Abstract
The matrix (M) protein of vesicular stomatitis virus (VSV) functions in virus assembly and inhibits host-directed gene expression independently of other viral components. Experiments in this study were carried out to determine the ability of M protein to inhibit transcription directed by each of the three host RNA polymerases (RNA polymerase I [RNAPI], RNAPII, and RNAPIII). The effects of wild-type (wt) VSV, v6 (a VSV mutant isolated from persistently infected cells), and tsO82 viruses on poly(A)+ and poly(A)− RNA synthesis were measured by incorporation of [3H]uridine. v6 and tsO82 viruses, which contain M-gene mutations, had a decreased ability to inhibit synthesis of both poly(A)+ and poly(A)− RNA. Nuclear runoff analysis showed that VSV inhibited transcription of 18S rRNA and α-tubulin genes, which was dependent on RNAPI and RNAPII, respectively, but infection with wt virus enhanced transcription of 5S rRNA by RNAPIII. The effect of M protein alone on transcription by RNAPI-, RNAPII-, and RNAPIII-dependent promoters was measured by cotransfection assays. M protein inhibited transcription from RNAPI- and RNAPII-dependent promoters in the absence of other viral gene products. RNAPIII-dependent transcription of the adenovirus VA promoters was also inhibited by M protein. However, as observed during wt VSV infection, M protein enhanced endogenous 5S rRNA transcription, indicating that the inhibition of transcription by RNAPIII was dependent on the nature of the promoter.
Infection of cells with vesicular stomatitis virus (VSV) results in a rapid and potent shutoff of host macromolecular synthesis, including the inhibition of host DNA, RNA, and protein synthesis. The ability of VSV to inhibit host transcription has been examined extensively and is known to occur at the level of initiation by host RNA polymerases (30). VSV infection inhibits activity of all three host RNA polymerases (RNA polymerase I [RNAPI], -II, and -III), but genes transcribed by RNAPII appear to be more sensitive to the effects of virus infection than those transcribed by RNAPI and RNAPIII (28, 30).
Previous experiments have revealed that viral transcription is essential for shutoff of host transcription by VSV (29) and have implicated leader RNA as being involved. However, more recently, the viral matrix (M) protein has been found to play a role in the cytopathic effects associated with VSV infection. M protein is a major structural protein that normally functions in viral assembly by binding the ribonucleoprotein core of the virus to the host plasma membrane during the budding process (reviewed in reference 21). However, M protein causes the cell rounding characteristic of VSV infection when expressed in the absence of other viral components (5, 23, 31). In addition, M protein is capable of inhibiting host-directed transcription of RNAPII-dependent promoters in vivo in the absence of other viral gene components (2, 3, 11, 25). The ability of M protein to inhibit host transcription is quite potent, as 1,000-fold-more M protein is produced in infected cells than is necessary for 50% inhibition of target gene expression by M protein (22). The mechanism by which M protein inhibits host transcription is not known.
Further evidence for the role of M protein in inhibition of host gene expression has been provided by the conditionally temperature-sensitive VSV mutant, tsO82 (9). tsO82 virus is defective in the ability to shut off host RNA synthesis and induce the cell rounding characteristic of VSV infection (1, 9, 23). The M gene of tsO82 virus contains a single point mutation leading to a methionine-to-arginine substitution at position 51 of the protein sequence. The M51R mutation is the same as that found recently in the M protein of the T1026R1 mutant virus, which was previously found to be defective in shutting off host RNA and protein synthesis (10–12, 27). This mutation renders tsO82 M protein defective in its ability to inhibit RNAPII-dependent transcription at all temperatures but does not affect its function in virus assembly, as determined by complementation analysis (3, 19). These results demonstrate that the role of M protein in inhibition of host gene expression is genetically distinct from its function in virus assembly. This conclusion is reinforced by the MN1 mutant, which was generated by deleting the M-protein region spanning amino acids 4 to 21. MN1 protein displayed a phenotype complementary to that of tsO82 M protein in that it demonstrated full activity in inhibition of host gene expression, but its ability to function in virus assembly was abolished (3). More recently, the M genes of viruses isolated from persistently infected cells have been analyzed. v6 virus was plaque purified from a culture supernatant of L cells persistently infected with VSV (1, 14, 32). When tested for its ability to inhibit total host RNA synthesis, v6 was approximately twofold less effective than wild-type (wt) VSV but not as defective as the tsO82 virus (1). The reduced ability of v6 virus to inhibit host RNA synthesis was linked to a mutation at position 163 of the M-protein sequence leading to an asparagine-to-aspartate (N163D) substitution (1). Therefore, data from both tsO82 and v6 mutants indicate that M-gene mutations contribute to a reduction in the cytopathic effects of virus infection.
There is little, if any, promoter specificity in M-protein-induced inhibition of host cell transcription by RNAPII (22). The RNAPII-dependent promoters that have been shown to be inhibited by M protein include promoters with a wide variety of activating sequences, such as the following: the simian virus 40 (SV40) early promoter (1–3); the adenovirus major late promoter (22); the herpes simplex virus thymidine kinase promoter (22); the long terminal repeat promoters of human immunodeficiency virus (25), Rous sarcoma virus (22), and mouse mammary tumor virus (unpublished results); cellular promoters for class I major histocompatibility complex (22) and beta interferon (11); and the TATA-independent promoter for dihydrofolate reductase (22). It has been reported previously that some RNAPII-dependent promoters are relatively resistant to the inhibitory effects of VSV infection, for example, genes that are responsive to stimulation by interferon (6). However, this resistance appears to require the expression of double-stranded RNA following virus infection. These promoters are not resistant to the inhibitory effects of M protein expressed in the absence of other viral components (22). All of the promoters that have been examined, including those with interferon-stimulated response elements, appear to be as susceptible to M-protein-induced inhibition as the SV40 promoter is (22 and unpublished results).
