In the December 1997 issue, a report by Desrosiers et al. (2) described a novel herpesvirus in rhesus macaques (named RRV for rhesus rhadinovirus) with similarity to human herpesvirus 8 (HHV-8 or KSHV for Kaposi’s sarcoma-associated herpesvirus). In this article the authors listed various diseases that occur in macaques, including lymphomas and lymphoproliferative diseases, and speculated that “a causative role for RRV in these or other diseases would greatly facilitate studies of pathogenesis and vaccine and drug development.” Earlier the same year another report, by Rose et al. (4), described molecular sequences of two gammaherpesviruses in macaques associated with a proliferative disease in macaques called retroperitoneal fibromatosis (RF). This article also described the close genetic similarities between these new viruses (called RFHV for RF-associated herpesvirus) and HHV-8. HHV-8 is strongly implicated as a necessary cofactor in human Kaposi’s sarcoma (reference 1 and references therein). Therefore, availability of a monkey model in which pathogenesis with related viruses can be studied could prove to be very valuable. It is important to properly describe the genetic and functional relationships between these gammaherpesviruses. A comparative analysis of the available sequences is presented here.
In comparing the published sequences from RFHVMn, RFHVMm (RFHV variants from Macaca nemestrina and Macaca mulatta, respectively) and RRV, we noticed that contrary to our expectations RRV is not identical to RFHVMm despite the fact that both are gammaherpesviruses found in rhesus macaques. This finding has prompted us to evaluate the phylogenetic relationships between these three gammaherpesviruses from macaques and other gammaherpesviruses. Tables 1 and 2 list nucleotide and amino acid sequence identities between fragments of the DNA polymerase and glycoprotein B gene fragments of primate gammaherpesviruses, including the newly described RFHVMm, RFHVMn, and RRV. Based on these comparisons, HHV-8 appears to be more closely related to RFHVMm and RFHVMn than to RRV.
TABLE 1.
Nucleotide and amino acid identities between DNA polymerase gene fragments of primate gammaherpesviruses
| Nucleotide (amino acid) identities
|
|||||
|---|---|---|---|---|---|
| HHV-8 | RFHVMm | RFHVMn | RRV | HVS | |
| RFHVMm | 69 (83) | ||||
| RFHVMn | 71 (84) | 83 (90) | |||
| RRV | 68 (74) | 66 (66) | 66 (68) | ||
| HVS | 62 (67) | 60 (63) | 59 (64) | 60 (66) | |
| EBV | 62 (62) | 57 (62) | 64 (63) | 63 (58) | 52 (59) |
TABLE 2.
Nucleotide and amino acid identities between glycoprotein B gene fragments (319 bp) of primate gammaherpesviruses
| Nucleotide (amino acid) identities
|
|||||
|---|---|---|---|---|---|
| HHV-8 | RFHVMm | RFHVMn | RRV | HVS | |
| RFHVMm | 75 (86) | ||||
| RFHVMn | 77 (86) | 92 (93) | |||
| RRV | 71 (80) | 67 (75) | 68 (79) | ||
| HVS | 59 (57) | 58 (53) | 59 (56) | 54 (54) | |
| EBV | 53 (41) | 49 (39) | 50 (42) | 52 (42) | 48 (46) |
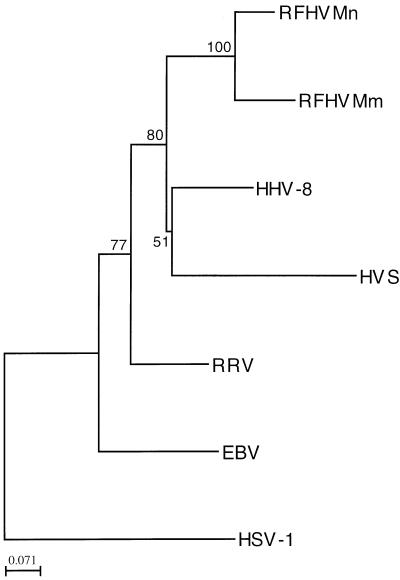
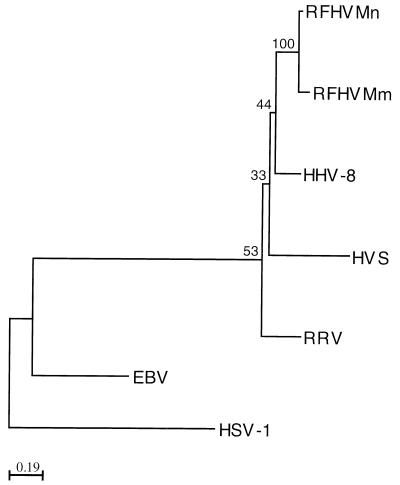
In Fig. 1 and 2 we present phylogenetic trees based on the same nucleotide sequences as used in Table 1, generated using the programs PROTDIST and NEIGHBOR (neighbor-joining method) from the Phylip package for computerized phylogenetic analysis (University of Washington, Seattle). In this tree, RFHVMm and RFHVMn cluster with HHV-8, whereas RRV branches off independently. Numbers at the branch points indicate bootstrap values obtained through implementation of the SEQBOOT and CONSENSE programs of the Phylip package. Despite the limited amount of sequence information, these values show that the branch points for RFHVMm and RFHVMm, HHV-8, and RRV can be reliably assigned, at least for the DNA polymerase gene sequences. The tree based on the glycoprotein B sequences has a topology similar to the DNA polymerase tree.
FIG. 1.
Phylogenetic tree of selected herpesvirus DNA polymerase gene fragments. Abbreviations: EHV-2, equine herpesvirus 2; HVS, herpesvirus saimiri; MHV68, murine herpesvirus 68; EBV, Epstein-Barr virus; HSV-1, herpes simplex virus type 1; CMV, human cytomegalovirus.
FIG. 2.
Phylogenetic tree of selected herpesvirus glycoprotein B gene fragments. See legend to Fig. 1 for abbreviations. GenBank accession numbers for RFHVMm and RFHVMn glycoprotein B gene sequences are AF061447 and AF061448 (5).
These data suggest the existence of two phylogenetically distinct groups of gammaherpesviruses in macaques, both of which are related to HHV-8. Based on the limited sequence comparisons presented here, the members of the RFHV group seem most closely related to HHV-8 and more sequence information in particular from the RFHV variants is needed to confirm this observation. Both RFHV variants were first detected in tissue sections from macaque RF. Similarities between RF and Kaposi’s sarcoma have been noted in the past (3), with the suggestion that RF in macaques could serve as a useful animal model to study Kaposi’s sarcoma pathogenesis. The high degree of similarity between HHV-8 and both RFHV variants supports that observation. At this point, no disease has been described in association with RRV and it will be interesting to see whether RRV infection in macaques is associated with pathogenesis related to that seen associated with HHV-8 infection in humans.
Footnotes
Ed. note: The author of the published article declined to reply.
REFERENCES
- 1.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Desrosiers R C, Sasseville V G, Czajak S Z, Zhang X, Mansfield K G, Kaur A, Johnson R P, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to the human Kaposi’s sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giddens W E, Jr, Tsai C-C, Morton W R, Ochs H D, Knitter G H, Blakley G A. Retroperitoneal fibromatosis and acquired immunodeficiency syndrome in macaques: pathologic observations and transmission studies. Am J Pathol. 1985;119:253–263. [PMC free article] [PubMed] [Google Scholar]
- 4.Rose T M, Strand K B, Schultz E R, Schaefer G, Rankin G W, Thouless M E, Tsai C-C, Bosch M L. Identification of two homologs of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strand, K., et al. Unpublished data.