Abstract
Sphingolipids are components of plant membranes, and their heterogeneous distribution gives different membrane systems distinct properties. For example, glycosyl inositol phosphoceramides (GIPCs), 1 major type of sphingolipids, aggregate in the outer layer of the plasma membrane (PM), as well as in extracellular vesicles (EVs), including the small (30 to 100 nm) EVs termed exosomes. How these sphingolipids are sorted and trafficked is not clear. In this work, we report that Arabidopsis thaliana TETRASPANIN8 (TET8) acts as a sphingolipid carrier and thus regulates the export of GIPCs from the Golgi apparatus. TET8 recognized the coat protein complex I (COPI) subunit γ2-COPI and moved to its proper location in the PM; this recognition required the TET8 C-terminal tail. Deleting the C-terminal tail of TET8 largely restricted its roles in GIPC transport and endosomal trafficking. Further, we show that TET8 affects EV secretion in association with GIPCs. Thus, our findings shed light on GIPC transport and the molecular machinery involved in EV biogenesis.
TETRASPANIN 8 transports glycosyl inositol phosphoceramide sphingolipids and participates in exosome biogenesis.
Introduction
Sphingolipids are a class of amphiphilic molecules that have a ceramide (Cer) backbone with a fatty acid and a sphingoid long-chain base (LCB; Carter et al. 1958). In plants, sphingolipids can be broadly classified as LCBs, Cers, hydroxyceramides (hCers), glucosylceramides (GlcCers), and glycosyl inositol phosphoceramides (GIPCs; Markham and Jaworski 2007). In Arabidopsis (Arabidopsis thaliana) leaf cells, Cers, hCers, GlcCers, and GIPCs account for ∼5%, ∼5%, ∼30%, and ∼60% of total sphingolipids, respectively, although the percentages vary in different membrane systems (Markham and Jaworski 2007; Liu et al. 2020a; Carmona-Salazar et al. 2021). GIPCs, for example, represent ∼30% of total sphingolipids in the vacuolar membrane, ∼67% in the plasma membrane (PM), and 99% in extracellular vesicles (EVs; Liu et al. 2020b; Carmona-Salazar et al. 2021). GIPCs tend to accumulate in membranes (PM and EVs) that are exposed to the extracellular (EC) environment (Liu et al. 2020b; Carmona-Salazar et al. 2021). GIPCs are important for normal plant growth and can act as signaling receptors in the PM for sodium (Na+) or fungal toxins (Rennie et al. 2014; Jiang et al. 2019; Lenarčič et al. 2017). Thus, investigating the mechanisms involved in GIPC sorting and trafficking may help us better understand plant responses to external stimuli that depend on this class of lipids.
In animals, the transmembrane protein p24 recruits coat protein complex I (COP I) vesicles that bud from the Golgi; these vesicles participate in the sorting and trafficking of the sphingolipid N-stearoyl sphingomyelin (Brügger et al. 2000; Contreras et al. 2012). The COPI complex has been reported to mediate lipid homeostasis (Contreras et al. 2012; Beller et al. 2008). Moreover, knocking out TRANSMEMBRANE 9 SUPERFAMILY MEMBER 2 in human bladder cancer cell (TM9SF2, encoding a transmembrane protein containing 9 transmembrane domains) impairs endosomal trafficking as well as the accumulation of the glycosphingolipid globotriaosylceramide (Tian et al. 2018). We previously showed that an Arabidopsis mutant defective in TETRASPANIN8 (TET8) contained lower levels of cellular GIPCs and secreted fewer EVs than the wild type (WT; Liu et al. 2020b), suggesting that GIPCs play roles in the secretion or function of plant EVs. TET8 is a member of the tetraspanin family and has been proposed to interact with GIPCs, as its fourth transmembrane domain (TMD4) contains a putative sphingolipid-binding site (Liu et al. 2020b; Contreras et al. 2012). However, how TET8 affects GIPCs is unknown.
EVs are lipid-bound vesicles that are released from the cell and carry proteins, nucleic acids, metabolites, and other biomolecules (Colombo et al. 2014). Plants and animals produce diverse EVs by different mechanisms. In mammalian cells, EVs are well characterized (Alenquer and Amorim 2015): apoptotic cells shed large apoptotic bodies (500 to 2,000 nm in diameter) and healthy cells release microvesicles (100 to 1,000 nm) and exosomes (30 to 100 nm; Harding et al. 1984). In general, microvesicles vary in size and are directly shed from the PM (Harding et al. 1984). In contrast, exosomes have a more uniform size distribution than microvesicles (Harding et al. 1984) and originate as intralumenal vesicles (ILVs) that form through inward budding of membrane-delimited multivesicular bodies (MVBs) within cells.
In mammalian cells, extensive research has been conducted on the mechanisms underlying exosome biogenesis (Harding et al. 1984; Hessvik and Llorente 2018). The endosomal sorting complex required for transport (ESCRT) machinery and a tetraspanin-dependent pathway function in MVB maturation (Escola et al. 1998; Huotari and Helenius 2011). Once mature, MVBs are absorbed by lysosomes for degradation or fuse with the PM to release exosomes (Huotari and Helenius 2011).
Additional work in mammalian cells has shown that soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), Rab GTPases, tethering factors, and lipids (including precursors) affect exosome release to the EC space from cells (Pfeffer 2007; Singh et al. 2014). Lipids that function in vesicular transport are vital for membrane deformation, fission, and fusion and thus affect exosome release (McMahon and Boucrot 2015). For example, in mammalian cells, a flotillin protein recruits cholesterol to MVBs and promotes the release of cholesterol-enriched exosomes into the intercellular space (Strauss et al. 2010).
Whether a similar mechanism exists in plants is not clear. Indeed, our understanding of plant exosome biogenesis and function remains rudimentary. Research on plants has primarily focused on the role of the Golgi in MVB biogenesis and regulating exosome maturation, both of which play an integral role in exosome production (Seguí-Simarro and Staehelin 2006; Scheuring et al. 2011). We previously proposed a link among TET8, GIPCs, and EV secretion (Liu et al. 2020b). In this study, we focus on the TET8-mediated pathway mediating GIPC trafficking and provide insight on GIPC–TET8-coupled exosome secretion.
Results
TET8 can bind to GIPCs and acts as a GIPC carrier
As TET8 mainly localized to the PM, we focused on GIPC contents in the PM, using sterols as an internal standard (campesterol, sitosterol, stigmasterol, and stigmastanol; Liu et al. 2020b; Supplemental Fig. S1). These sterols displayed no changes in leaf cell lipids between WT and tet8 (Liu et al. 2020b). However, the GIPCs/sterols ratio was significantly lower in the tet8 mutant relative to WT (Supplemental Fig. S1 and Data Set 1).
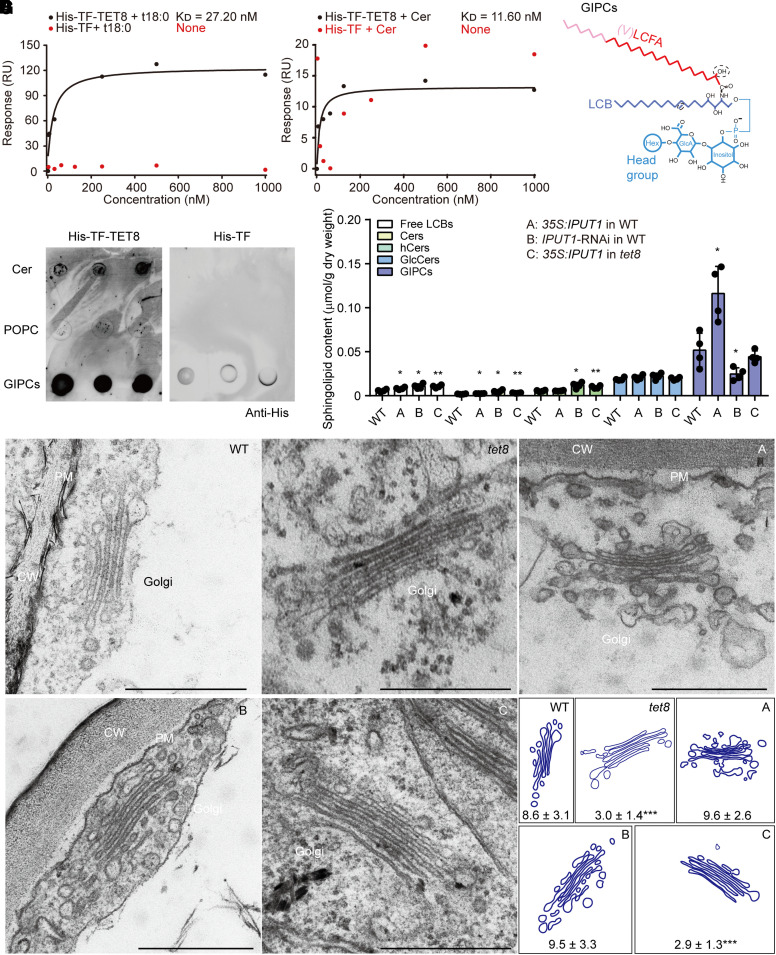
To specifically test for an interaction between TET8 and GIPCs, we conducted surface plasmon resonance (SPR) assays using purified recombinant His-TF-TET8 (a trigger factor [TF] chaperone) and sphingolipids (Supplemental Fig. S2; Fig. 1, A and B). The SPR analysis showed that recombinant His-TF-TET8 can bind strongly to phytosphinganine (t18:0, KD [affinity] = 27.2 nM) and Cer (t18:0-c24:0, KD [affinity] = 11.6 nM, backbone of GIPCs; Fig. 1, A to C). As a complementary approach, we spotted isolated GIPCs from Arabidopsis leaves onto a PVDF membrane for lipid strip tests (Fig. 1, C and D). Cer (t18:0-c24:0) and the phospholipid 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (16:0-18:1 PC, POPC) were used as positive and negative controls, respectively (Fig. 1D). After incubating the strips with recombinant His-TF-TET8, we detected a direct interaction between TET8 and leaf GIPCs (Fig. 1D). Therefore, we hypothesize that TET8 affects GIPC biosynthesis and/or participates in GIPC transport.
