Abstract
Colorectal cancer (CRC) is the third most common cancer affecting people. The discovery of new, non-invasive, specific, and sensitive molecular biomarkers for CRC may assist in the diagnosis and support therapeutic decision making. Exosomal miRNAs have been demonstrated in carcinogenesis and CRC development, which makes these miRNAs strong biomarkers for CRC. Deep sequencing allows a robust high-throughput informatics investigation of the types and abundance of exosomal miRNAs. Thus, exosomal miRNAs can be efficiently examined as diagnostic biomarkers for disease screening. In the present study, a number of 660 mature miRNAs were detected in patients diagnosed with CRC at different stages. Of which, 29 miRNAs were differentially expressed in CRC patients compared with healthy controls. Twenty-nine miRNAs with high abundance levels were further selected for subsequent analysis. These miRNAs were either highly up-regulated (e.g., let-7a-5p, let-7c-5p, let-7f-5p, let-7d-3p, miR-423-5p, miR-3184-5p, and miR-584) or down-regulated (e.g., miR-30a-5p, miR-99-5p, miR-150-5p, miR-26-5p and miR-204-5p). These miRNAs influence critical genes in CRC, leading to either tumor growth or suppression. Most of the reported diagnostic exosomal miRNAs were shown to be circulating in blood serum. The latter is a novel miRNA that was found in exosomal profile of blood serum. Some of the predicted target genes of highly expressed miRNAs participate in several cancer pathways, including CRC pathway. These target genes include tumor suppressor genes, oncogenes and DNA repair genes. Main focus was given to multiple critical signaling cross-talking pathways including transforming growth factor β (TGFβ) signaling pathways that are directly linked to CRC. In conclusion, we recommend further analysis in order to experimentally confirm exact relationships between selected differentially expressed miRNAs and their predicted target genes and downstream functional consequences.
Keywords: Exosomes, miRNAs, Colorectal cancer, Biomarkers, Deep sequencing
Subject terms: Biochemistry, Biological techniques, Biotechnology, Cancer, Chemical biology, Computational biology and bioinformatics, Genetics, Molecular biology, Biomarkers, Diseases, Gastroenterology, Health care, Medical research, Molecular medicine, Oncology, Pathogenesis, Risk factors
Introduction
Colorectal cancer (CRC) is result of the proliferation of cancerous cells from the mucosal layer in the colon and rectum1. CRC is a major public health concern in the world, as it is the third most common cause of cancer death2. Untreated CRC eventually progresses to systemic metastasis, which is the major cause of CRC mortality3. When first diagnosed, the stage of each individual patient’s disease needs to established, as it is the most important step in setting a treatment plan(see file for reference). Earlier detection of CRC and more accurate assessment of the cancer stage could potentially help physicians better tailor cancer therapy to the patients’ exact stage. Many risk factors contribute to the development of CRC, and individuals' poor lifestyles may also contribute to rise in number of CRC cases4. It was demonstrated that several tumor suppressor genes in CRC such as APC, TGFβR2, SMAD4, PTEN and TP53, were inactivated, while in contrast, oncogenes such as KRAS, BRAF, β-catenin, and MYC were overexpressed5–7. It was exhibited that 20–30% of CRC instances are caused by hereditary mutations, while the remaining cases of CRCs are sporadic in nature8.
MicroRNAs (miRNAs) are short non-coding RNAs around 18–23 nucleotides (nt) that regulate transcription and post-transcriptional modifications of their target genes9. Roles of miRNAs include inhibition or degradation of mRNA during binding to mRNAs in 3′-UTR termini10. Although most physiological functions of miRNAs are undefined, their expression profiles have been illustrated in various pathology fields11. Moreover, sensitivity and simplicity of sampling miRNAs contribute to production of appropriate non-invasive biomarkers11. Spotting properties of miRNAs could assist in comprehending carcinogenesis machinery, which could potentially lead to them becoming biomarkers that assist in establishing CRC diagnosis and prognosis12. However, biology of miRNAs is a massive area that is required for studying and understanding their mechanisms and applications in clinical cancer diagnosis and treatment.
Because exosomes play a critical role in transferring or exchanging materials between distant tissues and mediating intercellular communication13,14, they have been employed in many studies to assess their role as diagnostic and prognostic biomarkers for CRC15. Exosomes have been distinguished as significant transporters for miRNAs in serum fields16. Expression of serum miRNA is noted to be different in CRC patients, which needs more examination to identify link between expression of specific miRNAs in both serum and tissues of CRC patients. Because of insignificance of miRNA techniques, exosomal miRNAs might be conceivably utilized as indicative of biomarkers for different types of cancer13. Study of serum exosomal miR-92a-3p and miR-17-5p for diagnosis of primary and metastatic CRC suggested that up-regulation of these miRNAs directly correlated with stages of CRC17. Another study identified that low expression of circulating exosomal miR-4772-3p, isolated from serum, had been considered a predictive biomarker for cancer recurrence in colon cancer patients at stages II and III18. Next-generation sequencing (NGS) can provide a robust informatic analysis related to sorts and abundance of exosomal miRNAs. Thus, exosomal miRNAs can be efficiently examined as diagnostic biomarkers for several diseases19.
TGFβ gene family are normally regulated in epithelial cells, however, dysregulation of any member of this family can contribute to increasing tumorigenesis. Canonical TGFβ signaling is regulated by TGFβR1, and TGFβR2, which phosphorylate SMAD2 and SMAD3, after binding to TGFβ ligands. Then, activated SMAD2 and SMAD3 comprise a complex with SMAD4. SMAD complex is a nucleoprotein that modulates expression of several target genes and affects tumor growth by controlling cell proliferation, apoptosis, metastasis, and angiogenesis (Massagué et al.20; Feng and Derynck21). miRNAs have been shown to influence formation and activation of TGFβ signaling pathway, and miRNAs/TGF interaction acts in occurrence of various malignancies, including gastric, breast, lung, pancreatic, rectal and colon cancers (Soleimani et al.22,23).
In this study, deep sequencing approach was utilized for total blood RNAs to comparatively analyze circulating exosomal miRNAs expression in CRC patients at different cancer stages. The most strongly up-regulated exosomal miRNAs (e.g., let-7a-5p, let-7c-5p, let-7f.-5p, let-7d-3p, miR-423-5p, miR-3184-5p, and miR-584-5p) or down-regulated (e.g., miR-30a-5p, miR-99-5p, miR-150-5p, miR-26-5p, and miR-204-5p) were further investigated. These highly expressed miRNAs were predicted to target genes in cancer pathways, including CRC pathway. These target genes involve tumor-suppressor genes, oncogenes, and DNA repair genes. Many important signaling pathways such as transforming growth factors (TGFβ) were shown to directly associate with CRC. This work could support clarifying role of exosomal miRNAs in regulating expression of target genes that contribute to development of CRC progression. Exosomal miRNAs may, thus, distinguish CRC cases from healthy controls and serve as biomarkers for CRC diagnosis and prognosis.
Results
Morphological features of isolated exosomes
Exosome pellets obtained from membrane using spin column were examined firstly by TEM to prove isolation of exosomes on a morphological level of which they were shown as sphere-shaped extracellular vesicles harboring typical bilayer and their sizes were estimated to be between 100–200 nm (Fig. 1A). In addition, Zetasizer Nano (ZN) was used to detect physical properties of exosomes and size distribution of serum-derived extracellular vesicles (EVs) including exosomes with three replicates of a typical experiment, and similar findings were reached, where Z-Average was 232.1d.nm with Pdl of 0.157 indicating that most exosomes were collected with other additional EVs during extraction (Fig. 1B). These findings demonstrated that exosomes have been effectively isolated from serum, then, they were used further to detect new serum exosomal biomarkers for CRC.
Figure 1.
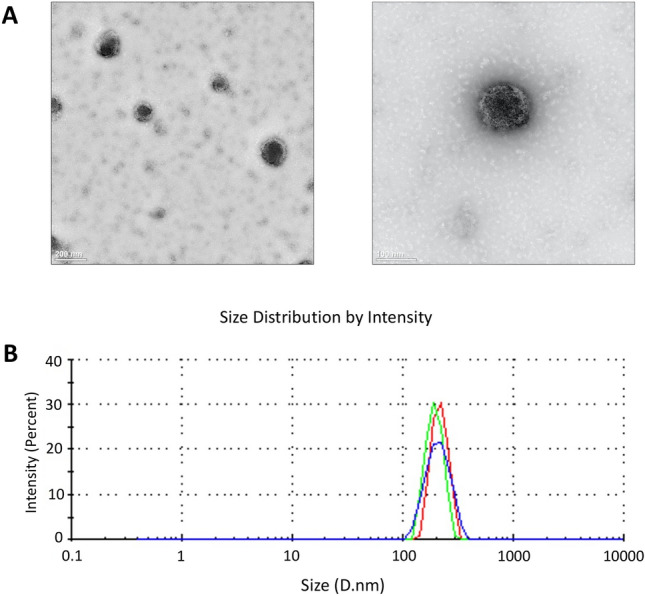
Characteristics of serum-derived exosomes from CRC patients. (A) TEM presented exosome morphology with scale bars are 200 nm (left row) and 100 nm (right row). (B) Physical properties of exosomes show size distribution of serum-derived EVs including exosomes, as assessed by Zetasizer Nano. The curve depicts means, and standard deviations (SD) from three experiment replicates that yielded similar results.
Exosomal miRNAs expression profiles
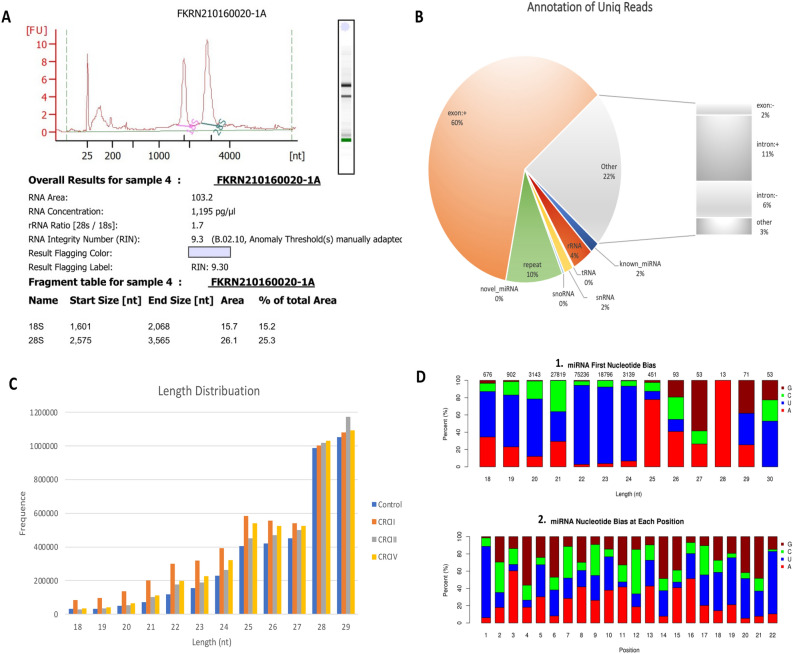
Small amounts of RNA were taken before approaching RNA sequencing using Agilent® 2100 Bioanalyzer (Agilent Technologies, United States) to test RNA integrity number (RIN), and results demonstrated high integrity of RNA (9.30) (Fig. 2A). Total RNA samples were taken from nine CRC patients at different stages (CRCII, CRCIII & CRCIV) and three healthy controls. miRNA sequencing libraries yielded 12,002,536, 12,850,941, 13,830,980 and 12,475,000 raw reads from control, CRCII, CRCIII, and CRCIV, respectively. Number of clean reads in libraries of control, CRCII, CRCIII and CRCIV were 11,798,483 (98.20%), 12,658,663 (98.48%), 13,641,029 (98.64%), and 12,316,277 (98.69%), respectively. miRNA annotation presented known miRNAs, rRNA, tRNAs, small nuclear (snRNA), small nucleolar (snoRNA), repeats, novel miRNAs, exonic RNAs, and intronic RNAs in serum-derived exosomes. Expectedly, most common reads in annotation referred to exon (60.50%) followed by others (22%), repeats (10%), rRNA (4%), known miRNAs (2%), snRNA (2%), snoRNA (0.35), tRNAs (0.20%), while the least was for novel miRNAs (0.01) (Fig. 2B & S1). Length distribution of clean reads of pre-miRNAs in control, CRCII, CRCIII, and CRCIV libraries were between 18–29 nt (longer reads were removed), while length of mature miRNA was between 21–22 nt (Fig. 2C). Previous reports indicated that seed region of conserved mature miRNAs is located at position between 2–8 bp. The results at first position demonstrated a negative bias against G nucleotide for mature miRNAs with 21–22 nt length, while bias was in favor of U nucleotide for miRNAs with 22–24 nt (Fig. 2D & S2). Within seed region (nt position 2–8), there was no bias towards or against a certain nucleotide. At nt position 22, there was a bias towards U nucleotide, while against C nucleotide (Fig. 2D).
Figure 2.
Overview of exosomal miRNAs analysis. (A) RNA integrity number (RIN) for total RNA (Agilent 2100 Bioanalyzer). (B) Pie chart showing summary of annotation of total RNA unique reads with percentages of several RNA classes detected in dataset. (C) Length distribution of exosomal miRNAs sequences in control and CRC groups (II, III and IV) library. (D) First nucleotide bias in resulted miRNAs with 18–30 nt length (1) and bias at different positions of miRNAs with 22 nt (2).
