Abstract
Introduction
Seed coat color is a significant agronomic trait in horticultural crops such as Brassica rapa which is characterized by brown or yellow seed coat coloration. Previous Brassica rapa studies have shown that BrTTG1 is responsible for seed coat proanthocyanidin formation, which is dependent on the MYB-bHLH-WD40 complex, whereas some studies have reported that TRANSPARENT TESTA GLABRA 1 (TTG1) directly interacts with the structural gene promoters of the flavonoid pathway.
Methods
Herein, the brown-seeded inbred B147 and ttg1 yellow-seeded inbred B80 mutants were used as plant materials for gene expression level analysis, gene promoter clone and transient overexpression.
Results
The analysis identified eleven structural genes involved in the flavonoid biosynthesis pathway, which are potentially responsible for BrTTG1- dependent seed coat proanthocyanidin formation. The promoters of these genes were cloned and cis-acting elements were identified. Yeast one-hybrid and dual-luciferase assays confirmed that BrTTG1 directly and independently interacted with proCHS-Bra008792, proDFR-Bra027457, proTT12-Bra003361, proTT19-Bra008570, proTT19-Bra023602 and proAHA10-Bra016610. A TTG1-binding motif (RTWWGTRGM) was also identified. Overexpression of TTG1 in the yellow-seed B. rapa inbred induced proanthocyanidin accumulation by increasing the expression levels of related genes.
Discussion
Our study unveiled, for the first time, the direct interaction between TTG1 and the promoters of the flavonoid biosynthesis pathway structural genes and glutathione S-transferases in Brassica rapa. Additionally, we have identified a novel TTG1-binding motif, providing a basis for further exploration into the function of TTG1 and the accumulation of proanthocyanidins in seed coats.
Keywords: BrTTG1, proanthocyanidins, flavonoid biosynthesis pathway, Brassica rapa L., promoters, gene overexpression
1. Introduction
Seed coat color is an important economic trait in Brassica crops and is closely associated with seed quality in terms of proanthocyanidins (PAs) (Ren et al., 2021), seed oil (Abbadi and Leckband, 2011), seed protein (Li et al., 2012), seed fiber (Wang et al., 2016), phenolic compound, and tannin contents (Zhai et al., 2020; Ding et al., 2021; Xie et al., 2020; Chao et al., 2022). Brassica rapa is a major rapeseed and vegetable crop in the Tibetan Plateau and northern China, with a short growth period and resistance to barren fields. B. rapa (AA = 20) is considered the original parent species of Brassica napus (AACC = 38) and Brassica juncea (AABB = 36) (Ren et al., 2021). Thus, the significance of studies on the seed coat color of B. rapa extends to other Brassica crops.
The pigment deposits responsible for seed coat color are attributed to the accumulation of PAs in the innermost cell layer of the testa (Debeaujon et al., 2000; Dixon et al., 2005; Lepiniec et al., 2006; Zhang et al., 2022). Several studies have shown that seed coat PA formation is directly controlled by structural genes involved in the flavonoid biosynthesis pathway (Akhov et al., 2009; Auger et al., 2010; Li et al., 2010; Lian et al., 2017; Ren et al., 2021; Xie et al., 2020). Early biosynthetic genes (EBGs) produce common precursors of PAs, and late biosynthetic genes (LBGs) are regulated by the ternary MYB-bHLH-WD40 complex (Xu et al., 2014). PA precursors are synthesized on the cytosolic surface of the endoplasmic reticulum and transported to the vacuole via glutathione S-transferases (GSTs) and membrane transporters regulated by genes such as including TT12, TT19, and AHA10 (Zhao, 2015).
TRANSPARENT TESTA GLABRA 1 (TTG1), a WD40 repeat protein involved in MYB-bHLH-WD40 (MBW) complexes, can regulate both the specific activity (i.e., interactions with other proteins or DNA) and quantity (e.g., stability and localization) of MBW complexes (Baudry et al., 2004; Koes et al., 2005; Thévenin et al., 2012; Xu et al., 2015) while being essential for preventing the effects of plant-specific negative regulators (Xu et al., 2015). Gonzalez et al. (2009) reported that the particular epidermal cell fate is specified by TTG1 in conjunction with a specific MYB and bHLH class transcription factor complex. Jia et al. (2021) showed that the tissue-specific pattern of PA biosynthesis relies more on R2R3-MYB factors, whereas TTG1 is an indispensable and constant regulator of MBW complexes.
Contrary to Ke et al. (2023) who showed that WD40 proteins lack transcriptional regulatory ability, we predicted and identified six TTG1-dependent structural genes involved in the flavonoid biosynthesis pathway. Yeast one-hybrid (Y1H) and dual-luciferase assays confirmed that TTG1 directly interacts with proCHS-Bra008792, proDFR-Bra027457, proTT12-Bra003361, proTT19-Bra008570, proTT19-Bra023602, and proAHA10-Bra016610 and induces their expression. Our study provides insights on the function of TTG1 and the PAs’ regulatory impact on seed coat color research.
2. Materials and methods
2.1. Plant materials and growth conditions
Brown-seeded inbred B147 and ttg1 yellow-seeded inbred B80 mutants were used as plant materials, and their seeds were sown in a greenhouse in the winter of 2016 for vernalization. Seedlings were subsequently transplanted and transferred to a plastic shed in the spring of 2017 for artificial pollination during blossom in Yangling, Shaanxi province, China. Seeds of different development stages were collected at 10, 14, 18, 22, 26, 30, and 34 days after flowering (DAF), frozen in liquid nitrogen, and immediately stored in a −80°C freezer (Sanyo, Japan). Three biological replicates were analyzed for each sample.
Leaves from tobacco (Nicotiana benthamiana) and yellow-seeded inbred B80 B. rapa ttg1 mutant plants were used for transient expression. Tobacco and B. rapa seeds were sown separately in the culture soil after soaking for 24 h. Fifteen-day-old tobacco seedlings were transplanted into plastic bowls (10 cm ×10 cm) and placed in the culture room under a 16-h light/8-h dark photoperiod, 22°C temperature and 60% humidity. Fourteen-day-old B. rapa seedlings were used for the transient expression analysis.
