Abstract
Molecular modifiers of KRAS G12C inhibitor (KRAS G12Ci) efficacy in advanced KRASG12C-mutant NSCLC are poorly defined. In a large unbiased clinico-genomic analysis of 424 NSCLC patients, we identified and validated co-alterations in KEAP1, SMARCA4 and CDKN2A as major independent determinants of inferior clinical outcomes with KRAS G12Ci monotherapy. Collectively, co-mutations in these three tumor suppressor genes segregated patients into distinct prognostic subgroups and captured ~50% of those with early disease progression (PFS≤3 months) with KRAS G12Ci. Pathway-level integration of less prevalent co-alterations in functionally related genes nominated PI3K/AKT/MTOR pathway and additional baseline RAS gene alterations, including amplifications, as candidate drivers of inferior outcomes with KRAS G12Ci, and revealed a possible association between defective DNA damage response/repair and improved KRAS G12Ci efficacy. Our findings propose a framework for patient stratification and clinical outcome prediction in KRASG12C-mutant NSCLC that can inform rational selection and appropriate tailoring of emerging combination therapies.
Keywords: KRAS, KRAS p.G12C, co-mutations, KEAP1, SMARCA4, CDKN2A, sotorasib, adagrasib, non-small cell lung cancer
Introduction
Activating mutations in the KRAS proto-oncogene are detected in 25%−30% of non-squamous non-small cell lung cancer (NSCLC), and most frequently (~42%) involve a glycine to cysteine substitution at residue 12 (G12C) as a result of a smoking-related G>T transversion (1). Replacement of glycine in codon 12 of KRAS is thought to sterically hinder insertion of the arginine finger (R-finger) of canonical GTPase activating proteins (GAPs, such as neurofibromin and p120RasGAP) into the GTPase active site and impairs GAP-stimulated GTP hydrolysis (2), thus shifting the KRAS nucleotide cycling equilibrium towards the active, GTP-bound state. For over 30 years since its initial discovery, KRAS remained an elusive therapeutic target due to: 1) picomolar binding affinity for its guanine nucleotide substrates coupled with high intracellular concentration of GTP, thus precluding the development of competitive inhibitors; 2) a featureless protein surface devoid of deep pockets suitable for docking of small-molecule inhibitors; 3) on-target toxicity from wild-type KRAS inhibition or concurrent targeting of the downstream effector RAF/MEK/ERK and PI3K/AKT/MTOR pathways; 4) paradoxical increase in RAS signaling with downstream pathway inhibitors due to release of negative feedback; 5) redundant prenylation pathways that control KRAS plasma membrane localization (3). The groundbreaking identification of compounds and subsequent development of covalent allosteric inhibitors that bind irreversibly to cysteine 12 and occupy a cryptic induced pocket in the switch II region of GDP-bound KRAS, trapping the oncoprotein in its inactive conformation, has enabled effective inhibition of KRAS G12C (4,5). Sotorasib (formerly AMG510), the first-in-class KRAS G12C inhibitor (KRAS G12Ci), and adagrasib (formerly MRTX849) both yielded robust single-agent clinical activity in previously treated patients with advanced KRASG12C-mutant NSCLC, producing objective response rates (ORR) of 37%−43% in single arm registration phase II studies (6,7). Based on these results, both sotorasib and adagrasib received FDA accelerated approval for previously treated patients with advanced KRASG12C-mutant NSCLC; furthermore, sotorasib improved progression-free survival (PFS) and ORR compared with docetaxel in the randomized phase III CodeBreaK 200 trial (8). Several additional KRAS G12C inhibitors are undergoing clinical development, with initial reports indicating comparable single-agent activity (9–12).
Despite promising ORR, KRAS G12Ci produce median PFS of approximately 6–7 months (6,7), which is inferior to what has been reported for targeted therapies in other oncogene addicted NSCLC subsets (e.g., EGFR mutations or ALK re-arrangements) (13,14). For individual patients, clinical outcomes with KRAS G12Ci vary widely from long-term durable responses and prolonged survival - with 2-year OS rate of 32.5% reported in CodeBreaK 100 - to early disease progression seen in ~5–16% of treated patients (6,7,15). De novo as well as adaptive and acquired resistance collectively curtail the efficacy of KRAS G12Ci monotherapy (7,15–20), and support the need for improved patient selection for sotorasib or adagrasib monotherapy and for combination regimens directed at treatment intensification. However, molecular or clinical determinants of distinct clinical outcomes with KRAS G12Ci are hitherto poorly defined and validated markers for patient stratification prior to treatment initiation are lacking. Co-occurring genomic alterations in key tumor suppressor genes underpin the molecular diversity of KRAS-mutant NSCLC and impact both tumor cell-intrinsic as well as non-tumor cell autonomous cancer hallmarks including shaping its immune contexture (21,22). Critically, co-mutations can impact responses to standard of care systemic therapies, including both chemotherapy and immunotherapy (22–26). Here, we systematically dissected the impact of genomic and clinical features on outcomes with KRAS G12Ci in the largest cohort to date of NSCLC patients treated with sotorasib or adagrasib, encompassing 424 patients from 21 centers in the U.S. and Europe. We demonstrate that prevalent co-alterations in KEAP1, SMARCA4 and CDKN2A are associated with inferior clinical outcomes with KRAS G12Ci therapy and collectively define a subgroup of patients with poor prognosis. In addition, we identify less prevalent candidate genomic modifiers of KRAS G12Ci efficacy and propose a framework for patient stratification with implications for treatment selection and clinical trial development for KRASG12C-mutant NSCLC.
Results
Clinical outcomes with KRAS G12Ci monotherapy in advanced NSCLC.
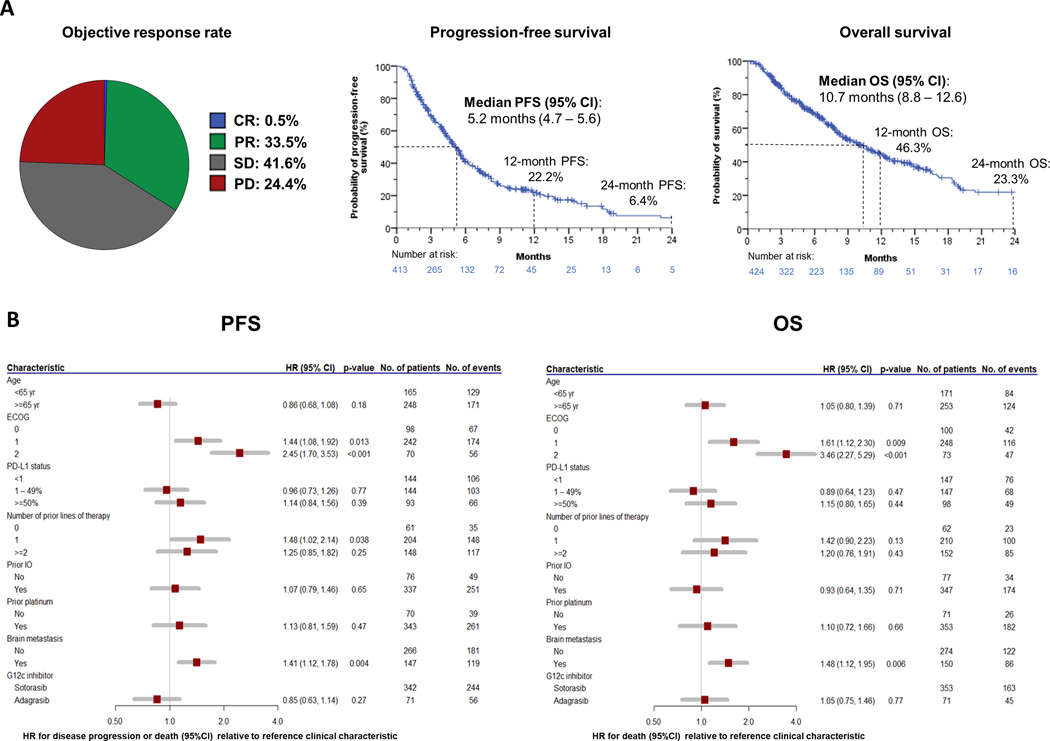
In order to comprehensively interrogate the impact of baseline clinico-genomic parameters on clinical outcomes with KRAS G12Ci, we assembled the largest cohort to date of patients with KRASG12C-mutant NSCLC that were treated with sotorasib or adagrasib, encompassing 424 unique evaluable patients across 21 centers in the U.S. and Europe (Supplementary Table S1). The study cohort was established by merging two independently collected retrospective cohorts [cohort A (N=330) and cohort B (N=94)], that were also analyzed separately to provide additional validation of key findings (Supplementary Tables S2 and S3, see Methods for detailed study eligibility criteria). In the overall cohort, median age was 68 years, patients were predominantly current or former smokers (96.9%), and most had ECOG performance status (PS) 0–1 (82.1%). Adenocarcinoma was the most common histology (92.7%). All patients had metastatic disease at start of KRAS G12Ci therapy, and 35.2% had history of brain metastases (26.2% previously treated, 9.0% untreated). The majority of patients received treatment with sotorasib (83.3%). Most patients received prior treatment with PD-1/PD-L1 inhibitors and platinum-based chemotherapy (75.9%). This cohort was overall representative of the general population of KRASG12C-mutant NSCLC patients (6,7). Most patients had genomic profiling performed on tumor tissue (62.3%), 18.2% had genomic profiling results from liquid biopsy, and 13.7% had both tumor and liquid biopsy profiling. 5.8% of patients had confirmed KRASG12C status from analysis of tumor DNA but did not undergo NGS-based profiling. Patient characteristics for the overall study cohort are summarized in Supplementary Table S1. In the overall cohort, ORR was 34.0% (95% CI 29.4 – 38.8), median PFS was 5.2 months (95% CI 4.7–5.6), and median OS was 10.7 months (95% CI 8.8–12.6) (Figure 1A). The estimated 12-month PFS and OS rates were 22.2% and 46.3% respectively, whereas the estimated 24-month PFS and OS rates were 6.4% and 23.3%, respectively. We observed similar results when analyzing the individual cohorts separately (Supplementary Figure S1A–B). PS of 1 or 2 was associated with shorter PFS and OS compared with PS 0, and patients with history of brain metastases had worse PFS and OS with KRAS G12Ci therapy compared to those without prior history of brain metastasis (Figure 1B). No difference in PFS and OS was observed depending on the KRAS G12Ci used (Figure 1B). When the analysis was limited to previously treated patients with ECOG PS 0–1 and either absent or treated and stable brain metastases at start of KRAS G12Ci therapy [comparable to the patient population enrolled in the registrational CodeBreaK 100 and KRYSTAL-1 clinical trials (6,7)], the ORR was 35.0% (95% CI 29.1 – 41.1), the median PFS was 5.5 months (95% CI 4.9–6.0) and the median OS was 11.4 months (95% CI 8.8–14.1) (Supplementary Figure S2A). Patients with untreated brain metastases had similar survival compared with those with previously treated brain metastases (PFS: 5.0 vs 4.3 months, log-rank p=0.964, multivariable [MV] hazard ratio [HR] 0.95 [95% CI 0.82–1.44]; OS: 8.8 vs 7.8 months, log-rank p=0.741, MV HR 1.13 [95% CI 0.68–1.88]) (Supplementary Figure S2B). Tumor cell PD-L1 expression and exposure to immune checkpoint inhibitors in prior line(s) of therapy was not associated with PFS or OS (Figure 1B, Supplementary Figure S2C–D).
