Abbreviations
- CXCL13
C‐X‐C Motif Chemokine Ligand 13
- CXCR2
C‐X‐C Chemokine Receptor 2
- E‐TLS
Early Tertiary Lymphoid Structures
- FL‐TLS
Follicle‐Like Tertiary Lymphoid Structures
- H&E
Hematoxylin‐and‐Eosin
- HEV
High Endothelial Venules
- HLA
Human Leukocyte Antigen
- ICB
Immune Checkpoint Blockade
- MHC‐II
Major Histocompatibility Complex II
- NSCLC
Non‐Small Cell Lung Cancer
- PD‐L1
Programmed Death‐Ligand 1
- PD1
Programmed Cell Death Protein 1
- PMN‐MDSCs
Polymorphonuclear Myeloid‐Derived Suppressor Cells
- PNAd
Peripheral Node Addressin
- ROI
Regions of Interest
- TANs
Tumor‐Associated Neutrophils
- TLS
Tertiary Lymphoid Structures
- Treg
Regulatory T
Dear Editor,
Tertiary lymphoid structures (TLSs) are organized clusters of immune cells found in non‐lymphoid tissues of chronic inflammation, including solid tumors [1]. High endothelial venules (HEVs) positive for peripheral node addressin (PNAd) are present in TLSs and may provide highway entry for lymphocytes. Tumor‐residing TLSs may be formed by the concerted actions of cytokines and chemokines, a finding supported by the relation between a 12‐chemokine signature and TLSs [2]. C‐X‐C motif chemokine ligand 13 (CXCL13) is central in TLS formation in tumors, including bladder cancer [3]. TLSs are associated with favorable prognoses in cancer and favorable responses to immune checkpoint blockade (ICB) independent of programmed death‐ligand 1 (PD‐L1) status [4]. TLSs may also contain immunosuppressive cells such as regulatory T cells and regulatory B cells. A prominent immunosuppressive population is polymorphonuclear myeloid‐derived suppressor cells (PMN‐MDSCs), which are essentially tumor‐associated neutrophils (TANs) that suppress effector T cells [5]. The relative distribution of TLSs and TANs/PMN‐MDSCs has not been studied in human cancers.
We designed a study to investigate the distribution of immune cells (especially TANs/PMN‐MDSCs) inside and near the TLSs of bladder cancer (the 10th most common cancer worldwide) and to evaluate the prognostic significance of TLSs and TANs/PMN‐MDSCs in bladder cancer patients treated with ICB therapy. We performed a retrospective study using formalin‐fixed paraffin‐embedded samples from 26 primary bladder cancers (Supplementary Table S1, Supplementary Methods). Samples were stained with hematoxylin‐and‐eosin (H&E) to recognize 58 TLS regions of interest (ROIs), which were further stained with 14 cell markers with MultiOmyx (Figure 1A, Supplementary Table S2). The co‐staining patterns allowed the identification of various immune populations, including TANs (Supplementary Figure S1A, Supplementary Table S3). The 58 TLSs were classified into 23 early TLSs (E‐TLSs) and 35 follicle‐like TLSs (FL‐TLSs) based on the morphology: E‐TLSs contained less organized B cells and T cells in an irregularly shaped region and very few (or no) PNAd+ HEVs, whereas FL‐TLSs had demarcated a B‐cell zone and a T‐cell zone in a follicular structure interspersed with several HEVs (Figure 1B). As expected, FL‐TLSs contained higher densities of B cells and extended larger areas than E‐TLSs (Figure 1C).
FIGURE 1.
