Abstract
Background
Chemoresistance is a major cause of treatment failure in gastric cancer (GC). Heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) is an N6‐methyladenosine (m6A)‐binding protein involved in a variety of cancers. However, whether m6A modification and hnRNPA2B1 play a role in GC chemoresistance is largely unknown. In this study, we aimed to investigate the role of hnRNPA2B1 and the downstream mechanism in GC chemoresistance.
Methods
The expression of hnRNPA2B1 among public datasets were analyzed and validated by quantitative PCR (qPCR), Western blotting, immunofluorescence, and immunohistochemical staining. The biological functions of hnRNPA2B1 in GC chemoresistance were investigated both in vitro and in vivo. RNA sequencing, methylated RNA immunoprecipitation, RNA immunoprecipitation, and RNA stability assay were performed to assess the association between hnRNPA2B1 and the binding RNA. The role of hnRNPA2B1 in maintenance of GC stemness was evaluated by bioinformatic analysis, qPCR, Western blotting, immunofluorescence, and sphere formation assays. The expression patterns of hnRNPA2B1 and downstream regulators in GC specimens from patients who received adjuvant chemotherapy were analyzed by RNAscope and multiplex immunohistochemistry.
Results
Elevated expression of hnRNPA2B1 was found in GC cells and tissues, especially in multidrug‐resistant (MDR) GC cell lines. The expression of hnRNPA2B1 was associated with poor outcomes of GC patients, especially in those who received 5‐fluorouracil treatment. Silencing hnRNPA2B1 effectively sensitized GC cells to chemotherapy by inhibiting cell proliferation and inducing apoptosis both in vitro and in vivo. Mechanically, hnRNPA2B1 interacted with and stabilized long noncoding RNA NEAT1 in an m6A‐dependent manner. Furthermore, hnRNPA2B1 and NEAT1 worked together to enhance the stemness properties of GC cells via Wnt/β‐catenin signaling pathway. In clinical specimens from GC patients subjected to chemotherapy, the expression levels of hnRNPA2B1, NEAT1, CD133, and CD44 were markedly elevated in non‐responders compared with responders.
Conclusion
Our findings indicated that hnRNPA2B1 interacts with and stabilizes lncRNA NEAT1, which contribute to the maintenance of stemness property via Wnt/β‐catenin pathway and exacerbate chemoresistance in GC.
Keywords: chemoresistance, gastric cancer, hnRNPA2B1, NEAT1, stemness
List of Abbreviations
- ADR
Adriamycin
- ASO
antisense oligonucleotide
- CCK‐8
Cell Counting Kit‐8
- CLIP‐seq
crosslinking immunoprecipitation‐high‐throughput sequencing
- CSC
cancer stem cell
- EMT
epithelial‐mesenchymal transition
- FPS
first progression survival
- GC
gastric cancer
- GCSC
gastric cancer stem cell
- hnRNPA2B1
heterogeneous nuclear ribonucleoprotein A2B1
- IC50
half‐maximal inhibitory concentration
- IF
immunofluorescence
- IgG
immunoglobulin G
- IHC
immunohistochemistry
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
mitogen‐activated protein kinase
- m6A
N6‐methyladenosine
- MeRIP
methylated RNA‐immunoprecipitation
- METTL3
methyltransferase 3
- mIHC
multispectral immunohistochemistry
- NEAT1
nuclear enriched abundant transcript 1
- OS
overall survival
- PFS
progression free survival
- PPS
post progression survival
- qPCR
quantitative real‐time PCR
- RBP
RNA binding proteins
- RIP
RNA binding protein immunoprecipitation
- SEM
standard error
- siRNA
small interfering RNA
- TCGA
The Cancer Genome Atlas
- TCF7L2
transcription factor 7 like 2
- VCR
Vincristine
- 5‐Fu
5‐fluorouracil
1. BACKGROUND
Gastric cancer (GC) is the fifth most common malignancy and the fourth leading cause of cancer‐related deaths worldwide [1]. Chemotherapy is the basic means and backbones of GC clinical treatment, and can significantly reduce tumor size and improve the progression‐free survival (PFS) and overall survival (OS) in patients with metastatic GC [2]. However, intrinsic and acquired resistance impedes efficacy of chemotherapy and plays a vital role in disease progression, leading to a 5‐year OS of less than 10% [3]. Therefore, it is important to understand how cancer cells resist chemotherapy‐induced cell death.
N6‐methyladenosine (m6A) is the most prevalent modification in eukaryotic RNAs and is implicated in diverse physiological processes, including drug resistance [4]. The m6A RNA modification process is regulated by a dynamic interaction of writers, erasers, and readers [5]. Heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1), an m6A reader, is a critical participant in multiple malignant behaviors, including tumorigenesis, angiogenesis, metastasis and chemoresistance [6]. We have previously found that long noncoding RNA (lncRNA) MIR100HG interacts with hnRNPA2B1 to facilitate the mRNA stability of transcription factor 7 like 2 (TCF7L2), which contributes to cetuximab resistance and metastasis in colorectal cancer (CRC) [7]. However, hnRNPA2B1 has been poorly studied in GC, and its detailed biological functions and corresponding molecular mechanisms, especially in drug resistance of GC, remain to be investigated.
Nuclear enriched abundant transcript 1 (NEAT1) is a widely expressed lncRNA involved in multiple physiological and pathological processes, such as the immune response [8], viral infection [9], tumorigenesis [10], and neurodegenerative diseases [11]. NEAT1 drives tumor initiation and progression through modulation of gene expression by associating with RNA‐binding proteins (RBPs), recruiting transcription factors, or acting as a competing endogenous RNA to alter target gene expression [12]. Upregulation of NEAT1 in digestive system carcinomas is significantly associated with tumor size, TNM stage, distant metastasis, and poor prognosis [12]. However, the relationship between hnRNPA2B1 and NEAT1 remains to be further investigated.
The Wnt/β‐catenin pathway is a crucial signaling cascade involved in embryonic development, tissue homeostasis, and cell proliferation [13, 14]. Studies have shown that aberrant activation of Wnt/β‐catenin pathway contributes to tumor initiation [15], progression [16], and metastasis [17] in GC. This dysregulation leads to increased stemness properties within cancer cells, which are characterized by self‐renewal capacity and differentiation potential [14]. However, whether hnRNPA2B1 is related to Wnt/β‐catenin pathway and stemness properties requires further exploration.
In this study, we aimed to investigate the role of hnRNPA2B1 and the downstream mechanism in GC chemoresistance. Investigating these factors can provide valuable insights into the mechanisms underlying chemoresistance and potentially lead to the development of novel therapeutic strategies for chemotherapy‐resistant GC.
2. MATERIALS AND METHODS
2.1. Human tissues and tissue collection
A GC tissue microarray (HStmA180Su20) was purchased from Outdo Biotech (Shanghai, China). The microarray contained samples for 75 cases of GC tissues and paired adjacent normal tissues, and 28 cases of unpaired GC tissues. Surgically resected specimens from 30 GC patients who received adjuvant chemotherapy and 20 paired specimens of primary GC tissues and adjacent normal tissues were obtained from Xijing Hospital of Digestive Diseases (Xi'an, Shaanxi, China). All samples were clinically and pathologically diagnosed. The clinical information of all patients is listed in Supplementary Tables S1‐S2. This study was approved by Xijing Hospital's Protection of Human Subjects Committee (KY20222241‐X‐1). Written informed consent was obtained from each patient.
2.2. Cells and cell culture
GC cell lines (SGC7901, MGC803, BGC823, MKN28, AGS, HGC27 and MKN45) and normal gastric epithelial cells (GES‐1) were obtained from the China Infrastructure of Cell Line Resources (Beijing, China). Adriamycin (ADR)‐resistant SGC7901 (SGC7901ADR) and vincristine (VCR)‐resistant SGC7901 (SGC7901VCR) cells had been previously established from SGC7901 cells at our lab [18, 19]. To maintain the multidrug‐resistant (MDR) phenotype, 0.5 µg/mL ADR (MCE, Monmouth Junction, NJ, USA) or 1 µg/mL VCR (MCE) was added to the culture medium of SGC7901ADR or SGC7901VCR cells, respectively. All cells were cultured in RPMI‐1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) at 37°C in a humidified atmosphere containing 5% CO2. All cell lines were confirmed to be free of contamination.
2.3. Mice
Female BALB/c nude mice aged 4‐6 weeks were provided by the Experimental Animal Center of the Fourth Military Medical University (Xi'an, Shaanxi, China). All animals were housed and maintained in pathogen‐free conditions. Mice were euthanized using carbon dioxide anesthesia suffocation method, either 4 weeks after the injection of tumor cells or when the tumor diameter reached over 15 mm, or when the tumor burden exceeded 10% of the mouse's body weight. All animal studies complied with the Fourth Military Medical University animal use guidelines, and the protocol was approved by the Fourth Military Medical University Animal Care Committee (IACUC20221013).
2.4. Constructs, oligonucleotides, transduction, and transfection
Lv‐hnRNPA2B1 sequences were amplified and cloned into the GV492 vector (GeneChem, Shanghai, China), and sh‐hnRNPA2B1 sequences were cloned into the GV493 vector (GeneChem). The empty vectors CON335 (GeneChem, Shanghai, China) and CON313 (GeneChem, Shanghai, China) were used as the negative controls respectively. Viral transfection was performed in the indicated cells according to the manufacturer's instructions. To generate stable cell lines, after 72 h of transfection, cells were subjected to puromycin (Sigma, St. Louis, MO, USA) selection for 2 weeks. NEAT1 overexpression plasmid, pcDNA3.1‐NEAT1, was constructed by Tsingke Biotechnology (Beijing, China), and empty pcDNA3.1 vector (Tsingke Biotechnology, Beijing, China) was used as the control. Antisense oligonucleotides (ASOs) specifically targeting NEAT1 were purchased from RiboBio (Guangzhou, Guangdong, China). NEAT1 overexpression plasmid and ASOs were transfected into the indicated cells using jetPRIME transfection reagent (Polyplus Transfection, IIIkirch, France) following the manufacturer's instructions. The sequences used in this study are listed in Supplementary Table S3.
2.5. Protein extraction and Western blotting
Proteins were extracted from cultured cells using RIPA buffer (Beyotime, Shanghai, China) supplemented with protease and phosphatase inhibitors (Beyotime, Shanghai, China). Denatured proteins (20‐30 µg) were fractionated by sodium dodecyl‐sulfate polyacrylamide gel electrophoresis and transferred to nitrocellulose or polyvinylidene fluoride membranes (Merck Millipore (Billerica, MA, USA)). After being blocked with the primary and secondary horseradish peroxidase (HRP)‐conjugated antibodies (Supplementary Table S4) in 5% non‐fat milk, the immunoreactive proteins were detected with enhanced chemiluminescence reagents (Beijing 4A Biotech, Beijing, China). The blots were scanned by ChemiDoc XRS+ Imaging System (Bio‐Rad, Hercules, CA, USA).
2.6. RNA extraction and quantitative PCR (qPCR)
Total RNA was extracted from cells using a GeneJET RNA Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA) and was used for cDNA synthesis with a Goldenstar RT6 cDNA Synthesis Kit (Tsingke Biotechnology). qPCR was performed using Master qPCR Mix (Tsingke Biotechnology) and a CFX96 Touch qPCR Detection System (Bio‐Rad). The primers were synthesized by Tsingke Biotechnology, and GAPDH was used as an internal control. The 2−ΔΔCT method was used to calculate relative gene expression. The primer sequences are listed in Supplementary Table S5.