2.2. DNA, RNA extraction, and gene expression
Genomic DNA was extracted from fresh young leaves using a modified version of the cetyltrimethylammonium bromide method (Porebski et al., 1997). Total RNA extraction and cDNA synthesis were performed as previously described (Ren et al., 2017). Gene expression analysis was performed using quantitative real-time PCR (RT-qPCR), and a housekeeping gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH; GO0048316) was used as a reference gene (Li et al., 2016). The RT-qPCR reaction was performed in 20-µL volume in a Roche LightCycler 480 Real-Time PCR Detection System with three replications as previously described (Ren et al., 2017). Relative expression level of genes was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The specificity of primers ( Supplementary Table 1 ) used for the amplifications was confirmed by melting curve analysis.
2.3. Cloning and analysis of gene promoters
Based on the location of the ATG initiation codon of each gene in the BRAD database, 1,500- to 2,000-bp sequences upstream the ATG were screened to design promoter cloning primer ( Supplementary Table 2 ) using Primer Premier 5.0. PCR amplification procedure was performed in a 25-µL volume according to previously described instructions. A tail A was added to the purified PCR amplification products, which were subsequently cloned to the pMD19-T simple vector and transformed to Escherichia coli strain DH5α for sequencing. Promoter function was predicted using PlantCARE database online tools. Visualization of the cis-acting elements was performed using the online software Gene Structure Display Server.
2.4. Yeast one-hybrid assays
For the Y1H assays, promoters were inserted into the pAbAi vector, and, then, pAbAi-promoter recombination plasmids were transformed into the Y1H Gold yeast strain for aureobasidin A (AbA; Clontech, USA) concentration screening. Confirmed transformants were spread on SD/-Ura, SD/-Ura/AbA (150 ng/mL), and SD/-Ura/AbA (300 ng/mL) culture media, and the optimal AbA concentration was determined on the basis of colony growth.
The TTG1-ORF was integrated into the pGADT7 vector, and the pGADT7-TTG1 recombinant plasmid was transformed into a recombinant yeast strain containing pAbAi promoters. Recombinant transformants were spread on SD/-Leu/AbA culture medium, whereas SD/-Leu without AbA was used as a control. The interaction between BrTTG1 and its promoter was assessed on the basis of the colony growth. The combination of pGADT7-p53 + pAbAi-p53 was used as a positive control, and the combination of empty pGADT7 + pAbAi-promoters was used as a negative control. All primers used for recombinant plasmid construction in the Y1H assay are listed in Supplementary Table 3 .
2.5. Dual-luciferase assays
For the dual-luciferase assays, BrTTG1-ORF and promoters were introduced into pGreenII 62-SK and pGreenII 0800-LUC vectors by homologous recombination. The recombinant plasmids were then separately transformed into GV3101. pGreenII 62-SK-BrTTG1 was used as an effector, and promoter-pGreenII 0800-LUC was used as a reporter. All primers used for the construction of recombinant plasmids in the dual-luciferase assay are listed in Supplementary Table 4 .
Six independent tobacco (Nicotiana benthamiana) leaves were transfected during the transient expression assays. The transfected plants were kept in the dark overnight and then transferred to the culture room for 2 days. Luciferase activity was evaluated using a TransDetect Double-Luciferase Reporter Assay Kit (Promega, USA), and the regulatory relationship between TTG1 and the promoters was determined on the basis of the ratio of firefly luciferase activity to sea Renilla luciferase activity.
2.6. Promoters activity assays
Based on the distribution of cis-acting elements in the promoter sequences, several fragments of different lengths containing HindIII and SaII restriction sites were cloned and introduced into the pcambia1391-GUS vector by homologous recombination individually. All primers used for recombinant plasmid construction in promoter activity assays are listed in Supplementary Table 5 .
Transient expression was conducted in tobacco leaves, and six independent leaves were transfected as described above. The promoter: GUS-transfected leaves were dyed by soaking in β-glucuronidase (GUS) staining buffer [0.05% m/v 5-bromo-4-chloro-3-indolyl-β-glucuronide, 100 mM NaH2PO4·H2O, 10 mM EDTA, 0.5 mM K4FE(CN)6·3H2O, 0.1% Triton] and vacuum penetration for 30 min followed by staining at 37°C for 24 h. The stained leaves were soaked in 70% alcohol several times until they were completely decolorized, and photographs were taken. The reaction of GUS with 4-methylumbelliferyl β-D-glucuronide (MUG) resulted in the production of the fluorescent substance 4-methylumbelliferone (MU), and its content was determined by a fluorospectrophotometer. Thus, following the method described by Jefferson et al. (1987), the GUS content was quantified on the basis of the MU content of the plant total protein per minute. The protein content was determined using the Bradford method (Bradford, 1976). Our strategy followed the ones reported by Debeaujon et al. (2003); Thévenin et al. (2012), and Xu et al. (2014) with minor modifications. TTG1-binding motif enrichment was predicted using the MEME-ChiP (Bailey and Machanick, 2012).
2.7. Transient overexpression assays in B. rapa
The CDS of TTG1 was inserted into the pVBG2307 vector by homologous recombination, and the recombinant plasmid pVBG2307-TTG1 was transformed into GV3101 for transient injection into 14-day-old B. rapa cotyledons. GV3101 cells with an optical density (OD600) set to approximately 0.600 using a UV spectrophotometer (Thermo Fisher Scientific, USA) were injected into the leaves. Transformed plants were kept in the dark for 12 h, and, then, the light conditions were changed to 16-h light/8-h dark photoperiod for 2 days. Three individually transformed cotyledons and their corresponding leaves were collected for gene expression analysis and PA content determination.
2.8. Total proanthocyanidin analysis
Total PA extraction and content analysis were performed using the Plant PA Content Detection kit (Solarbio, China, BC1355). A quantity of 100 mg of dried transformed leaves was collected and dissolved in 1 mL of extract buffer, and, then, the ultrasonic method (300 W, 25°C, 30 min) was used for extraction followed by a centrifugation at 12,000 rpm at 25°C for 10 min. The supernatant was collected and diluted in 1 mL of extraction buffer, and the absorbance was measured at 500 nm.
2.9. Data statistics
All data calculations were performed using Microsoft Excel 2019, and column diagrams were drawn using Origin 2021. Significance analysis was performed using SPSS version 20.0.
