Abstract
l-Chicoric acid is an inhibitor of human immunodeficiency virus type 1 (HIV-1) integrase in vitro and of HIV-1 replication in tissue culture. Following 3 months of selection in the presence of increasing concentrations of l-chicoric acid, HIV-1 was completely resistant to the compound. Introduction of the mutant integrase containing a single glycine-to-serine amino acid change at position 140 into the native, l-chicoric acid-sensitive virus demonstrated that this change was sufficient to confer resistance to l-chicoric acid. These results confirm through natural selection previous biochemical studies showing that l-chicoric acid inhibits integrase and that the drug is likely to interact at residues near the catalytic triad in the integrase active site.
With the recent success of combination therapies targeting human immunodeficiency virus (HIV) protease and reverse transcriptase (RT) (5, 15, 17), it seems apparent that multiple drug therapy, rather than monotherapy, will be required for long-term survival of HIV-infected individuals. However, due to the rapid turnover of HIV-infected cells and the likely repetitive rounds of HIV replication within an infected individual (18, 41), the emergence of resistant organisms is a likely sequela of drug therapy. Indeed, although it is slower to appear than with monotherapy, resistance of HIV to combination therapy has been demonstrated (15). Therefore, to improve upon existing multidrug therapies, anti-HIV agents that inhibit enzymes other than HIV protease and RT will likely be required. One such therapeutic target is HIV integrase.
Integration is absolutely required for the maintenance of stable and productive HIV infection (8, 21, 37, 38, 40); therefore, inhibition of integration is likely to profoundly impair HIV replication. To date, a large number of candidate inhibitors of HIV integrase have been reported based on the ability to inhibit HIV integrase enzyme in vitro. However, few have affected HIV replication in tissue culture (reviewed in reference 33). Recently, we described a group of compounds, the dicaffeoylquinic and dicaffeoyltartaric acids, that blocked HIV replication in tissue culture at nontoxic concentrations (34, 36). In vitro studies indicated that they were potent and selective inhibitors of HIV integrase (25, 34, 36). Indeed, these compounds exhibited 10- to 100-fold selectivity against HIV integrase over other HIV enzymes (25); nevertheless, definitive proof of their mechanism of anti-HIV activity has been lacking. Indeed, several findings by other investigators suggested that they might be acting through mechanisms other than the inhibition of integrase. For example, other bis-catechols do not inhibit HIV replication (3, 7, 9–11, 20, 24, 27, 42), and several previous reports indicated that compounds related to the dicaffeoyltartaric acids inhibited gp120 binding to CD4 (22) and RT (29, 30).
Although inhibition of integrase in vitro by small molecules has been relatively simple to demonstrate, inhibition of HIV integration within the cell has proven difficult to study. One method by which the mechanism of an antiviral compound can be deduced is via the isolation of drug-resistant variants and the demonstration that drug resistance maps to changes in the amino acid sequence of a viral enzyme. For example, the resistance of HIV to nucleoside and nonnucleoside RT inhibitors maps to amino acid changes in HIV RT (12–14, 23). In contrast, a G quartet oligonucleotide currently in clinical trials (1) was reported to inhibit HIV-1 replication via the inhibition of integrase (31). However, a recent report indicates that drug resistance maps to the HIV envelope, not integrase (4), indicating this putative integrase inhibitor acts via inhibition of virus penetration, not integration. Therefore, to address whether l-chicoric acid inhibited HIV replication at the level of integrase, we chose to isolate variants of HIV resistant to the antiviral effects of l-chicoric acid.
H9 and MT-2 are CD4+ T-lymphoblastoid cell lines that support replication of tissue culture-adapted and syncytium-inducing, lymphocytotropic clinical isolates of HIV-1. Both were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (Rockville, Md.). HIVNL4-3 plasmid (a gift from P. Krogstad, University of California, Los Angeles) was transfected in HeLa cells with Lipofectin (Gibco/BRL). Excess DNA was removed by washing, and the cells were cocultured with H9 cells for 18 h. The H9 cells were removed and recultured in growth medium. When the culture was 100% positive for HIV antigens by indirect immunofluorescence (35), the virus was inoculated onto H9 cells and incubated at 37°C for several weeks in the presence of 2 μM l-chicoric acid. When this culture was 100% positive, the virus was isolated and one aliquot was passaged in a similar manner in 4 μM l-chicoric acid. Finally, the virus was cultured in the presence of 8 μM l-chicoric acid, and the resultant virus was filter clarified, aliquoted, and stored at −70°C.