Differential expression levels of exosomal miRNAs
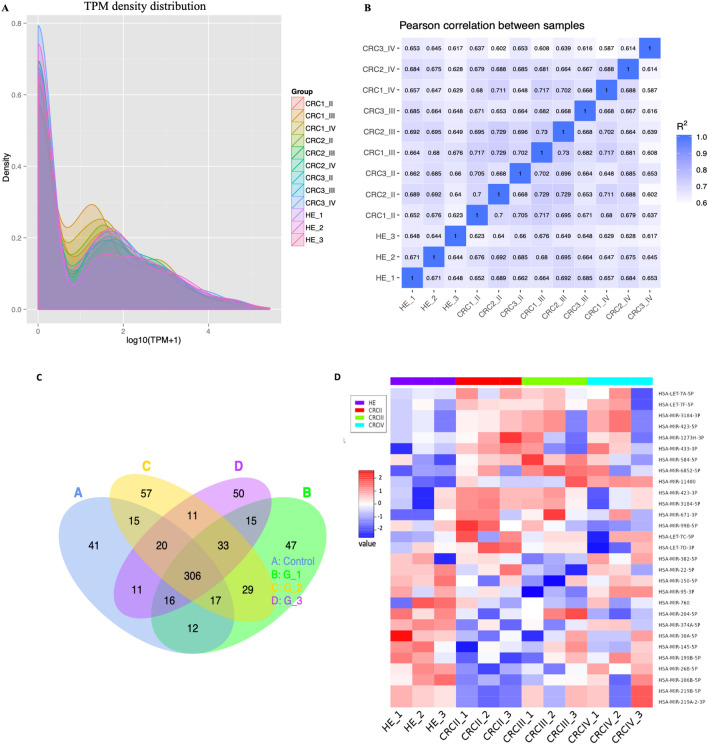
Known and unique miRNAs expression in each sample are statistically analyzed and normalized by transcript per million (TPM) (Zhou et al.25). The formula is normalized expression = (read count*1,000,000)/lib-size (miRNA read count of sample). Quality control of RNA sequencing data was shown in TPM density distribution and Pearson correlation coefficient. The patterns of miRNAs expression of each sample were presented in miRNA TPM density distribution as shown in Fig. 3A. While the RNA-Seq correlation shown in Fig. 3B presented the biological replicate for each sample. The results showed that the correlation between same samples is similar (1), but the correlation between each sample with its group, e.g., (CRC 1, 2 and 3 of group II) were (0.7), (CRC 1, 2 and 3 of group III) and (CRC 1, 2 and 3 of group IV), was 0.6.
Figure 3.
(A) The libraries of each group; control (HC), and CRC (II, III, and IV) are shown in different colors. The x-axis defines the log 10 (TPM + 1). The y-axis defines the probability value of the TPM distribution. TPM: transcripts per million. (B) Pearson's correlation coefficient of RNA-seq between each sample control and CRC (II, III, and IV). Pearson correlation coefficient square presented in shades of blue color, the ideal experiment settings is greater than 0.92, and the R2 (the square of the Pearson correlation coefficient) must be at least 0.8. (C) Venn diagram representing shared known miRNAs identified in each CRC group as well as healthy control. (D) Hierarchical clustering of 29 differentially expressed exosomal miRNAs shows colorectal cancer (CRC) patients in different stages and healthy control (HE).
To identify known miRNAs in serum exosomal miRNA libraries (control and CRCII, CRCIII and CRCIV), mapped sequences were aligned to miRNA sequences of Homo sapiens in miRBase database. Total mapped miRNAs were annotated in each group (control and CRCII, CRCIII and CRCIV) and a total of 660 known miRNAs were detected as shown in Venn diagram of Fig. 3C (S3). Of these, a number of 306 were shared among three CRC as well as control groups. Differential expression of exosomal miRNAs between control vs. groups CRCII (G1), CRCIII (G2), or CRCIV (G3) was established using statistical criteria of fold change ≥ 2.0 and P-value of ≤ 0.05. Results indicated occurrence of 29 miRNAs with significantly different expression levels. Of which, a number of 14 miRNAs were significantly up-regulated, while 15 miRNAs were significantly down-regulated (Fig. 3D). Hierarchical clustering based on 29 DEEMs showed unclear separation between CRC groups where only group of CRC II clustered clearly and separately from a healthy control. However, each group of CRC represents a different expression pattern (Fig. 3D, S4).
Predicted functions of differently expressed exosomal miRNAs
Target genes of differently expressed exosomal miRNAs (DEEMs) were predicted by using miRanda with present relationships between miRNAs and corresponding target genes. GO Enrichment Analysis is a technique of generating gene numbers 26. It begins by mapping all target gene candidates in database to Gene Ontology GO (http://www.geneontology.org/).
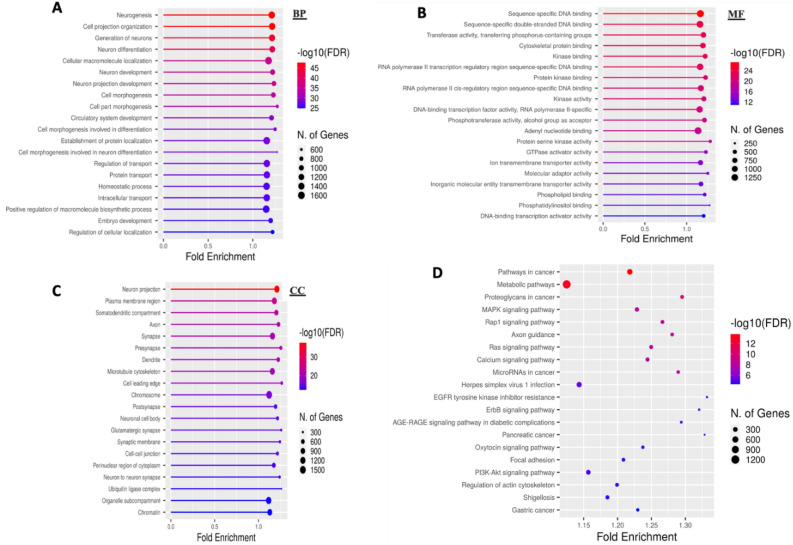
To find significant enriched GO terms in target gene candidates, Wallenius non-central hyper-geometric distribution was used in comparison with reference gene. From 30 DEEMs, around 14,782 predicted target genes were identified by miRanda database and by applying ShinyGO v0.75 which presents all top 30 in each three classes: biological process (BP), molecular function (MF) and cellular component (CC). Probable target genes in BP class, putative target genes were highly correlated to neurogenesis, cell projection organization, generation of neurons, and neuron differentiation. While MF class was largely associated with sequencing-specific DNA binding, sequencing-specific duple-stranded DNA binding, transferase activity, and transferring phosphorus-containing groups. In CC class, probable target genes were largely connected to neuron projection, plasma membrane region, and synapse (Fig. 4A–C, S5). KEGG analysis was applied to distinguish roles of 30 DEEMs based on function of their target genes. From KEGG pathways, the top 20 high pathways were selected.
Figure 4.
Histogram of target gene candidates. Three GO ontologies in next GO term are presented in x-axis, while number and percentage of target gene candidates annotated in this GO term are presented in y-axis (A–C). Biological process(BP), molecular function (MF) and cellular component (CC). (D) KEGG analysis for target genes of differently expressed miRNAs. Name of pathway is shown on y-axis, while Rich factor is shown on x-axis. Size of dots represents number of target genes, and color represents p-value.
Differentially expressed miRNA target genes were enriched in metabolic pathways, pathways in cancer, proteoglycans in cancer, miRNAs in cancer, MAPK, Rap1, RAS, and several cancers were involved, ex., gastric cancer, and pancreatic cancer (Fig. 4D, S6). It will be significant in the future to functionally confirm these differentially expressed miRNA target genes.
Identification of potential prognostic biomarkers for CRC
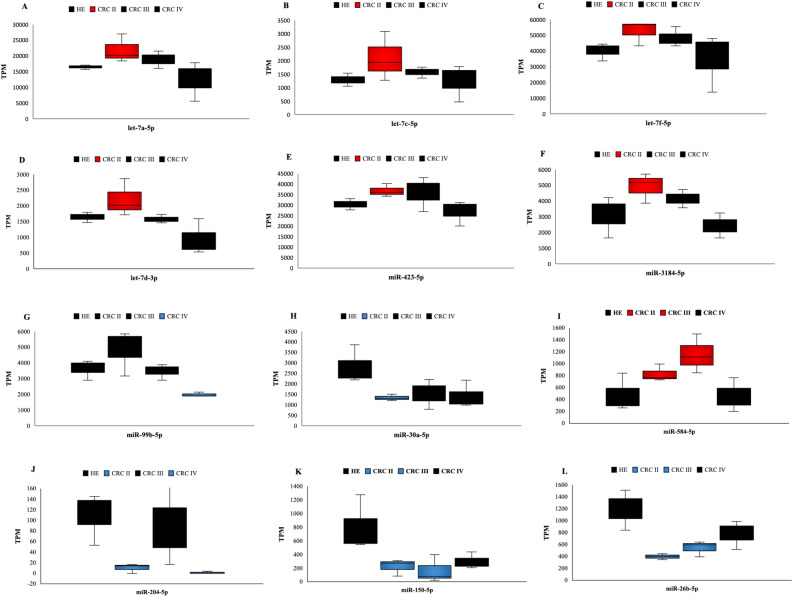
The 12 differently expressed exosomal miRNAs (DEEMs) showed high significance (P value < 0.05) (S7). Log2 FoldChange (FC) means effective size estimated and level of miRNAs expression of CRC samples in comparison with healthy control. Positive log2 (FC) indicates up-regulation of miRNAs, while the negative log2 (FC) refers to down-regulation. The six miRNAs that were up-regulated are let-7a-5p, let-7c-5p, let-7f.-5p, let-7d-3p, miR-423-5p, and miR-3184-5p, whereas those that were down-regulated (2) are miR-30a-5p, and miR-99-5p (Fig. 5A–H). Compared with control, there was a significant up-regulation of serum exosomal miRNAs with high abundance (let-7f.-5p, miR-423-5p, let-7a-5p, miR-3184, let-7c-5p, and let-7d-3p) (Fig. 5A–F). Results of let-7a-5p, let-7c-5p, let-7f.-5p, let-7d-3p, miR-423-5p, and miR-3184 were observed at a high level of miRNAs expression (P < 0.05) in CRC II, while these miRNAs were gradually down-regulated in CRC III and IV compared with healthy control (Fig. 5A–F).
Figure 5.
Top 12 regulated miRNAs in CRC compared with control. (A–F) miRNAs let-7a-5p, let-7c-5p, let-7f-5p, miR-423-5p, and miR-3184-5p, respectively, showed a high level of expression only in CRC stages II. The two lowest expressed in CRC. (G) miRNA miR-99B-5p was observed at low level of expression only in CRC stage IV compared with healthy control. (H) miRNAs miR-30a-5p was observed at low level of expression in CRC stages II. (I) miR-584-5p was up-regulated at CRC stages II and III. (J) miR-204-5p was down-regulated at CRC stages II and IV. (K, L) miR-150-5p and miR-26b-5p were down-regulated at CRC stages II and III. TPM; transcript per million. Red box: up-regulated miRNAs; blue box: down-regulated miRNAs; black box: miRNAs not differentially expressed.
Nevertheless, exosomal miR-99b-5p and miR-30a-5p were observed at a low expression level with high abundance compared with other down-regulated miRNAs. miR-99b-5p was down-regulated only in CRC IV, while miR-30a-5p was only identified in CRC II compared with healthy controls (Fig. 5G, H). Several miRNAs were shared among CRC stages II, III, and IV including miR-150-5p, miR-204-5p, miR-26b-5p, and miR-584-5p (Fig. 5I–L).
Expression level of miR-584-5p increased progressively in CRC II and III (827 to 1153) and decreased in CRC IV to (458 TPM) close to expression level of healthy control (476 TPM) (Fig. 5I). In contrast, expression level of miR-150-5p decreased gradually from healthy control until stage III (799, 221 and 164 TPM, respectively), then increased slightly at stage IV (296 TPM), which indicates that expression level of miR-150-5p was down-regulated from CRC II to III (Fig. 5K). While miR-26b-5p showed a sharp decrease in expression level of healthy control compared with CRC patients at stage II (1191–396 TPM), then gradually increased at stages III and IV (542 and 781 TPM, respectively). That indicates that miR-26b-5p was down-regulated in CRC II and III, and expression level between these stages was steady (Fig. 5L). On the other hand, expression level of miR-204-5p was different among stages, presenting a sharp decrease from healthy control to stage II (109 and 10 TPM, respectively) with negative log2 (FC) that is three times lower in stages II than in healthy control. Interestingly, in stage III, level of miR-204-5p expression was increased slightly then decreased sharply at stage IV (88 to 1 TPM) with also negative log2 (FC) that is four times lower than healthy control (Fig. 5J).
