Summary
Cigarette smoking adversely affects many aspects of human health, and epigenetic responses to smoking may reflect mechanisms that mediate or defend against these effects. Prior studies of smoking and DNA methylation (DNAm), typically measured in leukocytes, have identified numerous smoking-associated regions (e.g., AHRR). To identify smoking-associated DNAm features in typically inaccessible tissues, we generated array-based DNAm data for 916 tissue samples from the GTEx (Genotype-Tissue Expression) project representing 9 tissue types (lung, colon, ovary, prostate, blood, breast, testis, kidney, and muscle). We identified 6,350 smoking-associated CpGs in lung tissue (n = 212) and 2,735 in colon tissue (n = 210), most not reported previously. For all 7 other tissue types (sample sizes 38–153), no clear associations were observed (false discovery rate 0.05), but some tissues showed enrichment for smoking-associated CpGs reported previously. For 1,646 loci (in lung) and 22 (in colon), smoking was associated with both DNAm and local gene expression. For loci detected in both lung and colon (e.g., AHRR, CYP1B1, CYP1A1), top CpGs often differed between tissues, but similar clusters of hyper- or hypomethylated CpGs were observed, with hypomethylation at regulatory elements corresponding to increased expression. For lung tissue, 17 hallmark gene sets were enriched for smoking-associated CpGs, including xenobiotic- and cancer-related gene sets. At least four smoking-associated regions in lung were impacted by lung methylation quantitative trait loci (QTLs) that co-localize with genome-wide association study (GWAS) signals for lung function (FEV1/FVC), suggesting epigenetic alterations can mediate the effects of smoking on lung health. Our multi-tissue approach has identified smoking-associated regions in disease-relevant tissues, including effects that are shared across tissue types.
Keywords: smoking, epigenome, DNA, methylation, EWAS, multi-tissue
Introduction
Cigarette smoking has many detrimental effects on human health, including increased risk for cancer, cardiovascular diseases, and respiratory diseases.1 Tobacco smoke contains thousands of chemicals, dozens of which are known carcinogens, and the potential mechanisms underlying the adverse effects of these chemicals on health include DNA damage, inflammation, and oxidative stress.2 The effects of smoking on specific features of the human epigenome have been described previously, including studies that identify associations between smoking behaviors and epigenetic features, such as DNA methylation (DNAm) at cytosine-guanine (CpG) dinucleotides.3,4
The association between cigarette smoking and DNAm has been characterized in prior epigenome-wide association studies (EWASs).5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22 Gene regions where DNAm in leukocytes shows consistent association with smoking status include the aryl-hydrocarbon receptor repressor (AHRR),7,9,19,20,21,22 coagulation factor II (thrombin) receptor-like 3 (F2RL3),7,8,9,19,20,21 G protein-coupled receptor 15 (GPR15),7,19,21 2q37.1 (containing ALPPL2),7,9,19 and 6p21.33 (major histocompatibility complex) regions.7,9,19,20 These studies demonstrate that DNAm changes in blood can be used as biomarkers of tobacco exposure and smoking history,6 and subsequent studies have reported associations between smoking-related DNAm features and risk for smoking-related diseases, such as lung cancer.23,24
While the majority of prior studies on this topic focus on the effects of smoking on DNAm in leukocytes, there have been a small number of studies focusing on other tissue types, including lung,6,15 cord blood,25 placenta,17 and blood from newborns with prenatal exposure.11,16 These studies highlight regions affected by tobacco exposure in multiple tissue types (e.g., AHRR),10,16,18,22,25,26 as well as effects that are potentially tissue-specific (e.g., cg27402634 near LEKR1 and long noncoding RNA LINC00886, a hallmark of maternal smoking in placenta).17 However, the association of cigarette smoking with DNAm in non-blood tissue types has received relatively little attention, as most tissue types are typically inaccessible in human studies.
In this study, we generate genome-wide array-based DNAm data using human tissue samples from the Genotype-Tissue Expression (GTEx) project to assess the association of smoking and DNAm in the lung, colon, and seven additional tissue types.
Material and methods
Sample collection and ethics approval
As detailed in Oliva et al.,27 the GTEx research protocol was reviewed by Chesapeake Bay Review, Roswell Park Comprehensive Cancer Center’s Office of Research Subject Protection, and the institutional review boards at the University of Pennsylvania. Analyses of DNA samples from GTEx participants at the University of Chicago was not considered human subjects research by the university's institutional review board since only deidentified data on deceased individuals were utilized in this study.
The GTEx project
The GTEx project has established a biobank of human tissue samples from >950 postmortem multi-tissue donors for the study of molecular phenotypes.28 The GTEx v8 dataset consists of tissue-specific RNA-sequencing and genotyping data from 838 donors and 17,382 unique samples from 52 tissue types.29,30 GTEx also provides information on sex, age, and race/ethnicity based on questionnaire data, as well as measurements of ischemic time for all samples. For this project, we obtained DNAm measurements for 916 GTEx tissue samples representing nine tissue types (lung, colon, ovary, prostate, whole blood, breast, testis, kidney, and muscle), described previously.27 These nine tissue types were selected based on several criteria reflecting our research interests, including inclusion of cancer-relevant tissues (lung, colon, prostate, ovary, breast, and kidney), tissues with unique aging biology (testis and skeletal muscle), and tissues commonly used in epidemiological research (whole blood). With resources available to profile DNAm for ∼1,000 samples, we selected larger numbers of samples for some tissue types of strong public health interest (e.g., lung, colon, and ovary) and to assess the impact of sample size on power for DNAm quantitative trait loci (mQTL) detection.27
Determination of smoking status for GTEx donors
Smoking status was assigned “ever cigarette smoker” for GTEx donors with a reported history of cigarette smoking and “never cigarette smoker” for donors with no reported history of cigarette smoking. Assignment was based on the MHSMKSTS variable (smoking status: yes, no, unknown) and the MHSMKTP variable (smoke type: cigarette, cigar, pipe, others) provided by GTEx. We were able to assign ever/never status to 396 donors with DNAm data (269 cigarette smokers and 127 non-cigarette smokers), with 21 donors (46 samples) lacking data on cigarette smoking status. Ever cigarette smokers include both current and former smokers; however, the distinction between these two smoking categories was not assessed in our primary analyses due to incomplete information of prior smoking and smoking duration for many GTEx donors. However, we constructed a “current smoker” variable, which was used for secondary analyses. This variable was constructed using free text comments from family members of the tissue donors, recorded in the MHSMKCMT variable provided by GTEx.
DNAm data and quality control
DNA was extracted from GTEx tissue samples via the Qiagen Gentra Puregene method at GTEx Laboratory Data, Analysis and Coordinating Center (LDACC). The LDACC shipped DNA from 1,000 unique tissue samples to the University of Chicago. These 1,000 samples represent 424 unique GTEx donors and 9 unique GTEx tissue types. Genome-wide DNAm at >850,000 CpG sites was assessed using the Infinium MethylationEPIC array (Illumina, San Diego, CA, USA). All DNA samples were prepared and analyzed in accordance with the manufacturer’s guidelines and protocols. For sample quality control (QC), we excluded 3 samples with undetectable or missing methylation values (detection p > 0.01) in ≥5% of CpGs, 6 samples with mismatched sex, and 13 samples that did not show clear clustering with their corresponding tissue type. The EPIC array measures 59 high-frequency SNPs that can be used as a genetic fingerprint.31 Using these SNPs, we identified one sample that did not match the donor’s existing genotype data, and this sample was excluded. The 15 male samples from breast tissue were excluded from the DNA methylation data. The 46 samples lacking data on cigarette smoking status were also excluded from the analyses. In total, 84 samples were excluded. After excluding samples due to quality control or missing data issues, there were 916 samples used for analysis (representing 9 tissue types and 398 GTEx donors).
For CpG QC, we followed guidance from Pidsley et al.32 We excluded CpGs measured by probes with potential non-specific binding (n = 43,254), CpGs overlapping genetic variants or with variants that overlap single-base extension sites for type 1 probes (n = 7,708), CpGs mapping to the X and Y chromosomes (n = 16,037), and poorly performing CpGs according to Illumina (n = 169). We also excluded CpGs that had detection p > 0.01 in at least one sample (n = 44,135). A total of 754,119 CpGs passed QC and were retained for analyses. Genomic positions for all CpGs (and for all SNP and gene expression analyses described below) are based on human reference genome build hg19.
GTEx gene expression data
Gene-level expression data (v8) derived from RNA sequencing was obtained from the GTEx portal. The expression value for each gene was estimated as reads per kilobase of transcript per million mapped reads (RPKM) using RNA-SeQC on uniquely mapped, properly paired reads fully contained within exon boundaries and with alignment distances ≤6.33 Samples with <10 million mapped reads or with outlier expression measurements based on the D statistic were removed.34 A total of 56,200 genes in the v8 dataset had expression levels recorded in both read counts and transcripts per million (TPM). Read counts from these genes were normalized across samples using the trimmed mean of M-values (TMM) normalization method in the “edgeR” package to generate TMM-normalized TPM for each gene.35 Following TMM normalization, genes were selected based on the expression threshold of >0.1 TPM in at least 20% of samples and ≥6 reads in at least 20% of the samples. We then restricted to the fully processed, filtered, and normalized autosomal genes from the GTEx v8 dataset, which resulted in 25,272 genes expressed in lung (n = 541) and 24,580 genes expressed in colon (n = 382).
Association of cigarette smoking status with DNAm
The beta values for each CpG were logit transformed into M-values prior to analyses using the following formula: log2[beta/(1 − beta)]. For each tissue type, the association between smoking status (ever/never) and DNAm at each CpG site was estimated using a linear model implemented in the “limma” package36 in R, with age, sex, body mass index (BMI), race/ethnicity, ischemic time, batch/plate, and surrogate variables (SVs) included as covariates. For analyses of lung tissue, we also adjusted for common lung-related health conditions, including asthma (n = 24), chronic obstructive pulmonary disease (COPD) (n = 34), and pneumonia (n = 24). The R “sva” package37 was used to generate the SVs for each tissue type. We included the smoking variable in the full model matrix but omitted the smoking variable from the null model matrix to prevent the effects of smoking from being captured by SVs. The resulting SVs were used to control for technical variation and other biologic sources of variability (i.e., cell-type composition). As a general rule, we adjusted for 10 SVs for tissue types with n > 100 and 5 SVs for tissue types with n < 100. To ensure the SVs captured all variability due to cell-type composition (but not effects of smoking), we examined correlations of the first 20 SVs (per tissue type) with smoking status and cell-type composition estimates (derived using the EPISCORE method as described below). SVs showing clear association with EPISCORE cell-type estimates were typically among the top 5 SVs (Table S1). We considered the exclusion of SVs associated with smoking status. For example, for lung tissue, four of the top 10 SVs showed association with smoking status (p < 0.05); however, including these SVs as covariates resulted in only mild attenuation of associations observed, so all 10 SVs were retained. For colon DNAm data, no SVs were associated with smoking status, so all 10 SVs were retained. The false discovery rate (FDR) was estimated using the Benjamini-Hochberg method.38
Estimating power to detect smoking-related CpGs at varying sample sizes
We estimated the power to detect the effect sizes observed for CpGs identified in lung tissue (FDR 0.05) at sample sizes similar to other tissues by first generating 1,000 random subsamples of our lung tissue samples at sample sizes of 50, 100, and 150. We then performed an EWAS (described above) in each of these subsamples and determined the proportion of subsamples where we identified the smoking-associated CpG with the largest effect size magnitude (cg01584760), median effect size magnitude (cg20291548), and the smallest effect size magnitude (cg09138315) observed in lung.
