Abstract
The von Willebrand factor (VWF) propeptide (domains D1D2) is essential for the assembly of VWF multimers and its tubular storage in Weibel-Palade bodies. However, detailed molecular mechanism underlying this propeptide dependence is unclear. Here, we prepared Weibel-Palade body-like tubules using the N-terminal fragment of VWF and solved the cryo-electron microscopy structures of the tubule at atomic resolution. Detailed structural and biochemical analysis indicate that the propeptide forms a homodimer at acidic pH through the D2:D2 binding interface and then recruits 2 D'D3 domains, forming an intertwined D1D2D'D3 homodimer in essence. Stacking of these homodimers by the intermolecular D1:D2 interfaces brings 2 D3 domains face-to-face and facilitates their disulfide linkages and multimerization of VWF. Sequential stacking of these homodimers leads to a right-hand helical tubule for VWF storage. The clinically identified VWF mutations in the propeptide disrupted different steps of the assembling process, leading to diminished VWF multimers in von Willebrand diseases (VWD). Overall, these results indicate that the propeptide serves as a pH-sensing template for VWF multimerization and tubular storage. This sheds light on delivering normal propeptide as a template to rectify the defects in multimerization of VWD mutants.
Key Points
-
•
Cryo-electron microscopy structure of VWF tubule at 2.85 Å resolution.
-
•
The propeptide serves as a pH-sensing template for VWF multimerization and tubule formation.
Introduction
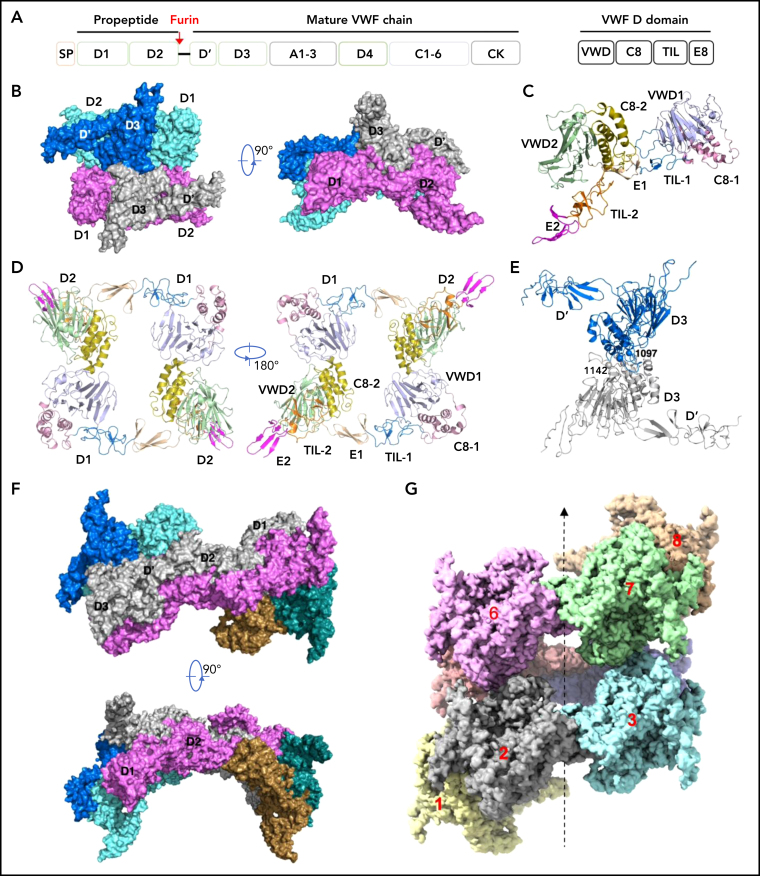
von Willebrand factor (VWF) is the largest known multimeric glycoprotein in the blood that controls platelet adhesion and aggregation. Its normal function requires the assembly of VWF into large, disulfide-linked multimers.1, 2, 3 Defects in VWF multimerization cause several forms of von Willebrand disease (VWD), the most common inherited bleeding disorder worldwide.4, 5, 6, 7 VWF assembly begins in the endoplasmic reticulum (ER) of endothelial cells and megakaryocytes, where it is synthesized as a precursor pro-VWF containing a signal peptide of 22-aa, a propeptide (domains D1D2) of 741-aa, and a mature subunit of 2050-aa of multiple domains (Figure 1A). The pro-VWF monomers form “tail-to-tail” homodimers through interchain disulfide linkages between cystine knot (CK) domains in the ER.8, 9, 10, 11 After being transported to the Golgi, these pro-VWF dimers assemble into “head-to-head” multimers through interchain disulfide linkages between D3 domains of 2 pro-VWF dimers. These multimers can condense into tubules in the storage granules, called Weibel-Palade bodies, which play a critical role in hemostasis by delivering VWF multimers into circulation.8
Figure 1.
Cryo-EM structures of VWF tubules. (A) Domain arrangement of pro-VWF. Furin cleaves pro-VWF between D2 and D' domains. Each VWF D domain has 4 modules (VWD, C8, TIL, and E8). (B) The tubule structure of 1 repeating unit shows a D'D3 dimer (marine and gray) docked in the central hole of a donut-shaped D1D2 dimer (cyan and pink). (C) The configuration of D1D2 fragment with each module colored differently. (D) The donut-shaped D1D2 dimer formed through D1:D2 interface. (E) The D'D3 dimers with 2 disulfide bonds (Cys1142-Cys1142' and Cys1097-Cys1097') in the interface. (F) Structure of tubule containing 2 repeating units. The 2 units are linked through the D2:D2 interface where D1D2 in 1 unit (gray) is expected to link with 1 D'D3 of the other unit (gray) in vivo. (G) Cryo-EM structure of tubule containing 8 repeating units packed as a right-hand helix with each unit colored differently. SP, signal peptide.
It has been shown that the propeptide is indispensable for VWF multimerization and tubular storage.12, 13 Both D domains, with each containing 4 modules (VWD, C8, TLE, and E8; Figure 1A), are required for multimerization, with deletion of either D1 or D2 resulting in complete loss of VWF multimerization. Interestingly, multimerization of mature VWF can be reestablished by coexpressing the propeptide “in trans” using 2 separate plasmids.12 VWF multimerization and storage are believed to be 2 independent intracellular processes because VWF multimerization is not a prerequisite for storage.9 As detailed molecular mechanism underlying this propeptide dependence is not clear, here we solved the cryo-electron microscopy (cryo-EM) structures of VWF tubules and analyzed the structural basis of VWF assembly. This shows how VWF tubule formation is initiated by a pH-sensing homodimer of the propeptide in a multistep process; how clinical VWD mutations disrupt this process, leading to diminished VWF multimers in plasma; and how these defects could potentially be rectified.