Figure 1.
TET8 interacts with sphingolipids and acts as a GIPC carrier. A) SPR analysis of recombinant His-TF-TET8 protein with t18:0. B) SPR analysis of recombinant His-TF-TET8 protein with Cer t18:0-c24:0. In A) and B), the KD (affinity) values were calculated based on the fitting curves and represent the binding affinity between protein and lipid. None, the molecule cannot bind to the target protein His-TF (negative control). C) Molecular structure of GIPC. The red line represents the long-chain fatty acid (LCFA), the pink part represents the very-long-chain fatty acid (VLCFA), the purple part represents long-chain base (LCB), and blue lines represent the head group of GIPC. D) Lipid strip assays showing that GIPCs can bind to His-TF-TET8. Same amounts of different lipids were spotted onto a PVDF membrane. Cer (t18:0-c24:0) and POPC (16:0-18:1) were used as a positive and negative control, respectively, for GIPCs. Recombinant His-TF protein was used as a negative control for His-TF-TET8. E) The sphingolipid profiles of WT, 35S:IPUT1 in WT (A), IPUT1-RNAi in WT (B), and 35S:IPUT1 in tet8 (C). Four independent repeats (see black dots) were conducted. Data are shown as means ± Sd. Statistical significance was determined using Student's t test. *P < 0.05; **P < 0.01. Details are listed in Supplemental Data Set 2. F, G) Golgi observations F) and analysis of leaf cells from 4-wk-old plants of WT, tet8, 35S:IPUT1 in WT (A), IPUT1-RNAi in WT (B), and 35S:IPUT1 in tet8 (G). CW, cell wall; PM, plasma membrane. Scale bars, 500 nm. The number of vesicles around the Golgi is represented as means ± Sd. Statistical significance was determined between WT and other plants using Student's t test. ***P < 0.001.
The Golgi-located GIPC glucuronosyltransferase INOSITOL PHOSPHORYLCERAMIDE GLUCURONOSYLTRANSFERASE 1 (IPUT1) is involved in the biosynthesis of GIPCs (Rennie et al. 2014). Compared to WT plants, the content of GIPCs was twice as high in IPUT1-overexpressing plants (35S:IPUT1) and decreased by half in an RNA interference (RNAi) line (IPUT1-RNAi) targeting the IPUT1 transcript (Fig. 1E; Supplemental Data Set 2 and Fig. S3). Overexpressing IPUT1 in the tet8 background also elevated the cellular content of GIPCs but altered sphingolipid homeostasis by leading to the accumulation of LCBs, Cers, and hCers compared to WT (Fig. 1E). These lipid profiling results indicate that the loss of TET8 function does not completely block the function of the GIPC synthase IPUT1; however, loss of TET8 function decreases the final concentration of GIPCs.
Biosynthesis of the sphingolipid backbone initiates in the endoplasmic reticulum (ER), but GIPCs form in the Golgi apparatus (Rennie et al. 2014). We thus investigated Golgi morphology and its surrounding vesicles in leaf cells from WT, tet8, 35S:IPUT1, and IPUT1-RNAi plants using transmission electron microscopy (TEM). We detected fewer vesicles around the Golgi apparatus in the tet8 mutant, whereas neither overexpressing nor knocking down IPUT1 affected the number of Golgi vesicles (Fig. 1, F and G). IPUT1 overexpression did not return the number of Golgi vesicles in the tet8 mutant back to WT levels (Fig. 1, F and G). Together, these data suggest that TET8 can bind to GIPCs and modulate their content through the Golgi, probably in parallel to GIPC biosynthesis.
The C-terminal tail of Arabidopsis TET8 may be responsible for COPI recognition and proper localization
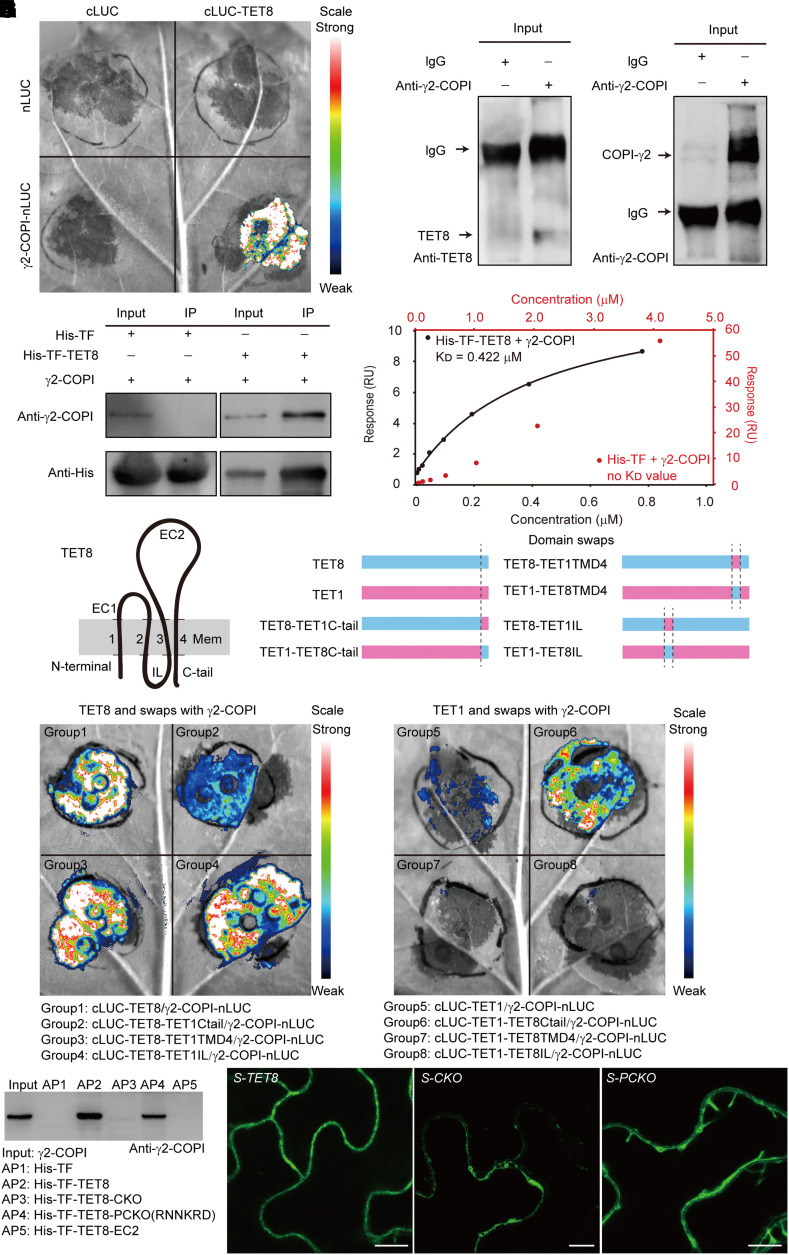
To examine how TET8 affects sphingolipids, we performed affinity purification coupled with mass spectrometry (AP–MS) using recombinant His-TF-TET8 as bait (Supplemental Data Set 3 and Fig. S2). Among the 286 candidate interactors, we noticed several subunits of the COPI complex. In Arabidopsis, the coatomer is composed of several subunits encoded by at least 11 genes (Cabada Gomez et al. 2020; Sánchez-Simarro et al. 2022). Split firefly luciferase (LUC) imaging (LCI) assays showed that only γ2-COPI (also known as Sec21p) strongly binds to TET8 (Supplemental Fig. S4; Fig. 2A). We confirmed the interaction of γ2-COPI and TET8 by co-immunoprecipitation (Co-IP) and pull-down assays (Fig. 2, B and C). We also quantified the binding affinity of TET8 for γ2-COPI by SPR, yielding KD (affinity) = 0.422 µM (Fig. 2D).
Figure 2.
The C-tail of TET8 can recognize γ2-COPI and determines proper TET8 localization. A) Bimolecular luminescence complementation (BiLC) assay showing that TET8 interacts with γ2-COPI strongly. Luminescence intensities indicate the degree of binding capacity. B) Co-IP assays showing that TET8 interacts with γ2-COPI in vivo. Beads incubated with IgG were used as negative control. C) Pull-down assays demonstrating that recombinant His-TF-TET8 binds to γ2-COPI in vitro. Beads incubated with recombinant His-TF were used as negative control. For B) and C), same amounts of total proteins or bait proteins were loaded. D) SPR assay showing that His-TF-TET8 can bind to γ2-COPI. The KD (affinity) value was calculated based on the fitting curves and represents the binding affinity. E) Topology of TET8 in the membrane. The transmembrane domains of TET8 are numbered 1 to 4. EC1, extracellular loop 1; EC2, extracellular loop 2, IL, internal loop; C-tail, C-terminal tail. F) Diagrams illustrating the domain swaps between TET1 and TET8. G, H) BiLC assays showing the interaction between recombinant TET proteins with γ2-COPI. Fluorescence intensities indicate the degree of binding capacity. I) Pull-down assays showing the direct interaction between TET8-C-tail and γ2-COPI. AP, affinity precipitate. The same amounts of proteins were loaded in each lane. J) Cellular distribution of EGFP-TET8, EGFP-TET8-CKO, and EGFP-TET8-PCKO in transgenic plants. Scale bars, 10 μm.
According to the topology of TET8 in membranes, the protein can be divided into 3 cytosolic domains (N-terminal part, internal loop [IL], and C-terminal tail [C-tail]), 2 EC domains (EC1 and EC2), and 4 transmembrane domains (Fig. 2E). Although we observed an interaction between TET8 and γ2-COPI when coinfiltrating the constructs cLUC-TET8 (encoding a fusion between the C-terminal half of LUC and TET8) and γ2-COPI-nLUC (encoding a fusion between γ2-COPI and the N-terminal half of LUC), we failed to detect an interaction when changing the positions of nLUC and cLUC with the constructs TET8-nLUC and cLUC-γ2-COPI (Supplemental Fig. S5, A to C; Fig. 2A). This result suggests that the interaction between γ2-COPI and TET8 mainly relies on the IL or the C-tail.