Gene target analysis of selected exosomal miRNAs
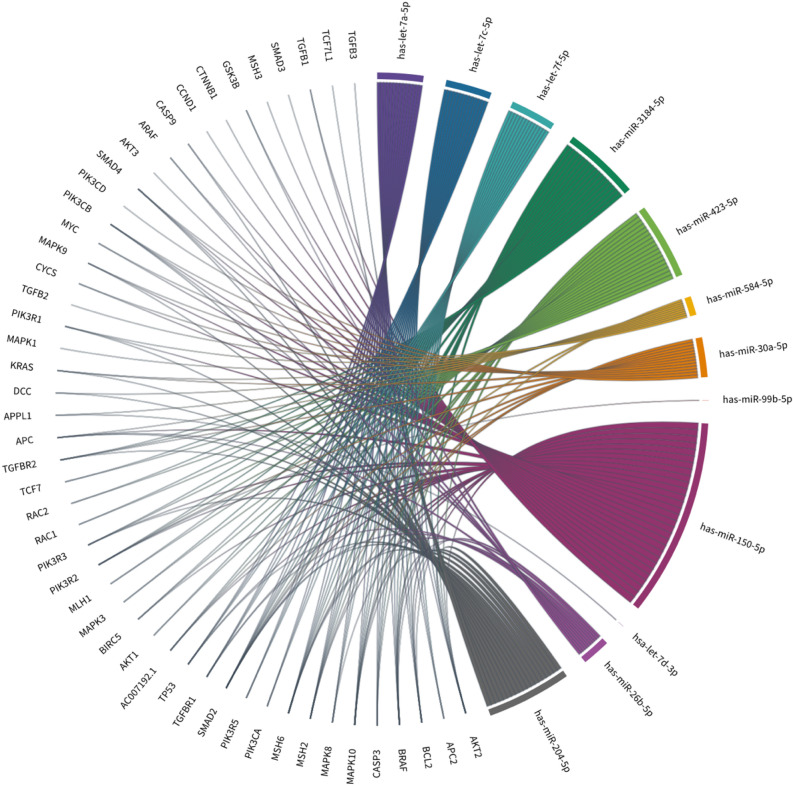
miRanda analysis was used to predict gene-targeted of 12 up/down-regulated miRNAs. To determine which of the 12 miRNAs could be related to CRC progression, target genes that were identified in miRanda were screened in colorectal cancer pathways, highlighting 50 genes previously associated with CRC. Results of Chord-diagram showed that all 12 exosomal miRNAs interacted with at least one gene in CRC (Fig. 6). miR-150-5p was detected to target a high number of CRC genes, around 29 out of 49 genes. While miR-9-5p, miR-30a-5p, and miR-584-5p were shown to interact with a total of 16, 13, 11, and 9 target genes. However, let-7a-5p, let-7c-5p, and let-7f.-5p were interacted with the same 14 CRC genes, including AKT2, APC2, BCL2, BRAF, CASP3, MAPK10, MAPK8, MSH2, MSH6, PIK3CA, PIK3R5, SMAD2, TGFBR1, and TP53. While let-7d-3p interacted with one gene, APPL1. In addition, miR-3184-5p and miR-423-5p were interacting with same 18 CRC genes including AC007192.1, AKT1, AKT2, BIRC5, BRAF, CASP3, MAPK10, MAPK3, MLH1, MSH2, MSH6, PIK3R2, PIK3R3, RAC1, RAC2, SMAD2, TCF7, TGFBR2. However, miR-99b-5p target only one gene, PIK3R2.
Figure 6.
Chord-diagram of 12 differentially expressed exosomal miRNAs targeting several genes expressed in CRC. Around 49 target genes interacted with the 12 differently expressed exosomal miRNAs. miRNAs are defined in a different color, while each gene target is defined in gray color.
Some of the known genes that altered in CRC were highly targeted by most of the 12 exosomal miRNAs. For example, SMAD2/3 and SMAD4 were highlighted and disrupted in sporadic CRC27 and our result identified 11 exosomal miRNAs were interacted with SMAD2, one exosomal miRNAs with SMAD3 and four exosomal miRNAs with SMAD4. Moreover, TGFβ signaling pathways including TGFβ1, TGFβ2, TGFβ3, TGFβR1, TGFβR2 were implicated among 10 exosomal miRNAs except let-7d-3p and miR-99b-5p. Furthermore, APC and TP53 are tumor suppressor genes that are inactivated in CRC28,29 and were identified to interact with seven and five exosomal miRNAs respectively. Overall, these miRNAs had the largest number of interactions with commonly changed genes in CRC, indicating that they may play a role in CRC development or progression through the regulation of key genes.
Pathway analysis for selected miRNAs
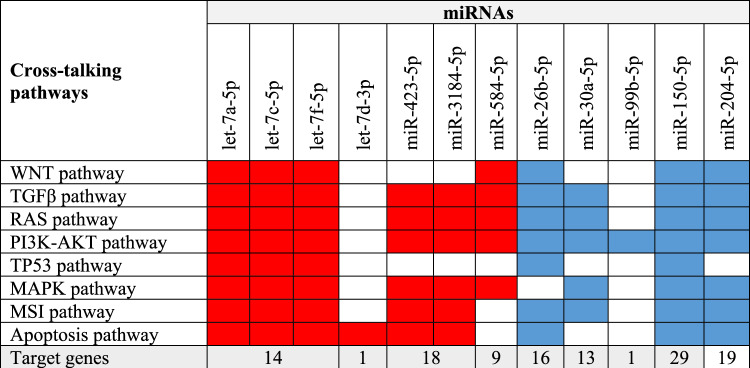
Pathways of 49 genes that were targeted by miRNAs were examined to determine involvement of the 12 DEEMs in CRC formation. Using KEGG database, enrichment pathway analysis indicated involvement of more than 100 pathways (P value > 0.05, Supplementary S3). The results showed that miRNAs play a critical role in KEGG pathways. The top highly DEEMs were identified as targeting genes related to "microRNAs in cancer” with P value = 4.87E−23 followed by “Pathways in Cancer" with a P value = 9.30E−19, MAPK signaling pathway with P value = 1.24E−14. RAS signaling pathway stood at position nine with P value = 3.06E−11, and PI3K-Akt signaling pathway stood at position 13 with P value = 1.59E−10. Also, several cancer pathways were involved in the top 30 pathways including breast cancer with P value = 5.70E−10, glioma with P value = 6.47E−10, gastric cancer with P value = 3.71E−08 and pancreatic cancer with P value = 6.84E−08. Nevertheless, colorectal cancer pathway represents a significant (p-value = 1.28E−06) but not included in the top 20 enriched KEGG pathways. Furthermore, additional signaling pathways were identified as crosstalk with CRC pathways such as WNT signaling pathway (P value = 7.39E−08), and TGFβ signaling pathway (P value = 9.96E−06). This is emphasizing that the 12 DEEMs play a significant role in CRC. These findings provide a strong connection between the 12 DEEMs and various changes that are frequently observed in CRC. Next step was to investigate where each miRNA interferes in often-changed pathways in CRC (e.g., WNT, TGFβ, RAS, PI3K-AKT, TP53, MAPK, and MSI signaling pathways)28,30–34. miRNA-target genes that are known to be associated with CRC were combined with genes engaged in each signaling pathway. According to these findings, majority of miRNAs were shown to be involved in at least one of these pathways. Intriguingly, various miRNAs appear to interfere in several pathways, and all 12 miRNAs appeared to target PI3K-AKT pathway. Moreover, four miRNAs (e.g., let-7a-5p, let-7c-5p, let-7f.-5p, and miR-150-5p) were shown to interact with the eight mentioned pathways. Interestingly, let-7d-3p was shown to interact with apoptosis pathway, only. Moreover, miR-204-5p presented targets in all of these pathways except for TP53 pathway. Furthermore, all up-regulated miRNAs were shown to communicate with MAPK pathway. However, TP53 pathway has lower interaction with these miRNAs. Interestingly, one miRNA (e.g., miR-99b-5p) targets PI3K-AKT pathway, only. As a result, it is possible that the changed expression of these miRNAs may participate in altering pathways in CRC through targeting several genes of these pathways (Fig. 7).
Figure 7.
Cross-talking KEGG pathways with Colorectal Cancer pathway showing association between frequently changed pathways in CRC and the nine differently expressed miRNAs. Through KEGG database, enrichment analysis was performed on the nine miRNA-target genes. The genes interfering in each signaling pathway interacting with the miRNA-target genes were found to be associated with CRC. Number of target genes for each miRNA presented in CRC pathways is shown. miRNAs' connection with each KEGG pathway is presented in red for up-regulation and blue for down-regulation.
Validation of miRNA Expression via RT-qPCR
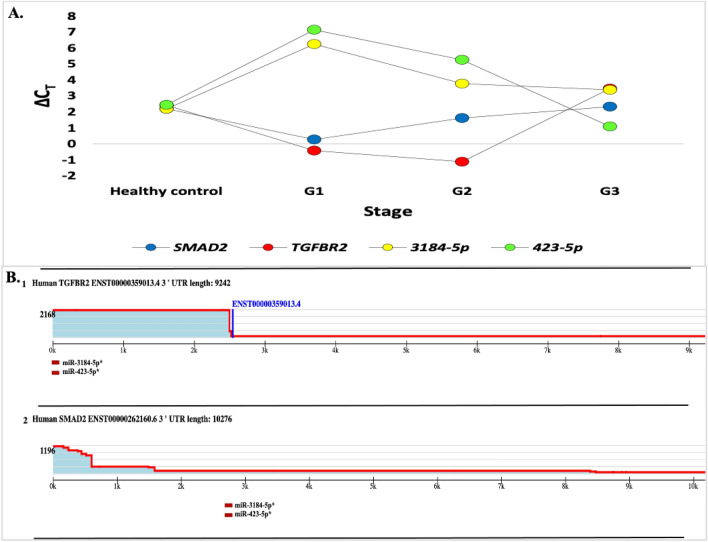
We analyzed RT-qPCR data derived from a study conducted to validate differentially expressed miRNAs in patients vs. healthy controls of the deep-sequencing data, we performed qRT-PCR on the identified. The validation was conducted for two miRNAs namely miR-3184-5p and miR-423-5p using the same RNA samples of the CRC patients except for one patient from stage IV (the result was undetermined). According to the present findings, these two miRNA were upregulated and significantly increased in both stage II (G1) and stage III (G2) when compared to the healthy control. In contrast, their target genes SMAD2 and TGFER2 were downregulated in stage II (G1) and stage III (G2) compared to healthy control. However, the expression of these genes gradually increased, and inversely with miR-3184-5p and miR-423-5p, which were decreased in stage VI (G3) (Fig. 8A).
Figure 8.
(A) Verification results of upregulated miRNA. The Fold Change bar graph shows expression of SMAD2, TGFBR2, 3184-5p, and 423-5p genes. (B) TargetScanHuman figure for the 3′ UTR of the TGFβ R2, and SMAD 2. The 3′-UTR profile on the top presents the relative expression of 3′-UTR isoforms and the total number of 3P-seq reads. The profile presents Gencode annotation (red line), the conserved sites of the predicted target of miRNAs (boxes). The predicted site of miRNA up-regulation is miR-423-5p and miR-3184-5p(red boxes).
It was previously reported that TGFβ signaling pathways are involved in multiple aspects of CRC progression; we hypothesized that the low expression of several genes in this pathway, such as TGFβRII, and SMAD2 in CRC, could be due to the up-regulation of miRNAs that target this pathway. The map of each gene in the TGFβ signaling pathway and a list of expressed miRNAs expected to target these genes based on TargetScan prediction are displayed in (Fig. 8B 1&2). The TargetScan database35 presented the TGFβRII and SMAD3 genes were targeted by both miR-423-5p and miR-3184-5p with one poorly conserved site.
Discussion
CRC initiation and progression are intimately linked to genetics and several environmental variables36. Because patients with advanced colorectal cancer (CRC) typically have a bad prognosis, detecting CRC early becomes is extremely important36,37. Development of useful biomarkers is critical to improve accuracy of CRC diagnosis and treatment. Researchers have discovered that miRNA plays a crucial role in the pathogenesis and progression of CRC38. Use of miRNAs properties that are recognized at early stages in blood circulation system, or early cancer detection, yielded highly encouraging results. However, based on differences in miRNA expression amongst populations, environments, nutrition, etc., findings of discovered miRNA expression may change39. Exosomal miRNA expression profiles have been reported in several studies to control various target genes that affect recipient cell function. Thus, exosomal miRNAs with specific properties are prospective biomarkers that can be employed as a diagnostic and prognostic tool for CRC or could be used as potential targets for therapeutic intervention of CRC metastasis.
In the present work, the importance of serum exosomal miRNAs in CRC patients compared with a healthy control group was examined using miRNA sequencing datasets. Detecting miRNAs in serum samples was discussed in this study to effectively discriminate malignancy from healthy controls and delivering panels of miRNAs as predictive biomarkers for improved patient classification, specifically for CRC stages II, III and IV. Towards this, deep sequencing was utilized to identify miRNA expression levels in serum samples at different stages of CRC. This method provides potential benefits in universal miRNA expression research, but it also introduces additional data analysis issues, since quantity of data obtained after deep sequencing involves millions of reads that must be mapped to genome and normalized36. The majority of miRNA expression alterations in CRC appears to begin during normal mucosa-to-cancer development and are sustained throughout metastatic transition40, which is consistent with our findings.
The sequencing data and bioinformatics analysis in our study provided a strong finding of mature exosomal miRNAs. A number of 660 mature miRNAs were found at different stages of CRC for patients that were successfully diagnosed. From the 29 differentially expressed miRNAs found in CRC patients compared with healthy controls, 12 regulated miRNAs were selected depending on their high abundance levels. The highly up-regulated miRNAs (e.g., let-7a-5p, let-7c-5p, let-7f.-5p, let-7d-3p, miR-423-5p, miR-3184-5p, and miR-584-5p) and down-regulated miRNAs (e.g., miR-30a-5p, miR-99-5p, miR-150-5p, miR-26-5p, and miR-204-5p) were analyzed further (Fig. 3A–D). We predicted that the 12 changed miRNAs may have influenced several critical target genes in CRC that lead to tumor occurrence and growth. Most of diagnostic exosomal miRNAs (e.g., let-7a-5p, let-7c-5p, let-7f.-5p, let-7d-3p, miR-423-5p, miR-584-5p, miR-30a-5p, miR-99-5p, miR-150-5p, miR-26-5p and miR-204-5p) have been discovered to be circulating in blood serum.