The lack of promoter specificity in M-protein-induced inhibition of RNAPII-dependent transcription suggests that M protein inactivates some component of the basal transcription machinery. However, it was not known whether M protein alone inhibits transcription directed by the other host RNA polymerases, RNAPI and RNAPIII. Alternatively, other viral components might be involved in inhibition of RNAPI- or RNAPIII-dependent transcription. Experiments presented here define the ability of M protein to suppress transcription directed by each of the host RNA polymerases both when expressed alone and in the context of a virus infection. It was found that M-protein mutations in tsO82 virus and the v6 virus from persistently infected cells decreased the ability of VSV to inhibit synthesis of both poly(A)+ and poly(A)− RNAs. Nuclear runoff analysis showed that VSV inhibited transcription of 18S rRNA and α-tubulin genes, which was dependent on RNAPI and RNAPII respectively, but infection with wt virus enhanced transcription of 5S rRNA by RNAPIII. Similarly, M protein inhibited transcription from RNAPI- and RNAPII-dependent promoters in the absence of other viral gene products, as shown by cotransfection experiments. However, the inhibition of transcription by RNAPIII appeared to be dependent on the nature of the promoter. Expression of M protein inhibited transcription from the RNAPIII-dependent adenovirus VA promoters but stimulated transcription of 5S rRNA.
Effects of VSV mutants on poly(A)+ and poly(A)− RNA synthesis.
The effects of wt, v6, and tsO82 viruses on poly(A)+ and poly(A)− RNA synthesis were determined to distinguish the ability of M protein to inhibit transcription by RNAPII compared to transcription by RNAPI and RNAPIII. BHK and mouse L cells were infected with wt, v6, and tsO82 viruses (or mock infected) at a multiplicity of infection of 20 PFU/cell. At 2, 4, and 6 h postinfection, cells were labeled with [3H]uridine (20 μCi/ml) for 30 min, which labels both host RNA and viral RNA. Parallel samples were incubated and labeled in the presence of actinomycin D to measure virus-specific RNA synthesis. Cells were harvested and resuspended in sodium dodecyl sulfate (SDS) lysis buffer, and lysates were incubated in the presence of oligo(dT) cellulose (InVitrogen) to separate RNA species into poly(A)+ and poly(A)− fractions. Aliquots of these separate fractions were precipitated with trichloroacetic acid, and acid-insoluble radioactivity was determined by scintillation counting. Values of samples incubated in the presence of actinomycin D were subtracted from the total counts to determine the rate of host RNA synthesis.
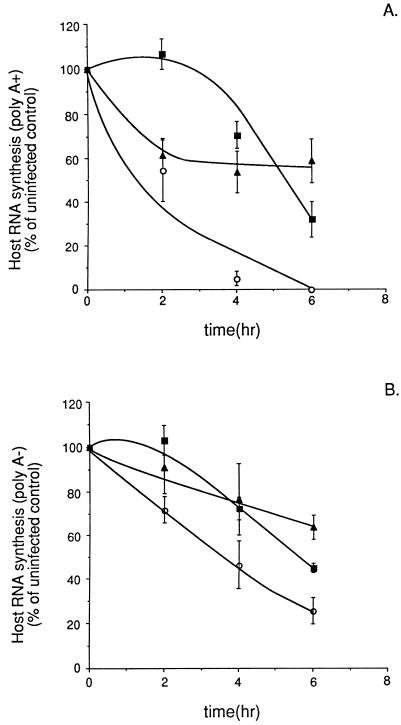
Data from a representative experiment (of four separate experiments) in infected L cells are shown in Table 1. L cells infected with wt VSV showed a progressive increase in actinomycin D-resistant viral RNA synthesis and a progressive decrease in host RNA synthesis for both poly(A)+ and poly(A)− RNA over the time course of the experiment. In contrast to previous data in other cell types (28, 30), poly(A)− RNA synthesis was at least as sensitive to VSV-induced inhibition as poly(A)+ RNA synthesis. In cells infected with tsO82 virus, which contains the M51R M-gene mutation, viral RNA was synthesized at a level similar to that of cells infected with wt VSV. tsO82 virus had decreased ability to inhibit both poly(A)+ and poly(A)− host RNA synthesis, supporting the idea that M protein is involved in the inhibition of synthesis of both poly(A)+ and poly(A)− RNA. Likewise, v6 virus, which contains the N163D M-gene mutation, failed to inhibit both poly(A)+ and poly(A)− host RNA synthesis as effectively as wt VSV did. Cells infected with v6 virus synthesized much less viral RNA than cells infected with wt VSV, despite the fact that viral proteins are synthesized at levels comparable to those of cells infected with wt VSV (1, 14). This is due to differences in control of translation in cells infected with viruses isolated from persistently infected cells, leading to more efficient translation of viral mRNAs (14).
TABLE 1.
Incorporation of [3H]uridine into poly(A)+ and poly(A)− RNA of VSV-infected L cells
| Virus | Time (h) postinfection |
3H cpm (103)a
|
Viral RNA synthesis (% of total)b
|
Host RNA synthesis (% of uninfected)c
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| − Act D
|
+ Act D
|
||||||||
| A+ | A− | A+ | A− | A+ | A− | A+ | A− | ||
| wt | 2 | 13.0 | 16.8 | 0.80 | 0.23 | 6.2 | 1.4 | 75 | 47 |
| 4 | 9.36 | 12.2 | 2.91 | 0.98 | 31 | 8.0 | 42 | 31 | |
| 6 | 8.20 | 4.55 | 4.15 | 0.76 | 51 | 16.7 | 25 | 8 | |
| tsO82 | 2 | 13.6 | 35.3 | 0.67 | 0.30 | 4.9 | 0.8 | 79 | 100 |
| 4 | 16.8 | 29.8 | 3.07 | 1.35 | 18 | 4.5 | 90 | 79 | |
| 6 | 13.3 | 20.3 | 3.66 | 1.30 | 28 | 6.4 | 60 | 41 | |
| v6 | 2 | 14.0 | 21.4 | 0.43 | 0.14 | 3.1 | 0.7 | 83 | 61 |
| 4 | 9.08 | 16.0 | 0.20 | 0.15 | 2.2 | 0.9 | 58 | 44 | |
| 6 | 8.59 | 10.3 | 0.20 | 0.12 | 2.3 | 1.2 | 51 | 22 | |
Abbreviations: − Act D, in the absence of actinomycin D; + Act D, in the presence of actinomycin D; A+, poly(A)+ RNA; A−, poly(A)− RNA.