Methods
Construction of expression vectors for VWF D1D2 and D1-D3 variants
The expression plasmids encoding VWF D1D2 (residues 1-763aa) and D1-D3 (residues 1-1270aa) were constructed by cloning the corresponding DNA sequence into the expression vector pCEP4 with a polyhistidine (His)-tag at the C terminus, respectively. All the related mutants including but not limited to D1D2, D1D2-Y87S, D1D2-E8del (p.Ser292_Glu333delinsLys),14 D1-D3-K762A-R763A, D1-D3-K762A-R763A-Y87S, and D1-D3-K762A-R763A-Y87K were generated by KOD-Plus-Mutagenesis Kit (SMK-101) and checked by direct sequencing.
Protein preparation and size-exclusion chromatography
All the recombinant proteins including D1D2-His variants and D1-D3-His variants, etc, were prepared from the expression medium of cultured human embryonic kidney 293 cells15 and purified by Ni-Chelating followed by an anion exchange column. The samples were then further purified by a Superdex200pg 16/60GL column (GE) in 10 mM Tris-HCl pH7.4 and 100 mM NaCl. The expression constructs and purification details of the main proteins are summarized in supplemental Figure 1 (available on the Blood Web site). The oligomerization of D1-D3-K762A-R763A variants was analyzed on a Superose 6 10/300GL column (GE) with buffer containing 10 mM Tris-HCl pH7.4, 100 mM NaCl, 5 mM EDTA or buffer containing 20 mM 4-morpholineethanesulfonic acid (MES), pH6.0, 100 mM NaCl, and 10 mM CaCl2, respectively. The binding interactions between D1D2 and D'D3 monomer were analyzed on a Superdex200pg 16/60GL column in 20 mM MES pH6.0 and 100 mM NaCl buffer with 10 mM CaCl2. All protein fractions from the columns were analyzed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE).
Cryo-EM sample preparation and data acquisition
Various protein samples including D1D2, mixture of D1D2 and D'D3 dimer (termed D1D2-D'D3 dimer mixture), D1-D3-K762A-R763A monomer, and D'D3-dimer complexed with D1D2 purified from the culture medium were all screened with pH (from 5.2 to 7.4), with or without 10 mM CaCl2, with a temperature from 0°C to 37°C and incubation time from 1 minute to overnight. This was followed by negative staining and imaging on a Tecnai Spirit TEM D1266 120 kV microscope (Thermo Fisher Scientific) to verify the sample quality.
The selected samples for cryo-EM were all in low pH buffer (20 mM MES pH6.0, 100 mM NaCl, 10 mM CaCl2). D1D2-D'D3 dimer mixture was incubated at 4°C for different time intervals (from 8 minutes to overnight) before cryo-EM sample preparation. Aliquots (4 μL) of the protein samples were placed on glow-discharged holey carbon grids (Quantifoil Au R1.2/1.3, 300 mesh). The grids were blotted and flash frozen in liquid ethane cooled by liquid nitrogen with Vitrobot (Mark IV; Thermo Fisher Scientific). The grids sample quality was verified with a Talos Arctica (Thermo Fisher Scientific) 200 kV electron microscope. The structures presented in this study were derived from the cryo-EM data of D1D2-D'D3 dimer mixture incubated overnight, which were collected using a 300 kV Titan Krios microscope equipped with Gatan K3 Summit detector (Gatan) (supplemental Figure 3) or from D1D2-D'D3 dimer mixture incubated for 8 minutes, which were collected using a 300 kV Titan Krios microscope equipped with Cs corrector, Gatan K3 Summit detector (Gatan), and GIF Quantum energy filter (slit width 20 eV) (supplemental Figure 2). Cryo-EM data of D1D2 dimer was collect on 200 kV Talos Arctica microscope equipped with Gatan K2 detector (Gatan).
In brief, SerialEM16 and AutoEMation17 were used for the automated data collection, and each movie had a total accumulate exposure of 50 e-/A2 fractionated in 32 frames exposure. Micrographs were recorded in the super-resolution mode with a calibrated pixel size of 0.485 Å, 0.54895 Å, and 0.468 Å, with binned 2-fold pixel size of 0.97 Å, 1.0979 Å, and 0.936 Å for final images. Motion correction (MotionCo2)18 and Ctf estimation (Gctf) function19 were automatically executed by TsinghuaTitan.py program (developed by Fang Yang) during data collection. Data collection statistics are summarized in supplemental Table 1.
Cryo-EM data processing
For D1D2-D'D3 dimer mixture datasets, the image processing procedures were presented in supplemental Figures 2 and 3. Initially, all the micrographs were inspected and selected using the TsinghuaTitan.py program, followed by particles autopicking using Gautomatch (developed by Kai Zhang, https://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/) or Relion 3.0.20, 21 Multiple rounds of 2-dimensional (2D) classification were performed by Relion3.0 to eliminate bad particles and ice contaminations and followed by 3D classification (Relion3.0) and autorefinement (Relion3.0 or CryoSPARC v3.222). All the D1D2-D'D3 dimer mixture particle datasets result in a helical or tube structure map after 3D classification or 3 dimensional (3D) autorefinement, but only 1 circular region containing 4 repeating units in the helix has decent electron density. To improve the map density of VWF filament building units, masks were applied on 1 unit or 2 units (radial direction) in Relion 3D Classification with further autorefinement resulting in higher resolutions (see supplemental Figures 2 and 3; supplemental Table 1). Further Ctf refinement and Bayesian polishing were performed in Relion3.123 for the final masked unit map. To separate different states during VWF D1-D3 filament formation, Relion 3D Classification and CryoSPARC heterogeneous refinement were performed. To obtain VWF filament structure, particles of polymer (with 8 or more units) were selected and subjected to autorefinement in Relion 3.0 or CryoSPARC v3.2. To improve map details of interface between units in longitudinal direction, particles belonging to classes with >6 building units were selected. Similarly, mask on 2 units of longitudinal direction also applied in Relion 3D Classification and autorefinement or Cryo-SPARC local refinement. All the detail particle numbers and map resolutions during image processing steps were indicated in supplemental Figures 2 and 3.
For the VWF D1D2 dimer dataset, particles were autopicked by Gautomatch or Relion 3.0 from 387 micrographs. All subsequent 2D and 3D classifications and autorefinement were performed using Relion 3.0. After several rounds of 2D classification and 3D classification, 199 496 particles were selected and applied for autorefinement, which resulted in a fly shuttle shape density map at the resolution of 11.4 Å (bin2 data, Apix = 1.872 Å). The reported resolutions were estimated with the gold-standard Fourier shell correlation cutoff of 0.143 criterion. Data processing statistics are summarized in supplemental Table 1.