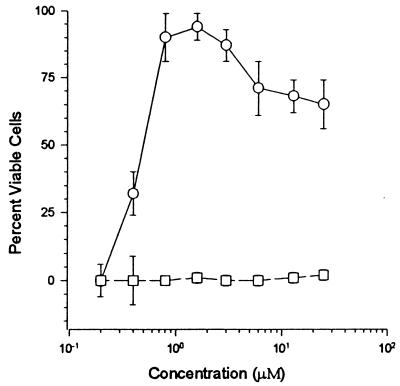
HIVNL4-3, following culture in 8 μM l-chicoric acid, was tested for resistance to the anti-HIV activity of the compound by using a cytopathicity-based assay (34, 36) first described by Montefiori et al. (26) (Fig. 1). This assay takes advantage of the lytic nature of T-cell-tropic clones of HIV, and decreased cell viability in the assay has been shown to correlate well with HIV replication (35). The 50% effective dose (ED50) of l-chicoric acid against HIVNL4-3 control virus was 400 nM, while HIVNL4-3 passaged in the presence of 8 μM l-chicoric acid was completely resistant to the compound (Fig. 1).
FIG. 1.
HIVNL4-3 passaged in l-chicoric acid develops drug resistance. HIVNL4-3 passaged in the presence (squares) or absence (circles) of increasing concentrations of l-chicoric acid was tested for sensitivity to l-chicoric acid. Each point is the mean of triplicate samples; the bars represent 1 standard deviation. HIV is a lytic virus, and increased levels of virus cause increased death of cells. Therefore, viability-based assays are good measures of HIV replication and anti-HIV activity. The viability of the cells is relative to that of cell controls (eight replicates; 100% viable) and virus controls (eight replicates; 0% viable) and was measured as first described by Montefiori et al. (26). Decreased cell viability in this assay correlates well with levels of HIV RNA, HIV protein expression, RT release, and numbers of infectious HIV particles (35).
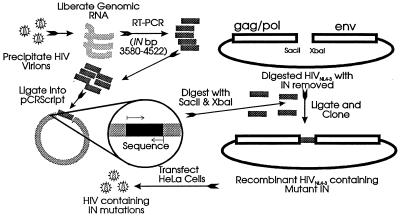
The overall cloning and sequencing strategy is illustrated in Fig. 2. For cloning and sequencing, HIV from 10 ml of culture was centrifuged at 33,000 × g for 4 h at 4°C. Virions were lysed, and RNA was isolated with Purescript (Gentra, Frederick, Md.). Primers used to amplify cDNA under these conditions recognize the 5′ and 3′ ends of integrase at nucleotide positions 3580 to 3605 (INS primer; 5′-GGTCTCCGCGGGAATCAGGAAAGTAC-3′) and 4497 to 4522 (INX primer; 5′-GCTTTTCTAGAAATATACATATGGTG-3′), respectively, and generate a 943-bp product. First-strand synthesis with INX primer and Superscript II, an avian myeloblastosis virus RT (Gibco/BRL), was performed at 42°C for 50 min according to the manufacturer’s instructions. Thirty-eight-cycle amplification was performed with thermostable Pfu DNA polymerase (Stratagene, La Jolla, Calif.) according to the manufacturer’s instructions. The optimum Mg2+ concentration for these studies was determined to be 1 mM. The conditions for PCR were 96°C for 1 min, 40°C for 30 s, and 72°C for 2 min for the first 2 cycles, followed by 96°C for 1 min, 55°C for 1 min, and 72°C for 3 min for 36 cycles. The final cycle included a 10-min, 70°C elongation step. The resulting RT-PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. Appropriately sized products were eluted from the gel and blunt end ligated into pCR-Script (Stratagene) for dideoxynucleotide sequencing with Sequenase II (U.S. Biochemical, Cleveland, Ohio) according to the manufacturer’s instructions. The entire integrase sequence was determined through the use of six oligonucleotide primers: INS, INX, Core 1 (5′-CAGCTGTGATAAATGTCAGCTA-3′ [nucleotides {nt} 3721 to 3741]), Core 2 (5′-CCATTTGTACTGCTGTCTTAA-3′ [nt 4122 to 4142]), INSPF (5′-GCAATTTCACCAGTACTACAGT-3′ [nt 3962 to 3983]), and INSPR (5′-GTAGGGAATGCCAAATTCCTG-3′ [nt 4016 to 4036]). Manual sequence analysis was confirmed by automated DNA sequencing.
