Abstract
Mouse adenovirus type 1 (MAV-1) early region 1A (E1A) viral mutants were used to determine the importance of this region in pathogenesis and establishment of a persistent infection in the natural host. Lethal dose analysis with adult male Swiss outbred mice revealed a significant reduction in virulence for all of the E1A mutants. During acute infections with 105 PFU of virus, an E1A null mutant, pmE109, was found in the same organs (brain, spleen, and spinal cord) and the same cell types (endothelial cells and mononuclear cells in lymphoid tissue) as wild-type virus. Another null mutant, pmE112, was detected in the same organs but in lower numbers. However, when mice were given a lower dose, 1 PFU, pmE109 and pmE112 reached none of the target organs analyzed by 14 days postinfection (p.i.). The absence of E1A did not hinder the ability of MAV-1 to establish a persistent infection. Viral nucleic acid was detected by PCR amplification or in situ hybridization in the kidneys, brains, spleens, and prefemoral lymph nodes of mice infected with wild-type or mutant virus up to 55 weeks p.i. The brain, spleen, and lymph node are recognized sites of acute viral infection but are previously unrecognized sites for MAV-1 persistence. Evidence for the potential reactivation of persistent MAV-1 infections is also presented.
The mouse adenovirus type 1 (MAV-1) early region 1A (E1A) produces a single message which encodes a single 30-kDa protein (3, 51). MAV-1 E1A has approximately 40% similarity to the larger, 289-amino-acid E1A protein from human adenovirus type 4 (hAd4), hAd5, and hAd7 within three conserved regions of the protein, with the strongest similarity being in conserved region 2 (CR2). MAV-1 E1A associates with both pRb and p107 through CR2 and CR1 (51). Removal of the E1A region from the MAV-1 genome does not appear to significantly hinder the ability of the virus to replicate in fibroblasts in cell culture (60). None of the early messages, except the E1A message, appears to be significantly reduced in expression during an infection of mouse 3T6 cells with a virus that produces no detectable E1A protein (60).
Pathogenesis studies done with MAV-1 in mice have shown that a widely disseminated systemic infection occurs upon intraperitoneal (i.p.) or intranasal infection (20, 32, 35). The primary targets of the virus are vascular endothelium and lymphoid tissue (32). Mice given a high dose of virus develop clinical signs of disease, with neurological compromise such as posterior paresis and flaccid paralysis (20, 35). These signs correlate with endothelial cells of the brain and spinal cord being the major sites of viral infection (32). Some mice that develop a neurological syndrome eventually succumb to the infection. Time to death ranges from 2 to 12 days postinfection (p.i.) and is dose dependent (35). However, many mice recover after 3 weeks p.i. and do not display any residual signs of disease. Evidence of chronic viruria and the presence of virus in the kidney long after virus has been cleared from other infected organs suggests that MAV-1 can establish a persistent infection in its natural host (16, 56).
Studies with human adenovirus have revealed many roles that E1A performs during an infection in cell culture, suggesting that E1A is important for virus-host interactions. These functions include transactivation of both cellular genes and the other early adenoviral genes (6, 15, 31, 41, 50), repression of enhanced transcription from a number of cellular genes (7, 22, 29, 53, 58), transformation of nonpermissive cells (47), and induction of cellular DNA synthesis and mitosis in growth-arrested cells (8, 49, 52, 61). The E1A protein also associates with a number of cellular proteins involved in cell cycle regulation, such as pRb, p107, p130, p300, p400, and p60 (cyclin A), which may explain its ability to perform many of its functions (reviewed in reference 40).
E1A also has immune modulatory functions, which can play a role in pathogenesis. In hAd12-transformed cells, the N terminus and/or CR2 of E1A is responsible for the down-regulation of transcription of the major histocompatibility complex class I (MHC-I) genes, resulting in decreased surface expression of MHC-I (42, 48). This may enable transformed cells to escape being recognized and killed by cytotoxic T lymphocytes (CTL) (42, 48, 57). CR1 of hAd5 E1A is required for repression of transcription of alpha interferon-induced genes (2, 21, 33, 44) and gamma interferon-induced genes (1, 38). Without the transcription of alpha interferon-induced genes, the cells can escape the antiviral activity of interferon.
Conversely, the E1A protein can also increase the recognition of infected cells by the immune system. Expression of hAd2/5 or hAd12 E1A renders nonpermissive mouse and rat cells susceptible to tumor necrosis factor alpha (TNF-α) killing (9, 13, 14). TNF-α is produced by activated macrophages and natural killer (NK) cells and can induce direct cytolysis of infected cells or activate other immune cells, leading to clearance of infected cells. Human adenoviruses have evolved means to counteract this E1A-induced susceptibility to TNF-α by producing other proteins (E3 14.7-kDa protein [14.7K], E1B 19K, and the E3 10.4K-14.5K complex) that can protect against TNF-α (17, 18, 23).
There is evidence that cellular immunity is important for the control of human adenovirus infections. Most fatal or severe adenovirus infections occur in infants, persons receiving bone marrow or renal transplants (and who are thus immunosuppressed), or persons with T-cell defects. hAd2/5 E1A induces susceptibility to both innate and acquired immune responses in nonpermissive cells as well as to nonspecific NK cell- and macrophage-mediated lysis in transformed hamster cells (12) and in hAd2/5-infected rodent cells (10, 11). However, E1A expression during hAd2/5 infection of human cells fails to induce susceptibility to NK cell-mediated killing (46). hAd5 E1A CR1, CR2, and the second exon encode immunodominant CTL epitopes on adenovirus-infected rat cells (5, 45, 55). Therefore, E1A peptides presented on the surface of infected cells by MHC-I molecules target infected cells for elimination by CTL.
Given the many immune-modulating functions described for human adenovirus E1A in cell culture, it is possible that MAV-1 E1A plays a role in dissemination of the virus as well as establishment of persistent infection in mice. MAV-1 provides an experimental system to study a virus in its natural host. In this study, we characterized E1A mutant viral infections in mice compared to wild type (wt) virus infection. We assessed the ability of these viruses to replicate and cause disease as well as the ability to establish a persistent infection in adult Swiss outbred mice. Dissemination of E1A mutant viruses was the same as that of the wt virus, although it occurred at a lower rate. E1A null mutant viruses were 105 times less virulent than wt MAV-1 but, like wt virus, were able to persistently infect adult Swiss outbred mice.
MATERIALS AND METHODS
Virus stocks.
pmE301, the parental wt virus for the E1A mutants (51), behaves like wt MAV-1 in all characteristics assayed to date and is referred to as wt MAV-1 throughout this work. The E1A mutants dlE102, dlE105, dlE106, and pmE109 have all been described previously (51). dlE102 contains a deletion of CR2, dlE105 contains a deletion of CR1, dlE106 contains a deletion of CR3, and pmE109 contains a point mutation changing the ATG initiator codon to TTG. pmE112 contains point mutations changing the ATG initiator codon to CAC (60).
LD50 experiments.
Virus stocks were diluted in conditioned tissue culture media, and 100 μl was injected i.p. into adult male NIH Swiss outbred mice obtained from Harlan Sprague-Dawley. For the 50% lethal dose (LD50) experiments, serial 10-fold dilutions were made of each virus and five mice were injected with each dilution. The mice were monitored for clinical signs of disease for 21 days. The LD50 for each virus was determined according to the method of Reed and Muench (43).
Acute mouse inoculations.
