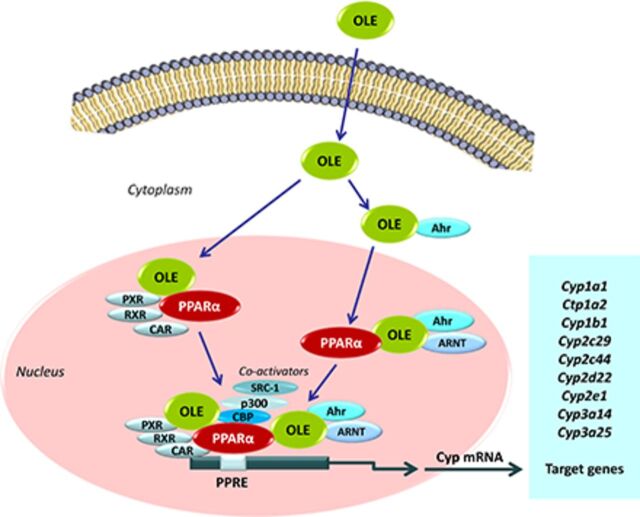
Abstract
Oleuropein (OLE), the main constituent of Olea europaea, displays pleiotropic beneficial effects in health and disease, which are mainly attributed to its anti-inflammatory and cardioprotective properties. Several food supplements and herbal medicines contain OLE and are available without a prescription. This study investigated the effects of OLE on the main cytochrome P450s (P450s) catalyzing the metabolism of many prescribed drugs. Emphasis was given to the role of peroxisome proliferator-activated receptor α (PPARα), a nuclear transcription factor regulating numerous genes including P450s. 129/Sv wild-type and Ppara-null mice were treated with OLE for 6 weeks. OLE induced Cyp1a1, Cyp1a2, Cyp1b1, Cyp3a14, Cyp3a25, Cyp2c29, Cyp2c44, Cyp2d22, and Cyp2e1 mRNAs in liver of wild-type mice, whereas no similar effects were observed in Ppara-null mice, indicating that the OLE-induced effect on these P450s is mediated by PPARα. Activation of the pathways related to phosphoinositide 3-kinase/protein kinase B (AKT)/forkhead box protein O1, c-Jun N-terminal kinase, AKT/p70, and extracellular signal-regulated kinase participates in P450 induction by OLE. These data indicate that consumption of herbal medicines and food supplements containing OLE could accelerate the metabolism of drug substrates of the above-mentioned P450s, thus reducing their efficacy and the outcome of pharmacotherapy. Therefore, OLE-induced activation of PPARα could modify the effects of drugs due to their increased metabolism and clearance, which should be taken into account when consuming OLE-containing products with certain drugs, in particular those of narrow therapeutic window.
SIGNIFICANCE STATEMENT
This study indicated that oleuropein, which belongs to the main constituents of the leaves and olive drupes of Olea europaea, induces the synthesis of the major cytochrome P450s (P450s) metabolizing the majority of prescribed drugs via activation of peroxisome proliferator-activated receptor α. This effect could modify the pharmacokinetic profile of co-administered drug substrates of the P450s, thus altering their therapeutic efficacy and toxicity.
Introduction
Globally, there is an upsurge in the use of medicines coming from herbs as many people claim that they are nontoxic based on their natural origin and their use in popular medicine for centuries. Nonetheless, herbs contain many active substances that could induce adverse effects, toxicity, or even cancer (Bensoussan et al., 2002; De Smet, 1995; Deng, 2002; Eisenberg et al., 1993; Ernst and Pittler, 2002a; Ernst and Pittler, 2002b; Greensfelder, 2000; Haller and Benowitz, 2000; Kennedy and Seely, 2010; Klepser and Klepser, 1999; Koh and Woo, 2000; Malliou et al., 2018; Mckenna et al., 2012; McRae et al., 2002; Stedman, 2002). When all foreign substances (xenobiotics) enter the body, they undergo biotransformation primarily in the liver, which accelerates their elimination. In particular, the metabolism of xenobiotics, such as drugs, precarcinogens, and toxic agents, during phase I is mainly catalyzed by cytochrome P450s (P450s) and can result either in activation of prodrugs or in inactivation of pharmacologically active drugs, or in activation of precarcinogens to carcinogens (Gonzalez and Gelboin, 1994; Ingelman-Sundberg, 2004a). It is estimated that P450s belonging to the CYP3A, CYP2C, CYP2D, and CYP1A subfamilies catalyze hepatic metabolism of more than 95% of the most widely prescribed drugs (Daskalopoulos et al., 2012a; Guengerich, 2003; Ingelman-Sundberg, 2004a; Konstandi, 2013). From a toxicological point of view, it is worth noting that CYP1A1/2 and CYP1B1 catalyze the bioactivation of the major groups of precarcinogens, the polycyclic aromatic hydrocarbons, polycyclic arylamines, and aflatoxin B1, to electrophilic DNA-binding derivatives (Cheng and Morgan, 2001; Flint et al., 2010; Harkitis et al., 2015; Kawajiri, 1999; Konstandi et al., 2006; Konstandi et al., 2005; Pasanen and Pelkonen, 1994). Interestingly, the biotransformation of steroids, fatty acids, and several other endogenous compounds is also catalyzed by P450s (Guengerich, 2003; Spatzenegger and Jaeger, 1995).
It is well documented that the traditional Mediterranean diet has various beneficial effects in health and longevity, and olive oil and olives are substantial ingredients of this diet (Impellizzeri et al., 2012). The main compounds found in the leaves and olive drupes of Olea europaea are oleuropein (OLE) and its hydrolysis product, hydroxytyrosol. There is accumulating evidence based on preclinical studies that OLE displays significant cardioprotective properties (Andreadou et al., 2006; Briante et al., 2001; Malliou et al., 2018; Oi-Kano et al., 2008; Tuck and Hayball, 2002; Visioli and Galli, 1994; Visioli et al., 2002a; Visioli et al., 2002b), which could be attributed to activation of peroxisome proliferator-activated receptor α (PPARα) (Harkitis et al., 2015; Malliou et al., 2018), a ligand-activated nuclear receptor that controls lipid homeostasis (Fruchart and Duriez, 2006; Hansen and Connolly, 2008; Kuusisto et al., 2007; Robillard et al., 2005). The anti-inflammatory properties of OLE may also contribute to the drug’s cardioprotective effects (Shimada et al., 1996; Spatzenegger and Jaeger, 1995). Preliminary data also indicated a pleotropic effect of OLE on several vital functions of the body including stimulation of neural plasticity and protection against neurodegenerating disorders, among others, effects potentially mediated by activation of PPARα (data not shown).
It is well defined that OLE activates PPARα (Malliou et al., 2018; Spatzenegger and Jaeger, 1995), which controls several genes participating in the regulation of inflammatory responses, the metabolism of lipids and glucose, as well as the adipose differentiation and cancer, among others (Malliou et al., 2018; Yang et al., 2008; YUMUK, 2006). It is of interest that PPARα apparently holds a substantial regulatory role for P450s (Choi and Waxman, 1999; Tauber et al., 2020). In particular, the hepatic sexual dimorphism of the P450 expression pattern is largely regulated by PPARs (Leuenberger et al., 2009).
Drug-drug interactions are of critical clinical significance because they markedly determine the outcome of pharmacotherapy, the side effects of the drugs, and pharmacotoxicity (Konstandi, 2013). They are usually dependent on the drugs’ effect on P450s acting either as inducers, inhibitors, or substrates. On one hand, induction of the most important P450s that catalyze the metabolism of the majority of prescribed drugs may accelerate the biotransformation of their drug substrates and, in most cases, result in their reduced pharmacological efficacy (Daskalopoulos et al., 2012a; Daskalopoulos et al., 2012b; Ingelman-Sundberg et al., 2007; Konstandi et al., 2020; Konstandi et al., 2005; Zhou et al., 2004) or, in other cases, result in perturbation of several endogenous regulatory circuits, often associated with pathophysiological states (Choi and Waxman, 1999). On the other hand, inhibition of P450s may lead to accumulation of their drug substrates in the blood, followed by adverse side effects of varying severity that may reach the level of toxicity; this is of particular clinical interest when drugs of low therapeutic index are administered (Daskalopoulos et al., 2012b; Konstandi et al., 2020; Spatzenegger and Jaeger, 1995). Therefore, it should be noted that food supplements or herbal medicines containing pharmacologically active compounds, such as OLE, which act either as substrates of P450s or even as their inducers or inhibitors (Zhou et al., 2004), may modify the efficacy of co-administered drugs in multidrug therapeutic schemes and, potentially, induce pharmacotoxicity (Daskalopoulos et al., 2012a; Daskalopoulos et al., 2012b; Gonzalez and Gelboin, 1994; Guengerich, 2003; Harkitis et al., 2015; Konstandi et al., 2014; Pelkonen et al., 2008).
In the light of the above considerations, the current study investigated the potential regulatory role of OLE for the main P450s that are involved in drug metabolism, emphasizing the role of PPARα in this regulation. To approach this issue, wild-type (WT) and Ppara-null mice were treated with OLE and P450 mRNA, and protein expressions were analyzed. OLE markedly upregulated several genes encoding the most significant drug-metabolizing P450 isozymes in the liver, a process profoundly mediated by PPARα activation.
Materials and Methods
Animals and treatment.