In addition, it is hypothesized that exosomes have a particular way for releasing nucleic acids into extracellular fluid and include miRNA species that have been carefully packed to migrate to other cells for knocking out cognate target genes41–43. Accordingly, gene target prediction software was used to suggest probable target genes based on sequence pairing. Predicted target genes of these highly expressed exosomal miRNAs strongly relate to cancer pathways that involve CRC pathway. The target genes in CRC pathway include tumor suppressor genes, oncogenes, DNA repair genes, and multiple critical signaling pathways such as TGFβ that are directly linked to CRC function in which this study focuses. KEGG analysis showed that differentially expressed miRNAs are included in cancer signaling pathways and gene ontology (GO) analysis specified the top expressed miRNAs involved in molecular functions, cellular components, and biological processes categories.
Transforming growth factor β (TGFβ) signaling pathway has been identified to promote growth inhibitory of tumorigenesis. However, dysregulation of TGFβ signaling have been reported in several studies to contribute to cancer proliferation, metastasis, and angiogenesis (Feng and Derynck21; Massagué et al.20). miRNAs have been shown to influence formation and activation of TGFβ signaling pathway, and miRNAs/TGF interaction was also shown to play a significant role in development of several malignancies, including lung, breast, pancreatic, gastric and CRC (Soleimani et al.22,23). Oncogenic miRNAs have been found in several cancer types to enhance proliferation, invasion, or resistance to chemotherapeutic treatments by targeting TGFβ receptors, particularly TGFβRII (Suzuki44).
By focusing on target gene sequences, let-7 family (let-7a-5p, let-7c-5p, and let-7f.-5p) has a highly similar sequence and shares the same seeds region (3’GAUGGAG’5), leading to targeting identical genes. Members of let-7 family (let-7a-5p, let-7c-5p and let-7f.-5p) share identical seed sequence, which represents highly conserved region that is used to recognize target mRNAs sequences45. This indicates that function and targets of let-7 family may be either similar or different46. Results of the present work showed that let-7 family members (let-7a, let-7c, let-7f) shared same target genes in CRC pathways. Let-7 family is considered as an essential tumor suppressor miRNA that regulates several oncogenes46. The study presented that through low level of let-7, several signaling pathways are enriched such as cell cycle and cellular proliferation, which promote cancer progression47. Down-regulation of let-7 negatively controls MYC and RAS expressions that are recognized to be important regulators for CRC progression47,48. Through direct binding, let-7 is able to inhibit RAS expression and hence control MAPK and PI3K/AKT pathways49. Kawata and colleagues determined that serum exosomal levels of let-7a were higher in primary CRC patients than in healthy controls, while much lower following tumor resection. In addition, authors demonstrated that colon cancer cell lines produced these miRNAs at considerably greater levels than normal colon cell lines50. Furthermore, a low level of let-7c expression in primary cancer tissues of CRC relates to metastases, progressed TNM stages, and poor prognosis of patients. Let-7c acts as a tumor suppressor for CRC development and metastasis by disrupting mRNAs of MMP11 and PBX351. While let-7f has been confirmed to be down-regulated in both plasma and stool samples of patients with early stages of CRC (I and II) compared with healthy control52. Co-expression of both let-7d-3p and let-7d-5p was found to contribute to colon cancer by exclusively inhibiting KRAS by let-7d-3p, and promoting IGF1R and THBS1 expression by let-7d-5p53. The study by Ohshima and colleagues demonstrated that cancer cell lines that produce exosomes load a high amount of let-7 family. It is proposed that these processes occur to release tumor suppressor miRNAs and support ongoing tumorigenesis and metastasis54,55. Referring to our results, where let-7 is a tumor suppressor miRNA, its abundance in serum primary CRC stage seems to be a predictor of enhanced carcinogenesis and metastatic action in malignant tissue, offering a novel perspective as to how these biomarkers are regulated (Fig. 5A–D).
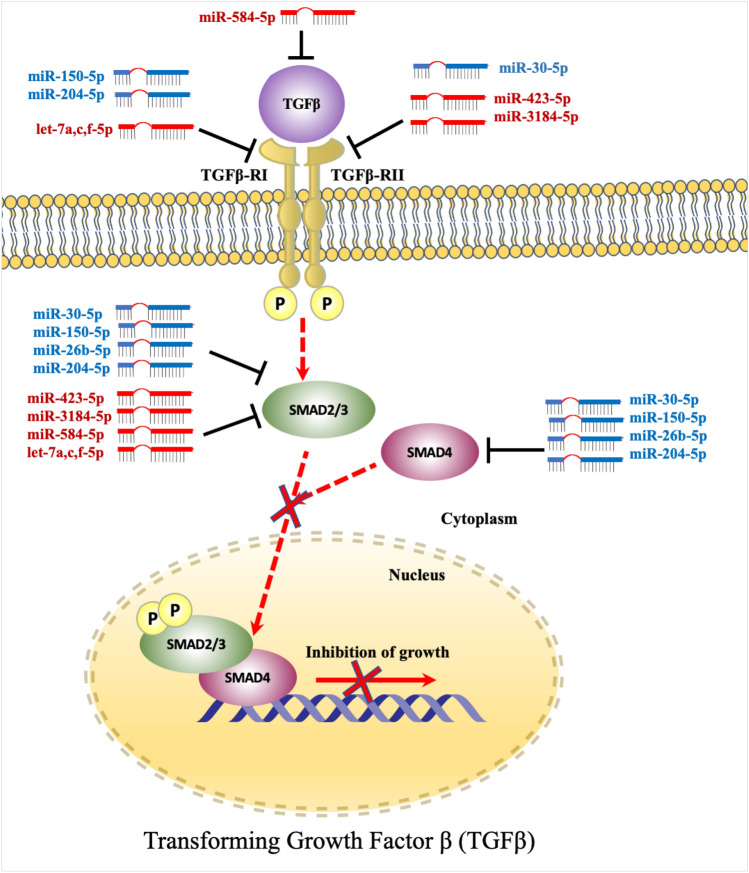
By bioinformatics analysis, we identified that TGFβ signaling pathways including TGFβRI and SMAD2 are targeted by let-7 family (Fig. 9). However, another study demonstrated that overexpression of let-7a interacts with TGF-β1 and SMAD4 and affects cervical cancer cells56. In addition, TGFβRI was regulated by let-7, which could play a significant role in TGFβ signaling activity at tumorigenesis stages57. However, no other gene targets for these miRNAs were detected. The results of miR-423-5p and miR-3184-5p indicate that they are significantly up-regulated in serum exosomal miRNAs at CRC II compared with control. However, these two miRNAs were down-regulated at CRC stage III and IV. miR-423-5p targets CRC pathways, particularly, tumor suppressor genes SMAD2/3 and TGFβR2. It has been identified that miR-423-5p and miR-3184-5p share same target genes because they have the same seed sequence (3’CGGGGAG’5)58. Morover, miR-423-5p and miR-3184-5p were significantly reported by several researches as oncogene and are involved in carcinogenesis, tumor development, therapeutic sensitivity, and prognosis in different types of cancer59–63. In glioblastomas, miR-423-5p acts as tumor promoter, leading to proliferation, invasion, and angiogenesis of cancer60. Plasma level of miR-423-5p has previously been identified as a possible biomarker for CRC occurrence60. While miR-3184-5p has been linked to formation of malignancies in a variety of human cancers. High expression level of miR-3184-5p was identified in hepatocellular carcinoma (HC) cell lines, according to miRNA array data64. Furthermore, exosomal miRNA-3184-5p was shown to be able to distinguish non-small-cell lung cancer (NSCLC) at early stages from normal controls65. miR-3184-5p was also shown to be highly elevated in breast cancer cells and operates as an oncogene in obesity-related breast cancer due to its influence on cell proliferation and invasion61. By predicting target genes for these miRNAs, it was demonstrated that certain target genes play a significant role in both activation and inhibition of CRC. Furthermore, up-regulation of miRNAs acts as oncogenic miRNA targeting certain tumor suppressor genes related to CRC initiation and development22. More specifically, TGFβR2, SMAD2, and SMAD3 genes are key components of TGFβ that regulate growth inhibitory, and abnormality of these genes contributes to tumorigenesis44. Inactivation of these genes results in dysregulation of TGFβ signaling, which leads to loss of growth inhibitory effect21. Our results suggest that miR-3184-5p and miR-423-5p may accumulate in these genes and disrupt transcriptional process of TGFβ signaling pathway at this stage (Fig. 5E, F). Also, up-regulation of exosomal miR-423-5p targeting GREM2 among TGFβ pathway enhances therapeutic resistance as well as contributes to malignant progression of prostate cancer (PC)66.
Figure 9.
TGFβ signaling pathway. TGFβRI, and TGFβRII are phosphorylate SMAD2 and SMAD3 after binding to TGFβ ligands. Activated SMAD2 and SMAD3 subsequently form a complex with SMAD4. SMAD complex is a nucleoprotein that controls cell proliferation, apoptosis, metastasis, and angiogenesis, as well as modulating target gene expression and tumor formation. MiRNAs target TGFβ, TGFβRI, TGFβRII, SMAD2/3 and SMAD4 leading to disrupt inhibition of growth in TGFβ signaling pathway. Overexpressed miRNAs (red color) and downexpressed miRNAs (blue color). Direct regulation is indicated by red dashed arrows and lines.
Moreover, miR-99b-5p and miR-30a were down-regulated in CRC in the present work and contributed to different cancer progression stages67–72. miR-99b-5p was down-regulated in CRC stage IV comparing to healthy control and was identified in our data to only target PIK3R2 gene in CRC pathway. The study by Li and colleagues showed that miR-99b-5p was overexpressed in primary tumors of patients with CRC and that suppressing this miRNA promotes cell migration and overexpression of mTOR of CRC cell lines. Also, authors suggested that miR-99b-5p is collaborated with more prolonged overall survival of patients and acts as a tumor suppressor in metastatic CRC by targeting mTOR. This indicates that miR-99b-5p may interact with mTOR and function as a predictive factor as well as a therapeutic target for anti-metastatic CRC patients72. Moreover, miR-99b-5p is associated with numerous malignancies, including colon cancer69. In gastric cancer, miR-99b-5p suppressed proliferation via negatively modulating insulin-like growth factor 1 receptor (IGF-1R) and stimulating AKT pathway73. It was identified that exosomal miR-99b-5p has been increased after tumor surgery, representing that exosomal miR-99b-5p expression level negatively influences tumor survival. As a result, miR-99b-5p (Fig. 5G) is identified as a possible clinical diagnostic biomarker capable of reliably and promptly distinguishing CRC patient69. It was identified that expression of TGF-β is affected by specific miR-99 family members such as miR-99a-5p, but not miR-99b-5p, and target SMAD2 and SMAD4 genes in acute megakaryoblastic leukemia (AMKL)67,74.
According to previous research, miR-30a expression is related to a variety of cancer types, such as thyroid, breast, and gastric cancers, as well as CRC68,70,75,76. miR-30a acts as a tumor suppressor in majority of these malignancies by modulating conserved target genes. miR-30a targets insulin receptor substrate (IRS2), phosphoinositide 3-kinase catalytic subunit delta (PIK3CD), and integrin β3 in CRC to inhibit proliferation, migration, and invasion24,71,77. Interestingly, low expression of miR-30a was in all stages but in stage II was more significant which indicates that this miRNA has a significant impact in this stage rather than other stages. It was demonstrated that miR-30a overexpression decreased migration and invasion and controlled E-cadherin and vascular endothelial growth factor (VEGF) expression by suppressing TM4SF1 gene. The findings indicate that miR-30a is related to progression stage and lymph node metastasis of CRC and that it might be a new promising source for diagnosing and therapy of CRC. Interestingly, our study identified that down-expression of miR-30a was represented in CRC stages III than in stages III and IV (Fig. 5H), which demonstrates that miR-30a plays a critical role in early stages of CRC development than in the later stages70. These findings suggest that miR-30a is negatively related to carcinogenesis and lymph node metastasis and may function as a tumor suppressor in CRC. Additionally, in CRC patients with severe clinical characteristics, low serum miR-30a-5p expression was more common. Moreover, CRC patients with high serum miR-30a-5p expression had substantially longer overall survival rate than those with low expression. Finally, analyses demonstrated that serum miR-30a-5p expression was a significant predictor of survival in CRC patients68.
Moreover, miR-30a-5p targets TGFβ signaling pathway, which could be disrupted by multiple processes related to tumor invasion. As described in anaplastic thyroid cancer (ATC), down-regulated miR-30a-5p correlates with inhibition of TGFβ signaling. This miRNA was associated with induced mesenchymal-epithelial transition (MET) and ATC cells invasion78. Expression of SMAD2 and TGFBR1 genes, which were elevated in most primary anaplastic thyroid cancer (ATC), was regulated by miR-30 in ATC-derived cells confirming involvement of TGF signaling in modifying MET/epithelial–mesenchymal transition (EMT)79. Based on these findings, miR-30a-5p is proposed to have a potential role as a novel biomarker for CRC diagnosis and/or prognosis. On the other hand, TGFβ has been reported as a tumor suppressor in the early stages but a tumor promoter in the later stages80. Some overexpressed miRNAs suppress anti-proliferative signals by targeting this pathway. In contrast, the TGFβ pathway, in collaboration with BRAF, causes EMT, migration, and invasion in later stages80. Our results presented that TGFBR2 has also interfered with miR-30a-5p, which could be a novel biomarker that can be studied and investigated further.