Adult male Swiss outbred mice obtained from Harlan Sprague-Dawley were inoculated i.p. with pmE301, pmE109, or pmE112 with a high dose (104 PFU) or a low dose (100 or 10−1 PFU), and three mice in each viral group were sacrificed on days 1 to 5 postinfection (p.i.) (high dose) or on days 5, 10, and 14 p.i. (low dose). Tissues (spleen, thymus, lung, liver, kidney, urinary bladder, intestine, prefemoral lymph nodes, brain, and spinal cord) were harvested immediately postmortem, fixed in 10% formalin for 24 h, and processed to paraffin. Sections were cut at 3-μm thicknesses for histological examination and in situ hybridization. Replicate tissues were snap frozen for DNA extraction for dot blot and PCR analyses.
Dot blot analysis.
DNA was extracted from ∼3-mm3 samples of brain and spleen by incubation overnight at 55°C in 500 μl of lysis buffer (37) plus 50 μg of proteinase K, followed by two phenol extractions and two ethanol precipitations. DNA quantitation was done with PicoGreen reagent (Molecular Probes) and a fluorescent plate reader (Molecular Dynamics Biolumin 960). Five micrograms of total DNA from each sample was spotted onto nitrocellulose and probed with a radioactively labeled probe corresponding to the E1B region of MAV-1. The dot blot was probed separately with an oligonucleotide complimentary to the mouse 18S rRNA gene (54). The E1B probe was made from random primer labeling of pHSP23 (nucleotides 1100 to 1647 of the MAV-1 E1B 21K cDNA cloned into pATH23) (34) digested with HinfI. Results were quantified with a phosphorimager and normalized to the 18S rRNA signal.
PCR of mouse organ DNA.
DNA (250 ng) extracted from brain, kidney, and spleen was used for PCR amplification of virus-specific sequences. Two consecutive rounds of amplification were done with two separate primer pairs spanning the E3 region of MAV-1 (nested PCR). The first-round primer pairs were MAVR851 (5′ CAT CAG CTA CAA CTA GCA GG 3′) and MAVR1816 (5′ AAA ATA GAC AGC ATT TAG CGC CTC TAC C 3′); the second-round primers were MAVR1508 (5′ ACG CTG CTG TTA GAA AC 3′) and MAVR1098 (5′ TGT GCC TGC TTC TAC TC 3′). PCR conditions for both rounds were 94°C for 1 min followed by 29 cycles of the following program: 92°C for 15 sec, 45°C for 15 sec, and 70°C for 1 min. One microliter of the first-round reaction mixture was used in the second-round amplification. PCR amplification was done in a total volume of 25 μl with 0.2 U of Taq polymerase, 1× PCR buffer (Promega), 1.2 mM MgCl2, 0.2 mM (each) deoxynucleoside triphosphates, and 50 ng of each primer in the first round and 250 ng of each primer in the second round. All reactions were carried out in a Perkin-Elmer GeneAmp PCR System 2400.
In situ hybridization.
In situ hybridization was performed on tissues harvested from acutely and persistently infected mice to detect the presence of viral nucleic acid. This was done as described previously, with an antisense digoxigenin-labeled riboprobe specific for the E3 region of MAV-1 (32).
Immunocapture PCR from urine.
A variation of a previously described immunocapture PCR method was used to detect MAV-1 viral particles being shed in the urine of infected mice (30). A 1:100 dilution of an anti-MAV-1 virion antibody (32) in 50 mM NaCO3 (pH 9.6) was incubated in 0.2-ml thin-wall tubes (Perkin-Elmer) for 4 h at 37°C and then blocked with 1% bovine serum albumin in 50 mM NaCO3 (pH 9.6) for 1 h at 37°C. The tubes were then washed three times with phosphate-buffered saline–0.05% Tween–0.02% sodium azide and kept at 4°C. Urine samples (200 μl) were added to the tubes and incubated at 4°C overnight to allow for virus capture. The urine samples were washed out, and the tubes were rinsed six times with 50 mM KCl, 10 mM Tris (pH 9), 0.1% Triton X-100, and 1.2 mM MgCl2 · 6H2O. PCR mix (48 μl of 1× PCR buffer [Promega]: 1.2 mM MgCl2, 0.2 mM [each] deoxynucleoside triphosphates, and 100 ng of each first-round primer) was then added to each tube. The tubes were incubated at 95°C for 5 min to denature the virus particles, and then 0.2 U of Taq polymerase in 0.2 μl was added to each reaction mixture. Nested PCR was carried out with primer pairs specific for the E1A region of MAV-1. One hundred micrograms of each of the first-round primers (MAVL170 [5′ GGT TTT TTA CTT TGC GGA GC 3′] and MAVL922 [5′ AAA ATG GCC CAG GTC AGC AGG TCC ATA AAA C 3′]) and 500 ng of each of the second-round primers (MAVL170 and MAVL892 [5′ AAA TCC TTG GCA GAC TCA TCA GGA ACT TC 3′]) were used under the same conditions as those described above for the E3 primers. The sensitivity of this method allows us to detect 1 PFU (1,000 particles) in 200 μl of urine (data not shown).
γ-Irradiation.
Adult Swiss outbred mice that had previously been infected with wt or mutant virus were subjected to a single dose (700 rads) of γ-irradiation from a 60Co source. Mice were subsequently maintained in sterile cages with sterilized food, water, and bedding.
RESULTS
The absence of E1A significantly decreases the virulence of MAV-1 in Swiss outbred mice.
We performed LD50 analyses to determine if the absence of the E1A region from MAV-1 would alter virulence in mice. Four- to 6-week-old adult Swiss outbred mice were inoculated i.p. with wt or mutant viruses, and LD50s were determined by the method of Reed and Muench (43). wt MAV-1 was compared to five different E1A mutant viruses (Table 1). Three mutants (dlE105, dlE102, and dlE106) contain deletions of discrete regions within the E1A coding region (CR1, CR2, and CR3, respectively) that are conserved in most adenovirus E1A proteins (51). The other two mutants possess point mutations which change the initiator methionine ATG codon to either TTG (pmE109) or CAC (pmE112) (51, 60). Each of the CR deletion mutants produces levels of E1A protein comparable to wt virus E1A levels. Neither of the two null mutants produces any detectable E1A protein, as assayed by Western blot analysis and immunoprecipitation with a MAV-1 E1A-specific antibody (51, 60). The mean LD50 for wt MAV-1 (pmE301) is 10−1.5 PFU (Table 1). wt MAV-1 has a particle-to-PFU ratio of approximately 1,000:1 (51a); therefore, 10−1.5 PFU is equivalent to 30 particles. All of the E1A mutants tested had higher LD50s than did the wt, indicating that each was less virulent than wt virus (Table 1). Of the mutants, the CR2 deletion virus (dlE102) had the lowest LD50, 100.9 PFU. The CR3 deletion virus (dlE106) had an intermediate LD50, 102.6 PFU. The dlE105 (CR1Δ) and pmE109 (ATG→TTG) mutants each had a mean LD50 of 103.5, and pmE112 (ATG→CAC) was the least virulent, with a mean LD50 of 103.9. Deletion of CR1 or elimination of translation from the entire E1A region had the most dramatic effect on the LD50, with a 5-log-unit increase over the wt LD50.
TABLE 1.
E1A mutant virus LD50s
| Virus | LD50 (log PFU)a
|
|||
|---|---|---|---|---|
| Trial 1 | Trial 2 | Trial 3 | Trial 4 | |
| pmE301 (wt) | <0 | −1.7 | −1.3 | ND |
| dlE102 (CR2Δ) | 1.1 | 1 | 0.5 | ND |
| dlE105 (CR1Δ) | 3.6 | 2.4 | ND | 4.6 |
| dlE106 (CR3Δ) | 3.3 | 1 | 2.9 | 3.1 |
| pmE109 (ATG→TTG) | 3.3 | 3.7 | ND | ND |
| pmE112 (ATG→CAG) | ND | 3.9 | 3.8 | ND |
Determined from four independent experiments with adult male Swiss outbred mice. Five mice per dose were injected with the indicated virus at 10-fold serial dilutions. LD50s were calculated by the method of Reed and Muench (43). Mice were monitored for 21 days p.i. for mortality. Time of death was dose dependent and ranged from 3 to 17 days p.i. for all mutants. ND, not determined.