Adult male 129/Sv WT and Ppara-null mice that were used in this study were bred in the Animal House of the University of Ioannina (Ioannina, Greece) and were housed up to five mice per cage. All animals (5–6 per group of treatment) had access to a standard chow diet for rodents (1324 TPF, Altromin Spezial futter GmbH & Co. KG) and water ad libitum. Throughout the experiments, all mice were housed in their cages under a standard 12-hour light/12-hour dark cycle (lights on at 6:00 AM). The experimental protocols employing procedures with animals received the approval of the Ethics Committee of the University of Ioannina–Faculty of Medicine. The procedure followed was in compliance with the European Commission ethical standards for the care and use of experimental animals (Directive 86/609-EEC). Both WT and Ppara-null mice were treated with food pellets containing OLE (100 mg/kg) daily for 6 consecutive weeks (Andreadou et al., 2006; Andreadou et al., 2014; Impellizzeri et al., 2012). For the isolation of OLE the leaves of O. europaea were used following a previously described method (Andreadou et al., 2014). The choice of the dose of OLE that was used in this study was based on information from the literature and our previous findings (Andreadou et al., 2006; Andreadou et al., 2014; Impellizzeri et al., 2012). It corresponds approximately to the average consumption of olive oil and olive drupes in the Mediterranean countries during a day (Abdel-aleem et al., 1997), represents total polyphenol consumption from these olive products, and is estimated to be approximately 100 mg/d (Abdel-aleem et al., 1997; Del Boccio et al., 2003). Mice were treated with OLE in food pellets because it is known that the drug is slightly absorbed in intestinal lumen even under normal iso-osmotic conditions. The absorption of OLE can be markedly improved by solvent flux through paracellular junctions, an effect that is facilitated by hypotonic luminal conditions (Edgecombe et al., 2000). A significant factor stimulating the water flux through the opening of paracellular junctions is the postprandial presence of glucose or amino acids in the intestinal lumen. This mechanism regulating the absorption of OLE in intestinal lumen appears to have similar effects with those of hypotonic solutions (Pappenheimer and Reiss, 1987). The pharmacokinetic profile of OLE in mice has not been determined, but only that in rats, when one dose of OLE (100 mg/kg per os) reached 200 ng/ml in the time of maximum concentration tmax of 2 hours (Del Boccio et al., 2003). Controls received regular rodent food for 6 weeks. Wild-type and Pparα-null mice also received intraperitoneally either fenofibrate (100 mg/kg), a selective PPARα agonist (Ghonem et al., 2015; Hu et al., 2019), or normal saline for 7 consecutive days. Upon completion of the experiments, all mice were euthanized using CO2 asphyxiation, and parts from the liver were dissected for the extraction of total RNA, nuclear/cytosolic proteins, and microsomal proteins. All liver tissue samples were preserved at −80°C until assayed.
Isolation of microsomal proteins.
Liver tissue was homogenized in homogenization buffer containing 0.15 M KCl, 10 mM K2EDTA, and 1 mM dithiothreitol (pH 7.4) at +4°C, for the isolation of microsomal fractions. The homogenates were then centrifuged for 20 minutes at 15,000 rpm (+4°C). The upper phase was transferred into clean vials followed by centrifugation at 27,500 rpm (+4°C) for 60 minutes. The formed microsomal pellet was resuspended by homogenization in the specific ice-cold homogenization buffer and centrifuged at 27,500 rpm for 45 minutes. Temperature was always kept at +4°C. Finally, the formed microsomal pellet was resuspended in the specific ice-cold storage buffer containing K2HPO4/KH2PO4 (pH 7.4), K2EDTA (1 mM), dithiothreitol (0.1 mM), and 20% glycerol. Aliquots of microsomal suspensions were stored at −80°C until assayed (Lang et al., 1981).
CYP2D activity, 1′-bufuralol hydroxylation.
1′-Bufuralol hydroxylation is mainly catalyzed by CYP2D isozymes (Matsunaga et al., 1990). Liver microsomal proteins (40 μg to ∼20 μl of sample) were preincubated in a 180 μl reaction mixture containing potassium phosphate (0.1 Μ, pH 7.4) at 37°C for 5 minutes, in the presence of 50 μM bufuralol (substrate) and NADPH (0.5 mM, Sigma-Aldrich). The duration of the reaction was 7.5 minutes and was terminated using 20 μl of perchloric acid (60%). After a 10-minute centrifugation at 14,075g, the supernatant containing the main metabolite of bufuralol (1′-hydroxy-bufuralol) was analyzed using a high-performance liquid chromatography (HPLC) method. The fluorescence detection was set at 252 nm (excitation wavelength) and 302 nm (emission wavelength), and a specific column (reverse-phase Luna C18, 5 μm, 150Χ 3 mm; Phenomenex, Torrance, CA) was used for this purpose. The mobile phase consisted of 30% acetonitrile/70% perchlorate buffer (20 mM, pH 2.5). The elution of each sample took place at a flow rate of 1 ml/min for 14 minutes. The recombinant rat CYP2D1 and CYP2D2, enriched with P450 reductase BD Supersomes, were used as a positive control of bufuralol 1′-hydroxylation (BD Gentest, Woburn, MA). Bufuralol (50 μM) was used as substrate, which was preincubated at 37°C for 2 minutes with approximately 200 μl 0.1 M potassium phosphate buffer (pH 7.4). Recombinant P450s (50 μM) along with NADPH (1 mM) were added in the mixture, which was incubated at 37°C for 30 minutes. The termination of the reaction was achieved with 20 μl acetonitrile. Then, all samples were placed on wet ice and left undisturbed for 10 minutes before their injection into the HPLC for analysis.
Western blot analysis.
Alterations in P450 apoprotein levels were assessed with immunoblot analysis using liver microsomes, whereas nuclear extracts were used for PPARα and hepatocyte nuclear factor 4α (HNF4α) analysis. Phosphorylated c-Jun N-terminal kinase (JNK), phosphorylated P38, phosphorylated extracellular signal-related kinase (ERK), phosphorylated P70, pregnane X receptor (PXR), constitutive androstane receptor (CAR), and phosphorylated forkhead box protein O1 (FOXO1) were analyzed using total cellular extracts. For the preparation of nuclear and cytosolic extracts, the corresponding cytosolic and nuclear extraction kit of Thermo Fisher Scientific (Waltham, MA) was used. The content of proteins in the sample was determined using the bovine serum albumin assay (Thermo Fisher Scientific). All proteins run on SDS-polyacrylamide gel electrophoresis followed by immunoblotting, and for this purpose the following antibodies were used: mouse/rat polyclonal CYP3A, CYP2C, and CYP2D IgGs; mouse/rat monoclonal HNF4α, CAR, and PXR IgGs; mouse polyclonal PPARα IgG (Santa Cruz Biotechnology); rabbit phosphorylated polyclonal IgGs cAMP responsive element-binding protein (Ser133), P38 (Thr180/Tyr182) (Santa Cruz Biotechnology), mitogen-activated protein kinase (Erk1/2) (Thr202/Tyr204) (Cell Signaling Technology), p70S6K (Thr389) (Cell Signaling Technology), FOXO1 (Ser256) (Santa Cruz Biotechnology), and protein kinase B (AKT) (Ser473) (Santa Cruz Biotechnology); and mouse phosphorylated monoclonal JNK (Thr183 and Tyr185) IgG (Santa Cruz Biotechnology). The secondary antibodies used in this study were conjugated with horseradish peroxidase (Santa Cruz Biotechnology). For the detection of proteins in the blot the enhanced chemiluminescence detection kit (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) was used. As a loading control, immunoblotting employing the specific antibodies, histone H3 or actin (Santa Cruz Biotechnology), and the secondary antibody, anti-mouse horseradish peroxidase–conjugated IgG, was used.
Quantitative real-time polymerase chain reaction assays.
TRIzol reagent (Invitrogen) was used for the isolation of total RNA from liver tissue following the instructions in the manufacturer’s protocol. Following a spectrophotometric method the concentration of total RNA in each sample was determined. Total RNA (1 μg) and a SuperScript II reverse transcriptase kit (Invitrogen) were used to generate cDNA, which was used in quantitative and reverse transcription–polymerase chain reaction (PCR) assays. Table 1 shows all sequences of the forward and reverse gene-specific primers that were used in this study. For the real-time reactions, the SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) was used, and these reactions were performed employing the C1000 Touch thermal cycler with a real-time detection system (Bio-Rad Laboratories, Hercules, CA). Relative mRNA expression was estimated using β-actin mRNA levels to normalize mRNA expression levels of each gene (QuantiTect primer assay; QIAGEN, Valencia, CA). The comparative threshold cycle method was used to quantify all values.
TABLE 1.
Oligonucleotide sequences used as primers for quantitation of gene mRNA levels through quantitative PCR assays
| Gene | Primer sequence |
|---|---|
| Ppara | F: CAGTGGGGAGAGAGGACAGA R: AGTTCGGGAACAAGACGTTG |
| Cyp2d22 | F: ACCGGTAAAGGTAGCTGGAGT R: CATAGGGCCTGGAGGGTAGT |
| Hnf4a | F: CGGAGCCCCTGCAAAGT R: ACTATCCAGTCTCACAGCCCATTC |
| Cyp2e1 | F: TGGTCCTGCATGGCTACAAG R: CGGGCCTCATTACCCTGTTT |
| Cyp2c29 | F: TGTTACAAACCCCCGTGACT R: GGATGTGGATAAAGACCTGAGAC |
| Cyp2c44 | F: CCTAAAGGCTCTGGTGGAGC R: GAAACAAATGCCCACGTGCT |
| Cyp3a14 | F: GGCCCAGTGGGGATAATGAG R: GGTGCCTTATTGGGCAGAGT |
| Cyp3a25 | F: TAGAAACCTGGGTGCTGCTG R: GGATGTGGATAAAGACCTGAGAC |
| Pxr | F: AAGAAGCAGACTCTGCCTTGGA R: GTGGTAGCCATTGGCCTTGT |
| Car | F: CCTCTTCTCCCCTGGTTTCTG R: TCATTGCCACTCCCAAGCTC |
| Rxra | F: CAGTACGCAAAGACCTGACCTACA R: GTTCCGCTGTCTCTTGTCGAT |
| Rxrb | F: AAGTGTCTGGAGCACCTGTTCTT R: CTCCATGAGGAAGGTGTCAATG |
| Cyp1a1 | F: GAAGTGGAAGGGCATAGGCAG R: GGCCAAAGCATATGGCACAG |
| Cyp1a2 | F: ACTTCGAACCAGTCAGCCAG R: GTGCTTGAACAGGGCACTTG |
| Cyp1b1 | F: CCAGCTTTTTGCCTGTCACC R: TGCACTGATGAGCGAGGATG |
| Ahr | F: TTCAGAACTGACTCCACCGC R: CCGGGTGTGATATCGGGAAG |
| Ahrr | F: AGTGTACATACGCCGGTAGG R: CAAGACTGGTGCCACAATGC |
| Hsp90 | F: CAGACCATGGTGAGCCCATT R: TCAACCACACCGCGGATAAA |
| eNos | F: GCAGAAGAGTCCAGCGAACA R: GGCAGCCAAACACCAAAGTC |
| iNos | F: GTGTTCCACCAGGAGATGTTG R: CTCCTGCCCACTGAGTTCGTC |
F, forward; R, reverse.