Some functions of these highly expressed miRNAs have been studied, where miR-584-5p targets some tumor suppressor genes including STAT1, PTEN, and cyclin D181. The miR-584 was found in the SW620 colorectal cancer cell line to interfere with binding of hnRNP A1 and CDK6. The study suggested that overexpression of miR-584 acts as a tumor suppressor, which can disrupt binding between hnRNP A1 and CDK6 mRNA, leading to apoptosis. This connection might be replicated for different miRs, which could lead to creation of new cancer treatment techniques82. Also, miR-584 was shown to be up-regulated in CRC and greater levels of this miRNA were only previously seen in colon adenoma, not carcinoma83. Another research revealed that miR-584 was overexpressed in the early stages of gastric cancer (GC) and then down-regulated in later stages. Their findings showed that high levels of miR-584 expression may be essential for primary tumor formation and gastric carcinogenesis, then, this miRNA undergoes low levels for tumor progression and invasiveness in the metastasis stage81. Furthermore, the latter study suggested that miR-584 has the potential to be a diagnostic biomarker for early identification of GC as well as a predictive factor for predicting patient clinical outcomes. Another study indicated that in the presence of FF/CAP18 treatment, which inhibits proliferation of colon cancer cell lines, exosomes release miR-584-5p at a high level84. Moreover, the latter study suggested that miR-584-5p may act as a tumor suppressor and can be a potential target to inhibit tumor progression. The study of Konishi and his team suggested that overexpression of miR-584-5p suppress the interaction between heterogeneous ribonucleoprotein A1 (hnRNP A1) and CDK6 mRNA, which induce apoptosis of CRC cell line in malignant transformation stage82. All the latter findings support our prediction that miR-584-5p is up-regulated in CRC stage (II and III), then, down-regulated at stage IV compared with healthy control, which indicated that miR-584-5p is highly expressed during carcinogenesis and, then, decreased to a lower level of expression during invasion and metastasis (Fig. 5I). Moreover, miR-584 targets SMAD2, and TGFβR2 genes leading to loss of growth inhibitory and induction of proliferation and angiogenesis of several malignancies, including CRC20,21,23.
MiR-150-5p was proven to be interfering in several cancers, including thyroid cancer cells (TCC), lung squamous cell carcinoma cells (LSCC), prostate cancer cells (PCC), lung, breast, pancreatic and esophageal cancer and CRC (Fig. 6)69,85–91. The miR-150-5p targets MAP3K12 gene that inhibits the proliferation and invasion of PCC and LSCC85,88. Moreover, following surgery, the expression of exosomal miR-150-5p was elevated, showing that tumor retention may have a detrimental impact on miR-150-5p expression69. Exosomal miR-150-5p has been reported to inhibit the proliferation of CRC cells, assisting as a potential novel biomarker for CRC diagnosis69. Another study found that a low expression level of miR-150 in colorectal tissue was associated with poor survival and responsiveness to chemotherapy, demonstrating that miR-150 expression levels were correlated to CRC. It is proposed that miR-150 be examined as a possible biomarker for CRC prognosis and treatment outcome90. On the other hand, in breast cancer cells, increased miR-150 expression has been shown to boost clonogenicity and cell proliferation, while reducing apoptosis92. Various studies have also shown that miR-150 functions as a tumor suppressor and a promising prospective biomarker by altering expression of several genes associated with CRC progression, including c-Myb, c-FLIP, and VEGFA90,93,94. A previous study was done to clarify the connection between miR-150 and β-catenin in CRC. It revealed that overexpression of miR-150 inhibited the viability, proliferation, and colony formation of SW480 and HT-29 cells by targeting the Wnt/β-catenin pathway95. The in vivo study presented that miR-150 inhibits CRC development through regulating β-catenin. Levels of β-catenin, c-MYC, and CCND1 were negatively correlated with miR-150 overexpression suggesting that miR-150 might decrease CRC growth and operate as a tumor suppressor by blocking β-catenin pathway95. Interestingly, a study of the TGFβ/SMAD signaling pathway in hepatic stellate cells (HSCs) revealed that inhibiting SMAD4 by the Smad4shRNA vector enormously improved miR-150 expression levels96. Overall, these findings support our result that found that down-regulation of miR-150 affects other downstream signaling pathways, especially TGFβ pathways (Fig. 5K). However, this miRNA was significantly down-regulated in stages II and III, not IV, suggesting that the down-regulation of miR-150 negatively regulates CRC progression and development.
Furthermore, miR-26b expression was found in various cancers, including neck squamous cell carcinoma, osteosarcoma, cell lung carcinoma, gastric cancer, ovarian cancer cell and CRC, which can be used as prognostic predictor97–102. The previous studies proved that miR-26b associates with CRC by interacting with different set of target genes82,102–106. It was recognized that miR-26b expression was elevated in serum and tissues during the progression of inflammatory bowel disease-related tumorigenesis107. Nevertheless, investigations have found that low level of miR-26b expression was detected in CRC tissue only, not in normal tissue, indicating that miR-26b reduction may contribute to CRC tumorigenesis102. Surprisingly in CRC cell lines, miR-26b correlates with increased invasion and metastasis and contributes to lymphatic metastasis. This result appears to be paradoxical although miR-26b expression is more expressed in normal cells than in malignant tissues. The study also determined that PTEN and WNT5A, as tumor suppressors, were inhibited by miR-26b, however, expression of miR-26b is a predictor of poor prognosis following carcinogenesis, which might be an indication of miRNA tissue-specificity103. The present study also suggests that instead of comparing the expression levels of miR-26b in CRC cells and normal colorectal mucosal epithelial cells, identifying the effect of various miR-26b expression levels on CRC cells could be more efficient. This miRNA may have a variety of roles depending on types of cells, and normal colorectal mucosal epithelial cells and CRC cells are the examples. Therefore, determining the function of miR-26b in CRC requires more than comparing the expression of miR-26b in CRC cells versus normal cells. Alternatively, tracking of miR-26b expression should simultaneously be in the same CRC cell lines103. Moreover, miR-26b was found to induce a stem cell-like phenotype in a fraction of CRC cells, which indicates that miR-26b acts as a regulator of CRC invasion and metastasis103. Interestingly, overexpression of miR-26b-5p plays a significant impact in chronic lymphocytic leukemia (CLL) development. It was determined that overexpression of miR-26b-5p disrupts the TGFβ/SMAD signaling pathway in CLL cells by targeting SMAD4 gene, leading to a reduction expression of p21-Cip1 kinase inhibitor and increased expression of c-MYC. This study identified miR-26b-5p as a novel molecular pathway relating CLL development to TGFβ regulation and a novel treatment method for CLL108. We can predict that miR-26b-5p could play a significant and key role in CRC. This includes low expression for progression in stages II and III, while for metastasis and invasion in stage IV. The current study presented that overexpression of miR-26b-5p contributes to preventing chondrocyte senescence and osteoarthritis (OA) by targeting asporin mRNA. Furthermore, asporin promotes chondrocyte senescence and exacerbates cartilage degradation by blocking the TGFβ1/SMAD2 pathway, and hence plays a role in OA. The miR-26b-5p/asporin/SMAD2 axis represents a prognostic biomarker and a possible therapeutic target in OA, according to these findings98. Our study demonstrated that miR-26b-5p was significantly decreased in stages II and III compered to control but still this miRNA has low expression in stage IV and no data for miR-26b-5p and TGF-β. In addition, our study supported prior research that found that miR-26b has biological impact on CRC development (Fig. 5L).
In addition, miR-204 was shown in the present study to be significantly down-regulated in CRC tissues compared with surrounding normal tissues. Previous research has found that miR-204 is often down-regulated in many different malignancies, indicating that miR-204 plays a common function in human carcinogenesis109–112. Moreover, previous research has found that miR-204 suppresses a variety of cancerous tumors via various signaling pathways, indicating that it may have distinct impacts on tumorigenesis. In the cases of pancreatic cancer and renal clear cell carcinoma, miR-204 might inhibit tumor growth111,113. Also, endometrial cancer, liver cancer, glioma, colon cancer, gastric cancer, intrahepatic cholangiocarcinoma, non-small cell lung cancer (NSCLC) and head and neck malignancies are all identified to be repressed by miR-204109–119. Furthermore, miR-204 plays a significant key role in the sensitivity of CRC to 5-Fu chemotherapeutic drugs by targeting high mobility group protein A2 (HMGA2). In a previous study, overexpression of miR-204 was detected to suppress proliferation of SW480 cell lines and increase response sensitivity with a 5-Fu dose, indicating that miR-204 might be used as a treatment for 5-Fu-resistant CRC120. Another study demonstrated that low expression of miR-204 correlates to colon cancer pathogenesis, CRC progression, and drug resistance through overexpression of angiopoietin-like protein 2 (ANGPTL2). However, up-regulation of miR-204 is accompanied with down-regulation of ANGPTL2 that led to induced apoptosis and inhibited cell proliferation121. Furthermore, this study determined that lower level of miR-204 expression correlates with higher TNM stages and elevated ANGPTL2 expression contributing to poor prognosis in CRC121. According to a previous study, overexpression of miR-204 in CRC cells significantly suppressed cell growth, lowered the ability of tumor progression of CRC cells in animals, impacted the intestinal cancer cell migration and invasion, and increased the responsiveness of cells to targeted therapies109. Prior studies have shown that miR-204 is also linked to CRC via interfacing with a broad set of target genes such as HMGA2, CXCL8, RAB22A, TPT1, PCAT6 and CREB1120,122–124. Other studies presented the role of miR-204-5p in interfering with TGFβ signaling pathways through regulating Six1 and SMAD4 genes in airway smooth muscle cells of asthma and human posterior capsule opacification, respectively125,126. These findings indicate that miR-204-5p expression was dramatically decreased in tumor tissues and cells of colorectal carcinoma patients in several studies, suggesting that miR-204-5p may function as a tumor suppressor in CRC. Our results, interestingly, align with these results, which proved that miR-204 -5p is valuable in malignancies, particularly CRC (Fig. 5J). Thus, it is an opportunity to study the role of miR-204-5p with TGFβ in the development or suppression of CRC.
The results also suggest that the highly expressed exosomal miRNAs, e.g., let-7a-5p, let-7c-5p, let-7f.-5p, miR-423-5p and miR-3184-5p, are connected to cancer and are potentially important in CRC by targeting TGFβ, which is the important signaling in the CRC pathway (Fig. 8). However, lower expressed exosomal miRNAs, e.g., miR-26b-5p, miR-150-5p, miR-204-5p, and miR-30a-5p, may contribute to the progression of CRC by missed targeting oncogenes. Only let-7d-3p and miR-99b-5p were not involved in this pathway. While an additional study is needed to validate the target genes and investigate the downstream implications. Exosomal miRNAs target mRNA species that are dysregulated in CRC patients, and functional pathway analysis revealed extremely significant connections with particular CRC molecular pathways. This builds confidence that miRNA biomarkers of CRC identified during our approach likely to be fully justified.
Significant correlation represents more than half of the statistically meaningful associations, indicating that miRNA and target gene expression are usually moving in opposite directions. Although an increase in miRNA levels should decrease target levels (or vice versa), it is widely known that miRNAs are dynamic gene expression regulators that inhibit protein translation rather than mRNA degradation127–133. The miRNAs' biological significance appears by area of binding to the conserved seed region, whether at 3p or 5p end. This indicates that miRNAs with -3p or -5p have a significant role in biological processes and can contribute to these processes in several ways and expression patterns53,134–137.
It is challenging to determine the relative relevance of high versus low miRNA expression since the absence or abundance of miRNAs might reflect equally relevant biological control signals. Over-expression of miRNAs might indicate that these miRNAs play vital roles as regulators of downstream targets of biological pathways. Furthermore, these miRNAs likely interfere with signaling pathways known to be aberrant in CRC, including TGFβ, TP53, KRAS, BRAF, MAPK, and BAX. Several investigations have also shown that miRNAs play a critical role in modulating these pathways22,30,57,138–141, which confirm our findings. Moreover, it was demonstrated that several dysregulated miRNAs in CRC are analytically mapped to targets implicated in cancer progression pathways142.
In conclusion, serum exosomal miRNAs profile may in the future become valuable biomarkers in the management of CRC. They represent one of the non-invasive methods that may have a potential role in the future diagnosis and screening of CRC. In addition, stage-specific expression of exosomal miRNAs might help establish a role for exosomal miRNAs in the staging, recurrence, and prognosis of CRC. Further experimentation is required before we recommend the use of such a molecular approach in controlling CRC.
Materials and methods
Ethical approval
The research experimental design conducted in this project followed the appropriate regulations and guidelines, including the Good Clinical Practice (GCP) Guidelines. The Biomedical Ethics Research Committee of King Abdulaziz University Hospital (KAUH), Jeddah, Saudi Arabia approved the study and assigned it registration number HA-02-J-008 with reference no. 510-20. All participants provided informed consent prior to their involvement. All blood samples were obtained at KAUH between years 2020 and 2021 after CRC patients as well as healthy individuals signed patient information and consent form. Participants were asked to complete a self-administered questionnaire regarding age, personal medical history and smoking. CRC was confirmed in patients based on monitored, diagnosed, and clinical data collected using standard clinical, endoscopic, radiographic, and histological criteria. Patients were qualified to participate in this study by surgeons and pathologists.
Inclusion criteria in the study healthy control arm was confirmed based on a recently completed normal colonoscopy with no previous history of CRC and no family history of cancer consistent with known dominant disorders. They also had no known genetic predisposition to the development of CRC. The exclusion criteria included CRC patients that have started chemotherapy or radiotherapy before blood collection and those identified to have hereditary non-polyposis CRC or familial adenomatous polyposis.
Samples were collected from CRC patients according to TNM staging system (American Joint Committee on Cancer AJCC). A total of 12 blood samples including nine CRC patients at stages II, III, and IV and three healthy individuals aged between 37 and 65 years old were utilized in this study. Samples were collected by a technician at endoscopy unit at KAUH. Clinicopathological characteristics of CRC patients are shown in Table 1.
Table 1.
