Abstract
We previously reported the unbound reversible (IC50,u) and time-dependent (KI,u) inhibition potencies of cannabidiol (CBD), delta-9-tetrahydrocannabinol (THC), and THC metabolites 11-hydroxy THC (11-OH THC) and 11-nor-9-carboxy-delta-9-THC (11-COOH THC) against the major cytochrome P450 (P450) enzymes (1A2, 2C9, 2C19, 2D6, and 3A). Here, using human liver microsomes, we determined the CYP2A6, 2B6, and 2C8 IC50,u values of the aforementioned cannabinoids and the IC50,u and KI,u of the circulating CBD metabolites 7-hydroxy CBD (7-OH CBD) and 7-carboxy CBD (7-COOH CBD), against all the P450s listed above. The IC50,u of CBD, 7-OH CBD, THC, and 11-OH THC against CYP2B6 was 0.05, 0.34, 0.40, and 0.32 μM, respectively, and against CYP2C8 was 0.28, 1.02, 0.67, and 3.66 μM, respectively. 7-COOH CBD, but not 11-COOH THC, was a weak inhibitor of CYP2B6 and 2C8. All tested cannabinoids except 11-COOH THC were weak inhibitors of CYP2A6. 7-OH CBD inhibited all P450s examined (IC50,u<2.5 μM) except CYP1A2 and inactivated CYP2C19 and CYP3A, with inactivation efficiencies (kinact/KI,u) of 0.10 and 0.14 minutes−1μM−1, respectively. Using several different static models, we predicted the following maximum pharmacokinetic interactions (affected P450 probe drug and area under the plasma concentration-time curve ratio) between oral CBD (700 mg) and drugs predominantly metabolized by CYP3A (midazolam, 14.8) > 2C9 (diclofenac, 9.6) > 2C19 (omeprazole, 7.3) > 1A2 (theophylline, 4.0) > 2B6 (ticlopidine, 2.2) > 2D6 (dextromethorphan, 2.1) > 2C8 (repaglinide, 1.6). Oral (130 mg) or inhaled (75 mg) THC was predicted to precipitate interactions with drugs predominately metabolized by CYP2C9 (diclofenac, 6.6 or 2.3, respectively) > 3A (midazolam, 1.8) > 1A2 (theophylline, 1.4). In vivo drug interaction studies are warranted to verify these predictions.
SIGNIFICANCE STATEMENT
This study, combined with our previous findings, provides for the first time a comprehensive analysis of the potential for cannabidiol, delta-9-tetrahydrocannabinol, and their metabolites to inhibit cytochrome P450 enzymes in a reversible or time-dependent manner. These analyses enabled us to predict the potential of these cannabinoids to produce drug interactions in vivo at clinical or recreational doses.
Introduction
Cannabis (Cannabis sativa) is used worldwide for both recreational and medicinal purposes. The major phytocannabinoids in cannabis are the nonpsychoactive cannabidiol (CBD) and the psychoactive delta-9-tetrahydrocannabinol (THC) (Fig. 1). CBD is approved to treat childhood epilepsy, namely the Lennox-Gastaut and the Dravet syndrome. THC is used as an antiemetic, analgesic, antispasmodic, and appetite stimulant (Amaral Silva et al., 2020). To date, cannabis has been legalized in 36 states for medicinal use and in 18 states for recreational use (https://www.ncsl.org/research/health/state-medical-marijuana-laws.aspx). By 2025, the estimated number of cannabis users in the United States is expected to reach 46.6 million (https://www.statista.com/statistics/1060216/us-total-estimated-cannabis-consumer-population/). Therefore, it is crucial to evaluate the risk of potential pharmacokinetic interactions between cannabinoids and pharmaceutical drugs. Indeed, two case reports have reported cannabinoid-drug interaction related adverse effects. In the first, a cannabis-warfarin (a CYP2C9 substrate) interaction was observed where the anticoagulant effect of warfarin was considerably increased (international normalized ratio of 12) to outside the therapeutic range (international normalized ratio of 2–3) in a patient who smoked cannabis frequently (Grayson et al., 2017). In the second, a CBD-tacrolimus (a CYP3A4 substrate) interaction resulted in increased serum creatinine concentration (a marker of tacrolimus renal toxicity) in a patient who consumed a high dose of CBD (1000 mg twice daily orally) with tacrolimus (Leino et al., 2019).
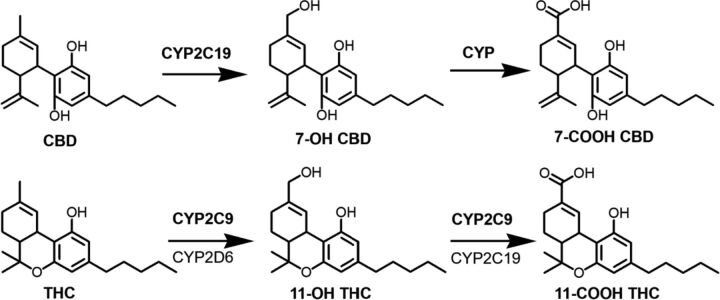
Fig. 1.
CBD and THC biotransformation pathways. Bolded enzymes are the predominant contributors (Patilea-Vrana and Unadkat, 2019; Beers et al., 2021). P450s involved in the metabolism of 7-OH CBD to 7-COOH CBD have not been identified.
CBD is metabolized to the pharmacologically active circulating metabolite 7-hydroxy CBD (7-OH CBD), which is further metabolized to the inactive circulating metabolite 7-carboxy CBD (7-COOH CBD) (Fig. 1). (https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/210365lbl.pdf). THC is metabolized to the psychoactive circulating metabolite 11-hydroxy THC (11-OH THC), which is further metabolized to the inactive circulating metabolite 11-nor-9-carboxy-delta-9-THC (11-COOH THC) (Fig. 1). The maximum systemic plasma concentration (Cmax) of these metabolites is reported to exceed that of CBD or THC except 7-OH CBD, for which the Cmax is approximately 50% of that of CBD (Frytak et al., 1984; Nadulski et al., 2005; Taylor et al., 2018). Therefore, as recommended by the US Food and Drug Administration, the effect of cytochrome P450 (P450) inhibition by these metabolites on the predicted magnitude of pharmacokinetic CBD- or THC-drug interactions should be evaluated (https://www.fda.gov/media/134582/download).
CBD or THC is known to reversibly inhibit cytochrome P450 1A, 2A6, 1B1, 2B6, 2C8, 2C9, 2C19, 2D6, 2J2, and 3A (Yamaori et al., 2010; 2011a; Arnold et al., 2018; Cox et al., 2019; https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/210365lbl.pdf). In addition, THC has been shown to be a time-dependent inhibitor (TDI) of CYP2A6 (Yamaori et al., 2011b). However, these studies did not consider the binding of CBD or THC to microsomal proteins or labware. We recently reported the unbound inhibition potency (IC50,u) of CBD, THC, 11-OH THC, and 11-COOH THC against CYP1A2, 2C9, 2C19, 2D6, and 3A (Bansal et al., 2020). In addition, we showed that CBD was a time-dependent inhibitor of CYP1A2, 2C19, and 3A. However, none of these studies determined the IC50,u of CBD, THC, 11-OH THC, or 11-COOH THC against CYP2A6, 2B6, and 2C8. Moreover, whether 7-OH CBD and 7-COOH CBD inhibits any of these P450s is not known.
The overall goal of the current study was to predict the magnitude of pharmacokinetic interactions between CBD or THC and drugs predominantly metabolized by CYP2A6, 2B6, or 2C8 and to determine the ability of the circulating CBD metabolites to inhibit the above mentioned CYPs. Specifically, our aims were to 1) determine the fraction unbound in the plasma (fu,p) and in the incubation mixture (fu,inc) of CBD, THC, and their metabolites; 2) determine the unbound inhibition or inactivation potency of CBD, THC, and their metabolites against CYP2A6, 2B6, or 2C8; 3) determine the potential of circulating CBD metabolites to inhibit or inactivate all the P450s listed above; and 4) predict, using a mechanistic static model, the magnitude of P450-mediated interactions between oral CBD or inhaled and oral THC and P450 probe drugs.