There were no differences in clinical signs of disease among the groups of mice infected with wt or E1A mutants during the LD50 experiments. Mice first developed a ruffled coat and lethargy, progressing to posterior paralysis and finally death or, in some instances, recovery. The amount of time required for the first signs of disease to appear or for death to occur was dose dependent for all of the viruses.
Reduced levels of virus are found in the brains and spleens of mice infected with an E1A null mutant virus.
Mice were infected with wt and E1A null mutant viruses (pmE109 and pmE112) to compare replication, dissemination, and pathogenesis. We investigated whether there were quantitative differences in the amount of viral DNA present in the brains and spleens of mice infected with wt and mutant viruses by dot blot analysis of total organ DNA. Three different doses of viruses were tested, due to the marked difference in the LD50s between the E1A null mutants and the wt. When the mice were infected with a high dose, 104 PFU, close to the LD50 of the E1A mutant viruses, tissues were harvested on days 1 to 5 p.i. This experiment could not progress beyond 5 days due to the virulence of the wt virus at this dose. No viral DNA was detected in the spleens of mice infected with wt virus or pmE109 at any day p.i., whereas by day 5 p.i. viral DNA was detected in the brains of mice infected with both wt and pmE109 virus (data not shown). The levels of viral DNA present at 5 days p.i. in the brains of mice infected with mutant pmE109 and wt virus were not significantly different, as determined by Student’s t test (t = 0.9; P > 0.10 [data not shown]). Surprisingly, the levels of viral DNA found in the brains of mice infected with pmE112, the other null mutant, at 5 days p.i. were significantly lower than the amount found in the brains of wt- and pmE109-infected mice (t = 26; P < 0.001 [data not shown]).
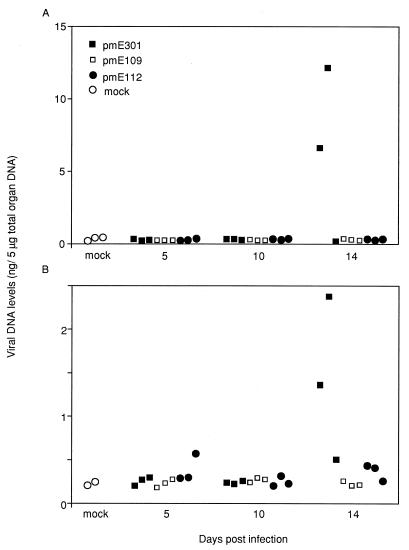
In order to allow for analysis for longer periods of time p.i., similar quantitative analyses were carried out with organs from mice infected with lower doses of viruses, 100 and 10−1 PFU, closer to the LD50 of wt MAV-1 (Fig. 1; Table 1). Tissues were harvested on days 5, 10, and 14 p.i. No viral DNA was detected by dot blot analysis or PCR amplification at 5, 10, and 14 days p.i. in either the brains or spleens of mice infected with the lowest dose, 10−1 PFU, of wt or E1A null mutant viruses (data not shown). For the mice infected with 100 PFU of wt virus on day 14 p.i., PCR amplification of DNA produced a virus-specific band from the brains and spleens. Similarly, by dot blot analysis, viral DNA was found only in those mice in brains and spleens at day 14 p.i. (Fig. 1). Two of the three mice succumbed to the wt virus infection on day 14 p.i. Dot blot analysis and quantitation with a phosphorimager indicated that brains and spleens of the wt-infected mice contained significantly higher amounts of viral DNA than those of mice infected with either pmE109 or pmE112 (Fig. 1). The Student t values for comparison of levels of viral DNA present in the brains of wt-infected mice to those of pmE109- or pmE112-infected mice were both 5.68 (P < 0.01). The Student t values for comparison of viral DNA levels in the spleens of wt- and either pmE109- or pmE112-infected mice were 2.23 and 1.94, respectively (P values of <0.10 and >0.10, respectively).
FIG. 1.
Viral DNA in organs of infected mice. Organs were harvested at 5, 10, and 14 days after i.p. infection with 1 PFU of wt (pmE301) or E1A mutant (pmE109 and pmE112) virus. At each time point, the organs were harvested from three mice from each virus group. Total DNA was extracted, and 5 μg was used for dot blot quantitation with a MAV-1 E1B viral probe. Each symbol represents results from a single mouse. (A) Brain samples. (B) Spleen samples.
Virus dissemination in mice is slower in the absence of E1A.
Tissues (spleen, thymus, lung, liver, kidney, urinary bladder, intestine, prefemoral lymph nodes, brain, and spinal cord) were harvested from mice in the acute infection experiments described above for histological examination. As has been reported for wt infections (32), there was a lack of any significant inflammation or tissue damage. This was also true for the mice infected with the E1A virus mutants pmE109 and pmE112.
The dissemination of wt, pmE109, and pmE112 viruses during the acute phase of infection in mice was compared by using in situ hybridization. At day 5 p.i. with a high dose of virus, 104 PFU, there were virtually no differences in viral tissue tropism between wt virus and the E1A mutants pmE109 and pmE112 (Table 2). Viral nucleic acid was detected by in situ hybridization on day 5 p.i. in the vascular endothelium of the brains and spinal cords and in the red pulp of the spleens of mice infected with either wt virus, pmE109, or pmE112. Rare endothelial cells in the lungs and macrophages in the prefemoral lymph nodes of mice infected with either wt, pmE109, or pmE112 also stained positive for the presence of viral nucleic acid (Table 2). Viral DNA from pmE112-infected mice was also detected in the thymus in two of the three mice sampled at 5 days p.i. (Table 2).
TABLE 2.
Localization of viral DNA in infected mice by in situ hybridization
| Dose (PFU)a | Virus | Day p.i.b | No. of mice positive/no. assayedc
|
|||||
|---|---|---|---|---|---|---|---|---|
| Brain | Spinal cord | Lung | Spleen | Lymph node | Thymus | |||
| 104 | pmE301 | 5 | 2/3 | 3/3 | 2/3 | 2/3 | 1/3 | 0/3 |
| pmE109 | 5 | 2/3 | 3/3 | 1/3 | 2/3 | 2/3 | 0/3 | |
| pmE112 | 5 | 1/3 | 1/3 | 2/3 | 2/3 | 2/3 | 2/3 | |
| 100 | pmE301 | 5 | 0/3 | 0/3 | 0/3 | 0/3 | 2/3 | 0/3 |
| 14 | 2/3 | 3/3 | 1/3 | 0/3 | 0/3 | 0/3 | ||
| pmE109 | 5 | 0/3 | 0/3 | 0/3 | 0/2 | 0/3 | 0/3 | |
| 14 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | ||
| pmE112 | 5 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | |
| 14 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | ||
Dose of virus injected i.p.
Day p.i. on which organs were harvested.
The in situ hybridization probe was a MAV-1 E3 riboprobe, as described in Materials and Methods.