Statistical analysis.
For the analysis of data, one-way analysis of variance followed by multiple comparisons employing Bonferroni’s and Tukey’s least honest significant difference methods was used. All values are presented as means ± S.E., and P values of <0.05 were considered significant.
Results
OLE-induced effect on CYP1s.
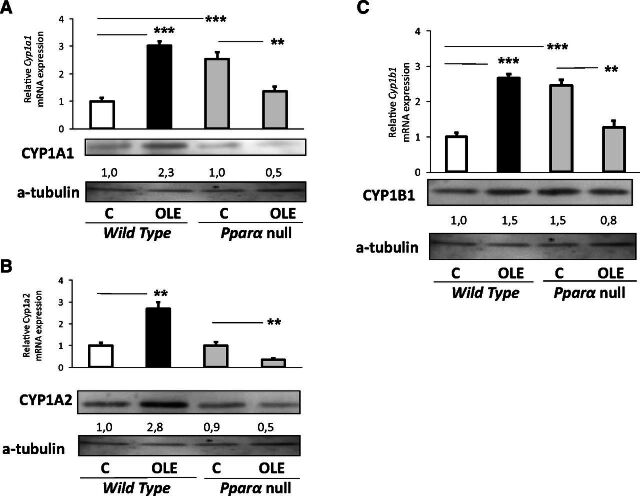
OLE markedly increased hepatic Cyp1a1 and Cyp1a2 mRNA expression and CYP1A protein expression in WT mice (Fig. 1A and B, respectively); Cyp1b1 mRNA levels were also increased by OLE (Fig. 1C).
Fig. 1.
PPARα-mediated regulation of hepatic CYP1A1, CYP1A2, and CYP1B1. Assessment of the effect of OLE, a PPARα agonist, on CYP1A1 (A), CYP1A2 (B), and CYP1B1 (C) protein levels employing Western blotting, and relative gene expression with quantitative PCR analysis. In wild-type and Ppara-null mice, comparisons were made between controls (designated C) and OLE-treated mice. Values are expressed as means ± S.E. Wild-type mice (n = 11); Ppara-null mice (n = 10). *P < 0.05; **P < 0.01; ***P < 0.001. Numbers below bands in Western blot captures show the ratio of the density of the sample band to that of a-tubulin.
Oleuropein-induced effect on CYP2 and CYP3s.
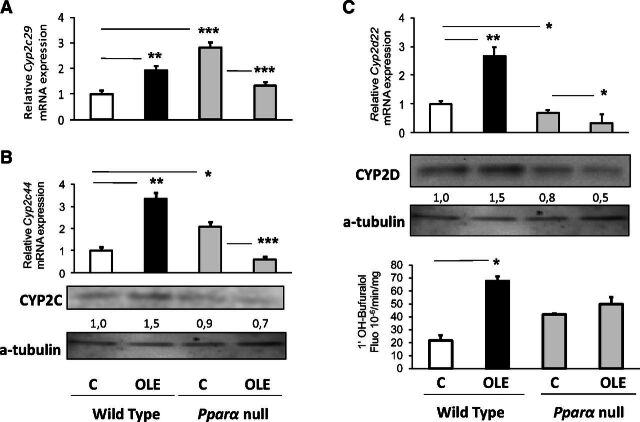
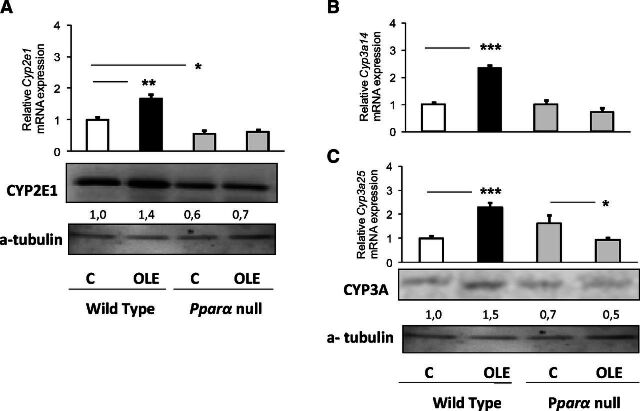
Oleuropein markedly increased hepatic Cyp2c29 and Cyp2c44 mRNA and CYP2C protein expression in WT mice (Fig. 2A and B, respectively). Similarly, OLE increased the hepatic Cyp2d22 mRNA, CYP2D protein and activity levels (Fig. 2C), and the hepatic Cyp2e1 (Fig. 3A), Cyp3a14, and Cyp3a25 expression (Fig. 3B) at mRNA and protein levels.
Fig. 2.
PPARα-mediated regulation of hepatic CYP2C29/2C44 and CYP2D22. Assessment of the effect of OLE, a PPARα agonist, on CYP2C29 (A), CYP2C44 (B), and CYP2D22 (C) protein levels employing Western blotting, and on relative gene expression with quantitative PCR analysis. The OLE-mediated effect on 1′-bufuralol hydroxylation was performed using HPLC. In wild-type and Ppara-null mice, comparisons were made between controls (designated C) and OLE-treated mice. Values are expressed as means ± S.E. Wild-type mice (n = 11); Pparα-null mice (n = 10). *P < 0.05; **P < 0.01; ***P < 0.001. Numbers below bands in Western blot captures show the ratio of the density of the sample band to that of a-tubulin.
Fig. 3.
PPARα-mediated regulation of hepatic CYP2E1 and CYP3A14/3A25. Assessment of the effect of OLE, a PPARα agonist, on CYP2E1 (A), CYP3A14 (B), and CYP3A25 (C) protein level employing Western blotting, and on relative gene expression with quantitative PCR analysis. In wild-type and Ppara-null mice, comparisons were made between controls (designated C) and OLE-treated mice. Values are expressed as means ± S.E. Wild-type mice (n = 11); Ppara-null mice (n = 10). *P < 0.05; **P < 0.01; ***P < 0.001. Numbers below bands in Western blot captures show the ratio of the density of the sample band to that of a-tubulin.
Assessment of OLE effect on transcription factors and signal transduction pathways related to P450 regulation.
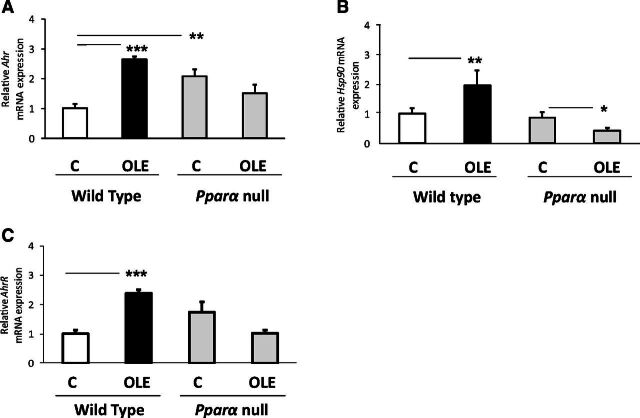
The OLE-induced effect on aryl hydrocarbon receptor (AHR), aryl hydrocarbon receptor repressor (AhrR), and heat shock protein 90 (HSP90) (Fig. 4A–C, respectively), critical transcription factors in Cyp1a1, Cyp1a2, and Cyp1b1 regulation (Nebert et al., 2013), is apparently followed by upregulation of these P450s, an effect profoundly associated with the OLE-induced PPARα activation (Malliou et al., 2018), because OLE did not increase Ahr, AhrR, and Hsp90 mRNAs in Ppara-null mice, but the drug rather slightly repressed the expression of these transcription factors (Fig. 4A–C).
Fig. 4.
Assessment of the effect of OLE, a PPARα agonist, on various factors involved in CYP1A and CYP1B regulation (Matsunaga et al., 1990). Ahr (A), AhRR (B), and HSP90 (C) relative mRNA expression was estimated with quantitative PCR analysis. In wild-type and Ppara-null mice, comparisons were made between controls (designated C) and OLE-treated mice. Values are expressed as means ± S.E. Wild-type mice (n = 11); Ppara-null mice (n = 10). *P < 0.05; **P < 0.01; ***P < 0.001.
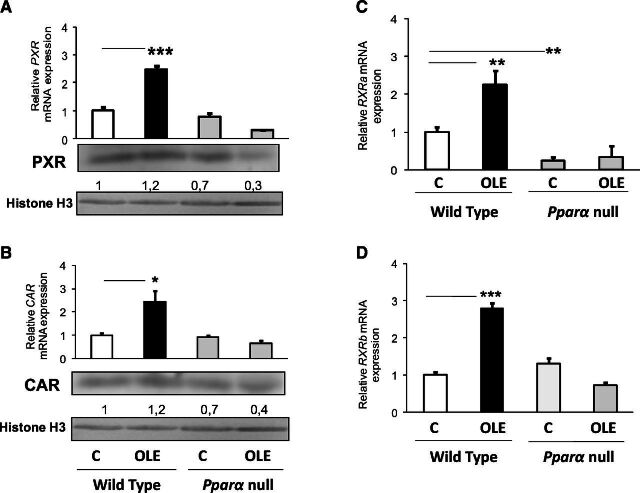
The OLE-induced effect on Cyp2c29, Cyp2c44, and Cyp3a25 mRNA expression in the liver of WT mice is apparently mediated by induction of Pxr, Car, retinoic X receptor (Rxr) a, and Rxrb, which encode critical transcription factors regulating the above P450 genes (Daskalopoulos et al., 2012a) (Fig. 5A–D). The drug’s upregulating effect on these transcription factors is probably mediated by PPARα because OLE had a repressing effect on Pxr, Car, and Rxrb in Ppara-null mice (Fig. 5A, B, and D, respectively).
Fig. 5.