To find a proper control, we compared all 17 Arabidopsis tetraspanins (Supplemental Fig. S5D). TET1, TET2, and TET9 have the same number of amino acids (aa) in their C-tail and about the same length for their IL as TET8. We tested the interaction between γ2-COPI and each tetraspanin using the LCI assay (Supplemental Fig. S5E). As we detected no interaction between γ2-COPI and TET1 (Supplemental Fig. S5E), we chose this tetraspanin as a negative control. We then swapped the IL, or the TMD4 (containing the sphingolipid-binding site), or the C-tail domain between TET1 and TET8 (Fig. 2F). The LCI results showed that the TET8 variant containing the TET1-C-tail no longer bind to γ2-COPI; conversely, the TET1 variant harboring the TET8-C-tail gained the ability to bind to γ2-COPI (Fig. 2, G and H). Notably, neither the IL nor TMD4 was responsible for the interaction (Fig. 2, G and H). We removed the entire C-tail of TET8 or part of the C-tail (retaining the RNNKRD sequence; Gao et al. 2012) to produce the variant recombinant proteins His-TF-TET8-C-tail-knockout (CKO) or His-TF-TET8-part of C-tail-knockout (PCKO) to test their interaction with γ2-COPI by pull-down assay (Supplemental Fig. S2; Fig. 2I). The presence of the RNNKRD motif in the C-tail of TET8 was necessary for the interaction with TET8, as we detected γ2-COPI by immunoblotting among the proteins pulled down with the TET8 variant PCKO, but not with CKO. The 139-aa fragment of TET8 comprising EC2 (His-TF-EC2) and His-TF showed no interaction with γ2-COPI (Fig. 2I). These data indicate that the C-tail of TET8 mediates the interaction with γ2-COPI.
We then examined the localization of these TET8 variants using the additional transgenic lines proTET8:EGFP-TET8 in tet8 (S-TET8), proTET8:EGFP-TET8-CKO in tet8 (S-CKO), and proTET8:EGFP-TET8-PCKO in tet8 (S-PCKO) (Supplemental Fig. S6; Fig. 2J). We detected full-length, intact TET8 mainly at the PM by confocal microscopy (Fig. 2J; Supplemental Fig. S7F) as well as a minor fraction in MVB–exosomes, as evidenced by immunogold TEM (Supplemental Fig. S7, A to D). In contrast, less of the TET8-CKO variant accumulated at the PM; instead TET8-CKO accumulated in well-defined puncta. The localization pattern for the variant TET8-PCKO was intermediate between that of intact TET8 and TET8-CKO (Fig. 2J; Videos 1 to 3). Thus, we conclude that the C-tail of TET8 is necessary for protein function, including providing a recognition signal for γ2-COPI and assisting in proper subcellular localization.
TET8 regulates the exit of GIPCs from the Golgi
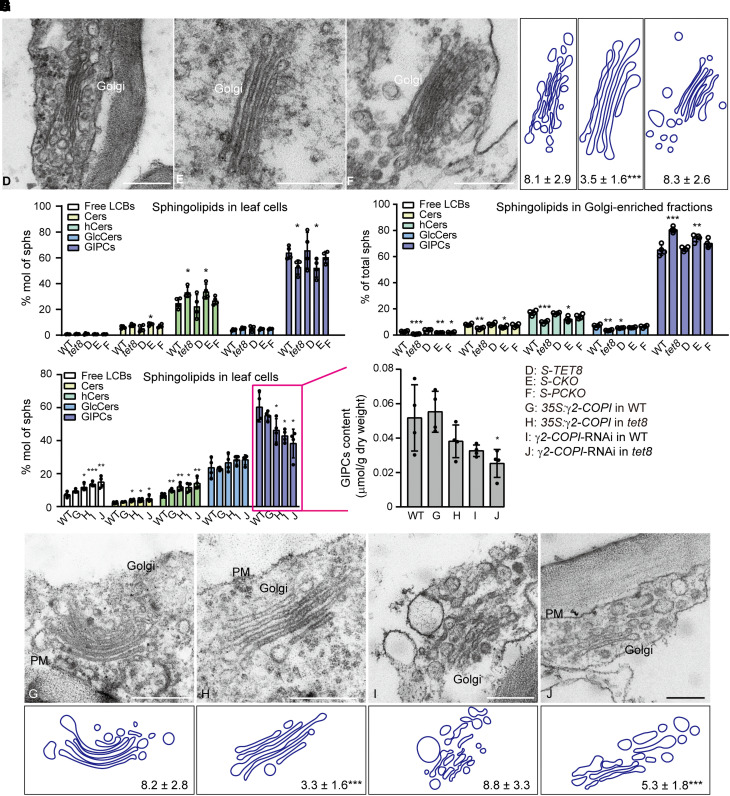
We further examined the structure of the Golgi in WT and these transgenic lines (Fig. 3A). Compared to the WT, S-CKO plants had fewer Golgi vesicles (Figs. 1F and 3, A and B), whereas the lines for the other variants (S-TET8 and S-PCKO) exhibited similar Golgi phenotypes as the WT (Figs. 1G and 3, A and B). Given that TET8 could bind GIPCs and regulate Golgi function, we explored its role in sphingolipid sorting or trafficking. Accordingly, we profiled leaf cell sphingolipids, which revealed that S-TET8 leaf cells have a similar fraction of GIPCs as WT leaves (∼65% of total sphingolipids), whereas the leaves from the tet8 mutant and the S-CKO lines displayed a substantially lower ratio of GIPCs (∼50% of total sphingolipids; Fig. 3C; Supplemental Data Set 4). The levels of GIPCs in S-PCKO appeared to be slightly lower than in WT, although this difference did not reach significance.
Figure 3.
The TET8 regulates Golgi vesicle budding and modulates GIPC exports. A) TEM observations of the Golgi in the leaves of 4-wk-old plants from S-TET8 (D), S-CKO (E), and S-PCKO (F). Scale bars, 500 nm. B) Number of Golgi vesicles. C) Sphingolipid profiles of leaf cells in WT, tet8, S-TET8 (D), S-CKO (E), and S-PCKO (F). D) Sphingolipid profiles of Golgi-enriched fractions extracted from WT, tet8, S-TET8 (D), S-CKO (E), and S-PCKO (F). E) Sphingolipid profiles of leaf cells in WT, 35S:γ2-COPI in WT (G), 35S:γ2-COPI in tet8 (H), γ2-COPI-RNAi in WT (I), and γ2-COPI-RNAi in tet8 (J). F) Content of leaf GIPCs in WT, 35S:γ2-COPI in WT (G), 35S:γ2-COPI in tet8 (H), γ2-COPI-RNAi in WT (I), and γ2-COPI-RNAi in tet8 (J). G) TEM observation of the Golgi in the leaves of 4-wk-old plants from 35S:γ2-COPI in WT (G), 35S:γ2-COPI in tet8 (H), γ2-COPI-RNAi in WT (I), and γ2-COPI-RNAi in tet8 (J). Scale bars, 500 nm. PM, plasma membrane. H) Number of Golgi vesicles. For B) and H), the number of vesicles around the Golgi is shown as means ± Sd. Statistical significance was determined using Student's t test. ***P < 0.001. For C to E), all data are represented as mol % of total sphingolipids. For C to F), 4 independent repeats (shown as black dots) were conducted. Data are presented as means ± Sd. Statistical significance was determined using Student's t test. *P < 0.05; **P < 0.01; ***P < 0.001. Details are given in Supplemental Data Sets 4 to 6.
As GIPCs are synthesized in the Golgi lumen (Rennie et al. 2014; Mortimer et al. 2013), and the loss of TET8 function resulted in fewer Golgi vesicles (Fig. 1, F and G), we isolated the crude Golgi fraction and analyzed their sphingolipid contents (Supplemental Fig. S8; Fig. 3D). Notably, the proportion of GIPCs in the Golgi fraction showed a reverse relationship with TET8 function. The tet8 Golgi fraction contained the highest percentage of GIPCs (more than 80% of total sphingolipids). The portion of GIPCs in S-CKO was ∼70%, followed by ∼66% in S-PCKO, which was slightly higher than that in S-TET8 and WT (∼60%; Fig. 3D; Supplemental Data Set 5). Together, these results indicate that loss of TET8 causes GIPCs to accumulate in the Golgi, at the expense of total GIPCs in the cell.
The COPI complex is important for lipid homeostasis (Contreras et al. 2012; Beller et al. 2008). In this study, we verified the interaction between TET8 and 1 COPI subunit (Fig. 2). We measured sphingolipid levels in transgenic lines overexpressing or knocked down for γ2-COPI: the knockdown of γ2-COPI decreased the accumulation of GIPCs while increasing the ratio of LCBs, Cers, and hCers (Fig. 3E). However, overexpressing γ2-COPI did not lead to an increase of GIPCs in total sphingolipids, nor did it rescue the lower level of GIPCs when we overexpressed γ2-COPI in the tet8 mutant. Overexpressing γ2-COPI in WT did not significantly alter the content of cellular GIPCs, while overexpressing γ2-COPI in the tet8 mutant and knocking down γ2-COPI expression in WT and tet8 resulted in lower cellular GIPC contents than that in the WT (Fig. 3, E and F; Supplemental Data Set 6).
The COPI complex is responsible for intra-Golgi trafficking and retrograde transport from the Golgi to the ER and is also involved in maintaining Golgi morphology and sorting function (Ahn et al. 2015; Gimeno-Ferrer et al. 2016). We thus examined Golgi structure in the γ2-COPI transgenic lines. Unlike in the tet8 mutant, knocking down γ2-COPI was associated with the conversion of Golgi ribbons into smaller, disconnected stacks (Supplemental Fig. S9; Fig. 3G). These ministacks were accompanied by increased Golgi fragmentation (Supplemental Fig. S9; Fig. 3G), suggesting that γ2-COPI has roles in material modification, trafficking, or sorting. Although overexpressing γ2-COPI in WT resulted in a WT Golgi phenotype (Fig. 3, G and H), overexpressing γ2-COPI in the tet8 mutant did not rescue the lack of vesicles typical of the mutant around the Golgi, and knocking down γ2-COPI in the tet8 mutant resulted in more severe fragmentation of the Golgi than in WT (Fig. 3, G and H). Our data are reminiscent of observations in the yeast (Saccharomyces cerevisiae) mutant sec21-3, which is defective in COPI function, as it displays decreased levels of inositol phosphoceramide (IPC, an intermediate in GIPC biosynthesis) together with Golgi remodeling of GPI-anchor proteins (Reggiori and Conzelmann 1998). Taking these results together, we propose that GIPCs might first undergo sorting via TET8 in Golgi membranes (possibly with the assistance of COPI) before being transported to the outer leaflet of the PM or the apoplastic space, in a process mediated by EVs, along with TET8.
