Abstract
Cannabidiol (CBD) is a naturally occurring nonpsychotoxic phytocannabinoid that has gained increasing attention as a popular consumer product and for its use in Food and Drug Administration–approved Epidiolex (CBD oral solution) for the treatment of Lennox-Gastaut syndrome and Dravet syndrome. CBD was previously reported to be metabolized primarily by CYP2C19 and CYP3A4, with minor contributions from UDP-glucuronosyltransferases. 7-Hydroxy-CBD (7-OH-CBD) is the primary active metabolite with equipotent activity compared with CBD. Given the polymorphic nature of CYP2C19, we hypothesized that variable CYP2C19 expression may lead to interindividual differences in CBD metabolism to 7-OH-CBD. The objectives of this study were to further characterize the roles of cytochrome P450 enzymes in CBD metabolism, specifically to the active metabolite 7-OH-CBD, and to investigate the impact of CYP2C19 polymorphism on CBD metabolism in genotyped human liver microsomes. The results from reaction phenotyping experiments with recombinant cytochrome P450 enzymes and cytochrome P450–selective chemical inhibitors indicated that both CYP2C19 and CYP2C9 are capable of CBD metabolism to 7-OH-CBD. CYP3A played a major role in CBD metabolic clearance via oxidation at sites other than the 7-position. In genotyped human liver microsomes, 7-OH-CBD formation was positively correlated with CYP2C19 activity but was not associated with CYP2C19 genotype. In a subset of single-donor human liver microsomes with moderate to low CYP2C19 activity, CYP2C9 inhibition significantly reduced 7-OH-CBD formation, suggesting that CYP2C9 may play a greater role in CBD 7-hydroxylation than previously thought. Collectively, these data indicate that both CYP2C19 and CYP2C9 are important contributors in CBD metabolism to the active metabolite 7-OH-CBD.
SIGNIFICANCE STATEMENT
This study demonstrates that both CYP2C19 and CYP2C9 are involved in CBD metabolism to the active metabolite 7-OH-CBD and that CYP3A4 is a major contributor to CBD metabolism through pathways other than 7-hydroxylation. 7-OH-CBD formation was associated with human liver microsomal CYP2C19 activity, but not CYP2C19 genotype, and CYP2C9 was found to contribute significantly to 7-OH-CBD generation. These findings have implications for patients taking CBD who may be at risk for clinically important cytochrome P450–mediated drug interactions.
Introduction
Cannabidiol (CBD) is one of several phytocannabinoids derived from the Cannabis sativa plant. CBD oral solution (Epidiolex) was approved by the Food and Drug Administration (FDA) in 2018 for the treatment of seizures associated with Lennox-Gastaut syndrome or Dravet syndrome in patients aged 2 years and older (www.epidiolex.com/sites/default/files/pdfs/0820/EPX-03645-0820_EPIDIOLEX_(cannabidiol)_USPI.pdf). CBD is also a popular consumer product marketed to the public in various dosage forms for unapproved indications such as anxiety, pain, seizures, appetite stimulation, and insomnia. Since 2015, the FDA has issued several warning letters to businesses for marketing CBD in violation of the Food, Drug, and Cosmetic Act (www.fda.gov/news-events/public-health-focus/warning-letters-and-test-results-cannabidiol-related-products).
The rise in popularity of CBD-containing consumer products has led to increased reports of CBD-related safety events. In addition, recent reports from Greenwich Pharmaceuticals have found that CBD causes dose-dependent hepatotoxicity (https://www.fda.gov/media/112565/download,https://www.fda.gov/media/112947/download); the risk of liver injury is increased when CBD is given concomitantly with the hepatotoxic antiepileptic drug valproate (Gaston et al., 2017; www.epidiolex.com/sites/default/files /pdfs/0820/EPX-03645-0820_EPIDIOLEX_(cannabidiol)_USPI.pdf). The toxicity observed with the FDA-approved formulation Epidiolex may be related to the high daily doses (up to 20 mg/kg per day) in patients prescribed CBD (www.epidiolex.com/sites/default/files/pdfs/0820/EPX-03645-0820_EPIDIOLEX_(cannabidiol)_USPI.pdf). The steady-state peak plasma concentration (Cmax) of CBD at therapeutic doses is approximately 2 μM (732 ng/ml, fasted), and the plasma concentrations are higher when taken with food (https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/210365Orig1s000OtherR.pdf). Doses ranging from <1 mg/kg per day to 50 mg/kg per day of CBD have been tested for several indications (Millar et al., 2019). Variations in CBD content among consumer products may pose further complications for risk assessment, as multiple reports have demonstrated discrepancies between reported and actual CBD content (www.fda.gov/news-events/public-health-focus/warning-letters-and-test-results-cannabidiol-related-products).
CBD-related safety events have also been tied to pharmacokinetic drug-drug interactions. CBD is extensively metabolized by cytochrome P450 enzymes, and adverse events have been reported in patients taking CBD concomitantly with cytochrome P450 substrates such as warfarin, clobazam, and methadone (Damkier et al., 2019; Morrison et al., 2019; Madden et al., 2020). Inhibition of cytochrome P450–mediated metabolism by CBD has been implicated as a causative mechanism for these drug interactions (Qian et al., 2019). Although multiple cases of CBD-related adverse events have been reported, the underlying mechanism(s) for these events remains unknown. It is possible that pharmacogenetic differences in cytochrome P450 expression and activity may influence the risk of CBD-related toxicities in diverse patient populations taking varying doses of CBD in combination with prescribed cytochrome P450 substrates.
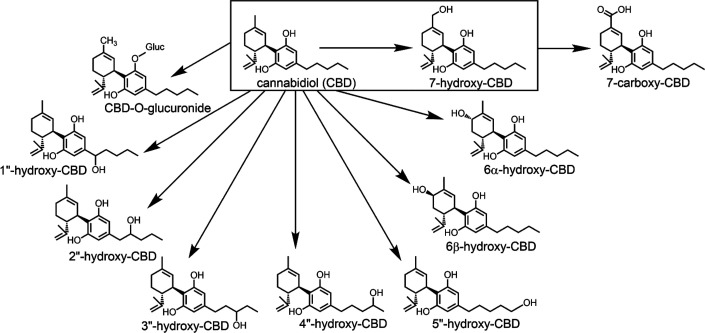
CBD undergoes hepatic oxidative and conjugative metabolism by cytochrome P450 and UDP-glucuronosyltransferase (UGT) enzymes, respectively (Fig. 1) (Mazur et al., 2009; Jiang et al., 2011). However, the individual enzyme contributions to CBD metabolism have not been fully characterized. Previous research by Jiang et al. (2011) indicated that CYP2C19 and CYP3A4 are the primary enzymes involved in oxidation of CBD to multiple mono-oxygenated metabolites. Specifically, CYP2C19 was reported to primarily form 7-hydroxy-CBD (7-OH-CBD), whereas CYP3A was involved in forming 6α-OH-CBD, 6β-OH-CBD, and side chain hydroxylation products such as 2″-OH- and 4″-OH-CBD (Jiang et al., 2011). 7-OH-CBD is the major pharmacologically active metabolite, which is further converted to the inactive metabolite 7-carboxy-CBD (7-COOH-CBD) (Whalley et al., 2017). Prior in vitro studies have identified direct CBD-glucuronides formed at the C1 and C5 hydroxyl groups by UGT1A7, UGT1A9, and UGT2B7 (Mazur et al., 2009). CBD and its metabolites predominantly undergo hepatobiliary elimination, with approximately 82% of the total dose recovered in feces after a single dose (https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/210365Orig1s000OtherR.pdf).
Fig. 1.
Metabolism of CBD by hepatic cytochrome P450 and UGT enzymes. Conversion to the active metabolite 7-OH-CBD has been outlined.
Although the role of CYP2C19 in 7-OH-CBD formation has been reported (Jiang et al., 2011), less is known about the effect of CYP2C19 polymorphism on generation of this active metabolite. Moreover, whether other cytochrome P450 enzymes contribute to 7-OH-CBD generation is not known. We hypothesized that variable CYP2C19 expression may lead to interindividual differences in CBD metabolism to 7-OH-CBD.
The objectives of this study were to 1) further characterize the roles of cytochrome P450 enzymes in CBD metabolism, specifically with respect to formation of the active metabolite 7-OH-CBD, and to 2) investigate the impact of CYP2C19 genotype on formation of 7-OH-CBD in human liver microsomes. Our findings demonstrate the involvement of CYP2C19 and CYP2C9 in the 7-hydroxylation of CBD, and the results indicate major contributions of CYP3A to other pathways of CBD oxidative metabolism.
Materials and Methods
Chemicals and Reagents.