Model building and structure refinement
Initially, 1 copy of VWF filament repeat unit (1 D1D2 dimer and 1 D'D3 dimer) was manually built in Coot.24 Two-unit models or multiple-unit models were generated by fitting the 1-unit models into target maps. The atomic models were refined using Phenix in real space25 and validated using Molprobity web application.26 UCSF Chimera,27 UCSF ChimeraX,28 and PyMol29 were used for map segmentation and figure generation. Model refinement statistics are summarized in supplemental Table 1.
Results
The structure of the VWF tubules
It has been shown that the smallest fragment of VWF that can direct tubular packing in storage granules consists of domains D1-D3 from the N terminus of the VWF precursor (Figure 1A).30, 31 The helical Weibel-Palade body-like tubules could be recreated in vitro by mixing D1D2 with D'D3 dimers at pH6 in the presence of calcium.32 Here, we prepared the VWF tubules using a similar procedure with optimization and analyzed the tubular samples by cryo-EM (supplemental Figures 2 and 3). Multiple cycles of 3D classifications and autorefinement yielded a reconstruction of an overall resolution at 2.85 Å for the tubule containing 1 repeating unit derived from the images of D1D2-D'D3 dimer mixture incubated for 8 minutes (supplemental Figure 2) and an overall resolution at 3.27 Å for the tubule with 2 repeating units derived from the images of D1D2-D'D3 dimer mixture incubated overnight (supplemental Figure 3). The models were built and refined with statistics summarized in supplemental Table 1.
Our structures show that each repeating unit, as designated in the previous EM structural study of the VWF tubules,32 is comprised of a donut-shaped D1D2 dimer and a D'D3 dimer (Figure 1B-E). The D1D2 homodimer is formed mainly through intermolecular D1:D2 interactions. Each D1D2 monomer is in an arched shape where D1 and D2 domains are linked by the extended disulfide-rich TIL1 and E1 modules of the D1 domain. This connecting region, also called the cradle region in a homologous protein mucin 2,33 was not resolved in the previous low-resolution EM structure.32 Two D'D3 molecules, with each cradled by 1 D1D2 with D' stacked on D2, are docked face-to-face in the central hole of the D1D2 dimer. There are 2 disulfide bonds (Cys1097-Cys1097' and Cys1142-Cys1142') in the D'D3 dimeric interface (Figure 1E), which resemble those observed in mucin 2.33 Notably, Cys1097 formed an intramolecular disulfide bond with Cys1091 in the crystal structure of monomeric D'D3 mutant (D'D3-C1099A-C1142A), indicating potential disulfide exchanges during D'D3 dimer formation as proposed by Dong et al.34, 35, 36 The interactions between the 2 neighboring repeating units are mainly mediated by the D2:D2 interface as seen in the tubule structure containing 2 repeating units (Figure 1F).
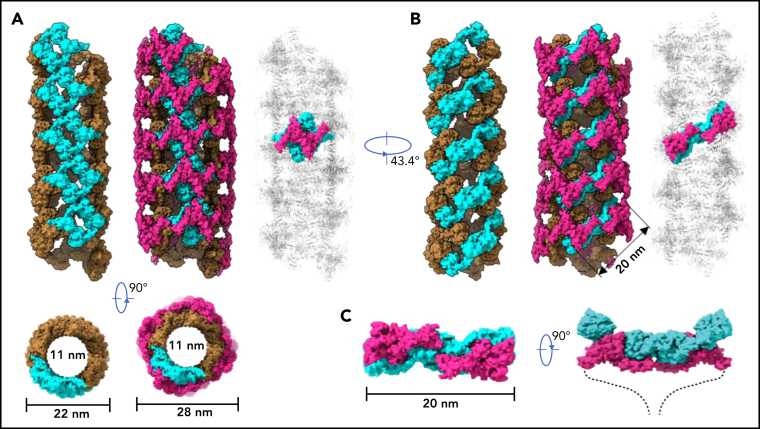
To further identify intermolecular interactions during tubule formation, we then solved a cryo-EM structure of tubule with 2 helical turns containing 8 repeating units at 4.3 Å resolution (Figure 1G). This shows that the repeating units in the tubule are packed in a right-hand helical conformation with an axial rise of 29.4 Å and twist of 86.8° per subunit, corresponding to ∼4.15 subunits per turn, largely consistent with previous EM studies32, 37 (Figure 2). The repeating units stack along the tubule extension axis mainly through the intermolecular interactions of D1 domains involving the C8-1 and TIL-1 modules (supplemental Figure 4). This results in D1D2s forming a tubule with an inside diameter of 11 nm and an outside diameter of 22 nm. The wall of the tubule is in a fishing net–like structure with holes of 2 different sizes. D'D3 dimers decorate the out surface of the tubule through docking in the smaller holes (Figure 2). The fully assembled tubule has an outside diameter of ∼28 nm. As the tails of neighboring D3 dimers along the tubule extension axis are close to each other, it is expected that the A1 domain linked after D3 in full-length VWF may form further stabilizing interactions on the tubule surface.32 The 2 C-terminal tails of mature VWF covalently linked by the CK domains will form a spike-like structure (dashed lines) pointing toward the outside of tubule, which will make a full VWF tubule like a round brush. Overall, these structures indicate that D1D2 functions as a template or framework allowing the docking of 2 D'D3 molecules in optimal positions for their intermolecular disulfide linkages and then serves as building blocks for tubular storage of mature VWF12, 38 (Figure 2).
Figure 2.
Model of VWF tubules. (A) The tubule structure of D1D2 with the small holes formed by a donut-shaped D1D2 dimer (cyan) for the docking of D'D3 dimers (left). D'D3 dimers (purple) decorate the out surface of the D1D2 tubule (middle). The conventional repeating unit with 1 D'D3 dimer and 1 D1D2 dimer is highlighted in the full VWF tubule (right). Scale bar, 22 and 28 nm. (B) An alternative view of the tubules formed by D1D2. The pH-sensitive D1D2 dimer is colored cyan (left). D'D3 dimers similarly docked in the smaller holes of the tubule (middle). A pH-sensitive D1D2 homodimer loaded with 2 D'D3 monomers in the full VWF tubule is highlighted with D1D2 in cyan and D'D3 in purple. (C) The distance between the 2 ends of D3 domains is ∼20 nm. The 2 C-terminal tails of mature VWF covalently linked by the CK domains will form a spike-like structure (dashed lines) pointing toward the outside of tubule, which makes the full VWF tubule look like a round brush. Scale bar, 20 nm.