Viral RNA synthesis calculated as a percentage of total RNA synthesis by dividing the 3H counts per minute (cpm) found in the presence of actinomycin D by 3H cpm found in the absence of actinomycin D.
Host RNA synthesis calculated as a percentage of uninfected control by subtracting the 3H cpm found in the presence of actinomycin D from 3H cpm found in the absence of actinomycin D and then dividing this value by the 3H cpm of mock-infected control.
Similar data were obtained for BHK cells. Host poly(A)+ RNA synthesis in infected BHK cells is shown in Fig. 1A as a percentage of the uninfected control value. wt VSV inhibited poly(A)+ RNA synthesis nearly completely by 6 h postinfection, while tsO82 and v6 viruses exhibited defective inhibition of poly(A)+ RNA synthesis. At 2 h postinfection, there was no detectable inhibition of poly(A)+ RNA synthesis by tsO82 virus and inhibition by v6 virus was intermediate between that of tsO82 virus and wt VSV. The ability of v6 virus to inhibit poly(A)+ RNA synthesis reached a constant level at 50 to 60% of uninfected controls by 6 h postinfection, whereas tsO82 virus continued to progressively inhibit poly(A)+ RNA synthesis over time, so that by 6 h postinfection, these two viruses inhibited host RNA synthesis to similar extents. A similar trend is shown in Fig. 1B demonstrating the effects of wt and mutant viruses on host poly(A)− RNA synthesis. Inhibition of poly(A)− RNA synthesis by wt VSV (Fig. 1B) was not as rapid as inhibition of poly(A)+ RNA synthesis was (Fig. 1A). M-gene mutations decreased the ability of the virus to inhibit poly(A)− RNA synthesis. However, poly(A)+ RNA synthesis mediated by RNAPII appeared to be more sensitive to the effect of M-gene mutations than did poly(A)− RNA synthesis by RNAPI and RNAPIII. The data from both L cells and BHK cells indicate that viruses with M-gene mutations are less effective than wt VSV in their ability to reduce both poly(A)+ and poly(A)− RNA synthesis, supporting the idea that M protein is involved in the inhibition of transcription of both poly(A)+ and poly(A)− RNA.
FIG. 1.
Poly(A)+ (A) and poly(A)− (B) RNA synthesis in BHK cells infected with wt, tsO82, and v6 viruses. BHK cells were infected with wt (open circles), tsO82 (closed squares), and v6 (closed triangles) viruses at a multiplicity of infection of 20 PFU/cell. Parallel samples were infected in the presence of actinomycin D. At 2, 4, and 6 h postinfection, cells were labeled with [3H]uridine (20 μCi/ml) for 30 min. Cells were harvested and lysed in SDS lysis buffer. To separate poly(A)+ and poly(A)− RNAs, lysates were incubated in the presence of oligo(dT) cellulose (Invitrogen), washed in high-salt buffer, and eluted in low-salt buffer. Samples were then precipitated with 7% trichloroacetic acid on ice and washed twice with 7% trichloroacetic acid. Acid-precipitable radioactivity was measured by scintillation counting. Values of samples incubated in the presence of actinomycin D were subtracted from the total counts to determine the rate of host RNA synthesis. Data shown are means ± standard deviations for four experiments.
Effect of M protein on transcription directed by RNAPI.
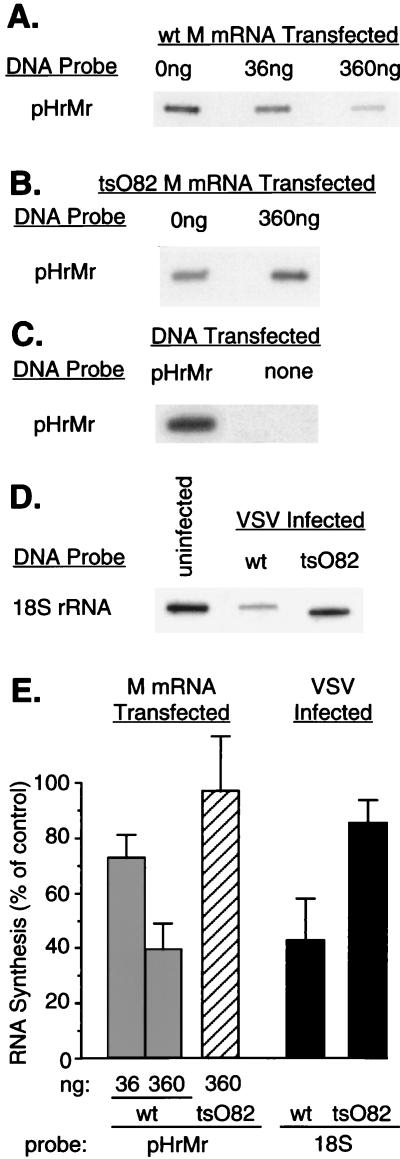
The effect of M protein on transcription directed by each of the host RNA polymerases in the absence of other viral components was tested by cotransfecting plasmid DNAs containing RNAPI-, RNAPII-, and RNAPIII-dependent promoters into BHK cells together with in vitro-transcribed M mRNA. M protein was expressed from in vitro-transcribed M mRNA instead of transfected plasmid DNA to avoid the M-protein-induced inhibition of its own synthesis from DNA vectors that require host transcriptional activity (2, 4). Transcriptional activity of these cotransfected cells was measured in a nuclear runoff assay, in which isolated nuclei were incubated in an in vitro transcription reaction mixture containing [α-32P]UTP. The basis of the nuclear runoff assay is that only transcripts that have initiated prior to the isolation of the nuclei are elongated in the runoff reaction, so that the amount of labeling reflects the number of polymerases actively transcribing in vivo. Labeled RNAs were then hybridized to DNA probes which had been fixed on nitrocellulose membrane filters in slots and analyzed by autoradiography. This is the method of choice for measuring in vivo transcription rates of individual RNAs, which is distinct from measurement of steady-state RNA levels by techniques such as Northern blotting (16). The transfected plasmid DNA containing the RNAPI promoter, pHrMr, encodes the mouse rRNA gene, which is recognized by the hamster polymerase in BHK cells (26). However, the transcript produced does not share enough sequence similarity with the endogenous hamster rRNA gene to cross-hybridize. Thus, only transcription in transfected cells, which also expressed M protein, was measured in these experiments.