2.7. Immunofluorescence (IF)
Cells were seeded on 4‐well glass chamber slides (Millipore, Billerica, MA, USA), left overnight to stably adhere, and then fixed in 4% paraformaldehyde (Beyotime, Shanghai, China) at room temperature for 30 min. Then, the cells were permeated with 0.1% Triton X‐100 (Beyotime, Shanghai, China) for 15 min and incubated with 5% bovine serum albumin (BSA; Sigma‐Aldrich, St. Louis, MO, USA) for 1 h at room temperature. Primary antibodies (Supplementary Table S4) diluted with 5% BSA to adequate concentrations were added, and the cells were incubated overnight at 4°C. Then, the cells were incubated with the corresponding fluorescent secondary antibodies (Supplementary Table S4) (Affinity Biosciences, Melbourne, Australia) at room temperature for 1 h. The nuclei were stained by DAPI (Beyotime, Shanghai, China) for 15 min. IF images were captured using a Nikon A1 Confocal Laser Microscope System (Nikon, Tokyo, Japan).
For spheroid IF, spheroids were fixed and permeabilized for 30 min at room temperature in phosphate‐buffered saline (PBS) containing 4% paraformaldehyde (PFA) and 0.5% Triton X‐100 and washed in PBS. Spheroids were then blocked in PBS containing 0.1% BSA, 0.2% Triton X‐100, and 10% FBS for 2 h at room temperature. Primary antibodies (Supplementary Table S4) diluted with prior block reagent were added, and the spheroids were incubated overnight at 4°C. Thereafter, spheroid IF was performed by as above‐mentioned.
2.8. Immunohistochemistry (IHC)
Paraffin‐embedded specimens were serially sectioned, deparaffinized by dimethylbenzene, 100% alcohol, 95% alcohol, 90% alcohol, 85% alcohol, 70% alcohol, and PBS. And then treated with 3% H2O2 to block endogenous peroxidase activity. Slides were immersed in an antigen retrieval buffer (EDTA antigen retrieval solution, 50×, pH 9.0, Proandy, Xi'an, Shaanxi, China) and heated to 120°C for 10 min and then allowed to cool to room temperature. Incubation with primary antibodies (Supplementary Table S4) was performed at 4°C overnight, and the samples were incubated with corresponding peroxidase‐conjugated secondary antibodies at room temperature for 1 h. Then, the proteins were visualized with DAB chromogenic substrate (ZSGB‐BIO, Beijing, China), which was followed by visualization with hematoxylin (Beyotime, Shanghai, China) and whole‐slide imaging by a pathological slice scanner (3DHISTECH, Budapest, Hungary).
2.9. Half‐maximal inhibitory concentration (IC50) and cell proliferation assay
For the IC50 assay, cells in the logarithmic growth phase were harvested and seeded in 96‐well plates (4,000 cells/well) and incubated overnight to allow cells to adhere. Then, different concentrations of ADR, VCR, and 5‐fluorouracil (5‐Fu; MCE) were then added to the medium. For Wnt inhibition, cells were additionally treated with ICG‐001 (25 µmol/L) (MCE, Monmouth Junction, NJ, USA). After 48 h of treatment, Cell Counting Kit‐8 (CCK‐8) reagent (GLPBIO, Montclair, CA, USA) mixed with RPMI‐1640 was used for cell viability assay, and the absorbance was measured at 450 nm with a microplate reader (Bio‐Rad).
For the cell proliferation assay, cells were plated into 96‐well plates (1,000 cells/well) and incubated overnight to adhere. Drugs (ADR (12 µg/mL), and 5‐Fu (20 µg/mL) for SGC7901ADR cells; VCR (15 µg/mL), and 5‐Fu (20 µg/mL) for SGC7901VCR cells; ADR (1 µg/mL), VCR (1 µg/mL), and 5‐Fu (1 µg/mL) for SGC7901 cells) were then added to the medium correspondingly. For Wnt inhibition, cells were additionally treated with ICG‐001 (25 µmol/L). After 0‐7 days of treatment, the absorbance values were measured via CCK‐8 assay.
2.10. Apoptosis analysis
The indicated cells (SGC7901ADR shNC and SGC7901ADR shA2B1 cells; SGC7901VCR shNC and SGC7901VCR shA2B1 cells; SGC790 lv‐NC and SGC790 lv‐A2B1 cells) were seeded in 6‐well plates at a concentration of 1,000,000 cells/well. After the cells had become adherent, culture medium containing corresponding drug (ADR [12 µg/mL], and 5‐Fu [20 µg/mL] for SGC7901ADR cells; VCR [15 µg/mL], and 5‐Fu [20 µg/mL] for SGC7901VCR cells; ADR [1 µg/mL], VCR [1 µg/mL], and 5‐Fu [1 µg/mL] for SGC7901 cells) was added. After 48 h, the cells were harvested, resuspended in staining buffer, and examined using an Annexin V‐PE/7AAD apoptosis detection kit (BD Biosciences, San Jose, CA, USA). After 48 h of treatment, the Annexin V‐PE+/7AAD− cells were detected by the flow cytometry (Beckman Coulter, Miami, FL, USA) and considered to have undergone apoptosis. The data were analyzed using CellQuest Pro software (BD Biosciences).
2.11. in vivo drug resistance assay
Suspensions of the cells were subcutaneously injected into the unilateral thighs of mice (1,000,000 cells/100 µL PBS per spot; 8 mice in each group), and tumor volume was measured every 3 days. When the tumor volume reached 100 mm3, 5‐Fu (20 mg/kg) or PBS was given intraperitoneally. After 4 weeks, the nude mice were euthanized, and the subcutaneous tumor tissues were fixed in 4% paraformaldehyde and embedded in paraffin. Then, serial sections made from xenograft tumor samples were stained with anti‐Ki‐67 and anti‐cleaved Caspase3 antibodies to explore the role of hnRNPA2B1 in cell proliferation and apoptosis in vivo. Besides, RNA was extracted from paraffin embedded specimens (TIANGEN, Beijing, China) to perform qPCR analysis of NEAT1.
2.12. Tumor sphere formation assay
A total of 1,000 Cells were seeded in ultra‐low attachment plates (Corning, Corning, NY, USA) and cultured in serum‐free RPMI‐1640 supplemented with 2% B27 supplement (STEMCELL, Vancouver, BC, Canada), 20 ng/mL EGF (MCE), and 10 ng/mL bFGF (MCE) in a humidified 5% CO2 at 37°C. Additionally, 25 µmol/L ICG‐001 or 100 ng/mL Wnt3A (MCE, Monmouth Junction, NJ, USA) was added in the culture medium. After one‐week incubation, tumor sphere numbers were counted under a phase‐contrast microscope using the 40× magnification lens (Olympus, Tokyo, Japan).
2.13. RNA sequencing and data analysis
To detect the changes in downstream genes after hnRNPA2B1 knockdown, total RNA was extracted from hnRNPA2B1‐knockdown SGC7901ADR cells (ADR_sh) and negative control cells (ADR_shNC). After that, mRNA was purified using Dynabeads Oligo (dT) (Thermo Fisher, CA, USA) and fragmented into short fragments. Then the cleaved fragments were reverse‐transcribed to create the cDNA, which were next used to synthesize U‐labeled second‐stranded DNAs. Then size selection and PCR amplification were performed. At last, the 2 × 150 bp paired‐end sequencing (PE150) were performed on an Illumina Novaseq™ 6000 (LC‐Bio Technology CO., Ltd., Hangzhou, Zhejiang, China) following the vendor's recommended protocol.
2.14. Methylated RNA‐immunoprecipitation (MeRIP)‐qPCR
The MeRIP assay was performed according to the manufacturer's instructions of riboMeRIP m6A Transcriptome Profiling Kit (Ribo Biotechnology, Guangzhou, Guangdong, China). Total RNA was fragmented into 200 nucleotides followed by magnetic immunoprecipitation with an m6A‐specific antibody (Ribo Biotechnology, Guangzhou, Guangdong, China). After washing with IP buffer, the RNA was eluted and precipitated by using MagenTM Hipure Serum/plasma miRNA kit (Magen Biotechnology, Guangzhou, Guangdong, China). Then, the isolated RNA fragments were reversely transcribed into cDNA by the PrimeScript RT Reagent Kit (TaKaRa, Tokyo, Japan) and subjected to qPCR.
2.15. RNA immunoprecipitation (RIP)
RIP assays were carried out using a Millipore EZ‐Magna Nuclear RIP (Native) Kit (Millipore) according to the manufacturer's protocol. In brief, 5 µg of hnRNPA2B1 antibody and corresponding control IgG were conjugated to protein A/G magnetic beads, and the beads were then incubated with precleared nuclear extracts to obtain the RNA‐binding protein‐RNA complex. According to the instructions, the immunoprecipitated RNAs were extracted and subjected to qPCR using total RNA as an input control.
2.16. RNA stability assay
Cells (300,000 cells/mL) were seeded into 6‐well plates to achieve 30%‐40% confluency after 24 h. The cells were then treated with 10 µg/mL actinomycin D (Sigma) for the indicated times. Total RNA was extracted every 2 h and subsequently subjected to qPCR. The half‐life of RNA was calculated as previously described [20]. Actinomycin D inhibited mRNA transcription and the degradation rate of RNA (Kdecay) was estimated by following equation: ln(C/C0) = ‐Kdecayt. C0 is the concentration of mRNA at time 0, t is the transcription inhibition time, and C is the mRNA concentration at the time t. The half‐life time (t1/2), which means C/C0 = 1/2, can be calculated by the following equation: t1/2 = ln2/Kdecay.
2.17. RNAscope assay
The expression of NEAT1 was detected by in situ hybridization (ISH) using an RNAscope Multiplex Fluorescent Kit V2 (Advanced Cell Diagnostics, Minneapolis, MN, USA) according to the manufacturer's instructions. NEAT1 probe (No. 411531) was purchased from Advanced Cell Diagnostics (Santa Clara, CA, USA). Nikon A1 confocal laser microscope (Tokyo, Japan) was used for image analysis. Three fields of each tissue section were selected for histology quantification. All tissues were categorized into NEAT1‐high and NEAT1‐low groups according to the median of the proportion of positive‐stained cells.
2.18. Multiplex immunohistochemistry (mIHC)
mIHC was performed using an Opal 7‐color IHC kit (Akoya Biosciences, Marlborough, MA, USA) as previously described [21]. First, the slices were baked, dewaxed, rehydrated, and then subjected to antigen retrieval in AR6 (Perkin Elmer, pH = 6.0) or EDTA (50×, pH = 9.0, Proandy, Xi'an, Shaanxi, China) antigen repair solution in a microwave oven for 10 min and then allowed to cool to room temperature. Then, the sections were blocked in blocking buffer for 10 min, incubated with primary antibody (Supplementary Table S4) for 1 h, and incubated with a polymer HRP‐conjugated secondary antibody for 10 min. Subsequently, the tissues were stained with the fluorophore‐4 tyramine signal amplification dye. After that, the paraffin sections were again subjected to antigen repair. The aforementioned steps were iteratively repeated until all antigens were labeled. Finally, the slides were stained with DAPI (BD Biosciences, San Jose, CA, USA) for 5 min and mounted with antifade mounting medium (Beyotime, Shanghai, China) after elution. Multispectral imaging and segmentation imaging analysis were performed using a Vectra multispectral imaging system (Perkin‐Elmer, Waltham, MA, USA).