The acidic pH-induced homodimer of VWF propeptide
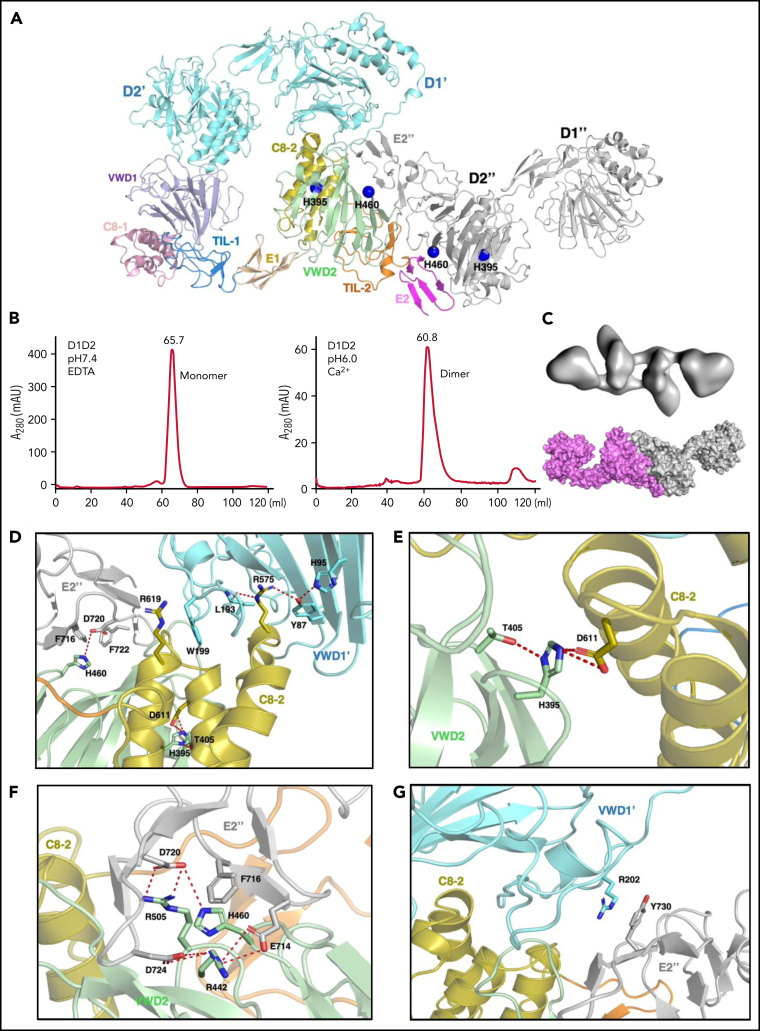
The above structures show the final state of the assembled VWF tubules; however, it is less clear how the assembling process is initiated and progressed in the cells. For example, the donut-shaped D1D2 dimeric interactions could form at the very early stage or a later stage of the assembling process, and D1D2 could form an alternative dimer with a neighboring molecule (Figure 2, Figure 3). The intact D1-D3 fragment without furin cleavage could similarly form alternative homodimers with different neighboring molecules, which may lead to different assembling steps (Figure 2). Because both VWF multimer assembling and packaging into Weibel-Palade bodies are regulated by the pH differences between ER (pH ∼7.4) and the relatively acidic Golgi (pH ∼5.8),32, 39, 40 and D1D2 in vitro forms noncovalent homodimers that bind tightly to multimeric VWF or D'D3 dimers at mild acidic pH but dissociate at pH7.4, this indicates that low pH is necessary for the intermolecular disulfide bonds formation during multimer assembly, after which low pH is also necessary for multimers to condense into tubules.41 So, we prepared the D1D2 homodimer by a gel filtration column at pH6 in the presence of calcium (Figure 3B), and analyzed its structure by cryo-EM using 200 kV Talos Arctica (supplemental Figure 5). The 2D averages and 3D reconstruction map of the D1D2 dimer show a flying-shuttle shape with C1 symmetry (Figure 3C; supplemental Figure 5B-C), which largely resembles the alternative D1D2 dimer linked by the D2:D2 interactions as seen in the tubule structure of 2 repeating units (Figure 1F). This indicates that dimerization through D2:D2 interface is the first step induced by the pH changes.
Figure 3.
The pH-induced D1D2 homodimer. (A) The binding interfaces of the neighboring D1D2 domains. His395 is located within the D2 domain, whereas His460 is in the D2:D2 interface. (B) D1D2 was assessed by a gel filtration Superdex S200 column at pH7.4 (left) or pH6 (right). (C) The overall shape of the pH-induced D1D2 dimer largely resembles that of D1D2 dimer formed through D2:D2 interface in the cryo-EM structure (Figure 1F). The flexible cradle region of D1D2 likely prevented the D1D2 dimer from forming larger complexes through D1:D2 interactions at pH6. (D) Both C8-2 and VWD2 module of D2 are involved in binding to the D1 domain. Protonated His395 would hold these 2 modules together. The connecting loop (Leu193-Trp199) of D1 is sandwiched by 2 Arginine residues from C8-2, with Arg575 stabilized by Tyr87 of VWD1' and Arg619 forming cation-π interaction with Trp199 of VWD1'. (E) His395 stabilized by hydrogen bonding with neighboring Thr405 forms intramolecular salt bridges with Asp611 of C8-2. (F) His460 forms intermolecular ionic integration with Asp720 and cation-π interaction with Phe716 of E2 module of the other D2 domain. (G) D1 domain also forms stabilization interaction with the other D2 domain of the D2:D2 interface as seen in panel A through the cation-π interaction between Arg202 and Tyr730. R202P and R202W mutations are associated with type 2A VWD52, 53 (supplemental Figure 6).