Clinicopathological characteristics of CRC patients. Tumor stages were detected according to staging system of AJCC, while location was detected according to proximal or distal classification.
| Variable | Frequency (N = 12) |
|---|---|
| Gender | |
| Male | 6 |
| Female | 6 |
| Median range (37–65) | |
| Age at diagnosis | |
| TNM stage | |
| II | 3 |
| III | 3 |
| IV | 3 |
| Nodal stage | |
| Positive | 5 |
| Negative | 4 |
| Tumor location | |
| Rectum | 3 |
| Distal colon | 4 |
| Proximal colon | 2 |
Sample collection and preparation
Blood samples were collected and divided into serum collection tubes (Hebei Xinle Sci. & Tech. Co., LTD, China). Samples were left for 60 min at room temperature to fully coagulate, then centrifuged for 10 min at 3000 rpm (1900×g) and 4°C by using a swinging bucket rotor. Additional centrifugation was applied on serum samples after being transferred into 2 ml tubes. Serum samples were centrifuged for 15 min at 3000×g and 4°C to remove additional cellular nucleic acids attached to cell debris. Collected serum samples were aliquoted into 1.5 ml microcentrifuge tubes, then labeled and stored in −80°C refrigerator. All samples were processed within 3 h of collection.
Exosome isolation
Exosomes were isolated using exoRNeasy Midi kit (Qiagen, cat. no. 77144) as described in manufacturer’s protocol except for adding buffer XE (45 ml) (Qiagen, cat. no.76214) instead of qiazol. Briefly, frozen serum (1 ml) was centrifuged for 5 min at 3000×g and 4°C after being completely thawed at room temperature to eliminate any remaining cell debris. Around 800 μl of clear supernatant was transferred to 2 ml new tube. By adding same volume of buffer XBP to sample and mixing gently. Mixture was added into exoEasy spin column and spun twice for 1 min at 500×g under cooling, and flow throw was discarded and then again spun for 1 min at 4000×g under cooling to remove liquid remaining on membrane. In this step, exosomes bound to exoEasy spin column. Around 3.5 ml washing buffer XWP was added onto spin column and centrifuged for 5 min at 4000×g under cooling. Then, spin column was transferred to a new collection tube. For eluting exosomes from membrane, 200 μl buffer XE was added and incubated for 1 min then centrifuged for 5 min at 500×g under cooling. For protein analysis, 1–2 ml buffer XE was added to membrane then ultrafiltration was applied to concentrate elution. To ensure all exosomes were collected, eluate was re-added onto exoEasy spin column and incubated for 1 min then centrifuged for 5 min at 5000×g under cooling.
Transmission electron microscopy (TEM)
Examining exosomes by transmission electron microscopy (TEM) was done at King Abdullah University of Science and Technology (KAUST), Thuwal, Saudi Arabia. Purified exosomes (4 μl) were carefully loaded onto surface of pre-glow-discharged carbon-coated 300-mesh Cu formvar grid and incubated for 1 min for adsorption. Samples were then removed using filter paper and stained negatively for 1 min with 1 drop of 2% uranyl acetate. After removing stain with filter paper, sample was left to dry. Sample was then ready for examination under microscope. After loading samples on single tilt holder, they were inserted into Titan CT (FEI Company, Thermo Fisher Scientific). Finally, screening and imaging of exosomes were performed using several magnifications Gatan US4000 CCD camera (Gatan, Inc., Pleasanton, CA). ImageJ software was used to compute radius of exosomes based on images of exosomes obtained from TEM. Exosomes were then diluted in 1 mL PBS and size examined by Zetasizer Nano ZS (Malvern instruments) at 25°C according to manufacturer’s instructions.
RNA extraction
For extraction of total RNAs, including miRNA, from exosomes and other extracellular vesicles (Evs), exoRNeasy Midi kit (Qiagen, cat. no. 77144) was used according to manufacturer’s protocol. To extract total RNA from vesicles, around 700 μl of lysate buffer ‘QIAzol’ was added into spin column and spun for 5 min at 4000 xg, then transferred to 1 ml tube. Lysate was incubated for 5 min at room temperature (15–25°C), which assists dissociation of nucleoprotein complexes. After incubation, 3.5 μl miRNeasy Serum/Plasma Spike-In Control was added as spike-in exogenous control and followed by adding 90 μl chloroform with shaking vigorously for 15 s to subsequent phase separation. Tube was incubated for 2–3 min at room temperature (15–25°C) and centrifuged for 15 min at 12,000 xg and 4°C. Through centrifugation, sample separates into 3 phases. An amount of 300 ul upper aqueous phase containing RNA was transferred to a new 2 ml collection tube. Then, two volumes of 100% ethanol were added and mixed thoroughly by pipetting slowly several times. Then, 650 μl of sample was transferred onto Rneasy MinElute spin column and centrifuged for 15 s. This step was repeated for remainder of sample. Then, 700 μl of buffer RWT was added to spin column and centrifuged for 15 s and 500 μl buffer RPE was added onto spin column and again centrifuged for 15 s. Then, 500 μl buffer RPE was added again onto spin column and centrifuged for 2 min. All centrifugations were done for each step at ≥ 8000×g (≥ 10,000 rpm) and in each time flow through was discarded except for last washing with buffer RPE. RNeasy MinElute spin column was transferred into a new 2 ml collection tube and centrifuged at high speed for 5 min with an open lid to dry membrane, then collection tube with flow through was discarded. Spin column was then transferred to a new 1.5 ml collection tube and 20–18 μl RNase-free water was directly added into center of membrane of spin column. Tube was incubated for 1 min and then centrifuged for 2 min with high speed to elute RNA. All processes of RNA extraction were done at room temperature (15–25°C) except phase separation process which was done at 4°C. Purified total RNA was measured by NanoDrop and Bioanalyzer, then frozen at −80°C until miRNA expression analysis was done.
RNA quantification and qualification
Novogene Experimental Department has performed RNA quantification and qualification processes, and 1% agarose gels were used to monitor RNA breakdown and protein contamination. NanoPhotometer spectrophotometer was used to assess purity of RNA (Implen, CA, USA). RNA Nano 6000 Assay kit of Agilent Bioanalyzer 2100 system was used to test RNA integrity and quantity (Agilent Technologies, CA, USA).
Small RNA library preparation and Illumina sequencing
Sequencing preparation was done by Novogene AIT Genomics (Singapore) utilizing NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB, United States). For miRNA library construction, 3 μg total RNA per sample was used as input material. NEBNext® Multiplex Small RNA Library Prep Set for Illumina®, NEB, USA) was used for generating sequencing libraries by following manufacturer’s recommendations. In summary, 3’ and 5’ adaptors were ligated to 3’ and 5’ ends of exososmal miRNA, respectively. Following hybridization with a reverse transcription primer, first strand cDNA was produced. PCR enrichment was used to create double-stranded cDNA library. Libraries containing insertions between 18 and 40 bp were ready for sequencing with SE50 after purification and size selection. Library was then tested for quantification using Qubit and real-time PCR, as well as size distribution detection using a Bioanalyzer 2100 system (Agilent Technologies, CA, USA). On Illumina platforms, quantified libraries were pooled and sequenced based on effective library concentration and data amount required.
miRNA alignments and prediction of target genes
miRNAs were mapped to recognize known from novel miRNAs. To get candidate miRNAs and design secondary structures, we utilized miRBase20.0 as a reference, along with customized software mirdeep2143, and srna-tools-cli. Then, miRNA counts, and base bias were calculated using custom scripts on first location of identified miRNA with a certain length and on each position of all identified miRNA. Then, Rfam 12.0 database was applied to analyze clean reads matching miRNAs in addition to rRNAs, tRNAs, snRNAs, snoRNAs, and other non-coding RNAs. For identification of known and novel miRNAs, miRbase database (version 21.0) was utilized by mapping specific sequences of obtained reads to reference genome. Transcriptome sequencing results from genome were used as reference genome to perform target gene prediction. miRanda software was performed to predict target genes of known and novel miRNA144. Transcript per million (TPM) was measured to assess miRNA expression levels using equation Normalized expression = (mapped read-count/total reads) × 1,000,000)25.
Differentially expressed exosomal miRNAs in CRC
Fold change (FC) values between CRC patients and healthy control were used to estimate miRNA differential expression. FC values were converted to base-2 logarithm of FC (log2 (FC)) once they were calculated. Exosomal miRNAs that were differentially expressed in dataset were then used to form Venn diagram and find miRNAs that showed statistical differences between CRC groups compared with healthy control. DESeq R package (1.8.3) was applied for differential expression analysis of two conditions or groups of biological replicates of samples. Benjamini and Hochberg method was used to adjust P values, while corrected P value of 0.05 was set as threshold for significant differential expression.
Bioinformatic analysis
All predicted target gene of differentially expressed (DE) miRNAs were categorized into functional classes through Gene Ontology (GO) enrichment analysis by GOseq software based on Wallenius non-central hyper-geometric distribution26. All target genes that bind to DE miRNAs were applied in Shiny GO to identify KEGG pathways and GO enrichment terms FM, CC, and BP. Heatmap designed by iDEP.951 (http://bioinformatics.sdstate.edu/idep/) and KOBAS145 (http://kobas.cbi.pku.edu.cn/home.do) softwares was performed to test statistical enrichment of all DE miRNAs target gene candidates in KEGG pathways (http://www.genome.jp/kegg/). Binding sites of miRNAs on target gene was predicted using a built-in function of TargetScan database (v7.0; targetscan.org)146. Predicted target genes were visualized by Flourish studio (Flourish Studio, Kiln Enterprises Ltd, London, UK). Sequence of pre-miRNA stem-loop secondary structure was taken from miRBase and designed consulting ViennaRNA Web Services147.
Verification of miRNA deep-sequencing results by qPCR
Freshly frozen serum exosomes were isolated to comprise the expression of miRNAs: miR-3184-5p and miR-423-5p at the qPCR level. Their respective target genes, SMAD2 and TGFBR2, were selected to validate expression of these miRNAs (miR-3184-5p and miR-423-5p) resulted from deep sequencing. The GAPDH gene was used as a housekeeping gene. The primers used in the validation were designed on the basis of miRNA mature sequence (listed in Supplementary S8). cDNA synthesis and qRT-PCR were performed using StepOne Real-Time PCR System (Applied Biosystems, USA) according to the manufacturer’s protocol148. All samples were run in three replications. qPCR data were analyzed by two methods (i) ∆∆CT or Livak and (ii) ∆CT. The average CT for the endogenous control and each gene was calculated from the raw data generated by RT-PCR StepOne System and Data Assist software. The outliers were either omitted from the analysis or substituted with means. The graph Pad PRISM software and MS Excel functions were used to perform the analysis and generate the statistical plots of the data. The mean CT and standard deviation for the reference gene (GAPDH), the two genes (SMAD2 and TGFER2), and 2 miRNAs (miR-3184-5p and miR-423-5p) genes were calculated. P-value and fold change were calculated to determine the statistical significance of the expression pattern of miR-3184-5p and miR-423-5p genes (P-value < 0.05 and fold change > ± 2).
Supplementary Information
Acknowledgements
We would like to thank Professor. Hani Choudhry for his invaluable input and support throughout the research process. We also thank authorities of the Cancer and Mutagenesis Unit at King Fahd Medical Research Center in King Abdulaziz University for providing lab facilities.
Author contributions
T.B. designed and executed the entire project. A.B. and S.A. assisted in the design, editing, and review of the project. A.F. was responsible for obtaining physician and ethical approval. A.Y., T.B., A.A., S.Q., and M.A. collaborated on sample collection and wet lab preparation. F.A. and O.E. provided recommendations and participated in editing the work. M.I. undertook the data analysis as a bioinformatician. All authors reviewed the manuscript.
Data availability
Exosomal miRNA raw data are deposited in the European Nucleotide Archive at EMBL-EBI under accession number PRJEB64383. The datasets produced and/or examined in the course of this study are not publicly accessible because they are still undergoing further investigation. However, they can be obtained from the corresponding author upon a reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tahani Bakhsh, Email: tabakhsh@uj.edu.sa.
Ahmed Bahieldin, Email: abmahmed@kau.edu.sa.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-58536-3.
References
- 1.Arvelo, F., Sojo, F., Cotte, C. Biology of colorectal cancer. Ecancermedicalscience9 (2015). [DOI] [PMC free article] [PubMed]
- 2.Ding, L., et al. The dual role of MicroRNAs in colorectal cancer progression.Int. J. Mol. Sci.19(9) (2018). [DOI] [PMC free article] [PubMed]
- 3.Wang LG, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2012;36(1):e61–e67. doi: 10.1016/j.canep.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Al-Sheikh YA, et al. Expression profiling of selected microRNA signatures in plasma and tissues of Saudi colorectal cancer patients by qPCR. Oncol Lett. 2016;11(2):1406–1412. doi: 10.3892/ol.2015.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aghabozorgi AS, et al. Role of adenomatous polyposis coli (APC) gene mutations in the pathogenesis of colorectal cancer; current status and perspectives. Biochimie. 2019;157:64–71. doi: 10.1016/j.biochi.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Guinney J, et al. The consensus molecular subtypes of colorectal cancer. Nature Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn M, et al. The genetic heterogeneity of colorectal cancer predisposition-guidelines for gene discovery. Cell. Oncol. 2016;39(6):491–510. doi: 10.1007/s13402-016-0284-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strubberg AM, Madison BB. MicroRNAs in the etiology of colorectal cancer: Pathways and clinical implications. Dis Model Mech. 2017;10(3):197–214. doi: 10.1242/dmm.027441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosseinahli N, et al. Treating cancer with microRNA replacement therapy: A literature review. J Cell Physiol. 2018;233(8):5574–5588. doi: 10.1002/jcp.26514. [DOI] [PubMed] [Google Scholar]
- 11.Igaz, P. Circulating Micrornas in Disease Diagnostics and their Potential Biological Relevance. vol. 106. 2015, Springer.
