Summary
Selenophosphate synthetase (SEPHS) plays an essential role in selenium metabolism. Two mammalian SEPHS paralogues, SEPHS1 and SEPHS2, share high sequence identity and structural homology with SEPHS. Here, we report nine individuals from eight families with developmental delay, growth and feeding problems, hypotonia, and dysmorphic features, all with heterozygous missense variants in SEPHS1. Eight of these individuals had a recurrent variant at amino acid position 371 of SEPHS1 (p.Arg371Trp, p.Arg371Gln, and p.Arg371Gly); seven of these variants were known to be de novo. Structural modeling and biochemical assays were used to understand the effect of these variants on SEPHS1 function. We found that a variant at residue Trp352 results in local structural changes of the C-terminal region of SEPHS1 that decrease the overall thermal stability of the enzyme. In contrast, variants of a solvent-exposed residue Arg371 do not impact enzyme stability and folding but could modulate direct protein-protein interactions of SEPSH1 with cellular factors in promoting cell proliferation and development. In neuronal SH-SY5Y cells, we assessed the impact of SEPHS1 variants on cell proliferation and ROS production and investigated the mRNA expression levels of genes encoding stress-related selenoproteins. Our findings provided evidence that the identified SEPHS1 variants enhance cell proliferation by modulating ROS homeostasis. Our study supports the hypothesis that SEPHS1 plays a critical role during human development and provides a basis for further investigation into the molecular mechanisms employed by SEPHS1. Furthermore, our data suggest that variants in SEPHS1 are associated with a neurodevelopmental disorder.
Keywords: SEPHS1, clinical exome sequencing, developmental delay, hypotonia, neurodevelopmental disorder, ROS production, selenophosphate synthetase, selenium metabolism
SEPHS1 is in the selenocysteine biosynthetic pathway. We report nine individuals from eight families with developmental delay, growth and feeding problems, hypotonia, and dysmorphic features, all with heterozygous missense variants in SEPHS1. Our findings provide insight into the molecular pathogenesis of this neurodevelopmental disorder.
Main text
Selenium (Se) is the sole essential trace element to be specified in the genetic code1 and is incorporated into proteins as the nonstandard amino acid selenocysteine (Sec) which is encoded by the UGA codon.2,3,4,5,6,7,8,9 The human genome encodes 25 selenoproteins, many of which are involved in metabolism, maintenance of the cellular redox environment, and oxidative stress management.10,11,12,13 The Sec biosynthesis machinery is responsible for generating Sec-tRNASec and delivering it to the ribosome for incorporation into the nascent polypeptide chain.2,3,4 The primary Se donor for Sec biosynthesis is provided by selenophosphate synthetase (SEPHS). Using ATP and inorganic Se as substrates, SEPHS cleaves and transfers the γ-phosphate of ATP onto selenide to form mono-selenophosphate (SeP). SeP is then delivered to O-phosphoseryl selenium transferase (SEPSECS) for the terminal reaction of Sec synthesis and subsequent incorporation into selenoproteins.7
Impairment in the Sec pathway has been associated with diverse human diseases including cardiovascular, central nervous, and endocrine system disorders, cancer, and pregnancy complications (miscarriage, premature birth, and preeclampsia).1,6,7,8,9,10,11,12,13 Currently, three genes in the Sec biosynthesis pathway have been identified to cause autosomal-recessive disease: SECISBP2 (MIM: 607693), SEPSECS (MIM: 613009), and TRU-TCA1-1 (MIM: 620198).10,12,14,15,16,17 Defects in SECISBP2 are associated with an abnormal thyroid hormone metabolism disorder (MIM: 609698),which is characterized by multiorgan defects including abnormal thyroid hormone metabolism, myopathy, hearing loss, and male infertility.15 SEPSECS pathogenic variants are associated with pontocerebellar hypoplasia type 2D (MIM: 613811) which is characterized by progressive microcephaly, postnatal onset of progressive atrophy of the cerebrum and cerebellum, profound intellectual delay, spasticity, and variable seizures.14,18 Pathogenic variants in TRU-TCA1-1 are associated with autosomal-recessive thyroid hormone metabolism, abnormal 3 (MIM: 165060) which is characterized by euthyroid hyperthyroxinemia, with elevated free T4 and reverse T3 levels and normal TSH (MIM: 188540) and free T3 levels.16,17 Affected individuals also show low plasma selenium levels and reduced levels of stress-related selenoproteins16,17
In the human Sec biosynthesis pathway, there are two SEPHS genes encoding SEPHS1 and SEPHS2 that both share a high degree of sequence identity and structural homology. However, it has been established that SEPHS2 catalyzes SeP synthesis.19,20 Due to the implications of the Sec biosynthesis pathway to human disease, variants in SEPHS1 (MIM: 600902) or SEPHS2 (MIM: 606218) may be linked to a genetic disorder. Interestingly, these two genes have yet to be implicated in human disease in OMIM.
SEPHS1 is an essential protein in mammals and is involved in promoting cell proliferation, differentiation, and survival.21,22,23,24 SEPHS1 has been found to modulate the cellular redox environment and promote cell migration, adhesion, and invasion.21,24 It is highly expressed in hepatocytes and thyroid glandular cells. The gene is ubiquitously expressed in the brain but has no region specificity.21,25 In Drosophila melanogaster, Sps1 is highly expressed during embryonic development and is prevalent in regions of the embryo undergoing rapid cell proliferation and differentiation.26
Given the fact that SEPHS1 does not directly participate in generating SeP, it is reasonable to propose that SEPHS1 plays a critical role in cellular processes outside of the Sec biosynthetic pathway. It has been found SEPHS1 modulates redox homeostasis and is responsible for cell proliferation and defense.19,20,25 However, the homeostatic mechanism by which this protein exerts its function is still poorly understood.
Sephs1 knockout mice embryos showed gradual increases in reactive oxygen species (ROS) resulting in growth retardation, structural brain abnormalities, and disrupted cardiac development.23,27 Therefore, it is likely that in humans, SEPHS1 is an essential gene and pathogenic variants may cause a human phenotype resembling other disorders in the Sec biosynthesis pathway.