3. Results
3.1. Prediction of structural genes involved in the BrTTG1-dependent flavonoid biosynthesis pathway
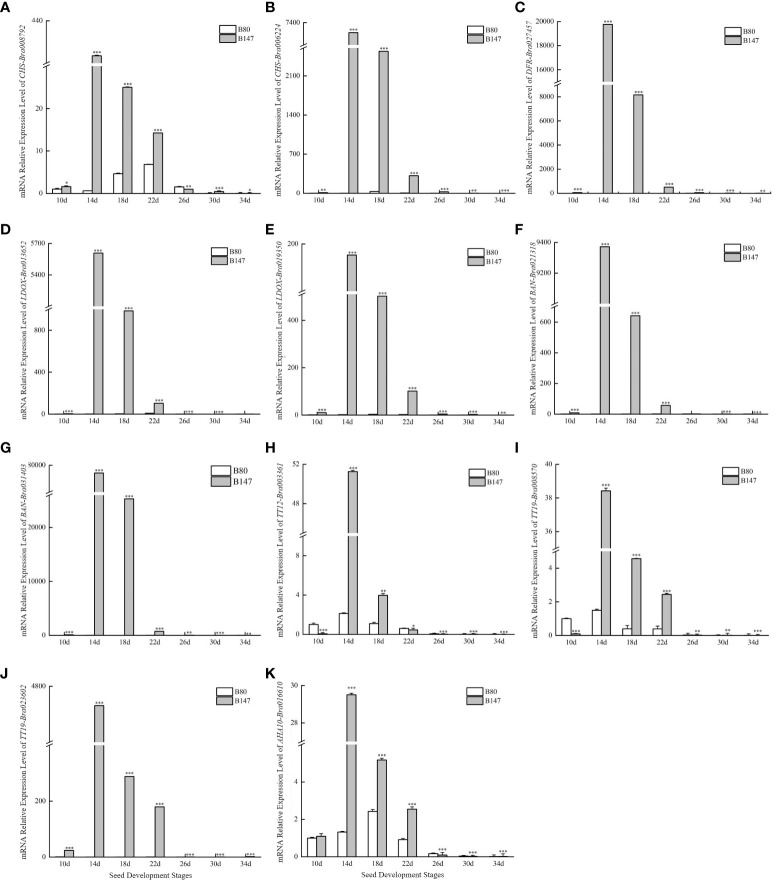
Seeds from brown-seeded inbred B147 and yellow-seeded inbred B80 plants collected at seven different developmental stages were used to predict the structural genes involved in the TTG1-dependent flavonoid biosynthesis pathway. Expression level analysis showed that 11 of the 22 structural genes had significant higher expression levels in B147 than in B80 at seven different developmental stages (10, 14, 18, 22, 26, 30, and 34 days after flowering) ( Figure 1 , Supplementary Figure 1 ). These were CHS-Bra008792, CHS-Bra006224, DFR-Bra027457, LDOX-Bra013652, LDOX-Bra019350, BAN-Bra021318, BAN-Bra031403, TT12-Bra003361, TT19-Bra008570, TT19-Bra023602, and AHA10-Bra016610. We speculated that these TTG1-dependent genes probably regulate seed coat PA formation in B. rapa.
Figure 1.
Expression levels of 11 structural genes involved in the flavonoid biosynthesis pathway dependent on TTG1 at seven different development stages seeds. (A) CHS-Bra008792; (B) CHS-Bra006224; (C) DFR-Bra027457; (D) LDOX-Bra013652; (E) LDOX-Bra019350; (F) BAN-Bra021318; (G) BAN-Bra031403; (H) TT12-Bra003361; (I) TT19-Bra008570; (J) TT19-Bra023602; (K) AHA10-Bra016610. ***represents significantly difference when p-value was 0.005.
3.2. Analysis of candidate target genes promoters
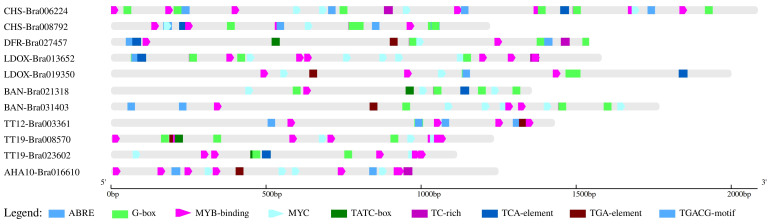
Based on the reference B. rapa Chiifu genome from the BRAD database, a total of 11 candidate target gene promoters of brown-seeded B147 plants were cloned and analyzed. These were proCHS-Bra008792, proCHS-Bra006224, proDFR-Bra027457, proLDOX-Bra013652, proLDOX-Bra019350, proBAN-Bra021318, proBAN-Bra031403, proTT12-Bra003361, proTT19-Bra008570, proTT19-Bra023602, and proAHA10-Bra016610 and had a length of 1,222 bp, 2,085 bp, 1,582 bp, 2,000 bp, 1,357 bp, 1,768 bp, 1,431 bp, 1,235 bp, 1,116 bp, and 1,249 bp, respectively. All promoter sequences are provided as Supplementary Materials . Analysis of these 11 promoter sequences revealed 98, 131, 77, 123, 140, 71, 102, 127, 100, 77, and 76 cis-acting elements, respectively. These elements are implicated in light response, circadian regulation, jasmonic acid response, salicylic acid response, regulation of zeolin metabolism, abscisic acid response, auxin-response, defense and stress responses, and anaerobic induction and constitute core elements of transcription initiation, promoter, and enhancer regions ( Figure 2 ).
Figure 2.
Cis-acting element analysis of 11 candidate target genes promoters.
Furthermore, considering that PA formation is regulated by the MYB and bHLH/MYC transcription factors, we analyzed their binding sites separately. Results revealed no MYC-binding sites in proTT12-Bra003361, whereas the remaining 10 promoters contained both MYB and MYC-binding sites.
3.3. Interaction analysis of TTG1 with the candidate target gene promoters
Eleven pAbAi promoter recombinant plasmid autoactivation assays were performed. Five recombinant yeast strains (pAbAi-CHS-Bra008792, pAbAi-BAN-Bra021318, pAbAi-BAN-Bra031403, pAbAi-LDOX-Bra019350, and pAbAi-TT19-Bra008570) failed to grow in the SD/-Ura/AbA (150 ng mL−1) culture medium. Six recombinant yeast strains, including pAbAi-CHS-Bra006224, pAbAi-DFR-Bra027457, pAbAi-LDOX-Bra013652, pAbAi-TT12-Bra003361, pAbAi-TT19-Bra023602, and pAbAi-AHA10-Bra016610, failed to grow in the SD/-Ura/AbA (300 ng mL−1) culture medium ( Supplementary Figure 2 ). Based on the autoactivation activity assay results, the interaction of TTG1 with the candidate target gene promoters was assessed using Y1H assays in two types of culture media. The results showed that TTG1 bound to proCHS-Bra008792, proDFR-Bra027457, proTT12-Bra003361, proTT19-Bra008570, proTT19-Bra023602, and proAHA10-Bra016610 ( Figures 3A, B ). A dual-luciferase assay was performed to confirm that TTG1 directly interacts with the above six promoters and induces their expression ( Figures 3C-I ).
