ABSTRACT
Bacteria are capable of withstanding large changes in osmolality and cytoplasmic pH, unlike eukaryotes that tightly regulate their pH and cellular composition. Previous studies on the bacterial acid stress response described a rapid, brief acidification, followed by immediate recovery. More recent experiments with better pH probes have imaged single living cells, and we now appreciate that following acid stress, bacteria maintain an acidic cytoplasm for as long as the stress remains. This acidification enables pathogens to sense a host environment and turn on their virulence programs, for example, enabling survival and replication within acidic vacuoles. Single-cell analysis identified an intracellular pH threshold of ~6.5. Acid stress reduces the internal pH below this threshold, triggering the assembly of a type III secretion system in Salmonella and the secretion of virulence factors in the host. These pathways are significant because preventing intracellular acidification of Salmonella renders it avirulent, suggesting that acid stress pathways represent a potential therapeutic target. Although we refer to the acid stress response as singular, it is actually a complex response that involves numerous two-component signaling systems, several amino acid decarboxylation systems, as well as cellular buffering systems and electron transport chain components, among others. In a recent paper in the Journal of Bacteriology, M. G. Gorelik, H. Yakhnin, A. Pannuri, A. C. Walker, C. Pourciau, D. Czyz, T. Romeo, and P. Babitzke (J Bacteriol 206:e00354-23, 2024, https://doi.org/10.1128/jb.00354-23) describe a new connection linking the carbon storage regulator CsrA to the acid stress response, highlighting new additional layers of complexity.
KEYWORDS: two-component regulatory systems, amino acid decarboxylation, acid stress response, SsrB, carbon storage regulation, CsrA, Salmonella pathogenicity island 2
INTRODUCTION
Acid stress mitigation systems have long been studied in bacteria, in part because of their importance in pathogenesis (1–3). After ingestion of contaminated food or water, bacteria have to survive the extreme acidic pH of the stomach and then migrate to the moderately acidic intestine. The ability of Gram-negative bacteria such as E. coli and Salmonella to tolerate acidic environments is attributable to a complex interplay of physiological, metabolic, and regulatory mechanisms. Amino acid decarboxylation systems play a central role in neutralizing the cytoplasm; secondary carbon metabolism and sugar derivatives come into play to produce fewer acid metabolites when compared to glucose. The cytoplasm is buffered by phosphates and polyamines, and changes in membrane lipid composition, as well as electron transport chain components, occur. Periplasmic chaperones are upregulated, and efflux through outer membrane porins is reduced. The development of new tools for measuring fluctuations in pH has aided in our understanding of acid stress responses.
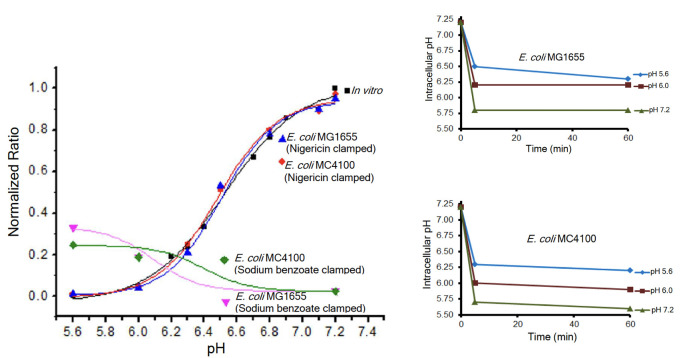
The commonly used pH indicator pHluorin is often expressed as an arabinose-inducible, plasmid-encoded, pH-sensitive GFP; it is heterogeneous with respect to its expression in single cells, and thus, it is not a good indicator for pH measurements in bacteria (2). In addition, sodium benzoate was frequently used as a clamping agent to equilibrate the internal pHi with the external pHe. A comparison of the standard curve of cells clamped using the ionophore nigericin and generated from the I-switch (a FRET-based DNA biosensor) or BCECF-AM with cells clamped using sodium benzoate, the salt of the weak acid benzoic acid, dramatically illustrated that sodium benzoate was a poor choice as a clamping agent (see Fig. 1) (2). It is immediately evident that the behavior of the BCECF-AM probe is quite different in the sodium benzoate-exposed strains compared to those containing nigericin. The nigericin-containing bacteria overlay perfectly with the probe alone in vitro (black curve). Furthermore, when we clamped cells with sodium benzoate at pH values 5.6, 6.0, and 7.2 and then measured the pHi using BCECF-AM, it was apparent that the cells were not clamped. At pHe = 5.6, the pHi was 6.3; at pHe = 6.0, the pHi was 6.2; and at pHe = 7.2, the pHi was 5.8. Thus, using sodium benzoate at neutral pHe, the intracellular pH would be presumed to have been neutralized, when it was actually quite acidic (pH 5.8). These values were not strain-dependent, i.e., they were similar for MC4100 and MG1655. However, important strain differences were evident when comparing E. coli strain MC4100 and its response to extracellular acid or osmotic stress compared to the probiotic Nissle strain (4, 5) and to MG1655, a sequenced strain (6).