Arabidopsis TET8 and GIPCs affect the secretion of plant EVs
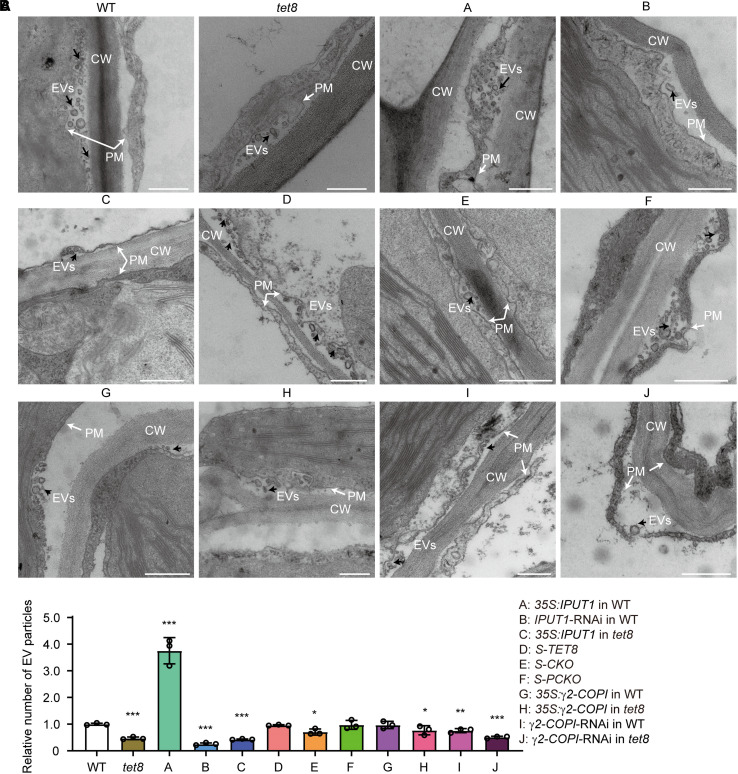
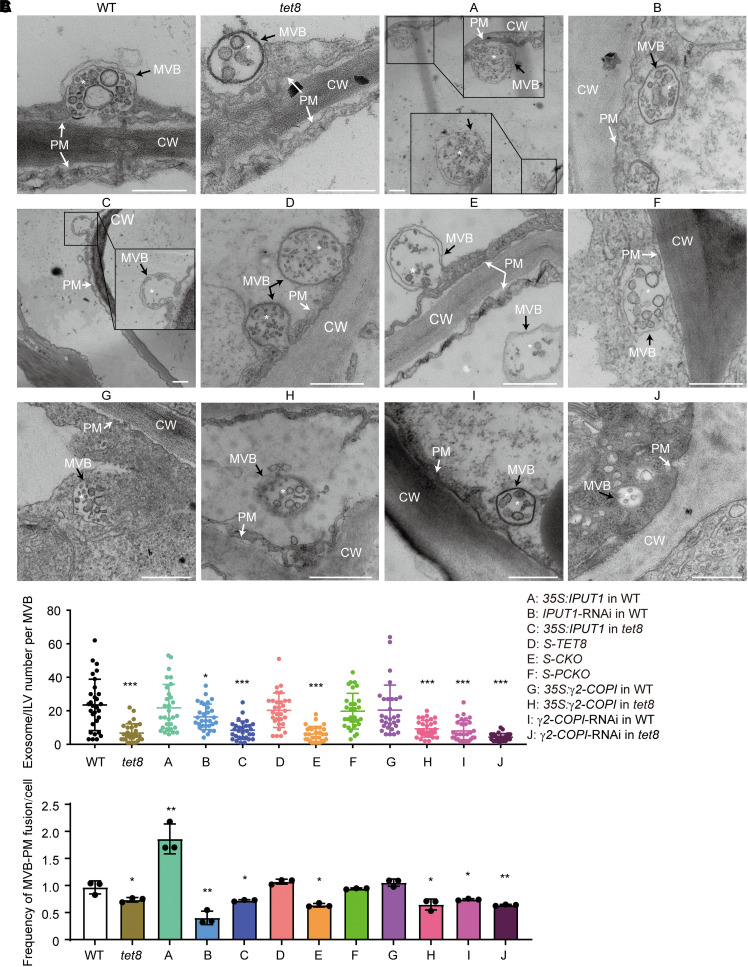
Arabidopsis secretes different subtypes of EVs decorated with distinct biomarkers (TET8-positive or PENETRATION1 [PEN1]-positive exosomes) and biogenesis pathways (He et al. 2021). We demonstrated the participation of TET8 in EV secretion and observed that TET8 was present only in some exosomes or ILVs (26%) undergoing MVB fusion with the PM (Supplemental Fig. S7; Liu et al. 2020b). We observed cellular membranes by TEM and performed a nanoparticle-tracking analysis (NTA) of EVs isolated from Arabidopsis leaves, which showed that all plants with lower GIPC contents or lacking functional TET8 release fewer EVs and contain significantly fewer ILVs or exosomes in MVBs than the WT (Figs. 4 and 5, A and B). It is worth noting that knocking down IPUT1 in WT significantly lowered the GIPC contents (Fig. 1E) while having no effect on the number of Golgi vesicles (Fig. 1G). Besides, compared to tet8 or S-CKO, IPUT1-RNAi plants in the WT background retained more ILVs in their MVBs. Overexpressing IPUT1 increased the secretion of plant EVs but not of MVB-ILVs (Figs. 4 and 5, A and B). We noticed more MVBs targeted to the PM in 35S:IPUT1 plants than in WT and fewer in the IPUT1-RNAi line (Fig. 5, A and C). Moreover, knocking down γ2-COPI in WT produced an abnormal Golgi morphology and fewer MVB-ILVs or exosomes, while overexpressing γ2-COPI resulted in the same phenotypes as the WT in terms of GIPC content, Golgi morphology, MVB-ILVs, MVB–PM fusion frequency, and exosome and EV secretion (Figs. 3, E to H, 4, and 5). These results suggest 2 possibilities: (i) COPI does not directly participate in the secretion of exosomes or EVs or (ii) EV secretion involving GIPCs, TET8, and COPI together is rate limited.
Figure 4.
TET8 and GIPCs together regulate the release of plant exosomes and EVs. A) TEM observation of EVs in WT, tet8, 35S:IPUT1 in WT (A), IPUT1-RNAi in WT (B), 35S:IPUT1 in tet8 (C), S-TET8 (D), S-CKO (E), S-PCKO (F), 35S:γ2-COPI in WT (G), 35S:γ2-COPI in tet8 (H), γ2-COPI-RNAi in WT (I), and γ2-COPI-RNAi in tet8 (J). CW, cell wall; EVs, extracellular vesicles (black arrows); PM, plasma membrane (white arrows). Scale bars, 500 nm. B) NTA of EVs isolated from the above plants. The value of WT EVs was set to 1.0. Three independent repeats (black dots) were conducted. Statistical significance was determined using Student's t test. *P < 0.05; **P < 0.01; ***P < 0.001.
Figure 5.
The TET8 and GIPCs together regulate plant MVB maturation. A) TEM observation of MVBs in WT, tet8, 35S:IPUT1 in WT (A), IPUT1-RNAi in WT (B), 35S:IPUT1 in tet8 (C), S-TET8 (D), S-CKO (E), S-PCKO (F), 35S:γ2-COPI in WT (G), 35S:γ2-COPI in tet8 (H), γ2-COPI-RNAi in WT (I), and γ2-COPI-RNAi in tet8 (J). CW, cell wall; MVBs, multivesicular bodies (black arrows); PM, plasma membrane (white arrows). White asterisks indicate ILV in the MVB. Scale bars, 500 nm. B) Number of ILVs in MVBs (n = 30 for each group, marked as dots in different colors). Statistical significance was determined using Mann–Whitney U test. C) Frequency of MVBs fusing with the PM. Three independent repeats (black dots) were conducted. Data are reported as means ± Sd. Statistical significance was determined using Student's t test. *P < 0.05; **P < 0.01; ***P < 0.001.
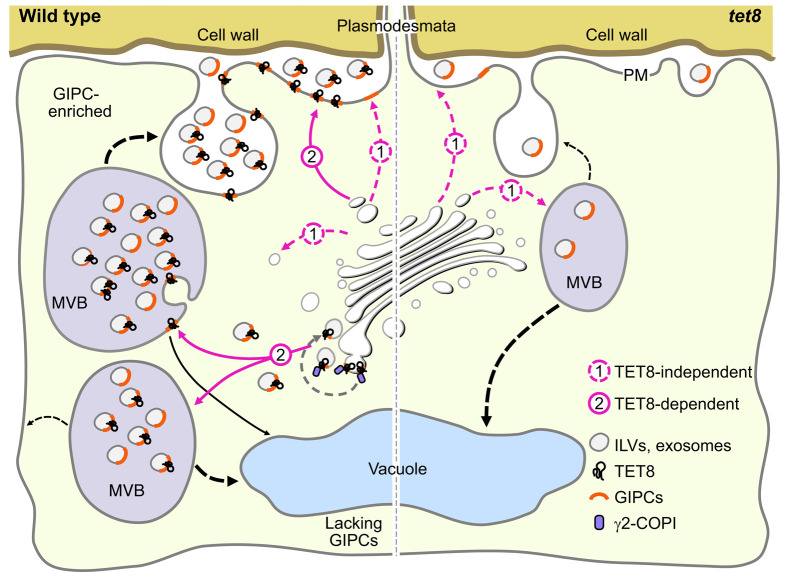
Finally, we propose a model for TET8-dependent transport of GIPCs that is associated with the biogenesis of exosomes and other EVs. In WT, TET8 and γ2-COPI sort and/or redistribute GIPCs, and TET8 promotes the generation of vesicles in the Golgi (Fig. 6). TET8 might reach the PM via a conventional secretion pathway involving membrane fusion or reach the PM by being transported to MVBs, entering ILVs by inward budding of MVB membranes in an endosomal sorting event, and then being released as exosomes, where it is secreted together with GIPCs. When GIPC levels are insufficient, MVBs cannot produce enough ILVs, resulting in a lower chance that ILVs will fuse with the PM. In the tet8 mutant and TET8-CKO variant line, the TET8-dependent transport of GIPCs does not occur, leading to impaired exosome release.
