Abstract
The legalization of cannabis in many parts of the United States and other countries has led to a need for a more comprehensive understanding of cannabis constituents and their potential for drug-drug interactions. Although (−)-trans-Δ9-tetrahydrocannabinol (THC), cannabidiol (CBD), and cannabinol (CBN) are the most abundant cannabinoids present in cannabis, THC metabolites are found in plasma at higher concentrations and for a longer duration than that of the parent cannabinoids. To understand the potential for drug-drug interactions, the inhibition potential of major cannabinoids and their metabolites on major hepatic cytochrome P450 (P450) enzymes was examined. In vitro assays with P450-overexpressing cell microsomes demonstrated that the major THC metabolites 11-hydroxy-Δ9-tetra-hydrocannabinol and 11-nor-9-carboxy-Δ9-THC-glucuronide competitively inhibited several major P450 enzymes, including CYP2B6, CYP2C9, and CYP2D6 (apparent Ki,u values = 0.086 ± 0.066 µM and 0.90 ± 0.54 µM, 0.057 ± 0.044 µM and 2.1 ± 0.81 µM, 0.15 ± 0.067 µM and 2.3 ± 0.54 µM, respectively). 11-Nor-9-carboxy-Δ9- tetrahydrocannabinol exhibited no inhibitory activity against any CYP450 tested. THC competitively inhibited CYP1A2, CYP2B6, CYP2C9, and CYP2D6; CBD competitively inhibited CYP3A4, CYP2B6, CYP2C9, CYP2D6, and CYP2E1; and CBN competitively inhibited CYP2B6, CYP2C9, and CYP2E1. THC and CBD showed mixed-type inhibition for CYP2C19 and CYP1A2, respectively. These data suggest that cannabinoids and major THC metabolites are able to inhibit the activities of multiple P450 enzymes, and basic static modeling of these data suggest the possibility of pharmacokinetic interactions between these cannabinoids and xenobiotics extensively metabolized by CYP2B6, CYP2C9, and CYP2D6.
SIGNIFICANCE STATEMENT
Major cannabinoids and their metabolites found in the plasma of cannabis users inhibit several P450 enzymes, including CYP2B6, CYP2C9, and CYP2D6. This study is the first to show the inhibition potential of the most abundant plasma cannabinoid metabolite, THC-COO-Gluc, and suggests that circulating metabolites of cannabinoids play an essential role in CYP450 enzyme inhibition as well as drug-drug interactions.
Introduction
Cannabis and cannabis-derived substances are the most widely consumed psychopharmaceuticals in the world (Atakan, 2012). The legal and ethical implications of their use continue to be the subject of intense global debate because of potential medical applications of cannabis-derived products and the progressive legalization of its use as a recreational drug. Modern clinical applications include treatment of multiple sclerosis, epilepsy, neuropathic pain, arthritis, nausea and vomiting due to chemotherapy, appetite stimulation in HIV/AIDS, depression, anxiety disorders, sleep disorders, psychosis, glaucoma, and Tourette syndrome among others (Solimini et al., 2017). However, concurrent usage of cannabis products (both recreational and medicinal) with conventional medications, nonprescription remedies, and other psychoactive substances can often lead to substance-use disorders (National Institute on Drug Abuse report, July 2020), suggesting that more information is needed to fully understand the effects of cannabis as a psychopharmaceutical (Weinberger et al., 2016).
More than 500 phytochemicals have been detected in commercial strains of the Cannabis sativa plant, with (-)-trans-Δ9-tetrahydrocannabinol (THC), cannabidiol (CBD), and cannabinol (CBN) being the three main active constituents (Aizpurua-Olaizola et al., 2016). THC is the most well described psychoactive constituent and interacts with the cannabinoid receptor type 1 (CB1) receptor and the complex network of neurologic transmitters to induce psychopharmacological effects (Pertwee, 2008). Guidelines from the Food and Drug Administration (FDA) classify any cannabis-derived substance that contains more than 0.3% THC as a schedule I controlled substance, whereas products with less than 0.3% THC are classified as the agricultural product hemp. CBD does not bind to the CB1 receptor and does not produce the same psychoactive responses. In fact, when CBD and THC are administered simultaneously, CBD appears to block some of the effects of THC by acting as an antagonist at the cannabinoid receptors (Pertwee, 2008). CBD administered without THC has been shown to reduce subjective anxiety in both animals and humans and is thought to produce a positive effect on conditions such as inflammation, diabetes, cancer, and neurodegenerative diseases (Maroon and Bost, 2018). CBN is only weakly psychoactive and appears to be formed primarily from the degradation of THC within the Cannabis plant itself (Russo and Marcu, 2017).
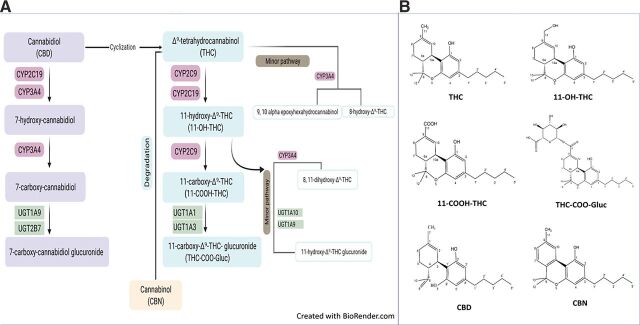
Upon consumption, the first step of metabolism of THC is hydroxylation mediated by CYP2C9, resulting in the active metabolite 11-hydroxy-Δ9-tetrahydrocannabinol (11-OH-THC; see Fig. 1). Although 11-OH-THC predominates as the first oxidation product, small amounts of 8β-hydroxy-THC (8β-OH-THC) and 8α-hydroxy-THC (8α-OH-THC) are also formed (Matsunaga et al., 1995). Much lower plasma 11-OH-THC concentrations (approximately 10% of THC concentrations) are found after cannabis smoking than after oral administration (Wall et al., 1983). Further hydroxylation by CYP2C9 leads to the inactive carboxylic acid derivative 11-nor-9-carboxy-Δ9- tetrahydrocannabinol (THC-COOH), which does not interact with the CB1 receptor. THC-COOH is a substrate for UDP-glucuronosyltransferase (UGT) enzymes that add a glucuronide moiety to form 11-nor-9-carboxy-Δ9-tetrahydrocannabinol-glucuronide (THC-COO-Gluc), the major end product of THC biotransformation in most species, including humans (Huestis, 2007). CBN is the nonenzymatic oxidation byproduct of THC and is most commonly found after prolonged storage, especially at higher temperatures.
Fig. 1.
Major metabolic pathways and metabolic structures of cannabinoids. (A) Metabolic pathways for THC and CBD. (B) Structure of cannabinoids and major THC metabolites.
CBD follows the same metabolic scheme as THC, with hydroxylation on the homologous 7 carbon via CYP2C19 and CYP3A4 to form 7-hydroxy-CBD (7-OH-CBD) and 7-carboxy-CBD (CBD-COOH), which is followed by glucuronidation to form 7-carboxy-CBD-glucuronide. However, 7-OH-CBD is only a minor biotransformation product in the plasma after cannabis inhalation, with unchanged CBD and, to a lesser extent, glucuronidated CBD as the main excretion products in urine (Harvey and Mechoulam, 1990; Huestis, 2007).
Cannabinoids are highly lipophilic, eventually concentrating in adipose tissue, liver, lung, and spleen, with slow release back into the bloodstream. This leads to varying plasma concentrations of active and inactive metabolites that persist in the bloodstream for a much wider timeframe than that of THC. Plasma concentrations of THC and its active metabolite 11-OH-THC change rapidly over time and vary widely depending on the mode of consumption and expertise of the user (occasional vs. frequent user, depth of inhalation, puff duration, and breath-hold) (Sharma et al., 2012). In contrast, the inactive metabolites THC-COOH and THC-COO-Gluc are present at relatively high levels regardless of the mode of consumption. Inhaled and ingested THC produces the same approximate level of THC-COOH. Once reaching maximal levels at approximately 80 minutes and 2 hours postinhalation, the level of THC-COOH and its glucuronide THC-COO-Gluc remain very stable in the bloodstream over many days, whereas THC is rapidly metabolized and cleared from the circulation (Huestis, 2007; Schwope et al., 2011; Sharma et al., 2012).
Although previous studies have shown that the cannabinoids THC, CBD, and CBN can inhibit several major hepatic P450s (Yamaori et al., 2010, 2011a,b,c; Jiang et al., 2013; Cox et al., 2019), no studies have focused on the inhibitory potential of the major abundant metabolites of THC. The present study is the first to perform a comprehensive analysis of the inhibition activities of major cannabinoids and THC metabolites against major human hepatic CYP450 enzymes.
Material and Methods
Chemicals and Reagents.