Figure 3.
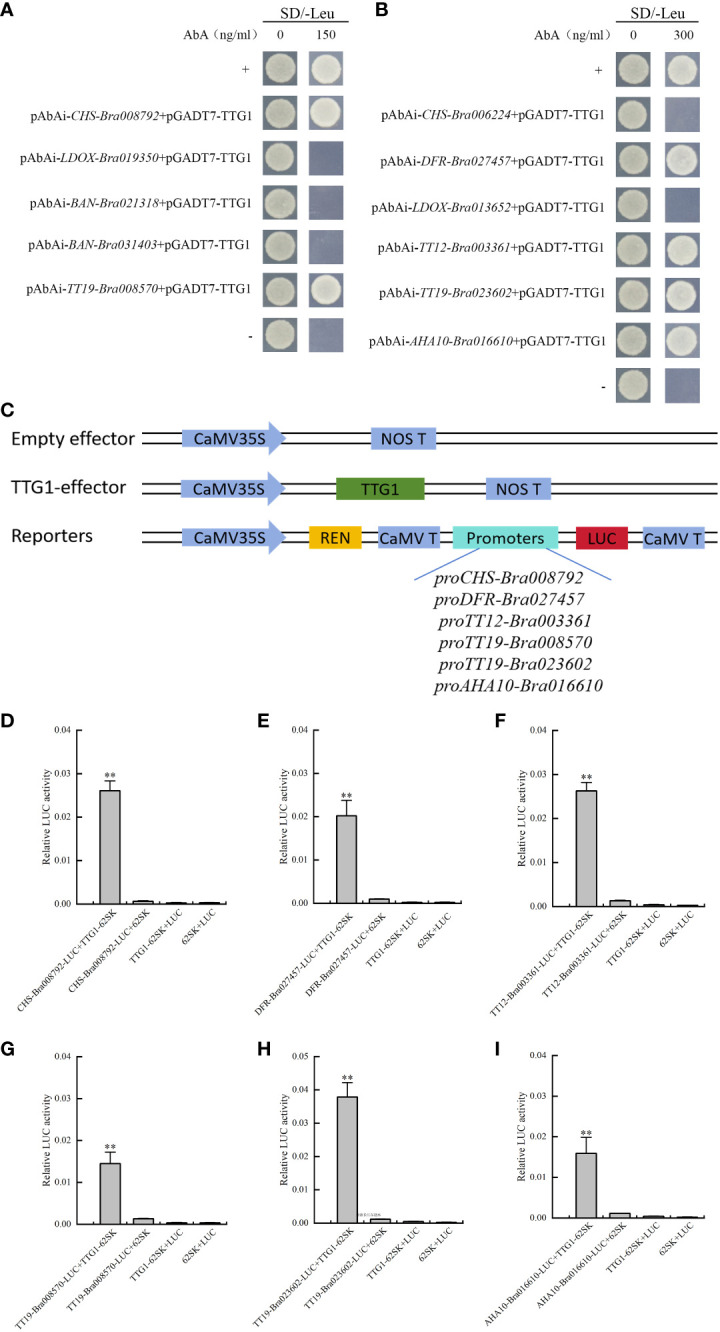
TTG1 directly regulates structural genes promoters of the flavonoid biosynthesis pathway and glutathione S-transferases. (A, B) TTG1 binding with proCHS-Bra008792, proDFR-Bra027457, proTT12-Bra003361, proTT19-Bra008570, proTT19-Bra023602, and proAHA10-Bra016610, separately. (C) Schematic diagrams of the effector and reporter plasmids used for the dual-luciferase assay. REN represents Renilla luciferase. LUC represents firefly luciferase. (D–I) TTG1 activates proCHS-Bra008792, proDFR-Bra027457, proTT12-Bra003361, proTT19-Bra008570, proTT19-Bra023602, and proAHA10-Bra016610 separately in dual-luciferase assay. The empty effector, empty reporters and empty effector with empty reporter were used as controls, separately. ** represents significantly difference when p-value was 0.01.
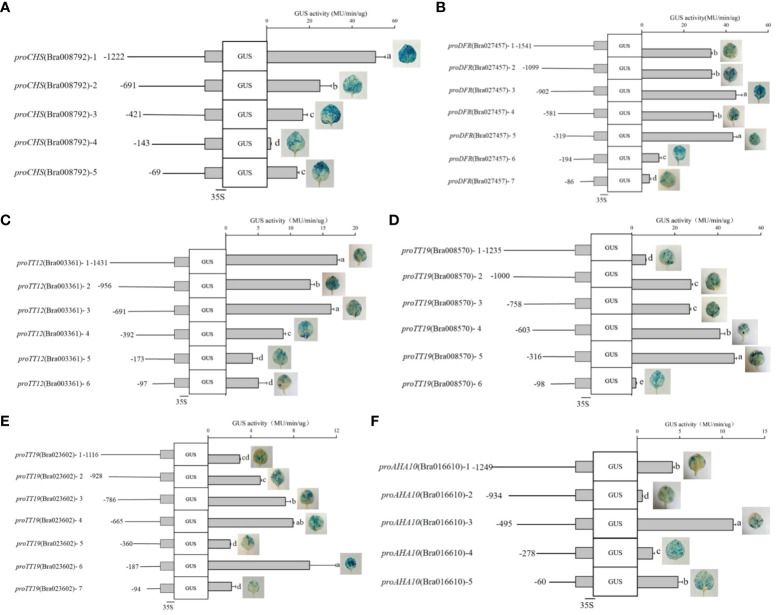
3.4. Analysis of the minimal active fragment of the promoter
To characterize the DNA regions involved in the regulation of proCHS-Bra008792, proDFR-Bra027457, proTT12-Bra003361, proTT19-Bra008570, proTT19-Bra023602, and proAHA10-Bra016610 activity by TTG1, transient transformation was performed in tobacco leaves. Based on the cis-acting element location in the promoter sequences, several fragments of different lengths were cloned into the pcambia1391-GUS vector for transient transformation, using GUS as a reporter. The key deletion fragments were characterized on the basis of GUS staining and the drastic changes in activity. GUS activity kept decreasing from proCHS-Bra008792-1 (−1,222 bp) to proCHS-Bra008792-4 (−143 bp) with the blue color of GUS staining gradually becoming lighter. Removal of an additional 278-bp fragment (proCHS-Bra008792-4) led to a strong decrease (~88.7%) in activity. These results indicate that this fragment contains the minimal information necessary to drive transcriptional activity. Intriguingly, proCHS-Bra008792-5 led to a stronger increase (~ 6.43-fold) compared to the GUS activity of proCHS-Bra008792-4, suggesting that some regions upstream of proCHS-Bra008792-5 have a negative impact on proCHS-Bra008792 activity ( Figure 4A ). Among the seven fragments of proDFR-Bra027457, GUS activity was higher than that of proDFR-Bra027457-1 (−1,541 bp) and proDFR-Bra027457-5 (−319 bp) and then decreased in proDFR-Bra027457-6 (−194 bp). Removal of an additional fragment of 125 bp (proDFR-Bra027457-6) led to a strong decrease (~81.2%) in the activity ( Figure 4B ), suggesting that this fragment contains the minimal information necessary to drive transcriptional activity. Analysis of six fragments of proTT12-Bra003361 led to the identification of a 219-bp promoter fragment from proTT12-Bra003361-4 (−392 bp) to proTT12-Bra003361-5 (−173 bp) ( Figure 4C ). Analysis of the proTT19-Bra008570 identified a 218-bp promoter fragment from proTT19-Bra008570-5 (−316 bp) to proTT19-Bra008570-6 (−98 bp), which led to a strong decrease (~95.9%) in activity ( Figure 4D ). Analysis of proTT19-Bra023602 identified a 219-bp promoter fragment from proTT19-Bra023602-4 (−665 bp) to proTT19-Bra023602-5 (−360 bp), which