Assessment of the effect of OLE, a PPARα agonist, on various factors involved in CYP3A and CYP2C regulation (Konstandi et al., 2006). Pxr (A), Car (B), Rxra (C), and Rxrb (D) relative mRNA expression with quantitative PCR analysis. In wild-type and Ppara-null mice, comparisons were made between controls (designated C) and OLE-treated mice. Values are expressed as means ± S.E. Wild-type mice (n = 11); Pparα-null mice (n = 10). *P < 0.05; **P < 0.01; ***P < 0.001. Numbers below bands in Western blot captures show the ratio of the density of the sample band to that of histone H3.
Long-term treatment of WT mice with OLE induced hepatic Ppara mRNA expression by 4–5-fold, a stimulating effect that was also evident at the protein level (Malliou et al., 2018; Supplemental Fig. 1). Further analysis employing molecular docking experiments suggests that OLE is a PPARα ligand, and the luciferase reporter gene assay revealed significant activation of this nuclear receptor and transcription factor by OLE (Malliou et al., 2018), followed by upregulation of various PPARα target genes including Acox1, Acot1, Cyp4a10, and Cyp4a14 (Malliou et al., 2018). Hnf4a, encoding a nuclear transcription factor, which holds a determinant role in the regulation of several P450 genes (Daskalopoulos et al., 2012a; Konstandi et al., 2020), was also upregulated by OLE in the liver of mice (Malliou et al., 2018; Supplemental Fig. 1). It is assumed that the OLE-upregulating effect on all P450s analyzed in this study is profoundly mediated by PPARα because no similar effects on these P450s were observed in Ppara-null mice (Fig. 1A–C, Fig. 2A–C, Fig. 3A–C). In particular, OLE either repressed Cyp1a1, Cyp1a2, Cyp1b1 (Fig. 1A–C) Cyp2c29, Cyp2c44, Cyp2d22 (Fig. 2A–C), and Cyp3a25 (Fig. 3C) or had no effect on Cyp2e1 (Fig. 3A) and Cyp3a14 (Fig. 3B) in the liver of Ppara-deficient mice. The determinant role of PPARα in P450 induction was also confirmed in mice treated with the selective PPARα agonist, fenofibrate (Ghonem et al., 2015; Hu et al., 2019), which increased CYP1A1, CYP1A2, CYP1B1, CYP2C, CYP2D, CYP2E1, and CYP3A protein expression in the liver of WT mice, whereas fenofibrate had no similar effects in the liver of PPARα-deficient mice (Fig. 6).
Fig. 6.
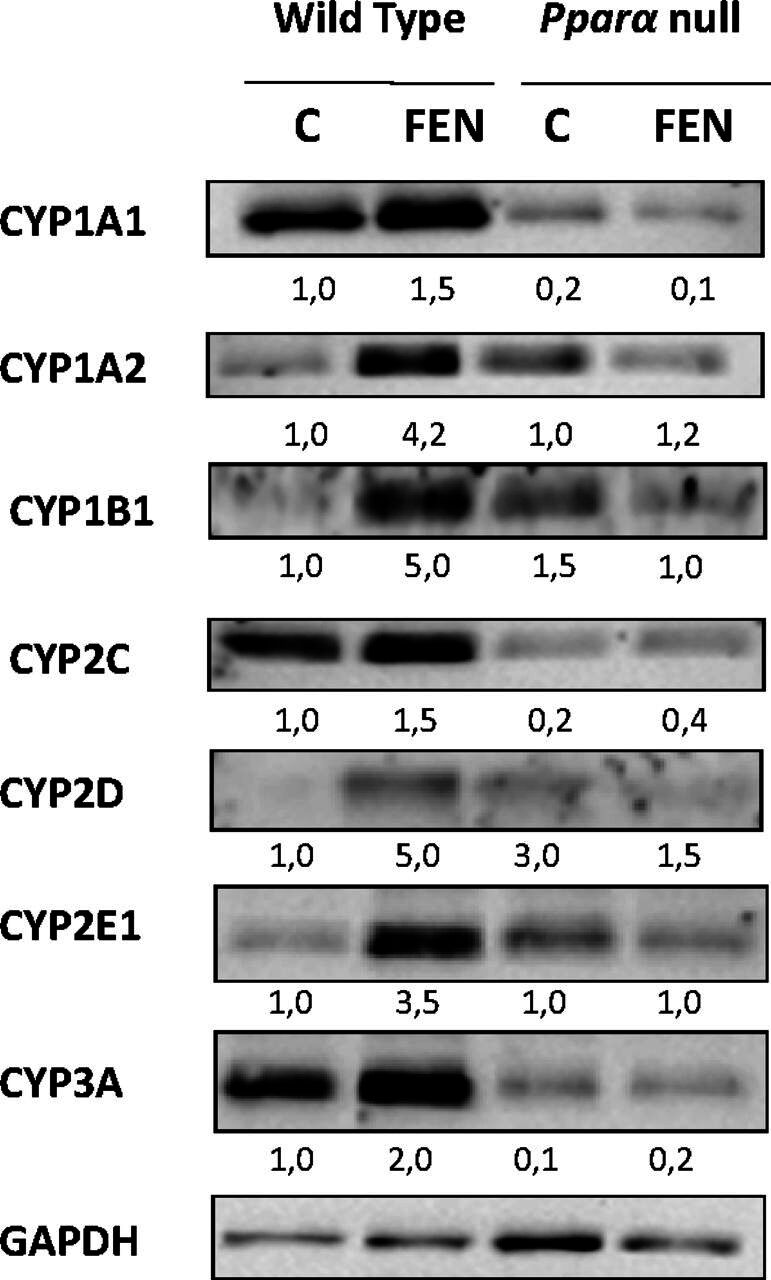
In vivo assessment of the effects of fenofibrate (FEN), a selective PPARα agonist, on P450 protein expression by Western blotting using microsomal proteins extracted from liver samples of wild-type and PPARα-null mice. FEN-induced hepatic CYP1A1, CYP1A2, CYP1B1, CYP2C, CYP2D, CYP2E1, and CYP3A protein expression. In wild-type and Ppara-null mice, comparisons were made between controls (designated C) and FEN-treated mice. The samples in the Western blot captures are representative of other three that were analyzed in separate blots. Numbers below bands in Western blot captures show the ratio of the density of the sample band to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
The role of the nitric oxide synthases (NOSs), endothelial NOS (eNOS) and inducible NOS (iNOS), in the regulation of hepatic Cyp1a, Cyp1b, Cyp3a, Cyp2c, Cyp2d, and Cyp2e1 by OLE in WT mice appears to be less significant because although OLE induced eNOS and iNOS mRNA expression (Fig. 7A and B), this effect was not followed by inhibition of the aforementioned P450s (Hara and Adachi, 2002). Instead, OLE induced the synthesis of these P450s (Fig. 1A–C, Fig. 2A–C and Fig. 3A–C), indicating that the NOS-inhibiting effect on P450s was probably overlapped by the inducing effect of other regulatory factors.
Fig. 7.
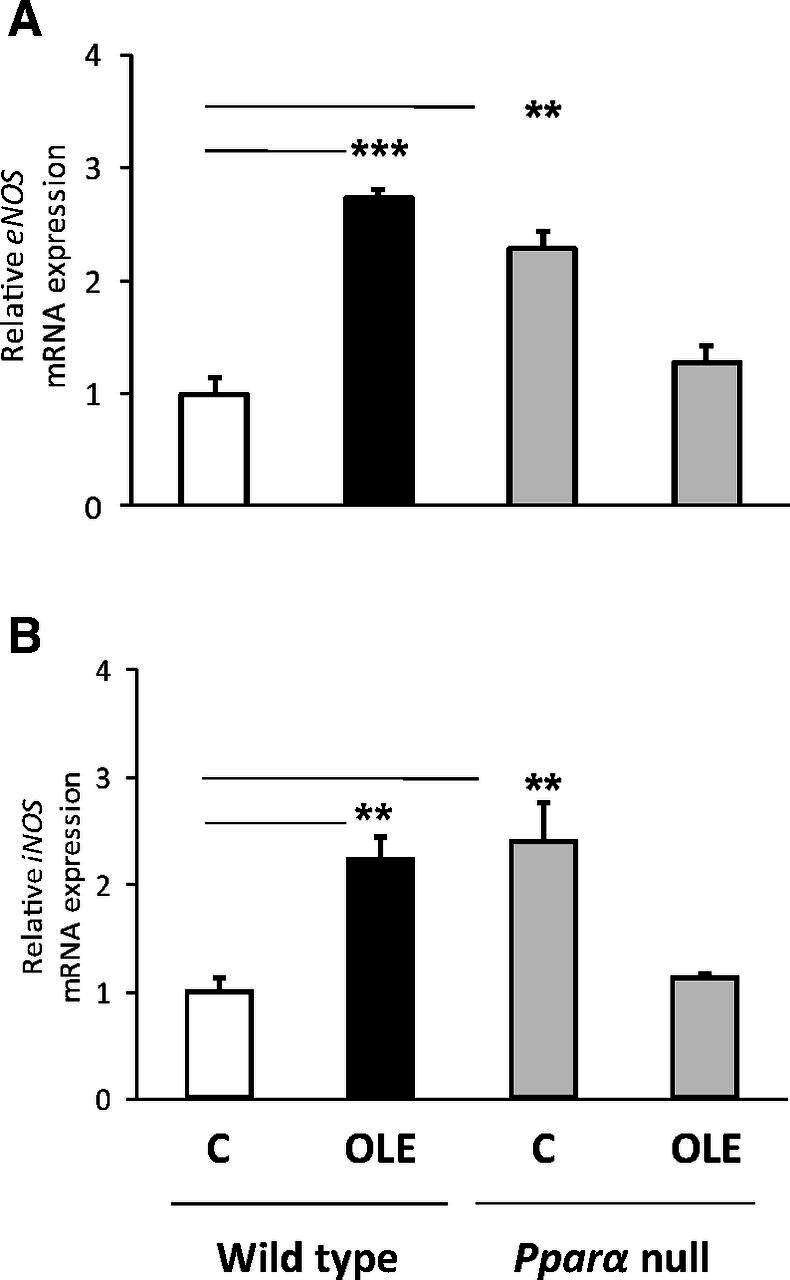
Assessment of the effect of OLE, a PPARα agonist, on eNOS (A) and iNOS (B) relative mRNA expression with quantitative PCR analysis. In wild-type and Ppara-null mice, comparisons were made between controls (designated C) and OLE-treated mice. Values are expressed as means ± S.E. Wild-type mice (n = 11); Pparα-null mice (n = 10). **P < 0.01; ***P < 0.001.