Figure 6.
TET8 mediates GIPC transport and affects exosome/EV biogenesis. In WT plants, sorting and trafficking of GIPCs are mediated in a TET8-dependent manner (with assistance of γ2-COPI) or in a TET8-independent manner. MVBs with enough GIPC-enriched ILVs prefer to fuse with the PM and release vesicles as EVs. In GIPC-deficient plants (such as IPUT1-RNAi in WT), the MVBs fuse less frequently with the PM, resulting in a decrease of EVs. In the tet8 mutant, GIPC accumulation is blocked, leading to impaired secretion of EVs. The red dotted lines represent the TET8-independent pathway. The red lines point to the TET8-dependent pathway. The black dotted lines are unknown pathways that mediate MVB fusion to the PM or vacuoles. CW, cell wall; EVs, extracellular vesicles; GIPCs, glycosyl inositol phosphoceramides; MVBs, multivesicular bodies; PD, plasmodesmata; PM, plasma membrane.
Discussion
In this study, we uncovered the coupled functions of TET8 and GIPCs in sphingolipid metabolism. The biosynthesis of simple sphingolipids occurs mainly in the ER, whereas GIPCs are produced in the Golgi apparatus (Rennie et al. 2014). We showed here that TET8 might regulate GIPC contents independently of GIPC biosynthesis. The Golgi in the tet8 mutant (or S-CKO line) accumulated GIPCs, which in turn resulted in decreased levels of Cers and hCers, the intermediate precursors of IPCs and then GIPCs, while the tet8 mutant largely accumulated LCBs, Cers, and hCers. Additionally, overexpressing or knocking down γ2-COP (affecting the Golgi) led to the accumulation of hCers in WT and tet8 plants. It is difficult to assess whether IPC synthase enzymes are somehow downregulated in the tet8 mutant, or whether the Golgi status affects sphingolipid homeostasis (as shown in Fig. 3). Therefore, we can only conclude that TET8 regulates the accumulation of GIPCs by affecting the Golgi status of plant cells, which might parallel GIPC biosynthesis.
TET8 might have the capacity to bind to most sphingolipids, as it interacted with the sphingolipid LCB phytosphinganine. However, the tet8 mutant only displayed decreased cellular contents of GIPCs, but not LCBs or Cers, relative to the WT (Liu et al. 2020b). Notably, GIPCs accumulated in the Golgi of the tet8 mutant (Fig. 3). These clues point to a role for TET8 as an outgoing transport carrier for GIPCs. TET8 might recruit γ2-COPI, which mainly mediates retrograde trafficking within the Golgi or from the Golgi to the ER. Brügger et al. (2000) demonstrated that COPI vesicles contained low amounts of sphingomyelin and cholesterol compared to their donor Golgi membranes and hinted at roles for COPI vesicles in lipid segregation or sorting. Lacking data for sphingolipids in plant COPI vesicles, we speculate that γ2-COPI might function in recycling GIPC-deficient TET8-decorated vesicles and participate in sphingolipid segregation with other lipids within the Golgi. The distribution of sphingolipids, especially complex ones such as GIPCs, is highly asymmetric across the PM as sphingolipids can form lipid raft domains (Riboni et al. 2010; Gronnier et al. 2016). In animals, COPI vesicles are believed to bud from liquid-disordered nonraft domains (Manneville et al. 2008). Thus, we suspect that COPI vesicles occur in non-GIPC membrane regions and reserve GIPCs in the Golgi, which enriches the level of GIPCs for further trafficking. Complex sphingolipids in membranes increase in content along the secretory pathway (Pinot et al. 2010). Depletion of the COPI complex in animal cells affected the sphingolipid ganglioside GM1, which accumulated in atypical compartments inside cells (Misselwitz et al. 2011). Removing the C-tail of TET8 not only blocked the recognition between TET8 and γ2-COPI but also led to TET8 retention inside cells, causing the retention of GIPCs in the Golgi and a decrease in the content of total GIPCs (Figs. 2 and 3). Retaining its interaction with γ2-COPI, the variant TET8-PCKO showed slight mislocalization and modestly lower GIPC contents relative to the WT (Fig. 3). These results hint at the function of TET8 in GIPC enrichment as well as GIPC escaping from the Golgi. Comparing the GIPCs between the PM of WT and tet8 plants showed that the loss of TET8 function eventually led to lower GIPC levels in its major storage location (Supplemental Fig. S1). In addition, the tet8 mutant released fewer EVs (also an important storage site for GIPCs) than the WT (Liu et al. 2020b). Together, these results suggest that TET8 acts as a GIPC carrier to modulate local GIPC concentration and packaging into some vesicles to the cell surface or EC space.
We also noted that knocking down γ2-COPI caused severe Golgi deformation (Fig. 3). The ɑ2-COPI knockout mutant has fewer cisternae per Golgi with abnormal vesicles around than WT (Gimeno-Ferrer et al. 2016), and silencing β’-COPI in Nicotiana benthamiana resulted in disassembly of the Golgi apparatus (Ahn et al. 2015). Loss of TET8 function also resulted in fewer Golgi vesicles (Figs. 1 and 3). Importantly, the Golgi apparatus is a dynamic organelle modulated via lipids and proteins, in turn affecting the sorting and trafficking of proteins and lipids. The COPI complex also functions in lipid homeostasis (Beller et al. 2008). It is, however, difficult to ascertain the identity of the main regulator within the network composed of γ2-COPI, TET8, GIPCs, and Golgi conditions.
We also elucidated the relationship between TET8−GIPCs and EV biogenesis. In our previous study, we reported that EVs were GIPC-enriched nanosized vesicles and that exogenous application of GIPCs promoted the release of EVs in both WT and the tet8 mutant (Liu et al. 2020b). In fact, EV biogenesis can be regarded as a type of cargo sorting that is subject to MVB maturation. Deficiency of TET8 or GIPCs led to impaired MVB–exosome production (Figs. 4 and 5, A and B). It was notable that excess amount of GIPCs did not further increase MVB-ILVs or exosomes but promoted the fusion of MVBs with the PM (Fig. 5, B and C). Plant MVBs likely can only contain a limited number of ILVs; if they are overloaded with ILVs, the MVBs will likely release the ILVs as exosomes. Artificially elevating the contents of GIPCs back to WT levels in tet8 by overexpressing IPUT1 did not fully rescue the decreased number of MVB-ILVs or exosomes of the mutant but did rescue the defect in MVB–PM fusion. MVBs may be destined to the vacuole for degradation or to the PM for exosome secretion. In the PM, GIPCs make up approximately 68% of total sphingolipids, but they make up only about ∼30% in the vacuole membrane (Carmona-Salazar et al. 2021). Most GIPCs are destined for the PM, rather than the vacuole membrane. Therefore, we propose that the GIPC contents of the MVBs (in the MVB or ILV membrane) might affect whether the MVB is sorted to the PM or vacuolar membrane.
Taken together, our results illustrate how GIPCs are sorted and transported and provide insight into how TET8 and GIPCs affect exosome biogenesis in plants.
Materials and methods
Plant materials and growth conditions
The Arabidopsis (A. thaliana) mutant tet8 (At2g23810, SALK_136039C) was obtained from the Eurasian Arabidopsis Stock Centre (uNASC). Homozygous lines were identified using the primers listed in Supplemental Table S1. The Arabidopsis lines S-TET8, S-PCKO, and S-CKO were generated by transforming the tet8 mutant with Agrobacterium (Agrobacterium tumefaciens) strain GV3101 carrying the pCAMBIA (myc-flag) vector comprising the Arabidopsis TET8 promoter and EGFP-TET8 (or its variants). The γ2-COPI-RNAi lines and IPUT1-RNAi lines were produced using the RNAi vector pCAMBIA-RNAi containing a specific 300-bp fragment from the coding sequence of γ2-COPI (At4g34450) or IPUT1 (At5g18480). The 35S:γ2-COPI lines and 35S:IPUT1 lines were produced using the pCAMBIA (myc-flag) vector containing the cauliflower mosaic virus (CaMV) 35S promoter followed by the full-length coding sequence of γ2-COPI or IPUT1. The transcript levels of γ2-COPI or IPUT1 in each line were verified by reverse transcription quantitative PCR (RT-qPCR). Arabidopsis UBIQUITIN5 (UBQ5) served as the internal standard. The primers used are listed in Supplemental Table S1.
The seeds of Arabidopsis accession Col-0 (WT) and mutants were sterilized with 75% (v/v) ethanol for 10 to 15 min and washed with sterile water 5 times and were cultivated on half-strength MS agar medium for about 2 wk before being transferred to soil in a controlled growth chamber under a condition of 16-h light/8-h dark photoperiod using LED bulbs (JIUPO-2835LED, China) at 100 µmo1/m2/s, 22 °C, and 70% relative humidity.
Sphingolipid extraction and LC–MS/MS
Sphingolipids were extracted from the leaves of 4-wk-old Arabidopsis plants as previously reported (Liu et al. 2020b). Briefly, ∼150 mg of freshly collected leaves was dried using freeze drier (at −80 °C, LABCONCO, United States) and weighted. Then the dried samples were pretreated with 400 μL deactivation buffer (0.01% [w/v] butylated hydroxytoluene in isopropanol) at 75 °C for 20 min. Next, 800 μL extraction solution I (chloroform:methanol = 1:1, v/v) was added to each sample with vortexing at room temperature. To obtain the Golgi-enriched membrane fraction, ∼200 μg (protein equivalent) of samples was extracted using extraction buffer II (chloroform:methanol:isopropanol = 1:1:1, v/v/v). The mixtures were incubated for 24 h with shaking at 150 rpm at room temperature before centrifugation at 3,000 × g for 20 min. Supernatants containing sphingolipids were dried in a SpeedVac rotary vacuum desiccator (Genevac, United Kingdom).