The wt and mutant viruses showed different patterns and kinetics of dissemination in mice when the dose was 100 PFU. At day 14 p.i., viral DNA was detected by in situ hybridization in the endothelium of the spinal cords of all three mice infected with wt virus but was not detected in the spinal cords of mice infected with either pmE109 or pmE112 (Table 2). Heavy staining was also detected in the brains of two mice that died from the wt infection on day 14 p.i., but no virus was detected in the brains of mice infected with pmE109 or pmE112 (Table 2). Prefemoral lymph nodes from two of the three wt-infected mice showed scattered staining for viral nucleic acid on day 5 p.i., and one of the three mice tested also had a few positive endothelial cells in the lung on day 14 p.i. In contrast with wt virus, which was present at day 5 p.i. in the lymph nodes and at day 14 p.i. in the brain and spinal cord at this dose, no viral nucleic acid was detected in any tissues from mice infected with the mutant viruses pmE109 and pmE112 (Table 2).
MAV-1 is shed in the urine of wt and E1A mutant virus-infected mice up to 55 weeks p.i.
An immunocapture PCR method was employed to detect the shedding of virus in the urine of infected mice and to determine whether the absence of E1A had an effect on the ability of the virus to persist. Urine was collected from individual mice infected with wt or mutant viruses from 12 to 22 weeks after the initial infection and tested for the presence of virus (Table 3). Urine was also collected after the mice had been infected with virus for 42 to 55 weeks, and samples were pooled according to the virus type initially used to infect the mice. Virus was detected in the urine samples of individual mice infected with wt and each of the viral mutants at various times p.i. Samples taken from a single mouse at different times were not always positive for virus, suggesting that there was intermittent shedding or that the amount of virus shed occasionally dropped below the level of detection. wt virus was detected in a sample pooled from two mice at 51 weeks p.i., but no virus was detected in pools of urine from dlE102-, dlE105-, dlE106-, pmE109-, or pmE112-infected mice at 42 to 55 weeks p.i. The dlE102 and pmE109 pools contained urine from mice that had previously shed virus at 12 and 22 weeks p.i., respectively. The variation in shedding may be due to different initial doses used to infect the mice or to the fact that the virus was not replicating at a high enough rate to produce detectable levels of virus in urine at the time of sampling.
TABLE 3.
Immunocapture of MAV-1 from urine of persistently infected mice
| Virus | Mouse no. | Result at wk p.i.a:
|
||||
|---|---|---|---|---|---|---|
| 12 | 15 | 17 | 22 | 42–55b | ||
| wt | 727 | + | − | + | − | ND |
| 728, 729c | ND | ND | ND | ND | + | |
| dlE105 | 752 | ND | + | − | − | ND |
| 755 | ND | ND | ND | + | ND | |
| 747, 749c | ND | ND | ND | ND | − | |
| dlE102 | 789 | + | − | − | ND | ND |
| 785, 786, 789c | ND | ND | ND | ND | − | |
| dlE106 | 815 | − | + | − | − | ND |
| 822 | ND | ND | ND | + | ND | |
| 808, 810c | ND | ND | ND | ND | − | |
| pmE109 | 835 | ND | ND | ND | + | ND |
| 840 | ND | ND | ND | + | ND | |
| 846 | − | − | + | − | ND | |
| 835, 836, 837c | ND | ND | ND | ND | − | |
| pmE112 | 869 | − | + | − | ND | ND |
| 862, 863c | ND | ND | ND | ND | − | |
+, virus detected in urine by immunocapture method described in Materials and Methods; −, no virus detected; ND, not determined.
Samples were collected on the same day, but the time p.i. depended on the original infection date for each mouse.
Urine samples from the indicated mice were pooled for the assay.
Because MAV-1 virus can be shed in urine, it was considered to be possible for it to be transmitted through urine aerosols or on food contaminated with urine from an infected mouse. We tested whether MAV-1 could be spread via mouse bedding to uninfected mice. Bedding that had been in contact with Swiss outbred mice acutely infected with 10−1 PFU of wt virus was removed every two days for 12 days and immediately put into a new cage with uninfected mice. None of the mice exhibited signs of disease or showed any evidence of virus shedding in the urine. Serum taken from these mice also tested negative for the presence of anti-MAV-1 antibody production, as determined by an enzyme-linked immunosorbent assay (4).
wt and E1A mutant viral DNA is found in the brains, spleens, and kidneys of mice for up to 55 weeks p.i.
With our evidence of virus being shed in the urine for up to 55 weeks p.i., as well as previous reports of chronic viruria (56) and MAV-1 persistence in the kidneys for up to 70 days p.i. (16), we wanted to determine if the kidney was the sole site for viral persistence and if the lack of E1A would affect the establishment of persistence by MAV-1. Brains, kidneys, and spleens from mice that had been infected for more than 42 weeks with wt or mutant viruses were harvested and assayed for the presence of viral DNA by PCR with MAV-1-specific primers. The results of this study are shown in Table 4 (0 weeks postirradiation). Viral DNA was detected in the brains and kidneys of all three mice and in the spleens of two of the three mice infected with wt MAV-1, indicating that a persistent infection had been established in these mice. A similar result was found in mice infected with an E1A null mutant, pmE109. Viral DNA was detected in the brains of almost half of the mice infected with the other E1A null mutant, pmE112. No viral DNA was detected in the organs of mice infected with either dlE105 or dlE106. Mice infected with dlE102 were not tested. In the presence or absence of E1A, MAV-1 DNA was found in the brains, kidneys, and spleens of infected mice for up to 42 weeks p.i.
TABLE 4.
Detection of viral DNA by PCR in organs of persistently infected mice
| Wk post- irradiation | Virus | No. of mice positive/no. assayed (%)a
|
||
|---|---|---|---|---|
| Brain | Kidney | Spleen | ||
| 0 | wt | 3/3 | 3/3 | 2/3 |
| dlE102 | ND | ND | ND | |
| dlE105 | 0/4 | 0/4 | 0/4 | |
| dlE106 | 0/3 | 0/3 | 0/3 | |
| pmE109 | 2/2 | 1/2 | 2/2 | |
| pmE112 | 3/7 | 2/7 | 2/7 | |
| Total | 8/19 (42) | 6/19 (32) | 6/19 (32) | |
| 1 | wt | 2/5 | 0/5 | 1/5 |
| dlE102 | 4/4 | 4/4 | 3/4 | |
| dlE105 | 4/5 | 5/5 | 2/5 | |
| dlE106 | 4/4 | 3/4 | 1/4 | |
| pmE109 | 4/4 | 3/4 | 0/4 | |
| pmE112 | 4/5 | 2/5 | 3/5 | |
| Total | 22/27 (81) | 17/27 (63) | 10/27 (37) | |
| 2 | wt | 0/5 | 0/5 | 0/5 |
| dlE102 | 3/5 | 1/5 | 1/5 | |
| dlE105 | 3/5 | 3/5 | 2/5 | |
| dlE106b | 4/5 | 4/5 | 1/5 | |
| pmE109 | 4/5 | 2/5 | 0/5 | |
| pmE112b | 2/5 | 2/5 | 2/5 | |
| Total | 16/30 (53) | 12/30 (40) | 6/30 (20) | |
| 3 | wt | 1/5 | 0/5 | 1/5 |
| dlE102 | 0/5 | 2/5 | 0/5 | |
| dlE105 | 1/5 | 2/5 | 0/5 | |
| dlE106 | 2/5 | 0/5 | 2/5 | |
| pmE109 | 4/5 | 1/5 | 1/5 | |
| pmE112 | 4/5 | 0/5 | 0/5 | |
| Total | 12/30 (40) | 5/30 (17) | 4/30 (13) | |
DNA was extracted from organs and used for PCR analysis with MAV-1 specific primers. ND, not determined.
Urine pool from this group of mice was positive by immunocapture PCR.
wt and E1A mutant viruses can persist in the kidney, spleen, and lymph nodes.