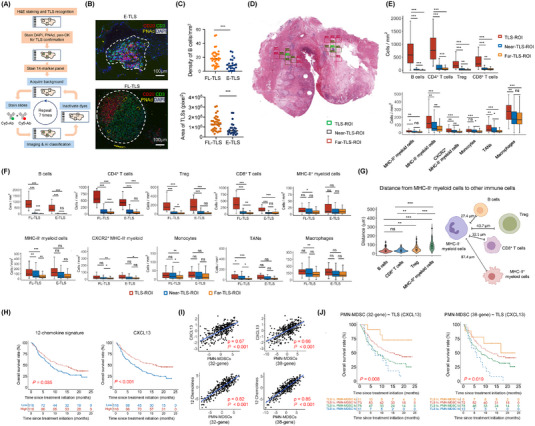
MultiOmyx and in silico analysis of TLSs and immune populations in bladder cancer. (A) Project workflow with MultiOmyx. (B) Representative MultiOmyx images of E‐TLS and FL‐TLS. TLS region was contoured with white dash lines. (C) Densities of B cells in E‐TLS and FL‐TLS in TLS‐containing ROIs (upper). TLS area sizes of E‐TLS and FL‐TLS in TLS‐ROIs (lower). P values were calculated with Mann Whitney U test. (D) Representative H&E section marked with two TLS‐ROIs (green) surrounded by multiple near‐TLS‐ROIs (purple) and far‐TLS‐ROIs (pink). (E) Comparison of the densities of various lymphocyte and myeloid populations in TLS‐ROIs, near‐TLS‐ROIs and far‐TLS‐ROIs. P values were calculated with Kolmogorov‐Smirnov test. The box limits are the lower and upper quantiles, and the whiskers extend to the most extreme values within 1.5 x IQR. (F) More refined comparison of the densities of lymphocyte and myeloid populations based on both ROI types (TLS, near, far) and TLS stages (E‐TLS, FL‐TLS). The box limits are the lower and upper quantiles, and the whiskers extend to the most extreme values within 1.5 x IQR. (G) Violin plots showing the distances of MHC‐II− myeloid cells to B cells, CD8+ T cells, Treg and MHC‐II+ myeloid cells in the TLS‐ROIs, with the schematic illustrating the median distances. P values were calculated with Kolmogorov‐Smirnov test. (H) Overall survival of the upper and lower tertiles of patients in IMvigor210 based on 12‐chemokine genes signatures or CXCL13 expression. Log‐rank test P values are marked. (I) Correlation of CXCL13 gene expression or 12‐chemokine gene signature expression with either of the two PMN‐MDSC gene signatures based on the IMvigor210 dataset. Spearman correlation coefficient ρ and P values are marked. (J) Kaplan‐Meier analysis for overall survival of patients in IMvigor210 classified into four groups based on CXCL13 expression (TLS marker) and either of the two PMN‐MDSC gene signatures. Upper and lower tertiles were classified as high (hi) and low (lo) for the signatures, respectively. P values based on two‐sided log‐rank tests are marked. In all the tests, * P < 0.05, ** P < 0.01, *** P < 0.001; ns, P > 0.05. AI, artificial intelligence, C‐X‐C chemokine receptor 2; CXCL13, C‐X‐C motif chemokine ligand 13; DAPI, 4′,6‐diamidino‐2‐phenylindole; E‐TLS, early tertiary lymphoid structure; FL‐TLS, follicle‐like tertiary lymphoid structure; H&E, hematoxylin‐and‐eosin; HLA‐DR, human leukocyte antigen DR CXCR2, IQR, interquartile range; MHC‐II, major histocompatibility complex class II; ns, not significant; PMN‐MDSC, polymorphonuclear myeloid‐derived suppressor cell; PNAd, peripheral node addressin; pan‐CK, pan‐cytokeratin; ROI, regions of interest; Treg, regulatory T; TAN, tumor‐associated neutrophil; TLS, tertiary lymphoid structure.
To examine the spatial distribution and interrelationship of immune cells relative to TLSs, we set the TLS‐ROIs as the center and selected ROIs 500 μm and 1,000 μm away as near‐TLS‐ROIs and far‐TLS‐ROIs (Figure 1D). We compared the immune cell densities in the three ROI types (Figure 1E). Lymphocytes, including B, CD4+ T, CD8+ T and regulatory T (Treg), were most abundant in the TLS‐ROIs and decreased as the distance from TLSs increased. Similar patterns were observed for human leukocyte antigen (HLA)‐DR− [major histocompatibility complex (MHC)‐II−] myeloid cells, C‐X‐C chemokine receptor 2 (CXCR2)+MHC‐II− myeloid cells, and TANs (Figure 1E). CXCR2 is a predominant chemokine receptor for PMN‐MDSCs in bladder cancer [6]. About 10%‐30% of TANs were CXCR2+ (Supplementary Figure S1B). For FL‐TLSs, the densities of lymphocytes, MHC‐II− myeloid cells and TANs followed the TLS>near>far pattern (Figure 1F). For E‐TLSs, lymphocyte densities remained highest in TLS‐ROIs, but showed no significant difference between near and far ROIs; the stepwise decreasing trend was present in myeloid cells but did not reach statistical significance (Figure 1F). Regarding cell‐cell distances, MHC‐II− myeloid cells were closest to B cells and CD8+ T cells, followed by Treg, and farthest from MHC‐II+ myeloid cells (Figure 1G, Supplementary Table S4). Distances between MHC‐II− myeloid cells and other immunocytes were shorter in TLS‐ROIs than in near‐ or far‐TLS‐ROIs (Supplementary Figure S1C).