Fig 1.
The problems with sodium benzoate as a clamping agent. (left graph) Cells were clamped with either 40 µM nigericin or 30 mM sodium benzoate, and the 488/440 ratio intensities were plotted as a function of pH and overlaid on the in vitro calibration curve. Intracellular pH values determined from BCECF-AM fluorescence of E. coli MG1655 (top right) and MC4100 (bottom right) clamped with sodium benzoate at pHe 5.6, pHe 6.1, and pHe 7.2 were plotted. It is evident that pHe was not equal to pHi (see text). Modified from reference 2.
Fluorescence methods involving pH-sensitive organic dyes such as BCECF-AM, SNARF, or fluorescein or plasmid-encoded fluorescent biosensors such as pHluorin and SypHer have been extensively applied to measure the intracellular pH (7–10). They each have their challenges and limitations. We have recently applied fluorescence lifetime imaging microscopy (FLIM) to a pH-sensitive mutant mCherry to quantitatively measure the pH in bacteria, as well as changes in pH that reflect fluctuating bacterial lifestyles, including in biofilms, in the intestines of Danio rerio and C. elegans, and in bacteria infecting HeLa and Thp-1 cells in tissue culture (M. K. Singh, M. Fernandez, R. Dalawari, and L. J. Kenney, unpublished data). Our results continue to demonstrate a sustained decrease in bacterial cytoplasmic pH in response to acid stress (1, 2, 11).
THE ROLE OF TWO-COMPONENT SYSTEMS
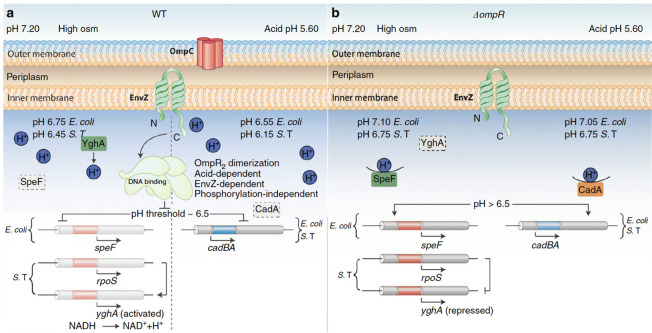
Two-component signaling systems are intimately involved in acid stress responses. A systems biology approach investigated the gene networks involved in acid resistance and discovered that the response regulator OmpR was a key regulator of the complex transcriptional program involved in acid adaptation (1). An ompR null strain was extremely acid-sensitive (12), and some direct targets of OmpR were later identified (2). We recently employed a FRET-DNA biosensor termed the I-switch and used it to measure the pH of the Salmonella cytoplasm while it was replicating within an acidic macrophage vacuole. The EnvZ/OmpR two-component system was essential in sensing the decrease in cytoplasmic pH and for repressing the cadCBA lysine decarboxylation system that would normally restore neutrality (1). Salmonella harnesses the acid stress response to convey that it is in a host, and it is time to turn on its virulence program (13), which allows it to replicate in an acidic vacuole and then disseminate. This process sets a series of events in motion, which lead to the up-regulation of the SsrA/B two-component system that is located on Salmonella pathogenicity island 2 (SPI-2) (14). The acid pH flips a molecular switch through a conserved histidine residue (His12) in the response regulator SsrB that drives a > 30 fold change in its affinity for DNA, stimulating the expression of virulence genes (3). Although it has not been studied, it is anticipated that other response regulators of the NarL/FixJ subfamily might also exhibit pH-dependent switches, as His12 is highly conserved (3). A summary of the complex regulation of acidification in response to osmotic and acid stress that requires OmpR is given in Fig. 2.
Fig 2.
