patients through unwanted drug-drug interactions, toxicities, and off-target effects. As seen from the IC50 values of THC, CBD, and CBN against the UGT2B7 probe substrate AZT in HLMs, the UGT2B7-mediated glucuronidation of AZT is moderately inhibited by these cannabinoids. In all three cases, the IC50 determined in HLMs is similar to the value determined in rUGT2B7, suggesting that UGT2B7 is inhibited by these cannabinoids and that this inhibition can be translated to the human liver, where the potential for unwanted DDIs may occur. Indeed, one such interaction has been observed when Epidolex (CBD) is prescribed as an antiseizure medication concurrently with the sedative midazolam (Patsalos et al., 2020). Although midazolam itself is not glucuronidated by UGT2B7, its active metabolite, 1-hydroxymidazolam, is a well documented UGT2B7 substrate (Seo et al., 2010). Administration of midazolam with steady state levels of Epidolex results in increased plasma concentrations of active 1-hydroxymidazolam (Cmax = ↑12%, area under the plasma concentration versus time curve from 0 to t = ↑68%), as well as a delay in tmax (difference in median of 2.2 hours) and an increase in t1/2 of 35%. Another study examined the disposition kinetics of the opioid morphine with and without concurrent inhaled vaporized cannabis (900 mg, 3.56% THC) (Abrams et al., 2011). Morphine is a well studied substrate for UGT2B7 (Osborne et al., 1990) and is glucuronidated to both the inactive 3-glucuronide and the highly active 6-glucuronide. A statistically significant decrease in steady state plasma levels of morphine was found when administered with vaporized cannabis (which was attributed to a decrease in the uptake of morphine), and a near significant decrease in the Cmax of inactive metabolite 3-glucuronide was also observed, indicating reduced UGT2B7 glucuronidation activity in the presence of the inhaled vaporized THC.
Although the role of renal metabolism is still an underexplored area compared with hepatic metabolism, mounting evidence from recent publications indicates that the human kidney has significant metabolic capacity. Renal metabolism by UGT enzymes plays a major role in clearance of many drugs including acetaminophen and furosemide (assayed in this study) as well as carbamazepine, codeine, gemfibrozil, morphine, and the commonly used over the counter nonsteroidal anti-inflammatory drugs ibuprofen, ketoprofen, and S-naproxen (Knights et al., 2013). Preferential inhibition of the renal UGTs may have a larger effect on drugs that are mainly excreted by renal glucuronidation, and interestingly, the two most highly expressed UGTs in human kidney (UGTs 1A9 and 2B7) were inhibited by CBD, THC, and CBN in the present study. Therefore, cannabinoids, and especially CBD, may significantly and disproportionately affect the 1.5 million people in the United States (Rein, 2020) who are diagnosed with chronic kidney disease and acute kidney injury. One-quarter to one-half of those patients also experience chronic symptoms such as pain, nausea, anorexia, sleep disturbance, anxiety, and depression (Rein, 2020), several of which are approved indications for medical cannabis (CBD). Additionally, chronic kidney disease is associated with decreased activity of drug metabolizing enzymes and transporters (Dreisbach and Lertora, 2008). Moreover, a recent study showed significant reduction in the glucuronidation capacity of drugs metabolized by UGT1A9 and UGT2B7 in patients with kidney tumors (Margaillan et al., 2015). AZT and propofol metabolism were decreased 96- and 7.6-fold, respectively, in a patient with neoplastic kidney when compared with normal kidney, suggesting that the use of Cannabis or CBD in these patients may be deleterious.