Materials and Methods
Biologic Materials, Chemicals, and Reagents
Pooled human liver microsomes (HLMs) (mixed gender; pool of 200 donors) and human plasma from a single healthy male donor were purchased from SEKISUI Xenotech, LLC (Kansas City, KS) and Bloodworks (Seattle, WA), respectively. THC, CBD, omeprazole, 5-hydroxy omeprazole, testosterone, 6β-hydroxy testosterone, dextromethorphan, dextrorphan, phenacetin, acetaminophen, diclofenac, and 4’-hydroxy diclofenac were purchased from Cayman Chemical (Ann Arbor, MI). 7-COOH CBD and 7-OH CBD were procured from BDG Synthesis (Lower Hutt, New Zealand). Methanolic solutions of 11-COOH THC, 11-OH THC, deuterated THC (THC-d3), and deuterated CBD (CBD-d3) were purchased from Cerilliant (Round Rock, TX). Acetonitrile, bovine serum albumin (BSA), and formic acid were bought from Fisher Scientific (Hampton, NH). Coumarin, 7-hydroxycoumarin, bupropion, 4-hydroxy bupropion, amodiaquine, N-desethylamodiaquine, tolbutamide, D-glucose 6-phosphate, β-nicotinamide adenine dinucleotide phosphate (NADP+), and glucose-6-phosphate dehydrogenase were purchased from Sigma-Aldrich (St. Louis, MO). Dulbecco’s phosphate-buffered saline was procured from Thermo Scientific (Asheville, NC). Micro ultracentrifuge polycarbonate tubes were purchased from Beckman Coulter (Brea, CA), and ultralow-binding microcentrifuge tubes were purchased from Genesee Scientific (EI Cajon, CA). All other chemicals and experimental reagents were obtained from reputable commercial sources.
Reversible Inhibition of P450s by Cannabinoids
As per our previous study (Bansal et al., 2020), low-binding microcentrifuge tubes were used to reduce the nonspecific binding and adsorption of cannabinoids to labware, and we included BSA (0.2%) in all incubations to increase cannabinoid solubility and reduce nonspecific binding.
The inhibition of CYP1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, and 3A by each cannabinoid was simultaneously evaluated using a previously validated P450 cocktail assay (Dinger et al., 2014). A metabolite of each probe substrate selectively formed by a single P450 was quantified using liquid chromatography-tandem mass spectrometry (LC-MS/MS) (described below). The probe substrate/metabolite pair and probe substrate concentration were as follows: phenacetin (Km = 1.7–152 μM)/acetaminophen and 50 μM (CYP1A2); coumarin (Km = 0.3–2.3 μM)/7-hydroxycoumarin and 2 μM (CYP2A6); bupropion (Km = 67–168 μM)/4-hydroxy-bupropion and 5 μM (CYP2B6); amodiaquine (Km = 1.9–2.4 μM)/N-desethylamodiaquine and 2 μM (CYP2C8); diclofenac (Km = 3.4–52 μM)/4’-hydroxydiclofenac and 5 μM (CYP2C9); omeprazole (Km = 17–26 μM)/5-hydroxyomeprazole and 10 μM (CYP2C19); dextromethorphan (Km = 0.44–8.5 μM)/dextrorphan and 5 μM (CYP2D6); and testosterone (Km = 52–94 μM)/6β-hydroxytestosterone and 10 μM (CYP3A) (Dinger et al., 2014; Spaggiari et al., 2014; Dahlinger et al., 2016). The probe substrates showed minimal interactions with each other (data not shown).
As an initial screen, each cannabinoid (10 μM) was added to an incubation mixture (200 μl) containing potassium phosphate buffer (100 mM; pH 7.4), HLMs (0.1 mg/ml), P450 cocktail, and BSA (0.2%). Stock solutions of probe substrates and cannabinoids were prepared in 100% DMSO. The final concentration of DMSO (v/v) in the incubation mixture was 0.2–0.5%. Each incubation mixture was prewarmed at 37°C with constant stirring for 10 minutes, and then the reaction was initiated with a NADPH regenerating system (1.3 mM NADP+, 3.3 mM D-glucose 6-phosphate, 3.3 mM magnesium chloride, and 0.4 units/ml glucose-6-phosphate dehydrogenase). After 15 minutes, the reaction was terminated with 200 μl ice-cold acetonitrile containing tolbutamide (internal standard; 25 nM final concentration) to precipitate the proteins. After vortexing, each sample was centrifuged at 18,000 g for 10 minutes. Then, an aliquot of the supernatant (50 μl) was stored at −20°C until analysis by LC-MS/MS (described below). Three independent experiments were conducted, each in duplicate.
Because 10 μM is much greater than circulating cannabinoid unbound plasma concentrations observed when cannabis or CBD products are used for medicinal or recreational purposes, IC50 values were determined only for those cannabinoids that likely inhibit P450s (in the screening assay) at pharmacologically relevant concentrations. Therefore, varying concentrations of CBD (0.003–100 μM), 7-OH CBD (0.003–50 μM), THC (0.003–100 μM), or 11-OH THC (0.01–100 μM) were used to determine their P450-inhibitory potencies. P450 inhibition assays were conducted as described above. Three independent experiments were conducted, each in duplicate. IC50 was determined by nonlinear least-squares regression analysis (GraphPad Prism 6.01, GraphPad Software Inc., San Diego, CA) using the following equation:
where [I] represents the inhibitor concentration, A0 and Amax represent minimum and maximum % P450 activity, respectively, and Hill Slope describes the steepness of the curve. The F-test and the Akaike information criterion were used as measures of goodness of fit. Residual plots were used to decide among standard, Poisson (1/y), or relative (1/y2) weighting.
Time-Dependent Inhibition of P450s by Cannabinoids
The TDI potential of each cannabinoid was screened at 10 μM. The previously reported TDI (Bansal et al., 2020) of CYP1A2, 2C19, and 3A by CBD (10 μM) was repeated as a positive control. Each primary incubation mixture (200 μl) consisted of HLMs (0.5 mg/ml protein), potassium phosphate buffer (100 mM; pH 7.4), BSA (0.2%), and cannabinoid. Blank DMSO (0.5% v/v) was used in place of cannabinoid as a negative control. The mixture was prewarmed at 37°C with constant stirring for 5 minutes, and then the reaction was initiated with the NADPH regenerating system and incubated at 37°C for 0 or 30 minutes. An aliquot (5 μl) of the primary incubation mixture was transferred to a prewarmed (37°C) secondary incubation mixture (195 μl) containing potassium phosphate buffer (pH 7.4), P450 cocktail (50 μM phenacetin, 2 μM coumarin, 5 μM bupropion, 2 μM amodiaquine, 5 μM diclofenac, 20 μM omeprazole, 5 μM dextromethorphan, and 20 μM testosterone), and the NADPH regenerating system. After 15 minutes, the reaction was terminated with 200 μl ice-cold acetonitrile containing tolbutamide (internal standard; 25 nM final concentration) to precipitate the proteins. Reaction mixtures were processed and analyzed as described for the reversible inhibition experiments. Three or four independent experiments were conducted, each singly.
Based on the screening results, cannabinoids that were likely to demonstrate TDI for P450s at pharmacologically relevant concentrations were studied in detail to determine the TDI kinetics. Briefly, varying concentrations of 7-OH CBD (1, 5, 10, 15, 20, 25, 30, 40, or 50 μM) or blank DMSO (0.5% v/v) were preincubated with HLMs (0.5 mg/ml protein) and BSA (0.2%) for 0, 4, 8, 12, or 16 minutes. The assay proceeded as described above. Four independent experiments were conducted, each in singlicate. The observed first-order rate constants for inactivation (kobs) were estimated as described previously (Bansal et al., 2020). The half-maximal inactivation concentration (KI) and maximal inactivation rate constant (kinact) were estimated by nonlinear least-squares regression analysis (GraphPad Prism 6.01, GraphPad Software Inc.) of the kobs versus inactivator concentration ([I]) data using the following equation:
Cannabinoid Plasma Protein Binding
The high nonspecific binding of cannabinoids to plastic and adsorption to filters or membranes preclude the use of ultrafiltration or equilibrium dialysis to determine their fraction unbound in plasma. Therefore, as previously described, the ultracentrifugation method and diluted plasma were used to measure the extent of plasma protein binding of the cannabinoids (Patilea-Vrana and Unadkat, 2019). Briefly, CBD (0.05, 5, or 50 μM), 7-OH CBD (0.25 or 1 μM), or THC (0.5 or 5 μM) was added to low-binding tubes and incubated with diluted plasma (10-fold dilution with Dulbecco’s phosphate buffer saline) (total volume = 200 μl) for 20 minutes at 37°C with shaking to facilitate nonspecific and protein binding. Two sets of these samples were prepared: one set (180 μl/sample) was subjected to ultracentrifugation at 37°C for 90 minutes, and the other (control) set (180 μl/sample) was incubated in the ultracentrifuge tubes at 37°C for 90 minutes without centrifugation. Samples undergoing ultracentrifugation were centrifuged at 435,000 g for 90 minutes at 37°C using the Sorval Discovery M150 SE centrifuge and S100-AT3 rotor (Thermo Scientific). The middle (aqueous) layer that was collected from the sample underwent centrifugation to determine unbound cannabinoid concentration. The total (bound and unbound) concentration of cannabinoid was quantified in the control incubations. Samples were analyzed using LC-MS/MS (described below). Because both sets of samples were prepared in the low-binding tubes and transferred into the ultracentrifuge tubes, the same degree of nonspecific binding was assumed for both sample types. The fraction unbound in undiluted plasma (fu,p) was estimated using the following equation (Schuhmacher et al., 2000), where fu,d represents the fraction unbound in the diluted plasma and DF represents the dilution factor (0.1):
Cannabinoid Protein Binding in the Incubation Mixtures
Although we and Nasrin and colleagues (2021) have previously determined fu,inc of CBD and THC by the tube adsorption method (Bansal et al., 2020), the ultracentrifugation method was used to determine fu,inc of CBD, 7-OH CBD, and THC. This method was used because the tube adsorption method requires determination of the partition ratio of unbound cannabinoid to the walls of the tube in protein-free aqueous buffer. Given the limited solubility of the cannabinoids in protein-free buffer, the ultracentrifugation method avoids this problem. Briefly, CBD (0.1, 1, or 5 μM), 7-OH CBD (1 or 10 μM), or THC (0.5 or 5 μM) was added to low-binding tubes and incubated with HLMs (0.1 or 0.5 mg/ml) in 0.1 mM potassium phosphate buffer (total volume, 200 μl) containing BSA (0.2%) for 15 minutes at 37°C with shaking to facilitate nonspecific and protein binding. Reaction mixtures were processed and analyzed as described for fu,p determination. The fu,inc of each tested cannabinoid was used to compute their corresponding unbound IC50 and KI values. Because 11-OH THC and 7-OH CBD have similar lipophilicity and have a similar chemical structure (ChemDraw Ultra, CambridgeSoft Corporation, Cambridge, MA), the fu,inc for 11-OH THC was assumed to be same as that for 7-OH CBD.