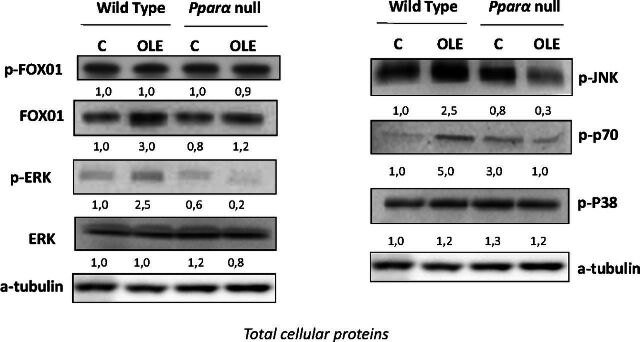
At signal transduction level, OLE increased FOXO1 and JNK phosphorylation, along with that of p70 in the livers of WT mice (Fig. 8). These findings indicate that the OLE-induced activation of phosphoinositide 3-kinase/AKT/FOXO1, JNK, and AKT/p70 pathways (Fig. 8) is profoundly related to the induction of P450s that belong to the CYP1A, CYP1B, CYP3A, CYP2C, CYP2D, and CYP2E subfamilies. Activation of ERK could also participate in the regulation of P450 induction by OLE (Fig. 8).
Fig. 8.
Inv vivo assessment of the effects of OLE, a PPARα agonist, on several signal transduction pathways involved in P450 regulation by Western blotting using total cellular proteins extracted from liver samples of mice. OLE induced activation of FOXO1, JNK, p70, and ERK but not to P38–mitogen-activated protein kinase. In wild-type and Ppara-null mice, comparisons were made between controls and OLE-treated mice. C, control; p-, phosphorylated. The samples in the Western blot captures are representative of other three that were analyzed in separate blots. Numbers below bands in Western blot captures show the ratio of the density of the sample band to that of a-tubulin.
Comparatively, OLE and fenofibrate at the doses given induced hepatic protein expression of CYP1A1, CYP2C, and CYP3A to a similar extent, whereas the induction of CYP1A2, CYP1B1, CYP2D, and CYP2E1 by fenofibrate was markedly higher than that by OLE (Figs. 1–3 and Fig. 6).
Discussion
There is an accumulating amount of evidence that supports the beneficial effects of OLE in preservation of good health and in the outcome of various disease states, including those related to the cardiovascular system (Ahamad et al., 2019; Andreadou et al., 2006; Araki et al., 2019; Lockyer et al., 2017; Vogel et al., 2014), which are mainly attributed to the drug’s anti-inflammatory properties (Malliou et al., 2018), to its effects on lipid homeostasis (Andreadou et al., 2014; Araki et al., 2019; Lockyer et al., 2017; Malliou et al., 2018), and to the protection of myocardium in conditions related to ischemia (Andreadou et al., 2014). Based on these OLE-related beneficial effects, the pharmaceutical industry produced several food supplements and herbal medicines containing the drug, which are available in the market without a prescription, a potentially high-risk practice for health and disease.
It is well documented that cytochromes belonging to P450 families 1–3 are involved in the metabolism of a plethora of diverse endogenous and exogenous compounds, such as drugs, precarcinogens, carcinogens, environmental pollutants, food additives, prostaglandins, fatty acids, lipid hydroperoxides, steroid hormones, biogenic amines, and numerous other xenobiotics (Cribb et al., 2005; Gonzalez, 2005; Gonzalez and Gelboin, 1994; Gonzalez and Yu, 2006; Guengerich, 2003; Ingelman-Sundberg, 2004b; Xu et al., 2005) to increase their water solubility and prepare them for the subsequent conjugation and elimination (Gonzalez, 2005). Their biotransformation takes place primarily in the liver and usually results in inactive metabolites or, in some cases, in active molecules, a process that usually depends on the structure of the parent compound. These active metabolic products may induce several serious toxic manifestations, such as teratogenesis, carcinogenicity, cell death, and oxidative stress, among others (Cribb et al., 2005; Gonzalez, 2005; Gonzalez and Gelboin, 1994; Gonzalez and Yu, 2006; Guengerich, 2003; Ingelman-Sundberg, 2004a; Konstandi, 2013; Xu et al., 2005). The findings of the current study clearly showed that OLE induced the expression of several P450s in the liver of WT mice, including CYP1A1, CYP1A2, CYP1B1, CYP3A14, CYP3A25, CYP2C29, CYP2C44, CYP2D22, and CYP2E1 via activation of major nuclear transcription and other cellular factors, such as AHR, CAR, RXR, and PXR (Dalton et al., 2000; Daskalopoulos et al., 2012a; Harkitis et al., 2015). Induction of P450s is a part of the regulatory mechanisms aimed at maintaining homeostasis. It is important for the adaptation of the organism to a modified biologic and chemical internal milieu and environment. Nonetheless, it is not always feasible to predict the extent of the effect of P450 induction on drug efficacy and toxicological risk assessment, determinant parameters in clinical drug therapy (Pelkonen et al., 2008). According to the regulatory standards of the U.S. Food and Drug Administration, a drug is considered a P450 inducer with clinical significance if the fold change of P450 mRNA expression relative to the vehicle control is ≥2-fold and then is efficient to accelerate the metabolism of P450 substrates (Fahmi and Ripp, 2010; https://www.fda.gov/drugs/drug-interactions-labeling/drug-development-and-drug-interactions-table-substrates-inhibitors-and-inducers). It is considered that induction of CYP1A2, CYP3A14, CYP3A25, CYP2C29, CYP2C44, CYP2D22, and CYP2E1 may result in reduced concentrations of their drug substrates in blood, potentially below their therapeutic levels, and therefore in failure of pharmacotherapy (Dalton et al., 2000; Kawajiri and Fujii-Kuriyama, 2007; Konstandi, 2013; Okey, 1990; Zhang et al., 1999), whereas induction of CYP1A1, CYP1A2, and CYP1B1 is followed by bioactivation of the major groups of precarcinogens, the polycyclic aromatic hydrocarbons, polycyclic arylamines, and aflatoxin B1, to DNA-binding metabolites (Cheng and Morgan, 2001; Kawajiri, 1999; Mulder et al., 2001; Pasanen and Pelkonen, 1994). In contrast to the present findings related to the induction of CYP3A14/25 by OLE, previous in vitro studies indicated that OLE and its metabolite hydroxytyrosol inactivated androstenedione 6beta-hydroxylase (CYP3A4-dependent) activity in microsomal preparations of human liver (Stupans et al., 2001; Stupans et al., 2000; Zhou et al., 2007). This discrepancy could be explained primarily on the basis of a species-specific regulation of the main P450s (Konstandi et al., 2020; Visioli et al., 2002a) and the different experimental approaches followed in these studies. The current in vivo study evaluated the OLE effect on CYP3A14/25 in mouse liver microsomes, whereas the in vitro studies mentioned above used human liver microsomes (Stupans et al., 2001; Stupans et al., 2000).
To investigate the mechanisms underlying the OLE-induced upregulating effect on P450s, the role of the nuclear transcription factor, PPARα, that regulates a variety of genes encoding P450s was assessed (Tauber et al., 2020). For this purpose, WT and Pparα-null mice were employed and received OLE in their diet for 6 weeks. The findings indicated that, in contrast to what happened in WT mice, OLE did not induce the expression of CYP1A1, CYP1A2, CYP1B1, CYP3A14, CYP3A25, CYP2C29, CYP2C44, CYP2D22, and CYP2E1 in the liver of Pparα-null mice. It is apparent that the OLE-mediated induction of these P450s in the liver of WT mice is mediated by the drug’s stimulating effect on PPARα activation. To further evaluate the involvement of PPARα in the regulation of P450s and, in particular, of P450 induction, WT mice were treated with the selective PPARα agonist fenofibrate (Ghonem et al., 2015; Hu et al., 2019), which induced CYP1A1, CYP1A2, CYP1B1, CYP2C, CYP2D, CYP2E1, and CYP3A protein expression in their livers. No similar effects were observed in the liver of Pparα-null mice after treatment with fenofibrate, and these findings confirm the role of PPARα in P450 induction. This hypothesis is also supported by other studies reporting the upregulating effects of PPARs on several P450s and, in particular, of PPARα on P450s displaying epoxygenase activities on polyunsaturated fatty acids, including isozymes that belong to the CYP1A, CYP3A, CYP2C, and CYP2E subfamilies (Tauber et al., 2020; Thomas et al., 2013). Interestingly, however, findings from the current and previous studies (Shi et al., 2017) showed that basal P450 expression levels in the liver of Pparα-null mice are markedly higher than those in WT mice. This observation indicates a distinct role of PPARα in the regulation of constitutive and inducible P450 expression. It is well established that P450 induction and constitutive expression are regulated by distinct mechanisms (Hahn et al., 2009; Nebert, 2000; Zanger and Schwab, 2013), including various transcription factors, coactivators, and corepressors, with both positive and negative regulatory roles that crosstalk between several regulatory pathways (Pelkonen et al., 2008). In this regard, the diverse role of PPARα in P450 regulation at basal and induced states may be attributed in the crosstalk between this transcription factor and AHR, CAR, PXR, and/or other transcription factors with significant regulatory roles in P450 induction by xenobiotics (Honkakoski and Negishi, 2000; Pelkonen et al., 2008; Zanger and Schwab, 2013). This hypothesis is supported by the finding that several functional PPARα-binding regions within the P450 promoters were detected (Makia and Goldstein, 2016; Oshida et al., 2016a; Thomas et al., 2013; Yao et al., 2007) (Oshida et al., 2016b). It should be noted that the mechanism underlying the PPARα-mediated P450 induction by OLE could also include modifications at PPARα phosphorylation status. It is well established that ligand-induced PPARα activation is mediated by increased expression of this nuclear receptor at transcriptional and protein level along with alterations at its phosphorylation state (Ning et al., 2016; Tamasi et al., 2008; Barger et al., 2001; Passilly et al., 1999).