LC–MS/MS analysis of sphingolipids was conducted using an Exion UPLC-QTRAP 6500 Plus (Sciex) LC–MS/MS system (curtain gas = 20, ion spray voltage = 5,500 V, temperature = 400 °C, ion source gas 1 = 35, and ion source gas 2 = 35; Lipidall Technologies Company, China) equipped with a Phenomenex Luna 3 μm silica column (internal diameter 150 × 2.0 mm). Buffer A (chloroform:methanol:ammonium hydroxide = 89.5:10:0.5, v/v/v) and buffer B (chloroform:methanol:ammonium hydroxide:water = 55:39:0.5:5.5, v/v/v/v) were used as mobile phases. The program was set as follows: 0 to 5 min, hold at 95% buffer A; 5 to 7 min, decrease gradient to 60% (v/v) buffer A; 7 to 11 min, hold at 60% (v/v) buffer A; 11 to 15 min, decrease gradient to 30% (v/v) buffer A; 15 to 30 min, hold at 30% (v/v) buffer A; and 30 to 35 min, increase gradient to 95% (v/v) buffer A. Internal standards were purchased from Avanti Polar Lipids: D-erythro-sphingosine (Sph-d17:1) for sphingoids, N-heptadecanoyl-D-erythro-sphingosine (Cer-C17, d18:1/17:0) for Cers, N-(dodecanoyl)-1-β-glucosyl-sphing-4-ene (GluCer-C12, d18:1/12:0) for glycosyl ceramides, and ganglioside (GM1-d18:1/18:0-d3) for GIPCs.
TEM and immunogold labeling TEM
Arabidopsis leaf tissues were cut into small pieces and fixed in a fixative solution (glutaraldehyde [GA] solution containing 5% [w/v] GA and 0.1 M phosphate buffer [50 mM Na2HPO4, 50 mM NaH2PO4]), pH 7.4. The leaf fragments together with the fixative solution were vacuumed for 2 h until all leaflets had sunk to the bottom of the container. To ensure that the samples were properly washed, the leaves were rinsed 6 times for 5 min each with 0.1 M phosphate buffer (pH 7.4). Next, the leaf fragments were dehydrated using a graded ethanol series (30%, 50%, 70%, 85%, 90%, 95%, and 100% ethanol, all v/v). These tissue fragments were then washed again with 0.1 M phosphate buffer (pH 7.4) and refixed in a solution of 2% (w/v) OsO4 and 1.5% (w/v) potassium ferrocyanide at room temperature. Before being embedded in Spurr's resin (SPI supplies), the samples were washed with phosphate buffer, then distilled H2O, and dehydrated through a graded ethanol series. The resin blocks were then processed with a Leica Microsystem UC7 ultramicrotome into 70-nm sections and mounted onto a copper grid before uranyl acetate/lead citrate staining.
Images of the Golgi, MVBs, and EVs were captured from leaf mesophyll cells. The outline of the Golgi was obtained using the Golgi stain plugin of ImageJ (Fiji; Vints et al. 2019). To quantify Golgi vesicles and ILVs in MVBs, a total of 30 images per sample were obtained from more than 3 blocks individually, taken from 3 different plant rosette leaves. To determine the MVB–PM fusion frequency, more than 100 cells per block and 3 independent blocks were manually counted and analyzed for each sample. Automated vesicle quantification was performed using a deep learning–based tool, Cellpose (Stringer et al. 2021). The images were preprocessed to a suitable size (∼512 × 512 pixels) via ImageJ (Fiji) before being labeled as masks. Ten typical images containing vesicles (50 to 100 nm, Golgi vesicles, ILVs) were manually labeled as masks to retrain the initial “cyto” model. The training parameters were set as follows: learning rate, 0.01; weight decay, 0.0001; and epoch, 100. The trained model was saved and reloaded for further detection. The value of region of interest (ROI) mask was recorded as the number of vesicles. Manual correction was also conducted to remove incorrectly labeled vesicles or add missing labeled vesicles.
For immunogold labeling, plant materials were fixed in 2.5% (w/v) paraformaldehyde solution and embedded in LR White resin. With the exception of the differences in the fixing solution and the embedding agent, all other steps of the TEM chemical fixation method were as described above. Resin blocks were cut into 70-nm sections and mounted onto nickel grids. Immunogold labeling was conducted using anti-TET8 antibody (1:50 dilution, PhytoAB; Supplemental Fig. S10) and goat anti-rabbit IgG antibody conjugated with 10-nm colloidal gold particles (1:50 dilution, Sigma-Aldrich) before uranyl acetate/lead citrate staining. TEM observation was conducted by using Tecnai G2 20 TWIN microscope (FEI) equipped with an Eagle 4k CCD at 120 kV, FEI. Eight images were used to calculate the labeling efficiency. The leaf cells of tet8 mutant were labeled with anti-TET8 antibody (1:50 dilution) and acted as negative control. Besides, the leaf cells of WT plant labeled with IgG (to replace the primary antibody) also acted as negative control.
Bimolecular luciferase complementation assay
N. benthamiana plants were grown under controlled conditions at 24 °C and 70% relative humidity, under a 14-h light/10-h dark photoperiod. The plants were grown for about 4 to 5 wk before being used for Agrobacterium-mediated infiltration. The appropriate constructs (encoding empty nLUC, empty cLUC, nLUC-target gene1, and cLUC-target gene2) were introduced into Agrobacterium strain GV3101 competent cells. For bacterial suspension, 1 mL of an overnight culture was diluted into 20 mL of LB culture medium with antibiotics and was grown at 28 °C until the optical density reached 0.5 to 0.6 at OD600. The bacteria were then collected by centrifugation at 6,000 × g for 10 min at room temperature. The pellet was fully resuspended in a suspension buffer containing 10 mM MgCl2, 10 mM MES, and 150 μM acetosyringone, pH 5.6. The optical density of the bacteria was adjusted to an OD600 of 1.0 and incubated for 2 to 3 h at room temperature. To introduce the target genes into N. benthamiana, equal volumes of 2 bacterial suspensions, each containing a different plasmid, were mixed well and infiltrated into the leaves of N. benthamiana plants. To detect luciferase activity, the infiltrated leaves were sprayed with luciferin solution (Yesen, diluted 1:1,000 [v/v] using sterile water with 0.1% Tween-20) using a 1-mL sprayer. The leaves were kept in the dark for 5 min to minimize chlorophyll phosphorescence and delayed fluorescence. Finally, a low-light cooled CCD imaging apparatus (Tanon 5200 Multi, China) was used to capture images.
Recombinant protein purification
The full-length coding sequences of Arabidopsis TET8 and its variants (CKO, PCKO) were cloned and assembled into the vector pCold-TF (with a 48-kD TF chaperone and a 6×His tag). The full-length coding sequence of γ2-COPI was cloned into the pGEX4T-1 vector (with a 26-kD glutathione transferase [GST] tag). Constructs were transformed in Escherichia coli strain BL21 (DE3) for protein production.
For soluble His-TF purification, whole cell lysates (in 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 20 mM imidazole) were centrifuged at 10,000 × g for 40 min at 4 °C. The suspension was collected and incubated with Ni resin (Transgen) for 1 h at 4 °C. The target proteins were eluted in His-elution buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 300 mM imidazole). Purification of the membrane protein fusion His-TF-TET8 was conducted as described in Liu et al. (2020a). Protein desalting was conducted with a desalting column (GE) in phosphate-buffered saline (PBS; 150 mM NaCl, 15 mM Na2HPO4, and 15 mM NaH2PO4, pH 7.4) with 0.02% (v/v) N-dodecyl-β-D-maltoside (DDM; for pull-down assay and immunoblotting assay assay) or 0.05% (v/v) Tween-20 (for SPR).
For GST-γ2-COPI protein purification, whole cell lysates (in PBS) were also centrifuged at 10,000 × g for 40 min at 4 °C to remove the precipitates. Soluble proteins were purified using GST resin (Transgen), and target proteins were eluted in GST elution buffer (PBS with 10 mM glutathione, pH 7.4). The GST tag was cleaved in digestion buffer (400 U/mL thrombin in PBS), and γ2-COPI protein was further separated via Millipore (50,000 MW) in PBS.
AP–MS, pull-down, and Co-IP
Total proteins from Arabidopsis leaves were extracted in IP buffer (50 mM Tris-HCl, pH 7.5, 10 mM EDTA, 150 mM NaCl, 15% [v/v] glycerol, 0.5% [v/v] NP40, 1 mM PMSF, and 1 × protease inhibitor cocktail). For AP–MS, His-TF-TET8 and His-TF (as negative control) were, respectively, preincubated with Ni resin beads before incubation with total proteins. Proteins bound on the resin were washed with IP buffer, and the proteins retained on the beads were identified by MS. The list of candidate proteins (with those present in the negative control removed) is given in Supplemental Data Set 3.
For pull-down assays, purified His-TF-TET8 and its variants were preincubated with Ni resin beads before being incubated with purified γ2-COPI. Mixtures were fully washed with PBS, and retained proteins were detected by immunoblotting with an anti-γ2-COPI polyclonal antibody (1:1,000 dilution, Agrisera) and secondary antibody horseradish peroxidase (HRP)–conjugated goat anti-rabbit (1:5,000 dilution, Transgen).
For Co-IP assay, IgG Dynabeads (Invitrogen) were blocked with anti-γ2-COPI antibody or IgG before being incubated with Arabidopsis total leaf protein extracts. Proteins retained on the beads were eluted with glycine buffer and detected by immunoblotting with an anti-TET8 polyclonal antibody (1:1,000 dilution, PhytoAB, PHY1490A) and HRP-conjugated goat anti-rabbit (1:5,000 dilution, Transgen). The specificity of the antibody to TET8 is shown in Supplemental Fig. S10.