LC-MS/MS Analysis
Each P450-mediated metabolite was quantified using an ultrahigh performance liquid chromatography (UPLC)-MS/MS system which consisted of an ACQUITY UPLC system (Waters, Milford, MA) coupled with a SCIEX 6500 QTRAP mass spectrometer (SCIEX, Framingham, MA) equipped with an electrospray ion source. Chromatography was performed on an ACQUITY UPLC BEH C18 column (2.1 × 50 mm, 1.7 μm) which was maintained at 45°C and was preceded by an ACQUITY UPLC BEH C18 VanGuard pre-column (2.1 × 5 mm, 1.7 μm). The autosampler compartment were set at 4°C to prevent the evaporation of the samples. The sample injection volume was 10 μl. Water containing 0.1% formic acid and acetonitrile containing 0.1% formic acid were used as the mobile phase A and B, respectively, with a flow rate of 0.4 ml/min. The gradient conditions were optimized over 4.5 minutes as follows: 5% B at 0.0–0.3 minutes, linear increase from 5% to 60% B at 0.3–3.0 minutes and from 60% to 90% B at 3.0–4.0 minutes, linear decrease from 90% to 5% B at 4.0–4.2 minutes, and 5% B at 4.2–4.5 minutes. The quantification was performed in the positive multiple reaction monitoring mode after optimizing the compound- and ion source-dependent parameters. Acetaminophen, 7-hydroxycoumarin, 4-hydroxy bupropion, N-desethylamodiaquine, 4’-hydroxy diclofenac, 5-hydroxy omeprazole, dextrorphan, 6β-hydroxy testosterone, and tolbutamide were quantified using the following mass transitions of mass-to-charge ratio: 152.0 → 110.0, 162.9 → 106.9, 256.2 → 238.1, 328.2 → 283.1, 312.0 → 231.0, 362.0 → 214.0, 258.1 → 157.0, 305.3 → 269.1, and 271.3 → 155.0, respectively. The optimized spray voltage, ion source temperature, curtain gas, ion source gas 1, and ion source gas 2 were 5500 V, 600°C, 30 psi, 50 psi, and 50 psi, respectively.
CBD, 7-OH CBD, CBD-d3 (internal standard for CBD and 7-OH CBD assay), THC, and THC-d3 (internal standard for THC assay) were quantified using the same LC-MS/MS system, column, and solvents as mentioned above. The flow rate was optimized to be 0.3 ml/min. The gradient conditions were optimized over 4.5 minutes as follows: 10% B at 0.0–0.5 minutes, linear increase from 10% to 95% B at 0.5–2.0 minutes, 95% B at 2.0–4.0 minutes, linear decrease from 95% to 10% B at 4.0–4.1 minutes, and 10% B at 4.1–4.5 minutes. CBD (or THC), 7-OH CBD, and CBD-d3, (or THC-d3) were quantified using the following mass transitions of mass-to-charge ratio: 315.2 → 193.3, 331.2 → 313.1, and 318.3 → 196.3, respectively. The optimized spray voltage, ion source temperature, curtain gas, ion source gas 1, and ion source gas 2 were 4000 V, 300°C, 30 psi, 50 psi, and 50 psi, respectively. After method development, the LC-MS/MS methods were validated. The calibration curves were linear over the calibrator concentration range used (r2 > 0.97) and the accuracy and precision of the quality control samples (low, middle, and high end of the calibration curves) were within 14% of the nominal concentrations and CV% of <12.5%, respectively.
Prediction of the Potential of the Cannabinoids to Precipitate Pharmacokinetic Drug Interactions
Unbound reversible inhibition (IC50,u) and inactivation (KI,u and kinact) kinetics of CBD, 7-OH CBD, and 11-COOH CBD or THC, 11-OH THC, and 11-COOH THC were incorporated into a previously described mechanistic static model (Fahmi et al., 2008; Cheong et al., 2017) to predict the magnitude of drug-drug interaction (eq. 4). Assuming competitive inhibition, the ki,u of the cannabinoids was assumed to be equal to IC50,u when the P450 inhibition experiments were performed at probe concentration <Km (CYP2B6, 2C19, and 3A) or half the IC50,u when the P450 inhibition experiments were performed at probe concentration ≅ Km (CYP1A2, 2C8, 2C9, and 2D6) (Cheng and Prusoff, 1973). AUCR (the magnitude of drug-drug interaction) represents the ratio of area under the plasma concentration-time curve of the object (probe) drug in the presence (AUC’PO) of the cannabinoid P450 inactivator/inhibitor to that of the probe drug AUC in the absence of the inactivator/inhibitor.
where A is the term that describes TDI of a P450 enzyme in the liver:
B is the term that describes reversible inhibition of a P450 enzyme in the liver:
X is the term that describes TDI of a P450 enzyme in the intestine:
Y is the term that describes reversible inhibition of a P450 enzyme in the intestine:
and n is the number of inactivators/inhibitors (parent and metabolites) present. [I]H and [I]G are the inhibitor/inactivator concentrations in the liver and intestine, respectively. kdeg,H and kdeg,G represent the degradation rates of the P450 in the liver and intestine, respectively. The fraction of the object drug metabolized by a given P450 is represented by fm, and the fraction of the object drug escaping intestinal metabolism is represented by FG. The fm and FG values of object drugs and kdeg values of P450 enzymes are provided in Supplemental Table 1.
A variation of the four different models (Models A–D) described by Tseng et al. (2021) was used to predict the magnitude of a pharmacokinetic cannabinoid-drug interaction in the liver or intestine after a single (THC) or multiple (CBD) doses of oral or inhaled cannabinoid (liver only) administration (Bansal et al., 2020). AUCR values were predicted using the estimated [I]H and [I]G concentrations (Supplemental Table 2). [I]H was estimated to be either the unbound maximum hepatic inlet concentration (Cmax,hepatic inlet,u, Model A), the unbound maximum systemic concentration (Cmax,sys,u, Model B) or, the unbound average systemic concentration (Cave,sys,u, Models C and D) (Tseng et al., 2021). [I]G of the cannabinoids, due to their poor aqueous solubility, was estimated as their maximum intestinal fluid solubility (Models A–C) (Bansal et al., 2020) or the unbound average portal vein concentration (Cave,portal,u, Model D) to predict any interaction in the intestine (Tseng et al., 2021).
To predict drug interactions precipitated by the cannabinoid metabolites 7-OH CBD, 7-COOH CBD, 11-OH THC, or 11-COOH THC, the Cmax,sys,u and Cave,sys,u of each was set equal to [I]H. The doses (and justification) of CBD and THC to predict in vivo hepatic and intestinal P450-mediated drug interactions were the same as described previously (Bansal et al., 2020). Briefly, the average low, average high, and maximum doses of CBD or THC used for recreational or medicinal purposes were estimated from those reported in clinical trials, case reports, social media, cannabinoid vendor websites, and newspapers (Bansal et al., 2020). The oral doses for CBD were 70 mg, 700 mg, and 2,000 mg and for THC were 20 mg, 130 mg, and 160 mg. The inhaled doses of THC were 25 mg, 75 mg, and 100 mg.