To determine the effect of M protein on transcription by RNAPI, BHK cells in 100-mm-diameter dishes (approximately 3 × 106 cells per culture) were cotransfected with M mRNA (36 or 360 ng) or yeast RNA (360 ng) as a negative control (1) together with a constant amount of pHrMr plasmid DNA. At 24 h posttransfection, nuclei were isolated and total RNA was elongated in a nuclear runoff reaction (2). Labeled RNA was hybridized to linearized plasmid DNA, an autoradiogram of which is shown in Fig. 2A. The dose-dependent inhibition of transcription from the RNAPI-dependent promoter in pHrMr by wt M protein is readily apparent. As a control, cotransfection of pHrMr plasmid DNA with tsO82 M mRNA did not lead to detectable inhibition of transcription (Fig. 2B). As an additional control, there was no hybridization with labeled RNA from cells that were not transfected with pHrMr DNA (Fig. 2C), indicating that only transcription in transfected cells, which also expressed M protein, was measured in these experiments. These data indicate that M protein of wt VSV inhibits RNAPI-dependent transcription in the absence of other viral gene products.
FIG. 2.
Effect of M protein on transcriptional activity of genes dependent on RNAPI. (A) BHK cells were cotransfected with pHrMr plasmid DNA and 0, 36, or 360 ng of in vitro-transcribed wt M mRNA. Cells that received no M mRNA were cotransfected with 360 ng of yeast RNA as a negative control. At 24 h posttransfection, nuclei were isolated and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to linearized pHrMr plasmid DNA fixed on nitrocellulose membrane filters. (B) BHK cells were cotransfected with pHrMr plasmid DNA and 0 or 360 ng of tsO82 M mRNA. Transcription of pHrMr DNA was assayed by nuclear runoff analysis as described above for panel A. (C) BHK cells were transfected with pHrMr DNA or no plasmid DNA as a control for the specificity of hybridization. Transcription of pHrMr DNA was assayed by nuclear runoff analysis as described above for panel A. (D) BHK cells were infected at a multiplicity of infection of 20 PFU/cell with wt or tsO82 virus. Mock-infected cells were used as a control. Nuclei were isolated 6 h postinfection, and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to a cDNA fragment of 18S rRNA immobilized on nitrocellulose membranes. (E) The data from four (A and D) or three (B) separate experiments were quantitated by densitometry and expressed as a percentage of the control without M mRNA for the transfection experiments and as a percentage of the uninfected control for the virus-infected cells. The data are means ± standard deviations.
The effect of M protein expressed from transfected mRNA on transcription by RNAPI in nuclear runoff experiments was compared quantitatively to the effect of virus infection. BHK cells were infected with wt or tsO82 virus at a multiplicity of infection of 20 PFU/cell or were mock infected. Nuclei were isolated at 6 h postinfection, and total RNA was elongated in a nuclear runoff reaction. Labeled RNA was hybridized to a 300-bp 18S rRNA cDNA probe generated by reverse transcription-PCR of total RNA isolated from BHK cells. The autoradiogram in Fig. 2D shows that wt VSV inhibited transcription of the 18S rRNA gene, whereas tsO82 virus did not inhibit transcription as effectively as wt VSV did. Results from four separate experiments similar to those in Fig. 2A, B, and D were quantitated by densitometry (Fig. 2E). Data were expressed as a percentage of the uninfected control for results from virus-infected cells or as a percentage of control cells transfected without M mRNA for the transfection experiments. wt VSV inhibited 18S rRNA synthesis to a degree similar to that exhibited by M protein when 360 ng of M mRNA was cotransfected together with pHrMr, indicating that M protein inhibits RNAPI-dependent transcription to a level comparable to the inhibition observed during virus infection. There was little or no inhibition by tsO82 M protein both when expressed in a virus infection and from transfected mRNA. The inhibition of 18S rRNA transcription by wt and tsO82 viruses observed in the nuclear runoff experiments at 6 h postinfection (Fig. 2E) was in good agreement with the extent of inhibition of [3H]uridine incorporation into poly(A)− RNA (Fig. 1B).
Effect of M protein on transcription directed by RNAPII.