2.19. Bioinformatic analysis
UALCAN (https://ualcan.path.uab.edu/index.html) was used to compare the expression of hnRNPA2B1, NEAT1, CD133 and CD44 in GC and normal tissues as previously described [22]. The prognostic value of hnRNPA2B1, NEAT1, CD133 and CD44 in GC patients, especially those who received chemotherapy, was analyzed by Kaplan‐Meier Plotter [23]. OS was analyzed in this study, which is the length of time from the date of diagnosis.
The POSTAR3 database generates a comprehensive map of RBPs using crosslinking immunoprecipitation high‐throughput sequencing (CLIP‐seq) data, allowing users to retrieve RNAs that bind to RBPs and their corresponding binding sites [24]. Using POSTAR3 database, we identified potential hnRNPA2B1‐binding RNAs. The SRAMP online tool could predict mammalian m6A sites based on sequence‐derived features, and was used to predict the potential m6A sites of NEAT1 [24].
RNA sequencing of hnRNPA2B1‐silenced SGC7901ADR cells and control cells was performed to identify the downstream RNAs regulated by hnRNPA2B1 (ǀlog2FCǀ > 2, P < 0.05, q < 0.05, FPKM average > 0.5). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis identified significantly enriched pathways in differentially expressed genes (DEGs) comparing with the whole genome background [25]. Pathways meeting this condition with P < 0.05 were defined as significantly enriched pathways in DEGs.
RNA sequencing expression profiles and corresponding clinical information were downloaded from The Cancer Genome Atlas Stomach Adenocarcinoma (TCGA‐STAD) (https://portal.gdc.cancer.gov/) dataset. Using the one‐class linear regression algorithm, we calculated the mRNA stemness index (mRNAsi) scores via R v4.0.3 [26]. We recruited 10 stemness‐related gene signatures from StemChecker, and the enrichment scores were quantified by the single‐sample gene set enrichment analysis (ssGSEA) algorithm via the GSVA R package (v1.34.0) [27, 28]. GSEA was performed to identify signaling pathways in hnRNPA2B1high NEAT1high group and hnRNPA2B1low NEAT1low group.
2.20. Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 (La Jolla, CA, USA) and SPSS 23.0 (IBM, Armonk, NY, USA). P < 0.05 was considered to indicate statistical significance. The data are presented as the mean ± standard error of the mean (SEM). Two‐tailed Student's t tests, ANOVA, χ2 tests, and Spearman's correlation analysis were used according to the type of experiment.
3. RESULTS
3.1. hnRNPA2B1 was highly expressed in GC cells and tissues and associated with poor clinical outcomes
Aberrant hnRNPA2B1 expression had been reported in a variety of tumors and was associated with patients’ poor prognosis [6]. However, the specific role of hnRNPA2B1 in GC warrants further exploration. Therefore, we investigated the expression of hnRNPA2B1 in GC. First, analysis of the TCGA‐STAD dataset indicated that hnRNPA2B1 expression was significantly higher in GC tissues than in noncancerous tissues (Figure 1A). Kaplan‐Meier survival analysis revealed that higher hnRNPA2B1 expression was related with poorer OS, first progression survival (FPS), and post progression survival (PPS) of GC patients, especially in those who had received 5‐Fu‐based adjuvant chemotherapy (Figure 1B).
FIGURE 1.
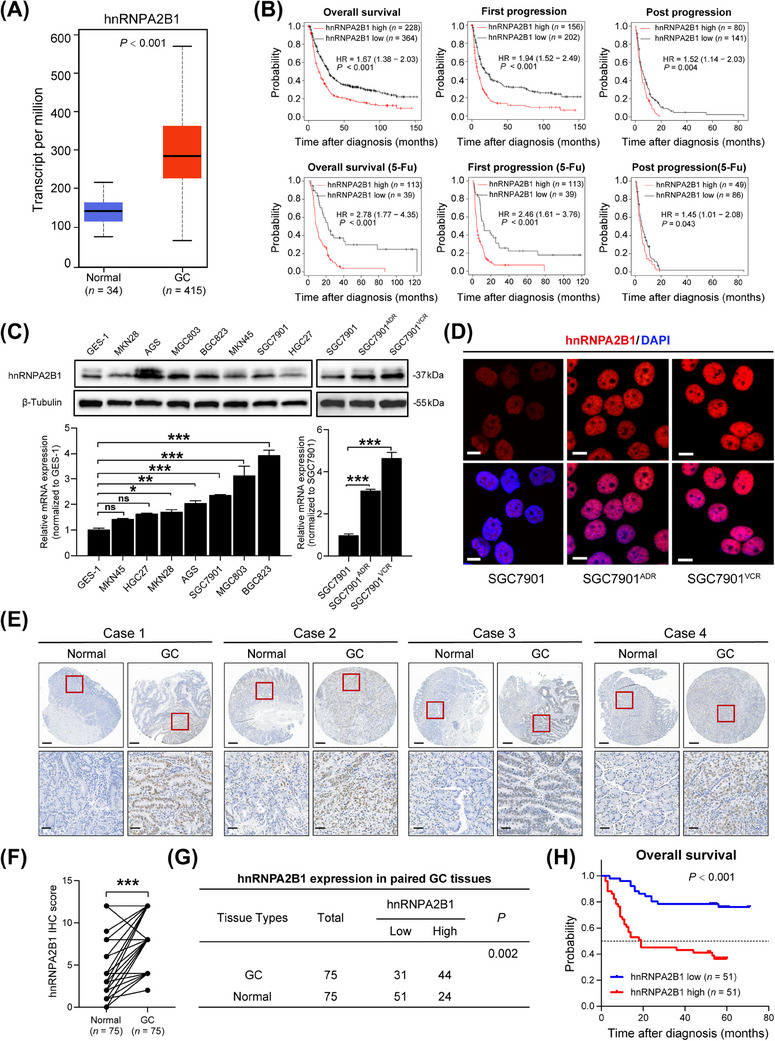
hnRNPA2B1 was highly expressed in GC and was associated with poor clinical outcomes. (A) mRNA expression of hnRNPA2B1 in the UALCAN database. (B) Kaplan‐Meier analyses of correlations between hnRNPA2B1 expression and OS, FPS, and PPS of GC patients and patients who received 5‐Fu treatment. (C‐D) Western blotting (C), qPCR (C) and representative IF images (D) of hnRNPA2B1 in GC cell lines. Scale bars, 10 µm. (E) Representative IHC staining of hnRNPA2B1 in matched GC and adjacent normal tissues. Scale bars, 400 µm (upper) and 50 µm (lower). (F) The IHC scores of hnRNPA2B1 in matched GC and adjacent normal tissues are shown. (G) The expression of hnRNPA2B1 in 75 paired GC and adjacent normal tissues. (H) Log‐rank test for overall survival of GC patients (n = 102). GC, gastric cancer; 5‐Fu, 5‐fluorouracil; IHC, immunohistochemistry. The data are presented as the mean ± SEM. The P value was calculated by paired t test (C), Wilcoxon's matched‐pairs signed‐rank test (F), χ2 test (G) and log‐rank test (H). *P < 0.05, ** P < 0.01, *** P < 0.001, ns, not significant. Abbreviations: 5‐Fu, 5‐fluorouracil; GC, gastric cancer; hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; HR, hazard ratio; IHC, immunohistochemistry.
Subsequently, Western blotting and qPCR verified that the expression of hnRNPA2B1 was elevated in most GC cell lines such as AGS, MGC803, BGC823, and SGC7901 compared with normal gastric epithelial cells (GES‐1). However, no significant increasement was observed in MKN45, HGC27, and MKN28 cells (Figure 1C). Particularly, hnRNPA2B1 was significantly higher in the MDR GC cell lines SGC7901ADR and SGC7901VCR than in the parental cell line SGC7901 (Figure 1C‐D). To further assess the clinical significance of hnRNPA2B1 in GC, we performed IHC staining in a GC tissue microarray (Figure 1E) and analyzed the relationships between hnRNPA2B1 expression levels and clinicopathological features. A marked increase in hnRNPA2B1 expression was observed in GC tissues compared with paired noncancerous tissues (Figure 1F‐G), and higher hnRNPA2B1 expression was related with poor clinicopathological features (such as differentiation grade, TNM stage, tumor size, microvascular invasion, and nerve invasion) and shorter OS (Figure 1H, Supplementary Table S6). Collectively, these results indicated that hnRNPA2B1 was overexpressed in GC tissues, especially in the chemoresistance setting, and its overexpression might contribute to the malignant progression of GC.
3.2. hnRNPA2B1 induced chemoresistance by inhibiting apoptosis and promoting proliferation both in vitro and in vivo
To ascertain the biological roles of hnRNPA2B1 in GC chemoresistance, we knocked down hnRNPA2B1 in MDR GC cells (SGC7901ADR cells and SGC7901VCR cells) (Figure 2A, Supplementary Figure S1A). We found that the downregulation of hnRNPA2B1 sensitized SGC7901VCR cells to VCR and 5‐Fu, as shown by decreased IC50 values (Figure 2B) and increased early apoptotic cell proportions (Figure 2C‐D). Knockdown of hnRNPA2B1 dramatically impaired the proliferative capacity of SGC7901VCR cells without or with chemotherapy (Figure 2E). Similar results were observed in SGC7901ADR cells in response to ADR and 5‐Fu (Supplementary Figure S1A‐E). In addition, we overexpressed hnRNPA2B1 in SGC7901 cells and found that ectopic expression of hnRNPA2B1 induced chemoresistance in SGC7901 cells, as demonstrated by elevated IC50 values and decreased early apoptotic cell proportions upon exposure to ADR, VCR, and 5‐Fu (Figure 2F‐I). Moreover, overexpression of hnRNPA2B1 caused an obvious increase in cell proliferation in the absence or presence of chemotherapy (Figure 2J). Downregulation of cleaved Caspase3 after hnRNPA2B1 overexpression and upregulation of cleaved Caspase3 after hnRNPA2B1 knockdown during chemotherapy treatment (Supplementary Figure S1F) also indicated that hnRNPA2B1 exerts an anti‐apoptosis effect in the chemo‐resistant setting.
FIGURE 2.
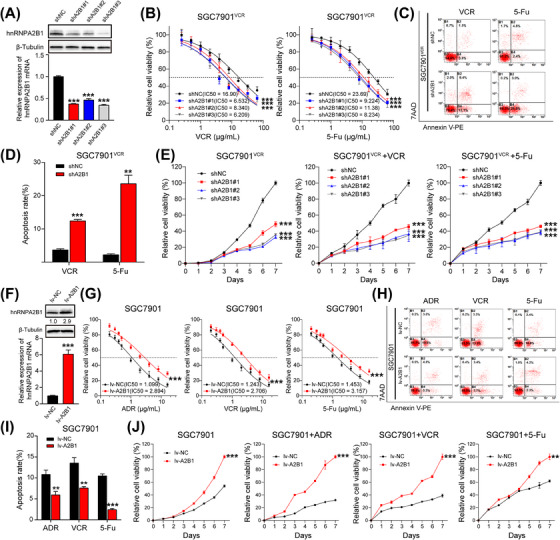
hnRNPA2B1 induced chemoresistance by promoting proliferation and inhibiting apoptosis in vitro. (A) Western blotting and qPCR were used to detect the knockdown efficiency of hnRNPA2B1 in SGC7901VCR cells. (B‐D) IC50 values (B) and apoptosis (C and D) of SGC7901VCR shNC and SGC7901VCR shA2B1 cells treated with VCR and 5‐Fu. (E) Growth curves of SGC7901VCR shNC and SGC7901VCR shA2B1 cells treated without or with chemotherapy. (F) Western blotting and qPCR were used to detect the overexpression efficiency of hnRNPA2B1 in SGC7901 cells. (G‐I) IC50 values (G) and apoptosis (H and I) of SGC7901 lv‐NC and SGC7901 lv‐A2B1 cells treated with ADR, VCR and 5‐Fu. (J) Growth curves of SGC7901 lv‐NC and SGC7901 lv‐A2B1 cells treated without or with chemotherapy. The data are presented as the mean ± SEM. ** P < 0.01, *** P < 0.001 by one‐way ANOVA test (A), paired t test (D, F and I) and one‐way ANOVA with Dunnett's multiple‐comparison test (B, E, G and J). Abbreviations: hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; IC50, half‐maximal inhibitory concentration; lv‐NC, empty overexpression control; lv‐A2B1, overexpression of hnRNPA2B1; shNC, negative control short hairpin RNA; shA2B1, short hairpin RNA of hnRNPA2B1.