In conclusion, the present study is the first to demonstrate that the major cannabinoids present in Cannabis are able to inhibit several of the primary UGT enzymes involved in phase II metabolism. CBD was shown to be the most potent cannabinoid inhibitor, exhibiting IC50 values 2–3-fold lower than that observed for THC. Although this is the first study to specifically address the inhibition of UGTs by CBD and other cannabinoids, previous reports indicate that CBD, THC, and several THC metabolites are potent inhibitors of several major P450 enzymes (Yamaori et al., 2011a; Yamaori et al., 2011b; Yamaori et al., 2011c; Bansal et al., 2020; Nasrin et al., 2021). Results from this study now show that two major hepatic UGTs and three of the most highly expressed UGTs present in kidney are strongly inhibited by these cannabinoids, suggesting that deleterious drug-drug interactions may be more likely to occur in patients in whom reduced hepatic or kidney function and cannabis use are occurring simultaneously. In light of the rising acceptance of cannabis use in the United States and internationally, further in vivo studies examining cannabinoid-drug interactions of both phase I and phase II are warranted.
Acknowledgments
The authors would like to thank Shelby M. Coates for her helpful contributions to the study. Figure 1 was created with BioRender.com.
Abbreviations
- AZT
azithromycin
- BSA
bovine serum albumin
- CBD
cannabidiol
- CBN
cannabinol
- DDI
drug-drug interaction
- HEK
human embryonic kidney
- HKM
human kidney microsome
- HLM
human liver microsome
- IC50,u
binding-corrected IC50
- 7-OH-CBD
7-hydroxy-cannabidiol
- 11-OH-THC
11-hydroxy-Δ9-tetrahydrocannabinol
- P450
cytochrome P450
- rUGT
recombinant UGT
- THC
(−)-trans-Δ9-tetrahydrocannabinol
- THC-COO-Gluc
11-nor-9-carboxy-Δ 9-tetrahydrocannabinol glucuronide
- THC-COOH
11-nor-9-carboxy-Δ9- tetrahydrocannabinol
- UDPGA
UDP glucuronic acid
- UGT
UDP-glucuronosyltransferase
- UPLC
ultra-high-performance liquid chromatography
- UPLC-MS/MS
UPLC–tandem mass spectrometry
Authorship Contributions
Participated in research design: Nasrin, Lazarus.
Conducted experiments: Nasrin, Watson, Bardhi, Fort.
Performed data analysis: Nasrin, Watson, Chen, Lazarus.
Wrote or contributed to the writing of the manuscript: Nasrin, Watson, Lazarus.
Footnotes
This work was supported by the National Institutes of Health National Institutes of Environmental Health Sciences [Grant R01-ES025460] to P.L., the Health Sciences and Services Authority of Spokane, WA [Grant WSU002292], and funds provided by the State of Washington Initiative Measure No. 502.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Abrams DI, Couey P, Shade SB, Kelly ME, Benowitz NL (2011) Cannabinoid-opioid interaction in chronic pain. Clin Pharmacol Ther 90:844–851. [DOI] [PubMed] [Google Scholar]
- Bansal S, Maharao N, Paine MF, Unadkat JD (2020) Predicting the potential for cannabinoids to precipitate pharmacokinetic drug interactions via reversible inhibition or inactivation of major cytochromes P450. Drug Metab Dispos 48:1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basit A, Neradugomma NK, Wolford C, Fan PW, Murray B, Takahashi RH, Khojasteh SC, Smith BJ, Heyward S, Totah RA, et al. (2020) Characterization of differential tissue abundance of major non-CYP enzymes in human. Mol Pharm 17:4114–4124. [DOI] [PubMed] [Google Scholar]
- Boychuk DG, Goddard G, Mauro G, Orellana MF (2015) The effectiveness of cannabinoids in the management of chronic nonmalignant neuropathic pain: a systematic review. J Oral Facial Pain Headache 29:7–14. [DOI] [PubMed] [Google Scholar]