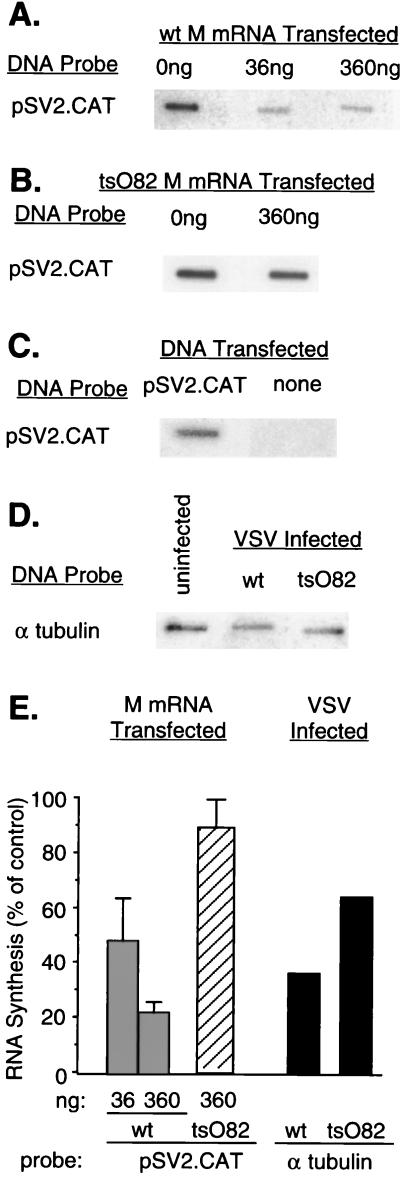
Nuclear runoff experiments similar to those in Fig. 2 compared the effect of M protein expressed from transfected mRNA with the effect of virus infection on transcription by RNAPII (Fig. 3). BHK cells were cotransfected with M mRNA together with pSV2.CAT plasmid DNA. pSV2.CAT contains the chloramphenicol acetyl transferase (CAT) reporter gene under control of the RNAPII-dependent SV40 early promoter (15). In the experiments shown in Fig. 3A, BHK cells were cotransfected with wt M mRNA (36 or 360 ng) or yeast RNA (360 ng; negative control) together with a constant amount of pSV2.CAT plasmid DNA. Cells were harvested 24 h posttransfection, and transcription of pSV2.CAT DNA was assayed by nuclear runoff analysis. Expression of wt M protein inhibited RNAPII-dependent transcription from pSV2.CAT plasmid DNA in a dose-dependent manner (Fig. 3A), while no inhibition was observed following cotransfection with tsO82 M mRNA (Fig. 3B). Since pSV2.CAT contains only viral or bacterial sequences, there was no detectable cross-hybridization with transcripts from untransfected cells in the nuclear runoff experiments (Fig. 3C). The extent of inhibition of pSV2.CAT-dependent transcription by wt M protein in these nuclear runoff experiments (Fig. 3E) was similar to the extent of inhibition of CAT expression measured by enzymatic activity in previously published experiments performed with the same ratios of M mRNA per cell (see, e.g., reference 22). These data are also in good agreement with those of previous nuclear runoff and Northern blot experiments in which M protein was expressed from plasmid DNA rather than transfected mRNA (2).
FIG. 3.
Effect of M protein on transcriptional activity of genes dependent on RNAPII. (A) BHK cells were cotransfected with pSV2.CAT plasmid DNA and 0, 36, or 360 ng of in vitro-transcribed wt M mRNA. Cells that received no M mRNA were cotransfected with 360 ng of yeast RNA as a negative control. At 24 h posttransfection, nuclei were isolated and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to linearized pSV2.CAT plasmid DNA fixed on nitrocellulose membrane filters. (B) BHK cells were cotransfected with pSV2.CAT plasmid DNA and 0 or 360 ng of tsO82 M mRNA. Transcription of pSV2.CAT DNA was assayed by nuclear runoff analysis as described above for panel A. (C) BHK cells were transfected with pSV2.CAT DNA or no plasmid DNA as a control for the specificity of hybridization. Transcription of pSV2.CAT DNA was assayed by nuclear runoff analysis as described above for panel A. (D) BHK cells were infected at a multiplicity of infection of 20 PFU/cell with wt or tsO82 virus. Mock-infected cells were used as a control. Nuclei were isolated 6 h postinfection, and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to a cDNA fragment of α-tubulin mRNA immobilized on nitrocellulose membranes. (E) The data from four (A and B) or two (D) separate experiments were quantitated by densitometry and expressed as a percentage of the control without M mRNA for the transfection experiments and as a percentage of the uninfected control in the case of the virus-infected cells. The data are means ± standard deviations.
Transcription of the cellular α-tubulin gene in virus-infected cells was used to compare quantitatively the effect of M protein expressed from transfected mRNA with the effect of virus infection on RNAPII-dependent transcription in nuclear runoff assays. The α-tubulin gene was chosen because it produces an abundant cellular transcript that was considered to be representative of RNAPII activity. BHK cells were infected with either wt VSV or tsO82 virus at a multiplicity of infection of 20 PFU/cell or were mock infected. At 6 h postinfection, nuclei were isolated and incubated in a nuclear runoff reaction. Labeled RNAs were hybridized to a 600-bp α-tubulin cDNA fragment amplified from hamster kidney cDNA made from poly(A) RNA that was primed with random hexamers and oligo(dT) (provided by Paul Dawson, Wake Forest University School of Medicine).
wt VSV inhibited transcription of the α-tubulin gene to a greater extent than tsO82 virus (Fig. 3D and E). However, the inhibition of α-tubulin gene transcription by wt VSV in the nuclear runoff assays (35% of control uninfected cells) was less pronounced than the virus-induced inhibition of [3H]uridine incorporation into poly(A)+ RNA (<10% of control uninfected cells, Fig. 1A). This contrasts with the good agreement between the nuclear runoff analysis of 18S rRNA transcription and [3H]uridine incorporation into poly(A)− RNA (Fig. 1B and 2D) and may reflect a resistance of α-tubulin gene transcription to the inhibitory effects of virus infection relative to other RNAPII-dependent transcripts. As a result, the inhibition of RNAPII-dependent transcription of pSV2.CAT by transfection of M mRNA (360 ng) was actually greater than the inhibition of α-tubulin gene transcription by infection with wt VSV (Fig. 3E).
Effect of M protein on transcription directed by RNAPIII.
