Abstract
Cytidine deaminase (CDA) is a determinant of in vivo gemcitabine elimination kinetics and cellular toxicity. The impact of CDA activity in pancreatic ductal adenocarcinoma (PDAC) cell lines has not been elucidated. We hypothesized that CDA regulates gemcitabine flux through its inactivation and activation pathways in PDAC cell lines. Three PDAC cell lines (BxPC-3, MIA PaCa-2, and PANC-1) were incubated with 10 or 100 µM gemcitabine for 60 minutes or 24 hours, with or without tetrahydrouridine, a CDA inhibitor. Extracellular inactive gemcitabine metabolite (dFdU) and intracellular active metabolite (dFdCTP) were quantified with liquid chromatography tandem mass spectrometry. Cellular expression of CDA was assessed with real-time PCR and Western blot. Gemcitabine conversion to dFdU was extensive in BxPC-3 and low in MIA PaCa-2 and PANC-1, in accordance with their respective CDA expression levels. CDA inhibition was associated with low or undetectable dFdU in all three cell lines. After 24 hours gemcitabine incubation, dFdCTP was highest in MIA PaCa-2 and lowest in BxPC-3. CDA inhibition resulted in a profound dFdCTP increase in BxPC-3 but not in MIA PaCa-2 or PANC-1. dFdCTP concentrations were not higher after exposure to 100 versus 10 µM gemcitabine when CDA activities were low (MIA PaCa-2 and PANC-1) or inhibited (BxPC-3). The results suggest a regulatory role of CDA for gemcitabine activation in PDAC cells but within limits related to the capacity in the activation pathway in the cell lines.
SIGNIFICANCE STATEMENT
The importance of cytidine deaminase (CDA) for cellular gemcitabine toxicity, linking a lower activity to higher toxicity, is well described. An underlying assumption is that CDA, by inactivating gemcitabine, limits the amount available for the intracellular activation pathway. Our study is the first to illustrate this regulatory role of CDA in pancreatic ductal adenocarcinoma cell lines by quantifying intracellular and extracellular gemcitabine metabolite concentrations.

Introduction
Gemcitabine (2′,2′-difluoro-2′-deoxycytidine [dFdC]) is a nucleoside analog used either alone or in combination with other cytostatic agents for treatment of inoperable pancreatic ductal adenocarcinoma (PDAC) and several other human cancers (https://www.legemiddelsok.no/). Following intracellular uptake, mainly by transmembrane equilibrative (hENT) and concentrative nucleoside transporter proteins (Wong et al., 2009), gemcitabine undergoes a stepwise phosphorylation process. Deoxycytidine kinase (dCK) catalyses the initial phosphorylation to gemcitabine monophosphate (dFdCMP) and is considered to be the rate-limiting step in the activation pathway (Wong et al., 2009). The main active metabolite is gemcitabine triphosphate (dFdCTP), which inhibits DNA replication. In tumor specimens from PDAC patients, high expression of hENT1 and dCK have been shown to favor the outcome of gemcitabine treatment (Maréchal et al., 2012).
Cytidine deaminase (CDA) catalyses the inactivation of gemcitabine to 2′,2′-difluoro-2′-deoxyuridine (dFdU) (Gusella et al., 2011; Simon et al., 2015; Cohen et al., 2019). CDA expression and activity in peripheral blood (Bowen et al., 2009) have been attributed both to lack of effect and increased toxicity of gemcitabine (Sugiyama et al., 2007; Ciccolini et al., 2010; Gusella et al., 2011). In PDAC tumor tissue, it has been found that CDA mRNA expression is higher compared with healthy tissues (Mameri et al., 2017). Bacteria and cells such as macrophages in the tumor microenvironment that express CDA might contribute to gemcitabine resistance (Vande Voorde et al., 2014; Weizman et al., 2014; Geller et al., 2017; Hessmann et al., 2018). However, the impact of intracellular CDA on gemcitabine metabolism in cancer cells is less studied (Morita et al., 2003; Vande Voorde et al., 2014).
Mameri et al. (2017) restored the expression of CDA in two a priori CDA-deficient cancer cell lines and showed that survival of these cells was higher than that of their CDA-deficient counterparts following in vitro incubation with gemcitabine. Indeed, similar results have also been achieved by others, indicating a reciprocal relationship between intracellular CDA activity and cellular gemcitabine sensitivity (Morita et al., 2003; Giovannetti et al., 2007; Yoshida et al., 2010; Peters et al., 2019). Thus, intracellular conversion of gemcitabine to dFdU is likely to be a mechanism contributing to gemcitabine resistance in this setting (Bardenheuer et al., 2005; Giovannetti et al., 2007; Ohmine et al., 2012; Vande Voorde et al., 2014; Mameri et al., 2017; Tibaldi et al., 2018).
In this study, we hypothesized that CDA plays a regulatory role in intracellular gemcitabine activation in PDAC cells. To test the hypothesis, we assessed intracellular and extracellular concentrations of gemcitabine and metabolites after exposure to gemcitabine with and without the use of the CDA inhibitor tetrahydrouridine (THU). We also determined basal mRNA and protein expression profiles of CDA and other main proteins involved in the transport and metabolism of gemcitabine.
Materials and Methods
Chemicals, Reagents, and Consumables.