We further confirmed that hnRNPA2B1 contributed to the chemoresistance of GC cells in vivo by establishing a subcutaneous xenograft model in athymic nude mice using SGC7901VCR shNC or SGC7901VCR shA2B1 cells followed by 5‐Fu or saline treatment. It was observed that tumor volumes and weights in the shA2B1 group with 5‐Fu treatment were markedly decreased compared to those in the shNC group with 5‐Fu treatment (Figure 3A). Ki‐67 proliferation index is often used to evaluate tumor proliferation, and cleaved Caspase3 is often used as an early indicator of apoptosis. Therefore, serial sections made from xenograft tumor samples were stained with anti‐Ki‐67 and anti‐cleaved Caspase3 antibodies to explore the role of hnRNPA2B1 in cell proliferation and apoptosis in vivo. IHC staining showed that shA2B1 groups treated with 5‐Fu presented fewer Ki‐67‐positive cells and more cleaved Caspase3‐positive cells (Figure 3B). In contrast, increased tumor volumes and weights were observed in hnRNPA2B1 overexpressi group (Figure 3C), with more Ki‐67‐positive cells and fewer cleaved Caspase3‐positive cells detected (Figure 3D). These in vivo observations confirmed that hnRNPA2B1 promoted GC chemoresistance by inhibiting apoptosis and promoting proliferation.
FIGURE 3.
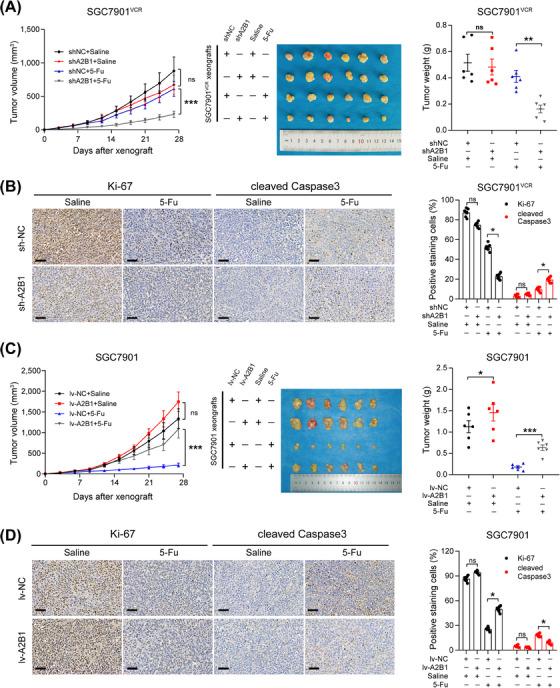
hnRNPA2B1 contributed to the chemoresistance of GC cells in vivo. (A) SGC7901VCR shNC and SGC7901VCR shA2B1 cells were subcutaneously implanted into nude mice, which were then treated with 5‐Fu or saline (20 mg/kg, i.p. injection). The tumor volumes and tumor weight were monitored, and growth curves were plotted every 3 days. (B) Ki‐67 and cleaved Caspase3 staining and percentages in the subcutaneous tumors of SGC7901VCR shNC and SGC7901VCR shA2B1 cells. (C) SGC7901 lv‐NC and SGC7901 lv‐A2B1 cells were subcutaneously implanted into nude mice, which were then treated with 5‐Fu or saline (20 mg/kg, i.p. injection). The tumor volumes and tumor weight were monitored, and growth curves were plotted every 3 days. (D) Ki‐67 and cleaved Caspase3 staining and percentages in the subcutaneous tumors of SGC7901 lv‐NC and SGC7901 lv‐A2B1 cells. The data are presented as the mean ± SEM, *P < 0.05, **P < 0.01, *** P < 0.001, ns, not significant by repeated‐measures ANOVA (A and C) or Student's t test (B and D). Scale bars, 50 µm. Abbreviations: hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; i.p., intraperitoneal; lv‐NC, empty overexpression control; lv‐A2B1, overexpression of hnRNPA2B1; shA2B1, short hairpin RNA of hnRNPA2B1; shNC, negative control short hairpin RNA.
3.3. lncRNA NEAT1 was a key molecule regulated by hnRNPA2B1
Subsequently, we investigated the molecular mechanisms through which hnRNPA2B1 participates in GC chemoresistance. As an RBP, hnRNPA2B1 participates in various malignant biological behaviors of tumors by affecting almost every stage of RNA synthesis [29]. Using POSTAR3 database, we identified 1,135 potential hnRNPA2B1‐binding RNAs. Meanwhile, we found that 127 genes were significantly upregulated, and 281 genes were significantly downregulated after hnRNPA2B1 knockdown. KEGG analysis revealed that the MAPK signaling pathway, PI3K‐Akt signaling pathway, Wnt signaling pathway, and Hippo signaling pathway were significantly enriched (Supplementary Figure S2A).
Accumulated evidence indicates that lncRNAs function critically in tumor progression, and our lab has been focusing on the functions and mechanisms of lncRNAs in gastrointestinal cancer chemoresistance [3, 12]. We found 640 downregulated lncRNAs in hnRNPA2B1‐silenced group, and the top 20 upregulated or down‐regulated lncRNAs are exhibited using heatmap (Figure 4A). By intersecting 640 downregulated lncRNAs with 1,135 potential hnRNPA2B1‐binding RNAs, we screened 3 lncRNAs: NEAT1, MALAT1, and CRNDE (Figure 4B). Among them, overexpression or downregulation of hnRNPA2B1 resulted in the corresponding changes of NAET1 both in vitro and in vivo (Figure 4C, Supplementary Figure S2B). Thus, we suspected that NEAT1 may be the potential target regulated by hnRNPA2B1.
FIGURE 4.
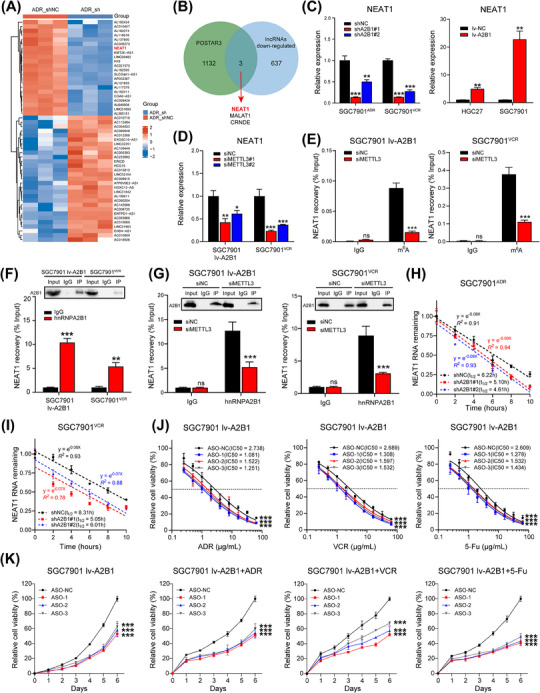
hnRNPA2B1 interacted with and stabilized NEAT1 in an m6A‐dependent manner. (A) Heatmap of differentially expressed lncRNAs (ǀlog2FCǀ > 1, P < 0.05, FPKM average > 0.5) in SGC7901ADR cells after hnRNPA2B1 knockdown. Red shades represent ADR_shNC, and blue shades represent ADR_sh. (B) Venn diagram of hnRNPA2B1‐binding lncRNAs. (C) hnRNPA2B1 knockdown or overexpression resulted in corresponding changes in NEAT1 levels. (D) qPCR analysis of NEAT1 expression in SGC7901 lv‐A2B1 and SGC7901VCR cells after METTL3 knockdown.(E) Analysis of MeRIP assays detecting NEAT1 retrieved by an m6A antibody in METTL3‐silenced SGC7901 lv‐A2B1 and SGC7901VCR cells. (F) Assessment of RIP assays detecting NEAT1 retrieved by an hnRNPA2B1 antibody or by IgG in SGC7901 lv‐A2B1 and SGC7901VCR cells. (G) Assessment of RIP assays detecting NEAT1 retrieved by an hnRNPA2B1 antibody or by IgG in METTL3‐silenced SGC7901 lv‐A2B1 and SGC7901VCR cells.(H) Assessment of the NEAT1 half‑life (t1/2) in hnRNPA2B1‑silenced SGC7901ADR cells. (I) Assessment of the NEAT1 half‑life (t1/2) in hnRNPA2B1‑silenced SGC7901VCR cells. (J‐K) IC50 values and growth curves of SGC7901 lv‐A2B1 cells transfected with ASO‐NEAT1 and treated with chemotherapy. The P value was calculated by one‐way ANOVA test (C left, D, H, and I), paired t test (C right and E, F, and G), one‐way ANOVA with Dunnett's multiple‐comparison test (H) and two‐way ANOVA with Dunnett's multiple‐comparison test (J and K). *P < 0.05, ** P < 0.01, *** P < 0.001, ns, not significant. Abbreviations: ADR_sh, hnRNPA2B1‐knockdown SGC7901ADR cell; ADR_shNC, negative control SGC7901ADR cell; ASO‐NC, negative control of antisense oligonucleotide; ASO‐NEAT1, antisense oligonucleotide of NEAT1; CRNDE, colorectal neoplasia differentially expressed; hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; IgG, immunoglobulin G; IP, immunoprecipitation; lncRNA, long noncoding RNA; METTL3, methyltransferase 3; lv‐A2B1, overexpression of hnRNPA2B1; lv‐NC, empty overexpression control; MALAT1, metastasis associated lung adenocarcinoma transcript 1; NEAT1, nuclear enriched abundant transcript 1; shNC, negative control short hairpin RNA; shA2B1, short hairpin RNA of hnRNPA2B1.
3.4. hnRNPA2B1 promoted cell proliferation and chemoresistance by interacting with NEAT1 in an m6A‐dependent manner
As hnRNPA2B1 is an m6A reader [6], we speculated that hnRNPA2B1 might regulate NEAT1 RNA stability in an m6A‐dependent manner. Firstly, we found abundant m6A modification sites were located in NEAT1 through the SRAMP database (Supplementary Figure S2C). Subsequently, silencing of the m6A Methyltransferase Like 3 (METTL3; Supplementary Figure S2D) in SGC7901VCR and hnRNPA2B1‐overexpressing SGC7901 cells led to decreased RNA levels of NEAT1 (Figure 4D). MeRIP assays confirmed the m6A modification of NEAT1, and also confirmed that silencing of METTL3 reduced the level of m6A modification in NEAT1 RNA (Figure 4E). Next, we determined whether there is a direct interaction between hnRNPA2B1 and NEAT1. RIP assays demonstrated that NEAT1 was significantly enriched by the hnRNPA2B1 antibody in SGC7901VCR and hnRNPA2B1‐overexpressing SGC7901 cells (Figure 4F), indicating a direct interaction between hnRNPA2B1 and NEAT1. Moreover, knockdown of METTL3 reduced this enrichment (Figure 4G), suggesting that the m6A modification on NEAT1 was necessary for its interaction with hnRNPA2B1. To ascertain the underlying mechanism of hnRNPA2B1‐mediated NEAT1 expression, we treated lentivirus‐transfected SGC7901ADR and SGC7901VCR cells with actinomycin D at 10 µg/mL for 0, 1, 2, 4, 8 or 10 h to halt new RNA synthesis and examined NEAT1 stability because hnRNPA2B1 has been implicated in RNA stability regulation [30]. The results showed that the decay of NEAT1 in the hnRNPA2B1‐silenced cells was faster than that in the control cells (Figure 4H‐I), indicating that hnRNPA2B1 can enhance NEAT1 RNA stability. Taken together, our results demonstrate that hnRNPA2B1 interacted with and stabilized NEAT1 in an m6A modification‐dependent manner.