Figure 1.
Clinical outcomes with KRAS G12Ci monotherapy in the overall study cohort. A) Objective response, progression-free survival and overall survival upon treatment with KRAS G12Ci in advanced KRASG12C-mutant NSCLC. B) Forest plot representation of clinical characteristics and their impact on progression-free survival and overall survival.
Co-alterations in KEAP1, SMARCA4, and CDKN2A are associated with early disease progression and poor clinical outcomes with KRAS G12Ci.
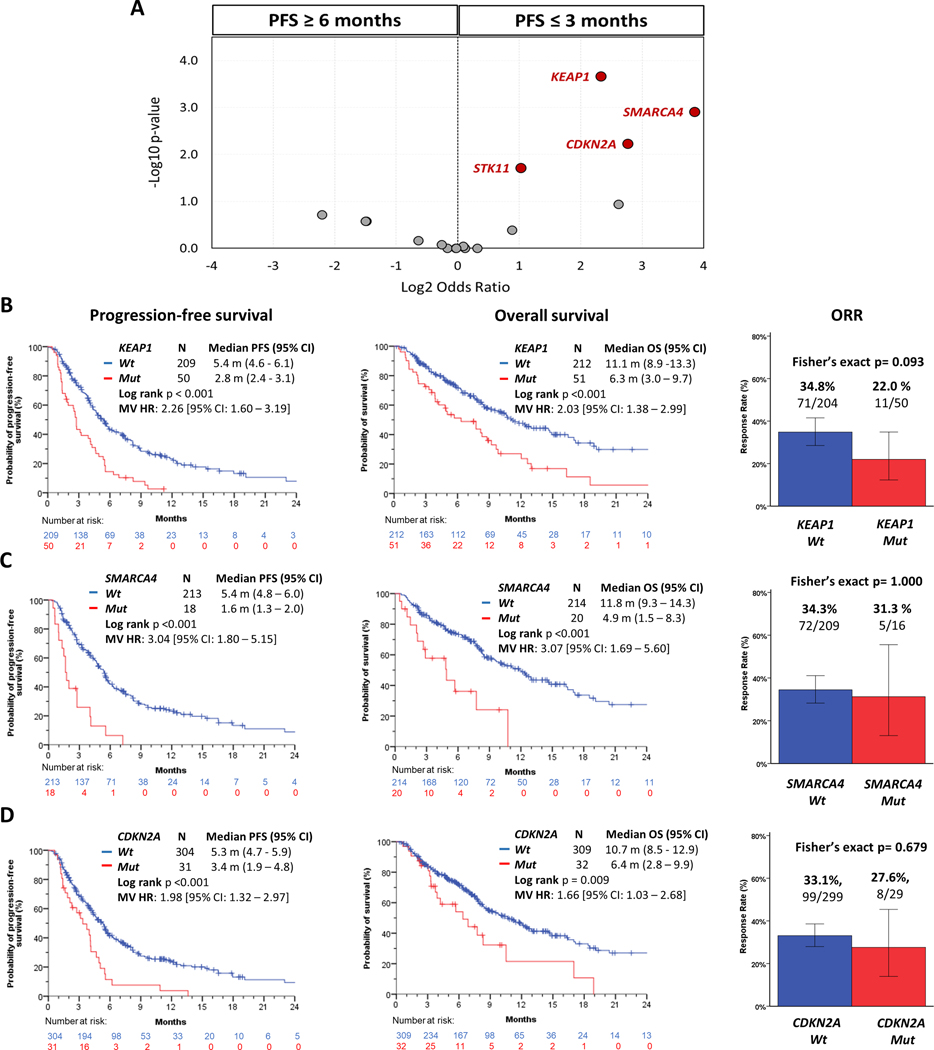
To dissect the impact of the tumor co-mutational landscape on clinical outcomes with KRAS G12Ci, we first classified patients into subgroups with durable clinical benefit (PFS ≥ 6 months; N=131) or early progression (PFS ≤ 3 months; N=124) (total N=255) (18). Patients censored with less than 3 months of follow-up were excluded from this analysis. We then performed an unbiased enrichment analysis of the most prevalent co-alterations (detected in at least 5% of patients) in the overall study cohort (see Methods section for additional details). We found that co-mutations in three tumor suppressor genes were significantly enriched in the early progression subgroup: KEAP1 (Fisher’s exact p<0.001, false discovery rate [FDR] q=0.004), SMARCA4 (Fisher’s exact p=0.001, FDR q=0.010), CDKN2A (Fisher’s exact p=0.006, FDR q=0.034) (Figure 2A). Patients bearing KEAP1 co-mutated tumors (KEAP1MUT) exhibited significantly shorter PFS (2.8 vs 5.4 months, log-rank p<0.001, MV HR 2.26 [95% CI 1.60 – 3.19]) and OS (6.3 vs 11.1 months, log-rank p<0.001, MV HR 2.03 [95% CI 1.38 – 2.99]) compared with those harboring KEAP1 wild-type (KEAP1WT) NSCLC (Figure 2B). SMARCA4 co-mutations were associated with markedly worse PFS and OS compared with SMARCA4 wild-type (SMARCA4MUT vs SMARCA4WT PFS: 1.6 vs 5.4 months, log-rank p<0.001, MV HR 3.04 [95% CI 1.80 – 5.15]; OS: 4.9 vs 11.8 months, log-rank p<0.001, MV HR 3.07 [95% CI 1.69 – 5.60]) (Figure 2C). Co-alterations in CDKN2A were also associated with worse PFS and OS upon treatment with KRAS G12Ci compared with CDKN2A wild-type (CDKN2AMUT vs CDKN2AWT PFS: 3.4 vs 5.3 months, log-rank p<0.001, MV HR 1.98 [95% CI 1.32 – 2.97]; OS: 6.4 vs 10.7 months, log-rank p=0.009, MV HR 1.66 [95% CI 1.03 – 2.68]) (Figure 2D). Similar findings were observed when cohorts A and B were analyzed separately (Supplementary Figure S3A–C), and when limiting the analysis only to patients that received prior immune checkpoint inhibitor therapy (Supplementary Figure S4A–C). KEAP1 co-mutations were associated with numerically lower ORR compared with KEAP1 wild-type, whereas there was no significant difference in ORR between patients with SMARCA4MUT vs SMARCA4WT and CDKN2AMUT vs CDKN2AWT NSCLC (Figures 2B–D).
Figure 2.
Co-mutations in KEAP1, SMARCA4, and CDKN2A are associated with inferior clinical outcomes with single-agent KRAS G12C inhibitor therapy. A) Volcano plot depicting relative enrichment of co-alterations in distinct clinical outcome subgroups [durable clinical benefit (PFS≥6 months) vs early disease progression (PFS≤3 months)]. Qualified genes were included based on p-value ≤ 0.05 (Fisher’s exact) and FDR q-value ≤ 0.10. Clinical outcomes in the overall study cohort according to co-mutation status of B) KEAP1, C) SMARCA4, and D) CDKN2A.