In this study, nine individuals were identified to have heterozygous missense variants in SEPHS1 by clinical exome sequencing (ES) (see supplemental material and methods). They were evaluated by clinical genetic centers in the United States and Canada via clinical exam, laboratory tests, and brain imaging. Clinical data were obtained from the providers via questionnaire. This study was approved by the institutional review board (IRB) (see supplemental material and methods). All parents or legal guardians provided written informed consent for their children to participate in the study and for publication of clinical information. Seven individuals (individuals 1–4 and 7–9) had ES performed by GeneDx. Individual 5 had ES performed at a pediatric medical center and was ascertained via personal communication. Individual 6 had ES performed through Care4Rare and was ascertained via GeneMatcher.28
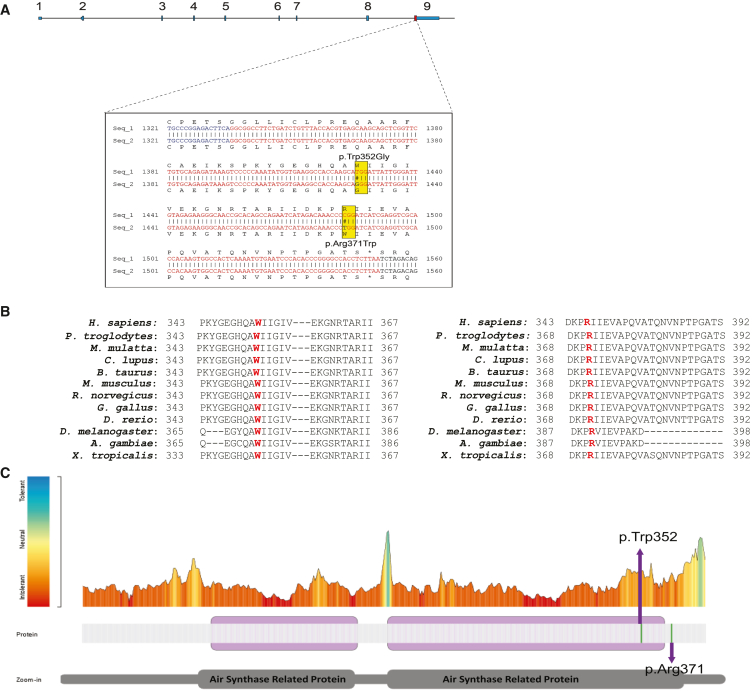
Eight individuals (individuals 1–8) had variants at the same amino acid position, p.Arg371 (Table 1). Individuals 1–3 shared a recurrent variant in SEPHS1 (GenBank: NM_012247.4: c.1111C>T [p.Arg371Trp]). This variant was de novo in individuals 1 and 2, but parental testing for individual 3 was not available (Figures S1A and S1B). Individuals 4–6 shared a different missense variant at the same residue in SEPHS1 (GenBank: NM_012247.4: c.1112G>A [p.Arg371Gln]). This variant was de novo in individuals 4–6 (Figures S1D and S1E). Individuals 7 and 8 were similarly affected siblings who shared a third unique missense variant at the p.Arg371 residue in SEPHS1 (GenBank: NM_012247.4: c.1111C>G [p.Arg371Gly]) (Figures S1F and S1G). This variant was de novo in the siblings, suggesting the possibility of parental germline mosaicism in this family. In addition, individual 7 had a de novo SRCAP (MIM: 611421) variant, GenBank: NM_006662.2 (c.3833C>A [p.Ser1278∗]), that was classified as a variant of uncertain significance (VUS).29 Individual 9 had a de novo missense variant at a different residue in SEPHS1 (GenBank: NM_012247.4: c.1054T>G [p.Trp352Gly]) (Figure S1H). All variants were located within exon 9 of SEPHS1 and located in/near the region encoding the AIR synthase-related protein, C-terminal domain of SEPHS1 (Figure 1A). None of these variants were reported in Genome Aggregation Database v.4.0.0 (gnomAD). SEPHS1 contains fewer missense variants than expected (Z score = 3.24) and is considered intolerant to loss of function (pLI = 0.98).30 The gene has a relatively %HI score implying that it is likely that variants within this gene operate through a mechanism of haploinsufficiency.31 The in silico prediction tool Provean predicts each of the identified missense changes to have a deleterious effect on SEPHS1 (Table 1). The amino acids at the two variant sites identified in the nine affected individuals (Trp352 and Arg371) are highly conserved throughout multiple species (Figure 1B) indicating that the tryptophan (Trp) and arginine (Arg) play an essential role in protein function. To identify the tolerance landscape of these missense variants, we utilized Metadome regional tolerance plot for genetic variation based on the ratio of observed missense and synonymous (dN/dS) variants that are included in gnomAD.32 Both altered positions in SEPHS1 are in regions that are intolerant to missense variants (Figure 1C).
Table 1.
Molecular findings in individuals with heterozygous missense SEPHS1 variants
| Individual 1 | Individual 2 | Individual 3 | Individual 4 | Individual 5 | Individual 6 | Individual 7 | Individual 8 | Individual 9 | |
|---|---|---|---|---|---|---|---|---|---|
| SEPHS1 variant (NM_012247.4) | c.1111C>T | c.1111C>T | c.1111C>T | c.1112G>A | c.1112G>A | c.1112G>A | c.1111C>G | c.1111C>G | c.1054T>G |
| Exon | exon 9 | exon 9 | exon 9 | exon 9 | exon 9 | exon 9 | exon 9 | exon 9 | exon 9 |
| Protein | p.Arg371Trp | p.Arg371Trp | p.Arg371Trp | p.Arg371Gln | p.Arg371Gln | p.Arg371Gln | p.Arg371Gly | p.Arg371Gly | p.Trp352Gly |
| Inheritance | de novo | de novo | unknown | de novo | de novo | de novo | de novo | de novo | de novo |
| PROVEAN (cutoff 2.5) | −3.75 | −3.75 | −3.75 | −1.15 | −1.15 | −1.15 | −3.65 | −3.65 | −10.49 |
| GnomAD frequency | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Reported CNV and SNVs | none reported | none reported | CMA: gain of chr13q14.1 (VUS, unknown inheritance)a | none reported | none reported | CMA: gain of chr3p25.3 (VUS, maternally inherited)a | de novo VUS in SRCAP, NM_006662.2: c.3833C>A (p.Ser1278∗)29 | none reported | heteroplasmic variant in MT-TL2; heterozygous VUS RAPSNa |
| ACMG criteria applied33 | internal criteria, PM2, PP3 | internal criteria, PM2, PP3 | internal criteria, PM2, PP3 | internal criteria, PS2, PM2, BP4 | internal criteria, PS2, PM2, BP4 | internal criteria, PS2, PM2, BP4 | internal criteria, PM2, PP3 | internal criteria, PM2, PP3 | PS2, PS3, PM2, PP3 |
| ACMG Class | PATH | PATH | PATH | PATH | PATH | PATH | PATH | PATH | PATH |
| ClinVar accession | SCV002103293 .1 | SCV002103293.1 | SCV002103293.1 | SCV002103295.1 | SCV002103295.1 | SCV002103295.1 | SCV002103294.2 | SCV002103294.2 | SCV002103296.1 |
CMA, chromosomal microarray
No HGVS notation available
Figure 1.
Identification of missense SEPHS1 variants in nine individuals
(A) Schematic of SEPHS1 organization. The blue boxes reflect exons. The yellow box depicts SEPHS1 variant position.
(B) Evolutionary conservation of missense variants observed in SEPHS1. Protein alignment of SEPHS1 orthologs was performed to determine protein sequence conservation of the region of interests. Residues impacted by variants within our cohort are highlighted in red. The Trp352 and Arg371 amino acids are highly conserved from human to frog.
(C) Tolerance Landscape of SEPHS1. The tolerance landscape of SEPHS1 was generated using Metadome (https://stuart.radboudumc.nl/metadome/). The color in the plot is an indication for the tolerance (red, intolerant; blue, tolerant). Graphical representation of the linear protein structure of SEPHS1 with two functional Air Synthase Related Protein domains is below the plot. Missense variants observed in this study are labeled, p.Arg371 and p.Trp352G. The missense variants in SEPHS1 are located in regions that are intolerable for variation (red).