Then we investigated whether NEAT1 participated in hnRNPA2B1‐mediated GC drug resistance. NEAT1 expression was elevated in GC cells, especially in MGC803 and BGC823 cells, as well as MDR cells (Supplementary Figure S2E). Knockdown of NEAT1 using ASOs (Supplementary Figure S2F) sensitized hnRNPA2B1‐overexpressing SGC7901 cells to ADR, VCR, and 5‐Fu, as indicated by reduced IC50 values (Figure 4J). ASO treatment also abolished the enhancement in proliferation of hnRNPA2B1‐overexpressing SGC7901 cells without or with chemotherapy (Figure 4K). These results suggested that hnRNPA2B1 could promote GC drug resistance via NEAT1.
3.5. hnRNPA2B1 promoted stemness‐like properties of GC cells
Cancer stem cells (CSCs) are potential culprits in GC chemotherapy resistance [31]. The mRNAsi describes the degree of similarity between tumor cells and stem cells, and higher mRNAsi scores are associated with higher CSC potential [26]. By analyzing mRNAsi scores in the TCGA‐STAD dataset, we found that the hnRNPA2B1 was positively correlated with the mRNAsi scores (Figure 5A). Additionally, ssGSEA was implemented based on our RNA sequencing results, and the 10 stemness‐related gene signatures were enriched in the ADR_shNC group compared with ADR_sh group (Supplementary Figure S3A), indicating that knockdown of hnRNPA2B1 may impair cancer stemness in GC. Then we further detected NEAT1 and CSC markers through qPCR and Western blotting analysis in MDR GC cell lines. The results illustrated that the expression of NEAT1 and CSC markers was significantly enhanced in the MDR GC cells compared with the parental cell SGC7901 (Supplementary Figure S3B‐C). In addition, qPCR indicated that downregulation of hnRNPA2B1 suppressed the expression of CSC surface markers including CD133, CD44, and LGR5, and pluripotent transcription factors including Nanog, Oct4, and Sox2 in SGC7901ADR and SGC7901VCR cells, while overexpression of hnRNPA2B1 in SGC7901 and HGC27 cells significantly elevated the expression of these genes (Figure 5B, Supplementary Figure S3D). Moreover, IF and Western blotting also illustrated that hnRNPA2B1 exerted positive effect on the expression of CD133 and CD44 (Figure 5C, Supplementary Figure S3E), indicating that hnRNPA2B1 may play a critical role in GC stemness acquisition.
FIGURE 5.
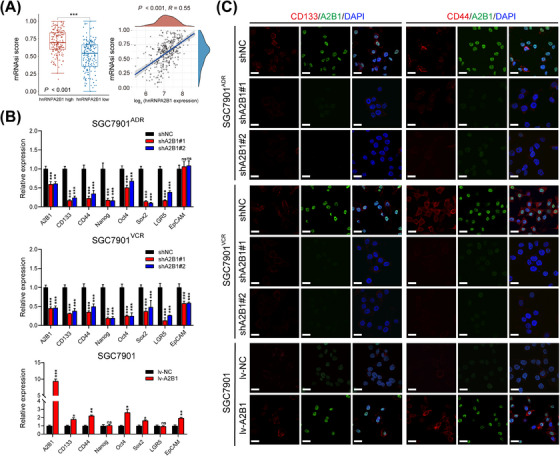
hnRNPA2B1 promoted cancer stemness‐like properties in vitro. (A) The expression of hnRNPA2B1 was correlated with mRNAsi scores in the TCGA GC dataset. (B‐C) qPCR and IF illustrated that regulation of hnRNPA2B1 resulted in corresponding changes in the expression of GCSC surface markers and pluripotent transcription factors in SGC7901ADR, SGC7901VCR, and parental SGC7901 cells. *P < 0.05, ** P < 0.01, *** P < 0.001, ns, not significant. Abbreviations: CD133, cluster of differentiation 133; CD44, cluster of differentiation 44; EpCAM, epithelial cell adhesion molecule; GCSC, gastric cancer stem cell; hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; lv‐NC, empty overexpression control; lv‐A2B1, overexpression of hnRNPA2B1; LGR5, leucine‐rich repeat‐containing G‐protein coupled receptor 5; Nanog, homeobox protein Nanog; Oct4, octamer‐binding protein 4; shA2B1, short hairpin RNA of hnRNPA2B1; shNC, negative control short hairpin RNA; Sox2, SRY‐box transcription factor 2.
To further explore the role of hnRNPA2B1 in stemness acquisition, we performed sphere formation assay in GC cells. The results showed that knockdown of hnRNPA2B1 significantly reduced the size and number of spheroids of SGC7901ADR and SGC7901VCR cells, while overexpression of hnRNPA2B1 improved the size and number of spheroids of SGC7901 cells (Figure 6A‐B). To exclude the effects of adhesive qualities on sphere formation, we performed secondary and tertiary sphere formation assays, similar results were also observed in SGC7901ADR, SGC7901VCR and SGC7901 cells (Figure 6A‐B). Furthermore, co‐expression of hnRNPA2B1, CD133, and CD44 was found in SGC7901ADR, SGC7901VCR and SGC7901 tumor spheroids, whereas knockdown of hnRNPA2B1 dramatically diminished CD133 and CD44 expression (Figure 6C). Collectively, these results demonstrate that hnRNPA2B1 enhances the stemness properties of GC cells.
FIGURE 6.
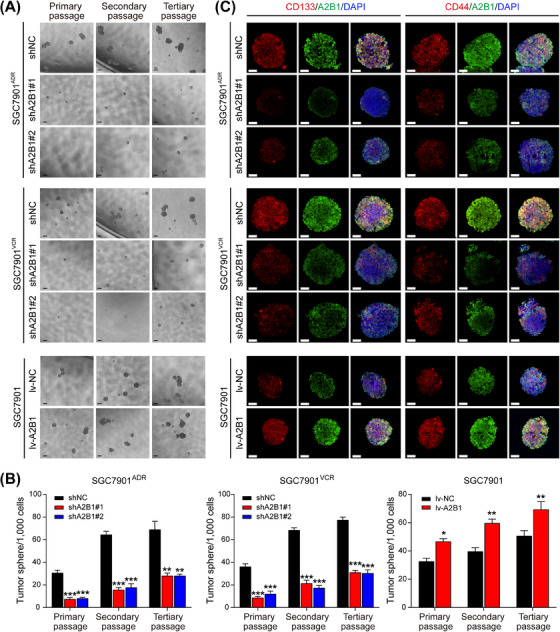
hnRNPA2B1 increased the self‐renewal and tumorigenic capacities of GCSCs in vitro. (A‐B) Representative images (A) and quantification (B) of the sphere‐forming abilities of SGC7901ADR, SGC7901VCR and SGC7901 at the indicated passages. Scale bar, 200 µm. (C) Representative images of CD133 /hnRNPA2B1 (left) and CD44/hnRNPA2B1 (right) immunostaining in SGC7901ADR, SGC7901VCR and SGC7901 tumor spheroids. Scale bars, 50 µm. The P value was calculated by one‐way ANOVA test (B left and middle) and paired t test (B right). *P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: A2B1, heterogeneous nuclear ribonucleoprotein A2B1; CD133, cluster of differentiation 133; CD44, cluster of differentiation 44; GCSC, gastric cancer stem cell; lv‐NC, empty overexpression control; lv‐A2B1, overexpression of hnRNPA2B1; shNC, negative control short hairpin RNA; shA2B1, short hairpin RNA of hnRNPA2B1.
3.6. hnRNPA2B1 and NEAT1 worked together to enhance stemness properties of GC cells and promote chemoresistance via Wnt/β‐catenin pathway
It was reported that NEAT1 confers chemoresistance and cancer stemness by activating various signaling pathways [32, 33]. Therefore, we supposed that hnRNPA2B1 and NEAT1 may regulate cancer stemness in GC through certain pathways. GC patients in TCGA‐STAD dataset with both hnRNPA2B1 and NEAT1 high expression (hnRNPA2B1highNEAT1high group, n = 105) and low expression (hnRNPA2B1lowNEAT1low group, n = 104) were selected and subjected to GSEA analysis. Results showed that the Wnt/β‐catenin pathway was significantly enriched in the hnRNPA2B1high NEAT1high group (Figure 7A), suggesting that hnRNPA2B1 and NEAT1 may activate the Wnt/β‐catenin signaling pathway in GC. Furthermore, downregulation of hnRNPA2B1 significantly reduced the expression of active β‐catenin, total β‐catenin, and Wnt/β‐catenin pathway downstream proteins such as c‐Myc and cyclin D1, while overexpression of hnRNPA2B1 exerted the opposite effect (Figure 7B). This suggested that hnRNPA2B1 could enhance the activity of Wnt/β‐catenin pathway in GC.
FIGURE 7.