Unless otherwise stated, chemicals and reagents were purchased from Merck KGaA (Darmstadt, Germany) and were of analytical grade. Horse serum and sodium pyruvate were bought from Thermo Fisher Scientific (Oslo, Norway), culture flasks and cryotubes from VWR (Oslo, Norway), centrifuge tubes from Sarstedt (Oslo, Norway), and THU from AH diagnostics (Oslo, Norway). All other reagents and equipment used for liquid chromatography tandem mass spectrometry methods have been described previously (Bjånes et al., 2015; Kamčeva et al., 2015).
Cell Culture.
Three human PDAC cell lines, BxPC-3, MIA PaCa-2, and PANC-1, authenticated and generously provided by Prof. Anders Molven (University of Bergen), were cultured in 75-cm2 flasks in a humidified atmosphere with 5% CO2 at 37°C and subcultured twice weekly. BxPC-3 cells were cultured in RPMI 1640 medium. MIA PaCa-2 and PANC-1 were cultured in Dulbecco’s modified Eagle’s medium D5671 (DMEM). All media were supplemented with 10% FBS, 4 mM sodium pyruvate, and 2 mM L-glutamine. The medium used for MIA PaCa-2 was additionally supplied with horse serum (2.5%) as recommended by the manufacturer. No antibiotics were used. Mycoplasma tests performed on a regular basis were negative.
Gemcitabine Incubation.
Cell-free media (RPMI, DMEM, and DMEM with horse serum) were spiked with 10 or 100 µM gemcitabine. Resulting spiked medium samples were aliquoted and stored in 1.5-ml Eppendorf tubes at 4°C, room temperature, and 37°C for up to 7 days and subsequently stored at −80°C until the entire batch was analyzed concurrently. The concentration ratios of dFdU over the sum of gemcitabine and dFdU in each sample, dFdU/(gemcitabine + dFdU) (%), was used as an indicator of CDA activity.
PDAC cell lines (0.25–0.4 × 106 cells per well in 2 ml culture medium) were seeded in 6-well plates 48 hours prior to gemcitabine incubation. Culture media was removed and replaced with freshly prepared drug-supplemented media at initiation of the experiments. The cells were incubated in quadruplicate for: 1) 24 hours with 10 or 100 µM gemcitabine with or without 200 µM THU or 2) 60 minutes with 10 or 100 µM gemcitabine with or without 200 µM THU. The two different durations of gemcitabine incubation were chosen based on: 1) that 24 hours is within a typical range applied in in vitro cytotoxicity experiments (Giovannetti et al., 2007; Yoshida et al., 2010; Mameri et al., 2017) and 2) that 60 minutes in vitro incubation reflects a comparable exposure to in vivo gemcitabine treatment (Gusella et al., 2011). Following gemcitabine incubation, media was collected, transferred to cryotubes, and stored at −80°C until quantification of extracellular gemcitabine and dFdU. Wells were rinsed twice with PBS, and cells were subsequently trypsinized for 5–8 minutes, harvested, and gently resuspended in cold culture medium. Manual cell counting was performed on a representative sample of the suspension. Cell suspensions were centrifuged for 5 minutes. Supernatant was discarded, and the cell pellets were dissolved in cold 60% methanol, transferred to cryotubes, vortexed for 20 seconds, and snap frozen on liquid nitrogen. All samples were stored at −80°C until quantification of intracellular dFdCTP.
Gemcitabine and Metabolite Quantification.
Quantification of gemcitabine and its metabolites was performed using an Agilent 1200 series HPLC system (Agilent Technologies, Waldbronn, Germany) for chromatographic separation and an Agilent 6410 triple-quad mass spectrometer for mass detection. Gemcitabine and dFdU in culture media samples were quantified as previously described (Bjånes et al., 2015) and optimized with lower limits of quantitation of 0.1 µM for both gemcitabine and dFdU. dFdCTP was analyzed in cell lysates with a modified version of our previously published method (Kamčeva et al., 2015). Modification consisted of shorter analysis time and the mass spectrometer operating in positive ionization mode because we were only interested in quantification of dFdCTP and not in the endogenous nucleosides that eluted later. dCTP was used as internal standard because of its similar structure and retention time with dFdCTP. Concentrations above the lower limit of quantitation of 0.05 µM were normalized to the cell count in each sample and expressed as picomoles per 106 cells (abbreviated to pmol/106 throughout the manuscript).
mRNA and Protein Expression.
Extraction of mRNA was performed on cell pellets from each cell line in quadruplicate by using the Qiagen column extraction kit. Two micrograms of mRNA was used for reverse transcription with Moloney Murine Leukemia Virus reverse transcriptase (InVitrogen). cDNA was diluted, and relative gene expression was determined by PCR in a final volume of 6.67 µl with Takyon NoRox SYBR MasterMix blue dTTP (Eurogentec). Triplicate runs were performed on a Lightcycler (LC480; Roche Life Science). Relative quantification was performed by the ΔΔCT method using 28S mRNA expression as a housekeeping gene and mean CT values as reference. Primers used for each gene are given in Supplemental Table 1A.