The clinical features of the nine individuals are summarized in Tables 2 and S1 and Figure S2. The cohort is comprised of 5 females and 4 males. The ages of the cohort ranged from two days to 16 years. Individual 7 expired at two days of age due to complications of persistent pulmonary hypertension of the newborn (PPHN); therefore, most of the phenotypic comparison was made among the other individuals. Despite the small sample size, there was a remarkably strong clinical overlap (Table 2; Figure S2). The most consistent feature observed in our cohort was developmental delay (8/8; 100%), with five of those individuals known or suspected to have intellectual disability. Two of the affected individuals also had neurobehavioral diagnoses (attention-deficit/hyperactivity disorder in two; obsessive-compulsive disorder in one). Hypotonia and/or muscle weakness was reported in all but one affected individual. Additional neurologic/neuromuscular findings present in single affected individuals include petit mal seizures, lower limb myalgia and fatigue, autoimmune encephalitis, and decreased muscle mass. None of the affected individuals who underwent brain MRI (n = 5) had a major malformation. Growth was also widely impacted. Five affected individuals had a history of both poor height growth and weight gain, four of whom had history of feeding difficulties. Two individuals of typical size also had a history of feeding problems; tube feedings were required in three affected individuals. Multiple facial dysmorphisms were observed in seven affected individuals (Table S1). Issues affecting other body systems were more variable within the cohort. Endocrine abnormalities reported in a single affected individual include Hashimoto’s thyroiditis (individual 5) and elevated B6 (individual 2). Two affected individuals had growth hormone deficiency (individuals 4 and 9) and the siblings had congenital hypocalcemia (individuals 7 and 8). Respiratory issues seen in multiple affected individuals include obstructive sleep apnea (n = 3), laryngomalacia (n = 2), recurrent pneumonia (n = 2), and asthma (n = 2). Gastrointestinal issues included constipation (n = 3) and anorectal malformation (n = 1). Two affected individuals had myopia, and ophthalmologic issues reported in single affected individuals included strabismus, astigmatism, ptosis, and congenital entropion. Cardiac/vascular anomalies were seen in three affected individuals (aberrant right subclavian artery in one affected individual; persistent pulmonary hypertension of the newborn in the siblings), with three affected individuals having normal echocardiograms. Overall, it is likely that de novo variants in SEPHS1 that disrupt SEPHS1 function result in a syndromic neurodevelopmental disorder. Suggestions for clinical care based on the medical and developmental histories of our cohort can be found in the supplemental note.
Table 2.
Clinical characteristics of individuals with SEPHS1 variants
| Demographics | Individual 1 | Individual 2 | Individual 3 | Individual 4 | Individual 5 | Individual6 | Individual 7 | Individual 8 | Individual9 |
|---|---|---|---|---|---|---|---|---|---|
| Age at last evaluation | 2.5 years | 23 months | 8 years | 11 years | 16 years | 7.6 years | 2 days | 3.5 years | 4 years |
| Neurocognitive features | |||||||||
| Developmental delay | + | + | + | + | + | + | NR | + | + |
| Intellectual disability | NR | – | + | NR | + | possiblea | NR | + | possiblea |
| Neurobehavioral diagnoses | NR | NR | NR | NR | + | + | NR | NR | – |
| Hypotonia and/or muscle weakness | + | + | + | – | + | + | + | NR | + |
| Clinically significant brain malformation on MRI | NR | NR | – | – | – | NR | NR | – | – |
| Other neurologic findings | – | – | + | – | + | + | NR | NR | – |
| Growth-related features | |||||||||
| History of poor height growth | + | + | – | + | + | – | – | – | + |
| History of poor weight gain | + | + | – | + | + | – | NR | – | + |
| Feeding difficulties | + | + | + | – | + | + | NR | NR | + |
| Additional features | |||||||||
| Dysmorphic craniofacial featuresb | + | + | + | + | + | + | NR | NR | + |
| Endocrine | – | + | NR | + | + | NR | + | + | – |
| Respiratory | + | – | + | + | + | – | + | + | – |
| Ophthalmologic | + | – | – | NR | + | + | NR | NR | + |
| Cardiac | – | – | – | – | + | NR | + | + | – |
| Gastrointestinal | + | – | + | + | – | – | NR | NR | NR |
NR, data not reported
No formal assessment of intellectual disability
Dysmorphic facial features are described in Table S1
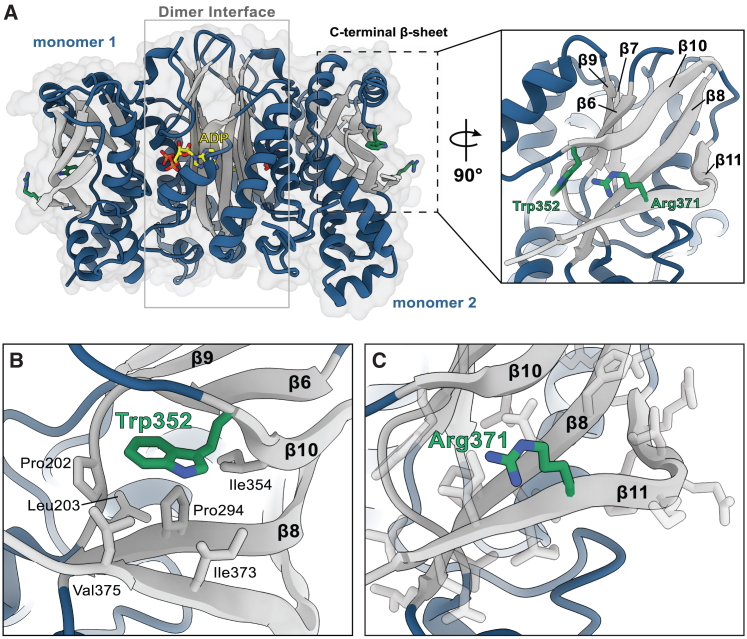
To further demonstrate whether these variants in SEPHS1 disrupt SEPHS1 function, we investigated the effect of the variants on protein function by using both biochemical assays and structural modeling. When we mapped the location of the altered residues, we observed that both residues Trp352 and Arg371 are situated within a six-stranded β-sheet in the C-terminal domain (Figure 2A). The architecture of this β-sheet is notated as β7↑ β9↓ β6↑ β10↓ β8↑ β11↑ with Trp352 located on the N-terminal tail of β10 and Arg371 found in the middle of β11 (Figure 2A). The side chain of Trp352 makes extensive contacts with Pro202, Leu203, Pro294, Ile354, Ile373, and Val375, which contribute to the formation of a hydrophobic pocket that stabilizes three antiparallel β-strands β8, β10, and β11 (Figure 2B). By contrast, the side chain of Arg371 is solvent-exposed and does not appear to participate in any interactions stabilizing the enzyme structure (Figure 2C).
Figure 2.
Structural modeling of SEPHS1
(A) Overall structural organization of SEPHS1 (PDB: 3FD6). Interactions between the N-terminal regions of monomers 1 and 2 form a β-barrel-like structure to stabilize the homodimer and active sites. ADP is shown in yellow. Residues Trp352 and Arg371 are situated in the C-terminal β-sheet and are highlighted in green.
(B) Trp352 resides in a hydrophobic pocket stabilized by interactions with neighboring hydrophobic residues.