The nuclear transcription factor, HNF4α, profoundly participates in the OLE-induced upregulation of the above-mentioned P450s because OLE induced hepatic Hnf4a expression in WT mice. It is known that the transcription factors HNF4α, RXR, and HNF1α (Weltman et al., 1998; Wiwi and Waxman, 2004), along with FOXO1 and the nuclear receptors CAR and PXR (Kodama et al., 2004), belong to a complex crosstalk mechanism displaying central regulatory roles in the expression of various P450s.
In P450 regulation there is also an interplay between PPARα and iNOS, efficient enough to modify P450 expression. In particular, it has been reported that PPARα ligands reduced the lipopolysaccharide-induced iNOS expression and subsequent nitric oxide synthesis in macrophages by increasing the proteasome pathway mediated iNOS protein degradation (Paukkeri et al., 2007). Notably, nitric oxide displays downregulating effects on several P450 genes, including CYP1A1/2, CYP2B1/2, CYP2D6, CYP2E1, and CYP3A4, but the underlying mechanism of this regulation remains blurred (Gergel et al., 1997; Hara and Adachi, 2002; Wink et al., 1993). Current findings indicated that the OLE-induced effect on NOS has a weak impact on P450s compared with other regulatory factors. It appears that the upregulating effect of OLE on NOS, which should be followed by P450 downregulation (Gergel et al., 1997; Hara and Adachi, 2002; Wink et al., 1993), was overlapped by the upregulating effects of other transcription factors. Further analysis indicated that at signal transduction level, activation of the signaling pathways related to phosphoinositide 3-kinase/AKT/FOXO1, JNK, AKT/p70S6K, and ERK profoundly participate in P450 induction by OLE (Kim and Novak, 2007). It is well documented that upon activation, AKT stimulates the phosphorylation of the nuclear transcription factor, FOXO1, which in turn translocates into the cytoplasm. This process is usually followed by termination of FOXO1 transcriptional activity in P450 genes. But then, the OLE-mediated activation of JNK promotes the nuclear localization of FOXO1, an effect that restores its transcriptional activity in P450 genes and counteracts the downregulating effect of AKT (Daskalopoulos et al., 2012a; Hay, 2011). Current findings indicated that OLE decreased JNK phosphorylation in the liver of Pparα-null mice, an effect that likely participates in the OLE-mediated downregulation of P450s in these mice.
Taken together, the above data clearly show the upregulating effect of OLE on various P450s encoding the main drug-metabolizing enzymes of phase I, an effect that is apparently mediated by PPARα activation. Current and previous findings indicate that diverse and distinct mechanisms participate in the regulation of constitutive and inducible P450 expression by PPARα agonists. Apparently, PPARα displays a negative regulatory role in constitutive P450 expression and a positive role in P450 induction by OLE. Although it is not always feasible to accurately predict the clinical impact of the OLE-induced effect on CYP3A14, CYP3A25, CYP2C29, CYP2C44, CYP2D22, CYP2E1, CYP1A1, CYP1A2, and CYP1B1, under certain conditions it could modify the pharmacokinetic profile of drug substrates of these P450s and subsequently affect the outcome of pharmacotherapy and the severity of drug-related adverse reactions. Although these data come from a preclinical study and cannot be extrapolated directly to humans, they underscore the necessity of taking into consideration the consumption of herbal medicines and food supplements containing drugs such as OLE that may affect the pharmacokinetic profiles of co-administered drugs. This parameter should be considered predominately in multiple drug therapeutic schemes, in particular those of vital significance for the patient, or in drugs with narrow therapeutic windows or with severe side effects. This concern is of particular importance because herbal medicines and food supplements are widely used and freely available in the market. All of these concerns indicate that further investigation, pharmacovigilance, better regulatory control, and advice from health professionals is essential to ensure safety when using herbal medicines and food supplements.
Abbreviations
- AHR
aryl hydrocarbon receptor
- AhrR
aryl hydrocarbon receptor repressor
- AKT
protein kinase B
- CAR
constitutive androstane receptor
- eNOS
endothelial NOS
- ERK
extracellular signal-related kinase
- FOXO1
forkhead box protein O1
- HNF4α
hepatocyte nuclear factor 4α
- HPLC
high-performance liquid chromatography
- HSP90
heat shock protein 90
- iNOS
inducible NOS
- JNK
c-Jun N-terminal kinase
- NOS
nitric oxide synthase
- OLE
oleuropein
- P450
cytochrome P450
- PCR
polymerase chain reaction
- PPARα
peroxisome proliferator-activated receptor α
- PXR
pregnane X receptor
- RXR
retinoic X receptor
- WT
wild type
Authorship Contributions
Participated in research design: Konstandi, Gonzalez.
Conducted experiments: Malliou, Andriopoulou, Konstandi, Skaltsounis.
Performed data analysis: Konstandi, Andriopoulou, Malliou.
Wrote or contributed to the writing of the manuscript: Konstandi, Gonzalez.
Footnotes
This study was funded by the European Union (European Regional Development Fund) and by Greek national funds coming from the operational program “THESSALY- MAINLAND GREECE AND EPIRUS-2007-2013” of the National Strategic Reference Framework (NSRF 2007-2013, Grant 346985/80753). We certify that there is no involvement of the funding sources in the research conduct and/or preparation of this article, in study design, in the collection, analysis and interpretation of data, in the writing of the article, and the decision to submit the article for publication in DMD.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Abdel-aleem S, el-Merzabani MM, Sayed-Ahmed M, Taylor DA, Lowe JE (1997) Acute and chronic effects of adriamycin on fatty acid oxidation in isolated cardiac myocytes. J Mol Cell Cardiol 29:789–797. [DOI] [PubMed] [Google Scholar]
- Ahamad J, Toufeeq I, Khan MA, Ameen MSM, Anwer ET, Uthirapathy S, Mir SR, Ahmad J (2019) Oleuropein: A natural antioxidant molecule in the treatment of metabolic syndrome. Phytother Res 33:3112–3128. [DOI] [PubMed] [Google Scholar]
- Andreadou I, Iliodromitis EK, Mikros E, Constantinou M, Agalias A, Magiatis P, Skaltsounis AL, Kamber E, Tsantili-Kakoulidou A, Kremastinos DT (2006) The olive constituent oleuropein exhibits anti-ischemic, antioxidative, and hypolipidemic effects in anesthetized rabbits. J Nutr 136:2213–2219. [DOI] [PubMed] [Google Scholar]
- Andreadou I, Mikros E, Ioannidis K, Sigala F, Naka K, Kostidis S, Farmakis D, Tenta R, Kavantzas N, Bibli S-I, et al. (2014) Oleuropein prevents doxorubicin-induced cardiomyopathy interfering with signaling molecules and cardiomyocyte metabolism. J Mol Cell Cardiol 69:4–16. [DOI] [PubMed] [Google Scholar]
- Araki R, Fujie K, Yuine N, Watabe Y, Nakata Y, Suzuki H, Isoda H, Hashimoto K (2019) Olive leaf tea is beneficial for lipid metabolism in adults with prediabetes: an exploratory randomized controlled trial. Nutr Res 67:60–66. [DOI] [PubMed] [Google Scholar]
- Barger PM, Browning AC, Garner AN, Kelly DP (2001) p38 mitogen-activated protein kinase activates peroxisome proliferator-activated receptor α: a potential role in the cardiac metabolic stress response. J Biol Chem 276:44495–44501. [DOI] [PubMed] [Google Scholar]
- Bensoussan A, Myers SP, Drew AK, Whyte IM, Dawson AH (2002) Development of a Chinese herbal medicine toxicology database. J Toxicol Clin Toxicol 40:159–167. [DOI] [PubMed] [Google Scholar]
- Briante R, La Cara F, Tonziello MP, Febbraio F, Nucci R (2001) Antioxidant activity of the main bioactive derivatives from oleuropein hydrolysis by hyperthermophilic beta-glycosidase. J Agric Food Chem 49:3198–3203. [DOI] [PubMed] [Google Scholar]
- Cheng PY, Morgan ET (2001) Hepatic cytochrome P450 regulation in disease states. Curr Drug Metab 2:165–183. [DOI] [PubMed] [Google Scholar]
- Choi HK, Waxman DJ (1999) Growth hormone, but not prolactin, maintains, low-level activation of STAT5a and STAT5b in female rat liver. Endocrinology 140:5126–5135. [DOI] [PubMed] [Google Scholar]
- Cribb AE, Peyrou M, Muruganandan S, Schneider L (2005) The endoplasmic reticulum in xenobiotic toxicity. Drug Metab Rev 37:405–442. [DOI] [PubMed] [Google Scholar]
- Dalton TP, Dieter MZ, Matlib RS, Childs NL, Shertzer HG, Genter MB, Nebert DW (2000) Targeted knockout of Cyp1a1 gene does not alter hepatic constitutive expression of other genes in the mouse [Ah] battery. Biochem Biophys Res Commun 267:184–189. [DOI] [PubMed] [Google Scholar]
- Daskalopoulos EP, Lang MA, Marselos M, Malliou F, Konstandi M (2012a) D2-dopaminergic receptor-linked pathways: critical regulators of CYP3A, CYP2C, and CYP2D. Mol Pharmacol 82:668–678. [DOI] [PubMed] [Google Scholar]
- Daskalopoulos EP, Malliou F, Rentesi G, Marselos M, Lang MA, Konstandi M (2012b) Stress is a critical player in CYP3A, CYP2C, and CYP2D regulation: role of adrenergic receptor signaling pathways. Am J Physiol Endocrinol Metab 303:E40–E54. [DOI] [PubMed] [Google Scholar]