SPR assays
To detect protein–protein interactions, a Biacore CM5 chip (GE) was first covered with purified γ2-COPI following the manufacturer’s instructions. His-TF-TET8 and His-TF were diluted to different concentrations. Each protein solution was applied onto the chip at a flow rate of 8 µL/min for 180 s followed by 180 s of dissociation time. The biosensor chip was regenerated using glycine solution (pH 1.5).
To detect protein–lipid interactions, a Biacore NTA chip was covered with purified His-TF-TET8 or His-TF protein. Lipids were diluted to different concentrations and applied to the chip at a flow rate of 10 µL/min for 180 s followed by 240 s of dissociation time.
All data were fit to 1:1 Langmuir adsorption model to calculate the dissociation constant (KD) using the BIA analysis software.
GIPC isolation and lipid strip assays
GIPCs were isolated as previously reported (Liu et al. 2020b; Buré et al. 2014). The purified GIPCs were dried under nitrogen gas and dissolved in methanol to a concentration of 0.1 mg/mL. Cer and POPC were also dissolved in methanol at a concentration of 0.1 mg/mL. About 100 ng of each lipid was spotted onto a PVDF membrane and dried at room temperature. Lipid strip assays were then conducted following the manufacturer's instructions, using Membrane Lipid Strips (Echelon Biosciences). Bound protein was then detected with an anti-His antibody (1:2,000 dilution, Transgene) and HRP-conjugated goat anti-mouse (1:5,000 dilution, Transgen).
Golgi-enriched fraction purification
The Golgi fraction was isolated from the leaves of 4-wk-old Arabidopsis plants as reported with modifications (Zeng et al. 2017). Briefly, mature leaves (∼10 g) were homogenized in precooled extraction buffer (50 mM HEPES-KOH, pH 6.8, 0.4 M sucrose, 1 mM DTT, 5 mM MnCl2, and 5 mM MgCl2). The homogenate was then filtered through 2 layers of gauze and centrifuged at 3,000 × g for 30 min at 4 °C. The supernatant (20 mL) was carefully transferred to a new centrifuge tube and preloaded with 6 mL Cushion Buffer (50 mM HEPES-KOH, 1.8 M sucrose, pH 6.8). Samples were ultracentrifuged at 100,000 × g for 1 h at 4 °C, and the crude microsomes at the bottom of the tube were collected. A continuous 25% to 40% sucrose gradient was prepared to separate the crude microsomes. Golgi-enriched layers (detected using anti-Arf1 [1:1,000 dilution, Agrisera] and anti-γ2-COPI antibody [1:1,000 dilution]) were collected and washed twice with washing buffer (50 mM HEPES-KOH, pH 6.8) for analysis.
Confocal microscopy and high-speed living cell rotary confocal microscopy
For confocal imaging, leaves from 4-wk-old transgenic plants were placed onto a glass slide and observed under a LEICA TCS SP8 STED (Germany) confocal microscope using a 40× or 63× oil immersion objective (excitation wavelengths: 488 nm for EGFP and 561 nm for RFP) or using a high-speed living cell rotary confocal microscope (Andor Revolution XD, Japan) equipped with a 40× water immersion objective. Sequential scanning was used to avoid potential spectral bleed-through (crosstalk) between the channels. Image analysis was performed using Fiji/ImageJ. The Pearson correlation coefficients were calculated for the whole image to analyze the cooccurrence of signals (pixels containing both fluorophores).
EV isolation and NTA
EVs from leaves were isolated using the method described by Rutter and Innes (2020). Briefly, 4-wk-old Arabidopsis rosettes were collected and precleaned with ultrapure water before being immersed in vesicle isolation buffer (VIB) containing 20 mM MES, 2 mM CaCl2, and 0.1 M NaCl, pH 6.0. The samples were then vacuum infiltrated in bottles until the buffer filled the whole leaves. The excess liquid on the leaves was gently wiped off with filter paper. The leaves were then mounted in modified centrifuge tubes and centrifuged at 700 × g at 4 °C for 20 min to obtain the crude apoplastic wash. Large insoluble particles in the crude apoplastic wash were removed by centrifugation at 10,000 × g at 4 °C for 30 min and filtered through a 0.22-μm filter. The suspension, named pretreated apoplastic wash, was continuously centrifuged at 40,000 × g at 4 °C for 1 h. The EVs were obtained from the pellet, which was then washed with 20 mM Tris-HCl, pH 7.0. The particles were quantified as concentration (particles/mL) using a NanoSight NS300 and analyzed with NanoSight NTA3.4 software. The initial weight of the plant leaves and the final volume of the EV suspension were recorded to calculate the number of EVs. The number of EVs in WT was set to 1.0, and the others were normalized to WT.
Statistical analysis
Statistical tests were applied as described in the text and figure legends. All statistical results are shown in Supplemental Data Set 7.
Accession numbers
Sequence data from this article can be found in the TAIR libraries under the following accession numbers: TET1 (At5g46700), TET2 (At2g19580), TET2 (At3g45600), TET4 (At5g60220), TET5 (At4g23410), TET6 (At3g12090), TET7 (At4g28050), TET8 (At2g23810), TET9 (At4g30430), TET10 (At1g63260), TET11 (At1g18520), TET12 (At5g23030), TET13 (At2g03840), TET14 (At2g01960), TET15 (At5g57810), TET16 (At1g18510), TET17 (At1g74045), ɑ1-COPI (At1g62020), ɑ2-COPI (At2g21390), β1-COPI (At4g31480), β2-COPI (At4g31490), β’1-COPI (At1g52360), β’2-COPI (At3g15980), β’3-COPI (At1g79990), γ1-COPI (At2g16200), γ2-COPI (At4g34450), ε1-COPI (At1g30630), ε2-COPI (At2g34840), and IPUT1 (At5g18480).
Supplementary Material
Acknowledgments
We thank Prof. Yuzhou Zhang from Northwest A&F University for helpful discussions. We thank Dr. Xiaoyan Gao, Zhiping Zhang, and Jiqin Li for technical assistance with electron microscopy, sample preparation, and image analysis and Dr. Sin-Man Lam, Prof. Guang-Hou Shui, and Zhiyang Zhou from Lipidall Technologies Company Ltd. for lipidomic analysis. We also thank Zhiyong Luo from Shanghai Xitu Biotechnology Co. Ltd. for vector construction.
Contributor Information
Ningjing Liu, State Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences/Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China; School of Life Sciences, East China Normal University, Shanghai 200241, China.
Lipan Hou, State Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences/Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China; University of Chinese Academy of Sciences, Shanghai 200032, China.
Xin Chen, State Key Laboratory of Protein and Plant Gene Research, College of Life Sciences, Peking University, Beijing 100871, China.
Jingjing Bao, State Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences/Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China; University of Chinese Academy of Sciences, Shanghai 200032, China.
Fangyan Chen, State Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences/Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Wenjuan Cai, State Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences/Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Huixian Zhu, State Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences/Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China; University of Chinese Academy of Sciences, Shanghai 200032, China.
Lingjian Wang, State Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences/Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Xiaoya Chen, State Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences/Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China; University of Chinese Academy of Sciences, Shanghai 200032, China.
Author contributions
X.-Y.C. and N.-J.L. conceived the research. N.-J.L. performed most of the experiments. L.-P.H. and X.-C. assisted with protein purification and confocal image analysis. X.-C., W.-J.C., and F.-Y.C. assisted with image capture by microscopy. J.-J.B. assisted with luciferase assays. H.-X.Z. and L.-J.W. assisted with plant mutant identification. N.-J.L. and X.-Y.C. wrote the manuscript.
Supplemental data
Supplemental Figure S1. Relative contents of PM GIPCs in WT and the tet8 mutant.
Supplemental Figure S2. Purification of recombinant TET8
Supplemental Figure S3. Relative IPUT1 expression levels in IPUT1-related transgenic plants.
Supplemental Figure S4. BiLC assays showing that TET8 hardly interacts with the other 10 COPI subunits.
Supplemental Figure S5. Detection of the interaction between different TET proteins and γ2-COPI.
Supplemental Figure S6. Relative TET8 expression levels in S-TET8, S-CKO, and S-PCKO transgenic lines.
Supplemental Figure S7. Subcellular localization of TET8.
Supplemental Figure S8. Immunoblot analysis of the crude Golgi fraction isolated from WT leaves.
Supplemental Figure S9. Relative expression level of γ2-COPI in γ2-COPI-related transgenic plants.
Supplemental Figure S10. Specificity test of the anti-TET8 antibody.
Supplemental Table S1. List of primers used in this study.
Supplemental Data Set 1. Sphingolipid profiling of the PM fraction from WT and tet8 leaves.
Supplemental Data Set 2. Sphingolipid profiling of leaves from WT and IPUT1-related transgenic lines.
Supplemental Data Set 3. Summary information of affinity-MS using His-TF-TET8 as bait.
Supplemental Data Set 4. Sphingolipid profiling of leaves from WT, tet8, S-TET8, S-CKO, and S-PCKO transgenic lines.
Supplemental Data Set 5. Sphingolipid profiling of the Golgi fraction isolated from the leaves of WT, tet8, S-TET8, S-CKO, and S-PCKO transgenic lines.
Supplemental Data Set 6. Sphingolipid profiling of leaves from WT and γ2-COPI-related transgenic lines.
Supplemental Data Set 7. Results of statistical analysis.
Funding
This research was supported by grants from the National Natural Science Foundation of China (32100275 and 32388201) and the Chinese Academy of Sciences (XDB27020207).