The purchase of THC, 11-OH-THC, THC-COOH, THC-COO-Gluc, CBD, and CBN was approved by the Drug Enforcement Agency, and they were obtained from Cayman Chemicals (Ann Arbor, MI) or Sigma-Aldrich (St. Louis, MO). The internal standards (ISs) D3-THC and D3-THC-COO-Gluc were purchased from Sigma-Aldrich. Pooled human liver microsomes (HLMs; n = 50 subjects, mixed sex) were obtained from Sekisui Xenotech, LLC (Lenexa, Kansas). NADPH-regenerating system (1.3 mM NADP, 3.3 mM glucose 6-phosphate, and 0.4 U/ml glucose 6-phosphate dehydrogenase) was obtained from Corning (Bedford, MA). CYP450 probe substrates (phenacetin, bupropion, (S)-bufuralol hydrochloride, amodiaquine, diclofenac, omeprazole, dextromethorphan, chlorzoxazone, and midazolam) and their corresponding metabolite standards (acetaminophen, hydroxybupropion, hydroxybupropion-D6, 4-hydroxydiclofenac, 4-hydroxydiclofenac-13C6, 5-hydroxyomeprazole, dextrorphan, dextrorphan-D3, 6-hydroxychlorzoxazone, and 1-hydroxymidazolam) along with probe inhibitors (furafylline, clopidogrel, montelukast, sulfaphenazole, tranylcypromine, quinidine, chlomethiazole, and ketoconazole) were all purchased from Sigma-Aldrich. (−)Warfarin and efavirenz were purchased from Cayman Chemicals (Ann Arbor, MI). LC-MS grade methanol, acetonitrile, and formic acid as well as ultra–low-binding microcentrifuge tubes were obtained from Fisher Scientific (Waltham, MA). All other chemicals and reagents were of analytical grade or the highest grade that was commercially available. Dulbecco’s modified Eagle’s medium, Dulbecco’s phosphate-buffered saline, fetal bovine serum, and geneticin (G418) were purchased from Gibco (Grand Island, NY). Bicinchoninic acid protein assays were purchased from Pierce (Rockford, Illinois).
In Vitro CYP450 Activity Assays and Metabolite Detection.
Human embryonic kidney (HEK) 293 cells individually overexpressing V5-tagged P450 isoforms 1A2, 3A4, 2B6, 2C8, 2C9, 2C19, 2D6, or 2E1 were developed and described previously (Peterson et al., 2017b). Microsomal membrane fractions of P450-overexpressing cell lines were prepared by differential centrifugation as previously described, with protein concentrations estimated using the bicinchoninic acid assay as per the manufacturer’s recommendations (Dellinger et al., 2006; Peterson et al., 2017a,b). An initial screen of the inhibition potential of individual cannabinoids and their metabolites against major P450s (1A2, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, and 3A4) were determined using microsomes (20–50 µg) from CYP450-overexpressing HEK293 cell lines in reactions containing either 1 µM or 10 µM of cannabinoid/THC metabolite, specific probe substrate (see Supplemental Table 1), 100 mM potassium phosphate buffer (pH 7.4), and magnesium chloride (3 mM) in a final reaction volume of 30 µl. The probe substrates were used at concentrations near or just below their respective Michaelis-Menten constant (Km) values to minimize unwanted interactions (Supplemental Table 1). As THC exhibits extensive nonspecific binding (70%–90%) to protein and labware (Garrett and Hunt, 1974), microsomal incubation conditions were optimized to prevent underestimation of inhibitory potency [IC50 or inhibitory constant (Ki)]. To reduce nonspecific binding and adsorption to labware, low-binding 0.6-ml microcentrifuge tubes were used for all reactions. Assays were preincubated for 5 minutes at 37°C, initiated by the addition of NADPH-regenerating system, and incubated for 5–30 minutes (Supplemental Table 1) at 37°C. Reactions were terminated by the addition of 30 µl of ice-cold stop solution (acetonitrile/methanol; 1:1). Samples were mixed on a vortex mixer and centrifuged at 17,000 × g for 15 minutes. The supernatant (∼50 µl) was transferred to an ultra–high-performance liquid chromatograph (UPLC) sample vial, and the specific probe substrate metabolite was detected using a UPLC (Waters Acquity; Waters Corp, Milford, MA) coupled to a triple-quadrupole mass spectrometer (Waters Xevo TQD; Waters Corp) by multiple reaction monitoring analysis (Supplemental Table 1). As a positive control for every inhibition experiment, 1 µM and 10 µM probe inhibitors (see Supplemental Table 1) were added instead of the cannabinoid compounds. As a negative control, only vehicle (3% methanol) was added (without inhibitor) to the reaction. Probe substrate without any inhibitor was used as a positive control for 100% activity. All analyses were performed in three independent experiments. Incubation conditions were optimized by incubations of the individual probe substrates with microsomes from overexpressing cell lines at 10, 20, 30, and 50 µg microsomal protein for different times (5, 15, 25, and 30 minutes). Remaining substrate and formed metabolite peaks for each reaction were analyzed, and conditions were chosen according to the following criteria: 1) metabolite formation was linear with time and enzyme concentration, 2) substrate consumption was no more than 20% of the initial amount, and 3) all metabolites could be easily detected by LC-MS/MS methods.
For UPLC-MS/MS, the mobile phase consisted of solvent A (0.1% formic acid in water) and solvent B (100% methanol). Samples (2–5 µl) were injected onto an Acquity UPLC column (BEH C18, 1.7 µM, 2.1 × 100 mm: Waters Corp). A 6-minute gradient elution was used as follows: 2 minutes at 95% A:5% B followed by a linear gradient for 2 minutes to 5% A:95% B, 1 minute at 5% A:95% B, and re-equilibration for 1 minute at 95% A:5% B. The flow rate was 0.4 ml/min, and the column temperature was 40°C.
Determination of IC50 and Ki Values.
For those cannabinoids/THC metabolites that exhibited a >50% decrease in relative P450 activity for any given probe substrate at ≤10 µM, IC50 determinations were performed in both microsomes from HEK293 CYP450-overexpressing cell lines (described above) and in HLMs using multiple concentrations (ranging between 0.5 and 100 µM) of cannabinoid/metabolite in incubations as described above, with all determinations performed for three independent experiments with three replicates per experiment.
To identify possible metabolism-dependent inhibition of the potential enzymes, IC50 shift studies (Parkinson et al., 2011) were performed using HEK-overexpressing cell lines. Reactions containing the test cannabinoid or THC metabolite (inhibitor) but no probe substrate were incubated with or without NADPH at 37°C for 30 minutes. After this 30-minute preincubation, probe substrate was added to the incubation mixture and incubated at 37°C for up to 30 minutes (Supplemental Table 1). Peak areas corresponding to the probe metabolite were determined, and the percentage of relative activity was calculated by comparing the peak area in incubations containing the inhibitors to incubations containing only the vehicle control, as described below. The differences between the IC50 values obtained with or without the NADPH preincubation period were compared, and the fold-IC50 shift was determined. The cannabinoid/THC metabolite with a fold-IC50 shift of ≥1.5 was considered a time-dependent inhibitor as recommended by the FDA guidelines.
IC50 data were used as a guide to generate appropriate probe substrate and test inhibitor concentrations for the determination of the Ki values for each isoform. The P450 enzyme-specific probe substrate concentrations used were 1–25 μM phenacetin for CYP1A2, 12.5–100 μM bupropion for CYP2B6, 2.5–30 μM diclofenac for CYP2C9, 0.5–5 μM omeprazole for CYP2C19, 1–10 μM dextromethorphan for CYP2D6, 12.5–100 μM chlorzoxazone for CYP2E1, and 1–25 μM midazolam for CYP3A4.
Data were exported and analyzed using an Excel spreadsheet (Microsoft). The amount of metabolite formed at each concentration relative to the control (percent relative activity) of specific enzyme in the presence and absence of probe inhibitor or test compound was calculated as:
% Relative activity = (Peak area with inhibitor/Peak area without inhibitor) × 100%
Percent Inhibition = [(Peak area without inhibitor − peak area with inhibitor)/peak area without inhibitor] × 100
The IC50 values were calculated by plotting the percent inhibition of P450 enzyme activities versus the log concentration of the test inhibitors using GraphPad Prism 7.04 software (GraphPad Software Inc., San Diego, CA).
To calculate Ki values, inhibition data were fit to different models of enzyme inhibition (competitive, uncompetitive, or noncompetitive) by nonlinear least-squares regression analysis using the GraphPad Prism 7.04 software. Ki values were calculated with the use of nonlinear regression according to the equations:
where I is compound concentration, Ki is the inhibition constant, S is the substrate concentration, and Km is the substrate concentration at half of the Vmax of the reaction (Cornish-Bowden, 1974).
Bound versus Unbound Cannabinoid Determinations.
Cannabinoid binding to proteins and tube walls in the incubation mixture (fu,inc) was measured using the tube adsorption method as described previously (Isoherranen et al., 2004; Patilea-Vrana et al., 2019). Briefly, in separate incubations, 50 µl of 50 µM THC, 11-OH-THC, THC-COO-Gluc, CBD, or CBN was added to each of two 0.6-ml low-binding centrifuge tubes containing buffer without protein, two 0.6-ml low-binding centrifuge tubes containing buffer with HLMs (25 µg), or two 0.6-ml low-binding centrifuge tubes containing buffer with HEK293 microsomal protein (30 µg). This was performed for each cannabinoid separately. Mixtures were incubated for 30 minutes at 37°C with occasional shaking. For one tube of the three incubation conditions (buffer alone, buffer + HLMs, and buffer + HEK293 microsomes), ice-cold acetonitrile (100 μl) containing D3-labeled cannabinoid IS was added (subgroup A). For the other tube of each condition, the entire 50-μl mixture was removed and added to a fresh tube that contained 100 μl ice-cold acetonitrile with D3-labeled cannabinoid IS (subgroup B). Samples were processed for UPLC-MS/MS as described above for the in vitro assay experiments. All analyses were performed in three independent experiments with four replicates per experiment.