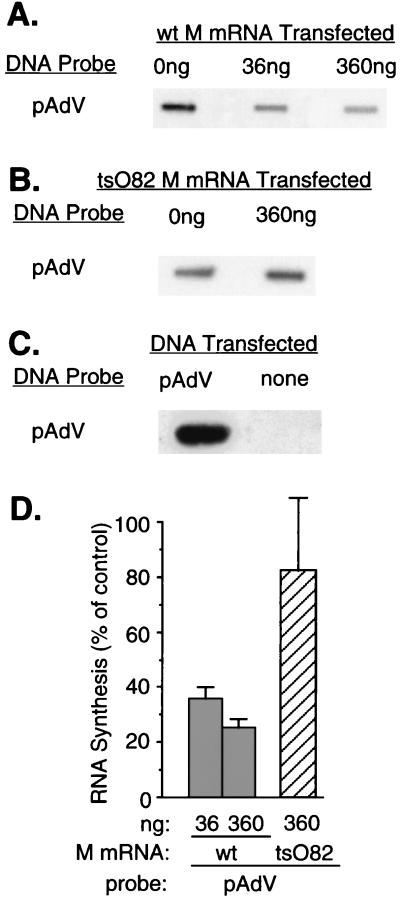
In the experiments shown in Fig. 4A, BHK cells were cotransfected with wt M mRNA (36 or 360 ng) or yeast RNA (360 ng; negative control) together with pAdVantage plasmid DNA (Promega), which contains RNAPIII-dependent promoters for the adenovirus VAI and VAII RNAs. At 24 h posttransfection, cells were harvested, and transcription of pAdVantage DNA was assayed by nuclear runoff analysis. Expression of wt M protein inhibited RNAPIII-dependent transcription from pAdVantage plasmid DNA in a dose-dependent manner (Fig. 4A), while no inhibition was observed following cotransfection with tsO82 M mRNA (Fig. 4B). Since pAdVantage contains only viral or bacterial sequences, there was no detectable cross-hybridization with transcripts from untransfected cells in the nuclear runoff experiments (Fig. 4C). The level of inhibition of RNAPIII-dependent transcription (Fig. 4D) was similar to the levels of inhibition of RNAPI- and RNAPII-dependent transcription observed in Fig. 2 and 3. Thus, transcription driven by the adenovirus VA promoters did not differ markedly from that of RNAPI- or RNAPII-dependent promoters in its sensitivity to M protein-induced inhibition. The promoters used in this study have been characterized previously to depend uniquely on RNAPI, -II, or -III for transcription. However, if M-protein expression allows an altered usage of polymerases, it is possible that the extent of inhibition of one of the polymerases is underestimated because of a contribution from transcription by another polymerase.
FIG. 4.
Effect of M protein on transcriptional activity of adenovirus VA genes dependent on RNAPIII. (A) BHK cells were cotransfected with pAdVantage (pAdV) plasmid DNA and 0, 36 or 360 ng of in vitro-transcribed wt M mRNA. Cells that received no M mRNA were cotransfected with 360 ng of yeast RNA as a negative control. At 24 h posttransfection, nuclei were isolated and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to linearized pAdV plasmid DNA fixed on nitrocellulose membrane filters. (B) BHK cells were cotransfected with pAdV plasmid DNA and 0 or 360 ng of tsO82 M mRNA. Transcription of pAdV DNA was assayed by nuclear runoff analysis as described above for panel A. (C) BHK cells were transfected with pAdV DNA or no plasmid DNA as a control for the specificity of hybridization. Transcription of pAdVantage DNA was assayed by nuclear runoff analysis as described above for panel A. (D) The data from four (A) or three (B) separate experiments were quantitated by densitometry and expressed as a percentage of the control without M mRNA. The data are means ± standard deviations.
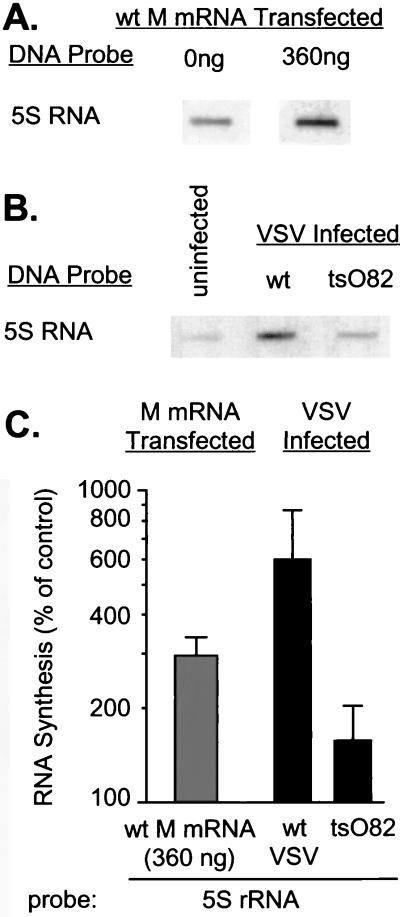
In contrast to the inhibition observed with the adenovirus VA promoters, the RNAPIII-dependent transcription of 5S rRNA was stimulated by expression of M protein. In the experiment in Fig. 5A, BHK cells were transfected with wt M mRNA (360 ng) or yeast RNA (360 ng; negative control) and were analyzed by nuclear runoff assay at 24 h posttransfection. For comparison, cells were infected with wt VSV or tsO82 virus or mock infected and then analyzed at 6 h postinfection (Fig. 5B). Labeled RNAs produced in the nuclear runoff reactions were hybridized to a hamster 5S rRNA cDNA probe (17) and analyzed by autoradiography and densitometry. 5S rRNA transcription in wt VSV-infected cells was stimulated about sixfold over that of uninfected cells, while synthesis by tsO82 virus-infected cells exhibited levels similar to those found in uninfected cells (Fig. 5C). Similarly, there was a threefold stimulation of 5S rRNA transcription in cells transfected with M mRNA. Under the conditions used in these experiments, approximately 40 to 60% of cells are transfected (23). Thus, the data in Fig. 5C underestimate the extent of stimulation by M protein, since the transcription rate measured contained contributions from both transfected and untransfected cells. This stimulation of 5S rRNA transcription contrasts markedly with the inhibition of transcription of the adenovirus VA RNAs.
FIG. 5.
Effect of M protein on transcriptional activity of 5S rRNA genes dependent on RNAPIII. (A) BHK cells were transfected with 0 or 360 ng of in vitro-transcribed wt M mRNA. At 24 h posttransfection, nuclei were isolated and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to linearized cDNA of 5S rRNA fixed on nitrocellulose membrane filters. (B) BHK cells were infected at a multiplicity of infection of 20 PFU/cell with wt or tsO82 virus. Mock-infected cells were used as a control. Nuclei were isolated 6 h postinfection, and RNA transcripts were elongated in the presence of [α-32P]UTP. Labeled RNAs were isolated and hybridized to a cDNA of 5S rRNA immobilized on nitrocellulose membranes. (C) The data from four separate experiments were quantitated by densitometry and expressed as a percentage of the control without M mRNA for the transfection experiments and as a percentage of the uninfected control in the case of the virus-infected cells. The data are means ± standard deviations and are plotted on a logarithmic scale to accommodate all of the values.