FIG. 2.
Cloning strategy for analyzing mutations in integrase. The diagram illustrates the general cloning scheme designed to insert integrases from drug-resistant organisms into the wild-type HIVNL4-3 background. Briefly, RNA was isolated from virions and subjected to RT-PCR with primers that introduced silent mutations (SacII and XbaI sites) upstream and downstream of the integrase gene. These RT-PCR products were ligated into pCR-Script for sequencing. Clones containing mutant integrases were digested with SacII and XbaI, and the integrase gene was ligated into a similarly digested HIVNL4-3 plasmid, allowing the entire integrase gene, and only the integrase gene, to be switched into a drug-sensitive HIVNL4-3 background.
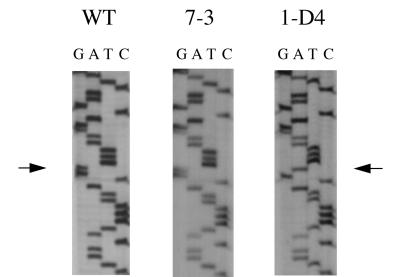
Sequencing the integrase genes from both drug-resistant and control HIVNL4-3 demonstrated several mutations. The control virus contained two silent mutations at nt 3832 and 4009. These silent mutations are believed to arise from a discrepancy in the published sequences of HIVNL4-3 (2, 39) and were likely not a result of the passage of HIV in the absence of inhibitor. Drug-resistant HIVNL4-3 had the same silent mutations as well as a single G-to-A transition at nucleotide position 4025 (Fig. 3), leading to an amino acid change from glycine to serine at amino acid 140.
FIG. 3.
Mutation of integrase at nt 4025 is associated with drug-resistance. Following the precipitation of virions, RNA was isolated and subjected to RT-PCR. The RT-PCR products were cloned into pCR-Script, and multiple clones were sequenced. WT, wild-type HIVNL4-3 sequence; 7-3, clone 7-3, a control sequence (HIVNL4-3 passaged in the absence of l-chicoric acid); 1-D4, clone 1-D4, a drug-resistant virus (HIVNL4-3 passaged in the presence of 8 μM l-chicoric acid). The arrows indicate the site of the mutation: the native sequence has a guanine at position 4025, while integrase from drug-resistant HIVNL4-3 contained an adenine at position 4025.