The separation and detection of individual cannabinoids was achieved using the same ACQUITY XEVO TQD (Waters Corporation, Millford, MA) chromatographic system described above using a Waters BEH C18 column 1.7 mm, 2.1 × 50 mm, at 40°C. UPLC-MS/MS was performed with a gradient elution using mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile) at a flow rate of 0.4 ml/min under the following conditions: 5% B for 1.0 minute, increasing to 95% B from 1.0 to 4.0 minutes, 95% B held for 2 minutes, which was was followed by a return to initial conditions for 1.5 minutes for a total time of 7.5 minutes. Detection of cannabinoids was performed in multiple reaction monitoring ES+ mode using the following mass transitions: m/z 315.1 → 193.2 for THC, m/z 331.3 → 313.1 for 11-OH-THC, m/z 315.2 → 181.1 for CBD, m/z 311.2 → 223.0 for CBN, and m/z 521.2 → 345.0 for THC-COO-Gluc. Cone voltages (V) of 40, 40, 50, and 30, respectively, and collision energies of 25, 18, 20, and 27, respectively, were used for THC, 11-OH-THC, CBD, CBN, and THC-COO-Gluc. Because of the nonavailability of deuterated CBD or CBN at the time of this study and because of the similar molecular weights and UPLC retention times between these cannabinoids, THC-D3 was used as the IS for the quantification of THC, CBD, and CBN; 11-OH-THC-D3 was used to quantify 11-OH-THC; and THC-COO-Gluc-D3 was used to quantify THC-COO-Gluc. The internal standards were detected using the same cone voltages and collision energies as their respective standards, with the following mass transitions: m/z 318.1 → 196.2 for THC-D3, m/z 334.3 → 316.1 for 11-OH-THC-D3, and m/z 523.2 → 348.0 for THC-COO-Gluc-D3.
The unbound fraction of cannabinoid in incubations with either HLMs or HEK293 microsomes (fu, inc) was calculated using the following equation:
where CBA is the total (adsorbed and unadsorbed) test compound in buffer only, and CBB is unadsorbed test compound in buffer. So, CBA − CBB is the test compound adsorbed to the tube wall, and CBB/CBA − CBB is the ratio of unbound test compound relative to the test compound adsorbed to the wall. CPA is the total (adsorbed and unadsorbed) test compound in the incubation mixture, and CPB is unbound test compound in presence of protein source (incubation mixture). CPA − CPB is the test compound adsorbed to the tube wall when mixed with microsomes. Calculations of were performed in three independent experiments with four replicates per group. IC50,u and Ki, u values were calculated using the following equations:
Prediction of Potential In Vivo Drug-Drug Interactions by Static Modeling.
To predict the risk of a clinical drug-drug interaction, basic static models of reversible inhibition were used following FDA recommended guidelines. The predicted hepatic and systemic area under the curve ratios (AUCRhep and AUCRsys, respectively) were calculated to determine the overall effect of cannabinoids and their metabolites on a given probe substrate. The basic static model is described by the following equations:
AUCRhep is calculated using eq. 1, in which is the unbound maximum hepatic inlet concentration, and the Ki,u is calculated for each inhibitor as described above.
The is calculated using eq. 2, in which is the unbound concentration of drug in plasma and was set at 0.03 based on the plasma protein binding of THC (Garrett and Hunt, 1974), which was suggested to be accurate for 11-OH-THC, CBD, and CBN as well as THC-COO-Gluc in previous studies (Skopp et al., 2002). is the maximum concentration of total drug in plasma (parameters described below), is the fraction of dose absorbed from the gut lumen [set to 1 as recommended by the FDA (Food and Drug Administration, 2020)], is the rate of intestinal absorption time [set to 0.02; (Cox et al., 2019)], is the ratio of drug concentration in blood (CB) to drug concentration in plasma [set to 0.4 as recommended by Schwilke et al. (2009)], and is the hepatic blood flow = 1500 ml/min (Food and Drug Administration, 2020). For THC, the was set to 65 nM for a 40-mg oral dose of THC and 254 nM for a 54-mg inhaled dose of THC (Schwope et al., 2011; Cox et al., 2019). For 11-OH-THC, the was set to 69.7 nM for a 20-mg oral dose of THC and 50 nM for a 46-mg inhaled dose of THC (Ménétrey et al., 2005; Hunault et al., 2008). For THC-COO-Gluc, the was set to 405 nM for an oral dose of 50 mg of THC and to 385 nM for a 54-mg inhaled dose of THC (Spindle et al., 2020). For CBD, the was set to 703 nM for an oral dose of 800 mg of CBD and to 10.3 nM for a 20-mg inhaled dose of CBD (Manini et al., 2015; Cox et al., 2019).
The is calculated using eq. 3, in which is equal to , which is the maximum concentration of total unbound drug in plasma (/).
An ≥ 1.25 or an ≥ 1.02 indicates a strong presystemic hepatic or systemic drug interaction potential.
Results
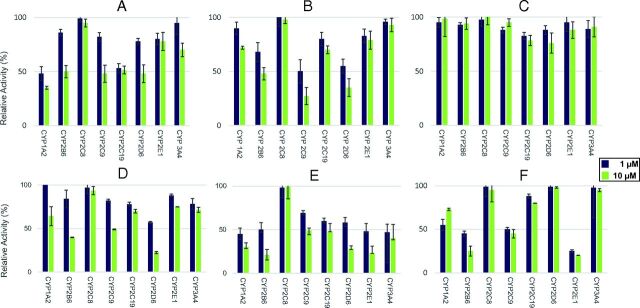
Prescreening of THC, 11-OH-THC, THC-COOH, THC-COO-Gluc, CBD, and CBN as potential CYP450 inhibitors showed that 10 µM THC decreased the relative activity of several P450 isoforms to approximately 50%, including CYP1A2 (35%), CYP2B6 (50%), CYP2C9 (48%), CYP2D6 (48%), and CYP2C19 (51%), as compared with reactions without added cannabinoid (Fig. 2A). Similar levels of inhibition were observed for 11-OH-THC against CYP2B6, CYP2C9, and CYP2D6, with 10 µM 11-OH-THC resulting in relative activity levels between 27% and 48% for the three enzymes (Fig. 2B). Although no significant inhibition was observed using up to 10 µM THC-COOH against any of the P450 enzymes tested (Fig. 2C), 10 µM THC-COO-Gluc resulted in 25%–49% relative activity for CYP2B6, CYP2C9, and CYP2D6 (Fig. 2D). In addition, decreases in the relative activity to less than 50% was observed for 10 µM CBD against several P450 enzymes (CYP1A2, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP2E1, and CYP3A4) and for 1 µM CBD against CYP1A2 (45%), CYP2B6 (50%), CYP2E1 (49%), and CYP3A4 (47%; Fig. 2E). Similar decreases in relative activity were observed for CBN against CYP2B6, CYP2C9, and CYP2E1, with 75%, 55%, and 80% decreases in activity observed for 10 µM CBN against all three enzymes and a 77% decrease observed for 1 µM CBN against CYP2E1 (Fig. 2F). Initial screening results were validated in assays using commercially prepared pooled HLMs (unpublished data).
Fig. 2.
Screening of cannabinoid inhibition of major hepatic P450s in microsomes from CYP450-overexpressing HEK293 cell lines. Probe substrates were phenacetin for CYP1A2, bupropion for CYP2B6, amodiaquine for CYP2C8, diclofenac for CYP2C9, omeprazole for CYP2C19, dextromethorphan for CYP2D6, chlorzoxazone for CYP2E1, and midazolam for CYP3A4. Incubations were performed using 1 or 10 µM of cannabinoid, with probe concentrations at or close to their known Km for their corresponding enzyme (see Supplemental Table 1). (A) THC; (B) 11-OH-THC; (C) THC-COOH; (D) THC-COO-Gluc; (E) CBD; (F) CBN. Three individual experiments were performed for each probe substrate. Data are expressed as a percentage of metabolite formation formed in assays with cannabinoid compared with assays without cannabinoids.
Using microsomes from both specific recombinant CYP450 (rCYP)-overexpressing HEK293 cell lines and HLMs, the inhibitory effects of THC, 11-OH-THC, THC-COOH, THC-COO-Gluc, CBD, and CBN were extended to establish IC50 values for each cannabinoid against the P450 enzymes shown to be inhibited in the rCYP screening assays (described above). The unbound fraction (fu,inc) of THC, 11-OH-THC, THC-COO-Gluc, CBD, and CBN in incubation mixtures (fu,inc) was calculated as 0.051 ± 0.033 and 0.043 ± 0.012, 0.094 ± 0.014 and 0.078 ± 0.042, 0.43 ± 0.071 and 0.45 ± 0.061, 0.098 ± 0.023 and 0.062 ± 0.011, and 0.052 ± 0.081 and 0.030 ± 0.052 in mixtures containing HLMs or microsomes from HEK293 cells, respectively. The range in fu,inc was from 0.030 (CBN) to 0.45 (THC-COO-Gluc) for HEK293 microsomes and 0.051 (THC) to 0.43 (THC-COO-Gluc) for HLMs. The values for HLMs were similar to those obtained for HEK293 microsomes for all cannabinoids tested. Interestingly, THC-COO-Gluc exhibited a fu,inc that was approximately 10-fold higher than the other cannabinoids, indicating less adsorption to the components of the incubation mixture.
CBD exhibited strong inhibition against every CYP450 tested, with a range in IC50,u values of 0.037–0.22 µM, with the strongest inhibition observed against rCYP2E1 microsomes (IC50,u = 0.037 ± 0.0081 µM; Table 1). Additionally, THC exhibited strong inhibition against rCYP1A2, rCYP2B6, rCYP2C9, rCYP2C19, and rCYP2D6 microsomes (IC50,u values of 0.12 µM ± 0.052 µM, 0.43 ± 0.28 µM, 0.19 µM ± 0.13 µM, 0.15 ± 0.030 µM, and 0.47 µM ± 0.077 µM, respectively). The THC metabolites 11-OH-THC and THC-COO-Gluc both exhibited slightly stronger inhibition against rCYP2D6 (IC50,u values of 0.34 ± 0.12 µM and 3.5 ± 0.72 µM, respectively) and rCYP2B6 (IC50,u values of 0.55 ± 0.26 µM and 2.5 ± 0.72 µM, respectively) than THC, while exhibiting similar or less effective inhibition against rCYP2C9 microsomes (IC50,u values of 0.30 ± 0.086 µM and 6.8 ± 2.0 µM, respectively). CBN exhibited strong inhibition against rCYP2B6 (IC50,u = 0.63 ± 0.26 µM), rCYP2C9 (IC50,u= 0.42 ± 0.13 µM), and rCYP2E1 (IC50,u= 0.28 ± 0.030 µM) microsomes. Similar IC50 values were obtained for HLMs using specific probe substrates for all CYP450s examined (Table 1).