(C) Arg371 is located on the solvent-exposed face of β11 and does not appear to form any significant interactions with surrounding residues. All modeling was performed using PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
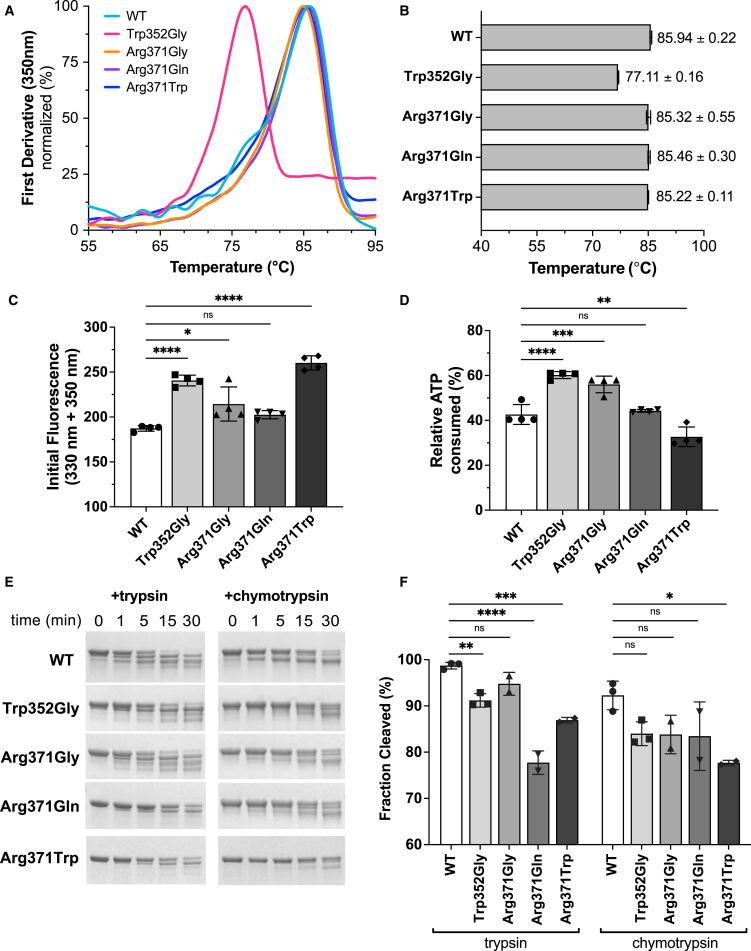
Given that affected residues are far removed from the dimer interface and active sites, we sought to determine how pathogenic SEPSH1 variants may affect overall protein stability and architecture, local conformational stability, and enzyme function. Using site-directed mutagenesis, we recapitulated the de novo variants (p.Trp352Gly, p.Arg371Gly, p.Arg371Gln, and p.Arg371Trp) and wild-type (WT) SEPHS1 enzymes in E. coli. All proteins were purified with a combination of affinity and size-exclusion chromatography. The native PAGE analysis confirmed all variants maintained the native dimer form, with the expected molecular weight of ∼85 kDa (Figure 3A). We then employed size-exclusion chromatography coupled with multi-angle light scattering (SEC-MALS) to accurately determine the molecular weight (MW) and polydispersity of all SEPHS1 samples. The average MW of all enzymes was calculated to be 85.41% ± 0.87% kDa with a polydispersity constant of 1.0% ± 0.001% (Figure S3), confirming that all recombinant SEPHS1 enzymes exist as dimers in solution.
Figure 3.
Analysis of SEPHS1 stability and enzymatic activity
(A) Protein thermal stability was measured using the Tycho NT.6 instrument. Intrinsic protein fluorescence was monitored as a thermal ramp was applied to each sample. The resulting curves are plotted as the first derivative and used to calculate the inflection temperature (Ti) for each sample.
(B) Calculated Ti values.
(C) Initial fluorescence values measured for each sample using the Tycho NT.6. All Ti and initial fluorescence values are reported with ±SD from 4 independent runs.
(D) SEPHS1-mediated ATP hydrolysis monitored using the CellTiter-Glo Assay 2.0 assay kit. Relative ATP consumption was measured after 18 h of incubation at +37°C. Assays were performed in triplicates and reported with ±SD.
(E) Trypsin or chymotrypsin was added to SEPHS1 samples and allowed to digest the protein for 30 min. Time points were taken at 0, 1, 5, 15, and 30 min. Digested products were analyzed on 4%–20% TGX gels. Representative gels are shown from 2 to 3 independent cleavage reactions.
(F) Densitometric quantification performed with ImageJ of total fraction cleaved after 30 min. Statistical significance for all panels was determined by one-way ANOVA with the Bonferroni correction, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Though not directly participating in the Sec biosynthetic pathway, SEPHS1 retains the ATPase activity. We investigated the possibility that pathogenic variants impact the active-site conformation and enzymatic activity. Using a luciferase-based ATPase assay, we monitored the relative change in ATP concentration over time. Our data show that WT, p.Trp352Gly, p.Arg371Gly, and p.Arg371Gln all consumed ∼50%–60% of the available ATP over the course of the assay (Figure 3D). The p.Arg371Trp exhibited a moderate reduction in activity consuming only ∼30% of the total ATP, suggestive of hypomorphic properties. Ultimately, we found all SEPHS1 variants are capable of hydrolyzing ATP in vitro, suggesting the dimer interface and active sites are not detrimentally impacted by the variants for this function.
To investigate whether the overall protein stability was altered by the variants, we used the Nanotemper instrument Tycho NT.6 to monitor intrinsic fluorescence changes of each enzyme sample during heat denaturation. When comparing the inflection temperatures (Ti) of the variants to WT SEPHS1, we observed a substantial decrease (∼10°C) in p.Trp352Gly while there was very little change with any of the Arg371 variants (Figure 3B). As this method is based on the intrinsic fluorescence of Trp residues, we expected the initial fluorescence of the p.Trp352Gly variant would decrease as a Trp residue was removed. Unexpectedly, the initial fluorescence for p.Trp352Gly was significantly higher than WT SEPHS1 (Figure 3C). This suggests that the variant causes other Trp residues, which are normally buried in the WT protein, to be exposed in the altered enzyme. Furthermore, p.Arg371Gly exhibited a small increase in initial fluorescence, suggesting slight changes in the local environment surrounding residue 371 but not enough to reduce thermal stability of the entire protein.
These results led us to speculate that pathogenic variants may elicit local structural changes in the C-terminal region of SEPHS1. To probe this, we used limited proteolysis with trypsin and chymotrypsin proteases (Figure 3E). In comparison to WT, SEPHS1 variants exhibited similar cleavage patterns for both proteases but exhibited varying degrees of proteolytic resistance. p.Trp352Gly, p.Arg371Gln, and p.Arg371Trp variants exhibited the highest resistance to proteolytic cleavage. Taken together, our results suggest that Trp352 stabilizes the C-terminal domain of SEPSH1 and, when replaced with Gly, induces local structural changes across the 6-stranded β-sheet. While exhibiting small changes in initial fluorescence and resistance to proteolysis, most of the Arg371 variants did not have a significant impact on protein stability. Thus, we suggest Arg371 variant does not greatly compromise the structural integrity of the C-terminal β-sheet and instead may play an integral role in modulating protein-protein interactions in the cell.