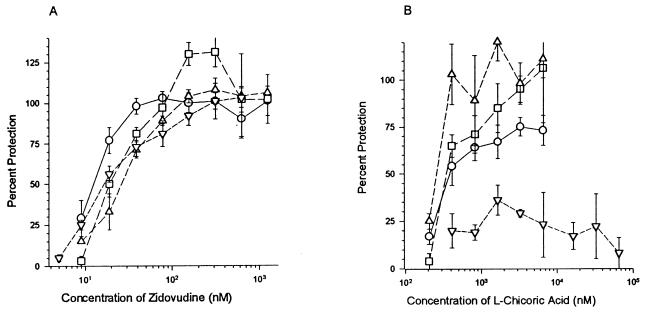
To determine whether this amino acid change was responsible for the observed resistant phenotype, the integrase genes from drug-resistant HIVNL4-3 were cloned into the native HIVNL4-3 plasmid (pNL4-3). This cloning was accomplished through site-directed mutagenesis, introducing several silent mutations immediately upstream and downstream of the integrase gene. These mutations generated two unique restriction sites (2, 39): an upstream SacII site and a downstream XbaI site (Fig. 2). Introduction of these mutations allowed the entire integrase gene, with only minimal upstream and downstream nucleotides, to be digested and “swapped” between drug-resistant and drug-sensitive clones. Two clones, 7-1 and 7-3, containing control integrase genes and wild type except for silent mutations generating the restriction sites, and clone 1-D4, containing drug-selected integrase with the G140S mutation, were chosen for further study. Once transfected into HeLa cells and amplified in H9 cells, the viruses from all three clones maintained the same sensitivity to zidovudine, a RT inhibitor, as the parental HIVNL4-3 (Fig. 4A). The three clones containing wild-type integrase (the control viruses, clones 7-1 and 7-3, and wild-type HIVNL4-3) maintained the l-chicoric acid-sensitive phenotype (Fig. 4B). Clone 1-D4, on the other hand, was resistant to the anti-HIV effects of l-chicoric acid. Although the drug-resistant clone retains some sensitivity to l-chicoric acid, the compound was unable to inhibit HIV replication by 50%, a requirement for determining activity (ED50). Therefore, the ED50 for the drug-resistant clone was >600-fold higher than that for the drug-sensitive clones (Table 1).
FIG. 4.
Resistance to l-chicoric acid but not zidovudine is conferred by G140S mutation in integrase. Wild-type HIVNL4-3 (circles), HIVNL4-3 control clones 7-1 (squares) and 7-3 (triangles), and clone 1-D4 (inverted triangles) were tested for sensitivity to zidovudine (A) or l-chicoric acid (B). The assays were performed as described in the legend to Fig. 1.
TABLE 1.
ED50s of l-chicoric acid, chlorogenic acid, and zidovudine against HIVLAI and HIVNL4-3 clones
| Virus | ED50 (μM)a
|
||
|---|---|---|---|
| Zidovudine | Chlorogenic acid | l-Chicoric acid | |
| HIVLAI | 0.031 | >700 | 8 |
| HIVNL4-3 | |||
| Wild type | 0.021 | >200 | 0.4 |
| Clone 7-1 | 0.024 | >200 | 0.11 |
| Clone 7-3 | 0.024 | >200 | 0.10 |
| Clone 1-D4 | 0.024 | >200 | >62.5 |
Amino acid 140 of integrase has not been mutated previously by site-directed mutagenesis. Furthermore, a search of the GenBank database does not indicate any naturally occurring mutations at this site. This amino acid is also highly conserved in integrases from other retroviruses, retrotransposons, and transposable elements of bacteria (19). Our data (Table 2) indicate that mutation at this site from the highly conserved glycine to serine has little effect on HIV replication but completely abrogates the anti-HIV activity of l-chicoric acid.
TABLE 2.
Characteristics of wild-type and drug-resistant variants of HIVNL4-3
| Virus | TCID50a (infectious particles/ml) | RTb
|
|
|---|---|---|---|
| cpm/ml | cpm/infectious particle | ||
| Wild-type NL4-3 | 640,000 | 1,091,040 | 1.7 |
| Clone 1-D4 | 640,000 | 575,008 | 0.9 |
Fifty-percent tissue culture infectious dose; determined by endpoint dilution on MT-2 cells in replicates of eight. Endpoints were measured after more than three replicative cycles (19 days).
l-Chicoric acid had been reported previously to have anti-HIV effects in tissue culture at nontoxic concentrations and to be a potent and selective inhibitor of HIV integrase in biochemical assays (25, 34, 36). Although one group has had difficulty demonstrating an anti-HIV effect of l-chicoric acid (27, 28), that may be due to their use of the tetrazolium dye, XTT, which is known to be incompatible with studies on bis-catechols (16), or because they used a different HIV isolate, HIVRF. Indeed, a previous report on the anti-HIV activity of a related compound, 3,4-dicaffeoylquinic acid (22), our reports of the anti-HIV activity of l-chicoric acid and the dicaffeoylquinic acids, and the independent confirmation by an outside source (18a) clearly show that these compounds inhibit HIV replication in tissue culture at nontoxic concentrations. By demonstrating that a single amino acid substitution at amino acid 140 in integrase confers resistance to l-chicoric acid, a potent inhibitor of HIV integrase, it is evident that the principal target for the anti-HIV activity of l-chicoric acid is integrase. Thus, there is now very strong evidence to suggest that small molecules can inhibit integration within cells and that inhibition of HIV integrase results in impaired HIV replication.