A previous study examined the synthesis of individual small RNA transcripts produced by RNAPII and RNAPIII during VSV infection and showed that there was a reduction in the synthesis of 5.8S, U1, and U2 RNAs, while synthesis of 5S and 4S RNAs was not reduced significantly (13). However, stimulation of 5S RNA transcription by VSV was not observed in this previous study. This difference from our results is probably related to their use of continuous labeling with [3H]uridine throughout infection versus the nuclear runoff assay at 6 h postinfection. Nonetheless, these results are all consistent with earlier work, which indicated that transcription by RNAPIII is the least sensitive to virus infection (28). However, the RNAPIII-dependent VAI and VAII promoters were at least as sensitive to M-protein-induced inhibition of host transcription as the RNAPI- and RNAPII-dependent promoters. Therefore, the inhibition of transcription by RNAPIII was dependent on the nature of the promoter.
The difference in the effect of M protein on transcription of the RNAPIII-dependent promoters is probably due to the fact that the 5S rRNA promoter has a structure that is distinct from that of the VA promoters. The VA promoters are similar to tRNA gene promoters in that they have two separated and variably space elements, box A and box B (20). Initiation complex formation occurs when TFIIIC recognizes box B, while box A orients the transcription factor on the start site. The bound TFIIIC allows association of the multisubunit complex TFIIIB, which is a pivotal step for RNAPIII recruitment and transcription initiation. The 5S rRNA gene promoter contains no A and B boxes and therefore, has no TFIIIC binding site. Transcription initiation is mediated by an intragenic control region called the box C element. This element is recognized by TFIIIA, which promotes the association of TFIIIC, thereby allowing subsequent binding of TFIIIB. However, there is some evidence suggesting that a preformed TFIIIA-TFIIIC complex exists in cells to facilitate RNAPIII transcription (20, 34). This difference between the 5S and VA RNAPIII promoters could account for the differential effects on transcription caused by virus infection as well as by M protein when expressed in the absence of other viral components.
Results from the experiments presented here are consistent with the idea that M protein plays a significant role in the VSV-mediated shutoff of transcription by all three host RNA polymerases. The hypothesis that M protein inhibits transcription by all three host RNA polymerases through a common mechanism is an attractive one. One possibility is that expression of M protein inactivates a cellular factor that is required by all three host RNA polymerases, such as TATA-binding protein (TBP), which is a subunit of transcription initiation factors for all three polymerases (34). Indeed, recent evidence indicates that the VSV-induced inhibition of RNAPII-dependent transcription involves inactivation of TBP (33). If inactivation of TBP is responsible for inhibition of all three host RNA polymerases, then the inactivation must not prevent interaction of the TBP-containing TFIIIB with TFIIIA in transcription of the 5S rRNA gene. The observed stimulation might result from an increased availability of TFIIIB due to inhibition of other RNAPIII-dependent promoters. Alternatively, M protein may act through different cellular targets to inhibit each host RNA polymerase. This would be analogous to the case with poliovirus, in which the viral 3C protease inhibits RNAPII-dependent transcription through inactivation of TBP (8) but inhibits RNAPIII-dependent transcription through inactivation of TFIIIC (7).
It has been demonstrated recently in Xenopus oocytes that M protein inhibits nuclear-cytoplasmic transport of RNA and protein mediated by the RAN GTPase and its guanine nucleotide exchange factor RCC1 (18). Thus, it is possible that inhibition of host transcription is an indirect effect of an M-protein-induced inhibition of nuclear-cytoplasmic transport, which could lead to a decrease in availability of transcription initiation factors for all three host RNA polymerases. This appears less likely, since inhibition of nuclear transport by a temperature-sensitive RCC1 mutation in BHK cells does not dramatically affect transcription rates as seen in virus-infected cells (24). Also, virus-induced inhibition of the activity of TBP does not appear to involve differences in the level of TBP in nuclear extracts (33). Future experiments to identify the cellular targets of M-protein action for each of the host RNA polymerases will resolve the question of whether there is a single mechanism or multiple mechanisms for inactivation of all three polymerases.
Acknowledgments
This work was supported by Public Health Service grant AI32983 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Ahmed M, Lyles D S. Identification of a consensus mutation in M protein of vesicular stomatitis virus from persistently infected cells that affects inhibition of host-directed gene expression. Virology. 1997;237:378–388. doi: 10.1006/viro.1997.8808. [DOI] [PubMed] [Google Scholar]
- 2.Black B L, Lyles D S. Vesicular stomatitis virus matrix protein inhibits host cell-directed transcription of target genes in vivo. J Virol. 1992;66:4058–4064. doi: 10.1128/jvi.66.7.4058-4064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black B L, Rhodes R B, McKenzie M O, Lyles D S. The role of vesicular stomatitis virus matrix protein in inhibition of host-directed gene expression is genetically separable from its function in virus assembly. J Virol. 1993;67:4814–4821. doi: 10.1128/jvi.67.8.4814-4821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black B L, Brewer G, Lyles D S. Effect of vesicular stomatitis virus matrix protein on host-directed translation in vivo. J Virol. 1994;68:555–560. doi: 10.1128/jvi.68.1.555-560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blondel D, Harmison G G, Schubert M. Role of matrix protein in cytopathogenesis of vesicular stomatitis virus. J Virol. 1990;64:1716–1725. doi: 10.1128/jvi.64.4.1716-1725.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bovolenta C, Lou J, Kanno Y, Park B K, Thornton A M, Coligan J E, Schubert M, Ozato K. Vesicular stomatitis virus infection induces a nuclear DNA-binding factor specific for the interferon-stimulated response element. Virology. 1995;69:4173–4181. doi: 10.1128/jvi.69.7.4173-4181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark M E, Hammerle T, Wimmer E, Dasgupta A. Poliovirus proteinase 3C converts an active form of transcription factor IIIC to an inactive form: a mechanism for inhibition of host cell polymerase III transcription by poliovirus. EMBO J. 1991;10:2941–2947. doi: 10.1002/j.1460-2075.1991.tb07844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark M E, Lieberman P M, Berk A J, Dasgupta A. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol Cell Biol. 1993;13:1232–1237. doi: 10.1128/mcb.13.2.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coulon P, Deutsch V, Lafay F, Martinet-Edelist C, Wyers F, Herman R C, Flamand A. Genetic evidence for multiple functions of the matrix protein of vesicular stomatitis virus. J Gen Virol. 1990;71:991–996. doi: 10.1099/0022-1317-71-4-991. [DOI] [PubMed] [Google Scholar]