Enrichment of smoking-associated CpGs in each tissue with previously reported CpGs
To assess whether smoking-associated CpGs in each of our tissues were significantly enriched for previously reported CpGs in whole blood,39 adipose,13 placenta,17 or CpGs identified in analyses of GTEx lung tissue, we calculated the proportion of all smoking-associated CpGs (p < 10−5) in each tissue that had been reported previously for each of these tissue types (and based on GTEx lung results). We performed a one-sided, two-sample z test of proportions to determine if CpGs previously reported were higher than the proportion of smoking-associated CpGs detected among all CpGs analyzed. We repeated these analyses using a p < 10−3 threshold for classifying smoking-associated CpGs.
Association of smoking status with gene expression
The association between smoking status and expression in lung and colon for each gene was estimated using a linear model implemented in “limma,” adjusting for age, sex, BMI, race/ethnicity, ischemic time, and 5 SVs (created using expression data). For lung tissue, we also adjusted for three lung-related diseases: asthma, COPD, and pneumonia (as described above).
Enrichment and pathway analyses for smoking-related CpGs
We compared the proportion of smoking-associated CpG sites (FDR 0.05) assigned to island, shore, shelf, and open sea (Illumina annotations) in lung and colon tissue to the distribution in the entire Infinium MethylationEPIC array (754,199 CpGs) using chi-square tests. We assessed enrichment of smoking-associated CpGs in chromatin segmentation features using reference data from the Roadmap Epigenomics project database40 (primary tissue colonic mucosa and lung). We used the R package “oddsratio” to calculate enrichment and Fisher’s exact test to compute p values. The above enrichment analyses were performed stratified by hypermethylated vs. hypomethylated smoking-associated CpG sites. Additionally, we performed a motif enrichment analysis to identify enrichment of smoking-associated CpGs in transcription factor binding sites (TFBS). We used annotations from the ENCODE version 2 and 3 chromatin immunoprecipitation sequencing experiments (1,256 experiments), representing 340 transcription factors (TFs) in 129 cell and tissue types. These annotations were obtained from the University of California, Santa Cruz (UCSC) table browser (encRegTfbsClustered, build hg38). We assessed enrichment via hypogeometric tests, using the phyper function in R. CpGs were additionally assigned to genes (based on annotations provided by Illumina), and genes were assigned to pathways and biologic processes using the hallmark gene set collections (n = 50 sets),41 as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations42,43,44 We conducted gene set enrichment analysis (GSEA) using the gometh function in “missMethyl”45 for lung and colon tissues using smoking-associated CpGs (FDR 0.05). This function accounts for the potential bias in GSEA due to the number of CpGs per gene by computing prior probabilities46 and evaluates enrichment using a hypergeometric test. Enriched gene sets were defined as those passing an FDR of 0.05. The motif enrichment analysis and pathway analysis were performed for all smoking-associated CpGs as well as stratified by hypomethylated vs. hypermethylated smoking-associated CpGs.
Identification of mQTLs for smoking-associated CpGs that co-localize with GWAS signals for tissue-relevant diseases
For the 2,478 smoking-associated CpGs observed in lung tissue (FDR < 0.01) and the 662 CpGs observed in colon tissue, we identified the CpGs previously shown to be affected by an mQTL in GTEx lung or colon tissue.27 For the 566 CpGs and 68 CpGs identified in lung and colon, respectively, we extracted the identifiers for the lead SNP for each corresponding mQTL (550 and 68 lead SNPs, respectively). We then searched for these lead SNPs of lung mQTLs in the genome-wide summary statistics from several large genome-wide association studies (GWASs) of lung-related diseases and phenotypes,27,47,48,49 including lung cancer, asthma, chronic obstructive pulmonary disease, as well as two spirometry-based phenotypes that have been clinically used to assess lung health: FVC (forced vital capacity) and FEV1/FVC (forced expiratory volume in the first 1 s divided by the forced vital capacity) (obtained from MR-Base).48 We additionally searched for these lead SNPs of colon mQTLs in several large GWASs of colorectal cancer and inflammatory bowel disease.50,51 We retained all mQTL lead SNPs showing association (p < 5 × 10−8) in these GWASs (corresponding to 10 mQTLs in lung). For each mQTL retained, we used mQTL results for SNPs within 250 kb of the lead mQTL SNP to test for colocalization with the corresponding GWAS association signal, using GWAS summary statistics for the same set of SNPs via the “coloc” package in R52 with the following default prior probabilities: the prior probability of a SNP is associated with either mQTL or GWAS = 1.0 × 10−4; the prior probability of a SNP is associated with both mQTL and GWAS = 1.0 × 10−5. We visualized evidence of colocalization using the LocusCompare software.53
Analyses of cell-type proportions
For lung and colon, we estimated cell-type composition using the methylation-based EPISCORE method (“wRPC” function) using a pan-tissue DNAm atlas54 as a reference dataset. We chose this method instead of expression-based deconvolution methods because (1) it provides cell-type estimates for all samples with DNAm data (some of which lack RNA-sequencing data) and (2) it provides cell-type estimates based on the same tissue samples used for DNAm measurement (as RNA was extracted from a different piece of tissue). The cell types estimated by EPISCORE and their distributions are shown in Figure S1. We observe inter-individual variability in cell-type proportions and correlation with DNAm-derived SVs.
For benchmarking purposes, we compared EPISCORE estimates of epithelial cell proportions to epithelial cell enrichment scores previously computed for GTEx samples using xCell, an expression-based method (Figure S2).55 EPISCORE epithelial cell estimates and xCell epithelial scores were not correlated in lung (Spearman rho = −0.093; p = 0.29) but showed a clear correlation in colon (rho = 0.55; p = 4.1 × 10−7). Thus, we have more confidence that our cell-type estimates accurately represent cell-type abundances in colon samples (as compared to lung). However, we utilize EPISCORE estimates for both tissue types for smoking-by-cell-type interaction analyses. To identify effects of smoking that vary by cell types in lung tissue, we identified CpGs involved in smoking and cell-type interactions (SxCT) by performing an interaction test between smoking and a given cell type for all CpGs and for each inferred cell type. For these SxCT analyses, we transformed cell-type proportion estimates obtained from EPISCORE to standard normal distributions. We performed a linear regression testing the association of DNAm with the smoking and cell-type proportion interaction term, adjusting for age, sex, BMI, race/ethnicity, and ischemic time. For analyses of lung tissue, we also adjusted for common lung-related health conditions mentioned above.
Results
Characteristics of GTEx tissue donors
We generated DNAm data for 917 unique tissue samples, obtained from 396 unique GTEx donors, representing 9 different GTEx tissue types (Table 1). Sample sizes analyzed for each tissue type ranged from 38 (breast) to 212 (lung). The number of tissues analyzed per donor ranged from 1 to 7 (average of 2.3). Among tissue types that are not sex specific, ∼70% of samples came from male donors. 85% of the 398 donors were reported to be white. Among all donors included, 70% were classified as smokers, and there were no clear differences in this percentage across age groups.
Table 1.
Summary of GTEx tissue samples used for DNA methylation analyses
|
Tissue types | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Lung (n = 212) | Colon (n = 209) | Ovary (n = 153) | Prostate (n = 111) | Whole blood (n = 52) | Breast (n = 38) | Testis (n = 48) | Kidney (n = 47) | Muscle (n = 46) | |
| Age (years) | 55.1 (11.1) | 55.7 (11.4) | 50.7 (13.5) | 54 (12.4) | 49.7 (12.7) | 50.0 (11.9) | 53.7 (12.2) | 59.3 (8.3) | 56.9 (10.5) |
| BMI (kg/m2) | 27.5 (3.9) | 27 (3.9) | 26.8 (4.2) | 27 (3.8) | 27.3 (4.2) | 25.4 (3.94) | 27.1 (3.8) | 26.2 (3.8) | 26.7 (4.4) |
| Sex | |||||||||
| Male | 150 (70.8) | 143 (68.4) | 0 | 111 (100) | 43 (82.7) | 0 | 48 (100) | 36 (76.6) | 27 (58.7) |
| Female | 62 (29.3) | 66 (31.6) | 153 (100) | 0 | 9 (17.3) | 38 (100) | 0 | 11 (23.4) | 19 (41.3) |
| Race | |||||||||
| White | 180 (84.9) | 183 (87.6) | 124 (81.1) | 101 (91) | 46 (88.5) | 32 (84.2) | 45 (93.8) | 42 (89.4) | 40 (87) |
| Af. Americans | 27 (12.7) | 21 (10) | 26 (17) | 8 (7.2) | 5 (9.6) | 6 (15.8) | 3 (6.3) | 5 (10.6) | 6 (13) |
| Others | 5 (2.4) | 5 (2.4) | 3 (2) | 2 (1.8) | 1 (1.9) | 0 | 0 (0) | 0 | 0 |
| Cigarette smoking | |||||||||
| Ever | 150 (70.8) | 152 (72.7) | 96 (62.8) | 73 (65.8) | 40 (76.9) | 27 (71.1) | 36 (75) | 37 (78.7) | 34 (73.9) |
| Current | 89 (42) | 91 (43.5) | 66 (43.1) | 42 (37.8) | 25 (48.1) | 19 (50) | 16 (33.3) | 19 (40.4) | 15 (32.6) |
| Former | 61 (28.7) | 61 (29.2) | 30 (19.6) | 31 (27.9) | 15 (28.8) | 8 (21.1) | 20 (41.7) | 18 (38.3) | 19 (41.3) |
| Never | 62 (29.3) | 57 (27.3) | 57 (37.3) | 38 (34.2) | 12 (23.1) | 11 (28.9) | 12 (25) | 10 (21.3) | 12 (26.1) |
Format of metrics in table is as follows (mean [SD] or n [%]). Report of the donor’s race was either reported by the donor, the donor’s family/next of kin, or abstracted from medical records. The classification categories were taken from the NIH and refer to geographically based categories that humans share (i.e., common history, nationality, or geographic distribution).