CBD (catalog number 90081, 1 mg/ml in methanol), 6α-hydroxy-cannabidiol (catalog number 27576, 1 mg), and cannabidiol-d9 (CBD-d9, internal standard; catalog number 16203, 1 mg/ml in methanol) certified reference compounds were purchased from Cayman Chemical (Ann Arbor, MI). 7-Carboxy-CBD (catalog number B140796-1, 1 mg/ml in methanol), 7-hydroxy-CBD (catalog number C-180-1ML, 1 mg/ml in methanol), 7-carboxy-CBD-d3 (catalog number C-224, 100 µg/ml in methanol), and 7-hydroxy-CBD-d3 (catalog number C-223, 100 µg/ml in methanol) standards were purchased from Cerilliant (Round Rock, TX). Purchased standards were formulated as Drug Enforcement Administration-exempt preparations of controlled substances. Stock solutions of CBD and metabolite standards were prepared in methanol and stored at −30°C. Chemical inhibitors ketoconazole, ticlopidine, α-naphthoflavone, sulfaphenazole, quinidine, clomethiazole, and 4-methylpyrazole were purchased from Sigma Aldrich (St. Louis, MO). Furafylline, 2-phenyl-2-(1- piperidinyl)propane, CYP3cide, and (+)-N-3-benzylnirvanol were purchased from Toronto Research Chemicals (Toronto, ON). Chemical inhibitors were prepared from stocks dissolved in DMSO and diluted in acetonitrile to make solutions in 1:9 DMSO:acetonitrile (v/v).
Human liver microsomes (HLM) pooled from 150 donors [mixed-gender (lots 38294, 38295, and 38296)], NADPH regenerating system solutions A and B, and UGT reaction solutions A and B were purchased from Corning Life Sciences (Woburn, MA). NADPH regenerating system solution A (catalog number 451220) contained 26 mM NADP+, 66 mM glucose-6-phosphate, and 66 mM magnesium chloride; NADPH regenerating system solution B (catalog number 451200) contained 40 U/ml glucose-6-phosphate dehydrogenase in 5 mM sodium citrate. UGT reaction mix solution A (catalog number 451300) contained 25 mM uridine 5′-diphospho-glucuronic acid (UDPGA) in water; UGT reaction mix solution B (catalog number 451320) contained 250 mM Tris-HCl buffer, 40 mM magnesium chloride, and 0.125 mg/ml alamethicin in water. CYP2C19-genotyped HLM were purchased from BioIVT (Baltimore, MD), XenoTech (Lenexa, KS), and Corning Life Sciences (Woburn, MA). Lot-specific demographic information, genotype, CYP3A4 activity, and CYP2C19 activity for individual CYP2C19-genotyped HLM are listed in Supplemental Table 1.
CBD Depletion in the Presence and Absence of Enzyme Cofactors.
Four reaction mixtures (1000 µl each) were prepared with 1 µM CBD and 150-donor pooled HLM (0.2 mg/ml protein) (Corning, lot 38294) in 100 mM potassium phosphate buffer (pH 7.4) with a final organic solvent composition of 1% v/v. These mixtures contained both NADPH regenerating system and UGT reaction mix, NADPH regenerating system alone, UGT reaction mix alone, or no cofactors (control) (final cofactor concentrations = 1.3 mM NADP+ and/or 2 mM UDPGA). Each mixture was prepared in triplicate for each experiment. Reactions were initiated with the addition of UDPGA and/or NADPH regenerating system. Mixtures were incubated in a 37°C shaking water bath over 0, 2, 5, 10, 20, 30, 45, and 60 minutes. At each time point, 100-µl aliquots of each reaction mixture were removed and added to 100 µl of ice-cold acetonitrile containing 100 ng/ml of CBD-d9 (internal standard). Samples were then mixed with a vortex device and centrifuged at 3400g and 4°C for 20 minutes. The clear supernatant was transferred to a separate liquid chromatography - mass spectrometry (LC-MS) vial and stored at −20°C prior to LC-MS/MS analysis.
Effect of Cytochrome P450–Selective Inhibitors on CBD and 7-OH-CBD Metabolism.
CBD (1 to 2 µM) and 7-OH-CBD (2 µM) was incubated with 150-donor pooled HLM (0.2 mg/ml protein) in the presence of cytochrome P450–selective chemical inhibitors to estimate relative cytochrome P450 contributions to metabolite formation (reaction phenotyping). Reaction mixtures (200 µl total volume) were coincubated with 1 µM α-naphthoflavone (CYP1A2 inhibitor), 25 µM furafylline (CYP1A2 inhibitor), 15 µM 2-phenyl-2-(1-piperidinyl)propane (CYP2B6 inhibitor), 5 µM ticlopidine (CYP2C19 and CYP2B6 inhibitor), 5 µM sulfaphenazole (CYP2C9 inhibitor), 5 µM (+)-N-3-benzylnirvanol (CYP2C19 inhibitor), 2 µM quinidine (CYP2D6 inhibitor), 100 µM 4-methylpyrazole (CYP2E1 inhibitor), or 1 µM ketoconazole (CYP3A4 and CYP3A5 inhibitor). Reactions occurred over 10 minutes in a 37°C water bath after addition of NADPH before quenching and sample preparation using the procedure described above. Samples were analyzed by LC-MS/MS for 6α-OH-CBD and 7-OH-CBD (when CBD was used as a substrate), and 7-COOH-CBD (when 7-OH-CBD was used as a substrate). In addition to the experimental procedures described above, in which all reaction conditions were initiated with NADPH as a direct, reversible inhibition assay, experiments were also performed with a preincubation step for the time-dependent inhibitors furafylline, clomethiazole (a time-dependent CYP2E1 inhibitor) (Stresser et al., 2016), ticlopidine, and CYP3cide. Procedures for time-dependent inhibition assays are provided in the Supplemental Material.
For CBD and 7-OH-CBD depletion and metabolite formation assays, reaction mixtures (1000 µl each) were prepared with 1 µM CBD or 1 µM 7-OH-CBD and 150-donor pooled HLM (0.2 mg/ml protein) (Corning, lots 38294 and 38295) in 100 mM potassium phosphate buffer (pH 7.4). To determine the relative roles of CYP2C9, CYP2C19, and CYP3A enzymes in CBD metabolism, mixtures each contained one of the following compounds: sulfaphenazole (a CYP2C9 inhibitor, 5 µM), (+)-N-3-benzylnirvanol (a CYP2C19 inhibitor, 5 µM), ketoconazole (a nonselective CYP3A inhibitor, 1 µM), or CYP3cide (a time-dependent CYP3A4-selective inactivator, 0.5 µM) (Walsky et al., 2012), or 1:9 DMSO:acetonitrile (vehicle control, 0.5% v/v). This experiment was also performed with 7-OH-CBD as a substrate in the presence of (+)-N-3-benzylnirvanol, quinidine (a CYP2D6 inhibitor, 2 µM), ketoconazole, CYP3cide, or vehicle control. Reactions were initiated with the addition of substrate and incubated for 2, 5, 10, 15, 20, and 30 minutes in a 37°C shaking water bath. For incubations containing the time-dependent inhibitor CYP3cide, substrate was added after a 10-minute preincubation using the method described by Towles et al. (2016). At each time point, 100-µl aliquots were removed and added to 100 µl of ice-cold acetonitrile (quench solution) containing 100 ng/ml CBD-d9, 7-OH-CBD-d3, and 7-COOH-CBD-d3 (internal standards). Samples were analyzed by LC-MS/MS as described below.
CBD and 7-OH-CBD Metabolism by Recombinant Cytochrome P450 Enzymes.
CBD (1 µM) and 7-OH-CBD (2 µM) were each added as substrates to reactions containing 20 pmol/ml each of individual P450 Supersomes consisting of CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, or CYP3A5. Briefly, reaction mixes (200 µl total) containing each enzyme supplemented with NADPH regenerating system were incubated for 10 minutes at 37°C and analyzed for formation of 7-OH-CBD (for reactions containing CBD as a substrate) or 7-COOH-CBD (for reactions containing 7-OH-CBD as a substrate) using LC-MS/MS. Metabolites were measured with the following mass transitions in negative ion mode: m/z 329 > 299 for 7-OH-CBD, and m/z 343 > 299 for 7-COOH-CBD. (See details below).
Kinetic Analysis of CBD 7-Hydroxylation in Pooled HLM, Recombinant CYP2C19, and Recombinant CYP2C9.
Prior to kinetic analysis, the linearity of metabolite formation with respect to time and protein concentration was measured in HLM, recombinant CYP2C19, and recombinant CYP2C9 to determine the appropriate reaction conditions for measuring the kinetic parameters (Km and Vmax) of metabolite formation. The slope of 7-OH-CBD formation was linear in HLM when incubated for 10 minutes with 0.2 mg/ml protein. Reaction mixtures (200 µl total volume containing a maximum of 1% organic solvent) containing 0, 0.1, 0.2, 1, 2, 5, 10, 20, 50, 100, and 200 µM CBD were prepared with either 150-donor pooled HLM (Corning, lot 38295) diluted to 0.2 mg/ml protein, 10 pmol/ml recombinant CYP2C19 (Corning, lot 0006001), or 10 pmol/ml recombinant CYP2C9 (Corning, lot 9241004). Reactions were initiated with the addition of NADPH and incubated in a 37°C shaking water bath for 10 minutes prior to addition of 400 µl of ice-cold acetonitrile containing 100 ng/ml CBD-d9 (quench solution). Samples were prepared and analyzed by LC-MS/MS as described below. Formation of 7-OH-CBD was quantified using an authentic chemical standard of 7-OH-CBD.