STK11 was the fourth most enriched somatically mutated gene in patients with early progression with KRAS G12Ci (Fisher’s exact p=0.019, FDR q=0.082) (Figure 2A). Patients with STK11MUT NSCLC had shorter PFS compared with patients that harbored STK11WT tumors (4.4 vs 5.5 months, log-rank p=0.010, MV HR 1.32 [95% CI 1.00 – 1.73]). No significant difference was observed between patients bearing STK11MUT and STK11WT tumors for OS (9.8 vs 10.5 months, log-rank p=0.167, MV HR 1.18 [95% CI 0.85 – 1.64]) or ORR (31.5% vs 34.3%, Fisher’s exact p=0.616) (Figure 3A). Because STK11 and KEAP1 mutations frequently overlap in KRAS-mutant NSCLC (21), we sought to de-convolute their individual impact by comparing outcomes with KRAS G12Ci in three distinct genomically defined subgroups: (1) KRASG12C/KEAP1WT/STK11WT; (2) KRASG12C/KEAP1WT/STK11MUT; (3) KRASG12C/KEAP1MUT/STK11MUT or WT. The KRASG12C/ KEAP1MUT/STK11MUT or WT subgroup exhibited significantly shorter PFS and OS compared with the KRASG12C/KEAP1WT/STK11WT subgroup (PFS: 2.8 vs 5.3 months, log-rank p<0.001, MV HR 2.30 [95% CI 1.60 – 3.30]; OS 6.3 vs 10.7 months, log-rank p<0.001, MV HR 2.13 [95% CI 1.41 – 3.20]), and numerically lower ORR (22.0% vs 34.9%, Fisher’s exact p=0.114). The KRASG12C/KEAP1WT/STK11MUT and KRASG12C/KEAP1WT/STK11WT subgroups had similar PFS (KRASG12C/KEAP1WT/STK11MUT vs KRASG12C/KEAP1WT/STK11WT PFS: 5.6 vs 5.3 months, MV HR 1.03 [95% CI 0.74 – 1.46]), OS (12.3 vs 10.7 months, MV HR 1.05 [95% CI 0.69 – 1.61]), and ORR (40.6% vs 34.9%) (Figure 3B). Similar results were observed when cohorts A and B were analyzed separately (Supplementary Figure S5A–B). Further deconvolution of patients with KEAP1MUT NSCLC based on STK11 mutation status yielded similar findings; each of the KRASG12C/KEAP1MUT/STK11MUT and KRASG12C/KEAP1MUT/STK11WT subgroups exhibited worse PFS and OS when compared with KRASG12C/KEAP1WT/STK11MUT or KRASG12C/KEAP1WT/STK11WT subgroups (Supplementary Figure S5C). We therefore conclude that STK11 co-mutations without concurrent KEAP1 mutations may not significantly influence outcomes with KRAS G12Ci monotherapy. This finding was also upheld when the analysis was limited to KEAP1WT, SMARCA4WT and CDKN2AWT (KSCWT) tumors (Supplementary Figure S5D).
Figure 3.
STK11 co-mutations may not impact clinical outcomes with KRAS G12Ci in the absence of concurrent KEAP1 alterations. A) Clinical outcomes with KRAS G12Ci according to STK11 co-mutation status in the overall cohort; B) De-convolution of clinical outcomes with KRAS G12Ci in the KRASG12C/KEAP1MUT/STK11MUT or WT, KRASG12C/KEAP1WT/STK11MUT and KRASG12C/KEAP1WT/STK11WT subgroups.
TP53 was the most frequently co-mutated gene in the overall cohort (45.7%), but TP53 mutations were not associated with clinical outcomes with KRAS G12Ci (Supplementary Figure S6A). This was further validated when cohorts A and B were analyzed separately (Supplementary Figure S6B–C).
Exploratory analysis identifies additional co-mutations associated with distinct clinical outcomes with KRAS G12Ci therapy.
Next, we interrogated our patient cohort to determine less prevalent functionally related co-mutations that are enriched in patients with early progression or durable clinical benefit. We focused this analysis on an expanded set of genes that were co-mutated in at least 3 patients. Due to the large-size dominant effects of KEAP1, SMARCA4, and CDKN2A (KSCMUT) this analysis was limited to patients whose tumor was KSCWT (N=128). CHEK2 and ATRX co-mutations were enriched in patients with durable clinical benefit with KRAS G12Ci (OR≤−2.0) whereas tumors harboring (i) KRAS amplification; (ii) co-mutations in TSC1, TSC2, MTOR or PTEN, encoding components of the PI3K/AKT/MTOR pathway, and (iii) co-mutations in some additional driver oncogenes (such as ALK, ROS1, NTRK3) were enriched in patients with early progression (OR≥2.0) (Figure 4A).
Figure 4.
Candidate low-prevalence genomic modifiers of clinical outcomes with KRAS G12Ci. A) Volcano plot of co-mutated genes enriched in the early progression (PFS≤3 months) or durable clinical benefit (PFS≥6 months) groups among patients with KSCWT tumors; B) Objective response rate according to co-mutation status of a group of established DDR genes – BRCA1/2, ATM, ATR, CHEK1/2, PALB2, RAD50/51/51B/51C/51D - in the overall mutation-evaluable population; C-F) Kaplan-Meier estimates of progression-free survival and overall survival with KRAS G12Ci depending on the mutational status of C) DDR genes (overall mutation-evaluable population); D) ATRX/DAXX (overall mutation-evaluable population); E) additional alterations (beyond KRASG12C) in RAS genes (KRAS/NRAS/HRAS; overall mutation-evaluable population); F) PI3K/AKT/MTOR pathway genes (mutation evaluable KSCWT population). Only cases with available comprehensive genomic profiling that included all functionally related genes within a group were considered wild-type for the grouped alterations.
We further examined the association of co-mutations in the identified candidate genes and clinical outcomes with KRAS G12Ci in the evaluable population for each individual gene. Patients whose tumor harbored co-mutations in CHEK2 had longer PFS compared with those whose tumor was CHEK2 wild-type, and median OS was not reached in CHEK2MUT patients (Supplementary Figure S7A). CHEK2 is a tumor suppressor gene that encodes a serine/threonine kinase involved in signal transduction in the cellular response to DNA double-strand breaks (DSBs) (27). We then further explored the impact of somatic genomic alterations in a group of well validated DDR genes – BRCA1/2, ATM, ATR, CHEK1/2, PALB2, RAD50/51/51B/51C/51D. Alterations in this group of DDR genes were present in 32.1% of patients. Patients whose tumor harbored DDR gene co-mutations had higher ORR (52.2% vs 27.7%, Fisher’s Exact test p=0.001) (Figure 4B), and significantly longer PFS with KRAS G12Ci compared with patients whose tumor was DDR gene wild-type (5.9 vs 4.6 months, log-rank 0.030, HR 0.68 [95% CI 0.48 – 0.97]), although there was no statistically significant difference in OS between patients harboring DDR gene-co-mutated and wild-type tumors (13.0 vs 8.4 months, log-rank p=0.075, HR 0.69 [95% CI 0.46 – 1.04]) (Figure 4C). Somatic mutations in ATRX were also enriched in patients with durable clinical benefit with KRAS G12Ci therapy (Figure 4A), and were associated with longer PFS and OS with KRAS G12Ci therapy when compared with patients bearing ATRX wild-type tumors (Supplementary Figure S7B). ATRX encodes an ATP-dependent chromatin remodeling protein, member of the SWI/SNF family, that interacts with the histone chaperone DAXX to deposit the variant histone H3.3 at sites of nucleosome turnover (28). The ATRX/DAXX complex has been implicated in transcriptional regulation and control of DNA replication, recombination and repair (28,29). Patients whose tumor harbored somatic mutations in the ATRX/DAXX genes had longer PFS and OS with sotorasib and adagrasib compared with patients whose tumor was ATRX/DAXX wild-type (Figure 4D).
Patients with NSCLC harboring additional (beyond the qualifying KRASG12C mutation) co-alterations in RAS genes (KRAS/NRAS/HRAS, including both somatic mutations and/or gene amplifications) prior to starting KRAS G12Ci therapy exhibited worse PFS and OS compared with those bearing tumors without additional RAS gene alterations in the mutation-evaluable population (Figure 4E) as well as in the KSCWT population (Supplementary Figure S8A). Presence of co-mutations in a group of functionally related PI3K/AKT/MTOR pathway genes (including AKT1, PIK3CA, MTOR, TSC1/2, PTEN) was not associated with survival in the overall mutation-evaluable population (Supplementary Figure S8B). This may be attributable to the large-size dominant effects of co-occurring KEAP1, SMARCA4, and CDKN2A mutations on clinical outcomes with KRAS G12Ci therapy (Figure 2B–D). Therefore, we tested the association of PI3K/AKT/MTOR pathway genes with survival in the KSCWT population, and observed that patients whose tumors harbored PI3K/AKT/MTOR co-mutations had significantly shorter PFS with sotorasib and adagrasib compared with patients harboring PI3K/AKT/MTOR wild-type tumors (Figure 4F). We also found that patients whose tumors harbored missense mutations in ROS1, ALK, and NTRK1–3 oncogenes - assessed together - had shorter PFS and OS with KRAS G12Ci therapy compared with patients whose tumors were ROS1, ALK, and NTRK1–3 wild-type (Supplementary Figure S8C–D). Additional co-mutated genes that were enriched in patients with early progression included LRP1B, KDM5C, FAT1, NOTCH2, NFE2L2, FLT1, and RAD50 (Figure 4A). However, none of these genes were associated with survival with KRAS G12Ci therapy (LRP1BMUT vs LRP1BWT: 3.0 vs 5.1 months, log-rank p=0.585, HR 1.24 [95% CI 0.57 – 2.67]; KDM5CMUT vs KDM5CWT: 2.2 vs 5.3 months, log-rank p=0.143, HR 1.83 [95% CI 0.80 – 4.19]; FAT1MUT vs FAT1WT: 4.1 vs 4.7 months, log-rank p=0.263, HR 1.66 [95% CI 0.68 – 4.09]; NOTCH2MUT vs NOTCH2WT: 1.9 vs 4.8 months, log-rank p=0.427, HR 1.39 [95% CI 0.61 – 3.16]; NFE2L2MUT vs NFE2L2WT: 5.5 vs 4.7 months, log-rank p=0.701, HR 1.17 [95% CI 0.52 – 2.65]; FLT1MUT vs FLT1WT: 2.8 vs 4.7 months, log-rank p=0.187, HR 1.93 [95% CI 0.71 – 5.24]; RAD50MUT vs RAD50WT: 2.7 vs 4.7 months, log-rank p=0.832, HR 1.13 [95% CI 0.36 – 3.59]). These findings collectively suggest that baseline co-alterations in RAS genes and PI3K/AKT/MTOR pathway genes, may exert a deleterious effect on clinical outcomes with KRAS G12Ci. It remains unclear if individual missense mutations in oncogenic drivers such as ALK, ROS1 and NTRK1–3 are functional and expressed in the absence of corresponding gene rearrangements. Meanwhile, somatic mutations in genes involved in DDR and chromatin remodeling/epigenetic regulation may have favorable impact on treatment outcomes with KRAS G12Ci. Due to the exploratory nature of this analysis, these findings warrant validation in subsequent preclinical and clinical studies.
Genomic landscape of early progression and durable clinical benefit with KRAS G12C inhibitors.