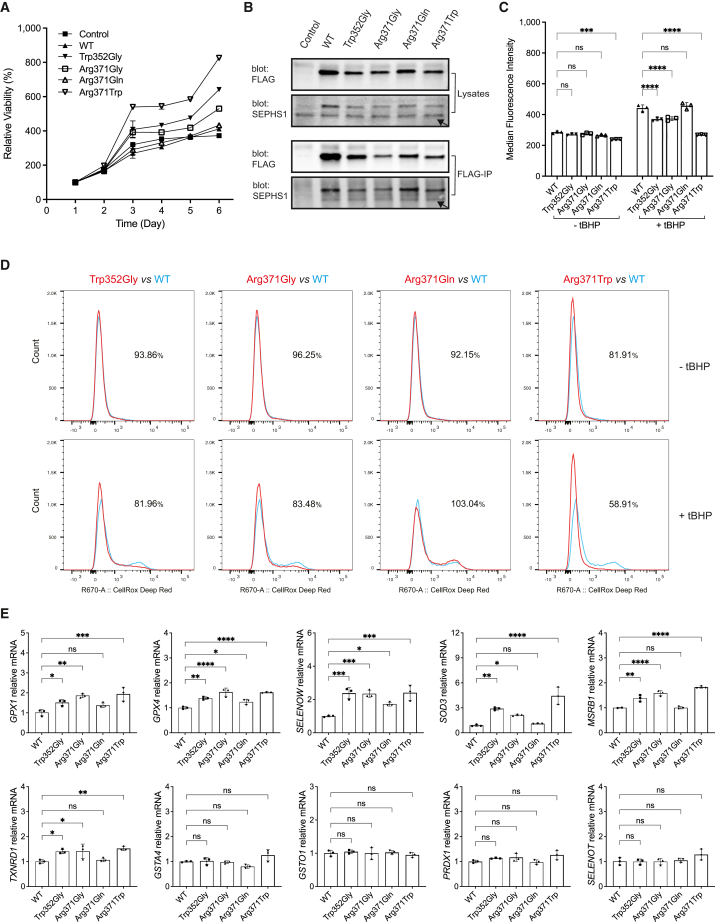
To investigate the physiological functions of the SEPHS1 variants, we introduced FLAG-tagged SEPHS1 variants into human neuroblastoma SH-SY5Y cells through lentiviral-mediated transduction. We initiated our assessment by examining the impact of SEPHS1 variants on cell proliferation using the alamarBlue viability assay (Figure 4A). The results indicated that the growth rate of p.Arg371Gln was comparable to that of the control and WT, while p.Trp352Gly, p.Arg371Gly, and p.Arg371Trp exhibited substantially increased cell proliferation, with p.Arg371Trp demonstrating the most pronounced effect among them. Furthermore, immunoprecipitation using the M2 anti-FLAG antibody revealed that the dimerization capability of FLAG-tagged SEPHS1 variants with endogenous SEPHS1 remained unaltered (Figure 4B). These findings align with our earlier observations from bacterial recombinant expression, indicating that the identified variants would not influence SEPHS1 functions by affecting SEPHS1 dimerization.
Figure 4.
Analysis of SEPHS1 cellular functions in SH-SY5Y cells
(A) Cell proliferation analysis of SH-SY5Y cells overexpressing SEPHS1 variants at the indicated days. The % live cell values were normalized to the day 1 cells (considered as 100% viable).
(B) Immunoblotting for immunoprecipitation of FLAG-tagged SEPHS1 variants. Arrows indicate the endogenous SEPHS1.
(C) Fluorescence-activated cell sorting (FACS) analysis of CellROX Deep Red fluorescence in SH-SY5Y cells with or without treatment with ROS inducer tBHP, showing the quantitative bar graphs and statistical analysis of the median fluorescence intensity (MFI). Error bars derived from three independent measurements.
(D) FACS histograms showing ROS production as described in (C). The numerical values accompanying each histogram signify the percentage of MFI for SEPHS1 variants relative to the wild-type (WT).
(E) Quantitative real-time PCR for genes encoding stress-related selenoproteins and ROS-scavenging enzymes in SH-SY5Y cells harboring SEPHS1 variants. mRNA levels were normalized to PolB. Data are represented as mean ± SEM, n = 3. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 and ns, not statistically significant.
Given SEPHS1’s pivotal role in regulating stress-related selenoproteins and maintaining redox homeostasis, we further investigated whether the identified variants influence cell proliferation through ROS production (Figure 4C). In the absence of oxidative stress stimulation, there were no significant differences in intracellular ROS production observed between the variants and WT (Figures 4C and 4D, top). Intriguingly, when the oxidant tBHP was employed in the cells to induce oxidative stress, p.Trp352Gly, p.Arg371Gly, and p.Arg371Trp exhibited a significant reduction in ROS production compared to WT. Among the variants, p.Arg371Trp displayed the most substantial reduction, while p.Arg371Gln had a marginal effect on tBHP-induced ROS production (Figures 4C and 4D, bottom).
The observed impact of SEPHS1 variants on ROS accumulation prompted us to investigate the mRNA expression levels of genes encoding stress-related selenoproteins, specifically glutathione peroxidase 1 (GPX1 [MIM: 138320]), glutathione peroxidase 4 (GPX4 [MIM: 138322]), thioredoxin reductase 1 (TXNRD1 [MIM: 601112]), selenoprotein T (SELENOT [MIM: 607912]), selenoprotein W (SELENOW [MIM: 603235]), and methionine sulfoxide reductase B1 (MSRB1 [MIM: 606216]). Additionally, we examined the expression of genes encoding ROS-scavenging enzymes, including peroxiredoxin 1 (PRDX1 [MIM: 176763]), glutathione S-transferase alpha 4 (GSTA4 [MIM: 605450]), glutathione S-transferase omega 1 (GSTO1 [MIM: 605482]), and superoxide dismutase 3 (SOD3 [MIM: 185490]). Quantitative real-time PCR analysis revealed that the mRNA expression of GPX1, MSRB1, SELENOW, SOD3, GPX4, MSBR1, and TXNRD1 was increased in the p.Trp352Gly, p.Arg371Gly, and p.Arg371Trp variants compared to the WT, whereas the expression of GSTA4, GSTO1, PRDX1, and SELENOT remained unaffected. Once again, the variant p.Arg371Gln exhibited little to no effect (Figure 4E).
In this study, we used ES to identify variants affecting the C-terminal region of SEPHS1 in nine individuals with an overlapping phenotype. The variants are classified as pathogenic based on the ACMG recommendations for interpretating and reporting sequence variants (Table 1).33
Functional characterization revealed that variant p.Trp352Gly significantly reduced the overall thermal stability of the protein, due to the disruption of a network of hydrophobic interactions. When compared to WT SEPHS1, we determined that p.Trp352Gly was more resistant to proteolytic cleavage and found that the initial fluorescence of p.Trp352Gly was drastically enhanced due to the exposure of Trp residues that are usually buried in the core of the WT protein. Replacement of Trp352 with a Gly residue destabilizes a C-terminal hydrophobic pocket containing residues spanning the entire β-sheet, leading to local structural changes which may modulate protein-protein interactions in the cell. With eight out of nine individuals having Arg371 variants, it was intriguing to us that they behaved differently than p.Trp352Gly and resembled WT SEPHS1. Arg371 variants did not significantly affect protein stability or proteolytic cleavage pattern. Thus, we propose that Arg371 does not contribute to stabilization of the C-terminal region but rather likely participates in recognition and/or regulation of SEPHS1 binding partners. Our findings in neuronal SH-SY5Y cells provide evidence that the identified SEPHS1 variants could enhance cell proliferation by modulating ROS homeostasis. However, the specific impact of the p.Arg371Gln variant on cellular function remains unclear. Further investigation is needed to elucidate the precise mechanism by which SEPHS1 variants influence the expression of genes encoding stress-related selenoproteins and ROS-scavenging enzymes.