CBD Metabolite Formation by Individual CYP2C19-Genotyped HLM.
CYP2C19-genotyped HLM from 19 individual donors were purchased from XenoTech, BioIVT, and Corning. Complete donor information, including lot numbers, genotype, enzyme activities, and demographics, are listed in Supplemental Table 1. Individual-donor HLMs consisted of seven CYP2C19*1/*1 donors (n = 7), two CYP2C19*1/*2 donors (n = 2), four CYP2C19*2/*2 donors (n = 4), three CYP2C19*1/*17 donors (n = 3), and three CYP2C19*17/*17 donors (n = 3). Donors included 17 adult males and two adult females. Donor CYP2C19 activity, measured as the rate of S-mephenytoin 4′-hydroxylation, was determined previously (Murray et al., 2020). Donor CYP3A4 activity, measured as the rate of testosterone 6β-hydroxylation, was provided by suppliers for the individual HLM (Supplemental Table 1). Reaction mixtures (200 µl total) were prepared with 2 µM CBD and individual-donor HLM (0.2 mg/ml protein) in 100 mM pH 7.4 potassium phosphate buffer. HLMs were diluted with potassium phosphate buffer to 20 mg/ml prior to addition to reaction mixes. Reactions were initiated with the addition of NADPH and incubated in a 37°C shaking water bath for 5 minutes prior to termination with 200 µl of ice-cold acetonitrile containing 100 ng/ml CBD-d9.
To further examine the role of CYP2C19 compared with other cytochromes P450 in 7-OH-CBD formation, CBD (2 µM) was incubated with a subset of individual HLMs with known CYP2C19 genotypes and activity in the presence of selective inhibitors for CYP2C9, CYP2C19, and CYP3A. Experiments were performed with HLMs from three CYP2C19*1/*1 donors (lots 710444, QLC, and IFF) and three CYP2C19*2/*2 donors (lots 810010, 689, and 863). Genotype data provided by BioIVT indicate that HLM lots QLC and IFF were both genotyped as CYP2C9*1/*1; however, the CYP2C9 genotypes for the other donors are not known. Reactions were prepared and incubated as described above for 10 minutes with vehicle control, 1 µM ketoconazole, 5 µM (+)-N-3-benzylnirvanol, or 5 µM sulfaphenazole (0.5% v/v final organic solvent concentration) prior to termination with 400 µl of ice-cold acetonitrile containing 100 ng/ml 7-OH-CBD-d3. Samples were prepared and analyzed as described previously.
LC-MS/MS Analysis.
Preliminary experiments, including initial substrate depletion and protein-time linearity experiments, were analyzed using an Agilent 1290 Infinity II ultraperformance liquid chromatography system with an AB Sciex triple quadrupole 6500 mass spectrometer. Samples were analyzed on this instrument with a 100 × 2.1 mm Thermo Hypersil GOLD C18 column with a 1.9-µm particle size. Mobile phase A was 5% methanol in water with 0.1% acetic acid, and mobile phase B was 5% methanol in acetonitrile with 0.1% acetic acid. The following LC gradient scheme (method 1) was used for compound separation at a flow rate of 0.5 ml/min: 50% A to 25% A from 0 to 1 minute, 25% A to 10% A from 1 to 5 minutes, 10% A from 5 to 5.5 minutes, 10% A to 50% A from 5.5 to 6 minutes, and 50% A from 6 to 7 minutes. For subsequent experiments, samples were analyzed using a Thermo TSQ Quantum Ultra triple quadrupole mass spectrometer coupled to a Waters Acquity ultraperformance liquid chromatography system. A 50 × 2.1 mm Phenomenex Kinetex EVO C18 column with a 2.6-µm particle size was used for analysis. A gradient elution scheme (method 2) was used with the same eluents and flow rate described above: 60% A from 0 to 0.7 minutes, 60% A to 10% A from 0.7 to 2.1 minutes, 10% A from 2.1 to 2.5 minutes, 10% A to 60% A from 2.5 to 2.6 minutes, and 60% A from 2.6 to 4 minutes. Selected reaction monitoring was employed for the analysis of CBD and CBD metabolites using the following precursor-to-product ion transitions: m/z 313 > 245 for CBD, m/z 322 > 254 for CBD-d9, m/z 332 > 302 for 7-OH-CBD-d3, m/z 346 > 302 for 7-COOH-CBD-d3, m/z 329 > 299 for 7-OH-CBD, m/z 343 > 299 for 7-COOH-CBD, and m/z 489 > 313 for CBD-glucuronide. Mass transitions were captured in the negative ion mode, [M − H]−. The m/z 329 > 311 mass transition corresponds to water loss and can identify not only 7-OH-CBD but also other monohydroxylated CBD metabolites. To separate and quantify both 7-OH-CBD and 6α-OH-CBD using this mass transition, a third gradient scheme with a longer run time (method 3) was used with the same mobile phases and flow rate: 60% A from 0 to 0.5 minutes, 60% A to 10% A from 0.5 to 4 minutes, 10% A from 4 to 4.4 minutes, 10% A to 60% A from 4.4 to 4.5 minutes, and 60% A from 4.5 to 6 minutes. A representative LC-MS/MS chromatogram for each analyte standard is shown in Supplemental Fig. 1A. Representative tandem mass spectrometry (MS/MS) product ion spectra for CBD, 7-OH-CBD, 6α-OH-CBD, and 7-COOH-CBD are shown in Supplemental Fig. 1B.
Quantitation of CBD and Metabolites.
Quantitation of CBD and metabolites was performed using freshly prepared standard curves in matrix matching the conditions for each experiment according to current recommendations for bioanalytical method development (https://www.fda.gov/files/drugs/published/Bioanalytical-Method-Validation-Guidance-for-Industry.pdf). Curves were fitted using Thermo Xcalibur 2.2 software with 1/X2 weighting for metabolites. Standard curves ranged between 1 and 1000 ng/ml with limits of quantitation between 1 and 25 ng/ml. Specific standard curve ranges and limits of quantitation for each experiment are listed in the legends for each figure.
Data Analysis.
Figure graph development and data analysis, including statistical tests, were performed using GraphPad Prism 8 software. Error bars on all figures represent the mean ± S.D. of three replicates for each sample unless otherwise indicated. Sample sizes for experiments containing individual CYP2C19-genotyped HLM were limited by the quantity of unique donors that were commercially available. The maximum number of lots available to our knowledge were used for these experiments.
The elimination rate constants for substrate depletion in the presence and absence of enzyme cofactors were calculated from the slope of the semilogarithmic plot of CBD remaining (measured as peak area ratio relative to internal standard) versus time.
Determination of the kinetic parameters (Km and Vmax) of CBD metabolite formation by recombinant cytochrome P450 enzymes was performed by graphically fitting the data to an enzyme kinetic model using GraphPad Prism 8 software. Graphical analysis of 7-OH-CBD formation was suggestive of substrate inhibition, a well known phenomenon wherein a substrate may inhibit its own metabolism at high concentrations (Lin et al., 2001). This phenomenon may occur by substrate binding at an additional site on the enzyme-substrate complex, forming an inactive ternary complex (Copeland, 2000); however, the actual mechanism has not been fully defined (Hutzler and Tracy, 2002). Since truncation of these data and fitting to the traditional Michaelis-Menten model may result in biased estimates of kinetic parameters, a substrate inhibition model with the following equation was used: Y = Vmax/(Km/X + 1 + X/Ki), where Vmax is the maximum enzyme velocity if the substrate did not inhibit enzyme activity, Km is the Michaelis constant, and Ki is the dissociation constant for additional substrate binding to the enzyme-substrate complex (Copeland, 2000).
For analysis of CBD and 7-OH-CBD metabolism in the presence of cytochrome P450–selective inhibitors, an estimated percent contribution of each cytochrome P450 enzyme was calculated for each reaction by determining the slopes of substrate depletion and metabolite formation and comparing these values to control reactions containing vehicle (1:9 DMSO:acetonitrile) without inhibitor. Slopes of substrate depletion were used to calculate intrinsic clearance as follows:
where kdeg is the negative slope of the semilogarithmic plot of the average peak area ratio versus time (Zientek et al., 2016).
Intrinsic clearance values were then converted to percentages of vehicle control by dividing the intrinsic clearance in the presence of inhibitor by the intrinsic clearance value of the vehicle control (without inhibitor). When the total estimated percent contribution was greater than 100%, the data were normalized to 100%, and the same proportional contribution by each cytochrome P450 was retained, according to the method described by Zientek et al. (2016). The relative percent contribution was calculated using the following equation (Zientek et al., 2016):
 |
Pearson’s r correlation analysis was performed using GraphPad Prism 8 software to assess the relationship between 7-OH-CBD generation and CYP2C19 enzyme activity for CYP2C19-genotyped HLM from 19 individual donors.