A summary of the role of OmpR in the acid stress response and acidification of the cytoplasm in response to osmolytes. Under acid and osmotic stress conditions, both S. Typhimurium and E. coli are acidified in an OmpR-dependent manner. OmpR requires n interaction with EnvZ, but not phosphorylation, to bring about cytoplasmic acidification. (A) In wild-type bacteria, under acid stress (right), OmpR represses the cad operon to eliminate proton consumption, resulting in acidification. At high osmolality, in S. Typhimurium, OmpR represses rpoS to relieve RpoS repression of yghA, producing protons (left). In E. coli, OmpR represses ornithine decarboxylase (speF), enabling cytoplasmic acidification. The pH optima of CadA is 6.1–6.5 (15); this optima contributes to a threshold of response. At pH 6.5 and below (achieved during acid stress), OmpR represses the cad operon, resulting in acidification. At high osmolality (pH 6.75, E. coli; 6.45, S. Typhimurium), acidification is less because the CAD system is working to restore neutrality. Acidification occurs through proton production (S. Typhimurium) or repression of a different amino acid decarboxylation system (E. coli). Intracellular acidification is required for activating SPI-2-dependent effector secretion. EnvZ senses and responds to cytoplasmic acidification via helix–coil transitions. A physical interaction of OmpR with EnvZ drives a conformational change, resulting in unphosphorylated OmpR dimer formation. EnvZ-dependent OmpR dimerization creates an active OmpR2 interface, favoring DNA binding and subsequent repression. (B) In the ompR null strain, during acid stress, CadC/BA is expressed and drives amino acid decarboxylation. This process consumes intracellular protons, restoring cytoplasmic pH and maintaining intracellular pH homeostasis. At high osmolality, in an ompR null S. Typhimurium strain, RpoS represses yghA, preventing proton release, resulting in a neutralized cytoplasm. In an ompR null E. coli strain, activated SpeF eliminates cytoplasmic protons during ornithine decarboxylation, maintaining pH homeostasis. Reprinted from reference 2.
The response regulator PhoP was identified in a two-dimensional SDS-PAGE analysis of acid-induced proteins. The phoQ/P two-component system was itself acid-induced, and phoQ/P mutants failed to induce some acid shock proteins (16). Direct targets of the PhoP-dependent response to acid shock were not identified. More recently, it was reported that PhoQ sensed cytoplasmic pH (17), presumably in a manner similar to EnvZ (18), although mechanistic studies are lacking.
Perhaps the most efficient acid resistance system involves glutamate decarboxylation by the GadA and GadB decarboxylases and the import of glutamate via the membrane protein GadC. GadE is the central transcriptional activator (19). In addition, RcsB, the response regulator of the Rcs system, is absolutely required for control of gadA/BC transcription, but its role is complex. In the presence of GadE, basal activity of RcsB stimulates gadA/BC expression, whereas activation of RcsB leads to general repression of the gad genes. Activation of gadA transcription involves binding of an RcsB/GadE heterodimer (20). In E. coli, EvgS/A upregulates a network of acid resistance genes, through a cascade of EvgA–YdeO–GadE regulators (21–23). In addition, the SafA–phoQ/phoP–IraM–RpoS network connects two-component systems EvgA/S and phoQ/P via the small connecting proteins SafA (formerly B1500) and IraM and the sigma factor RpoS (24, 25).
HOW DOES THE CARBON STORAGE REGULATOR (CsrA) INTERACT WITH THE ACID STRESS RESPONSE?
The answer lies in part through the RNA-binding protein CsrA. The Csr system plays a critical regulatory role in numerous cellular responses, including biofilm formation, stress response systems, motility, quorum sensing, virulence factor expression in pathogens, and central carbon metabolism [see References in (26)]. Disruption of csrA causes a pH-dependent growth defect. CsrA-mediated regulation involves its binding to sites containing a critical GGA motif, often found in the single-stranded loop of an RNA hairpin. These binding sites are usually located in the 5’ leader or early mRNA coding regions. CsrA binding can regulate acid stress response via multiple mechanisms, including transcription elongation, translation initiation, RNA stability, or modulating riboswitch activity. In a recent work by Gorelik et al. (26), the authors undertook a transcriptome-wide analysis of RNAs that bind to CsrA. Some of the mRNAs that were bound by the regulatory protein had roles in the response of E. coli to acidic conditions. To examine this connection in more detail, Gorelik and co-workers compared the sensitivity of wild-type and csrA mutant strains to both mild and extreme acidic conditions. CsrA binding to evgLA, gadA, gadB, gadE, and ydeO mRNAs was evident using mobility shift assays, as was CsrA repression of evgLA–, gadA–, gadB–, gadE–, ydeO-, and ydeP–lacZ fusions. Thus, CsrA binds directly in the absence of other factors to transcripts of the EvgA–YdeO–GadE circuit, with the highest affinity to evgA mRNA, where CsrA directly represses evgA translation. In vitro footprinting and toeprinting assays also elucidated where CsrA binds to the evgA, gadA, and gadB RNA fragments, identifying both repressive effects of CsrA on evgA translation as well as translational coupling through evgL.