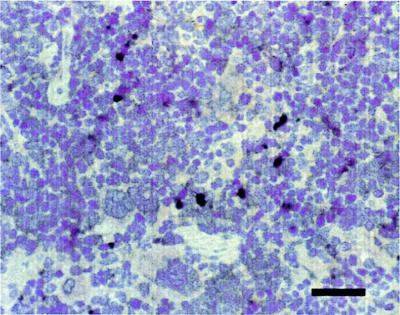
Along with the brain, spleen, and kidney, several other organs were sampled from the mice infected for 42 to 55 weeks to assay for the presence of viral nucleic acid by in situ hybridization. The results from this experiment are shown in Table 5. One of three mice infected with wt virus showed evidence of viral nucleic acid, which localized to a single endothelial cell in the cortex of the kidney. Mice infected with pmE109 and pmE112 also had evidence of viral nucleic acid in other organs. One mouse infected with pmE109 had abundant staining in the periarteriolar lymphoid sheaths in the white pulp of the spleen (Fig. 2). One mouse infected with pmE112 had staining in endothelium of four capillaries and one epithelial cell in the kidney, and the other mouse had distinct areas of staining in a prefemoral lymph node.
TABLE 5.
Localization of viral DNA in persistently infected mice by in situ hybridization
| Virus | No. of mice positive/no. assayed
|
||||
|---|---|---|---|---|---|
| Brain | Spinal cord | Spleen | Kidney | Lymph nodes | |
| pmE301 | 0/3 | 0/3 | 0/3 | 1/3 | 0/3 |
| pmE109 | 0/2 | 0/2 | 1/2 | 0/2 | 0/2 |
| pmE112 | 0/2 | 0/2 | 0/2 | 1/2 | 1/2 |
FIG. 2.
In situ hybridization with a MAV-1 E3 riboprobe of persistently infected spleen. Shown is a section of spleen from a mouse infected with E1A mutant pmE109 and sacrificed at 48 weeks p.i. The section was counterstained with hematoxylin, photographed on slide film, and digitized on a scanner. Positive viral staining appears brown. Bar = 100 μm.
Exposure to γ-irradiation may reactivate MAV-1 in persistently infected Swiss outbred mice.
We documented intermittent shedding of virus from mice persistently infected with either wt or mutant virus. We hypothesized that something was triggering transient increases in viral replication which allowed the levels of virus shed in the urine to increase to detectable levels. Reactivation of many viruses can occur in an infected individual during immunosuppression (19; reviewed in reference 39). We subjected persistently infected mice to a single sublethal dose of γ-irradiation. Such a dose would eliminate peripheral lymphocytes and some of the stem cell population and might lead to reactivation of the virus. Urine was collected at 1, 2, and 3 weeks postirradiation from groups of mice that had been infected with the same virus type for 42 to 55 weeks and tested for the presence of virus by the immunocapture PCR method described above. Urine pooled from dlE106- or pmE112-infected mice, taken 42 to 55 weeks p.i., contained detectable levels of virus when sampled at 2 weeks postirradiation (Table 4), although samples taken from mice prior to irradiation were negative (Table 3).
Exposure to γ-irradiation increases the levels of wt and E1A mutant virus in the brains, spleens, and kidneys of persistently infected mice.
The same groups of persistently infected, irradiated mice as described above were tested by PCR analysis for the presence of viral DNA in several organs. Brains, kidneys, and spleens were harvested, and DNA was extracted at 1, 2, and 3 weeks postirradiation. The results of the PCR analysis are shown in Table 4. One week after exposure to irradiation, 81% of the individuals tested (mice infected with both wt and mutant viruses) had detectable levels of viral DNA in the brain. This decreased to 53% after 2 weeks. After 3 weeks, the percentage of virus-positive brains was reduced to a level comparable to preirradiation levels, 40%. Similar results occurred in the kidneys and spleens. At 1 week postirradiation, 63% of the mice had detectable levels of viral DNA in the kidneys. After two weeks, 40% of the kidneys were virus positive, and after 3 weeks only 17% were positive. At 1 week after irradiation, 37% of the spleens tested were virus positive. At 2 weeks after irradiation, 20% of the spleens were virus positive, and after 3 weeks only 13% of the spleens were positive. Higher levels of virus detection in the organs at 1 week postirradiation correlated with a transitory atrophy of lymphoid tissue, as evidenced by the reduced size of lymph nodes, thymus, and spleen and the reduced number of observable Peyer’s patches (data not shown). Two and 3 weeks after irradiation, the spleens, lymph nodes, and thymus had regained their normal size, and Peyer’s patches were once again evident (data not shown).
DISCUSSION
Mutations were made in E1A of MAV-1 to determine the potential role of this region in pathogenesis and in establishment of persistence in the natural host. The growth pattern of these mutants in mouse fibroblasts in culture is not significantly different from that of the wt virus, and other early viral message levels are not reduced compared to a wt infection (60). These results suggest that E1A is not necessary for a productive infection in fibroblasts in culture. Recent studies have shown that the major targets of the virus in mice are the vascular endothelium and lymphoid tissue, not fibroblasts (32). Therefore, lack of a requirement for E1A in cultured fibroblasts may not accurately reflect its requirement for an in vivo infection.
The studies presented here of acute MAV-1 infection in mice with E1A null mutant viruses have revealed a requirement for E1A. One hundred thousand times more mutant virus was required initially to induce the same signs of disease and death in mice as induced by wt virus. When mice were injected with a low dose of virus, the E1A mutant viruses appeared to disseminate more slowly than wt virus, although they eventually reached the same sites (Table 2). Perhaps once the mutants reach an appropriate titer in the primary site of infection they are able to disseminate. Recent data from our laboratory indicates that in an i.p. infection the i.p. macrophages are infected and they harbor infectious virus (31a). This raises the possibility that i.p. macrophages are the primary site of viral replication upon i.p. infection. The macrophages then probably carry the virus to the secondary sites of infection, such as the lymph nodes, thymus, spleen, and brain.
The results of the LD50 experiments with the E1A mutants shown in Table 1 indicate that E1A plays a role during in vivo infection. The LD50s for the E1A mutant viruses were all higher than that of the wt, indicating that E1A is required for the virulence of this virus in adult male Swiss outbred mice. When CR2 of E1A was deleted (dlE102), the LD50 was almost 3 log units higher than that of wt virus. CR2 of MAV-1 E1A is required for E1A to associate with both mouse pRb and mouse p107 during an infection in cell culture (51). Association with these cellular proteins may allow infected cells to reenter the cell cycle and create a more favorable environment for viral replication (59). The increased LD50 in the CR2 mutant virus suggests that these interactions are important in establishing an infection in the mouse. Deletion of CR3 (dlE106) resulted in an LD50 4 log units higher than that of the wt. Deletion of CR1 (dlE105) or elimination of expression of the entire E1A region (pmE109 and pmE112) had the most dramatic effect, with a 5-log-unit increase in LD50 over the wt. Unlike the minimal effects that mutations of MAV-1 E1A have been shown to have in cultured fibroblasts (60), such E1A mutations significantly reduce the virulence of the virus in mice.
We compared the dissemination, replication, and pathogenesis of the E1A null viruses with the wt virus during an acute infection. For the mice injected with a high dose of virus, 104 PFU, there was no significant difference in the amount of viral DNA between wt- and pmE109-infected mice, as detected by dot blot analysis (Fig. 1). By in situ hybridization, pmE109 and pmE112 were found in the same target organs (brain, spleen, and spinal cord) and the same cell types (endothelial cells and macrophages) as was wt virus by day 5 p.i. (Table 2). wt virus, pmE109, and pmE112 were all detected in the endothelial cells in the brain and spinal cord as well as in the red pulp of the spleen. This implies that when mice were given a high initial dose of virus, the E1A null viruses (ATG→TTG [pmE109] and ATG→CAC [pmE112]) were able to replicate to comparable levels in the brains and spleens and to infect the same target cells as wt virus.