led to a strong decrease (~73.8%) in activity. The removal of an additional fragment of 173 bp of proTT19-Bra023602-5 led to a strong increase (~3.57-fold) in the GUS activity, suggesting that some regions upstream of proTT19-Bra023602-6 have a negative impact on proTT19-Bra023602-5 activity ( Figure 4E ). Five fragments were analyzed in proAHA10-Bra016610, and a 217-bp fragment deletion from proAHA10-Bra016610-3 (−495 bp) to proAHA10-Bra016610-4 led to a GUS activity decrease, whereas a 218-bp fragment deletion from proAHA10-Bra016610-4 (−316 bp) to proAHA10-Bra016610-5 led to a GUS activity increase ( Figure 4F ), indicating that the 218-bp fragment upstream of proAHA10-Bra016610-5 could possibly have an opposite effect with the 217-bp fragment upstream of proAHA10-Bra016610-4 in terms of driving the transcriptional activity.
Figure 4.
GUS staining and activity of the target gene promoters. (A) proCHS-Bra008792; (B) proDFR-Bra027457; (C) proTT12-Bra003361; (D) proTT19-Bra008570; (E) proTT19-Bra023602; (F) proAHA10-Bra016610. Left panels: The different promoters issued from the 5′-end deletion series were fused to the 35S cauliflower mosaic virus minimal promoter upstream of the GUS reporter gene. The transient expression assays was performed in tobacco leaves. Right panels: Column diagram represent GUS activity in transformed tobacco leaves driven by the different promoters fragment. Lowercase letters indicate significant difference.
Based on the GUS activity that was more than 5.0 MU/min/ug, motif enrichment analysis of promoter fragments was performed using MEME-ChiP to determine the sequence of TTG1-binding motifs. Five motifs were significantly enriched, including three novel motifs, a DOF-binding motif, and an NAC-binding motif ( Figure 5A ). Among these motifs, motif 2 (RTWWGTRGM) occurred one or two times per promoter; thus, novel motif 2 was considered the candidate TTG1-binding motif. The position of motif 2 in the promoters was analyzed and is shown in Figure 5B . Based on the combined results of GUS staining and GUS activity, fragments containing motif 2 exhibited higher GUS activity in proCHS-Bra008792-3 (−240 bp ~ −232 bp), proDFR-Bra027457-5 (−246 bp ~ −238 bp), proTT12-Bra003361-4 (−287 bp ~ −279 bp), and proTT19-Bra008570-5 (−215 bp ~ −207 bp). Interestingly, two motif 2 were detected in proTT19-Bra023602-5 (−287 bp to −279 bp and −166 bp to −158 bp) and proAHA10-Bra016610-3 (−456 bp to −448 bp and −148 bp to −140 bp), whose GUS activity differed. The proTT19-Bra023602-5 (−360 bp) containing two motif 2 showed lower GUS activity (2.07 ± 0.05 MU/min/ug), whereas proTT19-Bra023602-6 (−187 bp) containing one motif 2 showed higher GUS activity (9.48 ± 2.46 MU/min/ug). The proAHA10-Bra016610-3 (−495 bp) containing two motif 2 showed higher GUS activity (11.29 ± 0.32 MU/min/ug), whereas proAHA10-Bra016610-4 (-278 bp) containing one motif 2 showed lower GUS activity (1.82 ± 0.23 MU/min/ug).
Figure 5.
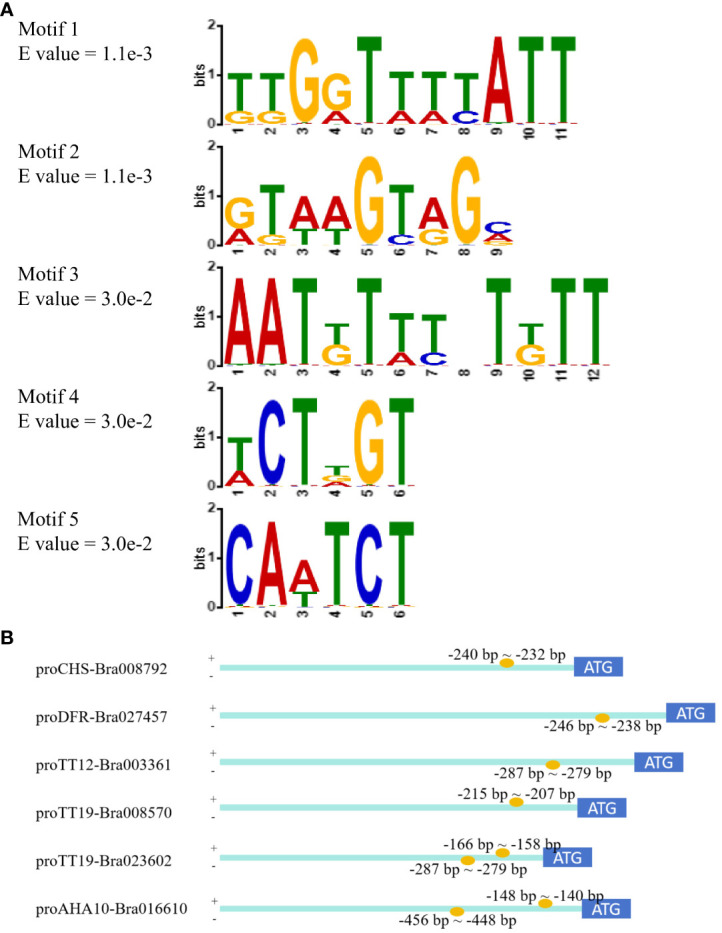
TTG1-binding motifs enrichment analysis by MEME-ChiP. (A) Five enriched motifs. (B) Positions of motif 2 in promoters.