Next, we aimed to determine the prevalence and overlap of enriched co-mutations in patients with either early progression or durable clinical benefit with KRAS G12Ci in order to further explore their clinical relevance and inter-relationships. For this purpose, we focused our analysis on patients whose tumor underwent comprehensive NGS profiling (≥400 covered genes). As expected, KEAP1, SMARCA4, and CDKN2A co-alterations were prevalent in patients with early disease progression (Figure 5A). In KSCWT tumors, additional alterations in RAS genes (KRAS, NRAS, HRAS), mutations in PI3K/AKT/MTOR pathway genes (AKT1, PIK3CA, MTOR, TSC1/2, PTEN), and somatic mutations in select driver oncogenes (ALK, ROS1, NTRK1/2/3) were identified in 11.1% (3/27), 18.5% (5/27), and 33.3% (9/27) of patients with early disease progression, respectively (Figure 5B). Co-mutations in the individual pathway genes PTEN, TSC1, TSC2 and MTOR were identified in 2%, 4%, 2% and 4% of patients with early progression, respectively, and were absent in patients with durable clinical benefit (Figure 5A). Meanwhile, co-mutations in CHEK2, PALB2, and ATRX were present in 8%, 2%, and 6% of patients with durable clinical benefit, and were lacking in those with early progression (Figure 5A). Co-mutations in ATRX/DAXX were present in 10%, and co-mutations in a group of well validated DDR genes – BRCA1/2, ATM, ATR, CHEK1/2, PALB2, RAD50/51/51B/51C/51D - were present in 40% of patients with durable clinical benefit (Figure 5C).
Figure 5.
Genomic landscape of early disease progression and durable clinical benefit with KRAS G12Ci. This analysis only included patients whose tumor underwent comprehensive NGS profiling (≥400 covered genes). A) OncoPrint illustrating co-alterations in patients with early disease progression (left) and durable clinical benefit (right); B) Pie chart representation of the prevalence of RAS co-alterations (left), co-mutations in PI3K/AKT/MTOR pathway genes (middle), and somatic mutations in ROS1/ALK/NTRK1–3 oncogenes (right) in patients with KSCWT NSCLC and early progression (PFS ≤3 months) with KRAS G12Ci; C) Pie chart representation of the prevalence of co-alterations in ATRX/DAXX (left) and DDR genes (BRCA1/2, ATM, ATR, CHEK1/2, PALB2, RAD50/51/51B/51C/51D) (right) in patients with durable clinical benefit (PFS ≥6 months) with KRAS G12Ci in the mutation-evaluable population.
Integration of KEAP1, SMARCA4 and CDKN2A co-mutations provides a framework for patient stratification and clinical outcome prediction with KRAS G12Ci monotherapy.
Through an unbiased approach, we identified genes that when co-mutated were associated with early progression with KRAS G12Ci therapy (Figure 2A). Prevalent alterations in KEAP1, SMARCA4, and CDKN2A (collectively identified in 32.0% of KRASG12C-mutant NSCLC in our overall cohort) were the most enriched in this group and captured 49.3% of patients with early disease progression with KRAS G12Ci (Figure 6A). Figures 6A and Supplementary Figure S9A show the overlap between KEAP1, SMARCA4, and CDKN2A co-mutations. The KSCMUT subgroup exhibited numerically lower ORR compared with the KSCWT subgroup (25.3% vs 38.1%, Fisher’s exact p=0.065) (Figure 6B). Despite approximately a quarter of patients achieving an early response, PFS and OS were significantly curtailed in the KSCMUT subgroup compared with the KSCWT subgroup (PFS: 2.8 vs 5.9 months, log-rank p<0.001, MV HR 2.51 [95% CI 1.79 – 3.52]; OS: 6.9 vs 13.0 months, log-rank p<0.001, MV HR 2.05 [95% CI 1.38 – 3.02]) (Figure 6C). Furthermore, the KSCMUT subgroup had markedly inferior 6- and 12-month PFS and OS compared with the KSCWT subgroup (estimated 6- and 12-month PFS rate: 15.7% vs 49.5%, and 3.3% vs 28.5%, respectively; estimated 6- and 12-month OS rate: 54.7% vs 75.2%, and 27.0% vs 54.6%, respectively) (Figure 6C). We also observed an incrementally detrimental effect based on co-mutation overlap of the KSC genes. Patients whose tumors harbored 2 or more co-mutations in any of the KSC genes exhibited significantly worse PFS and OS compared with patients with KSCWT NSCLC and with those with tumors bearing a single altered KSC gene upon treatment with KRAS G12Ci (Supplementary Figure S9B–C). Importantly, KEAP1, SMARCA4, and CDKN2A co-mutations were each independently associated with shorter PFS with KRAS G12Ci in a multivariable model that also incorporated key clinical characteristics (Supplementary Table S4). KEAP1 and SMARCA4 were also independently associated with shorter OS (Supplementary Table S5). Thus, co-mutations in KEAP1, SMARCA4 and CDKN2A are robust independent determinants of KRAS G12Ci efficacy that consistently segregate patients with advanced KRASG12C-mutant NSCLC into groups with markedly dissimilar clinical outcomes.
Figure 6.
Combined evaluation of KEAP1, SMARCA4 and CDKN2A co-mutations defines a subgroup of KRASG12C-mutant NSCLC (KSCMUT) with poor outcomes with KRAS G12Ci therapy. A) Pie chart depicting the prevalence of KEAP1, SMARCA4, and CDKN2A co-alterations in the mutation-evaluable population for all three genes (N=188) (left) and among patients with early disease progression with KRAS G12Ci (N=63) (right). B) Objective response to KRAS G12Ci in patients with KSCWT and KSCMUT NSCLC in the overall response-evaluable study population. C) PFS (left) and OS (right) with KRAS G12Ci according to KSC co-mutation status in the overall study population.
Discussion
In this study we identified genomic modifiers of KRAS G12Ci efficacy in advanced NSCLC through an unbiased clinico-genomic analysis of the largest cohort to date of patients treated with sotorasib or adagrasib. Prevalent co-alterations in KEAP1, SMARCA4 and CDKN2A were each associated with early disease progression and poor clinical outcomes with KRAS G12Ci monotherapy - independently of key clinical covariates - and collectively define subgroups of KRASG12C-mutant NSCLC patients with markedly divergent therapeutic response trajectories and overall prognosis. Furthermore, in an exploratory analysis we identified less frequent baseline somatic alterations in RAS genes and PI3K/AKT/MTOR pathway genes as candidate mediators of inferior clinical outcomes with KRAS G12Ci, whereas grouped alterations in DDR genes and components of the ATRX/DAX chromatin remodeling complex were associated with prolonged clinical benefit. These findings shed light on the molecular underpinnings of KRAS G12Ci clinical response heterogeneity in NSCLC and suggest a framework for patient stratification as well as for personalization of KRAS G12C inhibitor-anchored combination therapeutic strategies (Supplementary Figure S10).
Examined individually, co-alterations in KEAP1, SMARCA4 and CDKN2A were consistently associated with significantly shorter PFS and OS with KRAS G12Ci in two independently established cohorts of patients with advanced KRASG12C-mutant NSCLC, as well as in the overall merged cohort. In contrast, their impact on ORR was more heterogeneous and did not reach statistical significance, although a trend towards lower ORR was observed for KEAP1 mutations. KEAP1 co-mutations were associated with numerically lower ORR with both sotorasib and adagrasib in the phase II component of the CodeBreaK 100 and KRYSTAL-1 clinical trials respectively, but in both cases the confidence intervals overlapped (6,7); surprisingly, higher ORR was reported with adagrasib in patients with CDKN2AMUT compared with CDKN2AWT NSCLC in KRYSTAL-1 (7). Biologically, this discrepancy may underlie the emergence of adaptive - rather than primary - resistance, that can develop expeditiously in response to KRAS G12Ci (16) and manifest as rapid disease progression after initial radiological response. Therefore, assessment of the impact of co-mutations based on ORR alone may underestimate or fail to adequately capture their effect on the efficacy of KRAS G12Ci monotherapy. When assessed together, KSC alterations were identified in 32.0% of patients in the overall cohort and accounted for approximately half (49.3%) of patients that exhibited early disease progression (PFS ≤3 months) with sotorasib or adagrasib. The median PFS in patients with KSCMUT NSCLC was 2.8 months (compared with 5.9 months in KSCWT) and the estimated 12-month PFS rate was 3.3% (compared with 28.5% in KSCWT). Thus, co-mutations in key tumor suppressor genes delineate subsets of KRASG12C-mutant NSCLC with strikingly dissimilar clinical outcomes with KRAS G12Ci.