Identification of smoking-associated CpG sites across different tissue types
Analyses of smoking status in relation to genome-wide DNAm in lung (n = 212) identified 6,350 smoking-associated CpG sites at an FDR of 0.05 (p < 0.0004) (Table S2), including many reported in previous EWASs. However, the majority of the CpGs identified (6,209 CpGs) have not been reported in previous studies of DNAm in blood cells based on a recent review of smoking-related changes in DNAm and gene expression.39 Analyses of smoking and DNAm in colon (n = 210) resulted in 2,735 smoking-associated CpGs at an FDR of 0.05 (p < 0.0001). Smoking-associated CpGs identified in lung had effect sizes with magnitudes ranging from 0.073 to 1.459, with a mean effect size of 0.286; those identified in colon had effect sizes with magnitudes ranging from 0.072 to 1.075, with a mean effect size of 0.334. For all 7 other tissue types (with sample sizes ranging from 38 to 153), no clear associations with smoking status were observed (FDR of 0.05, Table S2 and Data S1. Lung tissue, Data S2. Colon tissue, Data S3. Ovary tissue, Data S4. Prostate tissue, Data S5. Whole blood, Data S6. Breast tissue, Data S7. Testis tissue, Data S8. Kidney tissue, Data S9. Muscle tissue).
Secondary analyses performed in lung and colon tissues using current vs. never smoking as the exposure variable produced a larger number of CpGs passing the FDR threshold (Table S2b), but the top CpGs were the same, and p values were similar (Data S10 and S11). Additionally, we observed strong correlation between association estimates from our primary EWAS in lung tissue (ever vs. never) and our secondary EWAS (current vs. never) (R = 0.87), with a slight bias toward stronger association in the secondary analysis (Figure S3); this correlation was even stronger for smoking-associated CpGs reaching an FDR of 0.05 (R = 0.99). However, our remaining analyses focus on our primary exposure variable (ever vs. never smoking).
The abundance of smoking-associated CpG sites observed for lung was clearly larger than those observed for colon (a tissue type with similar sample size). To assess the extent to which lung tissue showed more prominent effects of smoking than other tissue types, we randomly selected subsets of lung samples to produce sample sizes similar to those of the other tissue types studied (e.g., n = 111 for prostate). After this down-sampling, the number of smoking-related CpGs detectable in lung tissue was larger than the number detected in ovary (n = 153) in 1,000 of 1,000 subsamples and the number detected in prostate (n = 111) in 997 of 1,000 subsamples (Figure S4). However, for tissue types with sample sizes of ∼50, we had limited power to clearly demonstrate that lung had a larger number of smoking-associated CpGs (Figure S4). To assess power to detect the effect sizes observed in lung at lower sample sizes, we performed an EWAS in 1,000 random subsamples of lung tissue samples (at sample sizes of 50, 100, and 150) and determined the proportion of subsamples that included the smoking-associated CpG with the largest (cg01584760), median (cg20291548), and smallest (cg09138315) effect size magnitude observed in lung. In subsets with n = 50, none of the three CpGs were identified. For sample sizes of 100 and 150, the CpG with the largest effect size was detected in 54.0% and 98.2% of subsamples, respectively, and the CpG with the median effect size was discovered in 0.4% and 34.9% of subsamples, respectively (Table S3). Together, these results demonstrate the expected power for each tissue-specific analysis to detect effect sizes similar to those observed in lung.
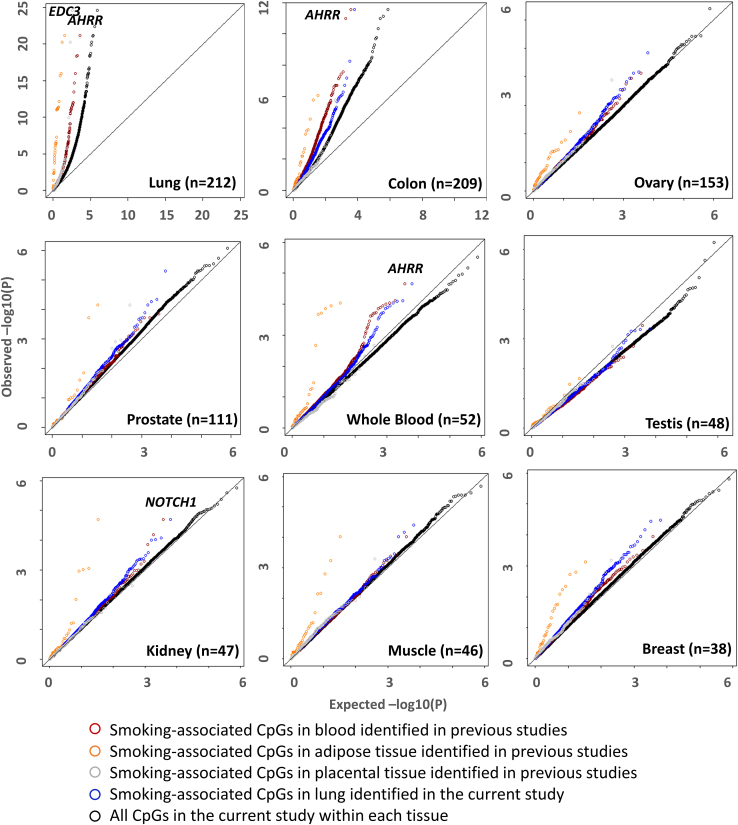
For each tissue type, we specifically examined the CpGs associated with smoking in lung (from analyses of GTEx reported here), blood cells (3,722 CpGs reported in a recent review of 30 studies focused on the association of active smoking with DNAm and gene expression),39 placenta tissue (443 CpGs),17 and adipose tissue (42 CpGs)13 (based on prior studies) to assess the evidence that some smoking-associated CpGs are shared across tissue types. Among the tissues examined, colon, ovary, whole blood, breast, and kidney showed the strongest evidence of enrichment for smoking-associated CpGs identified in GTEx lung tissue (Figure 1; Table S4). These tissue types were also enriched for CpGs identified previously in whole blood; in addition, muscle and prostate were enriched for CpGs previously identified in adipose tissue (Figure 1; Table S4). In contrast, with the exception of lung tissue, other tissue types were not enriched for smoking-associated CpGs identified previously in placental tissue (Figure 1; Table S4). The overlap of smoking-associated CpGs between tissue types and the overlap of genes annotated to these CpGs are characterized in Figures S5 and S6.
Figure 1.
Quantile-quantile plots of p values showing the association between smoking status and DNAm by tissue type
Results are shown for genome-wide analyses in each tissue (black circles), for the 6,350 CpG associated with smoking based on GTEx lung samples (blue circles), and for the 3,722 CpGs associated with smoking based on prior studies of DNAm in blood samples39 (red circles), adipose samples (orange circles),13 and placenta samples (gray circles).17 Several noteworthy genes with previously reported associations between smoking and DNAm are labeled including AHRR, NOTCH1, and EDC3.
Since the abundance of CpGs detected in each tissue is dependent on sample size, we compared association estimates of smoking-associated CpG sites in lung to estimates of the same of set CpG sites in the other 8 tissues (Figure S7). Association estimates for CpG sites identified in lung showed clear positive correlations (p < 1.9 × 10−7) with estimates from all other tissue types (except muscle). While correlations were generally weak, our results indicate that many smoking effects in lung are present in other tissues, suggesting the lack of signal in other tissues is due in part to limited power. Additionally, we performed stratified EWAS by sex for lung and colon tissues. We observed effect size estimates across smoking-associated CpGs identified in our primary analysis (FDR < 0.05) were strongly correlated between males and females in both lung (R2 = 0.70) and colon (R2 = 0.56) (Figure S8).
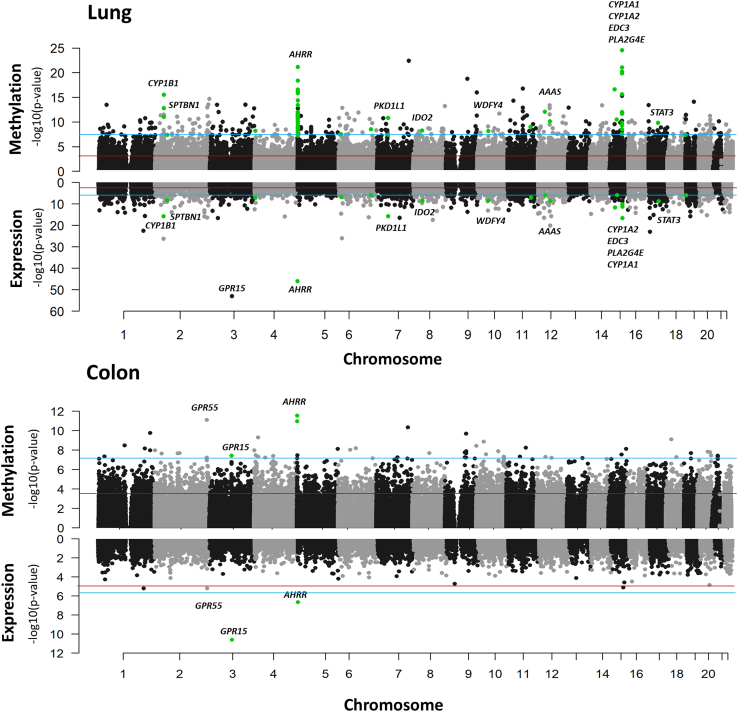
Regions in which smoking is associated with both DNAm and gene expression
Examining the association of smoking status with DNAm and gene expression (in lung and colon), we observe that smoking is associated with both DNAm and gene expression in some regions. In lung tissue, notable gene regions included CYP1B1, AHRR, and CYP1A1, with AHRR expression and methylation also showing clear association with smoking in colon tissue (Figure 2). The strongest smoking-related gene expression signal in both colon and lung was GPR15, a gene previously reported to be a biomarker of smoking in leukocytes (both expression and methylation).56,57 The CpG with the strongest evidence of association in this region in colon (cg19859270, p = 3.7 × 10−8) did not pass QC for lung. Overall, 994 loci in lung and 3 in colon showed association of smoking with both DNAm and gene expression (Tables S5 and S6).
Figure 2.
p values corresponding to the association of smoking status with DNAm and gene expression in lung (top) and colon (bottom) GTEx samples
Regions associated with both DNAm and gene expression based on a Bonferroni p value threshold are indicated with green dots. Gene expression analyses are based on 541 lung samples and 382 colon samples. Blue line represents the FDR threshold and red line represents the Bonferroni threshold.
The top 10 smoking-associated DNAm features/regions for lung and colon are shown in Table 2. For lung, 7 of the 10 regions have been previously reported in prior studies of DNAm, including blood cells,7,13,39,58,59,60 non-tumor lung tissue,6 adipose tissue,13 oral mucosa61 and/or non-small-cell lung cancer.62 Most of these regions contain multiple smoking-associated CpGs, including AHRR with 66 CpGs, CYP1A1 with 18 CpGs, and LRP5 with 15 CpGs. Six of these regions also showed evidence of association with smoking in colon (FDR 0.05): HIPK2, AHRR, CYP1A1, AOPEP, LRP5, and CYP1B1.
Table 2.