Results
Binding of CBD, 7-OH CBD, or THC to Plasma Proteins or in Microsomal Incubations
Because the fu,p of CBD, 7-OH CBD, and THC was concentration-independent, the data at all concentrations tested were pooled (Supplemental Table 3). The cannabinoids were highly bound to plasma proteins (>98%). THC demonstrated higher degree of binding to plasma proteins as compared with CBD or 7-OH CBD; the latter two had comparable protein binding (Supplemental Table 3). The fu,inc of CBD or 7-OH CBD was similar (P > 0.05) despite using different HLM concentrations (0.1 or 0.5 mg/ml) but the same BSA concentration (0.2%). THC behaved in a similar manner (Patilea-Vrana and Unadkat, 2019), indicating that the binding of these cannabinoids in the incubation mixture is driven by the BSA concentration (Supplemental Table 4). Because the fu,inc of CBD, 7-OH CBD, or THC was concentration-independent, values at all concentrations (for each cannabinoid) for a given HLM concentration were pooled (Supplemental Table 4). THC and CBD had similar degree of binding to proteins in the incubations, whereas that of 7-OH CBD was lower (Supplemental Table 4).
Reversible Inhibition of P450s by CBD, THC, and Their Metabolites
CBD and 7-OH CBD inhibited CYP2B6, 2C8, 2C9, and 2D6 by similar extents, whereas CBD was a more potent inhibitor of CYP2C19 and 3A than 7-OH CBD. 7-COOH CBD was a weak inhibitor of CYP2B6, 2C8, 2C9, and 2D6 (Supplemental Fig. 1). CBD and its metabolites were weak inhibitors of CYP2A6. THC and 11-OH THC (10 μM each) inhibited CYP1A2, 2B6, 2C8, 2C9, 2C19, and 2D6, but only THC inhibited CYP3A. Like CBD and its metabolites, THC and 11-OH THC were weak inhibitors of CYP2A6. 11-COOH THC did not inhibit any of the P450s tested.
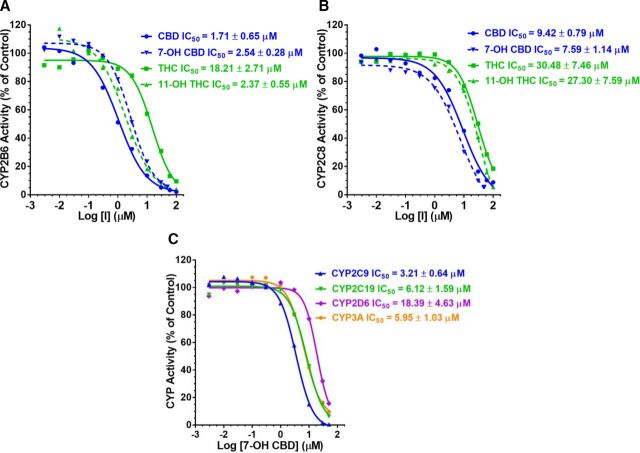
CBD, 7-OH CBD, THC, and 11-OH THC inhibited CYP2B6 and 2C8 in a concentration-dependent manner (Fig. 2). The CV% for IC50 estimates was <25%. Regarding the narrative below, potency refers to the unbound IC50 values (IC50,u) which were calculated from IC50 values (Supplemental Table 5) using fu,inc (Supplemental Table 4). Among the cannabinoids tested, CBD was the most potent inhibitor of CYP2B6 (Fig. 2, Table 1). CBD inhibited CYP2B6 with ∼8-fold greater potency than 7-OH CBD, THC, or 11-OH THC; the latter three inhibited CYP2B6 with comparable potency (Table 1). CBD was also the most potent inhibitor of CYP2C8, being 3.6- and 2.3-fold more potent than 7-OH CBD and THC, respectively; THC was a 5.5 times more potent inhibitor of CYP2C8 than 11-OH THC (Table 1).
TABLE 1.
IC50,u values of CBD, 7-OH CBD, THC, and 11-OH THC for P450s using HLMs
| Enzyme | CBD | 7-OH CBD | THC | 11-OH THC |
|---|---|---|---|---|
| IC50,ua (μM) | IC50,ua (μM) | IC50,ua (μM) | IC50,ua,b (μM) | |
| CYP2B6 | 0.05 ± 0.02 | 0.34 ± 0.04 | 0.40 ± 0.06 | 0.32 ± 0.07 |
| CYP2C8 | 0.28 ± 0.02 | 1.02 ± 0.15 | 0.67 ± 0.16 | 3.66 ± 1.02 |
| CYP1A2 | 0.11 ± 0.04 | >13.4 | 0.028 ± 0.007 | 1.81 ± 0.61 |
| CYP2C9 | 0.04 ± 0.01 | 0.43 ± 0.09 | 0.005 ± 0.001 | 0.36 ± 0.12 |
| CYP2C19 | 0.08 ± 0.01 | 0.82 ± 0.21 | 0.252 ± 0.098 | 1.76 ± 0.26 |
| CYP2D6 | 0.24 ± 0.12 | 2.46 ± 0.62 | 0.561 ± 0.108 | 5.25 ± 1.03 |
| CYP3A | 0.09 ± 0.03 | 0.80 ± 0.14 | 0.574 ± 0.149 | >6.7 |
aIC50,u values were calculated using the IC50 values provided in Supplemental Table 5.
bfu,inc for 11-OH THC was assumed to be same as 7-OH CBD.
Data shown are mean ± S.D. of three independent experiments, each conducted in duplicate.
Fig. 2.
Concentration-dependent inhibition of (A) CYP2B6 or (B) 2C8 activity in HLMs by CBD, 7-OH CBD, THC, or 11-OH THC and (C) concentration-dependent inhibition of CYP2C9, 2C19, 2D6, and 3A by 7-OH CBD. Of the cannabinoid tested, CBD was the most potent inhibitor of CYP2B6 and 2C8 (see Table 1). The order of 7-OH CBD inhibition potency was CYP2B6 ≈ 2C9 > 2C19 ≈ 3A > 2C8 > 2D6 (Table 1). Data shown are from a representative experiment conducted in duplicate. Solid lines represent model fit to the data.
7-OH CBD inhibited CYP2C9, 2C19, 2D6, and 3A with the order of potency CYP2C9 > 2C19 ≈ 3A > 2D6 (Fig. 2C, Table 1). The potency of 7-OH CBD to inhibit these enzymes was one-tenth that of CBD (Table 1). Like CBD, 7-OH CBD inhibited CYP2C9 with 50% less potency against CYP2C19 or 3A and inhibited 2C9 and 2B6 with similar potency (Table 1). The IC50 of 7-OH CBD against CYP1A2 and 7-COOH CBD against all the P450s could not be determined at the concentration range tested.
TDI of P450s by CBD, THC, and Their Metabolites
Of the P450s tested, 7-OH CBD (10 μM) demonstrated TDI of only CYP2C19 and 3A (Supplemental Fig. 2). Unlike CBD (Bansal et al., 2020), 7-OH CBD (10 μM) did not show TDI of CYP1A2 (Supplemental Fig. 2). TDI of P450s 1A2, 2C19, and 3A by CBD was repeated in the current study, and the data were consistent with our previously published findings (Bansal et al., 2020). CBD was not a time-dependent inhibitor of CYP2A6, 2B6, and 2C8 (data not shown). 7-COOH CBD was not a time-dependent inhibitor of any of the P450s tested (data not shown).
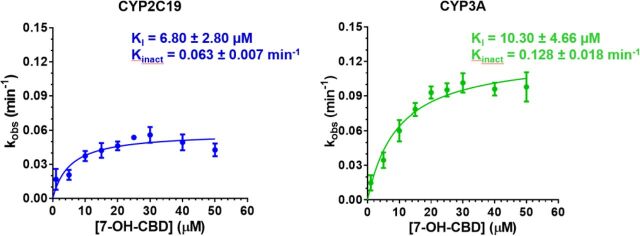
The inhibition of CYP2C19 and 3A activities by 7-OH CBD was concentration- and time-dependent (Supplemental Fig. 3). The KI for 7-OH CBD (Fig. 3, Supplemental Table 6) and the previously reported KI of CBD (Bansal et al., 2020) were converted to unbound KI (KI,u) using fu,inc (Supplemental Table 4). The CV% for KI or kinact estimates was <28%. 7-OH CBD inactivated CYP2C19 with similar efficiency (kinact/KI,u) as CYP3A, which was approximately one-sixth that of CBD (Table 2).
TABLE 2.