Based on the cellular data, the variants in SEPHS1 most likely act through a gain-of-function mechanism. Proper function of selenoproteins is shown to be crucial for neuronal function.6,9,11,13,18,34 It is conceivable that defective SEPHS1 could affect development of limbic system structure, including the hypothalamus, thalamus, basal ganglia, cingulate gyrus, hippocampus, and amygdala.19,20 In addition, individuals with impaired function of SEPHS1 may have neurodevelopmental deficits due to possible loss in the total number of pluripotent stem cells that survive during fetal development by an overactivation of the RAS/MAPK signaling pathway in these cells.23 Additionally, a deficiency of growth hormone-releasing hormone and thyroid-stimulating hormone secondary to diminished hypothalamic input to the anterior pituitary gland may exacerbate the neurodevelopmental and growth delays seen in our cohort.35,36,37
SEPHS1 has high evolutionary conservation across species, which allows for the study of the gene in various animal models.37 Based on findings from the in vitro, mouse, and fly models, SEPHS1 plays an essential role during cell development and proliferation and regulates cellular redox homeostasis and defense. Sephs1-targeted knockouts in the mice and the knockout of Sps1 mRNA in Drosophila show neurodevelopment being affected as well as embryonic lethality.23,26 Heterozygous Sps1 variants in Drosophila resulted in eye phenotypes, which could coincide with the variable eye phenotypes seen in our cohort.
In mouse and human cells, knockdown of SEPHS1 suppresses cell proliferation. Sephs1 knockout mice had embryos that were underdeveloped by day E8.5 and virtually resorbed by day E14.5.27 Knockdown of Sephs1 mRNA in a Drosophila SL2 immortalized cell line led to the inhibition of cell proliferation.26 In this model, megamitochondria were formed through the inhibition of pyridoxal phosphate which is the active form of vitamin B6 and necessary for SepSecS function, among other roles. Interestingly, one individual had elevated B6 levels. Taken together, our study helps support the role of SEPHS1 in human growth and development. In our study, individuals with SEPHS1 pathogenic variants exhibit hypotonia, muscle weakness, poor weight gain, growth delays, and short stature. The phenotypes point to the role of SEPHS1 in regulating growth via modulation of ROS. In the Ras/MAPK signaling pathway, ROS are messengers for growth and gain-of-function variants in this pathway are associated with a group of disorders termed RASopathies.38
To gain a deeper understanding of the potential link between SEPHS1 variants and cell proliferation via alterations in ROS homeostasis, it is imperative to conduct future experiments either in cell culture systems or animal models. These experiments will provide a more comprehensive and certain insight into the underlying mechanisms at play. By examining specific ROS homeostasis mechanisms and the signaling pathways affected by SEPHS1 variants, we can identify the precise molecular interactions responsible for the observed cellular changes. Additionally, a comparative analysis between cells with wild-type SEPHS1 and those with SEPHS1 variants will offer valuable insights into the differences in ROS regulation and cell proliferation rates, further confirming the role of these variants. Such investigations will not only advance our understanding of this neurodevelopmental disorder but also provide a foundation for the development of targeted therapeutic strategies aimed at restoring cellular homeostasis and improving patient outcomes.
Overall, individuals with pathogenic variants in SEPHS1 appear to have a neurodevelopmental and growth disorder, with variable effects on other body systems. We provide some considerations for immediate and long-term care of these individuals (see supplemental note). In conclusion, missense variants in exon 9 of SEPHS1 underlie a neurodevelopmental syndrome associated with developmental delay, growth problems, feeding difficulties, hypotonia, and dysmorphic features. Our findings provide insight into the molecular pathogenesis of this disorder.
Data and code availability
The sequence variants in SEPHS1 (NCBI ID 22929, transcript GenBank: NM_012247.4) reported in the paper have been deposited in ClinVar database (SCV002103293.1, SCV002103295.1, SCV002103294.2, SCV002103296.1) (https://www.ncbi.nlm.nih.gov/clinvar/) (Table 1). Exome sequencing data were generated in diagnostic setting and can not be shared due to privacy concerns.
Consortia
Members of the Undiagnosed Diseases Network are Maria T. Acosta, Margaret Adam, David R. Adams, Justin Alvey, Laura Amendola, Ashley Andrews, Euan A. Ashley, Mahshid S. Azamian, Carlos A. Bacino, Guney Bademci, Ashok Balasubramanyam, Dustin Baldridge, Jim Bale, Michael Bamshad, Deborah Barbouth, Pinar Bayrak-Toydemir, Anita Beck, Alan H. Beggs, Edward Behrens, Gill Bejerano, Hugo J. Bellen, Jimmy Bennet, Beverly Berg-Rood, Jonathan A. Bernstein, Gerard T. Berry, Anna Bican, Stephanie Bivona, Elizabeth Blue, John Bohnsack, Devon Bonner, Lorenzo Botto, Brenna Boyd, Lauren C. Briere, Elly Brokamp, Gabrielle Brown, Elizabeth A. Burke, Lindsay C. Burrage, Manish J. Butte, Peter Byers, William E. Byrd, John Carey, Olveen Carrasquillo, Thomas Cassini, Ta Chen Peter Chang, Sirisak Chanprasert, Hsiao-Tuan Chao, Gary D. Clark, Terra R. Coakley, Laurel A. Cobban, Joy D. Cogan, Matthew Coggins, F. Sessions Cole, Heather A. Colley, Cynthia M. Cooper, Heidi Cope, William J. Craigen, Andrew B. Crouse, Michael Cunningham, Precilla D'Souza, Hongzheng Dai, Surendra Dasari, Joie Davis, Jyoti G. Dayal, Matthew Deardorff, Esteban C. Dell'Angelica, Katrina Dipple, Daniel Doherty, Naghmeh Dorrani, Argenia L. Doss, Emilie D. Douine, Laura Duncan, Dawn Earl, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Cecilia Esteves, Marni Falk, Liliana Fernandez, Elizabeth L. Fieg, Paul G. Fisher, Brent L. Fogel, Irman Forghani, William A. Gahl, Ian Glass, Bernadette Gochuico, Rena A. Godfrey, Katie Golden-Grant, Madison P. Goldrich, Alana Grajewski, Irma Gutierrez, Don Hadley, Sihoun Hahn, Rizwan Hamid, Kelly Hassey, Nichole Hayes, Frances High, Anne Hing, Fuki M. Hisama, Ingrid A. Holm, Jason Hom, Martha Horike-Pyne, Alden Huang, Yong Huang, Wendy Introne, Rosario Isasi, Kosuke Izumi, Fariha Jamal, Gail P. Jarvik, Jeffrey Jarvik, Suman Jayadev, Orpa Jean-Marie, Vaidehi Jobanputra, Lefkothea Karaviti, Jennifer Kennedy, Shamika Ketkar, Dana Kiley, Gonench Kilich, Shilpa N. Kobren, Isaac S. Kohane, Jennefer N. Kohler, Deborah Krakow, Donna M. Krasnewich, Elijah Kravets, Susan Korrick, Mary Koziura, Seema R. Lalani, Byron Lam, Christina Lam, Grace L. LaMoure, Brendan C. Lanpher, Ian R. Lanza, Kimberly LeBlanc, Brendan H. Lee, Roy Levitt, Richard A. Lewis, Pengfei Liu, Xue Zhong Liu, Nicola Longo, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, Rachel Mahoney, Bryan C. Mak, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Rong