Top smoking-associated CpGs in GTEx lung and colon samples (based on p value)
| Tissue | Annotated genea(or region) | Chr:Position | CpG | p value | CpGs passing FDRb | Region identified previously | Other tissues (FDR 0.05) | Association withexpression (p value) |
|---|---|---|---|---|---|---|---|---|
| Lung | EDC3 | 15:74935742 | cg26843110 | 2.61E-25 | 4 | yes | no | up (5.63E-12) |
| HIPK2 | 7:139420300 | cg03224163 | 3.70E-23 | 4 | yes | colon | down (0.05) | |
| AHRR | 5:346695 | cg04135110 | 6.97E-22 | 66 | yes | colon | up (1.08E-46) | |
| CYP1A1c | 15:75015502 | cg23655854 | 8.26E-22 | 18 | yes | colon | up (3.08E-17) | |
| AOPEPd | 9:97544885 | cg21081352 | 1.79E-19 | 5 | no | colon | no | |
| 5p15.33 | 5:1128194 | cg03504128 | 3.76E-19 | 1 | no | no | N/A | |
| LRP5 | 11:68079135 | cg04840942 | 1.70E-17 | 15 | yes | colon | no | |
| PLA2G4E | 15:42313231 | cg16167478 | 2.47E-17 | 6 | no | no | up (2.61E-12) | |
| NOTCH1 | 9:139416102 | cg14120703 | 1.04E-16 | 2 | yes | no | down (0.04) | |
| CYP1B1 | 2:38296474 | cg01584760 | 2.83E-16 | 10 | yes | colon | up (1.90E-16) | |
| Colon | AHRR | 5:374252 | cg04141806 | 2.85E-12 | 17 | yes | lung | up (2.31E-7) |
| GPR55e | 2:231809610 | cg08840017 | 7.61E-12 | 1 | yes | no | up (6.31E-6) | |
| HIPK2 | 7:139366758 | cg25748521 | 4.40E-11 | 2 | yes | lung | no | |
| RHOU | 1:228871677 | cg27437294 | 1.73E-10 | 2 | no | no | no | |
| WNK2 | 9:95947164 | cg10281741 | 1.99E-10 | 5 | no | no | no | |
| FAM184B | 4:17783205 | cg01886556 | 4.88E-10 | 4 | yes | no | no | |
| LAMA3 | 18:21269793 | cg25009504 | 7.59E-10 | 3 | no | no | no | |
| NRP1 | 10:33624100 | cg09009410 | 1.35E-09 | 4 | no | lung | no | |
| NHLH2 | 1:116381475 | cg24106636 | 3.17E-09 | 2 | no | no | N/A | |
| DIP2C | 10:735472 | cg25488288 | 3.54E-09 | 2 | yes | no | no |
Based on Illumina’s annotation for the EPIC array. Cytoband is listed if there is no annotated gene.
The number CpGs passing FDR (0.05) that are annotated to the gene listed (based on Illumina’s annotation).
CYP1A1 and EDC3 are in the same region, separated by < 25 kb.
AOPEP is also known as C9ORF3.
CpG cg08840017 was assigned to GPR55 as it resides in a GRP55 isoform.
Among the top 10 smoking-associated regions in colon (Table 2), five have been previously reported in prior studies of non-tumor lung tissue6 and/or blood.25,58,63,64,65 However, among these 10 regions, only two (AHRR and GPR55) showed evidence of association with smoking in lung (FDR 0.05). The smoking-associated sites we identified in colon include CpGs annotated to RHOU and WNK2.
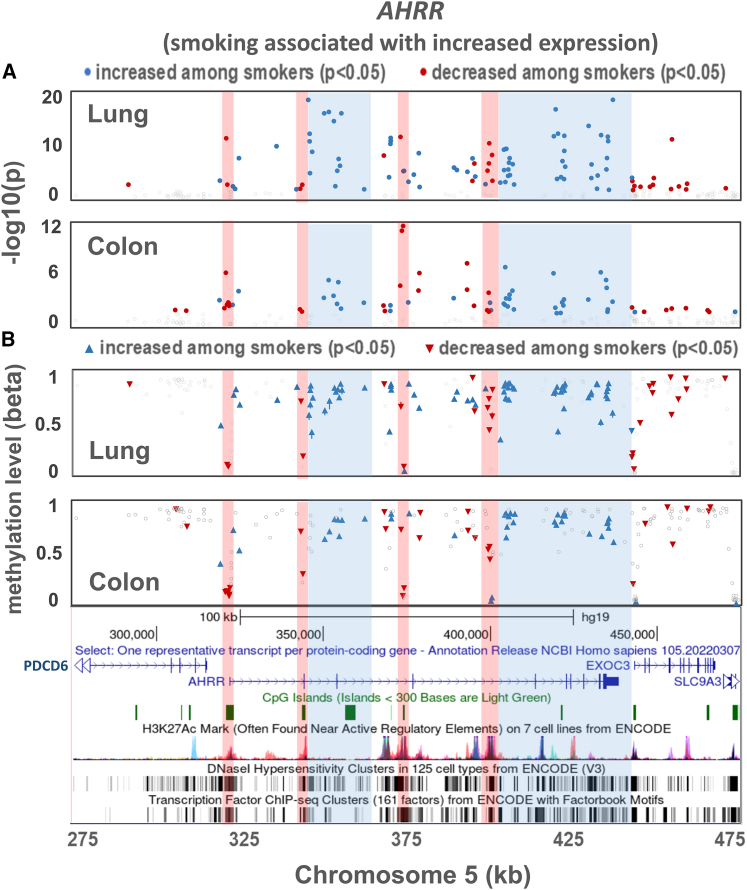
For several regions in which smoking was associated with both DNAm and gene expression (in both lung and colon), we examined the smoking and DNAm associations in detail. In the AHRR region (Figure 3), we observe clear differences between lung and colon with respect to the CpGs showing the clearest (based on p value) association with smoking. However, we observe some similarities between lung and colon with respect to patterns/clusters of increased and decreased methylation across the AHRR region (Figure S9). For example, decreased methylation among smokers is observed at CpG islands overlapping regulatory elements (based on ENCODE histone marks, DNase I hypersensitive sites (DHSs), and chromatin state), including the AHRR start site, for both lung and colon. These hypomethylated regions tend to have at least one site with a very low methylation level (beta value). In contrast, regions of increased methylation among smokers, in both lung and colon, tend to fall in the AHRR gene body, outside of regulatory elements coinciding with CpG islands (Figure 3). In both tissues, smoking is associated with increased expression of AHRR (lung p = 9.9 × 10−47; colon p = 2.4 × 10−7).
Figure 3.
Association between smoking and DNA methylation for CpG sites in the AHRR region for lung and colon tissues
(A) p values for association.
(B) Beta values for each CpG reflecting the level of DNA methylation at the CpG. Beta values represent the average across all individuals (smokers and non-smokers). Upward arrowheads indicate DNAm values were higher in smokers for a given CpG compared to in non-smokers, while downward arrowheads indicate DNAm values were lower in smokers.
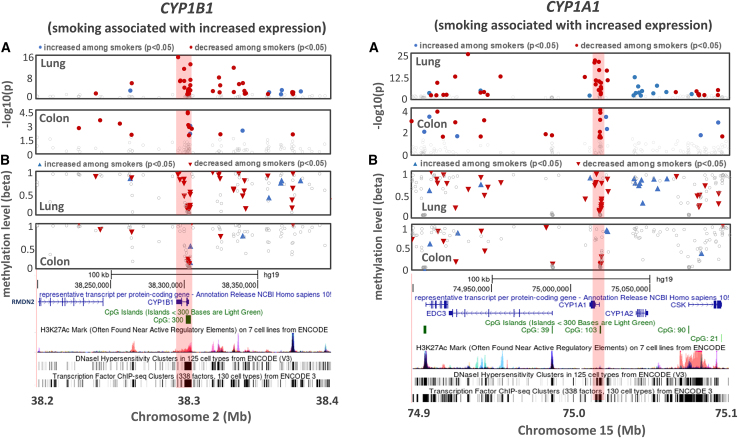
Sharper, more defined association signals (as compared to AHRR) were observed in both the CYP1B1 and CYP1A1 regions (Figure 4), for both lung and colon. While there are differences across tissues in terms of the specific CpGs showing the strongest association, these signals are located at CpG islands near the gene start site/promoter and show clear decreased methylation among smokers, with at least one smoking-associated CpG showing very low overall methylation levels (Figures 4B, S10, and S11). Smoking is associated with increased expression of CYP1B1 (lung p = 7.2 × 10−27; colon p = 0.002) and CYP1A1 (lung p = 3 × 10−17; colon p = 8.1 × 10−6).
Figure 4.
Association between smoking and DNA methylation for CpG sites in lung and colon for the CYP1B1 and CYP1A1 regions
Shown are p values for association (top) and beta values for each CpG reflecting the level of DNA methylation at the CpG (bottom). Beta values represent the average across all individuals (smokers and non-smokers). Upward arrows (blue) indicate DNAm values were higher in smokers for a given CpG compared to in non-smokers, while downward arrows (red) indicate DNAm values were lower in smokers.