Total proteins were extracted using cold buffer (20 mM Tris-HCl pH 6.8, 1 mM MgCl2, 2 mM EGTA, 0.5% NP40, and phosphatase inhibitor cocktails) with 60 minutes incubation on ice followed by centrifugation (15 minutes, 12,000g, 4°C). Proteins were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes using the iBlot system (Life Technologies). Membranes were incubated with specific antibodies, as shown in Supplemental Table 1B. Protein expression was visualized using the Odyssey infrared system (LI-COR Biosciences). Protein bands were quantified using the Odyssey system, subtracting background noise from a similarly sized area just below the band, and presented as ratio of the expression of proteins of interest versus beta-actin expression.
Data Processing and Statistics.
Quantitative data were analyzed with SPSS Statistics 24.0 (IBM Inc., Armon, NY) and GraphPad Prism 8 (San Diego, CA) for Windows. Results were expressed as means ± S.D. or as concentration ratios between analytes (%). A two-sided Student’s t test was used to compare results in individual cell lines under different experimental conditions. One-way ANOVA with Bonferroni post hoc test was used to compare results in different cell lines. A P value of less than 0.05 was considered significant.
Results
CDA Activity in Cell-Free Culture Media.
We investigated whether cell-free culture media had any CDA activity, which would be of importance in the subsequent interpretation of data from cell lines incubated with gemcitabine. We found CDA activity only in DMEM supplemented with horse serum used for culturing MIA PaCa-2 cells. Within the maximum duration of our cell experiments (24 hours), the highest dFdU/(gemcitabine + dFdU) ratio at both gemcitabine concentrations was 6.3% at 37°C (Supplemental Fig. 1). No CDA activity was found in either RPMI or DMEM media without horse serum.
Accumulation of Inactive Gemcitabine Metabolite in Culture Media.
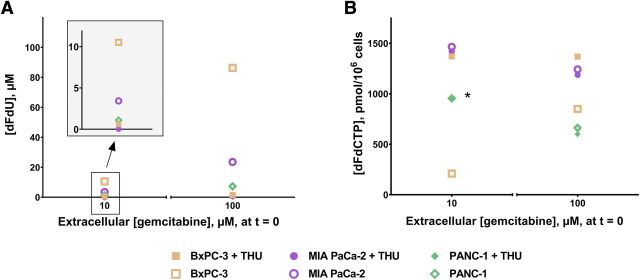
To quantify inactivation of gemcitabine in PDAC cells, we measured extracellular dFdU concentrations after incubation with 10 and 100 µM gemcitabine for 60 minutes or 24 hours, with or without inhibition of CDA. After 24 hours incubation of BxPC-3, MIA PaCa-2, and PANC-1 with 100 µM gemcitabine, mean dFdU concentrations were 86.3, 23.5, and 7.3 µM, respectively (Fig. 1A). After 60 minutes incubation with 100 µM gemcitabine, the corresponding dFdU concentrations were 17.7, 3.7, and 0.2 µM (Supplemental Fig. 2A). The percentage conversion of gemcitabine to dFdU was similar when cells had been incubated with 10 µM gemcitabine both after 60 minutes and 24 hours. After coincubation with gemcitabine and THU, dFdU was low or undetectable in medium from all three cell lines both after 60 minutes and 24 hours.
Fig. 1.
Concentrations of gemcitabine metabolites following 24 hours incubation with 10 or 100 µM gemcitabine ± 200 µM tetrahydrouridine (THU), a cytidine deaminase inhibitor. (A and B) show extracellular dFdU (micromolars) and intracellular dFdCTP (picomoles per 106), respectively. Insert in (A) shows data from 10 µM gemcitabine incubations in greater detail, with a differently scaled y-axis. Data are displayed as means (n = 4–8). Error bars excluded from view for clarity. Original data are shown in Supplemental Table 2. *dFdCTP concentrations in PANC-1 incubated with 10 µM gemcitabine with or without THU are overlapping and therefore appear as a single symbol.
Intracellular Accumulation of Active Gemcitabine Metabolite.
After 24 hours incubation of BxPC-3, MIA PaCa-2, and PANC-1 with 10 µM gemcitabine, mean dFdCTP concentrations were 210, 1466, and 955 pmol/106, respectively (Fig. 1B). After 24 hours incubation with 100 µM gemcitabine, dFdCTP concentrations in BxPC-3 were significantly higher (851 pmol/106; P < 0.001) than with 10 µM gemcitabine incubation. In MIA PaCa-2, dFdCTP concentrations were not significantly different between the two gemcitabine concentrations (P = 0.12), whereas in PANC-1, they were significantly lower at 100 µM gemcitabine (662 pmol/106; P < 0.05). CDA inhibition resulted in significantly higher dFdCTP concentrations in BxPC-3, with mean concentrations of 1370 (P < 0.01) and 1368 pmol/106 (P < 0.05) at 10 and 100 µM gemcitabine, respectively. In MIA PaCa-2 or PANC-1, dFdCTP concentrations were not significantly different with versus without CDA inhibition.
After 60 minutes incubation with 10 µM gemcitabine, mean dFdCTP concentrations were 92, 80, and 110 pmol/106 in BxPC-3, MIA PaCa-2, and PANC-1, respectively. Sixty minutes incubation with 100 µM gemcitabine did not result in significantly higher dFdCTP concentrations in any of the three cell lines. Also, CDA inhibition had no effect on dFdCTP concentrations at both gemcitabine concentrations under these experimental conditions (Supplemental Fig. 2B).