A limitation of the current study is that it does not allow for separation of predictive from prognostic effects of individual genomic alterations. However, both KEAP1 and CDKN2A loss were previously identified as drivers of improved cellular fitness under adagrasib selection in CRISPR/Cas9-based in vitro and in vivo knockout screens, thus supporting a causal - albeit context-dependent - role in mediating KRAS G12Ci insensitivity (5). KEAP1 encodes an adaptor protein that engenders substrate specificity for the CUL3/RBX E3 ubiquitin ligase complex and is critical for the ubiquitylation and proteasomal degradation of NRF2 (encoded by the NFE2L2 gene), a master regulator of cellular anti-oxidant, anti-inflammatory and cytoprotective signals (30). Importantly, NRF2 is involved in transcriptional control of genes encoding efflux transporters as well as several genes involved in xenobiotic detoxification (30). Inactivating KEAP1 somatic mutations have been associated with poor prognosis and inferior clinical outcomes with radiation therapy or chemo-radiation (31,32), platinum-doublet chemotherapy (22,24,33), PD-1 axis inhibitor monotherapy (24,33,34), and chemo-immunotherapy (25,26) in NSCLC, particularly in the context of KRAS-mutant tumors (22). Furthermore, KEAP1 depletion promoted resistance to multiple targeted therapies against components of the RTK/RAS/MAPK pathway in NSCLC cell lines by decreasing drug-induced generation of ROS and increasing glutathione synthesis (35). It should be noted that although NRF2 nuclear accumulation is considered the dominant molecular event downstream of KEAP1 loss in terms of carcinogenesis and therapeutic response, several NRF2-independent effects of KEAP1 inactivation have also been recognized (36). Inactivation of CDKN2A alone or in combination with the genetically and functionally related CDKN2B gene as a result of somatic mutation or bi-allelic deletion (frequently involving both genes as a result of an arm-level event in 9p21) can ostensibly promote KRAS G12Ci resistance by decoupling cell cycle progression from signaling downstream of KRASG12C. In this context, it is plausible that less prevalent alterations in other components of the cell cycle machinery may also influence individual responses to KRAS G12Ci as a result of dysregulated cell cycle control. Finally, deleterious somatic mutations in SMARCA4 encoding BRG1, one of two possible and mutually exclusive ATP-dependent core catalytic subunits of mammalian SWI/SNF ATP-dependent chromatin remodeling complexes, were previously linked with dedifferentiated histology and an atypical club cell lung cancer cell of origin in genetically engineered mouse models (37). In addition, SMARCA4 somatic mutations portend poor prognosis in patients with both early stage and advanced NSCLC - particularly among those that harbor KRAS-mutant tumors - although reports of their impact on immune checkpoint inhibitor efficacy have been conflicting (38–40). The mechanism(s) by which SMARCA4 loss may modulate response to KRAS G12Ci are currently unknown but previously reported pleiotropic functions in the regulation of cellular differentiation, DNA replication and repair as well as cell cycle progression are likely to be involved (41). The SMARCA4 genomic locus resides on the short arm of chromosome 19 (19p13.2), in topological proximity to KEAP1 and STK11, thus increasing susceptibility to co-deletion events that contribute to the frequent co-occurrence of alterations in the three genes.
Co-mutations in STK11, when present in the absence of concurrent alterations in KEAP1 (or KEAP1/SMARCA4/CDKN2A) did not impact ORR, PFS or OS with KRAS G12Ci. This finding has implications for clinical trial design and interpretation, because STK11 alterations are drivers of poor clinical outcomes with first-line PD-(L)1 inhibitor-encompassing chemo-immunotherapy regimens in advanced NSCLC (25,26) and constitute an eligibility criterion for clinical trials evaluating KRAS G12Ci in previously untreated patients (NCT04933695, NCT03785249). Furthermore, these results argue against a purely prognostic role for STK11 somatic mutations in NSCLC.
In order to identify additional, less prevalent, candidate mediators of diverse therapeutic outcomes with KRAS G12Ci, we adopted a pathway-level approach by initially surveying individual somatically mutated genes that were enriched in either the durable benefit (PFS ≥6 months) or early progression (PFS ≤3 months) subgroups and subsequently assessing their combined impact on KRAS G12Ci clinical outcomes. This analysis revealed association of mutations in genes implicated in DNA damage response and repair with improved clinical outcomes with KRAS G12Ci. Recurrent mutations in two distinct groups of genes were enriched in the durable benefit group including: a) DDR pathway genes, such as CHEK2, and b) ATRX and DAXX. The ATRX/DAXX complex has been implicated in the maintenance of genomic integrity through diverse effects in DNA repair, replication, methylation, gene expression and telomere homeostasis; accordingly, ATRX or DAXX-deficient tumors exhibit DNA repair defects and display genomic instability (28,29,42,43). Therefore, convergence on impaired DDR and genome maintenance pathways may underpin the increased KRAS G12Ci sensitivity of several low penetrance co-mutations. Notably, enrichment for DDR gene mutations in patients with durable clinical benefit was not uniform across individual genes and was not observed for ATM or RAD50; acquisition of secondary genomic alterations in this heavily chemotherapy-pretreated patient cohort may account for this discordant observation. Due to the exploratory nature of this analysis, these findings require further evaluation and validation in future studies.
Baseline co-alterations in RAS genes, including high level focal KRAS amplifications and co-existing oncogenic somatic mutations in KRAS/HRAS/NRAS, were enriched in patients with early progression, and were associated with worse PFS and OS with KRAS G12Ci. These results are aligned with prior pre-clinical and clinical work demonstrating that de novo and acquired RAS alterations are associated with and lead to resistance to single-agent KRAS G12Ci adagrasib and sotorasib (17,18). Co-occurring alterations in components of the PI3K/AKT/MTOR pathway were also associated with inferior PFS with KRAS G12Ci in KSCWT tumors; mutations in these genes can promote KRAS G12Ci insensitivity by establishing bypass signaling tracts, in agreement with direct effects in preclinical models (5). Finally, somatic mutations in some oncogenic kinase genes, including ROS1, ALK and NTRK1/2/3, were also associated with inferior PFS and OS with KRAS G12Ci in KSCWT tumors. Gradual expansion of subclonal mutations under the selective pressure imposed by KRAS G12Ci therapy may explain their more modest impact on clinical outcomes.
Taken together, our data establish co-mutations in KEAP1, SMARCA4 and CDKN2A as major independent determinants of inferior clinical outcomes with KRAS G12Ci monotherapy in advanced NSCLC. Additional granularity and accuracy in forecasting individual clinical response trajectories and patient stratification into distinct prognostic groups will likely be achieved by incorporation of less prevalent genomic as well as baseline and on-treatment transcriptomic and proteomic biomarkers (Supplementary Figure S10). For example, expression of RGS3, a non-canonical, mutant KRAS-inclusive GAP correlated with in vivo KRAS G12Ci sensitivity in a panel of NSCLC PDX models (44). Lineage- or cell state- specific as well as non-tumor cell intrinsic effects may also contribute to future integrated KRAS G12Ci efficacy predictive models. Finally, beyond patient stratification and individual clinical response prediction, our results are relevant for prioritization and precise tailoring of KRAS G12Ci-based combination therapeutic strategies – including currently ongoing and planned combinations with CDK4/6, mTOR, DNA repair, SHP2, EGFR and MEK/ERK inhibitors - to the co-mutation status of individual tumors in order to maximize therapeutic benefit.
Methods
Study population
Electronic medical record review was performed for two independently collected patient cohorts from 21 academic institutions in the U.S. and Europe. Cohort A includes MD Anderson Cancer Center, Cleveland Clinic, University of Chicago, Yale University, University of Cologne, University of Heidelberg, Columbia University Medical Center, Gustave Roussy, Henry Dunant Hospital Center, Johns Hopkins, Ohio State University, Instituto Nazionale Tumori Regina Elena-Rome, Stanford University, University of Torino–Orbassano, UC Davis, UCLA, UCSD, UCSF, Moffitt Cancer Center. Cohort B includes Dana Farber Cancer Institute and Massachusetts General Hospital. Patients with stage IV KRASG12C-mutant NSCLC who received treatment with single-agent KRAS G12Ci sotorasib or adagrasib, were alive for ≥14 days after start of treatment, had ECOG PS ≤2, and had genomic profiling results available from tumor or blood prior to starting KRAS G12Ci were eligible. Patients with acquired KRAS mutation in the context of other oncogene-addicted NSCLC (e.g., EGFR, ALK) were excluded. Patients were treated between November 2018 and October 2022, and the dataset was locked on October 01, 2022 for the outcome analysis. Patient information was collected through chart review. Cohorts A and B were analyzed separately and in combination (overall study cohort) for scientific rigor and transparency to provide further validation of key findings. Number of prior lines of therapy was defined as lines of systemic therapy received for metastatic disease. Tumor cell PD-L1 expression was determined with the Dako 22C3 (61.8%), E1L3N (23.6%), Ventana SP263 (12%), QR1 (1.2%), Ventana SP142 (0.6%) and IHC411 (0.6%) assays. The study was IRB approved at participating centers and included a waiver of patient informed consent. This study was conducted in accordance with ethical guidelines including the Declaration of Helsinki and U.S. Common Rule.
Genomic profiling
Patients must have had genomic profiling results from tumor and/or plasma prior to starting KRAS G12Ci to be included in the analysis. Tests performed through commercially approved assays or in a CLIA-certified laboratory were allowed (see Supplementary Table S6 for included assays). When available, we integrated results from tumor and plasma profiling for the analysis. Test results for each individual patient were curated and annotated for pathogenic somatic non-synonymous variants. Variants reported as germline were excluded. To be classified as pathogenic, a variant must meet at least one of four criteria: 1) be defined as pathogenic per Catalogue of Somatic Mutations in Cancer (COSMIC - RRID:SCR_002260) entry; 2) be defined as pathogenic on the ClinVar database (RRID:SCR_006169); 3) have PolyPhen (Polymorphism Phenotyping - RRID:SCR_013189) score ≥ 0.95 (45); 4) have SIFT (Sorting Intolerant From Tolerant - RRID:SCR_012813) score ≤ 0.05 (46). Biallelic (homozygous) copy number losses for tumor suppressor genes, amplifications for oncogenes, and gene rearrangements - where reported - were considered relevant alterations and were included in the analysis.
Statistical analysis
To determine genomic modifiers of clinical outcomes with KRAS G12C inhibitors, we first classified patients into two subgroups: durable clinical benefit (PFS ≥ 6 months) or early progression (PFS ≤ 3 months) with sotorasib and adagrasib, following similar methodology as previously reported (18). Patients censored with less than 3 months of follow-up were excluded. We then performed an unbiased enrichment analysis of the most prevalent co-alterations (detected in at least 5% of patients) in the overall study cohort. If a given patient underwent profiling (tumor or plasma) with a NGS panel that did not cover a specific gene, then that patient was removed from the analysis of that specific gene. Differences between durable clinical benefit and early progression subgroups were assessed with Fisher’s exact test adjusted for multiple comparisons using false discovery rate (Benjamini-Hochberg procedure). Significance was established at p ≤ 0.05 and FDR q ≤ 0.10.