TABLE 1.
IC50 values of cannabinoids and THC metabolites against major hepatic CYP450 enzymes in microsomes from recombinant CYP450-overexpressing cells or HLMs
| Probe Substrate | Microsomal Protein | THC | 11-OH-THC | THC-COO-Gluc | CBD | CBN | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IC 50 a | IC 50,u a , b | IC 50 | IC 50,u | IC 50 | IC 50,u | IC 50 | IC 50,u | IC 50 | IC 50,u | ||
| Phenacetin | rCYP1A2 | 2.7 ± 1.2 | 0.12 ± 0.052 | NAc | NA | 2.1 ± 0.93 | 0.13 ± 0.058 | NA | |||
| HLM | 5.2 ± 2.1 | 0.27 ± 0.11 | NA | NA | 6.8 ± 1.5 | 0.67 ± 0.15 | NA | ||||
| Midazolam | rCYP3A4 | NA | NA | NA | 3.0 ± 0.52 | 0.19 ± 0.032 | NA | ||||
| HLM | NA | NA | NA | 4.6 ± 1.3 | 0.45 ± 0.13 | NA | |||||
| Bupropion | rCYP2B6 | 10 ± 6.4 | 0.43 ± 0.28 | 7.1 ± 3.3 | 0.55 ± 0.26 | 5.5 ± 1.6 | 2.5 ± 0.72 | 2.1 ± 0.61 | 0.13 ± 0.038 | 2.1 ± 0.87 | 0.63 ± 0.26 |
| HLM | 19 ± 2.3 | 0.97 ± 0.12 | 8.2 ± 3.7 | 0.77 ± 0.35 | 5.3 ± 2.4 | 2.3 ± 1.0 | 2.7 ± 2.4 | 0.26 ± 0.24 | 1.7 ± 0.59 | 0.88 ± 0.31 | |
| Diclofenac | rCYP2C9 | 4.5 ± 3.0 | 0.19 ± 0.13 | 3.8 ± 1.1 | 0.30 ± 0.086 | 15 ± 4.5 | 6.8 ± 2.0 | 3.6 ± 1.6 | 0.22 ± 0.099 | 1.4 ± 0.44 | 0.42 ± 0.13 |
| HLM | 7.2 ± 2.6 | 0.37 ± 0.13 | 4.8 ± 2.6 | 0.45 ± 0.24 | 18 ± 2.5 | 7.7 ± 1.1 | 4.9 ± 1.6 | 0.48 ± 0.16 | 2.9 ± 0.51 | 1.51 ± 0.27 | |
| Omeprazole | rCYP2C19 | 3.6 ± 0.70 | 0.15 ± 0.030 | NA | NA | 2.5 ± .50 | 0.16 ± 0.031 | NA | |||
| HLM | 4.2 ± 1.1 | 0.21 ± 0.056 | NA | NA | 3.7 ± 1.5 | 0.36 ± 0.15 | NA | ||||
| Dextromethorphan | rCYP2D6 | 11 ± 1.8 | 0.47 ± 0.077 | 4.3 ± 1.5 | 0.34 ± 0.12 | 7.8 ± 1.6 | 3.5 ± 0.72 | 3.1 ± 0.64 | 0.19 ± 0.040 | NA | |
| HLM | 15 ± 5.3 | 0.77 ± 0.27 | 6.5 ± 3.4 | 0.61 ± 0.32 | 13 ± 3.6 | 5.6 ± 1.5 | 5.3 ± 1.0 | 0.52 ± 0.10 | NA | ||
| Chlorzoxazone | rCYP2E1 | NA | NA | NA | 0.59 ± 0.13 | 0.037 ± 0.0081 | 0.92 ± 0.10 | 0.28 ± 0.030 | |||
| HLM | NA | NA | NA | 1.4 ± 0.25 | 0.14 ± 0.025 | 1.1 ± 0.72 | 0.57 ± 0.37 | ||||
aValues are the mean ± S.D. in µM for four independent experiments with three replicates per group.
bIC50,u values represent IC50 values after correction for unbound fraction (fu,inc) of individual cannabinoids as described in Materials and Methods.
cNA, not analyzed.
To confirm the high level of inhibitory activity observed with cannabinoids against CYP2B6, CYP2C9, and CYP2D6, IC50 determinations were also performed for three additional probe substrates. Strong inhibition of activity against efavirenz, bufuralol, and S-warfarin in microsomes from rCYP2B6, rCYP2D6, and rCYP2C9 cell lines, respectively, was observed with THC, 11-OH-THC, THC-COO-Gluc, CBD, and CBN (Supplemental Table 2). Consistent with that observed for CBN with dextromethorphan as a probe substrate for CYP2D6, no inhibition was observed with this cannabinoid for bufuralol. These data suggest that the inhibition observed with these cannabinoids against these three enzymes is seen across multiple substrates.
IC50-shift assays were performed for microsomes from each CYP450-overexpressing cell line with the goal of evaluating the time-dependent inhibition (TDI) potencies of cannabinoid metabolites (Table 2). Using a value of 1.5 as the cutoff, none of the cannabinoids tested demonstrated TDI, with the possible exception of CBD for CYP2D6 with an exhibited shift of 1.5.
TABLE 2.
Effect of preincubation on inhibition of P450 activity by cannabinoids and THC metabolites in microsomes from CYP450-overexpressing cell lines
| THC | 11-OH-THC | THC-COO-Gluc | CBD | CBN | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme | (−)a | (+)b | Shiftc | (−) | (+) | Shift | (−) | (+) | Shift | (−) | (+) | Shift | (−) | (+) | Shift |
| CYP1A2 | 2.7 ± 1.0 | 1.8 ± 0.55 | 1.4 ± 0.12 | NAd | NA | 2.1 ± 0.25 | 1.8 ± 0.31 | 1.2 ± 0.20 | NA | ||||||
| CYP3A4 | NA | NA | NA | 3.1 ± 1.2 | 2.0 ± 1.1 | 1.1 ± 0.077 | NA | ||||||||
| CYP2B6 | 19 ± 5.1 | 18 ± 5.0 | 1.1 ± 0.049 | 7.3 ± 1.0 | 6.1 ± 1.0 | 1.2 ± 0.094 | 5.1 ± 1.6 | 4.1 ± 1.0 | 1.4 ± 0.16 | 1.9 ± 0.66 | 1.3 ± 0.45 | 1.3 ± 0.048 | 2.1 ± 0.83 | 2.0 ± 0.70 | 1.1 ± 0.061 |
| CYP2C9 | 3.4 ± 2.3 | 2.3 ± 1.7 | 1.4 ± 0.17 | 3.2 ± 0.61 | 2.2 ± 0.59 | 1.4 ± 0.18 | 15 ± 2.4 | 15 ± 2.7 | 1.0 ± 0.044 | 2.2 ± 1.2 | 1.8 ± 0.85 | 1.2 ± 0.12 | 3.0 ± 2.2 | 2.7 ± 2.1 | 1.1 ± 0.11 |
| CYP2C19 | 3.2 ± 2.5 | 2.5 ± 1.8 | 1.3 ± 0.13 | NA | NA | 2.5 ± 1.5 | 2.3 ± 1.3 | 1.1 ± 0.074 | NA | ||||||
| CYP2D6 | 13 ± 0.67 | 10 ± 0.92 | 1.3 ± 0.068 | 2.9 ± 0.86 | 2.1 ± 0.49 | 1.3 ± 0.14 | 8.1 ± 1.1 | 6.4 ± 2.5 | 1.4 ± 0.51 | 2.9 ± 1.1 | 2.0 ± 0.76 | 1.5 ± 0.33 | NA | ||
| CYP2E1 | NA | NA | NA | 1.1 ± 0.66 | 0.9 ± 0.40 | 1.2 ± 0.20 | 0.87 ± 0.15 | 0.73 ± 0.21 | 1.2 ± 0.16 | ||||||
a(−), IC50 ± S.D. (µM) value of three experiments with 30-min preincubation of test cannabinoid with enzyme without NADPH.
b(+), IC50 ± S.D. (µM) value of three experiments with 30-min preincubation of test cannabinoid with enzyme and NADPH.
cShift = (IC50 “minus NADPH”/IC50 “plus NADPH”) ± S.D. of three experiments.
dNA, not analyzed.
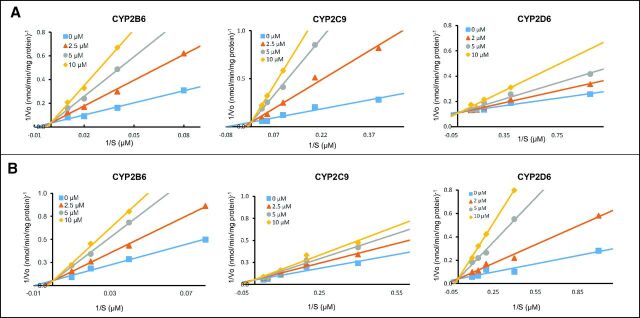
Additional experiments to determine Ki,u values were performed to further understand the strength and type of inhibition interactions occurring between the cannabinoids and the P450 enzymes. As shown in Lineweaver-Burk plots, the CYP2B6-catalyzed 4-hydroxylation of bupropion in microsomes from the rCYP2B6-overexpressing cell line was shown to be inhibited in a competitive manner by the THC metabolites 11-OH-THC and THC-COO-Gluc (Fig. 3) and the cannabinoids THC, CBD, and CBN (Supplemental Fig. 2), with Ki,u values similar for all inhibitors ranging from 0.068 ± 0.015 μM for CBD to 0.90 ± 0.054 μM for THC-COO-Gluc (Table 3). Slightly higher Ki,u values were observed for inhibition in HLMs, ranging from 0.22 ± 0.052 μM for CBD to 1.1 ± 0.47 μM for THC-COO-Gluc (Table 3).