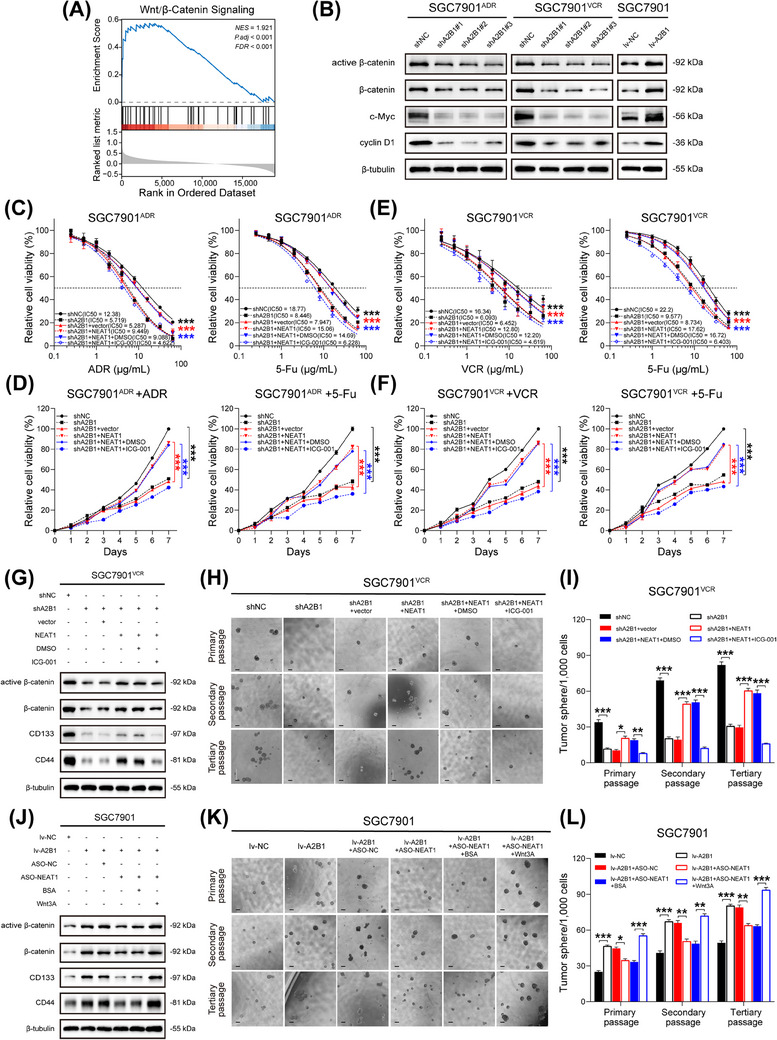
hnRNPA2B1 and NEAT1 enhanced stemness and promoted chemoresistance via Wnt/β‐catenin pathway in GC. (A) GSEA analysis between hnRNPA2B1high NEAT1high group (n = 105) and hnRNPA2B1low NEAT1low group (n = 104). (B) Immunoblots of active β‐catenin, β‐catenin, c‐Myc, and cyclin D1 in hnRNPA2B1‐silenced SGC7901ADR cells, hnRNPA2B1‐silenced SGC7901VCR cells and hnRNPA2B1‐overexpressed SGC7901 cells cell lines respectively. (C‐D) IC50 values and growth curves of SGC7901ADR cells after the indicated treatment (ICG‐001, 25 µmol/L for 24 h). (E‐F) IC50 values and growth curves of SGC7901VCR cells after the indicated treatment (ICG‐001, 25 µmol/L for 24 h). (G) Immunoblots of active β‐catenin, β‐catenin, CD133 and CD44 in SGC7901VCR cells after the indicated treatment (ICG‐001, 25 µmol/L for 24 h). (H‐I) Representative images (H) and quantification (I) of the sphere‐forming abilities of SGC7901VCR cells after the indicated treatment. Scale bar, 200 µm. (J) Immunoblots of active β‐catenin, β‐catenin, CD133 and CD44 in SGC7901 cells after the indicated treatment (Wnt3A, 100 ng/mL for 24 h). (K‐L) Representative images (K) and quantification (L) of the sphere‐forming abilities of SGC7901 cells after the indicated treatment. Scale bar, 200 µm. The P value was calculated by two‐way ANOVA test (C‐F), and paired t test (I and L). *P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: CD133, cluster of differentiation 133; CD44, cluster of differentiation 44; FDR, false discovery rate; GSEA, gene set enrichment analysis; hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; lv‐NC, empty overexpression control; lv‐A2B1, overexpression of hnRNPA2B1; NES, normalized enrichment score; P.adj, adjust P value; shNC, negative control short hairpin RNA; shA2B1, short hairpin RNA of hnRNPA2B1.
We then asked whether NEAT1 contributes to GC stemness properties. Interestingly, simultaneously silencing NEAT1 by ASO in hnRNPA2B1‐overexpressing SGC7901 cells hindered the expression of CD133, CD44, Nanog, Oct4, and Sox2, and similar results were observed in MGC803 and BGC823 cells (Supplementary Figure S3F‐G), while overexpression of NEAT1 in hnRNPA2B1‐silenced SGC7901ADR cells and SGC7901VCR cells exerted the opposite effect on stem cell markers (Supplementary Figure S3H). NEAT1 overexpression in hnRNPA2B1‐silenced SGC7901ADR cells showed drug resistance as revealed by increased IC50 values and faster proliferation rate. However, administration of ICG‐001, an inhibitor of β‐catenin/TCF mediated transcription, significantly reversed the effects of NEAT1 overexpression and resulted in increased drug sensitivity (Figure 7C‐D). Similar results were seen in hnRNPA2B1‐silenced SGC7901VCR cells (Figure 7E‐F). Moreover, treatment with ICG‐001 reversed the promoting effect of NEAT1 overexpression on the protein levels of active β‐catenin, total β‐catenin, CD133, and CD44 in hnRNPA2B1‐silenced SGC7901VCR cells (Figure 7G). Besides, treatment with ICG‐001 significantly decreased the size and number of spheroids which were previously enhanced by NEAT1 overexpression (Figure 7H‐I). In contrast, administration of recombinant Wnt3A, which activates Wnt/β‐catenin signaling, significantly reversed the inhibitory effect of NEAT1 silencing in hnRNPA2B1‐overexpressed SGC7901 cells (Figure 7J‐L). Taken together, the above results depicted that hnRNPA2B1 and NEAT1 worked together to enhance stemness properties of GC cells and exacerbate chemoresistance in GC via Wnt/β‐catenin pathway.
3.7. The clinical significance of cancer stemness‐associated hnRNPA2B1/NEAT1 axis in human GC tissues
To further assess the clinical significance of cancer stemness‐associated hnRNPA2B1/NEAT1 axis, we firstly analyzed the UALCAN database. The expression level of NEAT1 was significantly elevated in GC tissues compared with noncancerous tissues, and elevated NEAT1 expression was related with poor OS in GC patients, especially in those who had received 5‐Fu‐based adjuvant chemotherapy (Supplementary Figure S4A). Next, the expression NEAT1 was found significantly increased in 20 GC tissues compared with the paired adjacent normal tissues (Supplementary Figure S4B). A positive expression pattern between hnRNPA2B1 and NEAT1 was identified in GC tissues (Supplementary Figure S4C), and their co‐localization is shown in Supplementary Figure S4D. As for the cancer stemness, database analysis illustrated that the expression of CD133 and CD44 was significantly enhanced in GC tissues, and the elevated expression was related with poor OS in patients, especially in those who had received 5‐Fu treatment, whereas there was no significant difference between the expression of CD133 and OS in patient who had received 5‐Fu treatment (Supplementary Figure S4E). In addition, CD133, CD44, and hnRNPA2B1 were significantly overexpressed in a GC microarray containing 57 paired GC specimens, and a positive expression pattern was observed among them (Supplementary Figure S4F‐G). The above results illustrated that hnRNPA2B1, NEAT1, CD133, and CD44 were upregulated in GC tissues and related with poor prognosis, especially in patients with chemotherapy treatment.
Furthermore, 30 cases of surgically resected specimens were collected from GC patients who received postoperative adjuvant chemotherapy, and were divided into two groups, chemo‐sensitive vs. chemo‐resistant, based on if they derived or did not derive a survival benefit from chemotherapy (disease free survival [DFS] ≥ or < 2 years). We detected the expression of NEAT1 by RNAscope and the expression of hnRNPA2B1, CD133, and CD44 by mIHC. The results revealed that in clinical specimens from GC patients with chemotherapy treatment, the expression levels of hnRNPA2B1, NEAT1, CD133, and CD44 were markedly elevated in chemo‐resistant patients compared to chemo‐sensitive patients (Figure 8A‐B). Positive correlations between hnRNPA2B1 and NEAT1, CD133, or CD44 were observed (Figure 8C), suggesting that the cancer stemness‐associated hnRNPA2B1/NEAT1 axis may potentially contribute to the development of chemotherapy resistance in GC patients.
FIGURE 8.
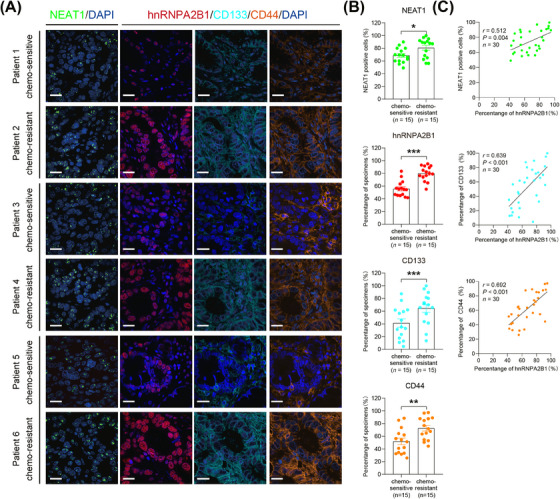
Coordinated expression of hnRNPA2B1, NEAT1, CD133 and CD44 in GC specimens. (A) Representative images of RNAscope ISH for NEAT1 in GC specimens (left panel). Scale bars: 20 µm. Multispectral staining and imaging for hnRNPA2B1, CD133 and CD44 in serial sections of GC tissues (right panel). Scale bars: 20 µm. (B) Semi‐quantification of the expression of NEAT1, hnRNPA2B1, CD133, and CD44 in chemo‐sensitive and chemo‐resistant GC tissues. *P < 0.05, ** P < 0.01, *** P < 0.001. (C) Association between hnRNPA2B1 levels and NEAT1, CD133 or CD44 levels in GC tissues. Spearman's rank correlation coefficients (r) and P values are shown. The P value was calculated by unpaired t test. Abbreviations: CD133, cluster of differentiation 133; CD44, cluster of differentiation 44; hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; NEAT1, nuclear enriched abundant transcript 1.
4. DISCUSSION
As an m6A reader, hnRNPA2B1 has been implicated in various malignant phenotypes, including therapy resistance, by recognizing and binding specific RNA substrates and DNA motifs [6]. In our previous study, we found that hnRNPA2B1 worked with MIR100HG and TCF7L2 to form a reciprocal positive feedback loop in the cetuximab resistance setting in CRC [7]. However, whether hnRNPA2B1 participates in GC chemoresistance has remained elusive. In this study, we observed that upregulation of hnRNPA2B1 in GC specimens was significantly associated with advanced TNM stage and shorter survival, which is also supported by the previous two studies [34, 35]. More importantly, hnRNPA2B1 expression was elevated in MDR GC cells, and high hnRNPA2B1 expression was associated with poor prognosis in patients receiving 5‐Fu chemotherapy, suggesting that high hnRNPA2B1 expression was associated with unfavorable clinical outcomes, particularly in patients undergoing chemotherapy. Subsequently, we identified that hnRNPA2B1 induced chemoresistance by inhibiting apoptosis and promoting proliferation both in vitro and in vivo.
With respect to the mechanism, we demonstrated that hnRNPA2B1 interacts with NEAT1 and regulates its stability in an m6A‐dependent manner. The collaboration between hnRNPA2B1 and NEAT1 enhances stemness properties through Wnt/β‐catenin pathway and exacerbates chemoresistance. The critical role of lncRNA NEAT1 in contributing cancer chemoresistance has been identified previously [32, 33]. However, whether NEAT1 is related to GC chemoresistance is unknown. In this study, we discovered that NEAT1 mainly accounts for the hnRNPA2B1‐mediated MDR phenotype in GC. Clinical data also confirmed that the expression of hnRNPA2B1 is positively associated with NEAT1. Moreover, we demonstrated that the RNA stability of NEAT1 was regulated by hnRNPA2B1 in an m6A‐dependent manner. Although the m6A modification on NEAT1 was reported previously, the functional role of m6A modification on NEAT1 is still controversial. For example, METTL3‐mediated m6A modification induced the aberrant expression of NEAT1, which promotes disease progression via miR‐766‐5p/CDKN1A axis in chronic myeloid leukemia [36]. However, METTL14 decreased the expression of NEAT1 in an m6A‐dependent manner and suppressed growth and metastasis of renal cell carcinoma [37]. In our study, we demonstrated that m6A modification facilitates NEAT1 expression through recognition and stabilization by hnRNPA2B1. This finding provided a comprehensive understanding of the impact of m6A modification on NEAT1.