Results
CBD Depletion in the Presence and Absence of Enzyme Cofactors
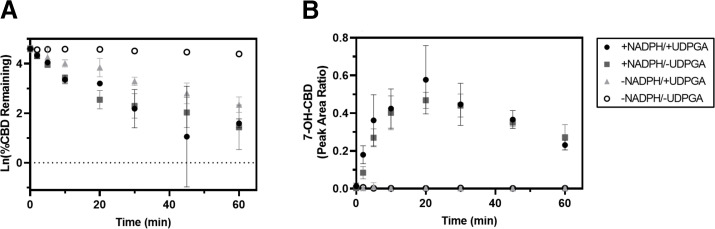
Depletion of CBD (1 μM) and formation of the active metabolite 7-OH-CBD in pooled HLM is shown in Fig. 2. Substrate depletion experiments confirmed that CBD remains relatively stable in the absence of cofactors for 60 minutes. The rate constant of CBD depletion for microsomal incubations supplemented with only NADPH was 0.119 minutes−1 versus 0.054 minutes−1 for incubations supplemented with only UDPGA (Table 1); thus, metabolism was approximately 2 times faster via oxidative pathways compared with conjugative metabolism, suggesting that oxidative metabolism by cytochromes P450 is a major pathway for CBD elimination. HLM supplemented with UDPGA alone depleted substrate to a lesser extent. Formation of the active metabolite 7-OH-CBD occurred rapidly in HLM supplemented with NADPH; 7-OH-CBD generation was detected within 2 minutes of initiating the reaction, and peak levels were observed at 20 minutes, followed by a gradual reduction in 7-OH-CBD levels, likely reflecting further metabolic clearance of 7-OH-CBD. In additional experiments with HLM supplemented with UDPGA, CBD-glucuronide conjugates were detected. Representative LC-MS/MS chromatograms of CBD metabolites formed in HLM in the presence and absence of NADPH and UDPGA are shown in Supplemental Fig. 2.
Fig. 2.
CBD substrate depletion and 7-OH-CBD formation in pooled HLM. CBD substrate depletion (A) and 7-hydroxylation (B) were measured in the presence and absence of the metabolic cofactors NADPH and UDPGA. CBD (1 µM) was incubated with 150-donor pooled HLM (0.2 mg/ml protein) for 0, 2, 5, 10, 20, 30, 45, and 60 minutes. (A) Rates of CBD depletion were calculated using natural log-transformed depletion data. Generation of 7-OH-CBD is expressed in (B) as the ratio of metabolite to internal standard, cannabidiol-d9. Data points represent the mean ± S.D. of duplicate experiments.
TABLE 1.
Cannabidiol depletion rate constants (min−1) in the presence and absence of metabolic cofactors
Rate constants for the depletion of CBD were calculated from linear range of the semilogarithmic slope of remaining CBD for each reaction condition vs. time in min.
| Cofactor(s) | CBD Depletion Rate Constant |
|---|---|
| min−1 | |
| +NADPH/+UDPGA | −0.122 |
| +NADPH/−UDPGA | −0.119 |
| −NADPH/+UDPGA | −0.054 |
| −NADPH/−UDPGA | −0.0006 |
TABLE 2.
Relative percent contribution of P450 enzymes to cannabidiol depletion
| Clint | Percentage of Vehicle Control | Est. % Contributionb | Relative % Contributionc | |
|---|---|---|---|---|
| μl/min/mg protein | % | |||
| Vehicle (control) | 460.6 | 100 | ||
| Sulfaphenazole (CYP2C9) | 362.9 | 79 | 21 | 15 |
| Benzylnirvanol (CYP2C19) | 262.6 | 57 | 43 | 31 |
| Ketoconazole (CYP3A) | 108.2 | 23 | 77 | 54 |
| CYP3cide (CYP3A4) | (14.79)a | (3)a | ||
aIntrinsic clearance was calculated according to Zientek et al. (2016): Clint (µl/min/mg protein) = kdeg × (µl of incubation)/(mg protein), where kdeg is the negative slope of the semilogarithmic plot of the average peak area ratio vs. time. Since the slope of ln(CBD remaining) vs. time was positive for incubations with CYP3cide, the resulting intrinsic clearance value was negative, and the apparent percentage (%) of vehicle control would be negative. The absolute value of Clint is therefore reported.
bPercent contributions of cytochrome P450 enzymes to CBD depletion were estimated by calculating the percent change in remaining CBD vs. time in reactions with cytochrome P450–selective inhibitors compared with vehicle control.
cBecause the total estimated % contribution was greater than 100%, the data were normalized to 100%, and the same proportional contribution by each cytochrome P450 was retained, according to the method described by Zientek et al. (2016).
Effect of Cytochrome P450–Selective Inhibitors on CBD and 7-OH-CBD Metabolism
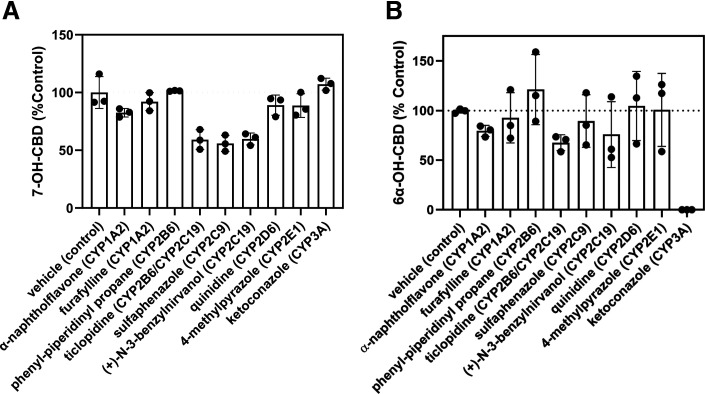
Reaction phenotyping studies were performed in pooled HLM with a panel of cytochrome P450–selective chemical inhibitors to explore the relative contributions of individual cytochrome P450 enzymes to CBD metabolism. These experiments were conducted at therapeutic concentrations of CBD (1 to 2 μM). Representative LC-MS/MS chromatograms of CBD metabolites formed in HLM in the presence and absence of cytochrome P450–selective chemical inhibitors are shown in Supplemental Fig. 3. The results from inhibition studies suggest that CYP2C9 and CYP2C19 were the primary enzymes to form 7-OH-CBD (Fig. 3A). A profound decrease in 6α-OH-CBD formation was observed in the presence of the CYP3A inhibitor ketoconazole (Fig. 3B). No significant differences were observed in 7-OH-CBD generation from CBD incubations with time-dependent cytochrome P450–selective inhibitors (Supplemental Fig. 4). When 7-OH-CBD (1 μM) was used as a substrate, minimal 7-COOH-CBD was formed in HLM, and 7-COOH-CBD formation was not significantly affected by the cytochrome P450 inhibitors tested (Supplemental Fig. 5). It is likely that the low levels of 7-COOH-CBD generated under the reaction conditions limited the ability to detect clear differences in 7-COOH-CBD formation in the presence and absence of cytochrome P450 inhibitors.
Fig. 3.
Effect of cytochrome P450–selective chemical inhibitors on CBD metabolism. CBD (1 to 2 µM) was incubated with pooled HLM (0.2 mg/ml protein) for 10 minutes. Formation of 7-OH-CBD (A) and 6α-OH-CBD (B) was measured in the presence of cytochrome P450–selective chemical inhibitors and compared with vehicle control incubations without inhibitor. The rate of 7-OH-CBD formation with vehicle control was 13.1 ± 2.30 pmol/min per milligram protein. Rates of metabolite formation were calculated using a standard curve in the range of 5–500 ng/ml (limit of quantitation = 25 ng/ml for 7-OH-CBD). The affected enzyme(s) for each inhibitor are listed in parentheses. Bars represent the mean ± S.D. of a single experiment performed in triplicate.