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Ahn HK, Kang YW, Lim HM, Hwang I, Pai HS. Physiological functions of the COPI complex in higher plants. Mol Cells. 2015:38(10):866–875. 10.14348/molcells.2015.0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenquer M, Amorim MJ. Exosome biogenesis, regulation, and function in viral infection. Viruses 2015:7(9):5066–5083. 10.3390/v7092862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller M, Sztalryd C, Southall N, Bell M, Jäckle H, Auld DS, Oliver B. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 2008:6(11):e292. 10.1371/journal.pbio.0060292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brügger B, Sandhoff R, Wegehingel S, Gorgas K, Malsam J, Helms JB, Lehmann W-D, Nickel W, Wieland FT. Evidence for segregation of sphingomyelin and cholesterol during formation of COPI-coated vesicles. J Cell Biol. 2000:151(3):507–518. 10.1083/jcb.151.3.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buré C, Cacas JL, Mongrand S, Schmitter JM. Characterization of glycosyl inositol phosphoryl ceramides from plants and fungi by mass spectrometry. Anal Bioanal Chem. 2014:406(4):995–1010. 10.1007/s00216-013-7130-8 [DOI] [PubMed] [Google Scholar]
- Cabada Gomez DA, Chavez MI, Cobos AN, Gross RJ, Yescas JA, Balogh MA, Indriolo E. COPI complex isoforms are required for the early acceptance of compatible pollen grains in Arabidopsis thaliana. Plant Reprod. 2020:33(2):97–110. 10.1007/s00497-020-00387-9 [DOI] [PubMed] [Google Scholar]
- Carmona-Salazar L, Cahoon RE, Gasca-Pineda J, González-Solís A, Vera-Estrella R, Treviño V, Cahoon EB, Gavilanes-Ruiz M. Plasma and vacuolar membrane sphingolipidomes: composition and insights on the role of main molecular species. Plant Physiol. 2021:186(1):624–639. 10.1093/plphys/kiab064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter HE, Gigg RH, Law JH, Nakayama T, Weber E. Biochemistry of the sphingolipides. XI. Structure of phytoglycolipide. J Biol Chem. 1958:233(6):1309–1314. 10.1016/S0021-9258(18)49332-5 [DOI] [PubMed] [Google Scholar]
- Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014:30(1):255–289. 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- Contreras F-X, Ernst AM, Haberkant P, Björkholm P, Lindahl E, Gönen B, Tischer C, Elofsson A, von Heijne G, Thiele C, et al. Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature 2012:481(7382):525–529. 10.1038/nature10742 [DOI] [PubMed] [Google Scholar]
- Escola J-M, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998:273(32):20121–20127. 10.1074/jbc.273.32.20121 [DOI] [PubMed] [Google Scholar]
- Gao C, Yu CKY, Qu S, San MWY, Li KY, Lo SW, Jiang L. The Golgi-localized Arabidopsis endomembrane protein12 contains both endoplasmic reticulum export and Golgi retention signals at its C terminus. Plant Cell 2012:24(5):2086–2104. 10.1105/tpc.112.096057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno-Ferrer F, Pastor-Cantizano N, Bernat-Silvestre C, Selvi-Martínez P, Vera-Sirera F, Gao C, Perez-Amador MA, Jiang L, Aniento F, Marcote MJ. α2-COP is involved in early secretory traffic in Arabidopsis and is required for plant growth. J Exp Bot. 2016:68(3):erw446. 10.1093/jxb/erw446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronnier J, Germain V, Gouguet P, Cacas JL, Mongrand S. GIPC: glycosyl inositol phospho ceramides, the major sphingolipids on earth. Plant Signal Behav. 2016:11(4):e1152438. 10.1080/15592324.2016.1152438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C, Heuser J, Stahl P. Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: demonstration of a pathway for receptor shedding. Eur J Cell Biol. 1984:35(2):256–263. [PubMed] [Google Scholar]
- He B, Cai Q, Qiao L, Huang C-Y, Wang S, Miao W, Ha T, Wang Y, Jin H. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat Plants. 2021:7(3):342–352. 10.1038/s41477-021-00863-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018:75(2):193–208. 10.1007/s00018-017-2595-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari J, Helenius A. Endosome maturation. EMBO J. 2011:30(17):3481–3500. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Zhou X, Tao M, Yuan F, Liu L, Wu F, Wu X, Xiang Y, Niu Y, Liu F, et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 2019:572(7769):341–346. 10.1038/s41586-019-1449-z [DOI] [PubMed] [Google Scholar]
- Lenarčič T, Albert I, Böhm H, Hodnik V, Pirc K, Zavec AB, Podobnik M, Pahovnik D, Žagar E, Pruitt R, et al. Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science 2017:358(6369):1431–1434. 10.1126/science.aan6874 [DOI] [PubMed] [Google Scholar]
- Liu N-J, Wang N, Bao J-J, Zhu H-X, Wang L-J, Chen X-Y. Lipidomic analysis reveals the importance of GIPCs in Arabidopsis leaf extracellular vesicles. Mol Plant. 2020b:13(10):1523–1532. 10.1016/j.molp.2020.07.016 [DOI] [PubMed] [Google Scholar]
- Liu N-J, Zhang T, Liu Z-H, Chen X, Guo H-S, Ju B-H, Zhang Y-Y, Li G-Z, Zhou Q-H, Qin Y-M, et al. Phytosphinganine affects plasmodesmata permeability via facilitating PDLP5-stimulated callose accumulation in Arabidopsis. Mol Plant. 2020a:13(1):128–143. 10.1016/j.molp.2019.10.013 [DOI] [PubMed] [Google Scholar]
- Manneville J-B, Casella J-F, Ambroggio E, Gounon P, Bertherat J, Bassereau P, Cartaud J, Antonny B, Goud B. COPI coat assembly occurs on liquid-disordered domains and the associated membrane deformations are limited by membrane tension. Proc Natl Acad Sci U S A. 2008:105(44):16946–16951. 10.1073/pnas.0807102105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham JE, Jaworski JG. Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2007:21(7):1304–1314. 10.1002/rcm.2962 [DOI] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. Membrane curvature at a glance. J Cell Sci. 2015:128(6):1065–1070. 10.1242/jcs.114454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz B, Dilling S, Vonaesch P, Sacher R, Snijder B, Schlumberger M, Rout S, Stark M, von Mering C, Pelkmans L, et al. RNAi screen of Salmonella invasion shows role of COPI in membrane targeting of cholesterol and Cdc42. Mol Syst Biol. 2011:7(1):474. 10.1038/msb.2011.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer JC, Yu X, Albrecht S, Sicilia F, Huichalaf M, Ampuero D, Michaelson LV, Murphy AM, Matsunaga T, Kurz S, et al. Abnormal glycosphingolipid mannosylation triggers salicylic acid-mediated responses in Arabidopsis. Plant Cell 2013:25(5):1881–1894. 10.1105/tpc.113.111500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR. Unsolved mysteries in membrane traffic. Annu Rev Biochem. 2007:76(1):629–645. 10.1146/annurev.biochem.76.061705.130002 [DOI] [PubMed] [Google Scholar]
- Pinot M, Goud B, Manneville JB. Physical aspects of COPI vesicle formation. Mol Membr Biol. 2010:27(8):428–442. 10.3109/09687688.2010.510485 [DOI] [PubMed] [Google Scholar]
- Reggiori F, Conzelmann A. Biosynthesis of inositol phosphoceramides and remodeling of glycosylphosphatidylinositol anchors in Saccharomyces cerevisiae are mediated by different enzymes. J Biol Chem. 1998:273(46):30550–30559. 10.1074/jbc.273.46.30550 [DOI] [PubMed] [Google Scholar]
- Rennie EA, Ebert B, Miles GP, Cahoon RE, Christiansen KM, Stonebloom S, Khatab H, Twell D, Petzold CJ, Adams PD, et al. Identification of a sphingolipid α-glucuronosyltransferase that is essential for pollen function in Arabidopsis. Plant Cell 2014:26(8):3314–3325. 10.1105/tpc.114.129171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riboni L, Giussani P, Viani P. Sphingolipid transport. Adv Exp Med Biol. 2010:688:24–45. 10.1007/978-1-4419-6741-1_2 [DOI] [PubMed] [Google Scholar]
- Rutter BD, Innes RW. Growing pains: addressing the pitfalls of plant extracellular vesicle research. New Phytol. 2020:228(5):1505–1510. 10.1111/nph.16725 [DOI] [PubMed] [Google Scholar]
- Sánchez-Simarro J, Selvi P, Bernat-Silvestre C, Minguet EG, Aniento F, Marcote MJ. Differential involvement of Arabidopsis β'-COP isoforms in plant development. Cells 2022:11(6):938. 10.3390/cells11060938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring D, Viotti C, Krüger F, Künzl F, Sturm S, Bubeck J, Hillmer S, Frigerio L, Robinson DG, Pimpl P, et al. Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 2011:23(9):3463–3481. 10.1105/tpc.111.086918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguí-Simarro JM, Staehelin LA. Cell cycle-dependent changes in Golgi stacks, vacuoles, clathrin-coated vesicles and multivesicular bodies in meristematic cells of Arabidopsis thaliana: a quantitative and spatial analysis. Planta 2006:223(2):223–236. 10.1007/s00425-005-0082-2 [DOI] [PubMed] [Google Scholar]
- Singh P, Jorgačevski J, Kreft M, Grubišić V, Stout RF, Potokar M, Parpura V, Zorec R. Single-vesicle architecture of synaptobrevin2 in astrocytes. Nat Commun. 2014:5(1):3780. 10.1038/ncomms4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss K, Goebel C, Runz H, Möbius W, Weiss S, Feussner I, Simons M, Schneider A. Exosome secretion ameliorates lysosomal storage of cholesterol in Niemann-Pick type C disease. J Biol Chem. 2010:285(34):26279–26288. 10.1074/jbc.M110.134775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer C, Wang T, Michaelos M, Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. Nat Methods. 2021:18(1):100–106. 10.1038/s41592-020-01018-x [DOI] [PubMed] [Google Scholar]
- Tian S, Muneeruddin K, Choi MY, Tao L, Bhuiyan RH, Ohmi Y, Furukawa K, Furukawa K, Boland S, Shaffer SA, et al. Genome-wide CRISPR screens for Shiga toxins and ricin reveal Golgi proteins critical for glycosylation. PLoS Biol. 2018:16(11):e2006951. 10.1371/journal.pbio.2006951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vints K, Vandael D, Baatsen P, Pavie B, Vernaillen F, Corthout N, Rybakin V, Munck S, Gounko NV. Modernization of Golgi staining techniques for high-resolution, 3-dimensional imaging of individual neurons. Sci Rep. 2019:9(1):130. 10.1038/s41598-018-37377-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Ebert B, Parsons HT, Rautengarten C, Bacic A, Heazlewood JL. Enrichment of Golgi membranes from Triticum aestivum (wheat) seedlings. Methods Mol Biol. 2017:1511:131–150. 10.1007/978-1-4939-6533-5_11 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.