Previous experiments had shown that the ED50 of l-chicoric acid for the uncloned, tissue culture-adapted strain of HIV, HIVLAI, was approximately 4 μM (34). Such a high ED50, coupled with questions regarding the intracellular (or extracellular) mechanism of action, had dampened enthusiasm for developing l-chicoric acid as an individual compound or the dicaffeoylquinic and dicaffeoyltartaric acids as a class of integrase inhibitors worth investigating in detail. Despite 50% inhibitory concentrations (IC50) of ∼200 to 400 nM against recombinant HIV integrase in biochemical assays (34, 36), the compounds lacked potencies suitable for clinical evaluation. However, the integrase used in biochemical assays was from the HIVNL4-3 molecular clone of HIV, not uncloned HIVLAI. The results here demonstrate that l-chicoric acid inhibits HIVNL4-3 replication (ED50) at a concentration that virtually matches the IC50 against purified HIVNL4-3 recombinant integrase. Furthermore, small changes in sequence, indeed, a single amino acid change in integrase, can significantly affect susceptibility to l-chicoric acid in tissue culture.
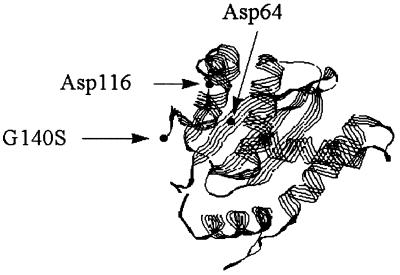
These data document that l-chicoric acid inhibits HIV replication, at least in part, through inhibition of HIV integrase. Furthermore, a single nonconservative amino acid change at a highly conserved amino acid, amino acid 140, is compatible with viral replication (Table 2) and confers resistance to an inhibitor of HIV-1 integrase. It is important to note that the packaging of virus and RT appears to be unaffected by the G140S mutation, as the RT-to-infectious particle ratio of the wild-type HIVNL4-3 and that of the drug-resistant variant were nearly equivalent (Table 2). This mutation occurs at a potentially important site in the integrase protein. First, this amino acid, glycine, is highly conserved throughout retroviral integrases (19). Second, this amino acid forms an anchor sequence in the integrase crystal structure (6): it was the last ordered amino acid prior to a disordered loop which contains one member of the catalytic triad of integrase (DD35E), a glutamine at residue 151. The location of this mutation in the catalytic core domain of integrase is indicated in Fig. 5.
FIG. 5.
HIV integrase catalytic core; diagram of the catalytic core domain containing the phenylalanine-to-lysine mutation at amino acid 185 (6). The location of the glycine-to-serine mutation at amino acid 140 is illustrated, as are the two catalytic residues, aspartate 64 and aspartate 116. The original figure, courtesy of D. Marcey (Kenyon College, Gambier, Ohio), was downloaded from http://www.kenyon.edu/depts/bmb/chime/integras/frames/intgrse.htm and is used with permission. The locations of the catalytic amino acids and the mutation were manually entered with Microsoft PowerPoint.
Thus, integrase inhibitors have promise as potential anti-HIV compounds. Powerful inhibition of integrase can impair virus replication, but any such inhibitors will likely need to be used in combination with existing anti-HIV agents as resistance, although difficult to achieve, requires only a single nucleotide change and results in only mild attenuation of viral replication. Now that an inhibitor of HIV replication that acts on integrase has been identified, it is possible to determine whether integrase is a suitable target in combination anti-HIV therapies. Ultimately, this class of anti-HIV agents may lead to the synthesis of clinically useful integrase inhibitors which can be used alone or in combination with existing anti-HIV agents to slow the progression to AIDS.