Basal mRNA and Protein Expression.
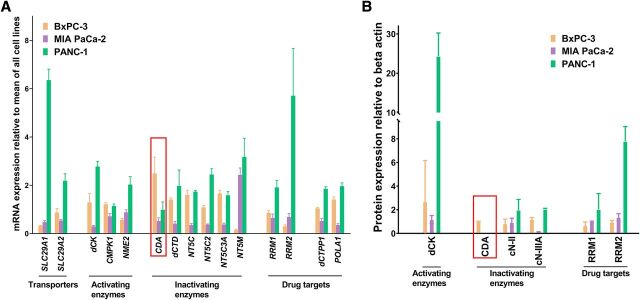
We assessed basal mRNA and protein expression of selected transporters and enzymes involved in gemcitabine uptake, metabolism, and activity in gemcitabine-untreated cell lines. Relative expression of mRNA and proteins are given in Fig. 2, respectively. Original Western blots can be seen in Supplemental Fig. 3. CDA showed highest mRNA and protein expression in BxPC-3. Lower CDA mRNA expression (Fig. 2A) and zero protein expression (Fig. 2B) was detected in both MIA PaCa-2 and PANC-1. The majority of the other transporters and enzymes revealed highest mRNA and protein expressions in PANC-1.
Fig. 2.
(A) Relative mRNA expression of selected proteins involved in the transport and metabolism of gemcitabine in BxPC-3, MIA PaCa-2, and PANC-1. Cytidine deaminase is highlighted (red rectangle). Data are displayed as means of four independent samples studied in triplicate, and error bars are S.D. SLC28A1*: hCNT1, concentrative nucleoside transporter 1; SLC29A1: hENT1, equlibrative nucleoside transporter 1; SLC29A2: CMPK1, uridine/cytosine monophosphate kinase; dCK, deoxycytidine kinase; hENT2, equlibrative nucleoside transporter 2; NME2: CDA, cytidine deaminase; dCTD, deoxycytidine monophosphate deaminase; NdPK, nucleoside diphosphate kinase; NT5C: cdN, cytosolic 5′(3′)-deoxyribonucleotidase; NT5C2: cN-II, cytosolic 5′-nucleotidase II; NT5C3: cN-IIIA, cytosolic 5′-nucleotidase III A; NT5M: CTPS1*, cytidine triphosphate synthase 1; DCTPP1, deoxycytidine triphosphate pyrophosphatase 1; mdN, mitochondrial 5′(3′)-deoxyribonucleotidase; POLA1, deoxyribonucleic acid polymerase alpha; RRM1, large subunit of ribonucleotide reductase; RRM2, small subunit of ribonucleotide reductase. mRNA expression of SLC28A1 not detectable. (B) Relative protein expression of selected proteins involved in the transport and metabolism of gemcitabine in BxPC-3, MIA PaCa-2, and PANC-1. Cytidine deaminase highlighted (red rectangle). Data are displayed as means of three independent samples, and error bars are S.D. Raw data are available in Supplemental Fig. 3. cN-II, cytosolic 5′-nucleotidase II (NT5C2); cN-IIIA, cytosolic 5′-nucleotidase III A (NT5C3); dCK, deoxycytidine kinase; RRM1, large subunit of ribonucleotide reductase; RRM2, small subunit of ribonucleotide reductase. *Antibodies against transporter proteins (hCNT and hENT) not available.
Discussion
Our overall finding was that intracellular cytidine deaminase plays a regulatory role for gemcitabine activation in PDAC cells, hence confirming our hypothesis.
Gemcitabine Inactivation.
Almost all gemcitabine added to the culture medium was converted to dFdU during 24 hours gemcitabine incubation of BxPC-3, highlighting the extensive CDA activity in this cell line. A comparable extent of gemcitabine conversion was reported by Bowen et al. (2009) in ex vivo whole blood from healthy volunteers, as 50% after 5 hours incubation and close to 100% after 24 hours. In accordance with other publications (Funamizu et al., 2012a,b), we also found that CDA displayed the highest mRNA (Fig. 2A) and protein expression (Fig. 2B) in BxPC-3 compared with MIA PaCa-2 and PANC-1.
Based on the pre-experimental stability assessments in cell-free culture media, all dFdU in BxPC-3 experiments was a result of cellular uptake, intracellular conversion, and subsequent efflux into the culture medium. In MIA PaCa-2 and PANC-1, respectively, the extent of gemcitabine conversion to dFdU was 20%–30% and <10% of BxPC-3 (Fig. 1A). This indicated that CDA activities were lower in MIA PaCa-2 and PANC-1. Gemcitabine was also to some extent converted to dFdU in the medium used for culturing MIA PaCa-2 (Supplemental Fig. 1). However, the conversion in cell-free medium only accounted for 20%–30% of the total amount found after 24 hours gemcitabine incubation of MIA PaCa-2 cells (Fig. 1A). The finding of no detectable CDA protein expression (Fig. 2B) in MIA PaCa-2 and PANC-1 did not fit with the appearance of dFdU following 24 hours gemcitabine incubation. These inconsistencies could preferably be explained by lack of sensitivity in the protein expression assay (Supplemental Fig. 3) because both cell lines expressed CDA mRNA (Fig. 2A). Moreover, it has been suggested that transcriptional, posttranscriptional (Mameri et al., 2017), and posttranslational (Frese et al., 2012) modulations could blur the relationship between mRNA and protein expression and the observed CDA phenotype.