TABLE 3.
Summary of Ki values and inhibition type for major cannabinoids and THC metabolites against hepatic CYP450 enzymes
| Microsomal Protein | THC | 11-OH-THC | THC-COO-Gluc | CBD | CBN | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kia | Ki,ua | Tb | K i | K i,u | T | K i | K i,u | T | K i | K i,u | T | K i | K i,u | T | |
| rCYP1A2c | 2.1 ± 0.62 | 0.090 ± 0.027 | C | NAd | NA | 1.9 ±1.2 | 0.12 ± 0.074 | M | NA | ||||||
| HLM | 2.0 ± 1.1 | 0.10 ± 0.056 | C | 2.1 ± 1.1 | 0.21 ± 0.11 | M | |||||||||
| rCYP3A4 | NA | NA | NA | 1.5 ± 0.59 | 0.093 ± 0.037 | C | NA | ||||||||
| HLM | 2.2 ± 0.45 | 0.22 ± 0.044 | C | ||||||||||||
| rCYP2B6 | 5.7 ± 1.0 | 0.25 ± 0.043 | C | 1.1 ± 0.85 | 0.086 ± 0.066 | C | 2.0 ± 1.2 | 0.90 ± 0.54 | C | 1.1 ± 0.24 | 0.068 ± 0.015 | C | 0.62 ± 0.19 | 0.19 ± 0.057 | C |
| HLM | 7.5 ± 0.56 | 0.38 ± 0.029 | C | 2.8 ± 0.44 | 0.26 ± 0.041 | C | 2.6 ± 1.1 | 1.1 ± 0.47 | C | 2.2 ± 0.53 | 0.22 ± 0.052 | C | 1.7 ± 0.42 | 0.88 ± 0.22 | C |
| rCYP2C9 | 1.7 ± 0.53 | 0.073 ± 0.023 | C | 0.73 ± 0.56 | 0.057 ± 0.044 | C | 4.7 ± 1.8 | 2.1 ± 0.81 | C | 1.5 ± 0.57 | 0.093 ± 0.035 | C | 1.0 ± 0.19 | 0.30 ± 0.057 | C |
| HLM | 3.4 ± 0.91 | 0.17 ± 0.046 | M | 2.2 ± 0.34 | 0.21 ± 0.032 | C | 7.8 ± 0.65 | 3.4 ± 0.28 | M | 1.9 ± 0.15 | 0.19 ± 0.015 | C | 1.9 ± 1.3 | 0.99 ± 0.68 | C |
| rCYP2C19 | 1.3 ± 0.42 | 0.056 ± 0.018 | M | NA | NA | 0.81 ± 0.092 | 0.050 ± 0.0057 | C | NA | ||||||
| HLM | 4.1 ± 1.6 | 0.21 ± 0.082 | M | 0.94 ± 0.063 | 0.092 ± 0.0062 | M | |||||||||
| rCYP2D6 | 2.5 ± 0.35 | 0.11 ± 0.015 | C | 1.9 ± 0.86 | 0.15 ± 0.067 | C | 5.0 ± 1.2 | 2.3 ± 0.54 | C | 1.2 ± 0.77 | 0.074 ± 0.048 | C | NA | ||
| HLM | 5.4 ± 0.58 | 0.28 ± 0.030 | C | 3.4 ± 2.5 | 0.32 ± 0.24 | C | 6.2 ± 1.7 | 2.7 ± 0.73 | C | 3.2 ± 1.4 | 0.31 ± 0.14 | C | |||
| rCYP2E1 | NA | NA | NA | 0.34 ± 0.098 | 0.021 ± 0.0061 | C | 0.21 ± 0.097 | 0.063 ± 0.029 | C | ||||||
| HLM | 0.59 ± 0.23 | 0.058 ± 0.023 | C | 0.33 ± 0.091 | 0.17 ± 0.047 | C | |||||||||
a,bValues are expressed as µM; shown are the mean ± S.D. for three independent experiments performed for the inhibition of each CYP450 enzyme by each cannabinoid.
bT, type of inhibition. C, competitive inhibition; M, mixed inhibition.
cThe probe substrate used were phenacetin, midazolam, bupropion, diclofenac, omeprazole, dextromethorphan, and chlorzoxazone for CYP1A2, CYP3A4, CYP2B6, CYP2C9, CYP2C19, CYP2D6, and CYP2E1, respectively.
dNA, not analyzed.
Fig. 3.
Lineweaver-Burk plots for the inhibition of CYP2B6, CYP2C9, and CYP2D6 in microsomes from recombinant CYP450-overexpressing cells by THC metabolites. (A) Inhibition by 11-OH-THC; (B) inhibition by THC-COO-Gluc.
Ki,u values for the inhibition of rCYP2C9 were determined by monitoring the hydroxylation of the CYP2C9 probe substrate diclofenac. Lineweaver-Burk plots for the inhibition of rCYP2C9 (Fig. 3; Supplemental Fig. 2) indicated a competitive type of inhibition for all cannabinoids tested in rCYP2C9-overexpressing cell microsomes, with Ki,u values ranging from 0.057 ± 0.044 μM for 11-OH-THC to 2.1 ± 0.081 μM for THC-COO-Gluc (Table 3). The inhibition of diclofenac hydroxylation in HLMs was similar, ranging from 0.17 ± 0.046 μM for THC to 3.4 ± 0.28 μM for THC-COO-Gluc. Slightly higher Ki,u values were observed for inhibition in HLMs, with THC and THC-COO-Gluc exhibiting mixed-type inhibition, whereas 11-OH-THC, CBD, and CBN exhibited competitive inhibition (Table 3).
For CYP2D6, Ki,u values were determined by examining the O-demethylation of dextromethorphan to dextrorphan. Lineweaver-Burk plots for the inhibition of CYP2D6 (Fig. 3; Supplemental Fig. 2) suggested a competitive type of inhibition for all inhibitors for both rCYP2D6 microsomes and HLMs. The Ki,u values for THC, THC-OH, THC-COO-Gluc, and CBD against rCYP2D6 were 0.11 ± 0.015 μM, 0.15 ± 0.067 μM, 2.3 ± 0.54 μM, and 0.074 ± 0.048 μM, respectively; similar Ki,u values were observed for dextromethorphan-O-demethylation activity for the four cannabinoids in HLMs (Table 3).
In addition to CYP2B6, CYP2C9, and CYP2D6, THC also inhibited CYP1A2-catalyzed phenacetin O-deethylation in rCYP1A2 microsomes, with a Ki,u value of 0.090 ± 0.027 μM; a similar level of inhibition (Ki,u = 0.12 ± 0.074 μM) was also observed for CBD. Inhibition by THC was determined to be competitive, whereas inhibition by CBD was mixed (Table 3). Highly similar Ki,u values were observed for the two cannabinoids in HLMs (Table 3).
Among the cannabinoid compounds tested, only CBD inhibited CYP3A4-catalyzed midazolam 1′-hydroxylation. This inhibition was competitive, with Ki,u values of 0.093 ± 0.037 μM and 0.22 ± 0.044 μM in rCYP3A4 microsomes and HLMs, respectively. CYP2C19-catalyzed omeprazole 5′-hydroxylation activity was inhibited by THC with a mixed-type inhibition mechanism for rCYP2C19 microsomes (Ki,u = 0.056 ± 0.018 μM) and HLMs (Ki,u = 0.21 ± 0.082 μM) and by CBD with a competitive mechanism in rCYP2C19 microsomes and a mixed-type in HLMs (Ki,u= 0.050 ± 0.0057 μM and 0.092 ± 0.0062 μM, respectively). CYP2E1-catalyzed chlorzoxazone 6-hydroxylation activity was competitively inhibited by CBD and CBN, with Ki,u values for rCYP2E1 of 0.021 ± 0.0061 μM and 0.063 ± 0.029 μM, respectively; the Ki,u values for the inhibition of chlorzoxazone 6-hydroxylation activity in HLMs were again similar to those observed for rCYP microsomes, with Ki,u values of 0.058 ± 0.023 μM and 0.17 ± 0.047 μM for CBD and CBN, respectively (Table 3).
A basic mechanistic static model populated with Ki,u data from the present study predicts AUCRhep and AUCRsys, with a strong potential for pharmacokinetic interactions between CBD (both oral and inhaled) and CYP1A2, CYP3A4, CYP2B6, CYP2C9, CYP2C19, CYP2D6, and CYP2E1 (Table 4). For oral dosing of THC, the potential for pharmacokinetic interactions was observed for CYP1A2, CYP2C9, CYP2C19, and CYP2D6, whereas for inhaled THC there is a potential for pharmacokinetic interactions with CYP1A2, CYP2B6, CYP2C9, CYP2C19, and CYP2D6. 11-OH-THC showed potential for pharmacokinetic interactions with CYP2B6, CYP2C9, and CYP2D6 for oral and inhaled dosing. THC-COO-Gluc showed potential for pharmacokinetic interactions with CYP2B6, CYP2C9, and CYP2D6 for both oral and inhaled dosing of THC.
TABLE 4.