Kinetics of time-dependent inhibition of CYPC19 and 3A by CBD or 7-OH CBD in HLMs
| Enzyme | CBDa | 7-OH CBDb | ||||
|---|---|---|---|---|---|---|
| KI,u (μM) | kinact (min−1) | kinact/KI,u (min−1 μM−1) | KI,u (μM) | kinact (min−1) | kinact/KI,u (min−1 μM−1) | |
| CYP2C19 | 0.07 ± 0.04 | 0.035 ± 0.006 | 0.62 ± 0.32 | 0.67 ± 0.27 | 0.063 ± 0.007 | 0.10 ± 0.02 |
| CYP3A | 0.11 ± 0.05 | 0.078 ± 0.022 | 0.79 ± 0.20 | 1.01 ± 0.46 | 0.128 ± 0.018 | 0.14 ± 0.05 |
aKI,u was calculated using the reported KI of CBD (Bansal et al., 2020).
bKI,u was calculated using the KI of 7-OH CBD provided in Supplemental Table 6.
Data are mean ± S.D. of four independent experiments, each conducted singly.
Fig. 3.
Kinetics of time-dependent inhibition of CYP2C19 and 3A by 7-OH CBD. Estimates of kinact and KI, obtained by fitting appropriate equations to the kobs data by nonlinear least-squares regression, showed that 7-OH CBD inactivated CYP2C19 and 3A with comparable efficiency (kinact/KI,u) but with lesser efficiency than CBD (positive control) (Table 2). Data shown are mean ± S.D. of four independent experiments, each conducted singly.
Prediction of In Vivo P450-Mediated Cannabinoid-Drug Interactions
The estimated maximum plasma concentrations of the cannabinoids and metabolites or the calculated maximum intestinal fluid solubility at various doses of the cannabinoids consumed for recreational or medicinal purposes were used to predict the magnitude of in vivo cannabinoid-drug interactions (Supplemental Table 2). Based on the predicted AUCR values using the Cmax,hepatic inlet,u (Model A), CBD at high oral doses (700 and 2000 mg) was predicted to precipitate pharmacokinetic interactions with drugs that are predominantly metabolized by CYP2B6 (ticlopidine AUCR > 2.1) or 2C8 (repaglinide AUCR > 1.5) (Table 3). However, except for a modest interaction with ticlopidine at the maximum dose (2000 mg), CBD was predicted not to produce these drug interactions using its unbound systemic plasma concentration (Models B–D) (Table 3). The predicted AUCR for CBD-ticlopidine or -repaglinide interactions was not affected by the inclusion of 7-OH CBD formed in vivo after CBD administration (data not shown). THC (oral or inhaled) and 11-OH THC were not predicted to precipitate interactions with ticlopidine or repaglinide, even at the highest doses consumed (Table 3).
TABLE 3.
Predicted magnitude of pharmacokinetic interactions between CBD or THC interaction and drugs extensively metabolized by P450 enzymes using different static models
| Precipitant | P450Enzyme | Object Drug | Predicted AUCR after Oral Administration of the Precipitant (Inhibition of Hepatic and Gut Metabolism) | Predicted AUCR after Inhalation of the Precipitant (Inhibition of Hepatic Metabolism) | ||||
|---|---|---|---|---|---|---|---|---|
| Model | Model | |||||||
| Aa | Bb | Cc | Dd | Be | C or Df | |||
| CBD (70 mg, Oral) | 1A2 | Theophylline | 3.9 | 3.2 | 2.7 | 2.7 | — | — |
| 2B6 | Ticlopidine | 1.2 | 1.0 | 1.0 | 1.0 | — | — | |
| 2C8 | Repaglinide | 1.1 | 1.0 | 1.0 | 1.0 | — | — | |
| 2C9 | Diclofenac | 2.4 | 1.6 | 1.6 | 1.1 | — | — | |
| 2C19 | Omeprazole | 5.3 | 1.9 | 1.5 | 1.5 | — | — | |
| 2D6 | Dextromethorphan | 1.1 | 1.0 | 1.0 | 1.0 | — | — | |
| 3A | Midazolam | 12.3 | 5.1 | 3.5 | 3.2 | — | — | |
| CBD (700 mg, Oral) | 1A2 | Theophylline | 4.0 | 3.9 | 3.8 | 3.8 | — | — |
| 2B6 | Ticlopidine | 2.2 | 1.2 | 1.1 | 1.1 | — | — | |
| 2C8 | Repaglinide | 1.6 | 1.1 | 1.0 | 1.0 | — | — | |
| 2C9 | Diclofenac | 9.6 | 2.3 | 1.9 | 1.6 | — | — | |
| 2C19 | Omeprazole | 7.3 | 5.1 | 3.9 | 3.9 | — | — | |
| 2D6 | Dextromethorphan | 2.1 | 1.1 | 1.0 | 1.0 | — | — | |
| 3A | Midazolam | 14.8 | 12.0 | 9.8 | 9.7 | — | — | |
| CBD (2000 mg, Oral) | 1A2 | Theophylline | 4.0 | 3.9 | 3.9 | 3.9 | — | — |
| 2B6 | Ticlopidine | 3.1 | 1.4 | 1.2 | 1.2 | — | — | |
| 2C8 | Repaglinide | 2.2 | 1.2 | 1.1 | 1.1 | — | — | |
| 2C9 | Diclofenac | 20.7 | 3.8 | 2.6 | 2.4 | — | — | |
| 2C19 | Omeprazole | 7.6 | 6.5 | 5.6 | 5.6 | — | — | |
| 2D6 | Dextromethorphan | 4.1 | 1.3 | 1.1 | 1.1 | — | — | |
| 3A | Midazolam | 15.0 | 13.9 | 12.6 | 12.6 | — | — | |
| THC (20 mg, Oral) | 1A2 | Theophylline | 1.1 | 1.0 | 1.0 | 1.0 | — | — |
| 2B6 | Ticlopidine | 1.0 | 1.0 | 1.0 | 1.0 | — | — | |
| 2C8 | Repaglinide | 1.0 | 1.0 | 1.0 | 1.0 | — | — | |
| 2C9 | Diclofenac | 2.1 | 1.6 | 1.6 | 1.1 | — | — | |
| 2C19 | Omeprazole | 1.0 | 1.0 | 1.0 | 1.0 | — | — | |
| 2D6 | Dextromethorphan | 1.0 | 1.0 | 1.0 | 1.0 | — | — | |
| 3A | Midazolam | 1.8 | 1.8 | 1.8 | 1.0 | — | — | |
| THC (130 mg, Oral) | 1A2 | Theophylline | 1.4 | 1.0 | 1.0 | 1.0 | — | — |
| 2B6 | Ticlopidine | 1.0 | 1.0 | 1.0 | 1.0 | — | — | |
| 2C8 | Repaglinide | 1.0 | 1.0 | 1.0 | 1.0 | — | — | |
| 2C9 | Diclofenac | 6.6 | 2.1 | 1.7 | 1.4 | — | — | |
| 2C19 | Omeprazole | 1.0 | 1.0 | 1.0 | 1.0 | — | — | |
| 2D6 | Dextromethorphan | 1.0 | 1.0 | 1.0 | 1.0 | — | — | |
| 3A | Midazolam | 1.8 | 1.8 | 1.8 | 1.0 | — | — | |
| THC (160 mg, Oral) | 1A2 | Theophylline | 1.5 | 1.1 | 1.0 | 1.0 | — | — |
| 2B6 | Ticlopidine | 1.0 | 1.0 | 1.0 | 1.0 | — | — | |
| 2C8 | Repaglinide | 1.0 | 1.0 | 1.0 | 1.0 | — | — | |
| 2C9 | Diclofenac | 7.6 | 2.3 | 1.7 | 1.4 | — | — | |
| 2C19 | Omeprazole | 1.0 | 1.0 | 1.0 | 1.0 | — | — | |
| 2D6 | Dextromethorphan | 1.1 | 1.0 | 1.0 | 1.0 | — | — | |
| 3A | Midazolam | 1.8 | 1.8 | 1.8 | 1.0 | — | — | |
| THC (20 mg, Inhaled) | 1A2 | Theophylline | — | — | — | — | 1.1 | 1.0 |
| 2B6 | Ticlopidine | — | — | — | — | 1.0 | 1.0 | |
| 2C8 | Repaglinide | — | — | — | — | 1.0 | 1.0 | |
| 2C9 | Diclofenac | — | — | — | — | 1.5 | 1.1 | |
| 2C19 | Omeprazole | — | — | — | — | 1.0 | 1.0 | |
| 2D6 | Dextromethorphan | — | — | — | — | 1.0 | 1.0 | |
| 3A | Midazolam | — | — | — | — | 1.0 | 1.0 | |
| THC (75 mg, Inhaled) | 1A2 | Theophylline | — | — | — | — | 1.2 | 1.0 |
| 2B6 | Ticlopidine | — | — | — | — | 1.0 | 1.0 | |
| 2C8 | Repaglinide | — | — | — | — | 1.0 | 1.0 | |
| 2C9 | Diclofenac | — | — | — | — | 2.3 | 1.2 | |
| 2C19 | Omeprazole | — | — | — | — | 1.0 | 1.0 | |
| 2D6 | Dextromethorphan | — | — | — | — | 1.0 | 1.0 | |
| 3A | Midazolam | — | — | — | — | 1.0 | 1.0 | |
| THC (100 mg, Inhaled) | 1A2 | Theophylline | — | — | — | — | 1.2 | 1.0 |
| 2B6 | Ticlopidine | — | — | — | — | 1.0 | 1.0 | |
| 2C8 | Repaglinide | — | — | — | — | 1.0 | 1.0 | |
| 2C9 | Diclofenac | — | — | — | — | 2.9 | 1.4 | |
| 2C19 | Omeprazole | — | — | — | — | 1.0 | 1.0 | |
| 2D6 | Dextromethorphan | — | — | — | — | 1.0 | 1.0 | |
| 3A | Midazolam | — | — | — | — | 1.0 | 1.0 | |
kapredicted AUCR using [I]H = Cmax,hepatic inlet,u in eq. 5 and 6 and [I]G = calculated maximum intestinal fluid solubility in eq. 7 and 8
bpredicted AUCR using [I]H = Cmax,sys,u in eq. 5 and 6 and [I]G = calculated maximum intestinal fluid solubility in eq. 7 and 8
cpredicted AUCR using [I]H = Cave,sys,u in eq. 5 and 6 and [I]G = calculated maximum intestinal fluid solubility in eq. 7 and 8
epredicted AUCR using only [I]H = Cmax,sys,u in eq. 6
fpredicted AUCR using only [I]H = Cave,sys,u in eq. 6
Dashes (—) in cells indicate not relevant.