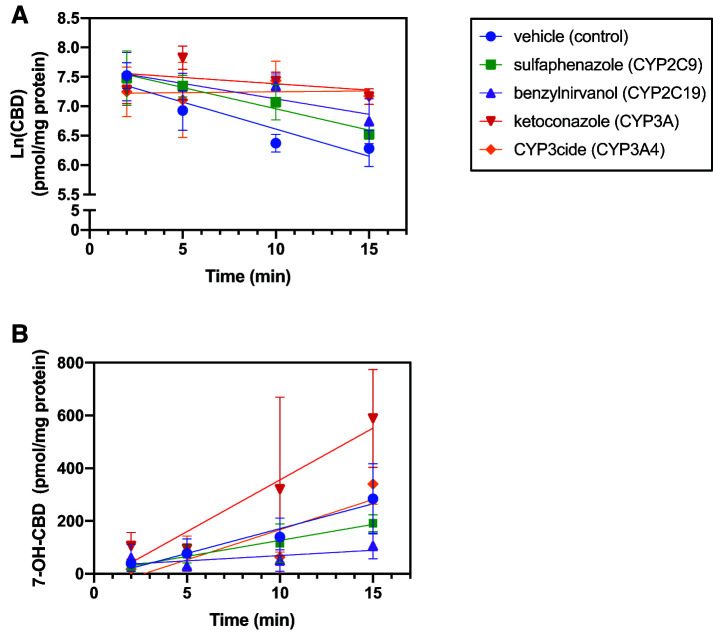
In a more detailed analysis, CBD (1 μM) depletion and metabolite formation were measured in the presence and absence of sulfaphenazole (a CYP2C9 inhibitor), benzylnirvanol (a CYP2C19 inhibitor), ketoconazole (an inhibitor of both CYP3A4 and CYP3A5), and CYP3cide (a time-dependent and selective CYP3A4 inhibitor). The results from these reactions are shown in Fig. 4, and the estimated relative cytochrome P450 contributions to each reaction are shown in Tables 2 and 3. Comparisons between the CYP3A4-selective inhibitor CYP3cide and the CYP3A nonspecific inhibitor ketoconazole were intended to estimate metabolic contributions of both CYP3A4 and CYP3A5 by calculating the differences between substrate depletion and metabolite formation with each inhibitor (Walsky et al., 2012; Tseng et al., 2014); however, no CBD depletion was observed in reactions preincubated with CYP3cide. The observation with CYP3cide contrasts with the results from coincubation with CYP3A inhibitor ketoconazole. The reason for this discrepancy is unknown. Thus, the results from experiments with CYP3cide were not used to estimate CYP3A4-specific contributions. CYP3A contributed approximately 54% to overall CBD depletion, whereas CYP2C19 and CYP2C9 were involved in an estimated 31% and 15%, respectively. CYP2C19 contributed the most to 7-OH-CBD formation (69%) in pooled HLM, followed by CYP2C9 (31%). Interestingly, coincubation with ketoconazole increased 7-OH-CBD formation by 2-fold, suggesting metabolic shunting from other oxidative pathways.
TABLE 3.
Percent contribution of P450 enzymes to 7-hydroxy-CBD formation
| Formation Rate | Percentage of vehicle control | Est. % Contributiona | Relative % Contributionb | |
|---|---|---|---|---|
| pmol/min/mg protein | % | |||
| Vehicle (control) | 18.81 | 100 | ||
| Sulfaphenazole (CYP2C9) | 12.12 | 64 | 36 | 31 |
| Benzylnirvanol (CYP2C19) | 4.013 | 21 | 79 | 69 |
| Ketoconazole (CYP3A) | 39.23 | 209 | ||
| CYP3cide (CYP3A4) | 22.84 | 121 | ||
aPercent contributions of cytochrome P450 enzymes to 7-OH-CBD formation were estimated by calculating the percent change in the slopes of 7-OH-CBD generation vs. time in reactions with cytochrome P450–selective inhibitors compared with vehicle control.
bBecause the total estimated % contribution was greater than 100%, the data were normalized to 100%, and the same proportional contribution by each cytochrome P450 was retained, according to the method described by Zientek et al. (2016).
Fig. 4.
Effect of CYP2C9, CYP2C19, and CYP3A inhibition on CBD depletion and metabolite formation. CBD (1 µM) was incubated with 150-donor pooled HLM (0.2 mg/ml protein) in the presence of cytochrome P450–selective chemical inhibitors (sulfaphenazole, 5 µM; (+)-N-3-benzylnirvanol, 5 µM; ketoconazole, 1 µM; and CYP3cide, 0.5 µM) and a vehicle control for 2, 5, 10, 15, 20, and 30 minutes. For reactions containing the time-dependent inhibitor CYP3cide, a 10-minute preincubation with CYP3cide and HLM was performed prior to addition of substrate. CBD depletion (A) and 7-OH-CBD formation (B) were quantified using a standard curve ranging from 5 to 500 ng/ml (limit of quantitation = 25 ng/ml) and measured as a peak area ratio with respect to internal standard (CBD-d9 and 7-OH-CBD-d3, respectively). Rates of CBD depletion were calculated using natural log-transformed depletion data, and rates of metabolite formation were calculated in the linear range of formation (over 15 minutes for 7-OH-CBD). Points represent the mean ± S.D. of a single experiment performed in triplicate.
7-OH-CBD (1 μM) depletion was also measured in pooled HLM in the presence of quinidine (a CYP2D6 inhibitor), benzylnirvanol, ketoconazole, and CYP3cide. The log-transformed rates of 7-OH-CBD depletion are represented in Supplemental Fig. 6A, and calculated relative cytochrome P450 contributions to 7-OH-CBD metabolism are shown in Table 4. Inhibition of CYP3A had the greatest impact on 7-OH-CBD depletion, suggesting further oxidation at sites other than the 7-position. Inhibition of CYP2D6, CYP2C19, and CYP3A4 had a modest effect on 7-COOH-CBD formation after 20 minutes of incubation (Supplemental Fig. 6B).
TABLE 4.
Percent contribution of P450 enzymes to 7-hydroxy-CBD depletion
Percent contributions of cytochrome P450 enzymes to 7-OH-CBD depletion were estimated by calculating the percent change in the slope of remaining 7-OH-CBD vs. time in reactions with cytochrome P450–selective inhibitors compared with vehicle control. 7-OH-CBD was quantitated using a standard curve (range = 1–1000 ng/ml; limit of quantitation = 5 ng/ml for 7-OH-CBD).
| Clint | Percentage of vehicle control | Est. % Contribution | |
|---|---|---|---|
| µl/min/mg protein | % | ||
| Vehicle (control) | 259.7 | 100 | |
| Benzylnirvanol (CYP2C19) | 233.8 | 90 | 10 |
| Quinidine (CYP2D6) | 300.6 | 116 | |
| Ketoconazole (CYP3A) | 91.75 | 35 | 65 |
| CYP3cide (CYP3A4) | 14.86 | 6 | |
CBD and 7-OH-CBD Metabolism by Recombinant Cytochrome P450 Enzymes
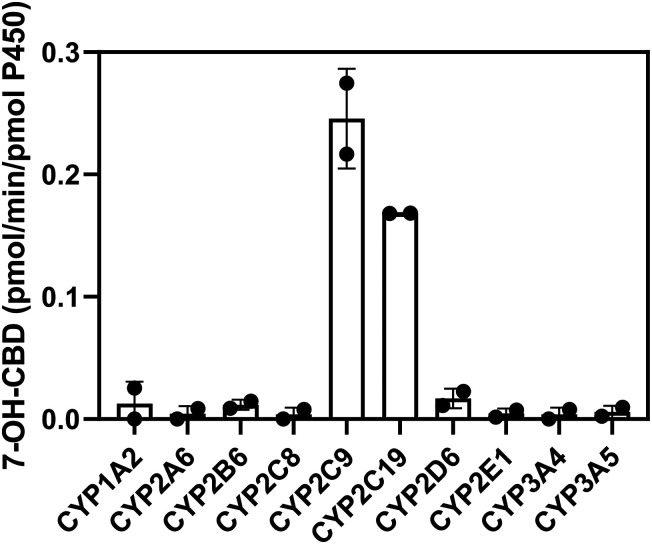
Screens with a panel of individual recombinant cytochrome P450 enzymes confirmed the involvement of CYP2C9 and CYP2C19 in formation of 7-OH-CBD (Fig. 5). Quantitatively the recombinant enzyme experiments suggest a greater role of CYP2C9 than expected from inhibition experiments with pooled HLM due to predicted higher average expression of CYP2C9 compared with CYP2C19 in HLM. Formation of 7-COOH-CBD from 7-OH-CBD as a substrate was catalyzed by recombinant CYP2C19 and CYP2D6, with minor contributions from CYP3A (Supplemental Fig. 7). The results for 7-COOH-CBD generation by recombinant cytochrome P450 enzymes were clearer than the findings from pooled HLM in the presence of cytochrome P450 inhibitors; this may be due to low turnover of 7-OH-CBD to 7-COOH-CBD in human liver microsomal assays.
Fig. 5.
Metabolism of CBD by recombinant cytochrome P450 enzymes. CBD (1 µM) was incubated with cytochrome P450 Supersomes (20 pmol/ml) for 10 minutes, and formation of 7-OH-CBD was measured by LC-MS/MS. Rates of formation were extrapolated from a 7-OH-CBD standard curve in the range of 5–500 ng/ml (limit of quantitation = 5 ng/ml). Bars represent the mean ± S.D. of two experiments performed in triplicate.
A summary table of CBD metabolites is provided in the Supplemental Table 2. The results from additional experiments and LC-MS/MS analysis of CBD metabolites formed by recombinant enzymes and HLM are shown in Supplemental Figs. 8–10.