- Bridgeman MB, Abazia DT (2017) Medicinal cannabis: history, pharmacology, and implications for the acute care setting. P&T 42:180–188. [PMC free article] [PubMed] [Google Scholar]
- Bushey RT, Lazarus P (2012) Identification and functional characterization of a novel UDP-glucuronosyltransferase 2A1 splice variant: potential importance in tobacco-related cancer susceptibility. J Pharmacol Exp Ther 343:712–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos AC, Fogaça MV, Sonego AB, Guimarães FS (2016) Cannabidiol, neuroprotection and neuropsychiatric disorders. Pharmacol Res 112:119–127. [DOI] [PubMed] [Google Scholar]
- Cimoch PJ, Lavelle J, Pollard R, Griffy KG, Wong R, Tarnowski TL, Casserella S, Jung D (1998) Pharmacokinetics of oral ganciclovir alone and in combination with zidovudine, didanosine, and probenecid in HIV-infected subjects. J Acquir Immune Defic Syndr Hum Retrovirol 17:227–234. [DOI] [PubMed] [Google Scholar]
- Court MH, Krishnaswamy S, Hao Q, Duan SX, Patten CJ, Von Moltke LL, Greenblatt DJ (2003) Evaluation of 3′-azido-3′-deoxythymidine, morphine, and codeine as probe substrates for UDP-glucuronosyltransferase 2B7 (UGT2B7) in human liver microsomes: specificity and influence of the UGT2B7*2 polymorphism. Drug Metab Dispos 31:1125–1133. [DOI] [PubMed] [Google Scholar]
- Cox EJ, Maharao N, Patilea-Vrana G, Unadkat JD, Rettie AE, McCune JS, Paine MF (2019) A marijuana-drug interaction primer: precipitants, pharmacology, and pharmacokinetics. Pharmacol Ther 201:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellinger RW, Fang J-L, Chen G, Weinberg R, Lazarus P (2006) Importance of UDP-glucuronosyltransferase 1A10 (UGT1A10) in the detoxification of polycyclic aromatic hydrocarbons: decreased glucuronidative activity of the UGT1A10139Lys isoform. Drug Metab Dispos 34:943–949. [DOI] [PubMed] [Google Scholar]
- Devinsky O, Marsh E, Friedman D, Thiele E, Laux L, Sullivan J, Miller I, Flamini R, Wilfong A, Filloux F, et al. (2016) Cannabidiol in patients with treatment-resistant epilepsy: an open-label interventional trial. Lancet Neurol 15:270–278. [DOI] [PubMed] [Google Scholar]
- Dreisbach AW, Lertora JJL (2008) The effect of chronic renal failure on drug metabolism and transport. Expert Opin Drug Metab Toxicol 4:1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Filippis DD, Cirillo C, Iuvone T, Capoccia E, Scuderi C, Steardo A, Cuomo R, Steardo L (2013) Cannabidiol in inflammatory bowel diseases: a brief overview. Phytother Res 27:633–636. [DOI] [PubMed] [Google Scholar]
- Garrett ER, Hunt CA (1974) Physiochemical properties, solubility, and protein binding of delta9-tetrahydrocannabinol. J Pharm Sci 63:1056–1064. [DOI] [PubMed] [Google Scholar]
- Harvey DJ, Mechoulam R (1990) Metabolites of cannabidiol identified in human urine. Xenobiotica 20:303–320. [DOI] [PubMed] [Google Scholar]
- Huestis MA (2007) Human cannabinoid pharmacokinetics. Chem Biodivers 4:1770–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L, King CD, Whitington PF, Green MD, Roy SK, Tephly TR, Coffman BL, Ratain MJ (1998) Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J Clin Invest 101:847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Yamaori S, Okamoto Y, Yamamoto I, Watanabe K (2013) Cannabidiol is a potent inhibitor of the catalytic activity of cytochrome P450 2C19. Drug Metab Pharmacokinet 28:332–338. [DOI] [PubMed] [Google Scholar]