In all cell lines, a long-lasting and strong inhibition of gemcitabine inactivation was achieved with 200 µM THU even at the highest gemcitabine concentrations and at both incubation durations. This is in line with previous studies in human blood performed by our own group (Bjånes et al., 2015) and other researchers (Bowen et al., 2009). dFdU could otherwise be assumed to be derived from the deamination of dFdCMP (Wong et al., 2009), but THU is not known to inhibit gemcitabine-inactivating enzymes other than CDA (Heinemann and Plunkett, 1989). The fact that coincubation of the cell lines with THU inhibited the formation of dFdU effectively underscores that direct gemcitabine deamination through CDA was the main source of dFdU in our experiments.
Gemcitabine Activation.
Without CDA inhibition, BxPC-3 accumulated significantly less dFdCTP over 24 hours compared with the two other cell lines (Fig. 1B). A probable explanation, in line with previous theories (Riva et al., 1992; Bardenheuer et al., 2005), was that the supply into the activation pathway was limited because of extensive conversion of gemcitabine to dFdU (Fig. 1A). This notion was supported by the observation that dFdCTP concentrations in BxPC-3 were significantly higher when gemcitabine exposure was increased, either by increasing gemcitabine concentrations from 10 to 100 µM (Fig. 1B) or by inhibiting CDA (Fig. 1B). No increase in dFdCTP concentrations was seen with increasing gemcitabine concentrations in MIA PaCa-2 or PANC-1, although baseline CDA activities were low. The same was true in BxPC-3 when CDA was inhibited. These findings were consistent with saturation kinetics of dCK, as previously described by others (Grunewald et al., 1991; Wong et al., 2009).
Despite the distinct effects after 24 hours incubation in BxPC-3, CDA inhibition had no effect on dFdCTP concentrations in any of the three cell lines when incubated for 60 minutes (Supplemental Fig. 2B). These findings could preferably be explained by sufficient concentrations of gemcitabine still available for the activation pathway in all three cell lines but with dCK operating close to its saturation limit. This view is supported by the fact that the mean percentage gemcitabine remaining in the medium after 60 minutes versus 24 hours incubation without THU was 77% versus <5% in BxPC-3, 92% versus 66% in MIA PaCa-2, and >98% versus 80% in PANC-1.
Overall Perspective.
Studies have highlighted the importance of CDA with respect to in vivo gemcitabine systemic pharmacokinetics (Sugiyama et al., 2007; Ciccolini et al., 2010; Gusella et al., 2011) and in vitro drug sensitivity (Yoshida et al., 2010; Funamizu et al., 2012b; Vande Voorde et al., 2014; Mameri et al., 2017), but the quantitative aspects of intracellular gemcitabine metabolism in PDAC cells has not previously been examined. We found that concentrations of both dFdU and dFdCTP after incubation with gemcitabine varied considerably between the PDAC cell lines, depending on CDA activity. As all three cell lines in this study are frequently used in in vitro PDAC studies (Funamizu et al., 2010, 2012a; Paproski et al., 2010; Mariglia et al., 2018), the observed metabolic variability may be important to take into account when interpreting results from gemcitabine incubation experiments. Moreover, the quantitative contribution of intracellular CDA in gemcitabine metabolism may provide a mechanistic explanation by which manipulating CDA activity could modify cellular gemcitabine sensitivity, as demonstrated by Bardenheuer et al. (2005) and Mameri et al. (2017).
By incubating the cell lines with gemcitabine with and without THU, we demonstrated that an extensive CDA-mediated gemcitabine conversion to dFdU in BxPC-3 was associated with less accumulation of the active metabolite dFdCTP. This was evident after 24 hours incubation but not after 60 minutes, indicating that a balanced substrate supply to dCK was an important factor for the accumulation of dFdCTP. In MIA PaCa-2 and PANC-1, no such effect of CDA inhibition on the gemcitabine activation pathway was seen, which was consistent with their a priori low CDA activities. This supports the idea that CDA activity may be a predictor for gemcitabine activation by regulating intracellular gemcitabine metabolism (Bardenheuer et al., 2005; Tibaldi et al., 2018). The observation that MIA PaCa-2 cells produced both more dFdU and dFdCTP than PANC-1 cells following 24 hours gemcitabine incubation could be explained by the higher expression of 5′-nucleotidases in PANC-1 (Fig. 2B), particularly cN-IIIA. Indeed, this enzyme has been suggested to dephosphorylate dFdCMP and thus oppose the accumulation of dFdCTP (Li et al., 2008; Aksoy et al., 2009). To decipher the exact mechanisms of these differences and the involvement of each of the other proteins shown in Fig. 2, it would be necessary to develop additional tools (protein-deficient cells, specific inhibitors, etc.) that are outside the scope of this work.