Acknowledgments
This work was supported in part by grants 1RO1-AI41360 and 5T32-GM-07311 from the Public Health Service.
We thank Suzanne Sandmeyer (University of California, Irvine), Hung Fan (University of California, Irvine), and Samson A. Chow (University of California, Los Angeles) for thoughtful suggestions on the manuscript and Brenda McDougall, Jean Kuan, Tracey Kim, and Keola Beale for their expert technical assistance. l-Chicoric acid was synthesized in the laboratory of, and kindly provided by, Manfred G. Reinecke (Texas Christian University, Fort Worth). The diagram of the integrase catalytic core was obtained from the Kenyon University website (http://www.kenyon.edu/depts/bmb/chime/integras/frames/intgrse.htm).
ADDENDUM IN PROOF
Recently, the X-ray crystal structure of the core domain of avian sarcoma virus (ASV) integrase in complex with an inhibitor of both ASV and HIV integrases was solved (J. Lubkowski, F. Yang, J. Alexandratos, A. Wlodawer, H. Zhao, T. R. Burke, Jr., N. Neamati, Y. Pommier, G. Merkel, A. M. Skalka, Proc. Natl. Acad. Sci. USA 95:4831–4836, 1998). This structure placed the glycine analogous to G140 of HIV integrase within close proximity to the inhibitor bound within the ASV integrase multimer interface. These findings support the interpretation that G140 is near an inhibitor-binding site within the HIV integrase core.
REFERENCES
- 1.Anonymous. AR-177 (zintevir) In: Abrams D, Cotton D, Markowitz M, Mayer M, editors. AIDS/HIV treatment directory. Vol. 8. New York, N.Y: American Foundation for AIDS Research; 1996. pp. 50–51. [Google Scholar]
- 2.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burke T R, Fesen M R, Mazumder A, Wang J, Carothers A M, Grunberger D, Driscoll J, Kohn K, Pommier Y. Hydroxylated aromatic inhibitors of HIV-1 integrase. J Med Chem. 1995;38:4171–4178. doi: 10.1021/jm00021a006. [DOI] [PubMed] [Google Scholar]
- 4.Cherepanov P, Este J A, Rando R F, Ojwang J O, Reekmans G, Steinfeld R, David G, DeClercq E, Debyser Z. Mode of interaction of G-quartets with the integrase of human immunodeficiency virus type 1. Mol Pharmacol. 1997;52:771–780. doi: 10.1124/mol.52.5.771. [DOI] [PubMed] [Google Scholar]
- 5.Collier A C, Coombs R W, Schoenfeld D A, Bassett R L, Timpone J, Baruch A, Jones M, Facey K, Whitacre C, McAuliffe V J, Friedman H M, Merigan T C, Reichman R C, Hooper C, Corey L. Treatment of human immunodeficiency virus infection with saquinavir, zidovudine, and zalcitabine. N Engl J Med. 1996;334:1011–1017. doi: 10.1056/NEJM199604183341602. [DOI] [PubMed] [Google Scholar]
- 6.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 7.Eich E, Pertz H, Kaloga M, Schulz J, Fesen M R, Mazumder A, Pommier Y. (−)-Arctigenin as a lead structure for inhibitors of human immunodeficiency virus type-1 integrase. J Med Chem. 1996;39:86–89. doi: 10.1021/jm950387u. [DOI] [PubMed] [Google Scholar]
- 8.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farnet C, Wang B, Lipford J R, Bushman F D. Differential inhibition of HIV-1 preintegration complexes and purified integrase protein by small molecules. Proc Natl Acad Sci USA. 1996;93:9742–9747. doi: 10.1073/pnas.93.18.9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fesen M R, Kohn K W, Leteurtre F, Pommier Y. Inhibitors of human immunodeficiency virus integrase. Proc Natl Acad Sci USA. 1993;90:2399–2403. doi: 10.1073/pnas.90.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fesen M R, Pommier Y, Leteurtre F, Hiroguchi S, Yung J, Kohn K W. Inhibition of HIV-1 integrase by flavones, caffeic acid phenethyl ester (CAPE) and related compounds. Biochem Pharmacol. 