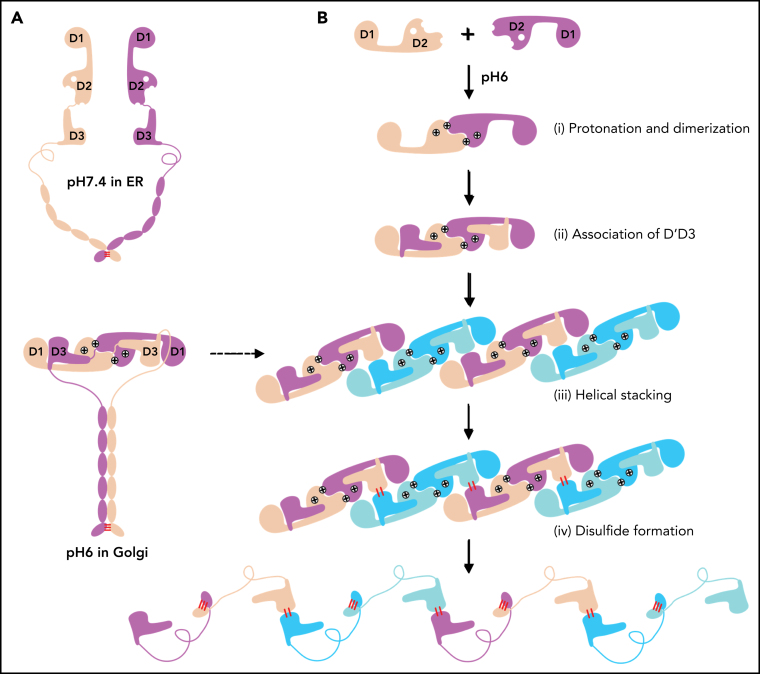
As the histidine sidechain is sensitive to pH changes between ER and Golgi and becomes positively charged upon protonation, previous histidine mapping studies have shown that His395 and His460 in the propeptide are indispensable for VWF multimerization.41 Here, we analyzed the ionic and cation-π interactions of all the histidine residues in the tubule structures. This showed that there are no direct involvements of histidine residues in the D1:D2 or D1D2:D3 interface (supplemental Figure 4A). His288 in C8-1 module of D1 is involved in the D1:D1 stacking interfaces along the tubule extension axis, and it likely forms stabilizing ionic interactions upon protonation for the stacking of the repeating units (supplemental Figure 4B). His395 from VWD2 module of the D2 domain forms intramolecular salt bridges with Asp611 from the C8-2 module upon protonation at pH6. These intramolecular interactions inside D2 would hold 2 modules together and provide a stable surface for D1 binding (Figure 3A,D-E). Replacements of this residue with Ala or Lys or Arg would disrupt its stabilizing interactions or the local packing, leading to the loss of disulfide linkages in the D'D3 dimeric interface.41 His460, also from the VWD2 module of D2 domain, can form intermolecular salt bridges with Asp720 and cation-π interactions with Phe716 from the E2 module of the other D2 domain (Figure 3F). Replacement of His460 with Ala would disrupt these interactions, leading to diminished VWF multimer formation, whereas Lys or Arg at position 460 could restore similar ionic and cation-π–stabilizing interactions with neighboring residues such as Phe722.41 The residues in D1 corresponding to these 2 histidines of D2 domain are Phe42 and Val98, respectively, indicating a unique feature of the D2 domain of VWF propeptide. These observations suggest that acidic pH would protonate histidine residues and generate the positively charged side chains to form key stabilizing interactions within the D2 domain and in the D2:D2 binding interface, which leads to the formation of a pH-sensitive D1D2 homodimer (Figure 3C). This points to an alternative repeating unit of the VWF tubule where 2 intact D1-D3 fragments of VWF would engage through the D2:D2 binding interactions first and then form an intertwined pH-induced homodimer by the intermolecular D1D2:D'D3 interactions (Figure 2B-C).
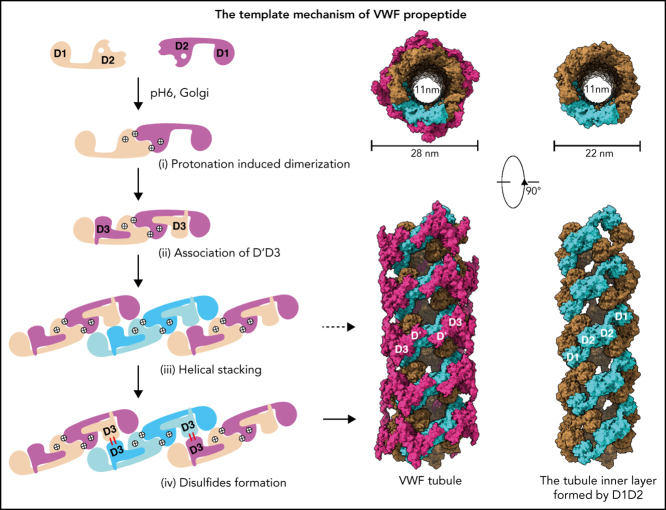
The template mechanism of VWF tubule formation
Similarly, the 2 D1-D3 fragments of a “tail-to-tail” pro-VWF homodimer linked by the disulfide bonds between their CK domains would engage noncovalently under acidic pH in the Golgi (Figure 4A). These closed pro-VWF dimers, further stabilized by the pH-dependent dimeric bouquet of the C-terminal tails of VWF,39 likely represent the true VWF assembly–starting units in the cell. They could then engage through the intermolecular D1:D2 interactions to align 2 D3 domains from 2 different closed pro-VWF dimers facilitating their disulfide formation (Figure 4Biii). Sequential stacking of these closed pro-VWF would lead to helical tubules formation, like that of mucin 2.33 This template mechanism does not require the propeptide being physically linked with the mature VWF chain. The D1D2 expressed “in trans” could form homodimer through the D2:D2 interface at acidic pH and then recruit 2 D'D3 domains forming a similar complex as the intertwined D1-D3 homodimer. They can stack through the D1:D2 interfaces to facilitate the intermolecular disulfide bonds formation between D3s and pack helically for VWF storage (Figure 4B). Thus, both multimerization and tubule formation of VWF are initiated by the pH-sensing D1D2 dimer and progressed by sequential stacking of the intertwined D1-D3 homodimers.
Figure 4.
The template mechanism of D1D2 in VWF multimerization and tubular storage. (A) Upon pH changes from ER to Golgi, the 2 histidine residues (His395 and His460) in D2 become protonated, and the N-terminal fragments of a pro-VWF dimer linked by the C-terminal CK domains would engage through the D2:D2 interface, forming a closed pro-VWF dimer in Golgi. (B) Two D1D2 molecules would form a pH-induced homodimer at pH6 (i) and then recruit 2 D'D3 monomers (ii). These D1D2 dimers loaded with 2 D'D3 molecules could then engage through the D1:D2 interface (iii) and align 2 D'D3 face-to-face, facilitating their intermolecular disulfide bond formation and helical packing of the repeating units (iv). The VWF multimers are then subject to tubular storage or release. The released VWF forms an extended head-to-head and tail-to-tail linear structure. All the D1D2 dimers loaded with D'D3 (ii-iv) could be transported into storage granules.
Clinical VWF mutations associated with the loss of VWF multimers
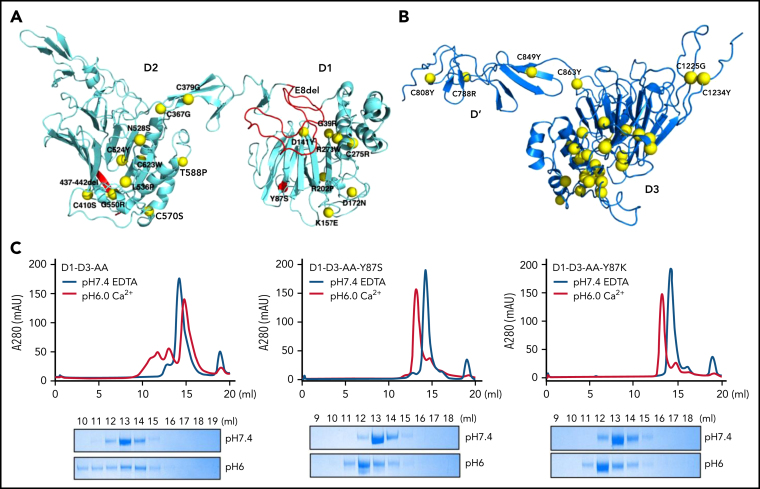
Many clinically identified VWF mutations are associated with qualitative or quantitative defects in VWF. Type 2A VWD often refers to limited or diminished VWF multimers in patients' plasma, which leads to defects in forming a stable platelet plug. Most of these type 2A mutations are in the D1D2 and D'D3 domains (Figure 5A-B). Detailed structural analysis of these mutants (supplemental Figure 6) indicated that these mutants often affect the local packing or the stability of the individual domain and subsequently impair the alignment of 2 D'D3 domains for their dimeric disulfide linkages, leading to diminished VWF multimers.