- Kasteel EEJ, Darney K, Kramer NI, Dorne JLCM, Lautz LS (2020) Human variability in isoform-specific UDP-glucuronosyltransferases: markers of acute and chronic exposure, polymorphisms and uncertainty factors. Arch Toxicol 94:2637–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights KM, Rowland A, Miners JO (2013) Renal drug metabolism in humans: the potential for drug-endobiotic interactions involving cytochrome P450 (CYP) and UDP-glucuronosyltransferase (UGT). Br J Clin Pharmacol 76:587–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen C, Shahinas J (2020) Dosage, efficacy and safety of cannabidiol administration in adults: a systematic review of human trials. J Clin Med Res 12:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie PI, Rogers A, Elliot DJ, Chau N, Hulin J-A, Miners JO, Meech R (2011) The novel UDP glycosyltransferase 3A2: cloning, catalytic properties, and tissue distribution. Mol Pharmacol 79:472–478. [DOI] [PubMed] [Google Scholar]
- Manini AF, Yiannoulos G, Bergamaschi MM, Hernandez S, Olmedo R, Barnes AJ, Winkel G, Sinha R, Jutras-Aswad D, Huestis MA, et al. (2015) Safety and pharmacokinetics of oral cannabidiol when administered concomitantly with intravenous fentanyl in humans. J Addict Med 9:204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaillan G, Rouleau M, Fallon JK, Caron P, Villeneuve L, Turcotte V, Smith PC, Joy MS, Guillemette C (2015) Quantitative profiling of human renal UDP-glucuronosyltransferases and glucuronidation activity: a comparison of normal and tumoral kidney tissues. Drug Metab Dispos 43:611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meech R, Hu DG, McKinnon RA, Mubarokah SN, Haines AZ, Nair PC, Rowland A, Mackenzie PI (2019) The UDP-glycosyltransferase (UGT) superfamily: new members, new functions, and novel paradigms. Physiol Rev 99:1153–1222. [DOI] [PubMed] [Google Scholar]
- Nasrin S, Watson CJW, Perez-Paramo YX, Lazarus P(2021) Cannabinoid metabolites as inhibitors of major hepatic CYP450 enzymes, with implications for cannabis-drug interactions. Drug Metab Dispos 49:1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin S, Watson CJ, Chen G, Lazarus P (2021) Cannabinoid metabolites as potential inhibitors of major CYP450 enzymes, with implications for cannabis-drug interactions. FASEB J DOI: 10.1096/fasebj.2020.34.s1.05863 [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne R, Joel S, Trew D, Slevin M (1990) Morphine and metabolite behavior after different routes of morphine administration: demonstration of the importance of the active metabolite morphine-6-glucuronide. Clin Pharmacol Ther 47:12–19. [DOI] [PubMed] [Google Scholar]
- Patilea-Vrana GI, Anoshchenko O, Unadkat JD (2019) Hepatic enzymes relevant to the disposition of (-)-Δ9-tetrahydrocannabinol (THC) and its psychoactive metabolite, 11-OH-THC. Drug Metab Dispos 47:249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsalos PN, Szaflarski JP, Gidal B, VanLandingham K, Critchley D, Morrison G (2020) Clinical implications of trials investigating drug-drug interactions between cannabidiol and enzyme inducers or inhibitors or common antiseizure drugs. Epilepsia 61:1854–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertwee RG (2008) The diverse CB1 and CB2 receptor pharmacology of three plant cannabinoids: delta9-tetrahydrocannabinol, cannabidiol and delta9-tetrahydrocannabivarin. Br J Pharmacol 153:199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein JL (2020) The nephrologist’s guide to cannabis and cannabinoids. Curr Opin Nephrol Hypertens 29:248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland A, Elliot DJ, Williams JA, Mackenzie PI, Dickinson RG, Miners JO (2006) In vitro characterization of lamotrigine N2-glucuronidation and the lamotrigine-valproic acid interaction. Drug Metab Dispos 34:1055–1062. [DOI] [PubMed] [Google Scholar]