Direct quantification of gemcitabine and its metabolites (Fig. 1), combined with CDA inhibition, provided insight into differential CDA activities that could not be revealed by expression analyses alone (Fig. 2). In a recent commentary by Peters et al. (2019), phenotyping with cytidine or gemcitabine was also recommended over genotyping for pretreatment assessment of in vivo CDA activity in patients. Hodge et al. (2011a,b) also demonstrated the value of applying different drug concentrations and duration of incubations, combined with enzyme inhibition, when studying cellular regulation of gemcitabine transport (Hodge et al., 2011b) and metabolic (Hodge et al., 2011a) pathways.
In our experiments, we measured the free dFdCTP concentrations and did not have a measure of the total intracellular amount comprising both free and DNA-bound gemcitabine that might correlate better with cytotoxicity (Gandhi et al., 1991). Indeed, using the AnnexinV-PI assay, no additional effect of CDA inhibition was observed in any of the cell lines (Supplemental Fig. 4), underscoring that free dFdCTP is not the only determinant of gemcitabine efficacy. The ratio between free and total dFdCTP is expected to change over time during and after gemcitabine incubation, and cell lines might also behave differently based on intracellular enzyme expressions, illustrated by our own results in Fig. 2. Based on in silico simulations, Battaglia and Parker (2011) suggested that the rate of DNA incorporation in general is a slow process compared with the production rate of dFdCTP. Hence, quantification of free dFdCTP could therefore be a better measure of cellular uptake and metabolism of gemcitabine following 60 minutes incubation compared with 24 hours incubation. Incubation for 60 minutes with 10–100 µM gemcitabine in vitro might also more accurately represent the in vivo drug exposure during and after clinically applied 30-minute gemcitabine infusions of 1000 mg/m2, with a comparable concentration-time product (area under the curve) of 41 ± 12 µM*h (Gusella et al., 2011). We calculated that 60 minutes or 24 hours in vitro incubation with 10 µM gemcitabine renders areas under the curve of 10 or 240 µM*h, respectively.
In general, data from in vitro experiments should be interpreted with caution in terms of in vivo relevance. However, our finding that increased gemcitabine exposure does not necessarily lead to an increase in the intracellular active metabolite concentrations are in line with observations from in vivo studies, as illustrated by Hessmann et al. (2018).
Conclusion
Our findings reveal quantitative aspects of gemcitabine intracellular metabolism in PDAC cell lines. The data support the notion that high CDA activity limits intracellular dFdCTP accumulation. However, low CDA activity may not necessarily result in increased dFdCTP accumulation and decreased cell viability. Both CDA activity and the cellular ability to synthesize active metabolites should be taken into consideration in future studies of gemcitabine delivery to pancreatic cancer cells.
Acknowledgments
The authors would like to thank Elisa Thodesen Murvold and Philip Webber for valuable technical support as well as Anders Molven for generously providing the PDAC cell lines.
Abbreviations
- CDA
cytidine deaminase
- dCK
deoxycytidine kinase
- dFdC
2′,2′-difluoro-2′-deoxycytidine
- dFdCMP
gemcitabine monophosphate
- dFdCTP
gemcitabine triphosphate
- dFdU
2′,2′-difluoro-2′-deoxyuridine
- DMEM
Dulbecco’s modified Eagle’s medium
- hENT1
equlibrative nucleoside transporter 1
- PDAC
pancreatic ductal adenocarcinoma
- THU
tetrahydrouridine
Authorship Contributions
Participated in research design: Bjånes, Jordheim, Schjøtt, Riedel.
Conducted experiments: Bjånes, Kamceva, Cros-Perrial.
Contributed new reagents or analytic tools: Bjånes, Jordheim, Schjøtt, McCormack.
Performed data analysis: Bjånes, Jordheim, Schjøtt, Cros-Perrial, Langer, Ruiz de Garibay.
Wrote or contributed to the writing of the manuscript: Bjånes, Jordheim, Schjøtt, Kamceva, Cros-Perrial, Langer, Ruiz de Garibay, Kotopoulis, McCormack, Riedel.