Kinetic Analysis of CBD 7-Hydroxylation with Pooled HLM and Recombinant CYP2C19
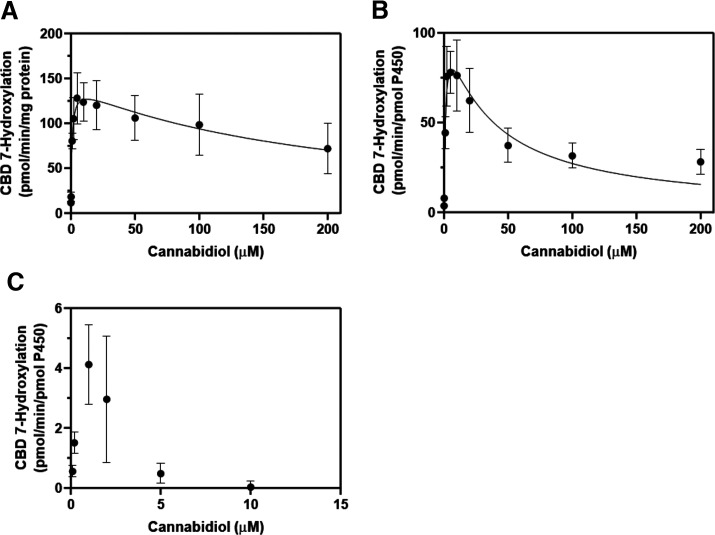
Based on the findings from reaction phenotyping studies, the kinetic parameters (Km and Vmax) of 7-OH-CBD formation were investigated with pooled HLM and recombinant CYP2C19 and CYP2C9. Initial incubations of CBD with pooled HLM for kinetic analysis generated an atypical curve for formation of 7-OH-CBD that did not follow typical Michaelis-Menten kinetics; rather, metabolite formation was instead suggestive of substrate inhibition, a phenomenon that may occur when a substrate inhibits its own metabolism at high concentrations through binding to a nonproductive inhibitory site on the drug-metabolizing enzyme (Fig. 6) (Lin et al., 2001). The exact mechanism of substrate inhibition remains unknown (Lin et al., 2001). The resultant kinetic parameters for pooled HLM and recombinant CYP2C19 are shown in Table 5. Kinetic parameter estimates for CYP2C9 could not be determined due to lack of 7-OH-CBD formation at CBD concentrations higher than 5 μM. This observation is indicative of anticipated cytochrome P450 substrate inhibition.
TABLE 5.
Kinetics of CBD 7-hydroxylation in pooled HLM and recombinant P450 enzymes
Kinetic parameters were calculated using a substrate inhibition model in GraphPad Prism 8 software.
| Kinetic Parameter | Estimate |
|---|---|
| Pooled HLM | |
| Apparent Vmax | 143.4 pmol/min/mg |
| Apparent Km | 0.8304 µM |
| Ki | 192.7 µM |
| CYP2C19 | |
| Apparent Vmax | 110.9 pmol/min/pmol P450 enzyme |
| Apparent Km | 1.279 µM |
| Ki | 32.56 µM |
Fig. 6.
Kinetic analysis of CBD 7-hydroxylation in pooled HLM (A), recombinant CYP2C19 (B), and recombinant CYP2C9 (C). CBD (0.1, 0.2, 1, 2, 5, 10, 20, 50, 100, and 200 µM) was incubated with 150-donor pooled HLM (0.2 mg/ml protein), rCYP2C19 (10 pmol/ml), and rCYP2C9 (10 pmol/ml) for 10 minutes, 2 minutes, and 5 minutes, respectively. Formation of 7-OH-CBD was measured by LC-MS/MS analysis using a standard curve in the range of 5–1000 ng/ml (limit of quantitation = 25 ng/ml for reactions with pooled HLM, 10 ng/ml for CYP2C19, and 25 ng/ml for CYP2C9). For pooled HLM and recombinant CYP2C19, Km and Vmax for 7-OH-CBD formation were calculated using GraphPad Prism 8 software using the following equation to describe substrate inhibition: Y= Vmax/(Km/X + 1 + X/Ki) (see Data Analysis). Data points represent mean ± S.D. of three experiments performed in triplicate.
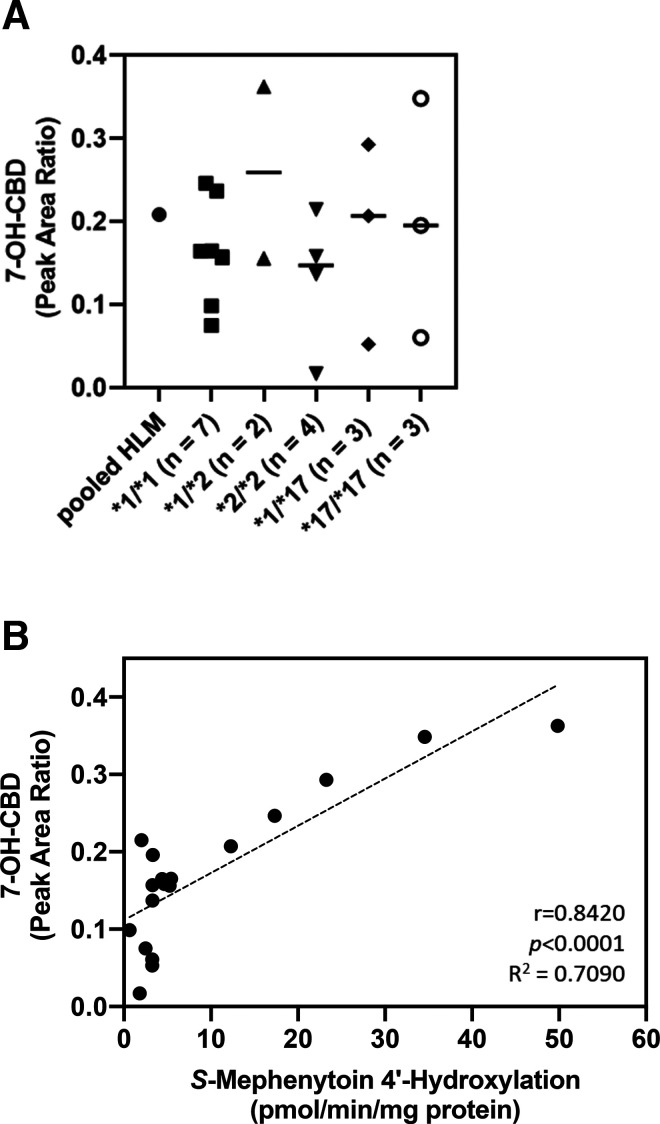
7-OH-CBD Formation by HLM from Individual CYP2C19-Genotyped Donors.
To explore the impact of CYP2C19 genetic variation on CBD active metabolite formation, CBD metabolism was studied in genotyped HLMs. HLM from 19 individual donors of varying CYP2C19 genotypes were used to evaluate 7-OH-CBD formation by genotype. Results are shown in Fig. 7. We found no association between 7-OH-CBD generation and CYP2C19 genotype. CYP2C19 activity, as measured by S-mephenytoin 4′-hydroxylation, was also not associated with CYP2C19 genotype among the donors tested (Supplemental Fig. 11). Pearson correlation analysis was also performed between CBD 7-hydroxylation and CYP2C19 activity. CBD 7-hydroxylation was significantly correlated with CYP2C19 activity measured by S-mephenytoin 4′-hydroxylation (Fig. 7B; Pearson r = 0.8420, P < 0.0001).
Fig. 7.
CBD metabolite formation in individual CYP2C19-genotyped HLMs. CBD (2 µM) was incubated with individual CYP2C19-genotyped HLM (0.2 mg/ml protein) for 5 minutes. (A) Formation of 7-OH-CBD was measured by LC-MS/MS analysis as a peak area ratio with respect to internal standard (CBD-d9). Data points represent the mean, and bars represent the median metabolite formation of three experiments performed in triplicate. (B) Correlation of 7-OH-CBD formation with CYP2C19 activity in individual CYP2C19-genotyped HLM. 7-OH-CBD formation was compared with CYP2C19 activity as measured by the rate of 4′-hydroxymephenytoin formation. Pearson r correlation coefficients, R2 values, and two-tailed P values were calculated using GraphPad Prism 8 software.
Additional experiments were conducted with a subset of individual-donor HLMs using selected chemical inhibitors to elucidate the discrepancies in the predicted contributions of CYP2C19 and CYP2C9 to 7-OH-CBD generation. When cytochrome P450 inhibitors were added to reactions with a subset of CYP2C19*1/*1 (homozygous wild-type) and CYP2C19*2/*2 (homozygous loss-of-function) genotyped HLMs, the CYP2C9 inhibitor sulfaphenazole inhibited CBD 7-hydroxylation in all donors regardless of CYP2C19 genotype (Fig. 8). Inhibition of CYP3A and CYP2C19 had minimal effects on 7-OH-CBD formation in single-donor HLMs. CYP2C19 inhibition was found to reduce 7-OH-CBD formation in 150-donor pooled HLM but not in the subset of individual donors tested.
Fig. 8.

Effect of cytochrome P450–selective chemical inhibitors on CBD metabolism by individual CYP2C19-genotyped HLMs. CBD (2 µM) was incubated with 150-donor pooled HLM and individual HLMs (diluted with 100 mM potassium phosphate buffer to 0.2 mg/ml protein) for 10 minutes. Formation of 7-OH-CBD was measured in the presence of sulfaphenazole (5 µM), (+)-N-3-benzylnirvanol (5 µM), or ketoconazole (1 µM) and compared with vehicle control incubations without inhibitor. Rates of metabolite formation were calculated using a standard curve in the range of 10–1000 ng/ml (limit of quantitation = 25 ng/ml for 7-OH-CBD; indicated by dotted line). The CYP2C19 genotype for each donor is shown in parentheses. Bars represent the mean ± S.D. of a single experiments performed in triplicate.