Bold values represent AUCR > 1.25 (cutoff recommended by US Food and Drug Administration).
Cmax,hepatic inlet,u and Cave,portal,u of CBD and THC and Cmax,sys,u and Cave,sys,u of CBD, THC, and their metabolites are provided in Supplemental Table 2. Maximum intestinal fluid solubility of CBD and THC is from Bansal et al. (2020).
Although the AUCR for CBD or THC (oral or inhaled) interactions with prototypic P450 substrates metabolized by CYP1A2 (theophylline), 2C9 (diclofenac), 2C19 (omeprazole), 2D6 (dextromethorphan), or 3A (midazolam) were predicted based on the protein binding data obtained from the current study (therefore revised IC50,u), these values were not different from those in our previous publication (Bansal et al., 2020) when predicted using the Cmax,hepatic inlet,u (Model A). That is, at the higher doses, oral CBD was predicted to precipitate drug interactions with drugs metabolized extensively by CYP1A2, 2C9, 2C19, 2D6, and 3A (Table 3). Oral THC was predicted to precipitate modest drug interactions with drugs metabolized extensively by CYP1A2 and 3A and much larger interaction with those extensively metabolized by CYP2C9. Inhaled THC was predicted to precipitate interactions with only CYP2C9-metabolized drugs (Table 3). However, compared with Model A, lower AUCR was predicted using Model B, C, or D. Moreover, Models C and D (using Cave,sys,u) did not predict any interaction between CBD and CYP2D6 substrate. Oral THC was predicted not to interact with CYP3A substrate when using Cave,portal,u as the enterocyte concentration (Model D vs. Model C) (Table 3).
Discussion
Using HLMs, we previously determined the unbound inhibition potency (IC50,u or KI,u) of CBD, THC, 11-OH THC, and 11-COOH THC toward CYP1A2, 2C9, 2C19, 2D6, or 3A (Bansal et al., 2020). To complete the assessment of the potential for CBD or THC (and their in vivo circulating metabolites) to precipitate P450-mediated cannabinoid-drug interactions, we determined for the first time the ability of CBD, THC, 7-OH CBD, 7-COOH CBD, 11-OH THC, and 11-COOH THC to reversibly and irreversibly inhibit (TDI) CYP2A6, 2B6, and 2C8 activities in HLMs. In addition, we determined the ability of the in vivo circulating metabolites of CBD to reversibly and irreversibly inhibit CYP1A2, 2C9, 2C19, 2D6, and 3A activity in HLMs. To mitigate cannabinoid nonspecific binding to labware and to increase their aqueous solubility, BSA (0.2%) was added to the incubation mixture. Incubations were conducted in low-binding Eppendorf tubes to further reduce cannabinoid nonspecific binding. In addition to determining unbound inhibition potency (IC50,u or KI,u), the fu,inc and fu,p of the cannabinoids were determined by ultracentrifugation, a method best used to determine the fraction unbound of highly bound drugs. The fu,inc of CBD and THC was 25% and 40%, respectively, of that determined by tube adsorption method (Bansal et al., 2020), and the fu,p of THC was 50% of that determined previously (Patilea-Vrana and Unadkat, 2019).
CBD and THC demonstrated potent reversible inhibition of CYP2B6 and weak reversible inhibition of 2A6, consistent with a previous report (Yamaori et al., 2011b; Nasrin et al., 2021). However, the previously reported CYP2B6 IC50 and IC50,u values were approximately 50- to 200-fold and 2.4- to 5-fold higher, respectively, than those determined in the current work, substantiating the necessity to consider the low aqueous solubility and nonspecific binding of cannabinoids when determining their inhibition potency (Bansal et al., 2020). The IC50 of CBD was comparable to that for the CYP2B6-selective inhibitor thio-tepa (IC50 = 1.75 μM), and was approximately 20% that of ticlopidine (IC50 = 0.32 μM), which to date is the most potent mixed-type (reversible and irreversible) inhibitor of CYP2B6 (Turpeinen et al., 2004). Ideally, these comparisons should be made using IC50,u, but the IC50,u values for these drugs are not available in the literature. Overall, these results suggest that CBD and THC are potent inhibitors of CYP2B6.
Here, we report for the first time that both CBD and THC are potent inhibitors of CYP2C8 (IC50,u < 0.7 μM). Nasrin and colleagues did not observe CYP2C8 inhibition by CBD or THC (Nasrin et al., 2021). Low aqueous solubility and nonspecific binding of these cannabinoids may be a plausible explanation for this discrepancy. As a comparison, the IC50 of CBD (IC50 = 9.42 μM) was approximately 500 times greater than that of montelukast (IC50 = 0.02 μM), the most potent inhibitor of CYP2C8 reported to date. However, this value was comparable to that of nifedipine, lovastatin, clopidogrel, irbesartan, and amlodipine (IC50 ∼10 μM) (Walsky et al., 2005). Again, IC50,u values of these drugs would provide the best comparison of their potencies, but these values are not available in the literature.
CBD, THC, and their metabolites did not demonstrate TDI of CYP2A6, 2B6, or 2C8 in HLMs. These results, except for the lack of TDI of CYP2A6 by THC, were consistent with a previous report in which Yamaori and colleagues (2011b) showed that preincubation of THC with recombinant CYP2A6 yielded weak to moderately potent inactivation of CYP2A6 (kinact/KI = 0.02 minutes−1 μM−1). A plausible explanation for this discrepancy could be the use of HLMs in the current study instead of recombinant CYP2A6. HLMs represent a complete P450 system that includes CYP2C9, which rapidly metabolizes THC (Patilea-Vrana and Unadkat, 2019). 7-OH CBD displayed TDI of CYP2C19 and 3A but showed 17% of the potency of CBD. COOH-CBD did not show TDI of these P450s. These results suggest that hydroxylation or carboxylation of CBD at the C7 position (Fig. 1) diminished or abolished the ability of the metabolite to inactivate CYP2C19 or CYP3A.
In this study, drug interaction (as measured by AUCR) was predicted using plasma concentrations observed after multiple doses of CBD or a single dose of THC. This is because CBD accumulates after multiple dosing (Taylor et al., 2018), likely due to autoinhibition of its metabolism (Bansal et al., 2020). In contrast, THC is predicted not to accumulate to a significant extent, as THC is not a TDI of any P450s and the terminal phase of THC concentration-time curve does not account for a significant portion of its AUC0–∞ (Patilea-Vrana and Unadkat, 2021).