Footnotes
This study was funded by the Western Health Board of Norway [Grant 912146].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Aksoy P, Zhu MJ, Kalari KR, Moon I, Pelleymounter LL, Eckloff BW, Wieben ED, Yee VC, Weinshilboum RM, Wang L (2009) Cytosolic 5′-nucleotidase III (NT5C3): gene sequence variation and functional genomics. Pharmacogenet Genomics 19:567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardenheuer W, Lehmberg K, Rattmann I, Brueckner A, Schneider A, Sorg UR, Seeber S, Moritz T, Flasshove M (2005) Resistance to cytarabine and gemcitabine and in vitro selection of transduced cells after retroviral expression of cytidine deaminase in human hematopoietic progenitor cells. Leukemia 19:2281–2288. [DOI] [PubMed] [Google Scholar]
- Battaglia MA, Parker RS (2011) Pharmacokinetic∕pharmacodynamic modelling of intracellular gemcitabine triphosphate accumulation: translating in vitro to in vivo. IET Syst Biol 5:34. [DOI] [PubMed] [Google Scholar]
- Bjånes T, Kamčeva T, Eide T, Riedel B, Schjøtt J, Svardal A (2015) Preanalytical stability of gemcitabine and its metabolite 2′, 2′-difluoro-2′-deoxyuridine in whole blood-assessed by liquid chromatography tandem mass spectrometry. J Pharm Sci 104:4427–4432. [DOI] [PubMed] [Google Scholar]
- Bowen C, Wang S, Licea-Perez H (2009) Development of a sensitive and selective LC-MS/MS method for simultaneous determination of gemcitabine and 2,2-difluoro-2-deoxyuridine in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci 877:2123–2129. [DOI] [PubMed] [Google Scholar]
- Ciccolini J, Dahan L, André N, Evrard A, Duluc M, Blesius A, Yang C, Giacometti S, Brunet C, Raynal C, et al. (2010) Cytidine deaminase residual activity in serum is a predictive marker of early severe toxicities in adults after gemcitabine-based chemotherapies. J Clin Oncol 28:160–165. [DOI] [PubMed] [Google Scholar]
- Cohen R, Preta LH, Joste V, Curis E, Huillard O, Jouinot A, Narjoz C, Thomas-Schoemann A, Bellesoeur A, Tiako Meyo M, et al. (2019) Determinants of the interindividual variability in serum cytidine deaminase activity of patients with solid tumours. Br J Clin Pharmacol 85:1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese KK, Neesse A, Cook N, Bapiro TE, Lolkema MP, Jodrell DI, Tuveson DA (2012) nab-Paclitaxel potentiates gemcitabine activity by reducing cytidine deaminase levels in a mouse model of pancreatic cancer. Cancer Discov 2:260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamizu N, Kamata Y, Misawa T, Uwagawa T, Lacy CR, Yanaga K, Manome Y (2012a) Hydroxyurea decreases gemcitabine resistance in pancreatic carcinoma cells with highly expressed ribonucleotide reductase. Pancreas 41:107–113. [DOI] [PubMed] [Google Scholar]
- Funamizu N, Lacy CR, Fujita K, Furukawa K, Misawa T, Yanaga K, Manome Y (2012b) Tetrahydrouridine inhibits cell proliferation through cell cycle regulation regardless of cytidine deaminase expression levels. PLoS One 7:e37424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamizu N, Okamoto A, Kamata Y, Misawa T, Uwagawa T, Gocho T, Yanaga K, Manome Y (2010) Is the resistance of gemcitabine for pancreatic cancer settled only by overexpression of deoxycytidine kinase? Oncol Rep 23:471–475. [PubMed] [Google Scholar]
- Gandhi V, Huang P, Xu YZ, Heinemann V, Plunkett W (1991) Metabolism and action of 2′,2′-difluorodeoxycytidine: self-potentiation of cytotoxicity. Adv Exp Med Biol 309A:125–130. [DOI] [PubMed] [Google Scholar]
- Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, et al. (2017) Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357:1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti E, Mey V, Loni L, Nannizzi S, Barsanti G, Savarino G, Ricciardi S, Del Tacca M, Danesi R (2007) Cytotoxic activity of gemcitabine and correlation with expression profile of drug-related genes in human lymphoid cells. Pharmacol Res 55:343–349. [DOI] [PubMed] [Google Scholar]
- Grunewald R, Abbruzzese JL, Tarassoff P, Plunkett W (1991) Saturation of 2′,2′-difluorodeoxycytidine 5′-triphosphate accumulation by mononuclear cells during a phase I trial of gemcitabine. Cancer Chemother Pharmacol 27:258–262. [DOI] [PubMed] [Google Scholar]
- Gusella M, Pasini F, Bolzonella C, Meneghetti S, Barile C, Bononi A, Toso S, Menon D, Crepaldi G, Modena Y, et al. (2011) Equilibrative nucleoside transporter 1 genotype, cytidine deaminase activity and age predict gemcitabine plasma clearance in patients with solid tumours. Br J Clin Pharmacol 71:437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann V, Plunkett W (1989) Modulation of deoxynucleotide metabolism by the deoxycytidylate deaminase inhibitor 3,4,5,6-tetrahydrodeoxyuridine. Biochem Pharmacol 38:4115–4121. [DOI] [PubMed] [Google Scholar]
- Hessmann E, Patzak MS, Klein L, Chen N, Kari V, Ramu I, Bapiro TE, Frese KK, Gopinathan A, Richards FM, et al. (2018) Fibroblast drug scavenging increases intratumoural gemcitabine accumulation in murine pancreas cancer. Gut 67:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge LS, Taub ME, Tracy TS (2011a) The deaminated metabolite of gemcitabine, 2′,2′-difluorodeoxyuridine, modulates the rate of gemcitabine transport and intracellular phosphorylation via deoxycytidine kinase. Drug Metab Dispos 39:2013–2016. [DOI] [PubMed] [Google Scholar]