Wnt/β‐catenin signaling pathway is a crucial signaling cascade involved in tumor growth, metastasis, chemoresistance, and stemness acquisition [38]. Our previous study found that hnRNPA2B1 interacts with lncRNA MIR100HG and controls the transcriptional activity of Wnt/β‐catenin signaling in CRC cetuximab resistance [7]. In this study, we found that hnRNPA2B1 and NEAT1 worked together to enhance stemness properties of GC cells and exacerbate chemoresistance in GC via Wnt/β‐catenin pathway. However, further investigation is still needed to explore the underlining mechanism of the dysregulation of Wnt/β‐catenin signaling. It was reported that hnRNPA2B1 could aggravate stemness in lung cancer and activate Wnt signaling pathway via inhibiting SFRP2 and promoting m6A‐mediated maturing of miR‐106b‐5p [39]. NEAT1 also promotes cancer stemness and chemoresistance via activation of Wnt/β‐catenin pathway. NEAT1 can bind to EZH2 and facilitate the trimethylation of H3K27 in the promoters of ICAT, GSK3β, and Axin2, which are factors that downregulate Wnt/β‐catenin signaling [40]. Furthermore, NEAT1 could indirectly activate the Wnt/β‐catenin signaling pathway via DDX5 and fulfilled its oncogenic functions in a DDX5‐mediated manner [41]. Consequently, we hypothesized a synergistic effect between hnRNPA2B1 and NEAT1 in activating the Wnt/β‐catenin pathway. Interestingly, Wnt/β‐catenin pathway could regulate m6A modification in multiple levels. For example, positive impact of β‐catenin on METTL3 was implicated in drug resistance of CRC cells with p53 R273H mutant protein [42]. Thus, whether there is a positive feedback loop between hnRNPA2B1 and Wnt/β‐catenin pathway in GC chemotherapy requires further exploration.
Recent research has revealed the existence of gastric cancer stem cells (GCSCs) and elimination of GCSCs could be focused on targeting cell surface markers that are essential for maintaining stemness properties [43]. Many CSC markers, such as CD133 [44], CD44 [45], CD24 [46], CD54 [47], CD90 [48], EpCAM [49], and LGR5 [50], have been employed to identify GCSCs. Among them, CD133 and CD44 are currently the most used cell surface markers [51]. CD133, also known as prominin‐1, belongs to the pentaspan transmembrane glycoprotein family [52]. CD133 expression is associated with tumorigenesis, invasion, metastasis, and drug resistance [53]. Notably, a cisplatin and anti‐CD133 chimeric antigen receptor‐T cell therapy (CAR‐T) combination strategy could simultaneously target normal and stem cell‐like GC cells to improve the treatment outcomes of GC patients [54]. CD44, a cell surface adhesion molecule and the receptor for the hyaluronan glycan, has been implicated in GC as well [55]. Aberrant expression of CD44 and CD44 variants was associated with lymph node metastasis, invasion, chemoresistance, and recurrence in GC [56]. In this study, systematic experiments on MDR cells demonstrated that higher hnRNPA2B1 or NEAT1 expression resulted in a stronger stemness phenotype indicated by CD133 and CD44. Correlation analyses also confirmed that hnRNPA2B1 expression was positively associated with CD133 or CD44 expression in GC specimens. This study has identified the hnRNPA2B1/NEAT1/Wnt pathway as a novel upstream regulator of cancer stemness, providing insight into the dysregulation of cancer stemness in cancer (Figure 9).
FIGURE 9.
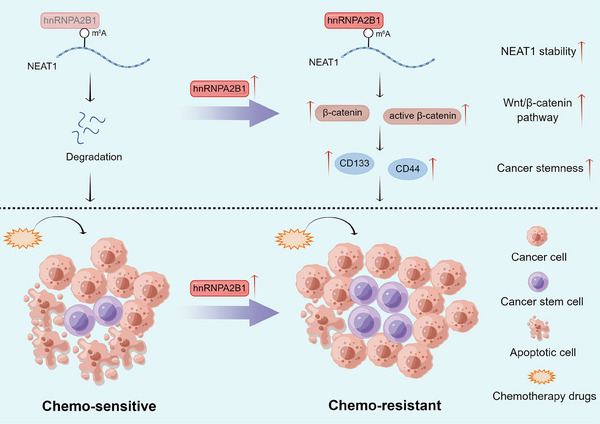
Proposed model for the effect of hnRNPA2B1‐mediated regulation of NEAT1 expression on GC chemoresistance. hnRNPA2B1 interacts with and stabilizes lncRNA NEAT1 in an m6A‐dependent manner. hnRNPA2B1 and NEAT1 collectively maintain cancer cell stemness property via Wnt/β‐catenin pathway and exacerbate chemoresistance in GC. Abbreviations: CD133, cluster of differentiation 133; CD44, cluster of differentiation 44; hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; m6A, N6‐methyladenosine; NEAT1, nuclear enriched abundant transcript 1.
m6A modification was found to be elevated in anti‐cancer drug resistance, and a great number of studies have focused on the underlying mechanisms [4, 57, 58]. Our conclusions are consistent with previous studies about the role of m6A modification in cancer, jointly verifying the detrimental effects of m6A modification in anti‐cancer drug resistance. Moreover, our findings have important therapeutic implications for targeting cancer stemness‐associated hnRNPA2B1/NEAT1 axis in chemotherapy‐resistant GC. For example, MO‐460 is a novel small‐molecule antagonist of hnRNPA2B1. MO‐460 inhibits the initiation of hypoxia‐inducible factor‐1α (HIF‐1α) translation by binding to the C‐terminal glycine‐rich domain of hnRNPA2B1 and inhibiting its subsequent binding to the 3’‐untranslated region of HIF‐1α mRNA, which offers an avenue for the development of novel anticancer therapies [59]. In addition, Wnt/β‐catenin signaling pathway has become a very attractive therapeutic target in recent years. ICG‐001, a preclinical agent which was used in this study, is efficacious to kill tumor cells in both in vitro experiments and mouse xenograft models by binding cyclic AMP response element‐binding protein (CBP) and disrupting its interaction with β‐catenin [60]. PRI‐724, an active enantiomer of ICG‐001, has already entered phase I clinical trials for treating colon cancer and advanced‐stage pancreatic adenocarcinoma [61]. Application of potential Wnt inhibitor seems to be promising in the GC patients with high hnRNPA2B1 expression.
The present study has some limitations as well. For instance, we didn't explore the mechanisms underlying the dysregulation of hnRNPA2B1 in MDR cells. Moreover, how hnRNPA2B1 and NEAT1 promote the activation of Wnt/β‐catenin signaling pathway also needs further investigation. Besides, as NEAT1 is bound by several RBPs, we could not exclude that other RBPs or signaling pathways may also contribute to drug resistance and stemness acquisition in GC.
5. CONCLUSIONS
This study investigated the potential role and mechanism of hnRNPA2B1 in GC chemoresistance. We found that upregulation of hnRNPA2B1 in GC is associated with poor clinical outcomes and that hnRNPA2B1 can induce chemoresistance by interacting with NEAT1 and increasing its stability in an m6A‐dependent manner. Moreover, hnRNPA2B1 and NEAT1 collaboratively enhance stemness properties via the Wnt/β‐catenin pathway and exacerbate chemoresistance. This study implies that the cancer stemness‐associated hnRNPA2B1/NEAT1 axis may represent a novel mechanism of GC chemoresistance. Targeting this axis may present a novel and efficacious therapeutic strategy for chemotherapy‐resistant GC.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Yuanyuan Lu, Xiaodi Zhao, and Daiming Fan designed the study. Jiayao Wang, Jiehao Zhang, Hao Liu, Lingnan Meng, Yihan Zhao, Chen Wang, Xiaoliang Gao, Ahui Fan, and Tianyu Cao performed the experiments, analyzed the data, and prepared the figures and tables. Xianchun Gao collected clinical samples and analyzed clinicopathological information. Jiayao Wang and Jiehao Zhang wrote the manuscript. Yuanyuan Lu, and Xiaodi Zhao conceived the project and supervised and coordinated all aspects of the work.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by Xijing Hospital's Protection of Human Subjects Committee (permit number: KY20222241‐X‐1). Written informed consent was obtained from each patient. All animal studies complied with the Fourth Military Medical University animal use guidelines, and the protocol was approved by the Fourth Military Medical University Animal Care Committee (permit number: IACUC20221013).
CONSENT FOR PUBLICATION
Not applicable.
Supporting information
Supporting Information
ACKNOWLEDGEMENTS
We appreciate the contributions of all participants who generously assisted with the study. We would also like to express our gratitude for the generation of the mechanism diagram by Figdraw. Special thanks to Professor Qiu Xiaoyan and Dr.Huang Xinmei from Peking University for their assistance with the spheroid immunofluorescence assay. This research was supported by National Natural Science Foundation of China (Grant Numbers: 82073197, 82273142, and 82222058).
Wang J, Zhang J, Liu H, Meng L, Gao X, Zhao Y, et al. N6‐methyladenosine reader hnRNPA2B1 recognizes and stabilizes NEAT1 to confer chemoresistance in gastric cancer. Cancer Commun. 2024;44:469–490. 10.1002/cac2.12534
Jiayao Wang, Jiehao Zhang, Hao Liu, and Lingnan Meng contributed equally to this work.
Contributor Information
Daiming Fan, Email: fandaim@fmmu.edu.cn.
Xiaodi Zhao, Email: leedyzhao@fmmu.edu.cn.
Yuanyuan Lu, Email: luyuandreamer@aliyun.com.
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study could be available from the corresponding author upon reasonable request.