- De Smet PA (1995) Health risks of herbal remedies. Drug Saf 13:81–93. [DOI] [PubMed] [Google Scholar]
- Del Boccio P, Di Deo A, De Curtis A, Celli N, Iacoviello L, Rotilio D (2003) Liquid chromatography-tandem mass spectrometry analysis of oleuropein and its metabolite hydroxytyrosol in rat plasma and urine after oral administration. J Chromatogr B Analyt Technol Biomed Life Sci 785:47–56. [DOI] [PubMed] [Google Scholar]
- Deng J-F (2002) Clinical and laboratory investigations in herbal poisonings. Toxicology 181-182:571–576. [DOI] [PubMed] [Google Scholar]
- Edgecombe SC, Stretch GL, Hayball PJ (2000) Oleuropein, an antioxidant polyphenol from olive oil, is poorly absorbed from isolated perfused rat intestine. J Nutr 130:2996–3002. [DOI] [PubMed] [Google Scholar]
- Eisenberg DM, Kessler RC, Foster C, Norlock FE, Calkins DR, Delbanco TL (1993) Unconventional medicine in the United States. Prevalence, costs, and patterns of use. N Engl J Med 328:246–252. [DOI] [PubMed] [Google Scholar]
- Ernst E, Pittler MH (2002a) Herbal medicine. Med Clin North Am 86:149–161. [DOI] [PubMed] [Google Scholar]
- Ernst E, Pittler MH (2002b) Risks associated with herbal medicinal products. Wien Med Wochenschr 152:183–189. [DOI] [PubMed] [Google Scholar]
- Fahmi OA, Ripp SL (2010) Evaluation of models for predicting drug-drug interactions due to induction. Expert Opin Drug Metab Toxicol 6:1399–1416. [DOI] [PubMed] [Google Scholar]
- Flint MS, Hood BL, Sun M, Stewart NA, Jones-Laughner J, Conrads TP (2010) Proteomic analysis of the murine liver in response to a combined exposure to psychological stress and 7,12-dimethylbenz(a)anthracene. J Proteome Res 9:509–520. [DOI] [PubMed] [Google Scholar]
- Fruchart JC, Duriez P (2006) Mode of action of fibrates in the regulation of triglyceride and HDL-cholesterol metabolism. Drugs Today (Barc) 42:39–64. [DOI] [PubMed] [Google Scholar]
- Gergel D, Misík V, Riesz P, Cederbaum AI (1997) Inhibition of rat and human cytochrome P4502E1 catalytic activity and reactive oxygen radical formation by nitric oxide. Arch Biochem Biophys 337:239–250. [DOI] [PubMed] [Google Scholar]
- Ghonem NS, Assis DN, Boyer JL (2015) Fibrates and cholestasis. Hepatology 62:635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez FJ (2005) Role of cytochromes P450 in chemical toxicity and oxidative stress: studies with CYP2E1. Mutat Res 569:101–110. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Gelboin HV (1994) Role of human cytochromes P450 in the metabolic activation of chemical carcinogens and toxins. Drug Metab Rev 26:165–183. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Yu A-M (2006) Cytochrome P450 and xenobiotic receptor humanized mice. Annu Rev Pharmacol Toxicol 46:41–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greensfelder L (2000) Alternative medicine. Herbal product linked to cancer. Science 288:1946. [DOI] [PubMed] [Google Scholar]
- Guengerich FP (2003) Cytochromes P450, drugs, and diseases. Mol Interv 3:194–204. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Allan LL, Sherr DH (2009) Regulation of constitutive and inducible AHR signaling: complex interactions involving the AHR repressor. Biochem Pharmacol 77:485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller CA, Benowitz NL (2000) Adverse cardiovascular and central nervous system events associated with dietary supplements containing ephedra alkaloids. N Engl J Med 343:1833–1838. [DOI] [PubMed] [Google Scholar]
- Hansen M, Connolly T (2008) Nuclear receptors as drug targets in obesity, dyslipidemia and atherosclerosis. Curr Opin Investig Drugs 9:247–255. [PubMed] [Google Scholar]
- Hara H, Adachi T (2002) Contribution of hepatocyte nuclear factor-4 to down-regulation of CYP2D6 gene expression by nitric oxide. Mol Pharmacol 61:194–200. [DOI] [PubMed] [Google Scholar]
- Harkitis P, Daskalopoulos EP, Malliou F, Lang MA, Marselos M, Fotopoulos A, Albucharali G, Konstandi M (2015) Dopamine D2-Receptor Antagonists Down-Regulate CYP1A1/2 and CYP1B1 in the Rat Liver. PLoS One 10:e0128708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N (2011) Interplay between FOXO, TOR, and Akt. Biochim Biophys Acta 1813:1965–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkakoski P, Negishi M (2000) Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochem J 347:321–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu DD, Zhao Q, Cheng Y, Xiao XR, Huang JF, Qu Y, Li X, Tang YM, Bao WM, Yang JH, Jiang T, Hu JP, Gonzalez FJ, Li F (2019) The protective roles of PPARα activation in triptolide-induced liver injury. Toxicol Sci 171:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impellizzeri D, Esposito E, Mazzon E, Paterniti I, Di Paola R, Bramanti P, Morittu VM, Procopio A, Perri E, Britti D, et al. (2012) The effects of a polyphenol present in olive oil, oleuropein aglycone, in an experimental model of spinal cord injury in mice. Biochem Pharmacol 83:1413–1426. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M (2004a) Human drug metabolising cytochrome P450 enzymes: properties and polymorphisms. Naunyn Schmiedebergs Arch Pharmacol 369:89–104. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M (2004b) Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends Pharmacol Sci 25:193–200. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C (2007) Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther 116:496–526. [DOI] [PubMed] [Google Scholar]
- Kawajiri K (1999) Cyp1a1. IARC Sci Publ (148):159–172. [PubMed] [Google Scholar]
- Kawajiri K, Fujii-Kuriyama Y (2007) Cytochrome P450 gene regulation and physiological functions mediated by the aryl hydrocarbon receptor. Arch Biochem Biophys 464:207–212. [DOI] [PubMed] [Google Scholar]
- Kennedy DA, Seely D (2010) Clinically based evidence of drug-herb interactions: a systematic review. Expert Opin Drug Saf 9:79–124. [DOI] [PubMed] [Google Scholar]
- Kim SK, Novak RF (2007) The role of intracellular signaling in insulin-mediated regulation of drug metabolizing enzyme gene and protein expression. Pharmacol Ther 113:88–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepser TB, Klepser ME (1999) Unsafe and potentially safe herbal therapies. Am J Health Syst Pharm 56:125–138, quiz 139–141. [DOI] [PubMed] [Google Scholar]
- Kodama S, Koike C, Negishi M, Yamamoto Y (2004) Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes. Mol Cell Biol 24:7931–7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh H-L, Woo S (2000) Chinese proprietary medicine in Singapore: regulatory control of toxic heavy metals and undeclared drugs. Drug Saf 23:351–362. [DOI] [PubMed] [Google Scholar]
- Konstandi M (2013) Psychophysiological stress: a significant parameter in drug pharmacokinetics. Expert Opin Drug Metab Toxicol 9:1317–1334. [DOI] [PubMed] [Google Scholar]
- Konstandi M, Andriopoulou CE, Cheng J, Gonzalez FJ (2020) Sex steroid hormones differentially regulate CYP2D in female wild-type and CYP2D6-humanized mice. J Endocrinol 245:301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstandi M, Johnson EO, Lang MA (2014) Consequences of psychophysiological stress on cytochrome P450-catalyzed drug metabolism. Neurosci Biobehav Rev 45:149–167. [DOI] [PubMed] [Google Scholar]
- Konstandi M, Kostakis D, Harkitis P, Johnson EO, Marselos M, Adamidis K, Lang MA (2006) Benzo(α)pyrene-induced up-regulation of CYP1A2 gene expression: role of adrenoceptor-linked signaling pathways. Life Sci 79:331–341. [DOI] [PubMed] [Google Scholar]
- Konstandi M, Kostakis D, Harkitis P, Marselos M, Johnson EO, Adamidis K, Lang MA (2005) Role of adrenoceptor-linked signaling pathways in the regulation of CYP1A1 gene expression. Biochem Pharmacol 69:277–287. [DOI] [PubMed] [Google Scholar]
- Kuusisto J, Andrulionyte L, Laakso M (2007) Atherosclerosis and cardiovascular risk reduction with PPAR agonists. Curr Atheroscler Rep 9:274–280. [DOI] [PubMed] [Google Scholar]
- Lang MA, Gielen JE, Nebert DW (1981) Genetic evidence for many unique liver microsomal P-450-mediated monooxygenase activities in heterogeneic stock mice. J Biol Chem 256:12068–12075. [PubMed] [Google Scholar]
- Leuenberger N, Pradervand S, Wahli W (2009) Sumoylated PPARalpha mediates sex-specific gene repression and protects the liver from estrogen-induced toxicity in mice. J Clin Invest 119:3138–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockyer S, Rowland I, Spencer JPE, Yaqoob P, Stonehouse W (2017) Impact of phenolic-rich olive leaf extract on blood pressure, plasma lipids and inflammatory markers: a randomised controlled trial. Eur J Nutr 56:1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makia NL, Goldstein JA (2016) CYP2C8 Is a Novel Target of Peroxisome Proliferator-Activated Receptor α in Human Liver. Mol Pharmacol 89:154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malliou F, Andreadou I, Gonzalez FJ, Lazou A, Xepapadaki E, Vallianou I, Lambrinidis G, Mikros E, Marselos M, Skaltsounis AL, et al. (2018) The olive constituent oleuropein, as a PPARα agonist, markedly reduces serum triglycerides. J Nutr Biochem 59:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga E, Zeugin T, Zanger UM, Aoyama T, Meyer UA, Gonzalez FJ (1990) Sequence requirements for cytochrome P-450IID1 catalytic activity. A single amino acid change (Ile380 Phe) specifically decreases Vmax of the enzyme for bufuralol but not debrisoquine hydroxylation. J Biol Chem 265:17197–17201. [PubMed] [Google Scholar]