- 12.Kumar, S., et al. MicroRNAs as peripheral biomarkers in aging and age-related diseases. In Progress in Molecular Biology and Translational Science, Elsevier. p. 47–94 (2017). [DOI] [PubMed]
- 13.Hibner, G., Kimsa-Furdzik, M., Francuz, T. Relevance of MicroRNAs as potential diagnostic and prognostic markers in colorectal cancer.Int. J. Mol. Sci.19(10) (2018). [DOI] [PMC free article] [PubMed]
- 14.Bakhsh T, et al. Exosomal miRNAs as a promising source of biomarkers in colorectal cancer progression. Int. J. Mol. Sci. 2022;23(9):4855. doi: 10.3390/ijms23094855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B, et al. Emerging microRNA biomarkers for colorectal cancer diagnosis and prognosis. Open Biol. 2019;9(1):180212. doi: 10.1098/rsob.180212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGough IJ, Vincent JP. Exosomes in developmental signalling. Development. 2016;143(14):2482–2493. doi: 10.1242/dev.126516. [DOI] [PubMed] [Google Scholar]
- 17.Fu F, et al. Circulating exosomal miR-17-5p and miR-92a-3p predict pathologic stage and grade of colorectal cancer. Translat. Oncol. 2018;11(2):221–232. doi: 10.1016/j.tranon.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C, et al. Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget. 2016;7(46):76250. doi: 10.18632/oncotarget.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schee K, et al. Deep sequencing the MicroRNA transcriptome in colorectal cancer. PLoS One. 2013;8(6):e66165. doi: 10.1371/journal.pone.0066165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massagué J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103(2):295–309. doi: 10.1016/S0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 21.Feng X-H, Derynck R. Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 22.Soleimani A, et al. The potential role of regulatory microRNAs of RAS/MAPK signaling pathway in the pathogenesis of colorectal cancer. J. Cell. Biochem. 2019;120(12):19245–19253. doi: 10.1002/jcb.29268. [DOI] [PubMed] [Google Scholar]
- 23.Soleimani A, et al. Role of TGF-β signaling regulatory microRNAs in the pathogenesis of colorectal cancer. J. Cell. Physiol. 2019;234(9):14574–14580. doi: 10.1002/jcp.28169. [DOI] [PubMed] [Google Scholar]
- 24.Zhong M, Bian Z, Wu Z. miR-30a suppresses cell migration and invasion through downregulation of PIK3CD in colorectal carcinoma. Cell. Physiol. Biochem. 2013;31(2–3):209–218. doi: 10.1159/000343362. [DOI] [PubMed] [Google Scholar]
- 25.Zhou, L., et al. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27. 3 associate with clear cell renal cell carcinoma. PloS one, 5(12): e15224 (2010). [DOI] [PMC free article] [PubMed]
- 26.Young MD, et al. goseq: gene ontology testing for RNA-seq datasets. R Bioconductor. 2012;8:1–25. [Google Scholar]
- 27.Fleming NI, et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013;73(2):725–735. doi: 10.1158/0008-5472.CAN-12-2706. [DOI] [PubMed] [Google Scholar]
- 28.Li X-L, et al. P53 mutations in colorectal cancer-molecular pathogenesis and pharmacological reactivation. World J. Gastroenterol.: WJG. 2015;21(1):84. doi: 10.3748/wjg.v21.i1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell SM, et al. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359(6392):235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 30.Slattery ML, et al. The p53-signaling pathway and colorectal cancer: Interactions between downstream p53 target genes and miRNAs. Genomics. 2019;111(4):762–771. doi: 10.1016/j.ygeno.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao M, Mishra L, Deng C-X. The role of TGF-β/SMAD4 signaling in cancer. Int. J. Biol. Sci. 2018;14(2):111. doi: 10.7150/ijbs.23230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamada T, Nowak JA, Ogino S. PIK3CA mutation and colorectal cancer precision medicine. Oncotarget. 2017;8(14):22305. doi: 10.18632/oncotarget.15724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zenonos K, Kyprianou K. RAS signaling pathways, mutations and their role in colorectal cancer. World J. Gastrointestinal Oncol. 2013;5(5):97. doi: 10.4251/wjgo.v5.i5.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Pasche B. TGF-β signaling alterations and susceptibility to colorectal cancer. Hum. Mol. Genet. 2007;16(R1):R14–R20. doi: 10.1093/hmg/ddl486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGeary, S.E., et al. The Biochemical Basis of MicroRNA Targeting Efficacy. Science, 2019. 366(6472): p. eaav1741. [DOI] [PMC free article] [PubMed]
- 36.Xie, Y., et al. RNA-seq profiling of serum exosomal circular RNAs reveals Circ-PNN as a potential biomarker for human colorectal cancer.Front. Oncol. p. 982 (2020). [DOI] [PMC free article] [PubMed]
- 37.Toiyama, Y., et al. MicroRNAs as potential liquid biopsy biomarkers in colorectal cancer: A systematic review.Biochim. et Biophys. Acta (BBA)-Reviews on Cancer, 1870(2), 274–282 (2018). [DOI] [PMC free article] [PubMed]
- 38.Ji D, et al. Serum-based microRNA signature predicts relapse and therapeutic outcome of adjuvant chemotherapy in colorectal cancer patients. EBioMedicine. 2018;35:189–197. doi: 10.1016/j.ebiom.2018.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ingenito F, et al. The role of exo-miRNAs in cancer: a focus on therapeutic and diagnostic applications. Int. J. Mol. Sci. 2019;20(19):4687. doi: 10.3390/ijms20194687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Świderska M, et al. The diagnostics of colorectal cancer. Contemp Oncol. 2014;18(1):1. doi: 10.5114/wo.2013.39995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ebrahimkhani S, et al. Deep sequencing of circulating exosomal microRNA allows non-invasive glioblastoma diagnosis. NPJ Precis. Oncol. 2018;2(1):1–9. doi: 10.1038/s41698-018-0071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sohel MH. Extracellular/circulating microRNAs: release mechanisms, functions and challenges. Achiev. Life Sci. 2016;10(2):175–186. [Google Scholar]
- 43.Palma J, et al. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012;40(18):9125–9138. doi: 10.1093/nar/gks656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki HI. MicroRNA control of TGF-β signaling. Int. J. Mol. Sci. 2018;19(7):1901. doi: 10.3390/ijms19071901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim LP, et al. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17(8):991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su J-L, et al. Function and regulation of let-7 family microRNAs. Microrna. 2012;1(1):34–39. doi: 10.2174/2211536611201010034. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H, et al. Silencing Lin28 promotes apoptosis in colorectal cancer cells by upregulating let-7c targeting of antiapoptotic BCL2L1. Mol. Med. Rep. 2018;17(4):5143–5149. doi: 10.3892/mmr.2018.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balzeau J, et al. The LIN28/let-7 pathway in cancer. Front. Genet. 2017;8:31. doi: 10.3389/fgene.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 50.Ogata-Kawata H, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PloS one. 2014;9(4):e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han HB, et al. Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J. Pathol. 2012;226(3):544–555. doi: 10.1002/path.3014. [DOI] [PubMed] [Google Scholar]
- 52.Ghanbari, R., et al. Simultaneous underexpression of let-7a-5p and let-7f-5p microRNAs in plasma and stool samples from early stage colorectal carcinoma: Supplementary issue: Biomarkers for colon cancer.Biomark. Cancer, 7: p. BIC. S25252 (2015). [DOI] [PMC free article] [PubMed]
- 53.Choo KB, et al. MicroRNA-5p and-3p co-expression and cross-targeting in colon cancer cells. J. Biomed. Sci. 2014;21(1):1–14. doi: 10.1186/s12929-014-0095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gmerek L, et al. MicroRNA regulation in colorectal cancer tissue and serum. PLoS One. 2019;14(8):e0222013. doi: 10.1371/journal.pone.0222013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohshima K, et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PloS one. 2010;5(10):e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu T, et al. Let-7a suppresses cell proliferation via the TGF-β/SMAD signaling pathway in cervical cancer. Oncol. Rep. 2016;36(6):3275–3282. doi: 10.3892/or.2016.5160. [DOI] [PubMed] [Google Scholar]
- 57.Butz H, et al. Crosstalk between TGF-β signaling and the microRNA machinery. Trends Pharmacol. Sci. 2012;33(7):382–393. doi: 10.1016/j.tips.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Miyamoto S, et al. Expression patterns of miRNA-423-5p in the serum and pericardial fluid in patients undergoing cardiac surgery. PLoS One. 2015;10(11):e0142904. doi: 10.1371/journal.pone.0142904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shang, Y., et al. Downregulation of miR-423-5p contributes to the radioresistance in colorectal cancer cells.Front. Oncol. p. 2990 (2021). [DOI] [PMC free article] [PubMed]
- 60.Lin H, et al. Inhibition of miR-423-5p suppressed prostate cancer through targeting GRIM-19. Gene. 2019;688:93–97. doi: 10.1016/j.gene.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 61.Rajarajan D, et al. Genome-wide analysis reveals miR-3184-5p and miR-181c-3p as a critical regulator for adipocytes-associated breast cancer. J. Cell. Physiol. 2019;234(10):17959–17974. doi: 10.1002/jcp.28428. [DOI] [PubMed] [Google Scholar]
- 62.Li H-T, et al. MiR-423-3p enhances cell growth through inhibition of p21Cip1/Waf1 in colorectal cancer. Cell. Physiol. Biochem. 2015;37(3):1044–1054. doi: 10.1159/000430230. [DOI] [PubMed] [Google Scholar]
- 63.Lin J, et al. MicroRNA-423 promotes cell growth and regulates G 1/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis. 2011;32(11):1641–1647. doi: 10.1093/carcin/bgr199. [DOI] [PubMed] [Google Scholar]
- 64.Morishita A, et al. MicroRNA profiles in various hepatocellular carcinoma cell lines. Oncol. Lett. 2016;12(3):1687–1692. doi: 10.3892/ol.2016.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin X, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin. Cancer Res. 2017;23(17):5311–5319. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 66.Shan G, et al. Cancer-associated fibroblast-secreted exosomal miR-423-5p promotes chemotherapy resistance in prostate cancer by targeting GREM2 through the TGF-β signaling pathway. Exp. Mol. Med. 2020;52(11):1809–1822. doi: 10.1038/s12276-020-0431-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eniafe J, Jiang S. MicroRNA-99 family in cancer and immunity. Wiley Interdiscip. Rev.: RNA. 2021;12(3):e1635. doi: 10.1002/wrna.1635. [DOI] [PubMed] [Google Scholar]
- 68.Sun Y, et al. Identification of serum miR-30a-5p as a diagnostic and prognostic biomarker in colorectal cancer. Cancer Biomark. 2019;24(3):299–305. doi: 10.3233/CBM-182129. [DOI] [PubMed] [Google Scholar]
- 69.Song, X., et al. Circulating exosomal miR-150-5p and miR-99b-5p as diagnostic biomarkers for colorectal cancer.Front. Oncol. p. 1129 (2019). [DOI] [PMC free article] [PubMed]
- 70.Park Y, et al. MicroRNA-30a-5p (miR-30a) regulates cell motility and EMT by directly targeting oncogenic TM4SF1 in colorectal cancer. J. Cancer Res. Clin. Oncol. 2017;143(10):1915–1927. doi: 10.1007/s00432-017-2440-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Q, et al. Role of microRNA 30a targeting insulin receptor substrate 2 in colorectal tumorigenesis. Mol. Cell. Biol. 2015;35(6):988–1000. doi: 10.1128/MCB.01242-14. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Li W, et al. miRNA-99b-5p suppresses liver metastasis of colorectal cancer by down-regulating mTOR. Oncotarget. 2015;6(27):24448. doi: 10.18632/oncotarget.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z, et al. MiR-99b-5p and miR-203a-3p function as tumor suppressors by targeting IGF-1R in gastric cancer. Sci. Rep. 2018;8(1):1–12. doi: 10.1038/s41598-018-27583-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Emmrich S, et al. LincRNAs MONC and MIR100HG act as oncogenes in acute megakaryoblastic leukemia. Mol. Cancer. 2014;13(1):1–12. doi: 10.1186/1476-4598-13-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soliman SE, et al. Down regulation of miR-30a-5p and miR-182–5p in gastric cancer: Clinical impact and survival analysis. Biochem. Biophys. Rep. 2021;27:101079. doi: 10.1016/j.bbrep.2021.101079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Visone R, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26(54):7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 77.Li, W., et al. Downregulation of β3 integrin by miR-30a-5p modulates cell adhesion and invasion by interrupting Erk/Ets‑1 network in triple-negative breast cancer Corrigendum in/10.3892/ijo.2021.5169.Int. J. Oncol.48(3): 1155–1164 (2016). [DOI] [PubMed]
- 78.Ramírez-Moya J, Santisteban P. miRNA-directed regulation of the main signaling pathways in thyroid cancer. Front. Endocrinol. 2019;10:430. doi: 10.3389/fendo.2019.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Braun J, et al. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene. 2010;29(29):4237–4244. doi: 10.1038/onc.2010.169. [DOI] [PubMed] [Google Scholar]
- 80.Riesco-Eizaguirre G, et al. The BRAFV600E oncogene induces transforming growth factor β secretion leading to sodium iodide symporter repression and increased malignancy in thyroid cancer. Cancer Res. 2009;69(21):8317–8325. doi: 10.1158/0008-5472.CAN-09-1248. [DOI] [PubMed] [Google Scholar]
- 81.Ghahnavieh LE, et al. Fluctuating expression of miR-584 in primary and high-grade gastric cancer. BMC Cancer. 2020;20(1):1–12. doi: 10.1186/s12885-020-07116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Konishi H, et al. microRNA-26a and-584 inhibit the colorectal cancer progression through inhibition of the binding of hnRNP A1-CDK6 mRNA. Biochem Biophys. Res. Commun. 2015;467(4):847–852. doi: 10.1016/j.bbrc.2015.10.055. [DOI] [PubMed] [Google Scholar]
- 83.Narasimhan, K., et al. Identification of unique miRNA biomarkers in colorectal adenoma and carcinoma using microarray: evaluation of their putative role in disease progression. Int. Scholarly Res. Notices. 2014 (2014).