- Hodge LS, Taub ME, Tracy TS (2011b) Effect of its deaminated metabolite, 2′,2′-difluorodeoxyuridine, on the transport and toxicity of gemcitabine in HeLa cells. Biochem Pharmacol 81:950–956. [DOI] [PubMed] [Google Scholar]
- Kamčeva T, Bjånes T, Svardal A, Riedel B, Schjøtt J, Eide T (2015) Liquid chromatography/tandem mass spectrometry method for simultaneous quantification of eight endogenous nucleotides and the intracellular gemcitabine metabolite dFdCTP in human peripheral blood mononuclear cells. J Chromatogr B Analyt Technol Biomed Life Sci 1001:212–220. [DOI] [PubMed] [Google Scholar]
- Li L, Fridley B, Kalari K, Jenkins G, Batzler A, Safgren S, Hildebrandt M, Ames M, Schaid D, Wang L (2008) Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res 68:7050–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mameri H, Bièche I, Meseure D, Marangoni E, Buhagiar-Labarchède G, Nicolas A, Vacher S, Onclercq-Delic R, Rajapakse V, Varma S, et al. (2017) Cytidine deaminase deficiency reveals new therapeutic opportunities against cancer. Clin Cancer Res 23:2116–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal R, Bachet JB, Mackey JR, Dalban C, Demetter P, Graham K, Couvelard A, Svrcek M, Bardier-Dupas A, Hammel P, et al. (2012) Levels of gemcitabine transport and metabolism proteins predict survival times of patients treated with gemcitabine for pancreatic adenocarcinoma. Gastroenterology 143:664–674.e6. [DOI] [PubMed] [Google Scholar]
- Mariglia J, Momin S, Coe IR, Karshafian R (2018) Analysis of the cytotoxic effects of combined ultrasound, microbubble and nucleoside analog combinations on pancreatic cells in vitro. Ultrasonics 89:110–117. [DOI] [PubMed] [Google Scholar]
- Morita T, Matsuzaki A, Kurokawa S, Tokue A (2003) Forced expression of cytidine deaminase confers sensitivity to capecitabine. Oncology 65:267–274. [DOI] [PubMed] [Google Scholar]
- Ohmine K, Kawaguchi K, Ohtsuki S, Motoi F, Egawa S, Unno M, Terasaki T (2012) Attenuation of phosphorylation by deoxycytidine kinase is key to acquired gemcitabine resistance in a pancreatic cancer cell line: targeted proteomic and metabolomic analyses in PK9 cells. Pharm Res 29:2006–2016. [DOI] [PubMed] [Google Scholar]
- Paproski RJ, Young JD, Cass CE (2010) Predicting gemcitabine transport and toxicity in human pancreatic cancer cell lines with the positron emission tomography tracer 3′-deoxy-3′-fluorothymidine. Biochem Pharmacol 79:587–595. [DOI] [PubMed] [Google Scholar]
- Peters GJ, Giovannetti E, Honeywell RJ, Ciccolini J (2019) Can cytidine deaminase be used as predictive biomarker for gemcitabine toxicity and response? Br J Clin Pharmacol 85:1213–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva C, Barra Y, Carcassonne Y, Cano JP, Rustum Y (1992) Effect of tetrahydrouridine on metabolism and transport of 1-beta-D-arabinofuranosylcytosine in human cells. Chemotherapy 38:358–366. [DOI] [PubMed] [Google Scholar]
- Simon N, Romano O, Michel P, Pinçon C, Vasseur M, Lemahieu N, Barthélémy C, Hebbar M, Décaudin B, Odou P (2015) Influence of infusion method on gemcitabine pharmacokinetics: a controlled randomized multicenter trial. Cancer Chemother Pharmacol 76:865–871. [DOI] [PubMed] [Google Scholar]
- Sugiyama E, Kaniwa N, Kim SR, Kikura-Hanajiri R, Hasegawa R, Maekawa K, Saito Y, Ozawa S, Sawada J, Kamatani N, et al. (2007) Pharmacokinetics of gemcitabine in Japanese cancer patients: the impact of a cytidine deaminase polymorphism. J Clin Oncol 25:32–42. [DOI] [PubMed] [Google Scholar]
- Tibaldi C, Camerini A, Tiseo M, Mazzoni F, Barbieri F, Vittimberga I, Brighenti M, Boni L, Baldini E, Gilli A, et al. Italian Oncological Group of Clinical Research (GOIRC) (2018) Cytidine deaminase enzymatic activity is a prognostic biomarker in gemcitabine/platinum-treated advanced non-small-cell lung cancer: a prospective validation study. Br J Cancer 119:1326–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Voorde J, Sabuncuoğlu S, Noppen S, Hofer A, Ranjbarian F, Fieuws S, Balzarini J, Liekens S (2014) Nucleoside-catabolizing enzymes in mycoplasma-infected tumor cell cultures compromise the cytostatic activity of the anticancer drug gemcitabine. J Biol Chem 289:13054–13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weizman N, Krelin Y, Shabtay-Orbach A, Amit M, Binenbaum Y, Wong RJ, Gil Z (2014) Macrophages mediate gemcitabine resistance of pancreatic adenocarcinoma by upregulating cytidine deaminase. Oncogene 33:3812–3819. [DOI] [PubMed] [Google Scholar]
- Wong A, Soo RA, Yong WP, Innocenti F (2009) Clinical pharmacology and pharmacogenetics of gemcitabine. Drug Metab Rev 41:77–88. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Endo Y, Obata T, Kosugi Y, Sakamoto K, Sasaki T (2010) Influence of cytidine deaminase on antitumor activity of 2′-deoxycytidine analogs in vitro and in vivo. Drug Metab Dispos 38:1814–1819. [DOI] [PubMed] [Google Scholar]