The E1A null mutant pmE112 (ATG→CAC) had no viral DNA detectable in the brain at 5 days p.i., a point at which it was detected in wt- and pmE109-infected mice by dot blot analysis. Although it was detectable by in situ hybridization in the brains, spleens, spinal cords, lungs, lymph nodes, and thymic medullas in some of the mice, the amount of viral nucleic acid staining in each organ was reduced in comparison to mice infected with either wt or pmE109. Possible explanations for this behavior may include the inability of this virus to replicate as efficiently as wt and pmE109 virus in mice. If pmE112 was replicating more slowly than wt virus and pmE109, then it may not have reached the titers necessary for dissemination to the other organs at the time when the mice were sampled. One explanation for the difference between the behavior of pmE109 and pmE112 may be a difference in E1A mRNA stability or expression. Both viruses have mutations in the methionine initiator codon, but perhaps the ATG→TTG mutation produces a more stable E1A mRNA than the ATG→CAC mutation or has some leaky (but still undetectable [60]) expression of the E1A protein.
When mice were infected with a low dose (10−1 PFU) of wt or mutant virus and monitored for 14 days, no detectable levels of viral DNA were found in either brains or spleens by dot blot analysis or by PCR amplification with virus-specific primers (data not shown). None of the mice in this experiment showed any signs of disease. When another group of mice was infected with 100 PFU of virus, two of the three wt-infected mice died on day 14 p.i. None of the mice infected with either pmE109 or pmE112 showed any signs of disease at this dose. By PCR amplification and dot blot analysis, we only detected viral DNA in the brains and spleens of the three mice infected with wt virus on day 14 p.i. (Fig. 1). In mice infected acutely with wt virus, the virus was detected at earlier times than mutants and in some additional sites not seen in mutant virus-infected mice. The wt virus reached the lymph nodes by day 5 p.i. and then was cleared from the lymph nodes, progressing to the brain and spleen and other sites by day 14 after i.p. infection (Table 2). Although the initial dose of virus injected into the mice was the same, the mutants took longer to replicate to detectable levels than did the wt virus.
The major sites of MAV-1 viral replication during the acute phase of a wt infection are the vascular endothelium and lymphoid tissue (32). However, virus is also detected in the epithelium lining the renal pelvis, which may explain viral excretion in the urine of infected mice. Reports indicate that MAV-1 can be detected in the kidneys of mice for up to 70 days p.i. (16) and is found in the urine of mice for up to 2 years after the initial infection (56). Our results are consistent with these findings: we detected both wt and mutant virus in the urine of mice for 12 to 55 weeks after infection (Table 3). Detection of virus in the urine was sporadic, which may be explained by only a small number of kidney epithelial cells being productively infected during a persistent infection.
We found evidence that the kidney, brain, and spleen are sites for viral persistence and that the lack of E1A does not affect the establishment of persistence by MAV-1. Organs from groups of mice which had been infected with wt or mutant viruses for 42 to 55 weeks were harvested and analyzed by PCR with virus-specific primers and by in situ hybridization with a virus-specific probe. In addition to the kidney, which had been previously reported as a site of MAV-1 persistence, we identified previously unrecognized target organs for persistence of MAV-1 (spleen, brain, and lymph node). Consistent with shedding of MAV-1 in urine, we detected virus (by in situ hybridization) in a single endothelial cell in the renal cortex at 42 weeks p.i. in one of the mice infected with wt virus and in endothelial cells of four capillaries and a single epithelial cell in the kidney of a mouse infected with pmE112. We also detected viral DNA by PCR amplification at 42 weeks p.i. in the kidneys of both wt and mutant virus-infected mice.
Other persistently infected organs were identified by either PCR or in situ hybridization from wt and mutant virus-infected mice. Viral DNA was detected by PCR in the brains and spleens at 42 weeks p.i. in mice infected with wt or E1A null mutant virus. Abundant staining by in situ hybridization was seen in the periarteriolar areas in the white pulp of the spleen of one mouse infected with pmE109. Distinct areas of staining were seen in a prefemoral lymph node of another mouse infected with pmE112. No viral DNA was detected by PCR in the organs of mice infected with either dlE105 or dlE106. There are at least two possible explanations for this result. CR1 and CR3 may be important for maintaining the virus in the host for long periods of time, or the mice used for this assay may have initially been infected with too low a dose to result in persistence of the virus. We favor the latter explanation since the null mutants (therefore lacking CR1 and CR3) were able to persist. Taken together, our immunocapture, in situ hybridization, and PCR results indicate that a persistent infection was established in multiple organs in wt and mutant virus-infected mice and that the absence of E1A did not affect the ability of the virus to persist in the mouse.
Down-regulation of the immune system may trigger a transient increase in viral replication during a persistent infection, which might allow detectable levels of virus to be made and shed. When mice persistently infected with either wt or mutant viruses were exposed to γ-irradiation, the percentage of mice possessing detectable levels of viral DNA in brains, kidneys, and spleens increased 1 week after irradiation (Table 4). The percentage of mice with viral DNA in the brains or kidneys 1 week after irradiation treatment was almost double the number before irradiation. The results with the brains and kidneys correlated with lower leukocyte counts and the presence of grossly visible atrophy of lymphoid organs, which are signs expected after γ-irradiation. Two weeks after irradiation, the percentage of mice with detectable levels of virus in the brains, kidneys, and spleens began to decrease and the lymphoid tissues began to return to normal size. Mice persistently infected with either dlE106 or pmE112 shed detectable levels of virus in urine at 2 weeks postirradiation. The percentage of viral DNA-positive organs continued to decrease through 3 weeks postirradiation, while the quantity of lymphoid tissue increased. At 3 weeks postirradiation, the percentage of viral DNA-positive organs was either comparable to or lower than preirradiation percentages. The results suggest that we successfully immunosuppressed the mice and that MAV-1 can undergo (at least transiently) increased replication in the brain, kidney, and spleen following immunosuppression. This may indicate that the cell-mediated immune response to mouse adenoviral infection (24–28) is important in establishing a persistent infection. These immunosuppression results also suggest that the brain, spleen, and kidney are sites of MAV-1 persistence in adult Swiss outbred mice infected i.p., which is consistent with our analysis of organs of long-term-infected mice.
It will be interesting to determine what factors allow the virus to persist in the host and what aspects of the immune system are important in maintaining this delicate balance. Previous studies have shown that MAV-1 does not decrease MHC-I surface expression of cultured infected cells (36). Down-regulation of MHC-I has been proposed as a possible mechanism for evading the immune system response by human adenoviruses. Except for the immunocapture method, which detects virus particles, in all of the studies described herein we assayed for the presence of viral nucleic acid rather than infectious virus production. Therefore, it is not known whether only the viral nucleic acid persists in the organs of MAV-1-infected mice (latent infection) or whether infectious virus is being produced (chronic infection).
ACKNOWLEDGMENTS
We thank Gwen Hirsch and Melissa Scott for technical assistance and the Animal Resources facility for maintenance of the mice. We also thank Lois Miller, Susan Kring Sullivan, Adriana Kajon, and Angela Cauthen for comments on the manuscript.
This work was supported by NIH grant AI23762 and American Cancer Society grant VM-176 to K.R.S. and by an NIH predoctoral traineeship (GM 07103) to K.S. K.R.S. is the recipient of an NIH Research Career Development Award.