- 84.Hayashi M, et al. Suppressive effect of an analog of the antimicrobial peptide of LL-37 on colon cancer cells via exosome-encapsulated miRNAs. Int. J. Mol. Med. 2018;42(6):3009–3016. doi: 10.3892/ijmm.2018.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suetsugu T, et al. Downregulation of matrix metalloproteinase 14 by the antitumor miRNA, miR-150-5p, inhibits the aggressiveness of lung squamous cell carcinoma cells. Int. J. Oncol. 2018;52(3):913–924. doi: 10.3892/ijo.2017.4232. [DOI] [PubMed] [Google Scholar]
- 86.Tang W, et al. MicroRNA-150 suppresses triple-negative breast cancer metastasis through targeting HMGA2. OncoTargets Ther. 2018;11:2319. doi: 10.2147/OTT.S161996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan R, et al. Retracted: MicroRNA-150-5p affects cell proliferation, apoptosis, and EMT by regulation of the BRAFV600E mutation in papillary thyroid cancer cells. J. Cell. Biochem. 2018;119(11):8763–8772. doi: 10.1002/jcb.27108. [DOI] [PubMed] [Google Scholar]
- 88.Yu J, et al. Aryl hydrocarbon receptor enhances the expression of miR-150-5p to suppress in prostate cancer progression by regulating MAP3K12. Arch. Biochem. Biophys. 2018;654:47–54. doi: 10.1016/j.abb.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 89.Lee KH, et al. Postoperative prognosis prediction of pancreatic cancer with seven microRNAs. Pancreas. 2015;44(5):764–768. doi: 10.1097/MPA.0000000000000346. [DOI] [PubMed] [Google Scholar]
- 90.Ma Y, et al. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61(10):1447–1453. doi: 10.1136/gutjnl-2011-301122. [DOI] [PubMed] [Google Scholar]
- 91.Yokobori T, et al. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB 1. Cancer Sci. 2013;104(1):48–54. doi: 10.1111/cas.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang S, et al. miR-150 promotes human breast cancer growth and malignant behavior by targeting the pro-apoptotic purinergic P2X7 receptor. PloS one. 2013;8(12):e80707. doi: 10.1371/journal.pone.0080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rong G, et al. miR-150-504-519d inhibits the growth of human colorectal cancer cell line SW48 and downregulates c-FLIP receptor. J Cell. Biochem. 2019;120(5):7962–7969. doi: 10.1002/jcb.28073. [DOI] [PubMed] [Google Scholar]
- 94.Feng J, et al. miR-150 functions as a tumour suppressor in human colorectal cancer by targeting c-Myb. J. Cell. Mol. Med. 2014;18(10):2125–2134. doi: 10.1111/jcmm.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.He Z, et al. The involvement of miR-150/β-catenin axis in colorectal cancer progression. Biomed. Pharmacother. 2020;121:109495. doi: 10.1016/j.biopha.2019.109495. [DOI] [PubMed] [Google Scholar]
- 96.Davoodian P, et al. Effect of TGF-β/smad signaling pathway blocking on expression profiles of miR-335, miR-150, miR-194, miR-27a, and miR-199a of hepatic stellate cells (HSCs) Gastroenterol. Hepatol. Bed Bench. 2017;10(2):112. [PMC free article] [PubMed] [Google Scholar]
- 97.Yi L, et al. MicroRNA-26b-5p suppresses the proliferation of tongue squamous cell carcinoma via targeting proline rich 11 (PRR11) Bioengineered. 2021;12(1):5830–5838. doi: 10.1080/21655979.2021.1969832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu, L., et al. Asporin regulated by miR-26b-5p mediates chondrocyte senescence and exacerbates osteoarthritis progression via TGF-β1/Smad2 pathway.Rheumatology, 2021. [DOI] [PubMed]
- 99.Wei Z, et al. MiR-26a/miR-26b represses tongue squamous cell carcinoma progression by targeting PAK1. Cancer Cell Int. 2020;20(1):1–14. doi: 10.1186/s12935-020-1166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koshizuka K, et al. Involvement of aberrantly expressed microRNAs in the pathogenesis of head and neck squamous cell carcinoma. Cancer Metastasis Rev. 2017;36(3):525–545. doi: 10.1007/s10555-017-9692-y. [DOI] [PubMed] [Google Scholar]
- 101.Li D, et al. MicroRNA-26b suppresses the metastasis of non-small cell lung cancer by targeting MIEN1 via NF-κB/MMP-9/VEGF pathways. Biochem. Biophys. Res. Commun. 2016;472(3):465–470. doi: 10.1016/j.bbrc.2016.01.163. [DOI] [PubMed] [Google Scholar]
- 102.Zhang C, Tong J, Huang G. Nicotinamide phosphoribosyl transferase (Nampt) is a target of microRNA-26b in colorectal cancer cells. PloS one. 2013;8(7):e69963. doi: 10.1371/journal.pone.0069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fan D, et al. Micro RNA 26b promotes colorectal cancer metastasis by downregulating phosphatase and tensin homolog and wingless-type MMTV integration site family member 5A. Cancer Sci. 2018;109(2):354–362. doi: 10.1111/cas.13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li Y, et al. Tumor-suppressive miR-26a and miR-26b inhibit cell aggressiveness by regulating FUT4 in colorectal cancer. Cell Death Dis. 2017;8(6):e2892–e2892. doi: 10.1038/cddis.2017.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De Robertis M, et al. Dysregulation of EGFR pathway in EphA2 cell subpopulation significantly associates with poor prognosis in colorectal cancer. Clin. Cancer Res. 2017;23(1):159–170. doi: 10.1158/1078-0432.CCR-16-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang Z, et al. MicroRNA-26b represses colon cancer cell proliferation by inhibiting lymphoid enhancer factor 1 expression. Mol. Cancer Therap. 2014;13(7):1942–1951. doi: 10.1158/1535-7163.MCT-13-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Benderska N, et al. miRNA-26b overexpression in ulcerative colitis-associated carcinogenesis. Inflammatory Bowel Dis. 2015;21(9):2039–2051. doi: 10.1097/MIB.0000000000000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Marquez ME, et al. TGF-β/SMAD pathway is modulated by miR-26b-5p: Another piece in the puzzle of chronic lymphocytic leukemia progression. Cancers. 2022;14(7):1676. doi: 10.3390/cancers14071676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yin Y, et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res. 2014;20(23):6187–6199. doi: 10.1158/1078-0432.CCR-14-1030. [DOI] [PubMed] [Google Scholar]
- 110.Bao W, et al. A TrkB–STAT3–miR-204-5p regulatory circuitry controls proliferation and invasion of endometrial carcinoma cells. Mol. Cancer. 2013;12(1):1–19. doi: 10.1186/1476-4598-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mikhaylova O, et al. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. 2012;21(4):532–546. doi: 10.1016/j.ccr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gong M, et al. MicroRNA-204 critically regulates carcinogenesis in malignant peripheral nerve sheath tumors. Neuro-oncology. 2012;14(8):1007–1017. doi: 10.1093/neuonc/nos124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen Z, et al. miR-204 mediated loss of Myeloid cell leukemia-1 results in pancreatic cancer cell death. Mol. Cancer. 2013;12(1):1–11. doi: 10.1186/1476-4598-12-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yu Y, et al. MiR-204 inhibits hepatocellular cancer drug resistance and metastasis through targeting NUAK1. Biochem. Cell Biol. 2019;97(5):563–570. doi: 10.1139/bcb-2018-0354. [DOI] [PubMed] [Google Scholar]
- 115.Huang G, et al. MiR-204 reduces cisplatin resistance in non-small cell lung cancer through suppression of the caveolin-1/AKT/Bad pathway. Aging (Albany NY) 2019;11(7):2138. doi: 10.18632/aging.101907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ying Z, et al. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 2013;73(2):990–999. doi: 10.1158/0008-5472.CAN-12-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Qiu Y-H, et al. miR-204 inhibits epithelial to mesenchymal transition by targeting slug in intrahepatic cholangiocarcinoma cells. Cell Physiol. Biochem. 2013;32(5):1331–1341. doi: 10.1159/000354531. [DOI] [PubMed] [Google Scholar]
- 118.Sacconi A, et al. miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis. 2012;3(11):e423–e423. doi: 10.1038/cddis.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee Y, et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput. Biol. 2010;6(4):e1000730. doi: 10.1371/journal.pcbi.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wu H, et al. MicroRNA-204 modulates colorectal cancer cell sensitivity in response to 5-fluorouracil-based treatment by targeting high mobility group protein A2. Biol. Open. 2016;5(5):563–570. doi: 10.1242/bio.015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang C, et al. Relationship between miR-204 and ANGPTL2 expression and diagnosis, pathological stage, and prognosis in patients with colon cancer. Translat. Cancer Res. 2021;10(8):3788. doi: 10.21037/tcr-21-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu H, et al. Long non-coding RNA PCAT6 targets miR-204 to modulate the chemoresistance of colorectal cancer cells to 5-fluorouracil-based treatment through HMGA2 signaling. Cancer Med. 2019;8(5):2484–2495. doi: 10.1002/cam4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li R, et al. Long noncoding RNA lncBRM promotes proliferation and invasion of colorectal cancer by sponging miR-204-3p and upregulating TPT1. Biochem. Biophys. Res. Commun. 2019;508(4):1259–1263. doi: 10.1016/j.bbrc.2018.12.053. [DOI] [PubMed] [Google Scholar]
- 124.Bian Z, et al. LncRNA—UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci. Rep. 2016;6(1):1–12. doi: 10.1038/srep23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang Z, et al. MiR-204-5p inhibits transforming growth factor-β1-induced proliferation and extracellular matrix production of airway smooth muscle cells by regulating Six1 in asthma. Int. Arch. Allergy Immunol. 2020;181(4):239–248. doi: 10.1159/000505064. [DOI] [PubMed] [Google Scholar]
- 126.Wang Y, et al. MicroRNA-204-5p regulates epithelial-to-mesenchymal transition during human posterior capsule opacification by targeting SMAD4. Invest. Ophthalmol. Visual Sci. 2013;54(1):323–332. doi: 10.1167/iovs.12-10904. [DOI] [PubMed] [Google Scholar]
- 127.Fonseca A, et al. Identification of colorectal cancer associated biomarkers: an integrated analysis of miRNA expression. Aging (Albany NY) 2021;13(18):21991. doi: 10.18632/aging.203556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nature Rev. Mol. Cell Biol. 2019;20(1):5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 129.Gao Z-J, et al. miR-486-5p functions as an oncogene by targeting PTEN in non-small cell lung cancer. Pathol.-Res. Pract. 2018;214(5):700–705. doi: 10.1016/j.prp.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 130.David P. Metazoan microRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Binefa G, et al. Colorectal cancer: From prevention to personalized medicine. World J. Gastroenterol.: WJG. 2014;20(22):6786. doi: 10.3748/wjg.v20.i22.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153(1):17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 133.Bardhan K, Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers. 2013;5(2):676–713. doi: 10.3390/cancers5020676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lebreton, M. D. P. C. A., Mammalian microRNAs and long noncoding RNAs in. Nucleic Acids Res. 41, 542–553 (2013).
- 135.Yang J-S, et al. Widespread regulatory activity of vertebrate microRNA* species. Rna. 2011;17(2):312–326. doi: 10.1261/rna.2537911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guo L, Lu Z. The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PloS One. 2010;5(6):e11387. doi: 10.1371/journal.pone.0011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Okamura K, et al. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nature Struct. Mol. Biol. 2008;15(4):354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Slattery ML, et al. The PI3K/AKT signaling pathway: associations of miRNAs with dysregulated gene expression in colorectal cancer. Mol. Carcinogenesis. 2018;57(2):243–261. doi: 10.1002/mc.22752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Therkildsen C, et al. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta Oncol. 2014;53(7):852–864. doi: 10.3109/0284186X.2014.895036. [DOI] [PubMed] [Google Scholar]
- 140.Nosho K, et al. Association of microRNA-31 with BRAF mutation, colorectal cancer survival and serrated pathway. Carcinogenesis. 2014;35(4):776–783. doi: 10.1093/carcin/bgt374. [DOI] [PubMed] [Google Scholar]
- 141.Miquel C, et al. Role of bax mutations in apoptosis in colorectal cancers with microsatellite instability. Am. J. Clin. Pathol. 2005;123(4):562–570. doi: 10.1309/JQ2X3RV3L8F9TGYW. [DOI] [PubMed] [Google Scholar]
- 142.Reid JF, et al. miRNA profiling in colorectal cancer highlights miR-1 involvement in MET-dependent proliferation. Mol. Cancer Res. 2012;10(4):504–515. doi: 10.1158/1541-7786.MCR-11-0342. [DOI] [PubMed] [Google Scholar]
- 143.Friedländer MR, et al. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40(1):37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Enright A, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;4(11):1–27. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mao X, et al. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21(19):3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 146.Agarwal, V., et al. Predicting effective microRNA target sites in mammalian mRNAs. elife, 4, p. e05005 (2015). [DOI] [PMC free article] [PubMed]
- 147.Kerpedjiev P, Hammer S, Hofacker IL. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics. 2015;31(20):3377–3379. doi: 10.1093/bioinformatics/btv372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yan J, et al. One-step real time RT-PCR for detection of microRNAs. Talanta. 2013;110:190–195. doi: 10.1016/j.talanta.2013.02.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Exosomal miRNA raw data are deposited in the European Nucleotide Archive at EMBL-EBI under accession number PRJEB64383. The datasets produced and/or examined in the course of this study are not publicly accessible because they are still undergoing further investigation. However, they can be obtained from the corresponding author upon a reasonable request.