Mao, Kenneth Maravilla, Ronit Marom, Gabor Marth, Beth A. Martin, Martin G. Martin, Julian A. Martínez-Agosto, Shruti Marwaha, Jacob McCauley, Allyn McConkie-Rosell, Alexa T. McCray, Elisabeth McGee, Heather Mefford, J. Lawrence Merritt, Matthew Might, Ghayda Mirzaa, Eva Morava, Paolo M. Moretti, Mariko Nakano-Okuno, Stan F. Nelson, John H. Newman, Sarah K. Nicholas, Deborah Nickerson, Shirley Nieves-Rodriguez, Donna Novacic, Devin Oglesbee, James P. Orengo, Laura Pace, Stephen Pak, J. Carl Pallais, Christina GS. Palmer, Jeanette C. Papp, Neil H. Parker, John A. Phillips III, Jennifer E. Posey, Lorraine Potocki, Barbara N. Pusey, Aaron Quinlan, Wendy Raskind, Archana N. Raja, Deepak A. Rao, Anna Raper, Genecee Renteria, Chloe M. Reuter, Lynette Rives, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Natalie Rosenwasser, Francis Rossignol, Maura Ruzhnikov, Ralph Sacco, Jacinda B. Sampson, Mario Saporta, Judy Schaechter, Timothy Schedl, Kelly Schoch, C. Ron Scott, Daryl A. Scott, Vandana Shashi, Jimann Shin, Edwin K. Silverman, Janet S. Sinsheimer, Kathy Sisco, Edward C. Smith, Kevin S. Smith, Emily Solem, Lilianna Solnica-Krezel, Ben Solomon, Rebecca C. Spillmann, Joan M. Stoler, Jennifer A. Sullivan, Kathleen Sullivan, Angela Sun, Shirley Sutton, David A. Sweetser, Virginia Sybert, Holly K. Tabor, Amelia L. M. Tan, Queenie K.-G. Tan, Mustafa Tekin, Fred Telischi, Willa Thorson, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Brianna M. Tucker, Tiina K. Urv, Adeline Vanderver, Matt Velinder, Dave Viskochil, Tiphanie P. Vogel, Colleen E. Wahl, Melissa Walker, Stephanie Wallace, Nicole M. Walley, Jennifer Wambach, Jijun Wan, Lee-kai Wang, Michael F. Wangler, Patricia A. Ward, Daniel Wegner, Monika Weisz-Hubshman, Mark Wener, Tara Wenger, Katherine Wesseling Perry, Monte Westerfield, Matthew T. Wheeler, Jordan Whitlock, Lynne A. Wolfe, Kim Worley, Changrui Xiao, Shinya Yamamoto, John Yang, Diane B. Zastrow, Zhe Zhang, Chunli Zhao, Stephan Zuchner.
Acknowledgments
We would like to thank all the families for participating in this study. This work was in part supported by The National Institute of General Medical Sciences, National Institutes of Health grant GM097042 (to M.S.). Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number(s) [U01HG010233]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Declaration of interests
S.V.M., E.T., H.G., T.B.P., A.T., D.A.C., M.M.M., I.M.W., K.G.M., and J.J. are employees of GeneDx., LLC. This article was prepared while M.S. was employed at the University of Illinois at Chicago. The opinions expressed in this article are the author’s own and do not reflect the view of the National Institutes of Health, the Department of Health and Human Services, or the United States government.
Published: March 25, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.02.016.
Contributor Information
Sureni V. Mullegama, Email: smullegama@genedx.com.
Jun Yang, Email: junyang@utmb.edu.
Web resources
Clustal Omega, https://www.ebi.ac.uk/Tools/msa/clustalo/
DGV, http://dgv.tcag.ca/dgv/app/home.
GeneMatcher, https://genematcher.org/
gnomAD, https://gnomad.broadinstitute.org/
MetaDome, https://stuart.radboudumc.nl/metadome/OMIM, https://www.omim.org/
The Human Protein Atlas, https://www.proteinatlas.org/
PROVEAN, http://provean.jcvi.org/
Supplemental information
References
- 1.Cardoso B.R., Cominetti C., Seale L.A. Editorial: Selenium, Human Health and Chronic Disease. Front. Nutr. 2021;8 doi: 10.3389/fnut.2021.827759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hilal T., Killam B.Y., Grozdanović M., Dobosz-Bartoszek M., Loerke J., Bürger J., Mielke T., Copeland P.R., Simonović M., Spahn C.M.T. Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon. Science. 2022;376:1338–1343. doi: 10.1126/science.abg3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palioura S., Sherrer R.L., Steitz T.A., Söll D., Simonovic M. The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science. 2009;325:321–325. doi: 10.1126/science.1173755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu X.M., Carlson B.A., Mix H., Zhang Y., Saira K., Glass R.S., Berry M.J., Gladyshev V.N., Hatfield D.L. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan J., Palioura S., Salazar J.C., Su D., O'Donoghue P., Hohn M.J., Cardoso A.M., Whitman W.B., Söll D. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc. Natl. Acad. Sci. USA. 2006;103:18923–18927. doi: 10.1073/pnas.0609703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellinger F.P., Raman A.V., Rueli R.H., Bellinger M.T., Dewing A.S., Seale L.A., Andres M.A., Uyehara-Lock J.H., White L.R., Ross G.W., Berry M.J. Changes in selenoprotein P in substantia nigra and putamen in Parkinson's disease. J. Parkinsons Dis. 2012;2:115–126. doi: 10.3233/JPD-2012-11052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillai R., Uyehara-Lock J.H., Bellinger F.P. Selenium and selenoprotein function in brain disorders. IUBMB Life. 2014;66:229–239. doi: 10.1002/iub.1262. [DOI] [PubMed] [Google Scholar]
- 8.Berry M.J., Banu L., Chen Y.Y., Mandel S.J., Kieffer J.D., Harney J.W., Larsen P.R. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3' untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 9.Boukhzar L., Hamieh A., Cartier D., Tanguy Y., Alsharif I., Castex M., Arabo A., El Hajji S., Bonnet J.J., Errami M., et al. Selenoprotein T Exerts an Essential Oxidoreductase Activity That Protects Dopaminergic Neurons in Mouse Models of Parkinson's Disease. Antioxidants Redox Signal. 2016;24:557–574. doi: 10.1089/ars.2015.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoenmakers E., Chatterjee K. Human Genetic Disorders Resulting in Systemic Selenoprotein Deficiency. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222312927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puppala A.K., French R.L., Matthies D., Baxa U., Subramaniam S., Simonović M. Structural basis for early-onset neurological disorders caused by mutations in human selenocysteine synthase. Sci. Rep. 2016;6 doi: 10.1038/srep32563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumitrescu A.M., Liao X.H., Abdullah M.S.Y., Lado-Abeal J., Majed F.A., Moeller L.C., Boran G., Schomburg L., Weiss R.E., Refetoff S. Mutations in SECISBP2 result in abnormal thyroid hormone metabolism. Nat. Genet. 2005;37:1247–1252. doi: 10.1038/ng1654. [DOI] [PubMed] [Google Scholar]