- 10.Dunigan D D, Baird S, Lucas-Lenard J. Lack of correlation between the accumulation of plus-strand leader RNA and the inhibition of protein and RNA synthesis in vesicular stomatitis virus infected mouse L cells. Virology. 1986;150:231–246. doi: 10.1016/0042-6822(86)90282-5. [DOI] [PubMed] [Google Scholar]
- 11.Ferran M C, Lucas-Lenard J M. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J Virol. 1997;71:371–377. doi: 10.1128/jvi.71.1.371-377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francoeur A M, Poliquin L, Stanners C P. The isolation of interferon-inducing mutants of vesicular stomatitis virus with altered viral P function for the inhibition of total protein synthesis. Virology. 1987;160:236–245. doi: 10.1016/0042-6822(87)90065-1. [DOI] [PubMed] [Google Scholar]
- 13.Fresco L D, Kurilla M G, Keene J D. Rapid inhibition of processing and assembly of small nuclear ribonucleoproteins after infection with vesicular stomatitis virus. Mol Cell Biol. 1987;7:1148–1155. doi: 10.1128/mcb.7.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frey T K, Youngner J S. Further studies of the RNA synthesis phenotype selected during persistent infection with vesicular stomatitis virus. Virology. 1984;136:211–220. doi: 10.1016/0042-6822(84)90260-5. [DOI] [PubMed] [Google Scholar]
- 15.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg M E, Bender T P. Identification of newly transcribed RNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. pp. 4.10.1–4.10.11. [DOI] [PubMed] [Google Scholar]
- 17.Hart R P, Folk W R. Structure and organization of a mammalian 5S gene cluster. J Biol Chem. 1982;257:11706–11711. [PubMed] [Google Scholar]
- 18.Her L-S, Lund E, Dahlberg J E. Inhibition of RAN GTPase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science. 1997;276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 19.Kaptur P E, McKenzie M O, Wertz G W, Lyles D S. Assembly functions of vesicular stomatitis virus matrix protein are not disrupted by mutations at major sites of phosphorylation. Virology. 1995;206:894–903. doi: 10.1006/viro.1995.1012. [DOI] [PubMed] [Google Scholar]
- 20.Lagna G, Kovelman R, Sukegawa J, Roeder R G. Cloning and characterization of an evolutionarily divergent DNA-binding subunit of TFIIIC. Mol Cell Biol. 1994;14:3053–3064. doi: 10.1128/mcb.14.5.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenard J. Negative-strand virus M and retrovirus MA proteins: all in a family? Virology. 1996;216:289–298. doi: 10.1006/viro.1996.0064. [DOI] [PubMed] [Google Scholar]
- 22.Lyles D S, McKenzie M O, Ahmed M, Woolwine S C. Potency of wild-type and temperature-sensitive vesicular stomatitis virus matrix protein in the inhibition of host-directed gene expression. Virology. 1996;225:172–180. doi: 10.1006/viro.1996.0585. [DOI] [PubMed] [Google Scholar]
- 23.Lyles D S, McKenzie M O. Activity of vesicular stomatitis virus M protein mutants in cell rounding is correlated with the ability to inhibit host gene expression and is not correlated with virus assembly function. Virology. 1997;229:77–89. doi: 10.1006/viro.1996.8415. [DOI] [PubMed] [Google Scholar]
- 24.Nishimoto T, Ellen E, Basilico C. Premature chromosome condensation in a ts DNA − mutant of BHK cells. Cell. 1978;15:475–483. doi: 10.1016/0092-8674(78)90017-x. [DOI] [PubMed] [Google Scholar]
- 25.Paik S-Y, Banerjea A C, Harmison G G, Chen C-J, Schubert M. Inducible and conditional inhibition of human immunodeficiency virus proviral expression by vesicular stomatitis virus matrix protein. J Virol. 1995;69:3529–3537. doi: 10.1128/jvi.69.6.3529-3537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudloff U, Eberhard D, Tora L, Stunnenberg H, Grummt I. TBP-associated factors interact with DNA and govern species specificity of RNA polymerase I transcription. EMBO J. 1994;13:2611–2616. doi: 10.1002/j.1460-2075.1994.tb06551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanners C P, Francoeur A M, Lam T. Analysis of VSV mutant with attenuated cytopathogenicity: mutation in viral function, P, for inhibition of protein synthesis. Cell. 1977;11:273–281. doi: 10.1016/0092-8674(77)90044-7. [DOI] [PubMed] [Google Scholar]
- 28.Weck P K, Wagner R R. Inhibition of RNA synthesis in mouse myeloma cells infected with vesicular stomatitis virus. J Virol. 1978;25:770–780. doi: 10.1128/jvi.25.3.770-780.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weck P K, Wagner R R. Transcription of vesicular stomatitis virus is required to shut off cellular RNA synthesis. J Virol. 1979;30:410–413. doi: 10.1128/jvi.30.1.410-413.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weck P K, Wagner R R. Vesicular stomatitis virus infection reduces the number of active DNA-dependent RNA polymerases in myeloma cells. J Biol Chem. 1979;254:5430–5434. [PubMed] [Google Scholar]
- 31.Ye Z, Wei S, Suryanarayana K, Justice P, Robinson D, Wagner R R. Membrane-binding domains and cytopathogenesis of the matrix protein of vesicular stomatitis virus. J Virol. 1994;68:7386–7396. doi: 10.1128/jvi.68.11.7386-7396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Youngner J S, Dubovi E J, Quagliana D O, Kelly M, Preble O T. Role of temperature-sensitive mutants in persistent infections initiated with vesicular stomatitis virus. J Virol. 1976;19:90–101. doi: 10.1128/jvi.19.1.90-101.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan, M., B. K. Yoza, and D. S. Lyles. Unpublished data.
- 34.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]