REFERENCES
- 1.Ackrill A M, Foster G R, Laxton C D, Flavell D M, Stark G R, Kerr I M. Inhibition of the cellular response to interferons by products of the adenovirus type-5 E1A oncogene. Nucleic Acids Res. 1991;19:4387–4393. doi: 10.1093/nar/19.16.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson K, Fennie E. Adenovirus early region 1A modulation of interferon antiviral activity. J Virol. 1987;61:787–795. doi: 10.1128/jvi.61.3.787-795.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball A O, Beard C W, Redick S D, Spindler K R. Genome organization of mouse adenovirus type 1 early region 1: a novel transcription map. Virology. 1989;170:523–536. doi: 10.1016/0042-6822(89)90444-3. [DOI] [PubMed] [Google Scholar]
- 4.Beard C W, Spindler K R. Analysis of early region 3 mutants of mouse adenovirus type 1. J Virol. 1996;70:5867–5874. doi: 10.1128/jvi.70.9.5867-5874.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellgrau D, Walker T A, Cook J L. Recognition of adenovirus E1A gene products on immortalized cell surfaces by cytotoxic T lymphocytes. J Virol. 1988;62:1513–1519. doi: 10.1128/jvi.62.5.1513-1519.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berk A J, Lee F, Harrison T, Williams J, Sharp P A. Pre-early adenovirus 5 gene product regulates synthesis of early viral messenger RNAs. Cell. 1979;17:935–944. doi: 10.1016/0092-8674(79)90333-7. [DOI] [PubMed] [Google Scholar]
- 7.Borelli E, Hen R, Chambon P. Adenovirus-2 E1A products repress enhancer-induced stimulation of transcription. Nature. 1984;312:608–612. doi: 10.1038/312608a0. [DOI] [PubMed] [Google Scholar]
- 8.Braithwaite A W, Cheetham B F, Li P, Parish C R, Waldron-Stevens L K, Bellett A J D. Adenovirus-induced alterations of the cell growth cycle: a requirement for expression of E1A but not of E1B. J Virol. 1983;45:192–199. doi: 10.1128/jvi.45.1.192-199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen M-J, Holskin B, Strickler J, Gorniak J, Clark M A, Johnson P J, Mitcho M, Shalloway D. Induction by E1A oncogene expression of cellular susceptibility to lysis by TNF. Nature. 1987;324:581–583. doi: 10.1038/330581a0. [DOI] [PubMed] [Google Scholar]
- 10.Cook J L, Lewis A M. Differential NK cell and macrophage killing of hamster cells infected with nononcogenic or oncogenic adenovirus. Science. 1984;224:612–615. doi: 10.1126/science.6710160. [DOI] [PubMed] [Google Scholar]
- 11.Cook J L, May D L, Lewis A M, Walker T A. Adenovirus E1A gene induction of susceptibility to lysis by natural killer cells and activated macrophages in infected rodent cells. J Virol. 1987;61:3510–3520. doi: 10.1128/jvi.61.11.3510-3520.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook J L, Walker T A, Lewis A M, Ruley H E, Graham F L. Expression of the adenovirus E1A oncogene during cell transformation is sufficient to induce susceptibility to lysis by host inflammatory cells. Proc Natl Acad Sci USA. 1986;83:6965–6969. doi: 10.1073/pnas.83.18.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duerksen-Hughes P, Wold W S M, Gooding L R. Adenovirus E1A renders infected cells sensitive to cytolysis by tumor necrosis factor. J Immunol. 1989;143:4193–4200. [PubMed] [Google Scholar]
- 14.Duerksen-Hughes P J, Hermiston T W, Wold W S M, Gooding L R. The amino-terminal portion of CD1 of the adenovirus E1A proteins is required to induce susceptibility to tumor necrosis factor cytolysis in adenovirus-infected mouse cells. J Virol. 1991;65:1236–1244. doi: 10.1128/jvi.65.3.1236-1244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaynor R B, Hillman D, Berk A J. Adenovirus early region 1A protein activates transcription of a nonviral gene introduced into mammalian cells by infection or transfection. Proc Natl Acad Sci USA. 1984;81:1193–1197. doi: 10.1073/pnas.81.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ginder D R. Increased susceptibility of mice infected with mouse adenovirus to Escherichia coli-induced pyelonephritis. J Exp Med. 1964;120:1117–1128. doi: 10.1084/jem.120.6.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gooding L R, Elmore L W, Tollefson A E, Brady H A, Wold W S M. A 14,700 MW protein from the E3 region of adenovirus inhibits cytolysis by tumor necrosis factor. Cell. 1988;53:341–346. doi: 10.1016/0092-8674(88)90154-7. [DOI] [PubMed] [Google Scholar]
- 18.Gooding L R, Ranheim T S, Tollefson A E, Aquino L, Duerksen-Hughes P, Horton T M, Wold W S M. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus function together to protect many but not all mouse cell lines against lysis by tumor necrosis factor. J Virol. 1991;65:4114–4123. doi: 10.1128/jvi.65.8.4114-4123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenlee J E, Dodd W K. Reactivation of persistent papovavirus K infection in immunosuppressed mice. J Virol. 1984;51:425–429. doi: 10.1128/jvi.51.2.425-429.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guida J D, Fejer G, Pirofski L-A, Brosnan C F, Horwitz M S. Mouse adenovirus type 1 causes a fatal hemorrhagic encephalomyelitis in adult C57BL/6 but not BALB/c mice. J Virol. 1995;69:7674–7681. doi: 10.1128/jvi.69.12.7674-7681.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutch M J, Reich N C. Repression of the interferon signal transduction pathway by the adenovirus E1A oncogene. Proc Natl Acad Sci USA. 1991;88:7913–7917. doi: 10.1073/pnas.88.18.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hen R, Borelli E, Chambon P. Repression of the immunoglobulin heavy chain enhancer by the adenovirus-2 E1A products. Science. 1985;230:1391–1394. doi: 10.1126/science.2999984. [DOI] [PubMed] [Google Scholar]
- 23.Horton T M, Ranheim T S, Aquino L, Kusher D I, Saha S K, Ware C F, Wold W S M, Gooding L R. Adenovirus E3 14.7K protein functions in the absence of other adenovirus proteins to protect transfected cells from tumor necrosis factor cytolysis. J Virol. 1991;65:2629–2639. doi: 10.1128/jvi.65.5.2629-2639.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inada T, Uetake H. Cell-mediated immunity assayed by 51Cr release test in mice infected with mouse adenovirus. Infect Immun. 1978;20:1–5. doi: 10.1128/iai.20.1.1-5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inada T, Uetake H. Cell-mediated immunity to mouse adenovirus infection: blocking of macrophage migration inhibition and T cell-mediated cytolysis of infected cells by anti-S antigen or anti-alloantigen serum. Microbiol Immunol. 1980;24:525–535. doi: 10.1111/j.1348-0421.1980.tb02856.x. [DOI] [PubMed] [Google Scholar]
- 26.Inada T, Uetake H. Cell-mediated immunity to mouse adenovirus infection: macrophage migration inhibition test. Microbiol Immunol. 1978;22:391–401. doi: 10.1111/j.1348-0421.1978.tb00385.x. [DOI] [PubMed] [Google Scholar]
- 27.Inada T, Uetake H. Nature and specificity of effector cells in cell-mediated cytolysis of mouse adenovirus-infected cells. Infect Immun. 1978;22:119–124. doi: 10.1128/iai.22.1.119-124.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inada T, Uetake H. Virus-induced specific cell surface antigen(s) on mouse adenovirus-infected cells. Infect Immun. 1977;18:41–45. doi: 10.1128/iai.18.1.41-45.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janaswami P M, Kalvakolanu D V R, Zhang Y, Sen G C. Transcriptional repression of interleukin-6 gene by adenoviral E1A proteins. J Biol Chem. 1992;267:24886–24891. [PubMed] [Google Scholar]
- 30.Jansen R W, Newbold J E, Lemon S M. Combined immunoaffinity cDNA-RNA hybridization assay for detection of hepatitis A virus in clinical specimens. J Clin Microbiol. 1985;22:984–989. doi: 10.1128/jcm.22.6.984-989.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones N C, Shenk T. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc Natl Acad Sci USA. 1979;76:3665–3669. doi: 10.1073/pnas.76.8.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Kajon, A., R. Ohira, and K. Spindler. Unpublished data.