- 13.Shao Z.Q., Zhang X., Fan H.H., Wang X.S., Wu H.M., Zhang L., Cheng W.H., Zhu J.H. Selenoprotein T Promotes Proliferation and G1-to-S Transition in SK-N-SH Cells: Implications in Parkinson's Disease. J. Nutr. 2019;149:2110–2119. doi: 10.1093/jn/nxz199. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Zeev B., Hoffman C., Lev D., Watemberg N., Malinger G., Brand N., Lerman-Sagie T. Progressive cerebellocerebral atrophy: a new syndrome with microcephaly, mental retardation, and spastic quadriplegia. J. Med. Genet. 2003;40:e96. doi: 10.1136/jmg.40.8.e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Çatli G., Fujisawa H., Kirbiyik Ö., Mimoto M.S., Gençpinar P., Özdemir T.R., Dündar B.N., Dumitrescu A.M. A Novel Homozygous Selenocysteine Insertion Sequence Binding Protein 2 (SECISBP2, SBP2) Gene Mutation in a Turkish Boy. Thyroid. 2018;28:1221–1223. doi: 10.1089/thy.2018.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoenmakers E., Carlson B., Agostini M., Moran C., Rajanayagam O., Bochukova E., Tobe R., Peat R., Gevers E., Muntoni F., et al. Mutation in human selenocysteine transfer RNA selectively disrupts selenoprotein synthesis. J. Clin. Invest. 2016;126:992–996. doi: 10.1172/JCI84747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geslot A., Savagner F., Caron P. Inherited Selenocysteine Transfer RNA Mutation: Clinical and Hormonal Evaluation of 2 Patients. Eur. Thyroid J. 2021;10:542–547. doi: 10.1159/000518275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavlidou E., Salpietro V., Phadke R., Hargreaves I.P., Batten L., McElreavy K., Pitt M., Mankad K., Wilson C., Cutrupi M.C., et al. Pontocerebellar hypoplasia type 2D and optic nerve atrophy further expand the spectrum associated with selenoprotein biosynthesis deficiency. Eur. J. Paediatr. Neurol. 2016;20:483–488. doi: 10.1016/j.ejpn.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 19.Kang D., Lee J., Jung J., Carlson B.A., Chang M.J., Chang C.B., Kang S.B., Lee B.C., Gladyshev V.N., Hatfield D.L., et al. Selenophosphate synthetase 1 deficiency exacerbates osteoarthritis by dysregulating redox homeostasis. Nat. Commun. 2022;13:779. doi: 10.1038/s41467-022-28385-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Na J., Jung J., Bang J., Lu Q., Carlson B.A., Guo X., Gladyshev V.N., Kim J., Hatfield D.L., Lee B.J. Selenophosphate synthetase 1 and its role in redox homeostasis, defense and proliferation. Free Radic. Biol. Med. 2018;127:190–197. doi: 10.1016/j.freeradbiomed.2018.04.577. [DOI] [PubMed] [Google Scholar]
- 21.Sjöstedt E., Zhong W., Fagerberg L., Karlsson M., Mitsios N., Adori C., Oksvold P., Edfors F., Limiszewska A., Hikmet F., et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science. 2020;367 doi: 10.1126/science.aay5947. [DOI] [PubMed] [Google Scholar]
- 22.Tobe R., Carlson B.A., Huh J.H., Castro N.P., Xu X.-M., Tsuji P.A., Lee S.-G., Bang J., Na J.-W., Kong Y.-Y., et al. Selenophosphate synthetase 1 is an essential protein with roles in regulation of redox homoeostasis in mammals. Biochem. J. 2016;473:2141–2154. doi: 10.1042/bcj20160393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bang J., Han M., Yoo T.-J., Qiao L., Jung J., Na J., Carlson B.A., Gladyshev V.N., Hatfield D.L., Kim J.-H., et al. Identification of Signaling Pathways for Early Embryonic Lethality and Developmental Retardation in Sephs1−/− Mice. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222111647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung J., Kim Y., Na J., Qiao L., Bang J., Kwon D., Yoo T.-J., Kang D., Kim L.K., Carlson B.A., et al. Constitutive Oxidative Stress by SEPHS1 Deficiency Induces Endothelial Cell Dysfunction. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222111646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tobe R., Carlson B.A., Huh J.H., Castro N.P., Xu X.M., Tsuji P.A., Lee S.G., Bang J., Na J.W., Kong Y.Y., et al. Selenophosphate synthetase 1 is an essential protein with roles in regulation of redox homoeostasis in mammals. Biochem. J. 2016;473:2141–2154. doi: 10.1042/BCJ20160393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shim M.S., Kim J.Y., Jung H.K., Lee K.H., Xu X.M., Carlson B.A., Kim K.W., Kim I.Y., Hatfield D.L., Lee B.J. Elevation of glutamine level by selenophosphate synthetase 1 knockdown induces megamitochondrial formation in Drosophila cells. J. Biol. Chem. 2009;284:32881–32894. doi: 10.1074/jbc.M109.026492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiao L., Dho S.H., Kim J.Y., Kim L.K. SEPHS1 is dispensable for pluripotency maintenance but indispensable for cardiac differentiation in mouse embryonic stem cells. Biochem. Biophys. Res. Commun. 2022;590:125–131. doi: 10.1016/j.bbrc.2021.12.091. [DOI] [PubMed] [Google Scholar]
- 28.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rots D., Chater-Diehl E., Dingemans A.J.M., Goodman S.J., Siu M.T., Cytrynbaum C., Choufani S., Hoang N., Walker S., Awamleh Z., et al. Truncating SRCAP variants outside the Floating-Harbor syndrome locus cause a distinct neurodevelopmental disorder with a specific DNA methylation signature. Am. J. Hum. Genet. 2021;108:1053–1068. doi: 10.1016/j.ajhg.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacDonald J.R., Ziman R., Yuen R.K.C., Feuk L., Scherer S.W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiel L., Baakman C., Gilissen D., Veltman J.A., Vriend G., Gilissen C. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum. Mutat. 2019;40:1030–1038. doi: 10.1002/humu.23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richards C.S., Bale S., Bellissimo D.B., Das S., Grody W.W., Hegde M.R., Lyon E., Ward B.E., Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y., Zhou Y., Schweizer U., Savaskan N.E., Hua D., Kipnis J., Hatfield D.L., Gladyshev V.N. Comparative analysis of selenocysteine machinery and selenoproteome gene expression in mouse brain identifies neurons as key functional sites of selenium in mammals. J. Biol. Chem. 2008;283:2427–2438. doi: 10.1074/jbc.M707951200. [DOI] [PubMed] [Google Scholar]
- 35.van Trotsenburg P., Stoupa A., Léger J., Rohrer T., Peters C., Fugazzola L., Cassio A., Heinrichs C., Beauloye V., Pohlenz J., et al. Congenital Hypothyroidism: A 2020-2021 Consensus Guidelines Update-An ENDO-European Reference Network Initiative Endorsed by the European Society for Pediatric Endocrinology and the European Society for Endocrinology. Thyroid. 2021;31:387–419. doi: 10.1089/thy.2020.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marr A., Yokubynas N., Tang K., Saleh D., Wherrett D.K., Stein R., Bassilious E., Chakraborty P., Lawrence S.E. Transient vs Permanent Congenital Hypothyroidism in Ontario, Canada: Predictive Factors and Scoring System. J. Clin. Endocrinol. Metab. 2022;107:638–648. doi: 10.1210/clinem/dgab798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santesmasses D., Mariotti M., Gladyshev V.N. Bioinformatics of Selenoproteins. Antioxidants Redox Signal. 2020;33:525–536. doi: 10.1089/ars.2020.8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hebron K.E., Hernandez E.R., Yohe M.E. The RASopathies: from pathogenetics to therapeutics. Dis. Model. Mech. 2022;15 doi: 10.1242/dmm.049107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence variants in SEPHS1 (NCBI ID 22929, transcript GenBank: NM_012247.4) reported in the paper have been deposited in ClinVar database (SCV002103293.1, SCV002103295.1, SCV002103294.2, SCV002103296.1) (https://www.ncbi.nlm.nih.gov/clinvar/) (Table 1). Exome sequencing data were generated in diagnostic setting and can not be shared due to privacy concerns.