Prediction of P450-mediated clinical drug interaction potential after oral or inhaled dose of THC or CBDa
| THC | 11-OH-THC | THC-COO-Gluc | CBD | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Oral | Inhaled | Oral | Inhaled | Oral | Inhaled | Oral | Inhaled | |||||
| Enzyme | AUCRhep | AUCRsys | AUCRsys | AUCRhep | AUCRsys | AUCRsys | AUCRhep | AUCRsys | AUCRsys | AUCRhep | AUCRsys | AUCRsys |
| CYP1A2 | 1.25 | 1.02 | 1.08 | NAb | NA | 7.38 | 1.20 | 1.09 | ||||
| CYP3A4 | NA | NA | NA | 9.24 | 1.26 | 1.11 | ||||||
| CYP2B6 | 1.09 | 1.01 | 1.03 | 1.32 | 1.03 | 1.02 | 1.08 | 1.03 | 1.03 | 12.26 | 1.35 | 1.15 |
| CYP2C9 | 1.31 | 1.03 | 1.10 | 1.44 | 1.04 | 1.03 | 1.05 | 1.02 | 1.02 | 9.24 | 1.26 | 1.11 |
| CYP2C19 | 1.40 | 1.03 | 1.13 | NA | NA | 16.32 | 1.48 | 1.21 | ||||
| CYP2D6 | 1.20 | 1.02 | 1.07 | 1.17 | 1.02 | 1.02 | 1.08 | 1.03 | 1.03 | 11.94 | 1.34 | 1.15 |
| CYP2E1 | NA | NA | NA | 37.47 | 2.14 | 1.49 | ||||||
aThe AUCRhep and AUCRsys were calculated for THC, 11-OH-THC and THC-COO-Gluc based on oral or inhaled dosing of THC; the AUCRhep and AUCRsys were calculated for CBD based on oral or inhaled dosing of CBD as described in Materials and Methods.
bNA, not analyzed.
Discussion
In the present study, the inhibitory effects of major cannabinoids and THC metabolites on major hepatic CYP450s were investigated. Consistent with previous studies (Jiang et al., 2011, 2013; Yamaori et al., 2011a,b,c; Cox et al., 2019), THC demonstrated reversible inhibition of CYP1A2, CYP2B6, CYP2C9, CYP2C19, and CYP2D6 activity; CBD demonstrated reversible inhibition of CYP1A2, CYP3A4, CYP2B6, CYP2C9, CYP2C19, and CYP2D6; and CBN demonstrated reversible inhibition of CYP2B6 and CYP2C9. Although not examined in previous studies, CYP2E1 was also inhibited by the major cannabinoids, with both CBD and CBN demonstrating strong, competitive inhibition of this P450. In ranking the magnitude of inhibition observed in CYP450-overexpressing cell lines, inhibition by THC was shown to be highest for CYP2C19, followed by 2C9 > 1A2 > 2D6 > 2B6; inhibition by CBD was highest for CYP2E1, followed by 2C19 > 2B6 > 2D6 > 3A4 ∼2C9 > 1A2; and inhibition by CBN was also highest for CYP2E1, followed by 2B6 > 2C9.
Prior to the present analysis, a comprehensive inhibition study of hepatic P450 enzymes by all major THC metabolites (11-OH-THC, THC-COOH, and THC-COO-Gluc) had not been previously performed. In the present study, 11-OH-THC and THC-COO-Gluc competitively inhibited CYP2B6, CYP2C9, and CYP2D6 activity, with no inhibition of any P450 enzyme observed with THC-COOH. Inhibition by 11-OH-THC was shown to be highest for CYP2C9, followed by CYP2B6 and then CYP2D6, whereas inhibition by THC-COO-Gluc was highest for CYP2B6, followed by CYP2C9 ≃ CYP2D6. In contrast to THC and its active metabolite THC-OH, THC-COOH and THC-COO-Gluc are present in the plasma at constant and relatively high levels and remain stable in the bloodstream over many days. THC-COOH and THC-COO-Gluc levels are approximately 2.0- and 7.6-fold higher, respectively, than THC after administration by cannabis inhalation, whereas after oral administration, 11-OH-THC and COOH-THC are 2.5- and 40-fold higher than the levels of THC (THCR-COO-Gluc levels have not been tested in plasma after oral administration) (Nadulski et al., 2005; Schwope et al., 2011; Bansal et al., 2020). These data suggest that 11-OH-THC and THC-COO-Gluc potentially play an important role in the inhibition of major hepatic P450 enzymes upon cannabis consumption.
All the cannabinoids and metabolites tested in the present study were reversible inhibitors of CYP2B6 bupropion hydroxylation activity. A strong inhibition was observed by the cannabinoid metabolites 11-THC-OH and THC-COO-Gluc acting in a competitive and reversible manner. In addition to the fact that CYP2B6 is responsible for the metabolism of numerous drugs and other agents, including bupropion, efavirenz, cyclophosphamide, ifosamide, pethidine, artemisinin, propofol, ketamine, and selegiline, current estimates indicate that CYP2B6 accounts for 2%–10% of total hepatic P450 content (Code et al., 1997; Hanna et al., 2000; Hesse et al., 2000; Hedrich et al., 2016). Two of the most well studied and better-understood drugs with respect to CYP2B6 metabolism are the anticancer drug cyclophosphamide and the anti-HIV medication efavirenz. Both of these widely used drugs have very narrow therapeutic indices, and variations in CYP2B6 expression along with enzyme inhibition lead to significantly altered drug plasma concentrations and associated toxicities (Hedrich et al., 2016). In recent years, cannabinoids have been extensively used for the treatment of various cancers to alleviate the side effects from anticancer agents, such as cyclophosphamide, which has been used as a treatment of cancer and autoimmune disorders for more than half a century. In a clinical study, cyclophosphamide did not demonstrate an improvement in nausea and vomiting when administered in combination with cannabis or oral THC as compared with placebo, which might be explained by the inhibition of CYP2B6 activity by THC and its metabolites and the subsequent increase of in vivo AUC of cyclophosphamide, which is primarily metabolized by CYP2B6 (Abrams, 2016). Additionally, there is growing recognition that individuals with HIV report greater cannabis use than the general population. Efavirenz is reported to carry a risk of side effects that includes adverse neuropsychiatric complications, such as depression, anxiety, hallucinations, and delusions. Several studies showed that the combination of EFV and THC might result in stronger hallucinating power and increased depression, which potentially could be caused by the inhibitory effects of cannabinoids and their metabolites (Mthembi et al., 2018). In addition, bupropion is an antidepressant that is often used as an aid to quit smoking and is almost exclusively metabolized by CYP2B6 (Hesse et al., 2000). Bupropion has been shown to cause increased adverse effects (irritability, restlessness, insomnia) in a clinical study when participants actively smoked cannabis (Haney et al., 2001). These increased adverse effects might also be caused by the inhibitory effects of cannabinoids on CYP2B6 when coadministered with bupropion. Lastly, ketamine is a medical-grade anesthetic that can be swallowed, snorted, injected, or smoked, often together with cannabis or tobacco. Although there are no studies exploring how ketamine and cannabis interact, co-users tend to report that cannabis increases the ketamine high and can, in some instances, exacerbate effects like drowsiness and dizziness (Garcia-Romeu et al., 2016).
The CYP2C9 hydroxylation of diclofenac was also strongly inhibited by the cannabinoid metabolites 11-THC-OH and THC-COO-Gluc in a reversible manner, with a competitive type in the rCYP and mixed in HLMs. Previous studies have also found potent inhibition of CYP2C9 by THC, CBD, and 11-OH-THC (Bansal et al., 2020). CYP2C9 is one of the most abundant P450 enzymes in the liver (about 20% of the hepatic total P450 content) (Ghodke-Puranik and Lamba, 2017). It metabolizes several important drugs, including anticancer agents, nonsteroidal anti-inflammatory agents, selective cyclooxygenase-2 inhibitors, antibiotics, antiepileptics, antihypertensives, anticoagulants, and antihyperlipidemics (Ghodke-Puranik and Lamba, 2017). Drug-drug interactions have been shown to arise when a CYP2C9 inhibitor is added to a therapeutic regimen, including low therapeutic index drugs like (S)-warfarin, tolbutamide, or phenytoin. In these cases, patients can risk life-threatening bleeding episodes, hypoglycemia, and neurotoxicity as a result of the diminished CYP2C9 enzyme activity. These in vitro predictions are largely consistent with many clinical CBD- or THC-drug interactions reported to date. In clinical studies, an increased anticoagulant effect of warfarin was previously observed in smokers of cannabis, potentially due to inhibition of CYP2C9-mediated warfarin metabolism by cannabinoids and THC metabolites (Yamreudeewong et al., 2009). In addition, a case report showed that phenytoin caused severe adverse effects when coadministered with cannabis (Jessen, 2004).
THC, 11-OH-THC, THC-COO-Gluc, and CBD also strongly inhibited the dextromethorphan-O-demethylase activity of CYP2D6 in a reversible and competitive manner. Although CYP2D6 comprises a relatively small percentage (2%–6%) of the total CYP450 protein in the liver, it is involved in the metabolism of 25% of the most commonly prescribed drugs (e.g., antidepressants, antipsychotics, opioids, and β blockers) (Shannon, 2007). Although there are no reports on in vivo drug interactions between cannabinoids and CYP2D6 substrates, the results from the present study can potentially explain the in vivo mechanism for increased adverse effects or potency of CYP2D6-metabolized agents. For example, dextromethorphan is a widely used antitussive agent metabolized by CYP2D6 into an active metabolite, dextrorphan. Using dextromethorphan together with cannabis has been shown to increase side effects, such as dizziness, drowsiness, confusion, and difficulty concentrating (Ziaee et al., 2005). In addition, the widely used anticancer drug and CYP2D6 substrate tamoxifen showed decreased tamoxifen metabolism in vivo, and the level of CYP2D6-mediated active metabolite endoxifen decreased in plasma in subjects also taking cannabinoids (Parihar et al., 2020).