Co-localization of cis-mQTLs and disease-related GWAS SNPs
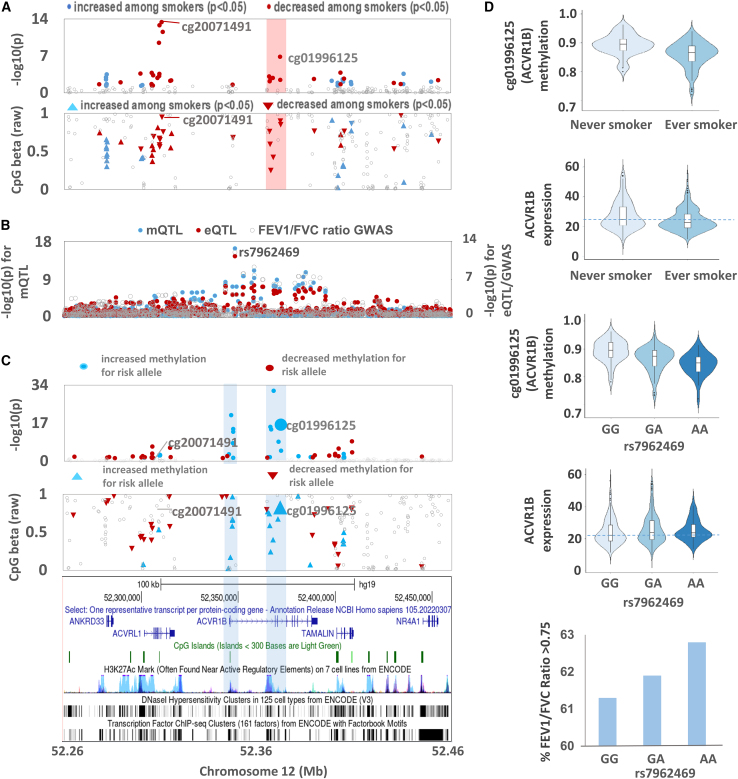
To identify CpGs in lung and colon that may mediate the effects of smoking on lung or colon health, we first identified smoking-associated CpGs that are affected by an mQTL (using existing mQTL results from GTEx passing an FDR of 0.01).27 For lung mQTLs, we determined if their lead SNPs were associated with lung-related phenotypes (FEV1/FVC, FVC, and lung adenocarcinoma) using GWAS summary statistics. Among our 2,478 smoking-associated CpGs in lung (FDR < 0.01), 566 are impacted by mQTLs in lung tissue. Among the 550 lead SNPs for these 566 lung mQTLs, 10 SNPs showed genome-wide significant associations with FEV1/FVC (p < 5 × 10−8) based on UK Biobank results47 (Table S7). We found evidence of co-localization (between the mQTL and a FEV1/FVC GWAS signals) for 4 of the 10 SNPs identified (PP4 < 0.99, Figure S12). The strongest co-localization detected was for an mQTL (lead SNP rs7962469) affecting cg01996125, within the gene body of ACVR1B on chromosome 12 (PP4 = 0.99). Smoking was associated with decreased DNAm at cg01996125 (Figures 5A, 5D, and S13). The co-localized GWAS and mQTL signals also co-localized with an eQTL for ACVR1B (Figure 5B). The FEV1/FVC risk allele (G) was associated with decreased FEV1/FVC, increased DNAm at cg01996125 (and several surrounding CpGs, Figure 5C), and decreased ACVR1B expression (Figure 5D). Smoking was associated with decreased DNAm at cg01996125 and decreased ACVR1B expression (Figure 5D). Given that the risk allele (G) and smoking were both associated with decreased ACVR1B expression, but with opposite effects on cg01996125 methylation (Figure 5D), these results suggest the epigenetic mechanism (or response) linking smoking to repression of ACVR1B may be different than from the mechanism of the FEV1/FVC risk allele. We also performed an interaction analysis regressing cg01996125 methylation on an interaction term between rs7962469 and smoking status (while adjusting for other covariates in the primary lung EWAS analysis) and observed an interaction p value of 0.28, corroborating the notion that these genetic and epigenetic mechanisms leading to ACVR1B repression may be distinct. Despite these differences in genetic and environmental effects on cg01996125, repression of ACVR1B expression represents a potential mediator by which smoking (and the risk allele at rs7962469) impacts lung health. Additional examples of co-localization between FEV1/FVC GWAS signals and mQTLs include SFTPA1 (PP4 = 0.99), PRSS23 (PP4 = 0.96), and MARCHF3/MARCH3 (PP4 = 0.96) (Figure S12). However, no eQTLs were observed for these genes.
Figure 5.
Co-localization of mQTL (for cg01996125), eQTL for ACVR1B, and lung function GWAS signal in the chromosome 12q13.13 region
(A) Plots of smoking EWAS (beta and p values) by effect direction for this region.
(B) Plot of p values for cg01996125 mQTL, ACVR1B eQTL, and FEV1/FVC ratio GWAS showing co-localization of all three association signals.
(C) Plot of the association of rs7962469 (FEV1/FVC ratio risk allele G) with all CpGs in the ACVR1B region.
(D) Distribution of cg01996125 methylation and ACVR1B expression by smoking status and rs7962469 genotype. Beta values represent the average across all individuals (smokers and non-smokers). Upward arrowheads (blue) indicate DNAm values were higher in smokers for a given CpG compared to in non-smokers, while downward arrowheads (red) indicate DNAm values were lower in smokers.
We did not find evidence of co-localization between colon mQTLs and GWAS signals of either colorectal cancer or inflammatory bowel disease. If we relax the mQTL discovery threshold from an FDR of 0.01 to 0.1, we find evidence of co-localization between a colon cis-mQTL (for smoking-associated CpG cg13616097 located in the gene body of WNT7B) and a colorectal cancer GWAS signal (PP4 = 0.98).50 We also find evidence of co-localization between a colon cis-mQTL (for smoking-associated CpG cg04048259 located downstream of ZNF831) and a GWAS signal for inflammatory bowel disease (PP4 = 0.99) (Table S8).51
Enrichment of smoking-associated CpGs within genomic features and biological pathways
Examining the distribution of hypermethylated and hypomethylated smoking-associated CpGs within genomic features, we observed that hypomethylated CpGs (FDR < 0.05) in colon were enriched in islands (p < 10−5). In contrast, in lung, hypomethylated CpGs were depleted in islands (p < 10−5) (Figure S14). Similarly, we observed different patterns of enrichment of smoking-associated CpGs sites in chromatin segmentation features between colon and lung. Both hypermethylated and hypomethylated lung CpGs showed enrichment in repressed polycomb states, whereas hypermethylated and hypomethylated colon CpGs showed enrichment in regions of active transcription (Figure S15). Finally, we observed clear enrichment of smoking-associated CpG sites across all TFBS in colon (p = 1.48 × 10−166) and lung (p = 1.59 × 10−105). The top enriched TFBS differed between the tissues and between hypermethylated and hypomethylated CpG sites within each tissue types (Figures S16 and S17).
We conducted pathway analyses of the 6,350 smoking-associated CpGs (from lung) assigned to 2,948 genes, which revealed 17 overrepresented biological pathways (FDR of 0.05). The top ten pathways identified using all 6,350 smoking-associated CpGs (Table 3) included xenobiotic metabolism (p = 9.3 × 10−4), a pathway that included several of our strongest signals already described (AHRR, CYP1A1, and CYP1B1), highlighting the response of biotransformation genes to the chemicals in cigarette smoke. Numerous cancer-related pathways also showed enrichment, including tumor necrosis factor alpha (TNF-α),66 signaling via nuclear factor kappa beta (NFKB), apoptosis,67 p53,68 IL6-JAK-STAT3 signaling,69 early estrogen response,70 ultraviolet (UV) radiation response,71 transforming growth factor β signaling,72 hypoxia,73,74 MTORC1 signaling,75 and cholesterol homeostasis.76 We additionally explored enrichment of gene sets related to human diseases among KEGG pathways and identified a pathway for lipids and atherosclerosis (Table 3). When examining enrichment separately for hypomethylated CpGs (n = 4,637), which comprised the majority of smoking-associated CpGs, we observed similar pathway enrichments (Table S9); the number of hypermethylated, smoking-associated CpGs was small and underpowered for pathway analysis. Similar analyses of the 2,735 smoking-associated CpGs from colon (assigned to 1,369 genes), of which 94.6% were hypomethylated, resulted in the detection of only two enriched Hallmark gene sets (FDR 0.05), epithelial to mesenchymal transition, and UV response (down regulation) (Table S10).
Table 3.
Pathway analysis of smoking-associated CpGs detected in lung tissue
| Description | Genes in gene set | Genes with smoking-associated CpGsa | Enrichment P | FDR-adjusted p value |
|---|---|---|---|---|
| Hallmark gene sets | ||||
| TNF-alpha signaling via NFKb | 199 | 56 | 8.40E-06 | 4.20E-04 |
| Apoptosis | 155 | 46 | 1.69E-04 | 4.22E-03 |
| P53 pathway | 196 | 53 | 3.35E-04 | 5.59E-03 |
| Xenobiotic metabolism | 197 | 49 | 9.01E-04 | 1.13E-02 |
| Early response to estrogen | 194 | 60 | 2.06E-03 | 1.69E-02 |
| IL6-JAK-STAT3 signaling | 81 | 23 | 2.21E-03 | 1.69E-02 |
| UV response (down regulated) | 142 | 52 | 2.36E-03 | 1.69E-02 |
| TGF-beta signaling | 53 | 21 | 3.59E-03 | 2.25E-02 |
| Hypoxia | 190 | 51 | 4.52E-03 | 2.51E-02 |
| Cholesterol homeostasis | 71 | 21 | 6.29E-03 | 3.14E-02 |
| Myogenesis | 195 | 55 | 7.08E-03 | 3.22E-02 |
| IL2-STAT5 signaling | 194 | 51 | 9.42E-03 | 3.93E-02 |
| Adipogenesis | 196 | 45 | 1.05E-02 | 4.02E-02 |
| Androgen response | 97 | 30 | 1.23E-02 | 4.17E-02 |
| Bile acid metabolism | 110 | 26 | 1.27E-02 | 4.17E-02 |
| MTORC1 signaling | 194 | 44 | 1.33E-02 | 4.17E-02 |
| KEGG Pathways | ||||
| Parathyroid hormone synthesis, secretion, and action | 105 | 42 | 9.85E-05 | 3.50E-02 |
| Lipid and atherosclerosis | 205 | 56 | 2.11E-04 | 3.75E-02 |
Genes with CpGs (as assigned by Illumina) that are associated with smoking.
While gene set enrichment analysis resulted in six Hallmark pathways that were shared between primary (ever vs. never) and secondary (current vs. never) analyses, there were also several pathways specific to either analysis with more pathways unique to the secondary analysis (Tables 3 and S11; Figure S18). When exploring enrichment of human diseases among KEGG pathways in our secondary analysis, we identified enrichment of a pathway for atherosclerosis, similar to our primary analysis. The secondary analysis further identified KEGG pathways for human health conditions including circadian entrainment and insulin resistance, thereby supporting the notion that dysregulation of the epigenome impacts these previously reported, smoking-associated health conditions.77,78 Together, these results suggest that epigenetic dysregulation, and the effects of this dysregulation on human health and disease, may be more pronounced between current vs. never smokers in comparison to ever vs. never smokers.
Cell-type-specific effects of smoking on DNAm
To search for evidence of cell-type-specific effects of smoking on DNAm, we tested the interaction between smoking and the EPISCORE-derived cell-type proportion estimates. In lung, cell types estimated, from most abundant to least, were endothelial cells, macrophages, epithelial cells, stromal cells, granulocytes, lymphocytes, and monocytes (Figure S1). In colon, cell types estimated, from most abundant to least, were lymphocytes, enterochromaffin cells, stromal cells, myeloid cells, and epithelial cells (Figure S1). The distribution of interaction p values (Figures S19 and S20) suggests that effects of smoking on methylation at certain CpG sites varies according to the abundance of the cell types present, including endothelial cells, lymphocytes, monocytes, and macrophages. The CpGs showing the strongest evidence of interaction between smoking and cell types (i.e., involved in SxCT) in lung tissue are listed in Table S12. Most CpGs involved in SxCT also show evidence of a residual “main effect” of smoking on the CpG in a direction consistent with the interaction effect. This observation suggests that a joint test of the main effect and the cell type interaction could potentially boost power for detecting environmental effects in DNAm/EWAS studies similar to methods developed for the GWAS context.79 Interestingly, our most significant EWAS signals from lung did not show clear evidence of interaction by cell type (Table S13).
Discussion
In this work, we generated and analyzed genome-wide DNAm data for 916 human tissue samples, representing 9 unique tissue types, and characterized the association of smoking status with genome-wide measures of DNAm. We detected >6,000 smoking-associated CpGs in lung, a tissue type that showed more prominent effects of smoking compared to other tissue types. Our results show that while DNAm in some regions is impacted by smoking in multiple tissue types, the specific CpGs affected (and the magnitude of those effects) can differ between tissues. Several mQTLs impacting smoking-associated CpGs in lung tissue were found to co-localize with association signals from GWASs of lung function, suggesting smoking-related epigenetic alterations may mediate the effects of smoking on lung health. Smoking-associated CpGs were enriched in pathways related to xenobiotic metabolism and cancer.