To identify less prevalent co-mutations that might be associated with clinical outcomes upon treatment with KRAS G12Ci, we performed an exploratory analysis focusing on 1) KSCWT tumors, 2) genes with co-mutations present in at least 3 patients. Genes of interest were selected based on Log2 OR ≥ 2.0 or ≤ −2.0 for early progression (patients with PFS≤3 months) relative to durable clinical benefit (patients with PFS≥6 months).
For the PFS analysis, patients who were alive and had no evidence of progression at the time of dataset lock or who were lost to follow-up were censored at the time of the last radiologic tumor assessment. For the OS analysis, patients who were alive or lost to follow-up at the time of dataset lock were censored at the time of the last documented patient contact. Kaplan-Meier method was used to estimate PFS and OS, and differences were assessed by log-rank test. Hazard ratios and corresponding confidence intervals were estimated with the use of stratified Cox proportional-hazards model adjusting for clinical variables (age, history of brain metastasis, prior lines of therapy for metastatic disease [0 vs ≥1], performance status [0–1 vs 2]). Univariate analysis was performed for the exploratory analysis of less prevalent candidate genes identified through the unbiased enrichment analysis and for gene groups established by biological significance. Best response was determined through investigator-assessed RECIST v 1.1 without central review. Patients who died ≥ 14 days after start of KRAS G12Ci, but prior to first restaging scan, were considered to have progressive disease. Differences in categorical variables were assessed by two-sided Fisher’s exact test. Significance was established at p ≤ 0.05. Statistical analysis was performed on IBM SPSS Statistics (RRID:SCR_002865), R (RRID:SCR_001905), Microsoft Excel (RRID:SCR_016137), and SAS 9.4 (RRID:SCR_008567).
Supplementary Material
Statement of significance.
In this work, we identify co-occurring genomic alterations in KEAP1, SMARCA4 and CDKN2A as independent determinants of poor clinical outcomes with KRAS G12C inhibitor monotherapy in advanced NSCLC and we propose a framework for patient stratification and treatment personalization based on the co-mutational status of individual tumors.
Acknowledgements
Dr Negrao acknowledges research funding from the Rexanna’s Foundation for Fighting Lung Cancer. Dr Hines acknowledges research funding from NIH Clinical Therapeutics Training Grant (T32-GM07019). Dr Awad was supported in part by the Elva J. and Clayton L. McLaughlin Fund for Lung Cancer Research, Team Stuie and LUNGSTRONG. Work in Dr Skoulidis’ laboratory was supported in part by NIH/NCI 1R01 CA262469-01 and the Tammi Hissom Grant from Rexanna’s Foundation for Fighting Lung Cancer. This work was supported by the generous philanthropic contributions to The University of Texas MD Anderson Lung Moon Shot Program and the MD Anderson Cancer Center Support Grant P30 CA016672. We acknowledge the MD Anderson GEMINI Team for their contributions to this manuscript.
Footnotes
Authors’ Disclosures
M.V.N reports consulting/advisory role from Mirati, Genentech, Merck, and Novartis; research funding to institution from Mirati, AstraZeneca, Pfizer, Novartis, Genentech, Alaunos, and Checkmate.
A.J.C. reports consulting honoraria from MJH Life Sciences.
J.K.H reports a consultant role for Jackson Laboratory Genomic Tumor Board.
G.M reports advisory role and lecture fees from AstraZeneca, BMS, Roche, Gilead.
M.A. reports consulting/advisory role from Viatris; travel grants from Sandoz.
P.B. reports consulting/advisory role from AstraZeneca, BeiGene, Bristol Myers Squibb, Roche, Takeda; research funding from Pfizer, Roche; Virtual Meetings Subscription from Daiichi Sankyo, Amgen.
S.C.S reports consulting/advisory role for Amgen, Genentech, Foundation Medicine.
M.S. reports consulting/advisory role from Amgen, AstraZeneca, Boehringer Ingelheim, Janssen, Novartis, Pfizer, Roche, Sanofi-Aventis, Siemens Healthineers, Takeda and research support from Amgen, Dracen Pharmaceuticals, Janssen, Novartis, Pfizer and Takeda.
P.C. reports advisory board honoraria from BMS, AstraZeneca, ThermoFisher, Novartis, speaker’s honoraria from BMS, Illumina, AstraZeneca, Novartis, Thermo-Fisher, MSD, Roche, and research funding from Chugai and BMS.
S.B.G. reports consulting/advisory board member for AstraZeneca, Boehringer Ingelheim, Bristol-Myers Squibb, Genentech, Amgen, Blueprint Medicine, Sanofi Genzyme, Daiichi-Sankyo, Regeneron, Takeda, and Janssen; Research funding from AstraZeneca, Boehringer Ingelheim, and Mirati.
B.R. reports consulting/advisory board role for Regeneron.
L.L reports consulting/advisory role from Pfizer, AstraZeneca, Roche, Takeda, BMS, MSD, Bayer, Lilly, Amgen, and Sanofi.
M.N. reports consulting/advisory role from Daiichi Sankyo, AstraZeneca; research funding from Merck, Canon.
G.R.B. reports honoraria for scientific advisory boards from Bristol Myers Squibb, Bayer, Celgene, Clovis Oncology, AbbVie, ARIAD, Genentech, Novartis, Xcovery, Adicet, Amgen, AstraZeneca, Roche, MedImmune, Maverick Therapeutics, Johnson & Johnson, Virogin Biotech, Gilead Sciences, Daiichi Sankyo Inc, Novartis, Tyme, Janssen Oncology, Lilly, Instil; research funding from Merck, Celgene, Genentech, Xcovery, Novartis, Bristol Myers Squibb, GlaxoSmithKline, Adaptimmune, Macrogenics, Kite Pharma, Immatics, Torque, Incyte, MedImmune, Exelixis, Immunocore, Roche, AstraZeneca, Bayer, Tmunity Therapeutics, Inc, Regeneron, BeiGene, Repertoire Immune Medicines, Daiichi Sankyo Inc, Verastem.
J.Z. reports consulting/advisory role from Johnson and Johnson, Novartis, Bristol Myers Squibb, AstraZeneca, GenePlus, Roche, Innovent and Hengrui; research grants from Merck.
D.H.O. reports institutional research funding from Merck, BMS, Genentech, Pfizer, Palobiofarma, Onc.Ai.
C.M.B reports consulting/advisory role/honoraria from Blueprint Medicines, Amgen, Oncocyte; research grants from AstraZeneca, Genentech, Takeda, Spectrum, Mirati, Erasca, Novartis.
G.M reports consulting/advisory role/honoraria from Roche Hellas, Novartis, BMS, MSD, AstraZeneca, Takeda Hellas, Janssen, GSK, Amgen Hellas, Sanofi, Boehringer.
C.A.S. reports consulting/advisory role from Arcus Biosciences, AstraZeneca, Genentech, Mirati Therapeutics, Janssen, Takeda.
C.M.B reports consulting/advisory role from AstraZeneca, BMS, CVS, Genentech, Jazz, JNJ, Novartis, Pfizer, Regeneron, Sanofi, Seattle Genetics, Takeda; Speakers Bureau from Merck; research funding from AstraZeneca, BMS.
M.C.G. reports consulting/advisory role/honoraria from: AstraZeneca, MSD International GmbH, BMS, Boehringer Ingelheim Italia S.p.A, Celgene, Eli Lilly, Ignyta, Incyte, Inivata, MedImmune, Novartis, Pfizer, Roche, Takeda, Seattle Genetics, Mirati, Daiichi Sankyo, Regeneron, Merck, Ose Immuno Therapeutics, Blueprint, Jansenn, Sanofi; institutional research funding from Eli Lilly, MSD, Pfizer (MISP); AstraZeneca, MSD International GmbH, BMS, Boehringer Ingelheim Italia S.p.A, Celgene, Eli Lilly, Ignyta, Incyte, MedImmune, Novartis, Pfizer, Roche, Takeda, Tiziana, Foundation Medicine, Glaxo Smith Kline GSK, Spectrum pharmaceuticals.
K.A.M. reports consulting/advisory role from: Amgen, AstraZeneca, BMS, Janssen, Regeneron; Research Funding (to institution) from: AstraZeneca, BMS, Mirati.
J.E.G. reports consulting/advisory role from Triptych Health Partners, Takeda Pharmaceuticals, Sanofi Pharmaceuticals, OncoCyte Biotechnology Company, Norvartis, Merck & Co., Loxo Oncology Inc, Jazz Pharmaceuticals, Janssen Scientific Affairs, LLC, Inivata, Genentech, EMD Serono – Merck KGaA, Daiichi Sankyo, Inc (DSI), Celgene Copr, Bristol-Myers Squibb, Blueprint Medicines, Axiom HC Strategies, AstraZeneca, AbbVie; research funding from Pfizer, Novartis, Merck & Co, Ludwig Institute of Cancer Research, G1 Therapeutics, Genentech, Bristol-Myers Squibb, Boehringer Ingelheim and AstraZeneca.
S.P.P reports consulting/advisory role from: Amgen, AstraZeneca, Bristol-Myers Squibb, Certis, Eli Lilly, Jazz, Genentech, Illumina, Merck, Pfizer, Rakuten, Tempus; receives institutional research funding from: Amgen, AstraZeneca/MedImmune, Bristol-Myers Squibb, Eli Lilly, Fate Therapeutics, Gilead, Iovance, Merck, Pfizer, Roche/Genentech, SQZ Biotechnologies.
A.L.C. reports consulting role from Tempus; research funding from Amgen, AstraZeneca, Genentech.
H.A.W. reports consulting/advisory boards from AstraZeneca, Blueprint, Mirati, Merck and Genentech/Roche; research funding from ACEA Biosciences, Arrys Therapeutics, AstraZeneca/Medimmune, BMS, Clovis Oncology, Genentech/Roche, Merck, Novartis, SeaGen, Xcovery, Helsinn.