For CYP1A2 and CYP2C19, a mixed-type inhibition was observed for CBD and THC, respectively, in both rCYPs and HLMs, suggesting that, in contrast to the competitive type inhibition observed for most of the cannabinoids and THC metabolites analyzed in this study, THC and CBD are not interacting with CYP1A2 and CYP2C19 in a similar fashion. In addition, CYP2C9 and CYP2C19 showed different types of inhibition when analyzing their corresponding rCYP microsomes versus HLMs; with THC, THC-COO-Gluc, and CBD all exhibiting competitive inhibition in the rCYP and mixed inhibition in HLMs, possibly due to a confounding effect of the presence of multiple P450s within HLMs.
Although cannabis can be consumed through oral dosing, smoking cannabis is still the most common way of consumption. These differences become important when analyzing metabolite levels, as the major THC metabolites will differ in plasma concentration based on the route of administration. After smoking a cannabis cigarette containing 10 mg THC, the plasma AUCs of THC, 11-OH-THC, and THC-COO-Gluc are 110 ug/l (0.35 µM), 19 ug/l (0.06 µM), and 840 ug/l (1.6 µM), respectively. In contrast, after oral dosing of a consumable containing 10 mg of THC, the AUCs of THC and 11-OH-THC are even higher at 360 ug/l (1.14 µM) and 14,600 ug/l (28 µM), respectively (Nadulski et al., 2005). THC-COO-Gluc levels were 139 ug/l (0.27 µM) after a controlled oral administration of 51 mg THC (Newmeyer et al., 2016). The daily dosage of cannabis varies widely among users, dependent on such factors as the THC:CBD ratio of the product, cannabis use history, and puff duration. THC is recreationally used at around 30–100 mg per joint or vaporizer, whereas the inhaled dose for medicinal use is lower at approximately 6–80 mg THC (Bansal et al., 2020). Based on these data, the plasma levels of THC and its metabolites will be at higher or similar levels compared with the Ki values and inhibitory concentrations calculated for the different CYP450 enzymes in the present study. In addition, since cannabinoids are readily distributed in various tissues because of their high lipophilicity, the tissue concentration of THC and its metabolites may be even higher than the plasma concentration.
Unlike that described by Bansal et al. (2020), TDI was not observed for CBD for the P450 enzymes tested in the present study, with the possible exception of CYP2D6. These differences are likely due to experimental conditions, with the present studies conducted using microsomes from CYP450-overexpressing cell lines, with each inhibition assay performed independently using individual probe substrates. In contrast, Bansal et al. (2020) performed their assays using a “cocktail method” with multiple probes added into one reaction with HLMs as the CYP450 protein source. In addition, although BSA was used in the reaction mixture in the Bansal studies, none was added to the assays within the present study.
In conclusion, cannabinoids and the major THC metabolites were shown to strongly inhibit several P450 enzymes, including CYP2B6, CYP2C9, and CYP2D6. Most importantly, the most abundant and previously untested THC metabolite in plasma, THC-COO-Gluc, is likely to play a major role in the inhibition of several hepatic P450 enzymes. Further investigations into the nature of inhibition of cannabinoids and their metabolites against specific agents will help to clarify the precise mechanism of in vivo cannabis-drug interactions.
Acknowledgments
The authors would like to thank Shelby Coates, Keti Bardhi, and Dr. Gang Chen for their helpful advice and contributions to the project.
Abbreviations
- AUC
area under the curve
- AUCRhep
hepatic AUC ratio
- AUCRsys
systemic AUC ratio
- CB1
cannabinoid receptor type 1
- CBD
cannabidiol
- 7-OH-CBD
7-hydroxy-cannabidiol
- CBD-COOH
7-carboxy-cannabidiol
- CBN
cannabinol
- FDA
Food and Drug Administration
- fu, inc
unbound fraction of cannabinoid in incubations with either HLMs or HEK293 microsomes
- HEK
human embryonic kidney
- HIV
human immunodeficiency virus
- HLM
human liver microsome
- IS
internal standard
- Ki
inhibitory constant
- Km
Michaelis-Menten constant
- LC
liquid chromatography
- MS
mass spectrometry
- MS/MS
tandem MS
- 11-OH-THC
11-hydroxy-Δ9-tetrahydrocannabinol
- P450
cytochrome P450
- TDI
time-dependent inhibition
- THC
(−)-trans-Δ9-tetrahydrocannabinol
- THC-COO-Gluc
11-nor-9-carboxy-Δ 9-tetrahydrocannabinol glucuronide
- THC-COOH
11-nor-9-carboxy-Δ9- tetrahydrocannabinol
- UGT
UDP-glucuronosyltransferase
- UPLC
ultra–high-performance LC
Authorship Contributions
Participated in research design: Nasrin, Watson, Lazarus.
Conducted experiments: Nasrin.
Contributed new reagents or analytic tools: Perez-Paramo.
Performed data analysis: Nasrin, Lazarus.
Wrote or contributed to the writing of the manuscript: Nasrin, Watson, Lazarus.
Footnotes
This work was supported by National Institutes of Health National Institutes of Environmental Health Sciences [Grant R01-ES025460] (to P. L.); the Health Sciences and Services Authority of Spokane, WA [Grant WSU002292]; and funds provided by the State of Washington Initiative Measure 502.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Abrams DI (2016) Integrating cannabis into clinical cancer care. Curr Oncol 23:S8–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizpurua-Olaizola O, Soydaner U, Öztürk E, Schibano D, Simsir Y, Navarro P, Etxebarria N, Usobiaga A (2016) Evolution of the cannabinoid and terpene content during the growth of cannabis sativa plants from different chemotypes. J Nat Prod 79:324–331. [DOI] [PubMed] [Google Scholar]
- Atakan Z (2012) Cannabis, a complex plant: different compounds and different effects on individuals. Ther Adv Psychopharmacol 2:241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S, Maharao N, Paine MF, Unadkat JD (2020) Predicting the potential for cannabinoids to precipitate pharmacokinetic drug interactions via reversible inhibition or inactivation of major cytochromes P450. Drug Metab Dispos 48:1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Code EL, Crespi CL, Penman BW, Gonzalez FJ, Chang TKH, Waxman DJ (1997) Human cytochrome P4502B6: interindividual hepatic expression, substrate specificity, and role in procarcinogen activation. Drug Metab Dispos 25:985–993. [PubMed] [Google Scholar]
- Cornish-Bowden A (1974) A simple graphical method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors. Biochem J 137:143–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox EJ, Maharao N, Patilea-Vrana G, Unadkat JD, Rettie AE, McCune JS, Paine MF (2019) A marijuana-drug interaction primer: precipitants, pharmacology, and pharmacokinetics. Pharmacol Ther 201:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellinger RW, Fang J-L, Chen G, Weinberg R, Lazarus P (2006) Importance of UDP-glucuronosyltransferase 1A10 (UGT1A10) in the detoxification of polycyclic aromatic hydrocarbons: decreased glucuronidative activity of the UGT1A10139Lys isoform. Drug Metab Dispos 34:943–949. [DOI] [PubMed] [Google Scholar]
- Food and Drug Administration (2020) In vitro drug interaction studies cytochrome P450 enzymes and transporter-mediated drug interactions. [Google Scholar]
- Garcia-Romeu A, Kersgaard B, Addy PH (2016) Clinical applications of hallucinogens: a review. Exp Clin Psychopharmacol 24:229–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett ER, Hunt CA (1974) Physiochemical properties, solubility, and protein binding of delta9-tetrahydrocannabinol. J Pharm Sci 63:1056–1064. [DOI] [PubMed] [Google Scholar]
- Ghodke-Puranik YA, Lamba JK (2017) Pharmacogenomics, in Innovative Approaches in Drug Discovery, chap 7. Patwardhan B, Chaguturu R, eds) pp 195–234, Academic Press, Boston. [Google Scholar]
- Haney M, Ward AS, Comer SD, Hart CL, Foltin RW, Fischman MW (2001) Bupropion SR worsens mood during marijuana withdrawal in humans. Psychopharmacology (Berl) 155:171–179. [DOI] [PubMed] [Google Scholar]
- Hanna IH, Reed JR, Guengerich FP, Hollenberg PF (2000) Expression of human cytochrome P450 2B6 in Escherichia coli: characterization of catalytic activity and expression levels in human liver. Arch Biochem Biophys 376:206–216. [DOI] [PubMed] [Google Scholar]
- Harvey DJ, Mechoulam R (1990) Metabolites of cannabidiol identified in human urine. Xenobiotica 20:303–320. [DOI] [PubMed] [Google Scholar]
- Hedrich WD, Hassan HE, Wang H (2016) Insights into CYP2B6-mediated drug-drug interactions. Acta Pharm Sin B 6:413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse LM, Venkatakrishnan K, Court MH, von Moltke LL, Duan SX, Shader RI, Greenblatt DJ (2000) CYP2B6 mediates the in vitro hydroxylation of bupropion: potential drug interactions with other antidepressants. Drug Metab Dispos 28:1176–1183. [PubMed] [Google Scholar]