Lung tissue had a much larger number of CpGs showing association with smoking status compared to colon and the other 7 tissue types examined. While this difference is in part due to the larger sample size for lung tissue, smoking effects in lung also appear to be more abundant after accounting for sample size differences. This is not unexpected, as lung tissue is exposed to tobacco combustion products directly via inhalation (as well as via the blood stream). In contrast, the other tissues examined are primarily exposed to tobacco combustion products via the blood stream, which carries chemicals that enter the pulmonary circulation (from the lungs) and then travel to other organs (although the colon could potentially be exposed to tobacco-derived chemicals through the gastrointestinal tract). Furthermore, we discovered that the number of CpGs passing an FDR of 0.05 between current vs. never smokers was around 2-fold that between ever vs. never smokers in both lung and colon (Table S2). Given that the ever-smokers category includes former smokers, these results suggest that smoking cessation may lead to a reduced impact of smoking on the epigenome compared to continued smoking. This conclusion aligns with studies reporting the benefits of smoking cessation related to reduced risk of adverse health conditions, including lung cancer80 and cardiovascular disease.81
Our results show that genomic regions affected by smoking can be shared across tissue types, consistent with prior studies,3,82 as we observe enrichment for smoking-related CpGs (identified from prior studies of blood) in other tissue types, including colon, ovary, and kidney. For example, NOTCH1 contained top CpGs for both lung and kidney (Figure 1), consistent with a prior study of adipose tissue.13 The enrichment observed for kidney (n = 47) suggests more signals are likely present (and observable at larger sample sizes), reflecting effects of smoking that may be consistent with the strong impact of smoking on the risk of renal cell carcinoma.83 However, for a given region, we observe that the specific CpGs associated with smoking and their relative magnitudes can vary substantially by tissue type (as observed for AHRR, CYP1A1, and CYP1B1). Thus, these findings suggest that while it is possible to assess exposure effects on DNAm within genes in accessible tissues to make inferences about effects in target tissues (for effects that are shared common across tissue types), it is more challenging to infer which specific CpGs may be impacted across tissues.
Our top smoking-associated regions in lung include several regions previously identified in blood, including the top three smoking-associated genes/regions involved in xenobiotic metabolism: AHRR, CYP1A1, and CYP1B1. Each of these regions has been identified in prior studies,7,9,19,20,21,22 and each region shows an association of smoking with increased gene expression. Each gene has a biologically plausible response to smoking, for example, AHRR encodes a transcription factor with key roles in sensing xenobiotics (including aromatic hydrocarbons) and regulation of metabolizing enzymes including CYP1A1. Our study has also discovered smoking associated CpGs, including the AOPEP, PLA2G4E, and PA2G4P4 gene regions in lung, as well as the RHOU and WNK2 gene regions in colon. Of note, PLA2G4E is part of the secretory phospholipase A2 family, a group of enzymes secreted during inflammation and involved in the cleavage of phospholipids during synthesis of eicosanoids, which are lipid mediators released by alveolar macrophages in response to toxic elements.84,85,86 Additionally, experimental evidence has shown that knockdown of RHOU, an atypical member of the RHO family, leads to higher proliferation and reduced apoptosis of colon cancer cells87; our discovery of smoking-associated CpGs in RHOU corroborates previous literature establishing smoking as a causal factor for colorectal carcinoma.1,88
Interestingly, we observed striking differences in the enrichment of hypermethylated and hypomethylated smoking-associated CpGs within genomic features (CpG islands and chromatin segmentation) for lung compared to colon. We additionally observed differences in the top enriched TFBS for lung and colon. Hypomethylated CpGs in colon were enriched in islands and active transcription regions, largely consistent with what we observe for our top smoking-associated regions involved in xenobiotic metabolism including AHRR, CYP1A1, and CYP1B1 (Figures 3 and 4). However, in lung, we observe depletion in islands and enrichment in repressed transcription states for both hypermethylated and hypomethylated CpGs. We speculate that these differences are due to the difference in the nature of exposure in lung versus colon. In lung, smoking effects appear larger and more pervasive, resulting in greater power to detect more subtle effects (e.g., transcriptionally repressed regions) beyond those related to response of xenobiotic metabolism genes.
Many of the smoking-associated CpGs identified in lung are also impacted by inherited genetic variation (i.e., mQTLs), including variants impacting lung health. Analyses of co-localization between FEV1/FVC GWAS hits and mQTLs of smoking-associated CpGs identified cg01996125 in ACVR1B as an epigenetic feature potentially involved in mediation of the effects of smoking on lung health. ACVR1B, expression of which is inversely associated with both smoking and the FEV1/FVC risk allele, is a part of the transforming growth factor beta (TGFR-β) superfamily contributing to inflammation and initiation of airway remodeling.89 Repression of ACVR1B (and any associated epigenetic alterations) may be a potential mediating pathway by which smoking (and the risk allele) have detrimental effects on lung health.
We estimated the proportions of individual cell types in lung and colon and observed substantial fractions of immune cells in both tissue types, including lymphocytes and myeloid cells (e.g., monocytes and macrophages). It is possible that the immune cell component of these tissues contributes to the observed overlap in smoking-associated regions between these tissues (and with regions previously reported in whole blood). To determine if effects of smoking on DNAm differ by cell type, we examined the interaction between smoking and the inferred cell-type proportions in lung tissue, identifying multiple CpGs potentially impacted by cell-type-specific effects. For example, we identified CpGs involved in SxCT, located upstream of COPS6 (SxCT: lymphocyte) and in the second intron of WASF2 (SxCT: monocyte and macrophage). COPS6 has been shown to promote tumor-infiltrating lymphocyte signaling in breast oncogenesis, facilitate tumor evasion,90 and promote the growth of various lung cancer cell lines.91 WASF2 mediates macrophage motility and phagocytosis by interacting with filamentous actin,92,93 with in vitro studies demonstrating immunoreactivity of WASF2 in many lung adenocarcinomas.94 Overall, our results suggest that analyses of SxCT can identify CpGs missed in analyses of marginal associations (i.e., main effects) alone, implicating genes with biologically plausible roles in cancer. Additional research is needed to explore mechanisms by which the effects of smoking are mediated by genes expressed in specific cell types. While the method we use for estimating cell-type composition (EPISCORE) is well established, it has not been validated specifically on GTEx samples. Additional work to further validate and characterize DNAm-based cellular deconvolution methods across diverse tissue types will improve our understanding of the shared cell-type-specific effects across tissues. Single-cell studies of DNAm in human tissues can also be leveraged to explore such mechanisms.
While we identified many previously unreported smoking-associated regions in disease-relevant tissues, including effects that are shared across tissues and tissue specific, this study is limited by the lack of whole-genome data on DNAm as the EPIC array is only able to capture a small fraction (∼2%) of all CpGs in the human genome. Additionally, we had small sample sizes for some tissues (e.g., kidney n = 48, muscle n = 46), which limited our power to detect associations. Therefore, larger studies of diverse tissues are needed to validate our results and generate additional data regarding the similarities and differences of DNAm across tissues. Overall, this work highlights the utility of using a multi-tissue approach to assess the effects of smoking on the human epigenome.
Data and code availability
Scripts to perform epigenome-wide association analyses are located at https://github.com/james-li-projects/SmokingEWAS.
Acknowledgments
This work was supported by grants U01 HG007601 (to B.L.P.), R35ES028379 (to B.L.P.), 2R01 GM108711 (to L.S.C), and U24 CA210993-SUB (to L.S.C) and was completed in part with computational resources provided by the Center for Research Informatics at the University of Chicago. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. We thank the donors and their families for their generous gifts of biospecimens to the GTEx research project; the Genomics Platform at the Broad Institute for data generation; F. Aguet, J. Nedzel, and K. Ardlie for sample-delivery logistics and data-release management. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
J.L.L. and N.J. performed analyses, interpreted the data, and wrote the main manuscript text. L.I.T. contributed to manuscript writing, data analysis, and interpreting results. L.T. performed analyses and prepared manuscript figures. M.G.K. and F.J. generated DNA methylation data. K.D. and M.O. performed data processing and quality control. L.S.C. advised on statistical analyses. B.L.P. conceived the project and contributed to writing/editing and data interpretation.
Declaration of interests
The authors declare no competing interests.
Published: March 14, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.02.012.