J.W. reports advisory boards and lecture fees from: Amgen, AstraZeneca, Bayer, Blueprint, BMS, Boehringer-Ingelheim, Chugai, Daiichi Sankyo, Janssen, Lilly, Loxo, Merck, MSD, Novartis, Nuvalent, Pfizer, Roche, Seattle Genetics, Takeda, Turning Point; and Research support (to institution) from BMS, Janssen Pharmaceutica, Novartis, Pfizer.
G.V.S reports consulting/advisory role from Eli Lilly, Beigene, Astrazeneca, Verastem, honoraria from AstraZeneca, Eli Lilly, MSD, Pfizer, Roche, Johnson & Johnson, Takeda and institutional research funding from Eli Lilly and MSD and Tesaro.
F.C. reports consulting/advisory role from Roche, AstraZeneca, BMS, Pfizer, Takeda, Lilly, Bayer, Amgen, Sanofi, Pharmamar, Novocure, Mirati,Galecto, OSE and MSD.
F.B. reports consulting/advisory role from Abbvie, ACEA, Amgen, Astra-Zeneca, Bayer, Bristol-Myers Squibb, Boehringer–Ingelheim, Eisai, Eli Lilly Oncology, F. Hoffmann–La Roche Ltd, Genentech, Ipsen, Ignyta, Innate Pharma, Loxo, Novartis, Medimmune, Merck, MSD, Pierre Fabre, Pfizer, Sanofi-Aventis and Takeda.
P.D.P reports consulting fees from Astrazeneca and Jazz pharmaceuticals.
L.D. is employee and stock holder of Guardant Health, Inc.
F.M.B. reports consulting/advisory role from Xencor, Debiopharm Group, Roche, PACT Pharma, eFFECTOR Therapeutics, Kolon Life Sciences, Tyra Biosciences, Zymeworks, Zentalis, Infinity Pharmaceuticals, AbbVie, Black Diamond Therapeutics, Eisai, OnCusp Therapeutics, Lengo Therapeutics, Tallac Therapeutics, Karyopharm Therapeutics, Biovica, AstraZeneca, Seattle Genetics, Loxo, Silverback Therapeutics; research funding from Novartis (Inst), AstraZeneca (Inst), Taiho Pharmaceutical (Inst), Genentech (Inst), Calithera Biosciences (Inst), Debiopharm Group (Inst), Bayer (Inst), Aileron Therapeutics (Inst), PUMA Biotechnology (Inst), CytomX Therapeutics (Inst), Jounce Therapeutics (Inst), Zymeworks (Inst), Curis (Inst), Pfizer (Inst), eFFECTOR Therapeutics (Inst), AbbVie (Inst), Boehringer Ingelheim (Inst), Guardant Health (Inst), Daiichi Sankyo (Inst), GlaxoSmithKline (Inst), Seattle Genetics (Inst), Klus Pharma (Inst), Takeda (Inst).
D.L.G. reports honoraria for scientific advisory boards from AstraZeneca, Sanofi, Alethia Biotherapeutics, Menarini, Eli Lilly, 4D Pharma and Onconova, and research support from Janssen, Takeda, Astellas, Ribon Therapeutics, NGM Biopharmaceuticals, Boehringer Ingelheim, Mirati Therapeutics and AstraZeneca.
J.V.H. reports honoraria for scientific advisory boards from AstraZeneca, Bristol Myers Squibb, Spectrum Pharmaceuticals, Guardant Health, Hengrui Pharmaceutical, GlaxoSmithKline, EMD Serono, Lilly, Takeda, Sanofi/Aventis, Genentech/Roche, Boehringer Ingelheim, Catalyst Biotech, Foundation medicine, Novartis, Mirati Therapeutics, BrightPath Biotheraputics, Janssen, Nexus Health Systems, Pneuma Respiratory, Kairos Ventures, Roche, Leads Biolabs; research support from AstraZeneca, Spectrum Pharmaceuticals, GlaxoSmithKline.
D.S.H reports consulting/advisory role from Adaptimmune, Alpha Insights, Acuta, Alkermes, Amgen, Aumbiosciences, Axiom, Baxter, Bayer, Boxer Capital, BridgeBio, COR2ed, COG, Cowen, ECOR1, Erasca, F. Hoffmann-La Roche, Gennao Bio, Genentech, Gilead, GLG, Group H, Guidepoint, HCW Precision, Immunogenesis, Janssen, Liberium, MedaCorp, Medscape, Numab, Oncologia Brasil, Orbi Captial, Pfizer, Pharma Intelligence, POET Congress, Prime Oncology, RAIN, SeaGen, ST Cube, Takeda, Tavistock, Trieza Therapeutics, Turning Point, Vertical, WebMD, YingLing Pharma, Ziopharm; and research funding from AbbVie, Adaptimmune, Aldai-Nortye, Amgen, Astra-Zeneca, Bayer, Bristol-Myers Squibb, Daiichi-Sankyo, Deciphera, Endeavor, Erasca, F. Hoffmann-La Roche, Fate Therapeutics, Genentech, Genmab, Immunogenesis, Infinity, Merck, Mirati, Navier, NCI-CTEP, Novartis, Numab, Pfizer, Pyramid Bio, Revolution Medicine, SeaGen, ST-Cube, Takeda, TCR2, Turning Point Therapeutics, VM Oncology; and has ownership interests in Molecular Match (Advisor), OncoResponse (Founder, Advisor) and Telperian (Founder, Advisor).
R.S.H reports consulting honoraria from Abbvie, Daichii Sankyo, EMD Serono, Lilly, Novartis, Regeneron, Sanofi; Research funding to institution from Abbvie, Agios, Corvus, Daichii Sankyo, Erasca, Lilly, Mirati, Novartis, Turning Point.
M.M.A. reports consulting/advisory role from: Bristol-Myers Squibb, Merck, Genentech, AstraZeneca, Mirati, Novartis, Blueprint Medicine, Abbvie, Gritstone, NextCure, EMD Serono; Research Funding (to institute) from: Bristol-Myers Squibb, Eli Lilly, Genentech, AstraZeneca, Amgen; Funding: V Foundation, Elva J. and Clayton L. McLaughlin Fund for Lung Cancer Research.
F.S reports consultant/advisory role/honoraria from AstraZeneca, Amgen, Novartis, Beigene, Guardant Health, BergenBio, Navire Pharma, Tango Therapeutics, Calithera Biosciences, Intellisphere LLC, Medscape LLC, PER LLC, Curio LLC, MI&T, IDEOlogy Health, MJH Life Sciences, VSPO McGill Universite de Montreal, RV Mais Promocao eventos LTDS; travel and lodging support from Dava Oncology, Tango Therapeutics, MJH Life Science, IDEOlogy Health, MI&T, PER LLC, Curio LLC; institutional research support from Amgen, Mirati therapeutics, Revolution Medicines, Pfizer, Novartis, Merck & Co; ownership of stock : BioNTech SE, Moderna Inc.
Data availability statement
The individual patient data generated in this study are governed by all participating institutions. To preserve patient confidentiality, to protect patient related information, and to remain compliant with each participating institution’s regulatory requirements, aggregate and/or summary de-identified data may be made available upon reasonable academic request to the corresponding author.