REFERENCES
- 1. Qiu H, Cao S, Xu R. Cancer incidence, mortality, and burden in China: a time‐trend analysis and comparison with the United States and United Kingdom based on the global epidemiological data released in 2020. Cancer Commun (Lond). 2021;41(10):1037‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang FH, Zhang XT, Li YF, Tang L, Qu XJ, Ying JE, et al. The Chinese Society of Clinical Oncology (CSCO): Clinical guidelines for the diagnosis and treatment of gastric cancer, 2021. Cancer Commun (Lond). 2021;41(8):747‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jim MA, Pinheiro PS, Carreira H, Espey DK, Wiggins CL, Weir HK. Stomach cancer survival in the United States by race and stage (2001‐2009): Findings from the CONCORD‐2 study. Cancer. 2017;123(Suppl 24):4994‐5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang F, Liu H, Duan M, Wang G, Zhang Z, Wang Y, et al. Crosstalk among m(6)A RNA methylation, hypoxia and metabolic reprogramming in TME: from immunosuppressive microenvironment to clinical application. J Hematol Oncol. 2022;15(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6‐methyladenosine and its role in cancer. Mol Cancer. 2019;18(1):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Y, Shi SL. The roles of hnRNP A2/B1 in RNA biology and disease. Wiley Interdiscip Rev RNA. 2021;12(2):e1612. [DOI] [PubMed] [Google Scholar]
- 7. Liu H, Li D, Sun L, Qin H, Fan A, Meng L, et al. Interaction of lncRNA MIR100HG with hnRNPA2B1 facilitates m(6)A‐dependent stabilization of TCF7L2 mRNA and colorectal cancer progression. Mol Cancer. 2022;21(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bu FT, Wang A, Zhu Y, You HM, Zhang YF, Meng XM, et al. LncRNA NEAT1: Shedding light on mechanisms and opportunities in liver diseases. Liver Int. 2020;40(11):2612‐26. [DOI] [PubMed] [Google Scholar]
- 9. Ghafouri‐Fard S, Khoshbakht T, Hussen BM, Taheri M, Arefian N. Regulatory Role of Non‐Coding RNAs on Immune Responses During Sepsis. Front Immunol. 2021;12:798713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li K, Yao T, Zhang Y, Li W, Wang Z. NEAT1 as a competing endogenous RNA in tumorigenesis of various cancers: Role, mechanism and therapeutic potential. Int J Biol Sci. 2021;17(13):3428‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li K, Wang Z. lncRNA NEAT1: Key player in neurodegenerative diseases. Ageing Res Rev. 2023;86:101878. [DOI] [PubMed] [Google Scholar]
- 12. Wang Z, Li K, Huang W. Long non‐coding RNA NEAT1‐centric gene regulation. Cell Mol Life Sci. 2020;77(19):3769‐79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nusse R, Clevers H. Wnt/β‐Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169(6):985‐99. [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y, Wang X. Targeting the Wnt/β‐catenin signaling pathway in cancer. J Hematol Oncol. 2020;13(1):165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flanagan DJ, Barker N, Costanzo NSD, Mason EA, Gurney A, Meniel VS, et al. Frizzled‐7 Is Required for Wnt Signaling in Gastric Tumors with and Without Apc Mutations. Cancer Res. 2019;79(5):970‐81. [DOI] [PubMed] [Google Scholar]
- 16. Yu S, Li L, Cai H, He B, Gao Y, Li Y. Overexpression of NELFE contributes to gastric cancer progression via Wnt/β‐catenin signaling‐mediated activation of CSNK2B expression. J Exp Clin Cancer Res. 2021;40(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao Q, Yang L, Shen A, Li Y, Li Y, Hu S, et al. A WNT7B‐m(6)A‐TCF7L2 positive feedback loop promotes gastric cancer progression and metastasis. Signal Transduct Target Ther. 2021;6(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cai XJZX, Fan DM. Establishment of multidrug resistant gastric cancer cell line and its biological characteristics. Zhongguo Zhongliu Linchuang. 1994(2):67‐71. [Google Scholar]
- 19. An HZZS, Fan DM. Establishment and characteristics of an adriamycin resistant human gastric carcinoma cell line. Xiandai Xiaohua Bing Ji Neijing Zazhi. 1997;(2):108‐10. [Google Scholar]
- 20. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R‐2HG Exhibits Anti‐tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172(1‐2):90‐105.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He Y, Han Y, Fan AH, Li D, Wang B, Ji K, et al. Multi‐perspective comparison of the immune microenvironment of primary colorectal cancer and liver metastases. J Transl Med. 2022;20(1):454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large‐scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556‐w60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jen J, Tang YA, Lu YH, Lin CC, Lai WW, Wang YC. Oct4 transcriptionally regulates the expression of long non‐coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol Cancer. 2017;16(1):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao W, Zhang S, Zhu Y, Xi X, Bao P, Ma Z, et al. POSTAR3: an updated platform for exploring post‐transcriptional regulation coordinated by RNA‐binding proteins. Nucleic Acids Res. 2022;50(D1):D287‐d94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanehisa M, Furumichi M, Sato Y, Ishiguro‐Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545‐d51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell. 2018;173(2):338‐54.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pinto JP, Kalathur RK, Oliveira DV, Barata T, Machado RS, Machado S, et al. StemChecker: a web‐based tool to discover and explore stemness signatures in gene sets. Nucleic Acids Res. 2015;43(W1):W72‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA‐seq data. BMC Bioinf. 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J. 2010;430(3):379‐92. [DOI] [PubMed] [Google Scholar]
- 30. Jiang F, Tang X, Tang C, Hua Z, Ke M, Wang C, et al. HNRNPA2B1 promotes multiple myeloma progression by increasing AKT3 expression via m6A‐dependent stabilization of ILF3 mRNA. J Hematol Oncol. 2021;14(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garcia‐Mayea Y, Mir C, Masson F, Paciucci R, ME LL. Insights into new mechanisms and models of cancer stem cell multidrug resistance. Semin Cancer Biol. 2020;60:166‐80. [DOI] [PubMed] [Google Scholar]
- 32. Jiang P, Xu H, Xu C, Chen A, Chen L, Zhou M, et al. NEAT1 contributes to the CSC‐like traits of A549/CDDP cells via activating Wnt signaling pathway. Chem Biol Interact. 2018;296:154‐61. [DOI] [PubMed] [Google Scholar]
- 33. Zhu Y, Hu H, Yuan Z, Zhang Q, Xiong H, Hu Z, et al. LncRNA NEAT1 remodels chromatin to promote the 5‐Fu resistance by maintaining colorectal cancer stemness. Cell Death Dis. 2020;11(11):962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peng WZ, Zhao J, Liu X, Li CF, Si S, Ma R. hnRNPA2B1 regulates the alternative splicing of BIRC5 to promote gastric cancer progression. Cancer Cell Int. 2021;21(1):281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dai P, Wang Q, Wang W, Jing R, Wang W, Wang F, et al. Unraveling Molecular Differences of Gastric Cancer by Label‐Free Quantitative Proteomics Analysis. Int J Mol Sci. 2016;17(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yao FY, Zhao C, Zhong FM, Qin TY, Wen F, Li MY, et al. m(6)A Modification of lncRNA NEAT1 Regulates Chronic Myelocytic Leukemia Progression via miR‐766‐5p/CDKN1A Axis. Front Oncol. 2021;11:679634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu T, Wang H, Fu Z, Wang Z, Wang J, Gan X, et al. Methyltransferase‐like 14 suppresses growth and metastasis of renal cell carcinoma by decreasing long noncoding RNA NEAT1. Cancer Sci. 2022;113(2):446‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. White BD, Chien AJ, Dawson DW. Dysregulation of Wnt/β‐catenin signaling in gastrointestinal cancers. Gastroenterology. 2012;142(2):219‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rong L, Xu Y, Zhang K, Jin L, Liu X. HNRNPA2B1 inhibited SFRP2 and activated Wnt‐β/catenin via m6A‐mediated miR‐106b‐5p processing to aggravate stemness in lung adenocarcinoma. Pathol Res Pract. 2022;233:153794. [DOI] [PubMed] [Google Scholar]
- 40. Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J, et al. Long Noncoding RNA NEAT1, Regulated by the EGFR Pathway, Contributes to Glioblastoma Progression Through the WNT/β‐Catenin Pathway by Scaffolding EZH2. Clin Cancer Res. 2018;24(3):684‐95. [DOI] [PubMed] [Google Scholar]
- 41. Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan C, et al. The lncRNA NEAT1 activates Wnt/β‐catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol. 2018;11(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Uddin MB, Roy KR, Hosain SB, Khiste SK, Hill RA, Jois SD, et al. An N(6)‐methyladenosine at the transited codon 273 of p53 pre‐mRNA promotes the expression of R273H mutant protein and drug resistance of cancer cells. Biochem Pharmacol. 2019;160:134‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Han J, Won M, Kim JH, Jung E, Min K, Jangili P, et al. Cancer stem cell‐targeted bio‐imaging and chemotherapeutic perspective. Chem Soc Rev. 2020;49(22):7856‐78. [DOI] [PubMed] [Google Scholar]
- 44. Soleimani A, Dadjoo P, Avan A, Soleimanpour S, Rajabian M, Ferns G, et al. Emerging roles of CD133 in the treatment of gastric cancer, a novel stem cell biomarker and beyond. Life Sci. 2022;293:120050. [DOI] [PubMed] [Google Scholar]
- 45. Takaishi S, Okumura T, Tu S, Wang SS, Shibata W, Vigneshwaran R, et al. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27(5):1006‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang C, Li C, He F, Cai Y, Yang H. Identification of CD44+CD24+ gastric cancer stem cells. J Cancer Res Clin Oncol. 2011;137(11):1679‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gómez‐Gallegos AA, Ramírez‐Vidal L, Becerril‐Rico J, Pérez‐Islas E, Hernandez‐Peralta ZJ, Toledo‐Guzmán ME, et al. CD24+CD44+CD54+EpCAM+ gastric cancer stem cells predict tumor progression and metastasis: clinical and experimental evidence. Stem Cell Res Ther. 2023;14(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fu L, Bu L, Yasuda T, Koiwa M, Akiyama T, Uchihara T, et al. Gastric Cancer Stem Cells: Current Insights into the Immune Microenvironment and Therapeutic Targets. Biomedicines. 2020;8(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Han ME, Jeon TY, Hwang SH, Lee YS, Kim HJ, Shim HE, et al. Cancer spheres from gastric cancer patients provide an ideal model system for cancer stem cell research. Cell Mol Life Sci. 2011;68(21):3589‐605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sigal M, Rothenberg ME, Logan CY, Lee JY, Honaker RW, Cooper RL, et al. Helicobacter pylori Activates and Expands Lgr5(+) Stem Cells Through Direct Colonization of the Gastric Glands. Gastroenterology. 2015;148(7):1392‐404.e21. [DOI] [PubMed] [Google Scholar]
- 51. Wang X, Gao J, Li C, Xu C, Li X, Meng F, et al. In situ gelatinase‐responsive and thermosensitive nanocomplex for local therapy of gastric cancer with peritoneal metastasis. Mater Today Bio. 2022;15:100305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Glumac PM, LeBeau AM. The role of CD133 in cancer: a concise review. Clin Transl Med. 2018;7(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yuan Z, Liang X, Zhan Y, Wang Z, Xu J, Qiu Y, et al. Targeting CD133 reverses drug‐resistance via the AKT/NF‐κB/MDR1 pathway in colorectal cancer. Br J Cancer. 2020;122(9):1342‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Han Y, Sun B, Cai H, Xuan Y. Simultaneously target of normal and stem cells‐like gastric cancer cells via cisplatin and anti‐CD133 CAR‐T combination therapy. Cancer Immunol Immunother. 2021;70(10):2795‐803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao S, Chen C, Chang K, Karnad A, Jagirdar J, Kumar AP, et al. CD44 Expression Level and Isoform Contributes to Pancreatic Cancer Cell Plasticity, Invasiveness, and Response to Therapy. Clin Cancer Res. 2016;22(22):5592‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zavros Y. Initiation and Maintenance of Gastric Cancer: A Focus on CD44 Variant Isoforms and Cancer Stem Cells. Cell Mol Gastroenterol Hepatol. 2017;4(1):55‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li B, Jiang J, Assaraf YG, Xiao H, Chen ZS, Huang C. Surmounting cancer drug resistance: New insights from the perspective of N(6)‐methyladenosine RNA modification. Drug Resist Updat. 2020;53:100720. [DOI] [PubMed] [Google Scholar]
- 58. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Soung NK, Kim HM, Asami Y, Kim DH, Cho Y, Naik R, et al. Mechanism of the natural product moracin‐O derived MO‐460 and its targeting protein hnRNPA2B1 on HIF‐1α inhibition. Exp Mol Med. 2019;51(2):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, et al. A small molecule inhibitor of beta‐catenin/CREB‐binding protein transcription. Proc Natl Acad Sci USA. 2004;101(34):12682‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ko AH, Chiorean EG, Kwak EL, Lenz H‐J, Nadler PI, Wood DL, et al. Final results of a phase Ib dose‐escalation study of PRI‐724, a CBP/beta‐catenin modulator, plus gemcitabine (GEM) in patients with advanced pancreatic adenocarcinoma (APC) as second‐line therapy after FOLFIRINOX or FOLFOX. J Clin Oncol. 2016;34(15_suppl):e15721‐e. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that supports the findings of this study could be available from the corresponding author upon reasonable request.