1994;48:595–608. doi: 10.1016/0006-2952(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 12.Gao Q, Gu Z, Parniak M A, Cameron J, Cammack N, Boucher C, Wainberg M A. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1993;37:1390–1392. doi: 10.1128/aac.37.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Q, Gu Z, Parniak M A, Li X, Wainberg M A. In vitro selection of variants of human immunodeficiency virus type 1 resistant to 3′-azido-3′-deoxythymidine and 2′,3′-dideoxyinosine. J Virol. 1992;66:12–19. doi: 10.1128/jvi.66.1.12-19.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu Z, Gao Q, Li X, Parniak M A, Wainberg M A. Novel mutation in the human immunodeficiency virus type 1 reverse transcriptase gene that encodes cross-resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine. J Virol. 1992;66:7128–7135. doi: 10.1128/jvi.66.12.7128-7135.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 16.Habtemariam S. Catechols and quercetin reduce through iron ions—a possible artifact in cell viability assays. Phytother Res. 1995;9:603–605. [Google Scholar]
- 17.Hammer S M, Squires K E, Hughes M D, Grimes J M, Demeter L M, Currier J S, Eron J J, Feinberg J E, Balfour H H, Jr, Deyton L R, Chodakewitz J A, Fischl M A AIDS Clinical Trials Group. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 18.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 18a.Hostomsky, Z. Personal communication.
- 19.Kulkosky J, Jones K S, Katz R A, Mack J P G, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaFemina R L, Graham P L, LeGrow K, Hastings J C, Wolfe A, Young S D, Emini E A, Hazuda D J. Inhibition of human immunodeficiency virus integrase by bis-catechols. Antimicrob Agents Chemother. 1995;39:320–324. doi: 10.1128/aac.39.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaFemina R L, Schneider C L, Robbins H L, Callahan P L, LeGrow K, Roth E, Schleif W A, Emini E A. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahmood N, Moore P S, De Tommasi N, De Simone F, Colman S, Hay A J, Pizza C. Inhibition of HIV infection by caffeoylquinic acid derivatives. Antiviral Chem Chemother. 1993;4:235–240. [Google Scholar]
- 23.Martin J L, Wilson J E, Haynes R L, Furman P A. Mechanism of resistance of human immunodeficiency virus type 1 to 2′,3′-dideoxyinosine. Proc Natl Acad Sci USA. 1993;90:6135–6139. doi: 10.1073/pnas.90.13.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazumder A, Gazit A, Levitzki A, Nicklaus M, Yung J, Kohlhagen G, Pommier Y. Effects of tyrphostins, protein kinase inhibitors, on human immunodeficiency virus type 1 integrase. Biochemistry. 1995;34:15111–15122. doi: 10.1021/bi00046a018. [DOI] [PubMed] [Google Scholar]
- 25.McDougall B, King P J, Wu B W, Hostomsky Z, Reinecke M G, Robinson W E., Jr Dicaffeoylquinic and dicaffeoyltartaric acids are selective inhibitors of human immunodeficiency virus type 1 integrase. Antimicrob Agents Chemother. 1998;42:140–146. doi: 10.1128/aac.42.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montefiori D C, Robinson W E, Jr, Schuffman S S, Mitchell W M. Evaluation of antiviral drugs and neutralizing antibodies against human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J Clin Microbiol. 1988;26:231–235. doi: 10.1128/jcm.26.2.231-235.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neamati N, Hong H, Mazumder A, Wang S, Sunder S, Nicklaus M C, Milne G W A, Proksa B, Pommier Y. Depsides and depsidones as inhibitors of HIV-1 integrase: discovery of novel inhibitors through 3D database searching. J Med Chem. 1997;40:942–951. doi: 10.1021/jm960759e. [DOI] [PubMed] [Google Scholar]