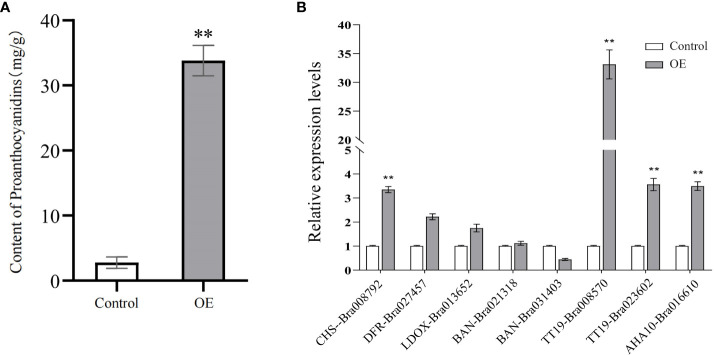
3.5. Overexpression of BrTTG1 enhanced proanthocyanidin formation
To clarify the effect of BrTTG1 on B. rapa seed coat color and PA formation, a BrTTG1 overexpression assay in B. rapa seedling cotyledons was carried out. The PAs in untransformed cotyledons were 2.78 ± 0.88 mg/g DW, whereas PAs in transformed cotyledons were 33.80 ± 3.30 mg/g DW. The PAs in the transformed seedling cotyledons were significantly higher than those in the control ( Figure 6A ). In addition, the expression levels of eight genes involved in the flavonoid biosynthesis pathway and Glutathione S-transferases were determined using qRT-PCR. Five target genes including CHS-Bra008792, DFR-Bra027457, TT19-Bra008570, TT19-Bra023602, and AHA10-Bra016610 exhibited a significantly increased expression than that in control, whereas LDOX-Bra13652, BAN-Bra021318, and BAN-Bra031403 exhibited no significant changes ( Figure 6B ).
Figure 6.
Overexpression of BrTTG1 enhanced proanthocyanidin formation. (A) The proanthocyanidin content in transformed seedling cotyledons. (B) Expression levels of eight genes involved in flavonoid biosynthesis pathway in transformed seedling cotyledons. ** represents significantly difference when p-value was 0.01.
4. Discussion
TTG1 was first reported almost half a century ago (Koornneef and Van der Veen, 1978) and has been demonstrated to be involved in the regulation of trichome initiation, seed coat mucilage biosynthesis (Koornneef, 1981), root hair formation (Galway et al., 1994; Zhang et al., 2009), seed development, and PA biosynthesis (Western et al., 2001; Debeaujon et al., 2003; Zhang et al., 2009; Chen et al., 2015; Ren et al., 2017; Yuan et al., 2019). PAs, the end products of the flavonoid biosynthetic pathway, are deposited in the innermost cell layer of the testa giving a characteristic seed coat pigment. Xu et al. (2014) quantified the mRNA accumulation levels of 12 characterized flavonoid biosynthetic genes, leading to PA accumulation between WT and ttg1 mutants in Arabidopsis and found significant decreases in F3H, DFR, LDOX, BAN, TT12, TT19, and AHA10 mRNA accumulation in ttg1 mutants compared with the WT, whereas the mRNA accumulation levels of CHS, CHI, and TT15 increased in ttg1 mutants. Ren et al. (2021) showed that the expression levels of EBGs in the flavonoid biosynthetic pathway were downregulated, whereas LBGs were hardly or not expressed at all in ttg1 mutant seeds at 10, 14, and 28 days after flowering in Brassica rapa. Here, seeds at seven different developmental stages were selected to analyze the mRNA accumulation levels of 22 characterized flavonoid biosynthetic genes in B. rapa, and 11 genes showed significantly higher expression levels in B147 than in B80 at seven different developmental stages. These results indicate that different copies of the same gene may perform different functions.
During PA formation, TTG1 interacts with R2R3 MYB transcription factors [TRANSPARENT TESTA 2 (TT2), MYB5, MYBPA1, and MYBPA2] and bHLH transcription factors (TT8, GL3, and EGL3) to regulate the expression of LBGs in the flavonoid biosynthesis pathway (Nesi et al., 2000; Baudry et al., 2004; Xu et al., 2014, 2015; Li et al., 2020; Tian and Wang, 2020; Rajput et al., 2022). Anthocyanin biosynthesis and PA biosynthesis possess the same EBGs and LBGs. Jia et al. (2021) showed that light-induced DcTTG1 regulates anthocyanin biosynthesis in Dendrobium candidum by binding to the promoters of DcCHS2, DcCHI, DcF3H, and DcF3′H and no direct binding of TTG1 to late anthocyanin biosynthetic gene promoters was observed. In this study, we found that BrTTG1 directly binds to the promoters of the flavonoid biosynthesis pathway and glutathione S-transferases, including CHS-Bra008792, DFR-Bra027457, TT12-Bra003361, TT19-Bra008570, TT19-Bra023602, and AHA10-Bra016610 and regulates PA formation in the presence of MYB and bHLH. Ke et al. (2023) showed that WD40 proteins stabilize the interaction between MYB and bHLH TFs but lack transcriptional regulatory ability. We showed that BrTTG1 directly bound to the promoters of the flavonoid biosynthesis pathway and promoted PA formation.