One of the most important steps in accurately predicting drug interaction is to estimate the hepatic and intestinal concentrations of the inhibitor. To do so, several models have been proposed (Bansal et al., 2020; Tseng et al., 2021). Here, we implemented a variation of these models. The magnitude of pharmacokinetic interactions between each cannabinoid and CYP2B6 or CYP2C8 probe substrates was predicted using Cmax,hepatic inlet,u, Cmax,sys,u, or Cave,sys,u as [I]H and the calculated maximum intestinal fluid solubility or Cave,portal,u as [I]G (Bansal et al., 2020; Tseng et al., 2021). Using Cmax,hepatic inlet,u as [I]H (Model A), a CBD oral dose of 700 mg was predicted to precipitate moderate (2 ≤ AUCR < 5) or weak (1.2 < AUCR < 2) interactions (https://www.fda.gov/media/134582/download) with drugs extensively metabolized (fm ∼0.75) by CYP2B6 or CYP2C8, respectively. If Cave,sys,u (vs. Cmax,hepatic inlet,u) is used as an estimate of [I]H and Cave,portal,u (vs. maximum cannabinoid solubility) as an estimate of [I]G (Model D), then CBD (700 mg) was predicted not to produce CYP2B6- or 2C8-mediated drug interactions. However, based on the in vitro data, the label for Epidiolex (CBD) recommends adjustment in dosage of substrates of CYP2C8 and CYP2B6 be considered, if clinically appropriate, when administered concomitantly with Epidiolex (https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/210365lbl.pdf).
Despite inhibition or inactivation by 7-OH CBD of the P450s tested, 7-OH CBD formed after oral administration of CBD was not predicted (based on 7-OH CBD Cmax,sys,u or Cave,sys,u) to enhance CBD-drug interactions. 7-OH CBD may be formed in the intestine by CYP3A (Jiang et al., 2011), and its Cmax,hepatic inlet,u could be higher than Cmax,sys,u. However, until the fraction of CBD metabolized to 7-OH CBD by intestinal CYP3A is known, it is not possible to correctly estimate 7-OH CBD Cmax,hepatic inlet,u. This fraction could be predicted using physiologically based pharmacokinetic modeling and simulation. 11-OH THC was also predicted to have no effect on the magnitude of THC-drug interactions after oral or inhalational administration.
As we predicted earlier (Bansal et al., 2020), CBD was predicted to precipitate drug interaction with CYP1A2, 2C9, 2C19, 2D6, or 3A substrates using Model A (Table 3). Drug interactions predicted with CYP1A2, 2C19, or 3A substrates were mostly due to TDI of these enzymes by CBD. Of the different static models used, as expected, Model A predicted the highest AUCR, whereas Model D predicted the lowest AUCR. Our predictions are largely consistent with the reported clinical CBD-drug interactions. For example, CBD (5–25 mg/kg per day or 750 mg twice daily orally) increases the AUC and Cmax (3- to 5-fold) of N-desmethylclobazam, which is metabolized predominantly by CYP2C19 (Geffrey et al., 2015; Gaston and Szaflarski, 2018; Morrison et al., 2019). CBD (750 mg twice daily) increases the AUC of caffeine, a CYP1A2-metabolized drug, by 2-fold (Thai et al., 2021). CBD (1000 mg twice daily) and CBD (400–800 mg daily) increase the Cmax of tacrolimus and citalopram, drugs cleared predominately by CYP3A enzymes, by 3-fold and 2-fold, respectively (Leino et al., 2019; Anderson et al., 2021). Surprisingly, CBD (dose unknown) does not affect midazolam plasma concentrations (https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/210365lbl.pdf), which could possibly be due to CYP3A induction (https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/210365Orig1s000ClinPharmR.pdf). However, it is puzzling as to why such induction would not affect the interaction with tacrolimus or citalopram. Due to the route of administration, inhaled THC-drug interactions were predicted using only Cmax,sys,u or Cave,sys,u and showed only CYP2C9-mediated drug interactions and not CYP1A2 or 3A interactions predicted for oral THC (Table 3). Except for the warfarin case study described in the introduction, well controlled THC-drug interaction studies (inhibition or induction) have not been reported in the literature. Although smoked cannabis appears to induce CYP1A2 (Jusko et al., 1978), it is not clear if this is due to THC or other constituents in the smoke (e.g., arylamines) that are known to induce this enzyme.
There are several uncertainties associated with the current static model predictions, which may affect the predicted magnitude of cannabinoid-drug interactions. First, the depletion of CBD, 7-OH CBD, THC, and 11-OH THC during incubations could result in the estimated IC50 or KI values that are higher than the true values. Second, the Cmax,hepatic inlet,u of the metabolites was not considered when predicting AUCRs for CBD- or THC-drug interactions after oral administration. Third, the predicted cannabinoid-drug interactions, based on static inhibitor concentrations, varied depending on the model used. Although Tseng et al. (2021) have proposed that Model D be used to predict such interactions, as to whether this model will accurately predict in vivo cannabinoid-drug interactions can only be confirmed by conducting such an interaction study. Fourth, static models do not consider the change in CBD, THC, or their metabolite concentrations with time in vivo. Therefore, we are developing a dynamic physiologically based pharmacokinetic model to capture these changes and predict the magnitude of drug interactions with CBD, THC, and their metabolites formed after oral or inhalational administration. Lastly, the TDI parameters (KI and kinact) obtained in HLMs might differ from those obtained in human hepatocytes due to proteins present in hepatocytes, but not in HLMs, that could neutralize the generated reactive metabolite(s) causing TDI of enzymes. Lastly, inhibition of non-P450 enzymes such as UGT1A9 and 2B7 by CBD or THC (Bansal et al., 2021) should be considered when predicting interactions with drugs metabolized by P450 or UGT1A9/2B7.
In conclusion, using mechanistic static models, we predicted modest or no (depending on the model used) pharmacokinetic interactions between CBD (>700 mg orally), but not THC, and drugs predominantly metabolized by CYP2B6 or 2C8. P450 inhibition exhibited by 7-OH CBD or 11-OH THC did not enhance the magnitude of drug interactions with CBD or THC after oral or inhalational administration. Overall, at higher oral doses (>700 mg) and for all the models used, CBD was predicted to precipitate interactions with drugs metabolized by all of the P450s examined except CYP2A6 and for some models CYP2D6. The order of magnitude of maximum drug interactions predicted at the highest dose (2000 mg) was CYP3A > 2C9 > 2C19 > 2D6 > 1A2 > 2B6 > 2C8. Oral or inhaled THC was predicted to precipitate interactions only with CYP2C9 substrates. In vivo drug interaction studies are needed to verify these predictions.
ABBREVIATIONS
- AUC
area under the plasma concentration-time curve
- AUCR
ratio of AUC of object drug in the presence to absence of inhibitor
- BSA
bovine serum albumin
- Cave, portal, u
unbound average portal vein concentration
- Cave, sys, u
unbound average systemic concentration
- CBD
cannabidiol
- CBD-d3
deuterated CBD
- Cmax, hepatic inlet, u
unbound maximum hepatic inlet concentration
- Cmax, sys, u
unbound maximum systemic concentration
- 7-COOH CBD
7-carboxy CBD
- 11-COOH THC
11-nor-9-carboxy-delta-9-THC
- fm
fraction metabolized
- fu, inc
fraction unbound in incubation containing HLM and BSA
- fu, p
fraction unbound in plasma
- HLM
human liver microsome
- IC50, u
unbound half-maximal inhibitory concentration
- [I]G
inhibitor/inactivator concentration in the intestine
- [I]H
inhibitor/inactivator concentration in the liver
- KI
half-maximal inactivation concentration
- kinact
maximum inactivation rate constant
- KI, u
unbound KI
- kobs
first-order rate constant for inactivation
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- 7-OH CBD
7-hydroxy CBD
- 11-OH THC
11-hydroxy THC
- P450
cytochrome P450
- TDI
time-dependent inhibition
- THC
(−)-trans-delta-9-tetrahydrocannabinol
- THC-d3
deuterated THC
- UPLC
ultrahigh performance liquid chromatography
Authorship Contributions
Participated in research design: Bansal, Unadkat.
Conducted experiments: Bansal.
Performed data analysis: Bansal.
Wrote or contributed to the writing of the manuscript: Bansal, Paine, Unadkat.
Footnotes
This work was supported by National Institutes of Health National Center for Complementary and Integrative Health and Office Dietary Supplements [Grant U54-AT008909] (to M.F.P.) and in part by National Institutes of Health National Institute on Drug Abuse [Grant P01-DA032507] (to J.D.U.).