- 28.Neamati N, Hong H, Sunder S, Milne G W, Pommier Y. Potent inhibitors of human immunodeficiency virus type 1 integrase: identification of a four-point pharmacophore and tetracyclines as novel inhibitors. Mol Pharmacol. 1997;52:1041–1055. doi: 10.1124/mol.52.6.1041. [DOI] [PubMed] [Google Scholar]
- 29.Nishizawa M, Yamagishi T, Dutschman G E, Parker W B, Bonder A J, Kilkuskie R E, Cheng Y-C, Lee K-H. Anti-AIDS agents, 1, isolation and characterization of four new tetragalloylquinic acids as a new class of HIV reverse transcriptase inhibitors from tannic acid. J Nat Prod. 1989;52:762–768. doi: 10.1021/np50064a016. [DOI] [PubMed] [Google Scholar]
- 30.Nonaka G-I, Nishioka I, Nishizawa M, Yamagishi T, Kashiwada Y, Dutschman G E, Bonder A, Kilkuskie R E, Cheng Y-C, Lee K-H. Anti-AIDS agents, 2, inhibitory effect of tannins on HIV reverse transcriptase and HIV replication in H9 lymphocyte cells. J Nat Prod. 1990;53:589–595. doi: 10.1021/np50069a008. [DOI] [PubMed] [Google Scholar]
- 31.Ojwang J O, Buckheit R W, Pommier Y, Mazumder A, De Vreese K, Esté J A, Reymen D, Pallansch L A, Lackman-Smith C, Wallace T L, De Clercq E, McGrath M S, Rando R F. T30177, an oligonucleotide stabilized by an intramolecular guanosine octet, is a potent inhibitor of laboratory strains and clinical isolates of human immunodeficiency virus type 1. Antimicrob Agents Chemother. 1995;39:2426–2435. doi: 10.1128/aac.39.11.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poiesz B J, Ruscetti F W, Gazder A F, Bunn B A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson W E., Jr HIV integrase: the next target? Infect Med. 1998;15:129–137. [Google Scholar]
- 34.Robinson W E, Jr, Cordeiro M, Abdel-Malek S, Jia Q, Chow S A, Reinecke M G, Mitchell W M. Dicaffeoylquinic acid inhibitors of human immunodeficiency virus (HIV) integrase: inhibition of the core catalytic domain of HIV integrase. Mol Pharmacol. 1996;50:846–855. [PubMed] [Google Scholar]
- 35.Robinson W E, Jr, Montefiori D C, Gillespie D H, Mitchell W M. Complement-mediated, antibody-dependent enhancement of human immunodeficiency virus type 1 (HIV-1) infection in vitro increases HIV-1 RNA and protein synthesis and infectious virus production. J Acquired Immune Defic Syndr. 1989;2:33–42. [PubMed] [Google Scholar]
- 36.Robinson W E, Jr, Reinecke M G, Abdel-Malek S, Jia Q, Chow S A. Inhibitors of HIV-1 replication that inhibit HIV integrase. Proc Natl Acad Sci USA. 1996;93:6326–6331. doi: 10.1073/pnas.93.13.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth M J, Schwartzberg P, Tanese N, Goff S P. Analysis of mutations in the integration function of Moloney murine leukemia virus: effect on DNA binding and cutting. J Virol. 1990;64:4709–4717. doi: 10.1128/jvi.64.10.4709-4717.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakai H, Kawamura M, Sakuragi J, Sakuragi S, Shibata R, Ishimoto A, Ono N, Ueda S, Adachi A. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J Virol. 1993;67:1169–1174. doi: 10.1128/jvi.67.3.1169-1174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salminen M, Koch C, Sanders-Buell E, Ehrenberg P, Michael N, Carr J, Burke D, McCutchan F. Recovery of virtually full-length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology. 1995;213:80–86. doi: 10.1006/viro.1995.1548. [DOI] [PubMed] [Google Scholar]
- 40.Shin C G, Taddeo B, Haseltine W A, Farnet C M. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J Virol. 1994;68:1633–1642. doi: 10.1128/jvi.68.3.1633-1642.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 42.Zhao H, Neamati N, Mazumder A, Sunder S, Pommier Y, Burke T R., Jr Arylamide inhibitors of HIV-1 integrase. J Med Chem. 1997;40:1186–1194. doi: 10.1021/jm960449w. [DOI] [PubMed] [Google Scholar]