During the analysis of the minimally active fragments of promoters, changes in GUS activity did not show an obvious pattern as the promoter fragment decreased, which was similar to the results of the MBW target gene promoter activity analysis reported by Xu et al. (2014). After TTG1-binding motif enrichment, fragments with two motif 2 in proTT19-Bra023602 and proAHA10-Bra016610 showed different GUS activities. We subsequently searched for cis-elements in the fragments with two motif 2 in proTT19-Bra023602 and proAHA10-Bra016610. The results showed that MYB-binding sites and bHLH recognition sites were found in proTT19-Bra023602-6 and proAHA10-Bra016610-3 fragments with higher GUS activity. No MYB-binding sites or bHLH recognition sites were found in the lower GUS activity fragments of proAHA10-Bra016610-4. This finding suggests that the GUS activity of these promoters may be affected by the presence or absence of MYB and bHLH or by other unknown elements.
Our findings indicate that BrTTG1 regulates seed coat PA formation through a direct interaction with structural gene promoters of the flavonoid biosynthesis pathway and glutathione S-transferases.
5. Conclusion
BrTTG1 directly interacts with structural gene promoters of the flavonoid pathway and glutathione S-transferases to regulate seed coat PA formation in B. rapa. A TTG1-binding motif (RTWWGTRGM) was identified. Overexpression of BrTTG1 in yellow seed B. rapa inbred plants induced PA accumulation by increasing the expression levels of target genes. Our study revealed, for the first time, a direct interaction between TTG1 and structural gene promoters of the flavonoid biosynthesis pathway and glutathione S-transferases in B. rapa and predicted a novel TTG1-binding motif. The above findings provide insights and could be the basis for future studies aiming to study the TTG1 function and PA accumulation in seed coats.
Data availability statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Author contributions
WZ: Conceptualization, Methodology, Software, Validation, Visualization, Writing – original draft. XL: Conceptualization, Methodology, Software, Writing – original draft. JW: Conceptualization, Writing – review & editing. QL: Writing – review & editing. SB: Methodology, Visualization, Writing – review & editing. YR: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing.
Acknowledgments
We would like to thank Shaanxi AUG Biotechnology Co., Ltd., for technological supporting and all the participants in this study.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (31960602), Project of Qinghai Provincial Science and Technology Department (2021-ZJ-763), and National Key Research and Development Program (2022YFD1602400).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1372477/full#supplementary-material
Expression levels of eleven structural genes involved in the flavonoid biosynthesis pathway independent on TTG1 at seven different development stages seeds. (A) CHS-Bra023441; (B) CHI-Bra007142; (C) CHI-Bra007145; (D) CHI-Bra003209; (E) F3H-Bra036828; (F) F3’H-Bra009312; (G) TT10-Bra037510; (H) TT15-Bra003021; (I) TT15-Bra035004; (J) TT15-Bra038445; (K) TT15-Bra023594.
AbA concentration screening of eleven pAbAi-promoters recombination plasmids.
References
- Abbadi A., Leckband G. (2011). Rapeseed breeding for oil content, quality, and sustainability. Eur. J. Lipid Sci. Tec. 113, 1198–1206. doi: 10.1002/ejlt.201100063 [DOI] [Google Scholar]
- Akhov L., Ashe P., Tan Y., Datla R., Selvaraj G. (2009). Proanthocyanidin biosynthesis in the seed coat of yellow-seeded, canola quality Brassica napus YN01-429 is constrained at the committed step catalyzed by dihydroflavonol 4-reductase. Botany 87, 616–625. doi: 10.1139/B09-036 [DOI] [Google Scholar]
- Auger B., Marnet N., Gautier V., Maia Grondard A., Leprince F., Renard M., et al. (2010). A detailed survey of seed coat flavonoids in developing seeds of Brassica napus L. J. Agri Food Chem. 58 (10), 6246–6256. doi: 10.1021/jf903619v [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Machanick P. (2012). Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 40, e128. doi: 10.1093/nar/gks433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A., Heim M. A., Debreucq B., Caboche M., Weisshaar B., Leprince F. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana . Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Chao H., Guo L., Zhao W., Li H., Li M. (2022). A major yellow-seed QTL on chromosome A09 significantly increases the oil content and reduces the fiber content of seed in Brassica napus. Theoret. Appl. Genet. 135, 1293–1305. doi: 10.1007/s00122-022-04031-0 [DOI] [PubMed] [Google Scholar]
- Chen M., Zhang B., Li C., Kulaveerasingam H., Chew F. T., Yu H. (2015). TRANSPARENT TESTA GLABRA1 regulates the accumulation of seed storage reserves in Arabidopsis . Plant Physiol. 169, 391–402. doi: 10.1104/pp.15.00943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I., Leon-Kloosterziel K. M., Koornneef M. (2000). Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis . Plant Physiol. 122, 403–413. doi: 10.1104/pp.122.2.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I., Nesi N., Perez P., Devic M., Grandjean O., Caboche M., et al. (2003). Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15 (11), 2514–2531. doi: 10.1105/tpc.014043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Yu S., Wang J., Li M., Qu C., Li J., et al. (2021). Comparative transcriptomic analysis of seed coats with high and low lignin contents reveals lignin and flavonoid biosynthesis in Brassica napus . BMC Plant Biol. 21, 1–16. doi: 10.1186/s12870-021-03030-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. A., Xie D. Y., Sharma S. B. (2005). Proanthocyanidins-a final frontier in flavonoid research. New Phytol. 165, 9–28. doi: 10.1111/j.1469-8137.2004.01217.x [DOI] [PubMed] [Google Scholar]
- Galway M. E., Masucci J. D., Lloyd A. M., Walbot V., Davis R. W., Schiefelbein J. W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166, 740–754. doi: 10.1006/dbio.1994.1352 [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Mendenhall J., Huo Y., Lloyd A. (2009). TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 325, 412–421. doi: 10.1016/j.ydbio.2008.10.005 [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/embj.1987.6.issue-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N., Wang J., Wang Y., Ye W., Liu J., Jiang J., et al. (2021). The light induced WD40-repeat transcription factor dcTTG1 regulates anthocyanin biosynthesis in dendrobium candidum. Front. Plant Sci. 12, 633333. doi: 10.3389/fpls.2021.633333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S., Jiang Y., Zhou M., Li Y. (2023). Genome-wide identification, evolution, and expression analysis of the WD40 subfamily in oryza genus . Int. J. Mol. Sci. 24, 15776. doi: 10.3390/ijms242115776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R., Verweij W., Quattrocchio F. (2005). Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242. doi: 10.1016/j.tplants.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Koornneef M. (1981). The complex syndrome of ttg mutants. Arab. Inf. Serv. 18, 45–51. [Google Scholar]