Given the density enrichment of TANs in TLSs and that many TANs function as PMN‐MDSCs, we assessed the clinical association between TLSs and PMN‐MDSCs using gene signatures based on the IMvigor210 phase 2 trial of atezolizumab (anti‐PD‐L1) on advanced urothelial carcinoma [7]. TLS signatures (12‐chemokine [2] or CXCL13 [3]) were associated with better survival (Figure 1H, Supplementary Table S5). Both TLS signatures were positively correlated with a 32‐gene PMN‐MDSC signature [8] or a 38‐gene PMN‐MDSC signature [9] (Figure 1I, Supplementary Table S5). When patients were stratified based on both TLS and PMN‐MDSC signatures, the survival from favorable to unfavorable followed the order TLShighPMN‐MDSClow > TLShighPMN‐MDSChigh > TLSlowPMN‐MDSClow > TLSlowPMN‐MDSChigh (Figure 1J). Note that the TLShighPMN‐MDSClow and TLSlowPMN‐MDSChigh groups had lower numbers of patients because the two signatures had opposite trends.
We tested the association of TLS signatures with two TAN signatures (Supplementary Table S5) in IMvigor210. Consistent with the PMN‐MDSC signatures, both TAN signatures were positively correlated with TLS signatures (Supplementary Figure S1D), and overall survival followed the order TLShighTANlow > TLShighTANhigh > TLSlowTANlow > TLSlowTANhigh from favorable to unfavorable outcomes (Supplementary Figure S1E). We also tested the prognostic significance of TLS signatures alone or in conjunction with the PMN‐MDSC signature in non‐small cell lung cancer (NSCLC) treated with anti‐PD‐1/PD‐L1 therapy [10]. TLS signatures were again associated with better overall survival (Supplementary Figure S1F) and with PMN‐MDSC signature (Supplementary Figure S1G). When both signatures were used to stratify NSCLC patients, overall survival also followed the order TLShighPMN‐MDSClow > TLShighPMN‐MDSChigh > TLSlowPMN‐MDSClow > TLSlowPMN‐MDSChigh from favorable to unfavorable outcomes (Supplementary Figure S1H). Therefore, the TLS/PMN‐MDSC prognostic association is also valid in anti‐PD‐1/PD‐L1 antibody‐treated NSCLC.
We stratified patients in IMvigor210 into basal and luminal subtypes based on a 47‐gene signature (Supplementary Figure S2A, Supplementary Table S5). The luminal subset survived better than the basal subset (Supplementary Figure S2B). TLShigh patients in both subtypes survived better than TLSlow patients (Supplementary Figure S2C), and TLS signatures were correlated with PMN‐MDSC signatures in both subtypes (Supplementary Figure S2D). TLS and PMN‐MDSC levels assessed by signature expressions were higher in the basal than in the luminal subtype (Supplementary Figure S2E). TLShighPMN‐MDSClow patients showed more favorable outcome than TLSlowPMN‐MDSChigh patients in the basal subtype (Supplementary Figure S2F). The luminal subtype was difficult to assess in the same manner due to very few PMN‐MDSChigh cases. Lastly, both TLS and PMN‐MDSC signatures were associated with PD‐1 or PD‐L1 levels (Supplementary Figure S2G) and T cell pathways (PD‐1‐ or exhaustion‐related) in IMvigor210 (Supplementary Figure S2H).
Our study made two main discoveries. First, lymphocytes and immunosuppressive myeloid cells (especially TANs) were most abundant in mature TLSs of bladder cancer, with densities decreasing as the distance from TLSs increased. While we speculated that TAN density follows this gradient if TANs enter TLSs through HEVs and diffuse radially, an alternative model where TANs extravasating into the tumor bed enrich through a chemokine gradient toward TLSs may lead to the same result. Future experiments with cell tracing may answer which mechanism prevails. Second, patients with bladder cancer characterized as TLShighPMN‐MDSClow and TLSlowPMN‐MDSChigh showed the best and worst prognosis with anti‐PD‐L1 therapy, respectively. These results may have the following clinical implications: (i) an immune score based on TLShighPMN‐MDSClow may help select patients who would benefit most from ICB therapy; (ii) for TLSlowPMN‐MDSChigh patients, strategies to induce TLS formation and debilitate PMN‐MDCSs may help overcome ICB therapy resistance.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Xuechun Wang, Anna Juncker‐Jensen: Conceptualization; investigation; methodology; data curation; formal analysis; validation; visualization; writing—original draft; writing—review & editing. Gang Huang, Mate Levente Nagy: Investigation; formal analysis. Xuemin Lu: Formal analysis; supervision. Liang Cheng, Xin Lu: Conceptualization; resources; supervision; funding acquisition; project administration; writing—original draft; writing—review & editing.