- Huestis MA (2007) Human cannabinoid pharmacokinetics. Chem Biodivers 4:1770–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunault CC, Mensinga TT, de Vries I, Kelholt-Dijkman HH, Hoek J, Kruidenier M, Leenders MEC, Meulenbelt J (2008) Delta-9-tetrahydrocannabinol (THC) serum concentrations and pharmacological effects in males after smoking a combination of tobacco and cannabis containing up to 69 mg THC. Psychopharmacology (Berl) 201:171–181. [DOI] [PubMed] [Google Scholar]
- Isoherranen N, Kunze KL, Allen KE, Nelson WL, Thummel KE(2004) Role of itraconazole metabolites in CYP3A4 inhibition. Drug Metab Dispos 32:1121–1131. [DOI] [PubMed] [Google Scholar]
- Jessen K (2004) Recreational use of phenytoin, marijuana, and alcohol: a case report. Neurology 62:2330. [DOI] [PubMed] [Google Scholar]
- Jiang R, Yamaori S, Okamoto Y, Yamamoto I, Watanabe K (2013) Cannabidiol is a potent inhibitor of the catalytic activity of cytochrome P450 2C19. Drug Metab Pharmacokinet 28:332–338. [DOI] [PubMed] [Google Scholar]
- Jiang R, Yamaori S, Takeda S, Yamamoto I, Watanabe K (2011) Identification of cytochrome P450 enzymes responsible for metabolism of cannabidiol by human liver microsomes. Life Sci 89:165–170. [DOI] [PubMed] [Google Scholar]
- Manini AFYiannoulos GBergamaschi MMHernandez SOlmedo RBarnes AJWinkel GSinha RJutras-Aswad DHuestis MA, et al. (2015) Safety and pharmacokinetics of oral cannabidiol when administered concomitantly with intravenous fentanyl in humans. J Addict Med 9:204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroon J, Bost J (2018) Review of the neurological benefits of phytocannabinoids. Surg Neurol Int 9:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga T, Iwawaki Y, Watanabe K, Yamamoto I, Kageyama T, Yoshimura H (1995) Metabolism of delta 9-tetrahydrocannabinol by cytochrome P450 isozymes purified from hepatic microsomes of monkeys. Life Sci 56:2089–2095. [DOI] [PubMed] [Google Scholar]
- Ménétrey A, Augsburger M, Favrat B, Pin MA, Rothuizen LE, Appenzeller M, Buclin T, Mangin P, Giroud C (2005) Assessment of driving capability through the use of clinical and psychomotor tests in relation to blood cannabinoids levels following oral administration of 20 mg dronabinol or of a cannabis decoction made with 20 or 60 mg Delta9-THC. J Anal Toxicol 29:327–338. [DOI] [PubMed] [Google Scholar]
- Mthembi PM, Mwenesongole EM, Cole MD (2018) Chemical profiling of the street cocktail drug ‘nyaope’ in South Africa using GC-MS I: stability studies of components of ‘nyaope’ in organic solvents. Forensic Sci Int 292:115–124. [DOI] [PubMed] [Google Scholar]
- Nadulski T, Pragst F, Weinberg G, Roser P, Schnelle M, Fronk EM, Stadelmann AM (2005) Randomized, double-blind, placebo-controlled study about the effects of cannabidiol (CBD) on the pharmacokinetics of delta9-tetrahydrocannabinol (THC) after oral application of THC verses standardized cannabis extract. Ther Drug Monit 27:799–810. [DOI] [PubMed] [Google Scholar]
- Newmeyer MN, Swortwood MJ, Barnes AJ, Abulseoud OA, Scheidweiler KB, Huestis MA (2016) Free and glucuronide whole blood cannabinoids’ pharmacokinetics after controlled smoked, vaporized, and oral cannabis administration in frequent and occasional cannabis users: identification of recent cannabis intake. Clin Chem 62:1579–1592. [DOI] [PubMed] [Google Scholar]
- Parihar V, Rogers A, Blain AM, Zacharias SRK, Patterson LL, Siyam MA-M (2020) Reduction in tamoxifen metabolites endoxifen and N-desmethyltamoxifen with chronic administration of low dose cannabidiol: a CYP3A4 and CYP2D6 drug interaction. J Pharm Pract 87-90. [DOI] [PubMed] [Google Scholar]
- Parkinson A, Kazmi F, Buckley DB, Yerino P, Paris BL, Holsapple J, Toren P, Otradovec SM, Ogilvie BW (2011) An evaluation of the dilution method for identifying metabolism-dependent inhibitors of cytochrome P450 enzymes. Drug Metab Dispos 39:1370–1387. [DOI] [PubMed] [Google Scholar]
- Patilea-Vrana GI, Anoshchenko O, Unadkat JD(2019) Hepatic Enzymes Relevant to the Disposition of (-)-Δ9-Tetrahydrocannabinol (THC) and Its Psychoactive Metabolite, 11-OH-THC. Drug Metab Dispos 47:249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertwee RG (2008) The diverse CB1 and CB2 receptor pharmacology of three plant cannabinoids: delta9-tetrahydrocannabinol, cannabidiol and delta9-tetrahydrocannabivarin. Br J Pharmacol 153:199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson A, Xia Z, Chen G, Lazarus P (2017a) Exemestane potency is unchanged by common nonsynonymous polymorphisms in CYP19A1: results of a novel anti-aromatase activity assay examining exemestane and its derivatives. Pharmacol Res Perspect 5:e00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson A, Xia Z, Chen G, Lazarus P (2017b) In vitro metabolism of exemestane by hepatic cytochrome P450s: impact of nonsynonymous polymorphisms on formation of the active metabolite 17β-dihydroexemestane. Pharmacol Res Perspect 5:e00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo EB, Marcu J (2017) Chapter Three - Cannabis Pharmacology: The Usual Suspects and a Few Promising Leads, in Adv Pharmacol (Kendall D, Alexander SPH, eds) pp 67–134, Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- Solimini R, Rotolo MC, Pichini S, Pacifici R (2017) Neurological disorders in medical use of cannabis: an update. CNS Neurol Disord Drug Targets 16:527–533. [DOI] [PubMed] [Google Scholar]
- Schwilke EW, Schwope DM, Karschner EL, Lowe RH, Darwin WD, Kelly DL, Goodwin RS, Gorelick DA, Huestis MA (2009) Delta9-tetrahydrocannabinol (THC), 11-hydroxy-THC, and 11-nor-9-carboxy-THC plasma pharmacokinetics during and after continuous high-dose oral THC. Clin Chem 55:2180–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwope DM, Karschner EL, Gorelick DA, Huestis MA (2011) Identification of recent cannabis use: whole-blood and plasma free and glucuronidated cannabinoid pharmacokinetics following controlled smoked cannabis administration. Clin Chem 57:1406–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon MW (2007) Drug Interactions, in Haddad and Winchester’s Clinical Management of Poisoning and Drug Overdose, Ed. 4th. chap 5. Shannon MW, Borron SW, Burns MJ, eds) pp 97–104, W.B. Saunders, Philadelphia. [Google Scholar]
- Sharma P, Murthy P, Bharath MMS (2012) Chemistry, metabolism, and toxicology of cannabis: clinical implications. Iran J Psychiatry 7:149–156. [PMC free article] [PubMed] [Google Scholar]
- Skopp G, Pötsch L, Mauden M, Richter B (2002) Partition coefficient, blood to plasma ratio, protein binding and short-term stability of 11-nor-Δ(9)-carboxy tetrahydrocannabinol glucuronide. Forensic Sci Int 126:17–23. [DOI] [PubMed] [Google Scholar]
- Spindle TR, Cone EJ, Goffi E, Weerts EM, Mitchell JM, Winecker RE, Bigelow GE, Flegel RR, Vandrey R (2020) Pharmacodynamic effects of vaporized and oral cannabidiol (CBD) and vaporized CBD-dominant cannabis in infrequent cannabis users. Drug Alcohol Depend 211:107937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall ME, Sadler BM, Brine D, Taylor H, Perez-Reyes M (1983) Metabolism, disposition, and kinetics of delta-9-tetrahydrocannabinol in men and women. Clin Pharmacol Ther 34:352–363. [DOI] [PubMed] [Google Scholar]
- Weinberger AH, Platt J, Goodwin RD (2016) Is cannabis use associated with an increased risk of onset and persistence of alcohol use disorders? A three-year prospective study among adults in the United States. Drug Alcohol Depend 161:363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaori S, Ebisawa J, Okushima Y, Yamamoto I, Watanabe K (2011a) Potent inhibition of human cytochrome P450 3A isoforms by cannabidiol: role of phenolic hydroxyl groups in the resorcinol moiety. Life Sci 88:730–736. [DOI] [PubMed] [Google Scholar]
- Yamaori S, Kushihara M, Yamamoto I, Watanabe K (2010) Characterization of major phytocannabinoids, cannabidiol and cannabinol, as isoform-selective and potent inhibitors of human CYP1 enzymes. Biochem Pharmacol 79:1691–1698. [DOI] [PubMed] [Google Scholar]
- Yamaori S, Maeda C, Yamamoto I, Watanabe K (2011b) Differential inhibition of human cytochrome P450 2A6 and 2B6 by major phytocannabinoids. Forensic Toxicol 29:117–124. [Google Scholar]
- Yamaori S, Okamoto Y, Yamamoto I, Watanabe K (2011c) Cannabidiol, a major phytocannabinoid, as a potent atypical inhibitor for CYP2D6. Drug Metab Dispos 39:2049–2056. [DOI] [PubMed] [Google Scholar]
- Yamreudeewong W, Wong HK, Brausch LM, Pulley KR (2009) Probable interaction between warfarin and marijuana smoking. Ann Pharmacother 43:1347–1353. [DOI] [PubMed] [Google Scholar]
- Ziaee V, Akbari Hamed E, Hoshmand A, Amini H, Kebriaeizadeh A, Saman K (2005) Side effects of dextromethorphan abuse, a case series. Addict Behav 30:1607–1613. [DOI] [PubMed] [Google Scholar]