Discussion
This study specifically investigated the 7-hydroxylation pathway of CBD because 7-OH-CBD is the primary active metabolite of CBD. 7-OH-CBD is reported to have equipotent activity compared with CBD, and the AUC of 7-OH-CBD is approximately 38% of CBD (https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/210365Orig1s000ClinPharmR.pdf). The major findings of this study were as follows: 1) both CYP2C19 and CYP2C9 are involved in CBD metabolism to 7-OH-CBD; 2) CYP3A4 is a major contributor to CBD metabolic clearance through pathways other than 7-hydroxylation; and 3) formation of 7-OH-CBD is positively associated CYP2C19 activity in human liver microsomes but not CYP2C19 genotype.
Complementary reaction phenotyping approaches were used to evaluate the cytochrome P450–mediated oxidative metabolism pathways of CBD in vitro. These experiments were conducted at clinically relevant concentrations of CBD (1 to 2 μM) to increase the translatability of the results to the in vivo situation. CYP3A had the highest contribution to the overall metabolic clearance of CBD in pooled HLM based on the effect of CYP3A-selective inhibitors on CBD depletion; however, CYP3A4 was not involved in 7-OH-CBD formation. The finding that 7-OH-CBD formation significantly increased in the presence of CYP3A inhibitors is consistent with the observation that CYP3A is involved in CBD metabolism through pathways other than 7-hydroxylation. Allosteric activation of cytochrome P450–mediated metabolism is also a possibility (Hutzler and Tracy, 2002). CBD can undergo hydroxylation at multiple sites on its alkyl side chain or monoterpenoid ring (Jiang et al., 2011). Prior studies indicate that CYP3A is the primary enzyme responsible for 6α-, 6β-, and 4″-OH-CBD formation (Jiang et al., 2011). Hydroxylation on the alkyl side chain of CBD is a minor pathway of metabolism in HLM (Jiang et al., 2011). In the present study, an authentic standard of 6α-OH-CBD was used to examine formation of this metabolite. CYP3A inhibition completely blocked 6α-OH-CBD formation. Our findings regarding the major role of CYP3A in overall CBD clearance are consistent with previous in vivo studies, in which CYP3A induction by rifampicin decreased the plasma Cmax of CBD by 52%, and CYP3A inhibition by ketoconazole increased the plasma Cmax of CBD by 89% (Stott et al., 2013).
Previous in vitro studies indicated that CYP2C19 is primarily responsible for formation of 7-OH-CBD (Jiang et al., 2011). Findings in the present study demonstrate that both CYP2C19 and CYP2C9 are involved in 7-OH-CBD formation. CYP2C19 and CYP2C9 inhibitors reduced CBD 7-hydroxylation in HLM, and recombinant CYP2C19 and CYP2C9 generated 7-OH-CBD. Jiang et al. (2011) previously reported no effect of CYP2C9 inhibitor sulfaphenazole on 7-OH-CBD generation. However, in the present study, sulfaphenazole decreased 7-OH-CBD formation in HLM compared with vehicle control, suggesting the involvement of CYP2C9. Kinetic analysis of CBD 7-hydroxylation revealed similar apparent Km values of 0.8 and 1.3 μM by pooled HLM and recombinant CYP2C19, respectively. However, the rate of 7-OH-CBD generation by recombinant CYP2C9 was negligible at CBD concentrations above 5 μM. The apparent substrate inhibition at higher concentrations of CBD is consistent with previous reports that CBD is a potent inhibitor of CYP2C9 and CYP2C19 (Jiang et al., 2013; Bansal et al., 2020).
Correlation analysis revealed a positive association between CYP2C19 activity, measured by S-mephenytoin 4′-hydroxylation, and 7-OH-CBD generation in HLM donors with high CYP2C19 activity, consistent with a previous report by Jiang et al. (2011). Further analysis of our results shown in Fig. 7B suggests that, although CYP2C19 activity may significantly affect 7-OH-CBD generation for some individuals, this correlation is not significant for the entire sample, as shown by the variation in 7-OH-CBD generation among individuals with low 4′-hydroxymephenytoin formation. These findings indicate that CYP2C19 may contribute to CBD 7-hydroxylation in individuals with high CYP2C19 activity, and other enzymes, such as CYP2C9, may be involved in individuals with lower CYP2C19 activity. No association was found between CYP2C19 genotype and 7-OH-CBD generation in HLM from CYP2C19-genotyped donors. It should be noted that CYP2C19 enzyme activity was also not associated with CYP2C19 genotype. This finding is consistent with our previous report demonstrating high variability in CYP2C19 activity even among donors of the same CYP2C19 genotype (Murray et al., 2020). These data suggest that factors besides CYP2C19 polymorphism influence CYP2C19 activity and CBD 7-hydroxylation.
To our knowledge, this is the first study to demonstrate a major role of CYP2C9 in CBD active metabolite formation. Herein, we found that inhibition of CYP2C9 (by sulfaphenazole) reduced 7-OH-CBD formation to a greater extent than CYP2C19 inhibition (by benzylnirvanol) in the subset of individual HLM donors (CYP2C19*1/*1 and CYP2C19*2/*2 donors) tested, suggesting a significant contribution by CYP2C9. This finding indicates that CYP2C9 may play a larger role in CBD 7-hydroxylation in some individuals, and the relative enzyme contributions may vary by patient. CYP2C9 is also highly polymorphic (Pharmacogene Variation Consortium https://www.pharmvar.org/gene/CYP2C9). A limitation to this analysis is that the CYP2C9 genotypes were not known for all of the HLM donors tested in this subset, except for two donors: QLC and IFF genotyped as CYP2C9*1/*1 (homozygous wild-type); the reported CYP2C9 activities were also not measured using the same probe substrate for all donors for comparison. Literature reports indicate that the average CYP2C9 protein abundance in HLMs is higher than CYP2C19 (Achour et al., 2014); however, relative cytochrome P450 enzyme expression and activity can vary widely between individuals and different genetic ancestry populations (Zhou et al., 2017). It is possible that CYP2C9 may play an increased role in CBD metabolism to 7-OH-CBD in some patients after multiple dosing with CBD due to time-dependent inhibition of CYP2C19; this requires future investigations.
When 7-OH-CBD was used as a substrate, recombinant CYP2C19 and CYP2D6 formed 7-COOH-CBD. However, low levels of 7-COOH-CBD were generated in HLM incubations with 7-OH-CBD. Cytochrome P450 inhibitors had minimal effects on 7-COOH-CBD formation, likely due to insufficient turnover of 7-OH-CBD to 7-COOH-CBD in HLM. When comparing the depletion data in Table 4 to the low 7-COOH-CBD formation observed in these experiments, the results suggest that 7-OH-CBD may be metabolized to products other than 7-COOH-CBD in HLM. Given the low levels of 7-COOH-CBD generated in liver microsomal incubations, it is conceivable that cytosolic enzymes may be involved in 7-COOH-CBD formation. This is relevant because 7-COOH-CBD is the most abundant circulating metabolite of CBD in vivo (GW Therapeutics Inc., 2017), and the enzymes that contribute to its formation will likely have a major impact on 7-COOH-CBD exposure in vivo. Therefore, future studies will focus on fully characterizing this pathway of CBD metabolism to identify the major enzymes involved.
CBD has been shown to be a reversible inhibitor of CYP2C9 and a time-dependent inhibitor of CYP2C19, CYP3A, and CYP1A2 (Bansal et al., 2020). These findings are supported by the substrate inhibition kinetics observed in HLM, recombinant CYP2C19, and CYP2C9 in this study. Clinically significant drug-drug interactions have been observed between CBD and the anticoagulant warfarin; S-warfarin is a CYP2C9 substrate with a narrow therapeutic index (Grayson et al., 2017; Damkier et al., 2019). Coadministration of CBD with the antiepileptic drug clobazam increased exposure (Cmax and AUCtau) to the active metabolite N-desmethylclobazam, a CYP2C19 substrate, by greater than 3-fold (Morrison et al., 2019), which may be due to CBD-mediated inhibition of CYP2C19. Less is known about clinically significant drug interactions with CYP3A substrates in which CBD is the perpetrator. For example, CBD did not significantly affect in vivo exposure to the sensitive CYP3A substrate midazolam (Morrison et al., 2018); however, concomitant use of CBD with CYP3A4/5 substrate tacrolimus was shown to increase dose-normalized trough concentrations of tacrolimus by 3-fold (Leino et al., 2019). Future investigations are needed to evaluate the interaction of CBD and its metabolites with other CYP2C9, CYP2C19, and CYP3A substrates in vivo. In addition, the effects of cytochrome P450 pharmacogenetic variations on CBD disposition, drug interaction potential, and toxicity in vivo have not been well defined, and these areas warrant further investigation.