No author has an actual or perceived conflict of interest with the contents of this article.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Amaral Silva D, Pate DW, Clark RD, Davies NM, El-Kadi AOS, Löbenberg R (2020) Phytocannabinoid drug-drug interactions and their clinical implications. Pharmacol Ther 215:107621. [DOI] [PubMed] [Google Scholar]
- Anderson LL, Doohan PT, Oldfield L, Kevin RC, Arnold JC, Berger M, Amminger GP, McGregor IS (2021) Citalopram and cannabidiol: in vitro and in vivo evidence of pharmacokinetic interactions relevant to the treatment of anxiety disorders in young people. J Clin Psychopharmacol 41:525–533. [DOI] [PubMed] [Google Scholar]
- Arnold WR, Weigle AT, Das A (2018) Cross-talk of cannabinoid and endocannabinoid metabolism is mediated via human cardiac CYP2J2. J Inorg Biochem 184:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S, Maharao N, Paine MF, Unadkat JD (2020) Predicting the potential for cannabinoids to precipitate pharmacokinetic drug interactions via reversible inhibition or inactivation of major cytochromes P450. Drug Metab Dispos 48:1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S, Paine M, Unadkat J (2021) Can cannabinoids precipitate UGT-mediated drug interactions? FASEB J 35:03854. [Google Scholar]
- Beers JL, Fu D, Jackson KD (2021) Cytochrome P450-catalyzed metabolism of cannabidiol to the active metabolite 7-hydroxy-cannabidiol. Drug Metab Dispos 49:882–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH (1973) Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 22:3099–3108. [DOI] [PubMed] [Google Scholar]
- Cheong EJY, Goh JJN, Hong Y, Venkatesan G, Liu Y, Chiu GNC, Kojodjojo P, Chan ECY (2017) Application of static modeling --in the prediction of in vivo drug-drug interactions between rivaroxaban and antiarrhythmic agents based on in vitro inhibition studies. Drug Metab Dispos 45:260–268. [DOI] [PubMed] [Google Scholar]
- Cox EJ, Maharao N, Patilea-Vrana G, Unadkat JD, Rettie AE, McCune JS, Paine MF (2019) A marijuana-drug interaction primer: precipitants, pharmacology, and pharmacokinetics. Pharmacol Ther 201:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlinger D, Duechting S, Nuecken D, Sydow K, Fuhr U, Frechen S (2016) Development and validation of an in vitro, seven-in-one human cytochrome P450 assay for evaluation of both direct and time-dependent inhibition. J Pharmacol Toxicol Methods 77:66–75. [DOI] [PubMed] [Google Scholar]
- Dinger J, Meyer MR, Maurer HH (2014) Development and validation of a liquid-chromatography high-resolution tandem mass spectrometry approach for quantification of nine cytochrome P450 (CYP) model substrate metabolites in an in vitro CYP inhibition cocktail. Anal Bioanal Chem 406:4453–4464. [DOI] [PubMed] [Google Scholar]
- Fahmi OA, Maurer TS, Kish M, Cardenas E, Boldt S, Nettleton D (2008) A combined model for predicting CYP3A4 clinical net drug-drug interaction based on CYP3A4 inhibition, inactivation, and induction determined in vitro. Drug Metab Dispos 36:1698–1708. [DOI] [PubMed] [Google Scholar]
- Frytak S, Moertel CG, Rubin J (1984) Metabolic studies of delta-9-tetrahydrocannabinol in cancer patients. Cancer Treat Rep 68:1427–1431. [PubMed] [Google Scholar]
- Gaston TE, Szaflarski JP (2018) Cannabis for the treatment of epilepsy: an update. Curr Neurol Neurosci Rep 18:73. [DOI] [PubMed] [Google Scholar]
- Geffrey AL, Pollack SF, Bruno PL, Thiele EA (2015) Drug-drug interaction between clobazam and cannabidiol in children with refractory epilepsy. Epilepsia 56:1246–1251. [DOI] [PubMed] [Google Scholar]
- Grayson L, Vines B, Nichol K, Szaflarski JP; UAB CBD Program (2017) An interaction between warfarin and cannabidiol, a case report. Epilepsy Behav Case Rep 9:10–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Yamaori S, Takeda S, Yamamoto I, Watanabe K (2011) Identification of cytochrome P450 enzymes responsible for metabolism of cannabidiol by human liver microsomes. Life Sci 89:165–170. [DOI] [PubMed] [Google Scholar]
- Jusko WJ, Schentag JJ, Clark JH, Gardner M, Yurchak AM (1978) Enhanced biotransformation of theophylline in marihuana and tobacco smokers. Clin Pharmacol Ther 24:405–410. [PubMed] [Google Scholar]
- Leino AD, Emoto C, Fukuda T, Privitera M, Vinks AA, Alloway RR (2019) Evidence of a clinically significant drug-drug interaction between cannabidiol and tacrolimus. Am J Transplant 19:2944–2948. [DOI] [PubMed] [Google Scholar]
- Morrison G, Crockett J, Blakey G, Sommerville K (2019) A phase 1, open-label, pharmacokinetic trial to investigate possible drug-drug interactions between clobazam, stiripentol, or valproate and cannabidiol in healthy subjects. Clin Pharmacol Drug Dev 8:1009–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadulski T, Pragst F, Weinberg G, Roser P, Schnelle M, Fronk E-M, Stadelmann AM (2005) Randomized, double-blind, placebo-controlled study about the effects of cannabidiol (CBD) on the pharmacokinetics of delta9-tetrahydrocannabinol (THC) after oral application of THC verses standardized cannabis extract. Ther Drug Monit 27:799–810. [DOI] [PubMed] [Google Scholar]
- Nasrin S, Watson CJW, Perez-Paramo YX, Lazarus P (2021) Cannabinoid metabolites as inhibitors of major hepatic CYP450 enzymes, with implications for cannabis-drug interactions. Drug Metab Dispos 49:1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patilea-Vrana GI, Unadkat JD (2019) Quantifying hepatic enzyme kinetics of (-)-Δ9-tetrahydrocannabinol (THC) and its psychoactive metabolite, 11-OH-THC, through in vitro modeling. Drug Metab Dispos 47:743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patilea-Vrana GI, Unadkat JD (2021) Development and verification of a linked Δ 9-THC/11-OH-THC physiologically based pharmacokinetic model in healthy, nonpregnant population and extrapolation to pregnant women. Drug Metab Dispos 49:509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher J, Bühner K, Witt-Laido A (2000) Determination of the free fraction and relative free fraction of drugs strongly bound to plasma proteins. J Pharm Sci 89:1008–1021. [DOI] [PubMed] [Google Scholar]
- Spaggiari D, Geiser L, Daali Y, Rudaz S (2014) Phenotyping of CYP450 in human liver microsomes using the cocktail approach. Anal Bioanal Chem 406:4875–4887. [DOI] [PubMed] [Google Scholar]
- Taylor L, Gidal B, Blakey G, Tayo B, Morrison G (2018) A phase I, randomized, double-blind, placebo-controlled, single ascending dose, multiple dose, and food effect trial of the safety, tolerability and pharmacokinetics of highly purified cannabidiol in healthy subjects. CNS Drugs 32:1053–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai C, Tayo B, Critchley D (2021) A phase 1 open-label, fixed-sequence pharmacokinetic drug interaction trial to investigate the effect of cannabidiol on the CYP1A2 probe caffeine in healthy subjects. Clin Pharmacol Drug Dev 10:1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng E, Eng H, Lin J, Cerny MA, Tess DA, Goosen TC, Obach RS (2021) Static and dynamic projections of drug-drug interactions caused by cytochrome P450 3A time-dependent inhibitors measured in human liver microsomes and hepatocytes. Drug Metab Dispos 49:947–960. [DOI] [PubMed] [Google Scholar]
- Turpeinen M, Nieminen R, Juntunen T, Taavitsainen P, Raunio H, Pelkonen O (2004) Selective inhibition of CYP2B6-catalyzed bupropion hydroxylation in human liver microsomes in vitro. Drug Metab Dispos 32:626–631. [DOI] [PubMed] [Google Scholar]
- Walsky RL, Gaman EA, Obach RS (2005) Examination of 209 drugs for inhibition of cytochrome P450 2C8. J Clin Pharmacol 45:68–78. [DOI] [PubMed] [Google Scholar]
- Yamaori S, Ebisawa J, Okushima Y, Yamamoto I, Watanabe K (2011a) Potent inhibition of human cytochrome P450 3A isoforms by cannabidiol: role of phenolic hydroxyl groups in the resorcinol moiety. Life Sci 88:730–736. [DOI] [PubMed] [Google Scholar]
- Yamaori S, Kushihara M, Yamamoto I, Watanabe K (2010) Characterization of major phytocannabinoids, cannabidiol and cannabinol, as isoform-selective and potent inhibitors of human CYP1 enzymes. Biochem Pharmacol 79:1691–1698. [DOI] [PubMed] [Google Scholar]
- Yamaori S, Maeda C, Yamamoto I, Watanabe K (2011b) Differential inhibition of human cytochrome P450 2A6 and 2B6 by major phytocannabinoids. Forensic Toxicol 29:117–124. [Google Scholar]