- Koornneef M., Van der Veen J. H. (1978). Gene localization with trisomics in Arabidopsis thaliana . Arab. Inf. Serv. 15, 38–43. [Google Scholar]
- Lepiniec L., Debeaujon I., Routaboul J. M., Baudry A., Pourcel L., Nesi N., et al. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57, 405–430. doi: 10.1146/annurev.arplant.57.032905.105252 [DOI] [PubMed] [Google Scholar]
- Li S. F., Allen P. J., Napoli R. S., Browne R. G., Pham H., Parish R. W. (2020). MYB-bHLH-TTG1 regulates Arabidopsis seed coat biosynthesis pathways directly and indirectly via multiple tiers of transcription factors. Plant Cell Physiol. 61, 1005–1018. doi: 10.1093/pcp/pcaa027 [DOI] [PubMed] [Google Scholar]
- Li Q. F., Zhao J., Zhang J., Dai Z. H., Zhang L. G. (2016). Ectopic expression of the Chinese cabbage malate dehydrogenase gene promotes growth and aluminum resistance in Arabidopsis . Front. Plant Sci. 7, 1180. doi: 10.3389/fpls.2016.01180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Chen L., Hong M. Y., Zhang Y., Zu F., Wen J., et al. (2012). A large insertion in bHLH transcription factor BrTT8 resulting in yellow seed coat in Brassica rapa . PloS One 7, e44145. doi: 10.1371/journal.pone.0044145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Westcott N., Links M., Gruber M. Y. (2010). Seed coat phenolics and the developing silique transcriptome of Brassica carinata . J. Agric. Food Chem. 58, 10918–10928. doi: 10.1021/jf102208a [DOI] [PubMed] [Google Scholar]
- Lian J., Lu X., Yin N., Ma L., Lu J., Liu X., et al. (2017). Silencing of BnTT1 family genes affects seed flavonoid biosynthesis and alters seed fatty acid composition in Brassica napus . Plant Sci. 254, 32–47. doi: 10.1016/j.plantsci.2016.10.012 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Nesi N., Debeaujon I., Jond C., Pelletier G., Caboche M., Lepiniec L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863–1878. doi: 10.1105/tpc.12.10.1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porebski S., Bailey L. G., Baum B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15. doi: 10.1007/BF02772108 [DOI] [Google Scholar]
- Rajput R., Naik J., Stracke R., Pandey. A. (2022). Interplay between R2R3 MYB-type activators and repressors regulates proanthocyanidin biosynthesis in banana (Musa acuminata). New Phytolo. 236 (3), 1108–1127. doi: 10.1111/nph.18382 [DOI] [PubMed] [Google Scholar]
- Ren Y. J., He Q., Ma X. M., Zhang L. G. (2017). Characteristics of color development in seeds of brown- and yellow-seeded heading Chinese cabbage and molecular analysis of Brsc the candidate gene controlling seed coat color. Front. Plant Sci. 8, 1410. doi: 10.3389/fpls.2017.01410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Zhang N., Li. R., Ma. X., Zhang L. (2021). Comparative transcriptome and flavonoids components analysis reveal the structural genes responsible for the yellow seed coat color of Brassica rapa L. PeerJ 9, e10770. doi: 10.7717/peerj.10770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenin J., Dubos C., Xu W., Le Gourrierec J., Kelemen Z., Charlot F., et al. (2012). A new system for fast and quantitative analysis of heterologous gene expression in plants. New Phytol. 193, 504–512. doi: 10.1111/j.1469-8137.2011.03936.x [DOI] [PubMed] [Google Scholar]
- Tian H., Wang S. (2020). TRANSPARENT TESTA GLABRA1, a key regulator in plants with multiple roles and multiple function mechanisms. Int. J. Mol. Sci. 21, 4881. doi: 10.3390/ijms21144881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. H., Xiao L., Guo S. M., An F. Y., Du D. Z. (2016). Fine mapping and whole-genome resequencing identify the seed coat color gene in Brassica rapa . PloS One 11, e0166464. doi: 10.1371/journal.pone.0166464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western T. L., Burn J., Tan W. L., Skinner D. J., Martin-McCaffffrey L., Moffatt B. A., et al. (2001). Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis . Plant Physiol. 127, 998–1011. doi: 10.1104/pp.010410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T., Chen X., Guo T., Rong H., Chen Z., Sun Q., et al. (2020). Targeted knockout of BnTT2 homologues for yellow-seeded Brassica napus with reduced flavonoids and improved fatty acid composition. J. Agric. Food Chem. 68, 5676–5690. doi: 10.1021/acs.jafc.0c01126 [DOI] [PubMed] [Google Scholar]
- Xu W., Dubos C., Lepiniec L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Xu W., Grain D., Bobet S., Le Gourrierec J., Thevenin J., kelemen Z., et al. (2014). Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytolo. 202, 132–144. doi: 10.1111/nph.12620 [DOI] [PubMed] [Google Scholar]
- Yuan F., Leng B., Zhang H., Wang X., Han G., Wang B. (2019). A WD40-repeat protein from the Recretohalophyte Limonium bicolor enhances trichome formation and salt tolerance in Arabidopsis . Front. Plant Sci. 10, 1456. doi: 10.3389/fpls.2019.01456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Yu K., Cai S., Hu L., Amoo O., Xu L., et al. (2020). Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 18, 1153–1168. doi: 10.1111/pbi.13281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Lu Y., Yuan Y., Geng J., Chen Y., Cloutier S., et al. (2009). Map-based cloning and characterization of a gene controlling hairiness and seed coat color traits in Brassica rapa . Plant Mol. Biol. 69, 553–563. doi: 10.1007/s11103-008-9437-y [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang H., Zhao H., Xia Y., Zheng X., Fan R., et al. (2022). Multi-omics analysis dissects the genetic architecture of seed coat content in Brassica napus . Genome Biol. 23, 86. doi: 10.1186/s13059-022-02647-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. (2015). Flavonoid transport mechanisms: how to go, and with whom. Trends Plant Sci. 20, 576–585. doi: 10.1016/j.tplants.2015.06.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression levels of eleven structural genes involved in the flavonoid biosynthesis pathway independent on TTG1 at seven different development stages seeds. (A) CHS-Bra023441; (B) CHI-Bra007142; (C) CHI-Bra007145; (D) CHI-Bra003209; (E) F3H-Bra036828; (F) F3’H-Bra009312; (G) TT10-Bra037510; (H) TT15-Bra003021; (I) TT15-Bra035004; (J) TT15-Bra038445; (K) TT15-Bra023594.
AbA concentration screening of eleven pAbAi-promoters recombination plasmids.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.