Figure 5.
Type 2A VWD associated mutations in D1D2 and D'D3. The clinically identified VWD mutations in D1D2 (A) and D'D3 (B) are mapped on the structures by yellow spheres. These mutations either disrupt the local packing of individual domains or interfere with the intermolecular interactions, leading to impaired alignment of 2 D'D3 domains for their dimeric disulfide linkages. Detailed structural analysis of each mutation is provided in supplemental Figure 6. The Y87S, E8del, and Δ437-442 mutation were highlighted in red. (C) The effect of Tyr87 mutations on oligomerization. The D1-D3 variants with the furin cleavage site mutated were analyzed by a Superose 6 gel filtration column with elution curves at pH7.4 with EDTA shown in blue and at pH6 with calcium shown in red. The corresponding fractions were analyzed by SDS-PAGE.
Notably, the previously well-characterized type 2A–associated Y87S mutation38 is located on the surface of the D1 domain. The side chain of Tyr87 is packed with the long side chain of Arg575 from the D2 domain in the D1:D2 binding interface (Figure 3D), and Y87S mutation is expected to impair the D1:D2 binding but not D'D3 binding of D1D2. Our previously reported type 2A VWD mutant VWF-E8del, which is derived from a splice site mutation c.875-5T>G leading to the skipping of the entire exon 8 of VWF gene with VWF synthesized in the proband having a substitution at 292 (S292K) and lacked Pro293_Glu333 in the propeptide (p.Ser292_Glu333delinsLys),14 has lost most of its TIL1 region of the D1 domain (Figure 5A). This mutation is expected to shorten the cradle region and affects the binding of D'D3 on D1D2. To confirm this, the D1D2 domains of both mutants were prepared and analyzed for their association with D'D3 at pH6 by a gel filtration column with the fractions analyzed by SDS-PAGE (supplemental Figure 7). Small amount of D'D3 could be seen in the early peak of the wild-type D1D2 (supplemental Figure 7A), consistent with previous observed weak association of D1D2 with D'D3 monomer at pH6.32 Small amount of D'D3 monomer could also be seen in the peak of D1D2-Y87S; however, little D'D3 could be seen in the D1D2-E8del mutant peak, indicating defects of D1D2-E8del in D'D3 binding as predicted (supplemental Figure 7B). To further evaluate the effect of Y87S mutation, the D1-D3-AA-Y87S and D1-D3-AA-Y87K mutants where furin cleavage site was mutated were prepared and analyzed by a gel filtration column (Figure 5C). Both mutants existed as monomers at pH7.4 as the wild-type fragment. At pH6, a large amount of wild-type D1-D3 fragment was eluted in early peaks from the column, indicating formation of high molecular weight oligomers; however, both mutants could form homodimers only. These results confirm that E8del mutation disrupts the D'D3 binding of D1D2 in the early stage of VWF assembling process (Figure 4Bii), whereas Y87S mutation affects the stacking of the closed pro-VWF dimers in the late stages (Figure 4Biii), with both defects leading to diminished VWF multimer formation. Overall, the phenotype of these clinical type 2A VWD mutants further supported the template mechanism of propeptide.
Discussion
In this study, we show that VWF tubule assembly is a multistep process initiated by a pH-sensitive homodimer of the propeptide (Figure 4). This homodimer could then serve as a template to promote the disulfide linkage in the dimeric interfaces of D'D3. Sequential packing of these dimers leads to helical tubule formation for VWF storage. This template mechanism is consistent with the notion that multimerization is not a prerequisite for tubular storage.9 Stacking of the intertwined homodimer of D1-D3 could occur sequentially without the need of disulfide linkages in the dimeric D3 interface (Figure 4Biii). The VWF-Y87S could not form multimers in Golgi due to weakened binding in the D1:D2 interfaces, but it could condense into Weibel-Palade body-like tubules in the storage granule, which is most likely due to its normal trafficking and high local concentration. This also indicates that the storage granules lack the redox components of Golgi to promote the disulfide linkages of the D3 dimeric interface in the VWF tubules of this mutant.
As the template mechanism of the propeptide readily explains how the separate and noncontiguous D1D2, when expressed “in trans,” mediates VWF multimers formation (Figure 4), it also implies that D1D2 expression “in trans” has the potential to restore the multimerization of defective type 2A VWD mutants. Careful review of literature revealed a delicate study on the type 2A VWD–associated VWF-Δ437-442 mutant.42 This mutant has lost the ability to form multimers and has defects in regulated storage, but coexpressed normal D1D2 completely restored multimerization of the mutant.42 Deletion of residues 437 to 442 is expected to disrupt the key stabilizing interactions near His460 in the D2:D2 interface (Figure 3F), leading to diminished VWF multimers. Through the same template mechanism as we speculate, the external expressed D1D2 could recruit D'D3 from the mutant protein and facilitate their dimerization and subsequent multimer formation of mutant VWF.
Despite current clinical management of VWD being considered effective with limited innovation in terms of treatment over the last 30 years, quality-of-life studies consistently reveal a higher-than-anticipated burden of VWD on patients, which is particularly true for women.5 As the VWD is an ideal target for gene therapy where a single treatment could potentially result in long-term correction of the disease. Even partial correction could have benefits in lowering the bleeding risks, like in the case of hemophilia.43 The preliminary study on VWD gene therapy showed limited yet promising prospect44, 45, 46, 47; however, one of the main obstacles of gene therapy approach for VWD treatment is the 8 kb size of VWF gene. This exceeds the packaging limit (5 kb) of the recombinant adeno-associated virus vectors that has been successfully used in gene therapy of hemophilia A and B and multiple other congenital disease.48, 49, 50 With the findings made in the current study on the template mechanism of propeptide, there is potential in introducing just the VWF propeptide for endothelium cell–targeted51 gene therapy to restore VWF multimer formation of type 2A VWD (supplemental Figure 8). For VWF mutants with defects in the D2:D2 interface, such as VWF-Δ437-442 and VWF-H460A, their multimerization could be restored by the external D1D2.42 However, it would likely require a larger amount of external D1D2 to restore the multimerization of other type 2A VWF mutants with normal D2:D2 interfaces, such as VWF-Y87S, which retains the ability to form closed pro-VWF dimers (supplemental Figure 8).