- 32.Kajon A E, Brown C C, Spindler K R. Distribution of mouse adenovirus type 1 in intraperitoneally and intranasally infected adult outbred mice. J Virol. 1998;72:1219–1223. doi: 10.1128/jvi.72.2.1219-1223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalvakolanu D V R, Bandyopadhyay S K, Harter M L, Sen G C. Inhibition of interferon-inducible gene expression by adenovirus E1A proteins—block in transcriptional complex formation. Proc Natl Acad Sci USA. 1991;88:7459–7463. doi: 10.1073/pnas.88.17.7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koerner T J, Hill J E, Myers A M, Tzagoloff A. High-expression vectors with multiple cloning sites for construction of trpE-fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- 35.Kring S C, King C S, Spindler K R. Susceptibility and signs associated with mouse adenovirus type 1 infection of adult outbred Swiss mice. J Virol. 1995;69:8084–8088. doi: 10.1128/jvi.69.12.8084-8088.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kring S C, Spindler K R. Lack of effect of mouse adenovirus type 1 infection on the cell surface expression of major histocompatibility complex class I antigens. J Virol. 1996;70:5495–5502. doi: 10.1128/jvi.70.8.5495-5502.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leonard G T, Sen G C. Effects of adenovirus E1A protein on interferon-signaling. Virology. 1996;224:25–33. doi: 10.1006/viro.1996.0503. [DOI] [PubMed] [Google Scholar]
- 39.McCance D J. Persistence of animal and human papovaviruses in renal and nervous tissues. In: Sever J L, Madden D L, editors. Polyomaviruses and human neurological diseases. New York, N.Y: Alan R. Liss, Inc.; 1983. pp. 343–357. [PubMed] [Google Scholar]
- 40.Moran E. Mammalian cell growth controls reflected through protein interactions with the adenovirus E1A gene products. Semin Virol. 1994;5:327–340. [Google Scholar]
- 41.Nevins J R. Induction of the synthesis of a 70,000 dalton mammalian heat shock protein by the adenovirus E1A gene product. Cell. 1982;29:913–919. doi: 10.1016/0092-8674(82)90453-6. [DOI] [PubMed] [Google Scholar]
- 42.Pereira D S, Rosenthal K L, Graham F L. Identification of adenovirus E1A regions which affect MHC class I expression and susceptibility to cytotoxic T lymphocytes. Virology. 1995;211:268–277. doi: 10.1006/viro.1995.1400. [DOI] [PubMed] [Google Scholar]
- 43.Reed L J, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 44.Reich N C, Pine R, Levy D, Darnell J E. Transcription of interferon-stimulated genes is induced by adenovirus particles but is suppressed by E1A gene products. J Virol. 1988;62:114–119. doi: 10.1128/jvi.62.1.114-119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Routes J M, Bellgrau D, McGrory W J, Bautista D S, Graham F L, Cook J L. Anti-adenovirus type 5 cytotoxic T lymphocytes: immunodominant epitopes are encoded by the E1A gene. J Virol. 1991;65:1450–1457. doi: 10.1128/jvi.65.3.1450-1457.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Routes J M, Cook J L. Adenovirus persistence in man: defective E1A gene product targeting of infected cells for elimination by natural killer cells. J Immunol. 1989;142:4022–4026. [PubMed] [Google Scholar]
- 47.Ruley H E. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983;304:602–606. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- 48.Schrier P I, Bernards R, Vaessen R T M J, Houweling A, van der Eb A J. Expression of class I major histocompatibility antigens switched off by highly oncogenic adenovirus 12 in transformed rat cells. Nature. 1983;305:771–775. doi: 10.1038/305771a0. [DOI] [PubMed] [Google Scholar]
- 49.Shimojo H, Yamashita T. Induction of DNA synthesis by adenoviruses in contact-inhibited hamster cells. Virology. 1968;36:422–433. doi: 10.1016/0042-6822(68)90167-0. [DOI] [PubMed] [Google Scholar]
- 50.Simon M C, Kitchener K, Kao H-T, Hickey E, Weber L, Voellmy R, Heintz N, Nevins J R. Selective induction of human heat shock gene transcription by the adenovirus E1A gene products, including the 12S E1A product. Mol Cell Biol. 1987;7:2884–2890. doi: 10.1128/mcb.7.8.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith K, Ying B, Ball A O, Beard C W, Spindler K R. Interaction of mouse adenovirus type 1 early region 1A protein with cellular proteins pRb and p107. Virology. 1996;224:184–197. doi: 10.1006/viro.1996.0520. [DOI] [PubMed] [Google Scholar]
- 51a.Spindler, K. R. Unpublished data.
- 52.Spindler K R, Eng C Y, Berk A J. An adenovirus early region 1A protein is required for maximal viral DNA replication in growth-arrested human cells. J Virol. 1985;53:742–750. doi: 10.1128/jvi.53.3.742-750.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stein R W, Ziff E B. Repression of insulin gene expression by adenovirus type 5 E1A proteins. Mol Cell Biol. 1987;7:1164–1170. doi: 10.1128/mcb.7.3.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torczynski R, Bollon A P, Fuke M. The complete nucleotide sequence of the rat 18S ribosomal RNA gene and its comparison with the respective yeast and frog genes. Nucleic Acids Res. 1983;11:4879–4890. doi: 10.1093/nar/11.14.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urbanelli D, Sawada Y, Raskova J, Jones N C, Shenk T, Raska K., Jr C-terminal domain of the adenovirus E1A oncogene product is required for induction of cytotoxic T lymphocytes and tumor-specific transplantation immunity. Virology. 1989;173:607–614. doi: 10.1016/0042-6822(89)90572-2. [DOI] [PubMed] [Google Scholar]
- 56.van der Veen J, Mes A. Experimental infection with mouse adenovirus in adult mice. Arch Gesamte Virusforsch. 1973;42:235–241. doi: 10.1007/BF01265648. [DOI] [PubMed] [Google Scholar]
- 57.Vasavada R, Eager K B, Barbanti-Brodano G, Caputo A, Ricciardi R P. Adenovirus type 12 early region 1A proteins repress class I HLA expression in transformed human cells. Proc Natl Acad Sci USA. 1986;83:5257–5261. doi: 10.1073/pnas.83.14.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Velcich A, Ziff E. Adenovirus E1A proteins repress transcription from the SV40 early promoter. Cell. 1985;40:705–716. doi: 10.1016/0092-8674(85)90219-3. [DOI] [PubMed] [Google Scholar]
- 59.Whyte P, Buchkovich K J, Horowitz J M, Friend S H, Raybuck M, Weinberg R A, Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 60.Ying, B., K. Smith, and K. R. Spindler. Mouse adenovirus type 1 early region 1A is dispensable for growth in cultured fibroblasts. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 61.Younghusband H B, Tyndall C, Bellett A J D. Replication and interaction of virus DNA and cellular DNA in mouse cells infected by a human adenovirus. J Gen Virol. 1979;45:455–467. doi: 10.1099/0022-1317-45-2-455. [DOI] [PubMed] [Google Scholar]