- Rowland A, Knights KM, Mackenzie PI, Miners JO (2008) The “albumin effect” and drug glucuronidation: bovine serum albumin and fatty acid-free human serum albumin enhance the glucuronidation of UDP-glucuronosyltransferase (UGT) 1A9 substrates but not UGT1A1 and UGT1A6 Activities. Drug Metab Dispos 36:1056–1062. [DOI] [PubMed] [Google Scholar]
- Rowland A, Miners JO, Mackenzie PI (2013) The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int J Biochem Cell Biol 45:1121–1132. [DOI] [PubMed] [Google Scholar]
- Russo EB, Marcu J (2017) Chapter Three - Cannabis Pharmacology: The Usual Suspects and a Few Promising Leads, in Adv Pharmacol (Kendall D, Alexander SPH, eds) pp 67–134, Academic Press, New York. [DOI] [PubMed] [Google Scholar]
- Seo K-A, Bae SK, Choi Y-K, Choi CS, Liu K-H, Shin J-G (2010) Metabolism of 1′- and 4-hydroxymidazolam by glucuronide conjugation is largely mediated by UDP-glucuronosyltransferases 1A4, 2B4, and 2B7. Drug Metab Dispos 38:2007–2013. [DOI] [PubMed] [Google Scholar]
- Sharma P, Murthy P, Bharath MMS (2012) Chemistry, metabolism, and toxicology of cannabis: clinical implications. Iran J Psychiatry 7:149–156. [PMC free article] [PubMed] [Google Scholar]
- Sun L, Li M, Zhang L, Teng X, Chen X, Zhou X, Ma Z, Qi L, Wang P (2017) Differences in UGT1A1 gene mutations and pathological liver changes between Chinese patients with Gilbert syndrome and Crigler-Najjar syndrome type II. Medicine (Baltimore) 96:e8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallman MN, Miles KK, Kessler FK, Nielsen JN, Tian X, Ritter JK, Smith PC (2007) The contribution of intestinal UDP-glucuronosyltransferases in modulating 7-ethyl-10-hydroxy-camptothecin (SN-38)-induced gastrointestinal toxicity in rats. J Pharmacol Exp Ther 320:29–37. [DOI] [PubMed] [Google Scholar]
- Vergara AG, Watson CJW, Chen G, Lazarus P (2020) UDP-glycosyltransferase 3A metabolism of polycyclic aromatic hydrocarbons: potential importance in Aerodigestive tract tissues. Drug Metab Dispos 48:160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, Peterkin V, Koup JR, Ball SE (2004) Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab Dispos 32:1201–1208. [DOI] [PubMed] [Google Scholar]
- Yamaori S, Ebisawa J, Okushima Y, Yamamoto I, Watanabe K (2011a) Potent inhibition of human cytochrome P450 3A isoforms by cannabidiol: role of phenolic hydroxyl groups in the resorcinol moiety. Life Sci 88:730–736. [DOI] [PubMed] [Google Scholar]
- Yamaori S, Koeda K, Kushihara M, Hada Y, Yamamoto I, Watanabe K (2012) Comparison in the in vitro inhibitory effects of major phytocannabinoids and polycyclic aromatic hydrocarbons contained in marijuana smoke on cytochrome P450 2C9 activity. Drug Metab Pharmacokinet 27:294–300. [DOI] [PubMed] [Google Scholar]
- Yamaori S, Kushihara M, Yamamoto I, Watanabe K (2010) Characterization of major phytocannabinoids, cannabidiol and cannabinol, as isoform-selective and potent inhibitors of human CYP1 enzymes. Biochem Pharmacol 79:1691–1698. [DOI] [PubMed] [Google Scholar]
- Yamaori S, Maeda C, Yamamoto I, Watanabe K (2011b) Differential inhibition of human cytochrome P450 2A6 and 2B6 by major phytocannabinoids. Forensic Toxicol 29:117–124. [Google Scholar]
- Yamaori S, Okamoto Y, Yamamoto I, Watanabe K (2011c) Cannabidiol, a major phytocannabinoid, as a potent atypical inhibitor for CYP2D6. Drug Metab Dispos 39:2049–2056. [DOI] [PubMed] [Google Scholar]