References
- 1.Karachaliou N, Mayo C, Costa C, Magrí I, Gimenez-Capitan A, Molina-Vila MA, et al. KRAS Mutations in Lung Cancer. Clin Lung Cancer [Internet]. Elsevier; 2013;14:205–14. Available from: 10.1016/j.cllc.2012.09.007 [DOI] [PubMed] [Google Scholar]
- 2.Simanshu DK, Nissley D v., McCormick F. RAS Proteins and Their Regulators in Human Disease. Cell [Internet]. Elsevier; 2017;170:17–33. Available from: 10.1016/j.cell.2017.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gimple RC, Wang X. RAS: Striking at the Core of the Oncogenic Circuitry [Internet]. Front Oncol. 2019;9:1–16. Available from: https://www.frontiersin.org/article/10.3389/fonc.2019.00965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–23. [DOI] [PubMed] [Google Scholar]
- 5.Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRAS G12C Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov [Internet]. 2020;10:54 LP–71. Available from: http://cancerdiscovery.aacrjournals.org/content/10/1/54.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. New England Journal of Medicine [Internet]. Massachusetts Medical Society; 2021;384:2371–81. Available from: 10.1056/NEJMoa2103695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jänne PA, Riely GJ, Gadgeel SM, Heist RS, Ou S-HI, Pacheco JM, et al. Adagrasib in Non–Small-Cell Lung Cancer Harboring a KRAS G12C Mutation. New England Journal of Medicine. 2022;1–12. [DOI] [PubMed] [Google Scholar]
- 8.de Langen AJ, Johnson ML, Mazieres J, Dingemans A-MC, Mountzios G, Pless M, et al. Sotorasib versus docetaxel for previously treated non-small-cell lung cancer with KRASG12C mutation: a randomised, open-label, phase 3 trial. The Lancet [Internet]. Elsevier; 2023;401:733–46. Available from: 10.1016/S0140-6736(23)00221-0 [DOI] [PubMed] [Google Scholar]
- 9.Nichols RJ, Yang YC, Cregg J, Schulze CJ, Wang Z, Dua R, et al. Abstract 3595: RMC-6291, a next-generation tri-complex KRASG12C(ON) inhibitor, outperforms KRASG12C(OFF) inhibitors in preclinical models of KRASG12C cancers. Cancer Res [Internet]. 2022;82:3595. Available from: 10.1158/1538-7445.AM2022-3595 [DOI] [Google Scholar]
- 10.Waizenegger IC, Lu H, Thamer C, Savarese F, Gerlach D, Rudolph D, et al. Abstract 2667: Trial in progress: Phase 1 study of BI 1823911, an irreversible KRASG12C inhibitor targeting KRAS in its GDP-loaded state, as monotherapy and in combination with the pan-KRAS SOS1 inhibitor BI 1701963 in solid tumors expressing KRASG12C mut. Cancer Res [Internet]. 2022;82:2667. Available from: 10.1158/1538-7445.AM2022-2667 [DOI] [Google Scholar]
- 11.Weiss A, Lorthiois E, Barys L, Beyer KS, Bomio-Confaglia C, Burks H, et al. Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C. Cancer Discov [Internet]. 2022;12:1500–17. Available from: 10.1158/2159-8290.CD-22-0158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sacher A, Patel MR, Miller WH Jr., Desai J, Garralda E, Bowyer S, et al. OA03.04 Phase I A Study to Evaluate GDC-6036 Monotherapy in Patients with Non-small Cell Lung Cancer (NSCLC) with KRAS G12C Mutation. Journal of Thoracic Oncology [Internet]. Elsevier; 2022;17:S8–9. Available from: 10.1016/j.jtho.2022.07.023 [DOI] [Google Scholar]
- 13.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med. 2018;378:113–25. [DOI] [PubMed] [Google Scholar]
- 14.Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim DW, et al. Alectinib versus crizotinib in untreated ALK-positive non–small-cell lung cancer. New England Journal of Medicine. 2017;377:829–38. [DOI] [PubMed] [Google Scholar]
- 15.Dy GK, Govindan R, Velcheti V, Falchook GS, Italiano A, Wolf J, et al. Abstract CT008: Long-term outcomes with sotorasib in pretreated KRASp.G12C-mutated NSCLC: 2-year analysis of CodeBreaK100. Cancer Res [Internet]. 2022;82:CT008–CT008. Available from: 10.1158/1538-7445.AM2022-CT008 [DOI] [Google Scholar]
- 16.Xue JY, Zhao Y, Aronowitz J, Mai TT, Vides A, Qeriqi B, et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature [Internet]. 2020;577:421–5. Available from: 10.1038/s41586-019-1884-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, Murciano-Goroff YR, Xue JY, Ang A, Lucas J, Mai TT, et al. Diverse alterations associated with resistance to KRAS(G12C) inhibition. Nature. Springer US; 2021;599:679–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Awad MM, Liu S, Rybkin II, Arbour KC, Dilly J, Zhu VW, et al. Acquired Resistance to KRASG12C Inhibition in Cancer. New England Journal of Medicine [Internet]. Massachusetts Medical Society; 2021;384:2382–93. Available from: 10.1056/NEJMoa2105281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solanki HS, Welsh EA, Fang B, Izumi V, Darville L, Stone B, et al. Cell type-specific adaptive signaling responses to KRASG12Cinhibition. Clinical Cancer Research. 2021;27:2533–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka N, Lin JJ, Li C, Ryan MB, Zhang J, Kiedrowski LA, et al. Clinical Acquired Resistance to KRAS G12C Inhibition through a Novel KRAS Switch-II Pocket Mutation and Polyclonal Alterations Converging on RAS–MAPK Reactivation. Cancer Discov [Internet]. 2021;11:1913 LP–1922. Available from: http://cancerdiscovery.aacrjournals.org/content/11/8/1913.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:861–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricciuti B, Arbour KC, Lin JJ, Vajdi A, Vokes N, Hong L, et al. Diminished Efficacy of Programmed Death-(Ligand)1 Inhibition in STK11 and KEAP1-Mutant Lung Adenocarcinoma Is Affected by KRAS Mutation Status. Journal of Thoracic Oncology [Internet]. Elsevier; 2022;17:399–410. Available from: 10.1016/j.jtho.2021.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8:822–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arbour KC, Jordan E, Kim HR, Dienstag J, Yu HA, Sanchez-Vega F, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clinical Cancer Research. 2018;24:334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skoulidis F, Arbour KC, Hellmann MD, Patil PD, Marmarelis ME, Awad MM, et al. Association of STK11/LKB1 genomic alterations with lack of benefit from the addition of pembrolizumab to platinum doublet chemotherapy in non-squamous non-small cell lung cancer. Journal of Clinical Oncology [Internet]. American Society of Clinical Oncology; 2019;37:102. Available from: 10.1200/JCO.2019.37.15_suppl.102 [DOI] [Google Scholar]
- 26.West HJ, McCleland M, Cappuzzo F, Reck M, Mok TSK, Jotte RM, et al. Clinical efficacy of atezolizumab plus bevacizumab and chemotherapy in KRAS-mutated non-small cell lung cancer with STK11, KEAP1, or TP53 comutations: subgroup results from the phase III IMpower150 trial. J Immunother Cancer [Internet]. 2022;10:e003027. Available from: http://jitc.bmj.com/content/10/2/e003027.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartek J, Falck J, Lukas J. Chk2 kinase — a busy messenger. Nat Rev Mol Cell Biol [Internet]. 2001;2:877–86. Available from: 10.1038/35103059 [DOI] [PubMed] [Google Scholar]
- 28.Clynes D, Higgs DR, Gibbons RJ. The chromatin remodeller ATRX: a repeat offender in human disease. Trends Biochem Sci [Internet]. Elsevier; 2013;38:461–6. Available from: 10.1016/j.tibs.2013.06.011 [DOI] [PubMed] [Google Scholar]
- 29.Truch J, Downes DJ, Scott C, Gür ER, Telenius JM, Repapi E, et al. The chromatin remodeller ATRX facilitates diverse nuclear processes, in a stochastic manner, in both heterochromatin and euchromatin. Nat Commun. Springer US; 2022;13:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu WL, Papagiannakopoulos T. The Pleiotropic Role of the KEAP1/NRF2 Pathway in Cancer. Annu Rev Cancer Biol. 2020;4:413–35. [Google Scholar]
- 31.Binkley MS, Jeon YJ, Nesselbush M, Moding EJ, Nabet BY, Almanza D, et al. KEAP1/NFE2L2 mutations predict lung cancer radiation resistance that can be targeted by glutaminase inhibition. Cancer Discov. 2020;10:1826–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sitthideatphaiboon P, Galan-Cobo A, Negrao M v, Qu X, Poteete A, Zhang F, et al. STK11/LKB1 Mutations in NSCLC Are Associated with KEAP1/NRF2-Dependent Radiotherapy Resistance Targetable by Glutaminase Inhibition. Clinical Cancer Research [Internet]. 2021;27:1720–33. Available from: 10.1158/1078-0432.CCR-20-2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh A, Daemen A, Nickles D, Jeon SM, Foreman O, Sudini K, et al. NRF2 Activation promotes aggressive lung cancer and associates with poor clinical outcomes. Clinical Cancer Research. 2021;27:877–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science (1979). 2018;362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krall EB, Wang B, Munoz DM, Ilic N, Raghavan S, Niederst MJ, et al. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. McMahon M, editor. Elife [Internet]. eLife Sciences Publications, Ltd; 2017;6:e18970. Available from: 10.7554/eLife.18970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marzio A, Kurz E, Sahni JM, di Feo G, Puccini J, Jiang S, et al. EMSY inhibits homologous recombination repair and the interferon response, promoting lung cancer immune evasion. Cell [Internet]. Elsevier; 2022;185:169–183.e19. Available from: 10.1016/j.cell.2021.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Concepcion CP, Ma S, LaFave LM, Bhutkar A, Liu M, DeAngelo LP, et al. Smarca4 Inactivation Promotes Lineage-Specific Transformation and Early Metastatic Features in the Lung. Cancer Discov [Internet]. 2022;12:562–85. Available from: 10.1158/2159-8290.CD-21-0248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alessi J v, Ricciuti B, Spurr LF, Gupta H, Li YY, Glass C, et al. SMARCA4 and Other SWItch/Sucrose NonFermentable Family Genomic Alterations in NSCLC: Clinicopathologic Characteristics and Outcomes to Immune Checkpoint Inhibition. Journal of Thoracic Oncology [Internet]. Elsevier; 2021;16:1176–87. Available from: 10.1016/j.jtho.2021.03.024 [DOI] [PubMed] [Google Scholar]
- 39.Fernando TM, Piskol R, Bainer R, Sokol ES, Trabucco SE, Zhang Q, et al. Functional characterization of SMARCA4 variants identified by targeted exome-sequencing of 131,668 cancer patients. Nat Commun [Internet]. Springer US; 2020;11:1–13. Available from: 10.1038/s41467-020-19402-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoenfeld AJ, Bandlamudi C, Lavery JA, Montecalvo J, Namakydoust A, Rizvi H, et al. The Genomic Landscape of SMARCA4 Alterations and Associations with Outcomes in Patients with Lung Cancer. Clinical Cancer Research [Internet]. 2020;26:5701–8. Available from: 10.1158/1078-0432.CCR-20-1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hodges C, Kirkland JG, Crabtree GR. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb Perspect Med. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clynes D, Jelinska C, Xella B, Ayyub H, Scott C, Mitson M, et al. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat Commun [Internet]. 2015;6:7538. Available from: 10.1038/ncomms8538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juhász S, Elbakry A, Mathes A, Löbrich M. ATRX Promotes DNA Repair Synthesis and Sister Chromatid Exchange during Homologous Recombination. Mol Cell [Internet]. Elsevier; 2018;71:11–24.e7. Available from: 10.1016/j.molcel.2018.05.014 [DOI] [PubMed] [Google Scholar]
- 44.Li C, Vides A, Kim D, Xue JY, Zhao Y, Lito P. The G protein signaling regulator RGS3 enhances the GTPase activity of KRAS. Science (1979) [Internet]. American Association for the Advancement of Science; 2021;374:197–201. Available from: 10.1126/science.abf1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ricciuti B, Recondo G, Spurr LF, Li YY, Lamberti G, Venkatraman D, et al. Impact of DNA Damage Response and Repair (DDR) Gene Mutations on Efficacy of PD-(L)1 Immune Checkpoint Inhibition in Non–Small Cell Lung Cancer. Clinical Cancer Research. 2020;26:4135–42. [DOI] [PubMed] [Google Scholar]
- 46.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The individual patient data generated in this study are governed by all participating institutions. To preserve patient confidentiality, to protect patient related information, and to remain compliant with each participating institution’s regulatory requirements, aggregate and/or summary de-identified data may be made available upon reasonable academic request to the corresponding author.