Web resources
AnVIL, https://anvil.terra.bio/#workspaces/anvil-datastorage/AnVIL_GTEx_V9_hg38
dbGaP, GTEx data, https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000424.v9.p2
epitools: Epidemiology Tools, R package, https://cran.r-project.org/web/packages/epitools/
GEO, DNAm normalized data, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE213478
GTEx Portal, https://gtexportal.org/home/
Supplemental information
References
- 1.Lushniak B.D., Samet J.M., Pechacek T.F., Norman L.A., Taylor P.A. 2014. The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General.https://stacks.cdc.gov/view/cdc/21569/cdc_21569_DS1.pdf [Google Scholar]
- 2.Akhmetova D.A., Kozlov V.V., Gulyaeva L.F. New Insight into the Role of AhR in Lung Carcinogenesis. Biochemistry. 2022;87:1219–1225. doi: 10.1134/s0006297922110013. [DOI] [PubMed] [Google Scholar]
- 3.Lee K.W.K., Pausova Z. Cigarette smoking and DNA methylation. Front. Genet. 2013;4:132. doi: 10.3389/fgene.2013.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mingay M., Chaturvedi A., Bilenky M., Cao Q., Jackson L., Hui T., Moksa M., Heravi-Moussavi A., Humphries R.K., Heuser M., Hirst M. Vitamin C-induced epigenomic remodelling in IDH1 mutant acute myeloid leukaemia. Leukemia. 2018;32:11–20. doi: 10.1038/leu.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terry M.B., Delgado-Cruzata L., Vin-Raviv N., Wu H.C., Santella R.M. DNA methylation in white blood cells. Epigenetics. 2011;6:828–837. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stueve T.R., Li W.Q., Shi J., Marconett C.N., Zhang T., Yang C., Mullen D., Yan C., Wheeler W., Hua X., et al. Epigenome-wide analysis of DNA methylation in lung tissue shows concordance with blood studies and identifies tobacco smoke-inducible enhancers. Hum. Mol. Genet. 2017;26:3014–3027. doi: 10.1093/hmg/ddx188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao X., Jia M., Zhang Y., Breitling L.P., Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin. Epigenetics. 2015;7 doi: 10.1186/s13148-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breitling L.P., Yang R., Korn B., Burwinkel B., Brenner H. Tobacco-Smoking-Related Differential DNA Methylation: 27K Discovery and Replication. Am. J. Hum. Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambatipudi S., Cuenin C., Hernandez-Vargas H., Ghantous A., Le Calvez-Kelm F., Kaaks R., Barrdahl M., Boeing H., Aleksandrova K., Trichopoulou A., et al. Tobacco smoking-associated genome-wide DNA methylation changes in the EPIC study. Epigenomics. 2016;8:599–618. doi: 10.2217/epi-2016-0001. [DOI] [PubMed] [Google Scholar]
- 10.Zeilinger S., Kühnel B., Klopp N., Baurecht H., Kleinschmidt A., Gieger C., Weidinger S., Lattka E., Adamski J., Peters A., et al. Tobacco Smoking Leads to Extensive Genome-Wide Changes in DNA Methylation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ringh M.V., Hagemann-Jensen M., Needhamsen M., Kular L., Breeze C.E., Sjöholm L.K., Slavec L., Kullberg S., Wahlström J., Grunewald J., et al. Tobacco smoking induces changes in true DNA methylation, hydroxymethylation and gene expression in bronchoalveolar lavage cells. EBioMedicine. 2019;46:290–304. doi: 10.1016/j.ebiom.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siemelink M.A., van der Laan S.W., Haitjema S., van Koeverden I.D., Schaap J., Wesseling M., de Jager S.C.A., Mokry M., van Iterson M., Dekkers K.F., et al. Smoking is Associated to DNA Methylation in Atherosclerotic Carotid Lesions. Circ. Genom. Precis. Med. 2018;11:e002030. doi: 10.1161/circgen.117.002030. [DOI] [PubMed] [Google Scholar]
- 13.Tsai P.-C., Glastonbury C.A., Eliot M.N., Bollepalli S., Yet I., Castillo-Fernandez J.E., Carnero-Montoro E., Hardiman T., Martin T.C., Vickers A., et al. Smoking induces coordinated DNA methylation and gene expression changes in adipose tissue with consequences for metabolic health. Clin. Epigenetics. 2018;10 doi: 10.1186/s13148-018-0558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barcelona V., Huang Y., Brown K., Liu J., Zhao W., Yu M., Kardia S.L.R., Smith J.A., Taylor J.Y., Sun Y.V. Novel DNA methylation sites associated with cigarette smoking among African Americans. Epigenetics. 2019;14:383–391. doi: 10.1080/15592294.2019.1588683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koo H.-K., Morrow J., Kachroo P., Tantisira K., Weiss S.T., Hersh C.P., Silverman E.K., DeMeo D.L. Sex-specific associations with DNA methylation in lung tissue demonstrate smoking interactions. Epigenetics. 2021;16:692–703. doi: 10.1080/15592294.2020.1819662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joubert B.R., Felix J.F., Yousefi P., Bakulski K.M., Just A.C., Breton C., Reese S.E., Markunas C.A., Richmond R.C., Xu C.J., et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. Am. J. Hum. Genet. 2016;98:680–696. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everson T.M., Vives-Usano M., Seyve E., Cardenas A., Lacasaña M., Craig J.M., Lesseur C., Baker E.R., Fernandez-Jimenez N., Heude B., et al. Placental DNA methylation signatures of maternal smoking during pregnancy and potential impacts on fetal growth. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-24558-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shenker N.S., Polidoro S., van Veldhoven K., Sacerdote C., Ricceri F., Birrell M.A., Belvisi M.G., Brown R., Vineis P., Flanagan J.M. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum. Mol. Genet. 2013;22:843–851. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- 19.Park S.L., Patel Y.M., Loo L.W.M., Mullen D.J., Offringa I.A., Maunakea A., Stram D.O., Siegmund K., Murphy S.E., Tiirikainen M., Le Marchand L. Association of internal smoking dose with blood DNA methylation in three racial/ethnic populations. Clin. Epigenetics. 2018;10 doi: 10.1186/s13148-018-0543-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Elgizouli M., Schöttker B., Holleczek B., Nieters A., Brenner H. Smoking-associated DNA methylation markers predict lung cancer incidence. Clin. Epigenetics. 2016;8:127. doi: 10.1186/s13148-016-0292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haase T., Müller C., Krause J., Röthemeier C., Stenzig J., Kunze S., Waldenberger M., Münzel T., Pfeiffer N., Wild P.S., et al. Novel DNA Methylation Sites Influence GPR15 Expression in Relation to Smoking. Biomolecules. 2018;8:74. doi: 10.3390/biom8030074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monick M.M., Beach S.R.H., Plume J., Sears R., Gerrard M., Brody G.H., Philibert R.A. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012;159B:141–151. doi: 10.1002/ajmg.b.32021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fasanelli F., Baglietto L., Ponzi E., Guida F., Campanella G., Johansson M., Grankvist K., Johansson M., Assumma M.B., Naccarati A., et al. Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nat. Commun. 2015;6 doi: 10.1038/ncomms10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baglietto L., Ponzi E., Haycock P., Hodge A., Bianca Assumma M., Jung C.H., Chung J., Fasanelli F., Guida F., Campanella G., et al. DNA methylation changes measured in pre-diagnostic peripheral blood samples are associated with smoking and lung cancer risk. Int. J. Cancer. 2017;140:50–61. doi: 10.1002/ijc.30431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joubert B.R., Håberg S.E., Nilsen R.M., Wang X., Vollset S.E., Murphy S.K., Huang Z., Hoyo C., Midttun Ø., Cupul-Uicab L.A., et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ. Health Perspect. 2012;120:1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markunas C.A., Xu Z., Harlid S., Wade P.A., Lie R.T., Taylor J.A., Wilcox A.J. Identification of DNA methylation changes in newborns related to maternal smoking during pregnancy. Environ. Health Perspect. 2014;122:1147–1153. doi: 10.1289/ehp.1307892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliva M., Demanelis K., Lu Y., Chernoff M., Jasmine F., Ahsan H., Kibriya M.G., Chen L.S., Pierce B.L. DNA methylation QTL mapping across diverse human tissues provides molecular links between genetic variation and complex traits. Nat. Genet. 2023;55:112–122. doi: 10.1038/s41588-022-01248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium G. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carithers L.J., Ardlie K., Barcus M., Branton P.A., Britton A., Buia S.A., Compton C.C., DeLuca D.S., Peter-Demchok J., Gelfand E.T., et al. A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv. Biobank. 2015;13:311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siminoff L.A., Wilson-Genderson M., Gardiner H.M., Mosavel M., Barker K.L. Consent to a Postmortem Tissue Procurement Study: Distinguishing Family Decision Makers' Knowledge of the Genotype-Tissue Expression Project. Biopreserv. Biobank. 2018;16:200–206. doi: 10.1089/bio.2017.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heiss J.A., Just A.C. Identifying mislabeled and contaminated DNA methylation microarray data: an extended quality control toolset with examples from GEO. Clin. Epigenetics. 2018;10 doi: 10.1186/s13148-018-0504-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pidsley R., Zotenko E., Peters T.J., Lawrence M.G., Risbridger G.P., Molloy P., Van Djik S., Muhlhausler B., Stirzaker C., Clark S.J. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17 doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.GTEx Consortium. Laboratory, Data Analysis &Coordinating Center LDACC—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx eGTEx groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.GTEx Consortium. Ardlie K.G., Deluca D.S., Segrè A.V., Sullivan T.J., Young T.R., Gelfand E.T., Trowbridge C.A., Maller J.B., Tukiainen T., et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyth GK. limma: Linear Models for Microarray Data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor: Springer-Verlag. p. 397-420.
- 37.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demanelis K., Argos M., Tong L., Shinkle J., Sabarinathan M., Rakibuz-Zaman M., Sarwar G., Shahriar H., Islam T., Rahman M., et al. Association of Arsenic Exposure with Whole Blood DNA Methylation: An Epigenome-Wide Study of Bangladeshi Adults. Environ. Health Perspect. 2019;127 doi: 10.1289/ehp3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva C.P., Kamens H.M. Cigarette smoke-induced alterations in blood: A review of research on DNA methylation and gene expression. Exp. Clin. Psychopharmacol. 2021;29:116–135. doi: 10.1037/pha0000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M., Furumichi M., Sato Y., Kawashima M., Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51:D587–D592. doi: 10.1093/nar/gkac963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phipson B., Maksimovic J., Oshlack A. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics. 2016;32:286–288. doi: 10.1093/bioinformatics/btv560. [DOI] [PubMed] [Google Scholar]
- 46.Geeleher P., Hartnett L., Egan L.J., Golden A., Raja Ali R.A., Seoighe C. Gene-set analysis is severely biased when applied to genome-wide methylation data. Bioinformatics. 2013;29:1851–1857. doi: 10.1093/bioinformatics/btt311. [DOI] [PubMed] [Google Scholar]
- 47.Shrine N., Guyatt A.L., Erzurumluoglu A.M., Jackson V.E., Hobbs B.D., Melbourne C.A., Batini C., Fawcett K.A., Song K., Sakornsakolpat P., et al. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat. Genet. 2019;51:481–493. doi: 10.1038/s41588-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hemani G., Zheng J., Elsworth B., Wade K.H., Haberland V., Baird D., Laurin C., Burgess S., Bowden J., Langdon R., et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7 doi: 10.7554/elife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elsworth B., Lyon M., Alexander T., Liu Y., Matthews P., Hallett J., Bates P., Palmer T., Haberland V., Smith G.D., et al. The MRC IEU OpenGWAS data infrastructure. bioRxiv. 2020 doi: 10.1101/2020.08.10.244293. Preprint at. [DOI] [Google Scholar]