In summary, the VWF propeptide functions as a pH-sensing switch by forming a homodimer once transported into Golgi and then serves as a template to facilitate VWF multimer formation and the packaging of mature VWF multimers into tubules for storage. This unique chaperon-like function of propeptide allows externally introduced propeptide to rectify the defective VWF multimerization process and sheds light on a novel approach for the treatment of type 2A VWD.
Acknowledgments
The authors thank the Core Facilities of Basic Medical Sciences, Shanghai Jiaotong University School of Medicine for support. They thank Lina Ma for performing the initial characterization and D1D2-mediated multimer recovery of VWF-E8del mutant. They acknowledge the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for providing the cryo-EM facility and the cluster of Bio-Computing Platform support. They thank Jianlin Lei, Xiaomin Li, Fan Yang, Jie Wen, Xiangjie Ge, and Tao Liu for cryo-EM technical support. They thank Tao Yang, Yakun Wang, Anbao Jia, and Jinhong Chen for computational support. They thank Jiawei Wang from Tsinghua University for kind help and the cryo-EM access, and thank Jinsong Duan for collecting some of the cryo-EM data.
This study was funded by the National Natural Science Foundation of China (grants 81572090, 32171190, and 82070137), Ministry of Science and Technology of China (grant 2016YFA0501103), and Shanghai Science and Technology Commission (grant 20JC1410100).
Footnotes
The cryo-EM density maps and atomic coordinates of VWF tubules have been deposited in Electron Microscopy and Protein Data Bank with accession codes EMD32687 and PDB 7WPP for 1 repeating unit, EMD-32688 and PDB 7WPQ for 2 repeating units, and EMD-32713 and PDB 7WQT for 8 repeating units, respectively.
For original data, please contact awz20@shsmu.edu.cn.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
The release of von Willebrand factor (VWF) into the blood controls platelet adhesion and aggregation. Zeng and colleagues used cryo-electron microscopy to reveal how VWF tubule formation is initiated by a pH-sensing homodimer of the propeptide in a multistep process. From this procedure, they showed how clinical Type 2A von Willebrand disease mutations disrupt this process, leading to diminished VWF multimers in plasma, and how these defects could potentially be rectified.
Contributor Information
Jianwei Zeng, Email: ceng-jw13@tsinghua.org.cn.
Xuefeng Wang, Email: wxf63@shsmu.edu.cn.
Aiwu Zhou, Email: awz20@shsmu.edu.cn.
Authorship
Contribution: A.Z. conceived the project; J. Zeng, Z.S., Q.L., and J. Zhang performed the experiments; A.Z., X.W., and W.W. supervised the study; A.Z. J. Zeng, and W.W. wrote the paper; and all authors analyzed the data and contributed to the paper preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for J. Zeng is Department of Biochemistry and Molecular Biophysics, Washington University in St. Louis, School of Medicine, St. Louis, MO.
Supplementary Material
REFERENCES
- 1.Sadler JE. von Willebrand factor assembly and secretion. J Thromb Haemost. 2009;7:24–27. doi: 10.1111/j.1538-7836.2009.03375.x. suppl 1. [DOI] [PubMed] [Google Scholar]
- 2.Springer TA. Biology and physics of von Willebrand factor concatamers. J Thromb Haemost. 2011;9:130–143. doi: 10.1111/j.1538-7836.2011.04320.x. suppl 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenting PJ, Christophe OD, Denis CV. von Willebrand factor biosynthesis, secretion, and clearance: connecting the far ends. Blood. 2015;125(13):2019–2028. doi: 10.1182/blood-2014-06-528406. [DOI] [PubMed] [Google Scholar]
- 4.Leebeek FW, Eikenboom JC. von Willebrand's disease. N Engl J Med. 2016;375(21):2067–2080. doi: 10.1056/NEJMra1601561. [DOI] [PubMed] [Google Scholar]
- 5.Denis CV, Susen S, Lenting PJ. von Willebrand disease: what does the future hold? Blood. 2021;137(17):2299–2306. doi: 10.1182/blood.2020008501. [DOI] [PubMed] [Google Scholar]
- 6.Springer TA. von Willebrand factor, Jedi knight of the bloodstream. Blood. 2014;124(9):1412–1425. doi: 10.1182/blood-2014-05-378638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montgomery RR, Flood VH. What have we learned from large population studies of von Willebrand disease? Hematology Am Soc Hematol Educ Program. 2016;2016:670–677. doi: 10.1182/asheducation-2016.1.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vischer UM, Wagner DD. von Willebrand factor proteolytic processing and multimerization precede the formation of Weibel-Palade bodies. Blood. 1994;83(12):3536–3544. [PubMed] [Google Scholar]
- 9.Haberichter SL, Fahs SA, Montgomery RR. von Willebrand factor storage and multimerization: 2 independent intracellular processes. Blood. 2000;96(5):1808–1815. [PubMed] [Google Scholar]
- 10.Haberichter SL. von Willebrand factor propeptide: biology and clinical utility. Blood. 2015;126(15):1753–1761. doi: 10.1182/blood-2015-04-512731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen S, Abuzenadah AM, Hinks J, et al. A novel von Willebrand disease-causing mutation (Arg273Trp) in the von Willebrand factor propeptide that results in defective multimerization and secretion. Blood. 2000;96(2):560–568. [PubMed] [Google Scholar]
- 12.Wise RJ, Pittman DD, Handin RI, Kaufman RJ, Orkin SH. The propeptide of von Willebrand factor independently mediates the assembly of von Willebrand multimers. Cell. 1988;52(2):229–236. doi: 10.1016/0092-8674(88)90511-9. [DOI] [PubMed] [Google Scholar]
- 13.Wagner DD, Saffaripour S, Bonfanti R, et al. Induction of specific storage organelles by von Willebrand factor propolypeptide. Cell. 1991;64(2):403–413. doi: 10.1016/0092-8674(91)90648-i. [DOI] [PubMed] [Google Scholar]
- 14.Liang Q, Qin H, Ding Q, et al. Molecular and clinical profile of VWD in a large cohort of Chinese population: application of next generation sequencing and CNVplex® technique. Thromb Haemost. 2017;117(8):1534–1548. doi: 10.1160/TH16-10-0794. [DOI] [PubMed] [Google Scholar]
- 15.L'Abbé D, Bisson L, Gervais C, Grazzini E, Durocher Y. Transient gene expression in suspension HEK293-EBNA1 cells. Methods Mol Biol. 2018;1850:1–16. doi: 10.1007/978-1-4939-8730-6_1. [DOI] [PubMed] [Google Scholar]
- 16.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Lei J, Frank J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J Struct Biol. 2005;150(1):69–80. doi: 10.1016/j.jsb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14(4):331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang K. Gctf: real-time CTF determination and correction. J Struct Biol. 2016;193(1):1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zivanov J, Nakane T, Forsberg BO, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheres SH. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012;415(2):406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14(3):290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 23.Zivanov J, Nakane T, Scheres SHW. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ. 2020;7(Pt 2):253–267. doi: 10.1107/S2052252520000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Afonine PV, Poon BK, Read RJ, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol. 2018;74(Pt 6):531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams CJ, Headd JJ, Moriarty NW, et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27(1):293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 28.Pettersen EF, Goddard TD, Huang CC, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30(1):70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeLano WL,. The PyMOL molecular graphics system. http://www.pymol.org. Accessed 13 March 2018.