In summary, we have shown that both CYP2C19 and CYP2C9 play a role in metabolism of CBD to the active metabolite 7-OH-CBD. Although CYP3A demonstrated the greatest contribution to overall CBD clearance through other oxidative pathways, CYP3A did not contribute significantly to 7-OH-CBD generation. Human liver microsomal formation of 7-OH-CBD was positively associated CYP2C19 activity but not CYP2C19 genotype, suggesting that factors besides CYP2C19 polymorphism influence CBD metabolism through this pathway. Reaction phenotyping studies at clinically relevant concentrations of CBD suggest that CYP2C9 may have a more significant contribution to CBD 7-hydroxylation than previously recognized.
Acknowledgments
The authors thank Dr. Kim L. R. Brouwer (University of North Carolina at Chapel Hill) and Dr. Paavo Honkakoski (University of North Carolina at Chapel Hill and University of Eastern Finland) for scientific discussions and feedback during manuscript preparation. We also thank Dr. Nina Isoherranen (University of Washington) for valuable scientific discussions during the revision of the manuscript. LC-MS/MS analysis was conducted at the UNC Biomarker Mass Spectrometry Core Facility, which is supported by NIH NIEHS award number P30ES010126. The authors also thank Leonard Collins and Peter Hans Cable for LC-MS/MS support.
Abbreviations
- CBD
cannabidiol
- FDA
Food and Drug Administration
- HLM
human liver microsome
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- 7-OH-CBD
7-Hydroxy-CBD
- 7-COOH-CBD
7-carboxy-CBD
- UDPGA
uridine 5′-diphosphate-glucuronic acid
- UGT
UDP-glucuronosyltransferase
Authorship Contributions
Participated in research design: Beers, Fu, Jackson.
Conducted experiments: Beers.
Performed data analysis: Beers.
Wrote or contributed to the writing of the manuscript: Beers, Fu, Jackson.
Footnotes
The authors have no financial conflicts of interest to disclose.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Achour BBarber J, Rostami-Hodjegan A (2014) Expression of hepatic drug-metabolizing cytochrome p450 enzymes and their intercorrelations: a meta-analysis. Drug Metab Dispos 42:1349–1356. [DOI] [PubMed] [Google Scholar]
- Bansal S, Maharao N, Paine MF, Unadkat JD (2020) Predicting the potential for cannabinoids to precipitate pharmacokinetic drug interactions via reversible inhibition or inactivation of major cytochromes p450. Drug Metab Dispos 48:1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland RA (2000) Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis, Ed. 2nd. Wiley-VCH, Inc., New York. [Google Scholar]
- Damkier P, Lassen D, Christensen MMH, Madsen KG, Hellfritzsch M, Pottegård A(2019) Interaction between warfarin and cannabis. Basic Clin Pharmacol Toxicol 124:28–31. [DOI] [PubMed] [Google Scholar]
- Gaston TE, Bebin EM, Cutter GR, Liu Y, Szaflarski JP; UAB CBD Program (2017) Interactions between cannabidiol and commonly used antiepileptic drugs. Epilepsia 58:1586–1592. [DOI] [PubMed] [Google Scholar]
- Grayson L, Vines B, Nichol K, Szaflarski JP; UAB CBD Program (2017) An interaction between warfarin and cannabidiol, a case report. Epilepsy Behav Case Rep 9:10–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA Center for Drug Evaluation and Research https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/210365Orig1s000ClinPharmR.pdf.
- Hutzler JM and Tracy TS (2002) Atypical kinetic profiles in drug metabolism reactions. Drug Metab Dispos 30:355–362. [DOI] [PubMed] [Google Scholar]
- Jiang R, Yamaori S, Okamoto Y, Yamamoto I, Watanabe K (2013) Cannabidiol is a potent inhibitor of the catalytic activity of cytochrome P450 2C19. Drug Metab Pharmacokinet 28:332–338. [DOI] [PubMed] [Google Scholar]
- Jiang R, Yamaori S, Takeda S, Yamamoto I, Watanabe K (2011) Identification of cytochrome P450 enzymes responsible for metabolism of cannabidiol by human liver microsomes. Life Sci 89:165–170. [DOI] [PubMed] [Google Scholar]
- Leino AD, Emoto C, Fukuda T, Privitera M, Vinks AA, Alloway RR (2019) Evidence of a clinically significant drug-drug interaction between cannabidiol and tacrolimus. Am J Transplant 19:2944–2948. [DOI] [PubMed] [Google Scholar]
- Lin Y, Lu P, Tang C, Mei Q, Sandig G, Rodrigues AD, Rushmore TH, Shou M (2001) Substrate inhibition kinetics for cytochrome P450-catalyzed reactions. Drug Metab Dispos 29:368–374. [PubMed] [Google Scholar]
- Madden K, Tanco K, Bruera E (2020) Clinically significant drug-drug interaction between methadone and cannabidiol. Pediatrics 145:e20193256. [DOI] [PubMed] [Google Scholar]
- Mazur A, Lichti CF, Prather PL, Zielinska AK, Bratton SM, Gallus-Zawada A, Finel M, Miller GP, Radomińska-Pandya A, Moran JH (2009) Characterization of human hepatic and extrahepatic UDP-glucuronosyltransferase enzymes involved in the metabolism of classic cannabinoids. Drug Metab Dispos 37:1496–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar SA, Stone NL, Bellman ZD, Yates AS, England TJ, O’Sullivan SE (2019) A systematic review of cannabidiol dosing in clinical populations. Br J Clin Pharmacol 85:1888–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison G, Crockett J, Blakey G, Sommerville K (2019) A phase 1, open-label, pharmacokinetic trial to investigate possible drug-drug interactions between clobazam, stiripentol, or valproate and cannabidiol in healthy subjects. Clin Pharmacol Drug Dev 8:1009–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison G, Taylor L, Crockett J, Critchley D, Tayo B (2018) A phase 1 investigation into the potential effects of cannabidiol on CYP3A4-mediated drug-drug interactions in healthy volunteers. American Epilepsy Society Annual Meeting, 2018. [Google Scholar]
- Murray JL, Mercer SL, Jackson KD (2020) Impact of cytochrome P450 variation on meperidine N-demethylation to the neurotoxic metabolite normeperidine. Xenobiotica 50:209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Gurley BJ, Markowitz JS (2019) The potential for pharmacokinetic interactions between cannabis products and conventional medications. J Clin Psychopharmacol 39:462–471. [DOI] [PubMed] [Google Scholar]
- Stott C, White L, Wright S, Wilbraham D, Guy G (2013) A phase I, open-label, randomized, crossover study in three parallel groups to evaluate the effect of Rifampicin, Ketoconazole, and Omeprazole on the pharmacokinetics of THC/CBD oromucosal spray in healthy volunteers. Springerplus 2:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stresser DM, Perloff ES, Mason AK, Blanchard AP, Dehal SS, Creegan TP, Singh R, Gangl ET (2016) Selective time- and NADPH-dependent inhibition of human CYP2E1 by clomethiazole. Drug Metab Dispos 44:1424–1430. [DOI] [PubMed] [Google Scholar]
- Towles JK, Clark RN, Wahlin MD, Uttamsingh V, Rettie AE, Jackson KD (2016) Cytochrome P450 3A4 and CYP3A5-catalyzed bioactivation of lapatinib. Drug Metab Dispos 44:1584–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng E, Walsky RL, Luzietti RA Jr, Harris JJ, Kosa RE, Goosen TC, Zientek MA, Obach RS (2014) Relative contributions of cytochrome CYP3A4 versus CYP3A5 for CYP3A-cleared drugs assessed in vitro using a CYP3A4-selective inactivator (CYP3cide). Drug Metab Dispos 42:1163–1173. [DOI] [PubMed] [Google Scholar]
- Walsky RL, Obach RS, Hyland R, Kang P, Zhou S, West M, Geoghegan KF, Helal CJ, Walker GS, Goosen TC, et al. (2012) Selective mechanism-based inactivation of CYP3A4 by CYP3cide (PF-04981517) and its utility as an in vitro tool for delineating the relative roles of CYP3A4 versus CYP3A5 in the metabolism of drugs. Drug Metab Dispos 40:1686–1697. [DOI] [PubMed] [Google Scholar]
- Whalley B, Stott C, Gray R, Jones N (2017) The human metabolite of cannabidiol, 7- hydroxyl cannabidiol, but not 7-carboxy cannabidiol, is anticonvulsant in the maxi-mal electroshock seizure threshold test (MEST) in mouse. American Epilepsy Society meeting abstract 1.435, 2017. [Google Scholar]
- Zhou Y, Ingelman-Sundberg M, Lauschke VM (2017) Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin Pharmacol Ther 102:688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientek MA, Goosen TC, Tseng E, Lin J, Bauman JN, Walker GS, Kang P, Jiang Y, Freiwald S, Neul D, et al. (2016) In vitro kinetic characterization of axitinib metabolism. Drug Metab Dispos 44:102–114. [DOI] [PubMed] [Google Scholar]