- Mckenna DJ, Jones K, Hughes K, Tyler VM (2012) Botanical Medicines: The Desk Reference for Major Herbal Supplements, 2nd ed, Taylor & Francis, Milton Park, UK. [Google Scholar]
- McRae CA, Agarwal K, Mutimer D, Bassendine MF (2002) Hepatitis associated with Chinese herbs. Eur J Gastroenterol Hepatol 14:559–562. [DOI] [PubMed] [Google Scholar]
- Mulder H, Breure AM, Rulkens WH (2001) Prediction of complete bioremediation periods for PAH soil pollutants in different physical states by mechanistic models. Chemosphere 43:1085–1094. [DOI] [PubMed] [Google Scholar]
- Nebert DW (2000) Drug-metabolizing enzymes, polymorphisms and interindividual response to environmental toxicants. Clin Chem Lab Med 38:857–861. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Wikvall K, Miller WL (2013) Human cytochromes P450 in health and disease. Philos Trans R Soc Lond B Biol Sci 368:20120431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning LJ, He AY, Li JM, Lu DL, Jiao JG, Li LY, Li DL, Zhang ML, Chen LQ, Du ZY (2016) Mechanisms and metabolic regulation of PPARα activation in Nile tilapia (Oreochromis niloticus). Biochim Biophys Acta 1861 (9 Pt A):1036–1048. [DOI] [PubMed] [Google Scholar]
- Oi-Kano Y, Kawada T, Watanabe T, Koyama F, Watanabe K, Senbongi R, Iwai K (2008) Oleuropein, a phenolic compound in extra virgin olive oil, increases uncoupling protein 1 content in brown adipose tissue and enhances noradrenaline and adrenaline secretions in rats. J Nutr Sci Vitaminol (Tokyo) 54:363–370. [DOI] [PubMed] [Google Scholar]
- Okey AB (1990) Enzyme induction in the cytochrome P-450 system. Pharmacol Ther 45:241–298. [DOI] [PubMed] [Google Scholar]
- Oshida K, Waxman DJ, Corton JC (2016a) Chemical and Hormonal Effects on STAT5b-Dependent Sexual Dimorphism of the Liver Transcriptome. PLoS One 11:e0150284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshida K, Waxman DJ, Corton JC (2016b) Correction: Chemical and Hormonal Effects on STAT5b- Dependent Sexual Dimorphism of the Liver Transcriptome. PLoS One 11:e0161519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappenheimer JR, Reiss KZ (1987) Contribution of solvent drag through intercellular junctions to absorption of nutrients by the small intestine of the rat. J Membr Biol 100:123–136. [DOI] [PubMed] [Google Scholar]
- Pasanen M, Pelkonen O (1994) The expression and environmental regulation of P450 enzymes in human placenta. Crit Rev Toxicol 24:211–229. [DOI] [PubMed] [Google Scholar]
- Passilly P, Schohn H, Jannin B, Cherkaoui Malki M, Boscoboinik D, Dauça M, Latruffe N (1999) Phosphorylation of peroxisome proliferator-activated receptor α in rat Fao cells and stimulation by ciprofibrate. Biochem Pharmacol 58:1001–1008. [DOI] [PubMed] [Google Scholar]
- Paukkeri EL, Leppänen T, Sareila O, Vuolteenaho K, Kankaanranta H, Moilanen E (2007) PPARalpha agonists inhibit nitric oxide production by enhancing iNOS degradation in LPS-treated macrophages. Br J Pharmacol 152:1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkonen O, Turpeinen M, Hakkola J, Honkakoski P, Hukkanen J, Raunio H (2008) Inhibition and induction of human cytochrome P450 enzymes: current status. Arch Toxicol 82:667–715. [DOI] [PubMed] [Google Scholar]
- Robillard R, Fontaine C, Chinetti G, Fruchart J-C, Staels B (2005) Fibrates, in Atherosclerosis: Diet and Drugs (von Eckardstein A ed) pp 389–406, Springer, Berlin. [DOI] [PubMed] [Google Scholar]
- Shi C, Min L, Yang J, Dai M, Song D, Hua H, Xu G, Gonzalez FJ, Liu A (2017) Peroxisome Proliferator-Activated Receptor α Activation Suppresses Cytochrome P450 Induction Potential in Mice Treated with Gemfibrozil. Basic Clin Pharmacol Toxicol 121:169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Hayes CL, Yamazaki H, Amin S, Hecht SS, Guengerich FP, Sutter TR (1996) Activation of chemically diverse procarcinogens by human cytochrome P-450 1B1. Cancer Res 56:2979–2984. [PubMed] [Google Scholar]
- Spatzenegger M, Jaeger W (1995) Clinical importance of hepatic cytochrome P450 in drug metabolism. Drug Metab Rev 27:397–417. [DOI] [PubMed] [Google Scholar]
- Stedman C (2002) Herbal hepatotoxicity. Semin Liver Dis 22:195–206. [DOI] [PubMed] [Google Scholar]
- Stupans I, Murray M, Kirlich A, Tuck KL, Hayball PJ (2001) Inactivation of cytochrome P450 by the food-derived complex phenol oleuropein. Food Chem Toxicol 39:1119–1124. [DOI] [PubMed] [Google Scholar]
- Stupans I, Stretch G, Hayball P (2000) Olive oil phenols inhibit human hepatic microsomal activity. J Nutr 130:2367–2370. [DOI] [PubMed] [Google Scholar]
- Tamasi V, Miller KK, Ripp SL, Vila E, Geoghagen TE, Prough RA (2008) Modulation of receptor phosphorylation contributes to activation of peroxisome proliferator activated receptor alpha by dehydroepiandrosterone and other peroxisome proliferators. Mol Pharmacol 73:968–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber Z, Koleckova M, Cizkova K (2020) Peroxisome proliferator-activated receptor α (PPARα)-cytochrome P450 epoxygenases-soluble epoxide hydrolase axis in ER + PR + HER2− breast cancer. Med Mol Morphol 53:141–148. [DOI] [PubMed] [Google Scholar]
- Thomas M, Burk O, Klumpp B, Kandel BA, Damm G, Weiss TS, Klein K, Schwab M, Zanger UM (2013) Direct transcriptional regulation of human hepatic cytochrome P450 3A4 (CYP3A4) by peroxisome proliferator-activated receptor alpha (PPARα). Mol Pharmacol 83:709–718. [DOI] [PubMed] [Google Scholar]
- Tuck KL, Hayball PJ (2002) Major phenolic compounds in olive oil: metabolism and health effects. J Nutr Biochem 13:636–644. [DOI] [PubMed] [Google Scholar]
- Visioli F, Galli C (1994) Oleuropein protects low density lipoprotein from oxidation. Life Sci 55:1965–1971. [DOI] [PubMed] [Google Scholar]
- Visioli F, Galli C, Galli G, Caruso D (2002a) Biological activities and metabolic fate of olive oil phenols. Eur J Lipid Sci Technol 104:677–684. [Google Scholar]
- Visioli F, Poli A, Gall C (2002b) Antioxidant and other biological activities of phenols from olives and olive oil. Med Res Rev 22:65–75. [DOI] [PubMed] [Google Scholar]
- Vogel P, Kasper Machado I, Garavaglia J, Zani VT, de Souza D, Morelo Dal Bosco S (2014) Polyphenols benefits of olive leaf (Olea europaea L) to human health. Nutr Hosp 31:1427–1433. [DOI] [PubMed] [Google Scholar]
- Weltman MD, Farrell GC, Hall P, Ingelman-Sundberg M, Liddle C (1998) Hepatic cytochrome P450 2E1 is increased in patients with nonalcoholic steatohepatitis. Hepatology 27:128–133. [DOI] [PubMed] [Google Scholar]
- Wink DA, Osawa Y, Darbyshire JF, Jones CR, Eshenaur SC, Nims RW (1993) Inhibition of cytochromes P450 by nitric oxide and a nitric oxide-releasing agent. Arch Biochem Biophys 300:115–123. [DOI] [PubMed] [Google Scholar]
- Wiwi CA, Waxman DJ (2004) Role of hepatocyte nuclear factors in growth hormone-regulated, sexually dimorphic expression of liver cytochromes P450. Growth Factors 22:79–88. [DOI] [PubMed] [Google Scholar]
- Xu M, Nelson GB, Moore JE, McCoy TP, Dai J, Manderville RA, Ross JA, Miller MS (2005) Induction of Cyp1a1 and Cyp1b1 and formation of DNA adducts in C57BL/6, Balb/c, and F1 mice following in utero exposure to 3-methylcholanthrene. Toxicol Appl Pharmacol 209:28–38. [DOI] [PubMed] [Google Scholar]
- Yang Q, Nagano T, Shah Y, Cheung C, Ito S, Gonzalez FJ (2008) The PPAR alpha-humanized mouse: a model to investigate species differences in liver toxicity mediated by PPAR alpha. Toxicol Sci 101:132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Zhu M, Sinz MW, Zhang H, Humphreys WG, Rodrigues AD, Dai R (2007) Development and full validation of six inhibition assays for five major cytochrome P450 enzymes in human liver microsomes using an automated 96-well microplate incubation format and LC-MS/MS analysis. J Pharm Biomed Anal 44:211–223. [DOI] [PubMed] [Google Scholar]
- Yumuk VD (2006) Targeting components of the stress system as potential therapies for the metabolic syndrome: the peroxisome-proliferator-activated receptors. Ann N Y Acad Sci 1083:306–318. [DOI] [PubMed] [Google Scholar]
- Zanger UM, Schwab M (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–141. [DOI] [PubMed] [Google Scholar]
- Zhang Q-Y, Dunbar D, Ostrowska A, Zeisloft S, Yang J, Kaminsky LS (1999) Characterization of human small intestinal cytochromes P-450. Drug Metab Dispos 27:804–809. [PubMed] [Google Scholar]
- Zhou S-F, Xue CC, Yu X-Q, Li C, Wang G (2007) Clinically important drug interactions potentially involving mechanism-based inhibition of cytochrome P450 3A4 and the role of therapeutic drug monitoring. Ther Drug Monit 29:687–710. [DOI] [PubMed] [Google Scholar]
- Zhou S, Chan E, Pan S-Q, Huang M, Lee EJD (2004) Pharmacokinetic interactions of drugs with St John’s wort. J Psychopharmacol 18:262–276. [DOI] [PubMed] [Google Scholar]