CONFLICTS OF INTERESTS STATEMENT
Anna Juncker‐Jensen and Mate Levente Nagy are employees of NeoGenomics Laboratories. The remaining authors declare no competing financial interest.
FUNDING INFORMATION
Xin Lu was supported by National Institutes of Health grants (R01CA248033 and R01CA280097) and Department of Defense Congressionally Directed Medical Research Programs grants (W81XWH2010312, W81XWH2010332 and HT94252310010).
CONSENT FOR PUBLICATION
Not applicable.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The experiments were approved by the Institutional Review Board of Indiana University School of Medicine and University of Notre Dame (#1808872882).
Supporting information
Supporting information
ACKNOWLEDGMENTS
We would like to thank the Lu lab members for helpful comments and suggestions during this work.
Xuechun Wang and Anna Juncker‐Jensen are co‐first authors with equal contributions
Contributor Information
Liang Cheng, Email: liang_cheng@brown.edu.
Xin Lu, Email: xlu@nd.edu.
DATA AVAILABILITY STATEMENT
The data presented in this study are available for request from the corresponding authors.
REFERENCES
- 1. Schumacher TN, Thommen DS. Tertiary lymphoid structures in cancer. Science. 2022;375(6576):eabf9419. [DOI] [PubMed] [Google Scholar]
- 2. Coppola D, Nebozhyn M, Khalil F, Dai H, Yeatman T, Loboda A, et al. Unique ectopic lymph node‐like structures present in human primary colorectal carcinoma are identified by immune gene array profiling. Am J Pathol. 2011;179(1):37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Groeneveld CS, Fontugne J, Cabel L, Bernard‐Pierrot I, Radvanyi F, Allory Y, et al. Tertiary lymphoid structures marker CXCL13 is associated with better survival for patients with advanced‐stage bladder cancer treated with immunotherapy. Eur J Cancer. 2021;148:181–189. [DOI] [PubMed] [Google Scholar]
- 4. Fridman WH, Meylan M, Petitprez F, Sun CM, Italiano A. Sautès‐Fridman C. B cells and tertiary lymphoid structures as determinants of tumour immune contexture and clinical outcome. Nat Rev Clin Oncol. 2022;19(7):441–457. [DOI] [PubMed] [Google Scholar]
- 5. Veglia F, Sanseviero E, Gabrilovich DI. Myeloid‐derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol. 2021;21(8):485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang H, Ye YL, Li MX, Ye SB, Huang WR, Cai TT, et al. CXCL2/MIF‐CXCR2 signaling promotes the recruitment of myeloid‐derived suppressor cells and is correlated with prognosis in bladder cancer. Oncogene. 2017;36(15):2095–2104. [DOI] [PubMed] [Google Scholar]
- 7. Balar AV, Galsky MD, Rosenberg JE, Powles T, Petrylak DP, Bellmunt J, et al. Atezolizumab as first‐line treatment in cisplatin‐ineligible patients with locally advanced and metastatic urothelial carcinoma: a single‐arm, multicentre, phase 2 trial. Lancet. 2017;389(10064):67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Condamine T, Dominguez GA, Youn JI, Kossenkov AV, Mony S, Alicea‐Torres K, et al. Lectin‐type oxidized LDL receptor‐1 distinguishes population of human polymorphonuclear myeloid‐derived suppressor cells in cancer patients. Sci Immunol. 2016;1(2):aaf8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang G, Lu X, Dey P, Deng P, Wu CC, Jiang S, et al. Targeting YAP‐Dependent MDSC Infiltration Impairs Tumor Progression. Cancer Discovery. 2016;6(1):80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ravi A, Hellmann MD, Arniella MB, Holton M, Freeman SS, Naranbhai V, et al. Genomic and transcriptomic analysis of checkpoint blockade response in advanced non‐small cell lung cancer. Nat Genet. 2023;55(5):807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
The data presented in this study are available for request from the corresponding authors.