- 30.Michaux G, Abbitt KB, Collinson LM, Haberichter SL, Norman KE, Cutler DF. The physiological function of von Willebrand's factor depends on its tubular storage in endothelial Weibel-Palade bodies. Dev Cell. 2006;10(2):223–232. doi: 10.1016/j.devcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 31.Voorberg J, Fontijn R, Calafat J, Janssen H, van Mourik JA, Pannekoek H. Biogenesis of von Willebrand factor-containing organelles in heterologous transfected CV-1 cells. EMBO J. 1993;12(2):749–758. doi: 10.1002/j.1460-2075.1993.tb05709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang RH, Wang Y, Roth R, et al. Assembly of Weibel-Palade body-like tubules from N-terminal domains of von Willebrand factor. Proc Natl Acad Sci USA. 2008;105(2):482–487. doi: 10.1073/pnas.0710079105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Javitt G, Khmelnitsky L, Albert L, et al. Assembly mechanism of mucin and von Willebrand factor polymers. Cell. 2020;183(3):717–729.e16. doi: 10.1016/j.cell.2020.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purvis AR, Gross J, Dang LT, et al. Two Cys residues essential for von Willebrand factor multimer assembly in the Golgi. Proc Natl Acad Sci USA. 2007;104(40):15647–15652. doi: 10.1073/pnas.0705175104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong X, Leksa NC, Chhabra ES, et al. The von Willebrand factor D'D3 assembly and structural principles for factor VIII binding and concatemer biogenesis. Blood. 2019;133(14):1523–1533. doi: 10.1182/blood-2018-10-876300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong X, Springer TA. Disulfide exchange in multimerization of von Willebrand factor and gel-forming mucins. Blood. 2021;137(9):1263–1267. doi: 10.1182/blood.2020005989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berriman JA, Li S, Hewlett LJ, et al. Structural organization of Weibel-Palade bodies revealed by cryo-EM of vitrified endothelial cells. Proc Natl Acad Sci USA. 2009;106(41):17407–17412. doi: 10.1073/pnas.0902977106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenberg JB, Haberichter SL, Jozwiak MA, et al. The role of the D1 domain of the von Willebrand factor propeptide in multimerization of VWF. Blood. 2002;100(5):1699–1706. doi: 10.1182/blood-2002-03-0789. [DOI] [PubMed] [Google Scholar]
- 39.Zhou YF, Eng ET, Nishida N, Lu C, Walz T, Springer TA. A pH-regulated dimeric bouquet in the structure of von Willebrand factor. EMBO J. 2011;30(19):4098–4111. doi: 10.1038/emboj.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayadas TN, Wagner DD. In vitro multimerization of von Willebrand factor is triggered by low pH. Importance of the propolypeptide and free sulfhydryls. J Biol Chem. 1989;264(23):13497–13503. [PubMed] [Google Scholar]
- 41.Dang LT, Purvis AR, Huang RH, Westfield LA, Sadler JE. Phylogenetic and functional analysis of histidine residues essential for pH-dependent multimerization of von Willebrand factor. J Biol Chem. 2011;286(29):25763–25769. doi: 10.1074/jbc.M111.249151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haberichter SL, Allmann AM, Jozwiak MA, Montgomery RR, Gill JC. Genetic alteration of the D2 domain abolishes von Willebrand factor multimerization and trafficking into storage. J Thromb Haemost. 2009;7(4):641–650. doi: 10.1111/j.1538-7836.2009.03290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nathwani AC, Davidoff AM, Tuddenham EGD. Advances in gene therapy for hemophilia. Hum Gene Ther. 2017;28(11):1004–1012. doi: 10.1089/hum.2017.167. [DOI] [PubMed] [Google Scholar]
- 44.De Meyer SF, Vanhoorelbeke K, Chuah MK, et al. Phenotypic correction of von Willebrand disease type 3 blood-derived endothelial cells with lentiviral vectors expressing von Willebrand factor. Blood. 2006;107(12):4728–4736. doi: 10.1182/blood-2005-09-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Rosenberg JB, De BP, et al. In vivo gene transfer strategies to achieve partial correction of von Willebrand disease. Hum Gene Ther. 2012;23(6):576–588. doi: 10.1089/hum.2011.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park SW, Choi SY. Long-term expression of von Willebrand Factor by a VSV-G pseudotyped lentivirus enhances the functional activity of secreted B-Domain-deleted Coagulation Factor VIII. Mol Cells. 2007;24(1):125–131. [PubMed] [Google Scholar]
- 47.Barbon E, Kawecki C, Marmier S, et al. Development of a dual hybrid AAV vector for endothelial-targeted expression of von Willebrand factor. Gene Ther. [DOI] [PMC free article] [PubMed]
- 48.Nichols TC, Dillow AM, Franck HW, et al. Protein replacement therapy and gene transfer in canine models of hemophilia A, hemophilia B, von willebrand disease, and factor VII deficiency. ILAR J. 2009;50(2):144–167. doi: 10.1093/ilar.50.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arruda VR, Doshi BS. Gene therapy for hemophilia: facts and quandaries in the 21st century. Mediterr J Hematol Infect Dis. 2020;12(1):e2020069. doi: 10.4084/MJHID.2020.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosen S, Tiefenbacher S, Robinson M, et al. Activity of transgene-produced B-domain-deleted factor VIII in human plasma following AAV5 gene therapy. Blood. 2020;136(22):2524–2534. doi: 10.1182/blood.2020005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abel T, El Filali E, Waern J, et al. Specific gene delivery to liver sinusoidal and artery endothelial cells. Blood. 2013;122(12):2030–2038. doi: 10.1182/blood-2012-11-468579. [DOI] [PubMed] [Google Scholar]
- 52.Baronciani L, Federici AB, Punzo M, et al. Type 2A (IIH) von Willebrand disease is due to mutations that affect von Willebrand factor multimerization. J Thromb Haemost. 2009;7(7):1114–1122. doi: 10.1111/j.1538-7836.2009.03457.x. [DOI] [PubMed] [Google Scholar]
- 53.Schneppenheim R, Budde U, Beutel K, et al. Response to DDAVP in children with von Willebrand disease type 2. Hamostaseologie. 2009;29(2):143–148. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.