- 50.Fernandez-Rozadilla C., Timofeeva M., Chen Z., Law P., Thomas M., Schmit S., Díez-Obrero V., Hsu L., Fernandez-Tajes J., Palles C., et al. Deciphering colorectal cancer genetics through multi-omic analysis of 100,204 cases and 154,587 controls of European and east Asian ancestries. Nat. Genet. 2023;55:89–99. doi: 10.1038/s41588-022-01222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Lange K.M., Moutsianas L., Lee J.C., Lamb C.A., Luo Y., Kennedy N.A., Jostins L., Rice D.L., Gutierrez-Achury J., Ji S.G., et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu B., Gloudemans M.J., Rao A.S., Ingelsson E., Montgomery S.B. Abundant associations with gene expression complicate GWAS follow-up. Nat. Genet. 2019;51:768–769. doi: 10.1038/s41588-019-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu T., Liu J., Beck S., Pan S., Capper D., Lechner M., Thirlwell C., Breeze C.E., Teschendorff A.E. A pan-tissue DNA methylation atlas enables in silico decomposition of human tissue methylomes at cell-type resolution. Nat. Methods. 2022;19:296–306. doi: 10.1038/s41592-022-01412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim-Hellmuth S., Aguet F., Oliva M., Muñoz-Aguirre M., Kasela S., Wucher V., Castel S.E., Hamel A.R., Viñuela A., Roberts A.L., et al. Cell type–specific genetic regulation of gene expression across human tissues. Science. 2020;369:eaaz8528. doi: 10.1126/science.aaz8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bauer M., Linsel G., Fink B., Offenberg K., Hahn A.M., Sack U., Knaack H., Eszlinger M., Herberth G. A varying T cell subtype explains apparent tobacco smoking induced single CpG hypomethylation in whole blood. Clin. Epigenetics. 2015;7 doi: 10.1186/s13148-015-0113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Obeidat M., Ding X., Fishbane N., Hollander Z., Ng R.T., McManus B., Tebbutt S.J., Miller B.E., Rennard S., Paré P.D., Sin D.D. The Effect of Different Case Definitions of Current Smoking on the Discovery of Smoking-Related Blood Gene Expression Signatures in Chronic Obstructive Pulmonary Disease. Nicotine Tob. Res. 2016;18:1903–1909. doi: 10.1093/ntr/ntw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imboden M., Wielscher M., Rezwan F.I., Amaral A.F.S., Schaffner E., Jeong A., Beckmeyer-Borowko A., Harris S.E., Starr J.M., Deary I.J., et al. Epigenome-wide association study of lung function level and its change. Eur. Respir. J. 2019;54 doi: 10.1183/13993003.00457-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuemmeler B.F., Dozmorov M.G., Do E.K., Zhang J.J., Grenier C., Huang Z., Maguire R.L., Kollins S.H., Hoyo C., Murphy S.K. DNA Methylation in Babies Born to Nonsmoking Mothers Exposed to Secondhand Smoke during Pregnancy: An Epigenome-Wide Association Study. Environ. Health Perspect. 2021;129 doi: 10.1289/ehp8099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joehanes R., Just A.C., Marioni R.E., Pilling L.C., Reynolds L.M., Mandaviya P.R., Guan W., Xu T., Elks C.E., Aslibekyan S., et al. Epigenetic signatures of cigarette smoking. Circ. Cardiovasc. Genet. 2016;9:436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richter G.M., Kruppa J., Munz M., Wiehe R., Häsler R., Franke A., Martins O., Jockel-Schneider Y., Bruckmann C., Dommisch H., Schaefer A.S. A combined epigenome- and transcriptome-wide association study of the oral masticatory mucosa assigns CYP1B1 a central role for epithelial health in smokers. Clin. Epigenetics. 2019;11 doi: 10.1186/s13148-019-0697-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang R., Lai L., Dong X., He J., You D., Chen C., Lin L., Zhu Y., Huang H., Shen S., et al. SIPA1L3 methylation modifies the benefit of smoking cessation on lung adenocarcinoma survival: an epigenomic–smoking interaction analysis. Mol. Oncol. 2019;13:1235–1248. doi: 10.1002/1878-0261.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song N., Sim J.A., Dong Q., Zheng Y., Hou L., Li Z., Hsu C.W., Pan H., Mulder H., Easton J., et al. Blood DNA methylation signatures are associated with social determinants of health among survivors of childhood cancer. Epigenetics. 2022;17:1389–1403. doi: 10.1080/15592294.2022.2030883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee M.K., Hong Y., Kim S.Y., Kim W.J., London S.J. Epigenome-wide association study of chronic obstructive pulmonary disease and lung function in Koreans. Epigenomics. 2017;9:971–984. doi: 10.2217/epi-2017-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cardenas A., Ecker S., Fadadu R.P., Huen K., Orozco A., McEwen L.M., Engelbrecht H.R., Gladish N., Kobor M.S., Rosero-Bixby L., et al. Epigenome-wide association study and epigenetic age acceleration associated with cigarette smoking among Costa Rican adults. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-08160-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang D., Tao D., Fang Y., Deng C., Xu Q., Zhou J. TNF-Alpha Promotes Invasion and Metastasis via NF-Kappa B Pathway in Oral Squamous Cell Carcinoma. Med. Sci. Monit. Basic Res. 2017;23:141–149. doi: 10.12659/msmbr.903910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lowe S.W., Lin A.W. Apoptosis in cancer. Carcinogenesis. 2000;21:485–495. doi: 10.1093/carcin/21.3.485. [DOI] [PubMed] [Google Scholar]
- 68.Whibley C., Pharoah P.D.P., Hollstein M. p53 polymorphisms: cancer implications. Nat. Rev. Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 69.Johnson D.E., O'Keefe R.A., Grandis J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oshi M., Tokumaru Y., Angarita F.A., Yan L., Matsuyama R., Endo I., Takabe K. Degree of Early Estrogen Response Predict Survival after Endocrine Therapy in Primary and Metastatic ER-Positive Breast Cancer. Cancers. 2020;12:3557. doi: 10.3390/cancers12123557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim K.-H., Kim H.J., Lee T.R. Epidermal long non-coding RNAs are regulated by ultraviolet irradiation. Gene. 2017;637:196–202. doi: 10.1016/j.gene.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 72.Ikushima H., Miyazono K. TGFβ signalling: a complex web in cancer progression. Nat. Rev. Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 73.Wilson W.R., Hay M.P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 74.Brahimi-Horn M.C., Chiche J., Pouysségur J. Hypoxia and cancer. J. Mol. Med. 2007;85:1301–1307. doi: 10.1007/s00109-007-0281-3. [DOI] [PubMed] [Google Scholar]
- 75.Tian T., Li X., Zhang J. mTOR signaling in cancer and mTOR inhibitors in solid tumor targeting therapy. Int. J. Mol. Sci. 2019;20:755. doi: 10.3390/ijms20030755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mok E.H.K., Lee T.K.W. The Pivotal Role of the Dysregulation of Cholesterol Homeostasis in Cancer: Implications for Therapeutic Targets. Cancers. 2020;12:1410. doi: 10.3390/cancers12061410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hwang J.-W., Sundar I.K., Yao H., Sellix M.T., Rahman I. Circadian clock function is disrupted by environmental tobacco/cigarette smoke, leading to lung inflammation and injury via a SIRT1-BMAL1 pathway. Faseb. J. 2014;28:176–194. doi: 10.1096/fj.13-232629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Artese A., Stamford B.A., Moffatt R.J. Cigarette smoking: an accessory to the development of insulin resistance. Am. J. Lifestyle Med. 2019;13:602–605. doi: 10.1177/1559827617726516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aschard H., Hancock D.B., London S.J., Kraft P. Genome-wide meta-analysis of joint tests for genetic and gene-environment interaction effects. Human Heredity. 2011:292–300. doi: 10.1159/000323318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tindle H.A., Stevenson Duncan M., Greevy R.A., Vasan R.S., Kundu S., Massion P.P., Freiberg M.S. Lifetime Smoking History and Risk of Lung Cancer: Results From the Framingham Heart Study. J. Nat. Can. Inst. 2018;110:1201–1207. doi: 10.1093/jnci/djy041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duncan M.S., Freiberg M.S., Greevy R.A., Jr., Kundu S., Vasan R.S., Tindle H.A. Association of Smoking Cessation With Subsequent Risk of Cardiovascular Disease. JAMA. 2019;322:642–650. doi: 10.1001/jama.2019.10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bakulski K.M., Dou J., Lin N., London S.J., Colacino J.A. DNA methylation signature of smoking in lung cancer is enriched for exposure signatures in newborn and adult blood. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-40963-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chow W.-H., Dong L.M., Devesa S.S. Epidemiology and risk factors for kidney cancer. Nat. Rev. Urol. 2010;7:245–257. doi: 10.1038/nrurol.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Granata F., Frattini A., Loffredo S., Del Prete A., Sozzani S., Marone G., Triggiani M. Signaling events involved in cytokine and chemokine production induced by secretory phospholipase A2 in human lung macrophages. Eur. J. Immunol. 2006;36:1938–1950. doi: 10.1002/eji.200535567. [DOI] [PubMed] [Google Scholar]
- 85.Saiga A., Uozumi N., Ono T., Seno K., Ishimoto Y., Arita H., Shimizu T., Hanasaki K. Group X secretory phospholipase A2 can induce arachidonic acid release and eicosanoid production without activation of cytosolic phospholipase A2 alpha. Prostaglandins Other Lipid Mediat. 2005;75:79–89. doi: 10.1016/j.prostaglandins.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 86.Serhan C.N. Resolution Phase of Inflammation: Novel Endogenous Anti-Inflammatory and Proresolving Lipid Mediators and Pathways. Annu. Rev. Immunol. 2007;25:101–137. doi: 10.1146/annurev.immunol.25.022106.141647. [DOI] [PubMed] [Google Scholar]
- 87.Slaymi C., Vignal E., Crès G., Roux P., Blangy A., Raynaud P., Fort P. The atypical RhoU/Wrch1 Rho GTPase controls cell proliferation and apoptosis in the gut epithelium. Biol. Cell. 2019;111:121–141. doi: 10.1111/boc.201800062. [DOI] [PubMed] [Google Scholar]
- 88.Habits P., Combustions I. 2012. France: IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. [Google Scholar]
- 89.Spitz M.R., Gorlov I.P., Amos C.I., Dong Q., Chen W., Etzel C.J., Gorlova O.Y., Chang D.W., Pu X., Zhang D., et al. Variants in Inflammation Genes Are Implicated in Risk of Lung Cancer in Never Smokers Exposed to Second-hand Smoke. Cancer Discov. 2011;1:420–429. doi: 10.1158/2159-8290.cd-11-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Du W.-Q., Zhu Z.M., Jiang X., Kang M.J., Pei D.S. COPS6 promotes tumor progression and reduces CD8+ T cell infiltration by repressing IL-6 production to facilitate tumor immune evasion in breast cancer. Acta Pharmacol. Sin. 2023;44:1890–1905. doi: 10.1038/s41401-023-01085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mahajan S., Majumder A., Stewart P.A., Chen Y.A., Adhikari E., Fang B., Yang Y., Lawrence H., Kinose F., Koomen J.M., Haura E.B. Deubiquitinase Vulnerabilities Identified through Activity-Based Protein Profiling in Non-Small Cell Lung Cancer. ACS Chem. Biol. 2022;17:776–784. doi: 10.1021/acschembio.2c00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kheir W.A., Gevrey J.C., Yamaguchi H., Isaac B., Cox D. A WAVE2-Abi1 complex mediates CSF-1-induced F-actin-rich membrane protrusions and migration in macrophages. J. Cell Sci. 2005;118:5369–5379. doi: 10.1242/jcs.02638. [DOI] [PubMed] [Google Scholar]
- 93.Cui H., Liu Y., Zheng Y., Li H., Zhang M., Wang X., Zhao X., Cheng H., Xu J., Chen X., Ding Z. Intelectin enhances the phagocytosis of macrophages via CDC42-WASF2-ARPC2 signaling axis in Megalobrama amblycephala. Int. J. Biol. Macromol. 2023;236 doi: 10.1016/j.ijbiomac.2023.124027. [DOI] [PubMed] [Google Scholar]
- 94.Semba S., Iwaya K., Matsubayashi J., Serizawa H., Kataba H., Hirano T., Kato H., Matsuoka T., Mukai K. Coexpression of actin-related protein 2 and Wiskott-Aldrich syndrome family verproline-homologous protein 2 in adenocarcinoma of the lung. Clin. Cancer Res. 2006;12:2449–2454. doi: 10.1158/1078-0432.CCR-05-2566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Scripts to perform epigenome-wide association analyses are located at https://github.com/james-li-projects/SmokingEWAS.