CsrA repression of the acid stress response is critical for managing the trade-off between growth and survival. Overexpression of acid stress genes caused by csrA disruption enhanced survival under extreme acidity, but was detrimental for growth under mildly acidic conditions. Thus, the authors have identified a new facet of the global role played by CsrA in balancing the opposing physiological demands of stress resistance with the capacity for growth and ultimately in modulating host interactions. It would be of interest to determine (i) what the changes in cytoplasmic pH (pHi) are during the stress conditions imposed, (ii) as well as the pHi range over which CsrA is active, and (iii) whether or not CsrA contains a molecular switch as we reported in SsrB (3), which dramatically affects its binding activity. It is also imperative to have a broader understanding of how the many acid stress responses are coordinated, i.e., how are signals integrated to coordinate a response to environmental stress? Does CsrA play a role in other amino acid decarboxylation systems and what determines specificity? Is it merely the absence of GGA motifs in mRNAs from the other decarboxylation systems or some other CsrA function?
ACKNOWLEDGMENTS
I acknowledge the following funding sources that have supported our work on the acid stress response: NIH R21AI-123640; GM-058746; VA IO1BX-000372; a Regional Centre of Excellence in Mechanobiology supported by the Ministry of Education, Singapore; a Texas STAR award and start-up funds from UTMB.
I am grateful to the Arnold family for the Chair in Gastroenterology. I thank E. Pete Greenberg for discussions on nigericin vs sodium benzoate clamping. Thanks also to Michael Galperin for his comments and gentle prodding. I owe a huge debt to the members of my laboratory and collaborators who contributed intellectually and with sweat equity to our understanding of acid stress in bacteria: Ganesh Anand, Smarajit Chakraborty, Soham Deolankar, Stuti K. Desai, Marion Fernandez, Yong Hwee Foo, Mike Heilemann, Andrew Liew, Hideaki Mizusaki, Khin K.Z. Mon, Dasvit Shetty, Moirangthem Kiran Singh, Christoph Spahn, Don Walthers, Parisa Zangoui, and Leslie K. Morgan, my dear lab manager of 20 years.
The views expressed in this article do not necessarily reflect the views of the journal or of ASM.
Contributor Information
Linda J. Kenney, Email: likenney@utmb.edu.
Michael Y. Galperin, NCBI, NLM, National Institutes of Health, Bethesda, Maryland, USA
REFERENCES
- 1. Chakraborty S, Mizusaki H, Kenney LJ. 2015. A FRET-based DNA biosensor tracks OmpR-dependent acidification of Salmonella during macrophage infection. PLoS Biol 13:e1002116. doi: 10.1371/journal.pbio.1002116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chakraborty S, Winardhi RS, Morgan LK, Yan J, Kenney LJ. 2017. Non-canonical activation of OmpR drives acid and osmotic stress responses in single bacterial cells. Nat Commun 8:1587. doi: 10.1038/s41467-017-02030-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shetty D, Kenney LJ. 2023. A pH-sensitive switch activates virulence in Salmonella. Elife 12:e85690. doi: 10.7554/eLife.85690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adediran J, Leatham-Jensen MP, Mokszycki ME, Frimodt-Møller J, Krogfelt KA, Kazmierczak K, Kenney LJ, Conway T, Cohen PS. 2014. An Escherichia coli Nissle 1917 missense mutant colonizes the streptomycin-treated mouse intestine better than the wild type but is not a better probiotic. Infect Immun 82:670–682. doi: 10.1128/IAI.01149-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Möndel M, Schroeder BO, Zimmermann K, Huber H, Nuding S, Beisner J, Fellermann K, Stange EF, Wehkamp J. 2009. Probiotic E. coli treatment mediates antimicrobial human beta-defensin synthesis and fecal excretion in humans. Mucosal Immunol 2:166–172. doi: 10.1038/mi.2008.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453 [DOI] [PubMed] [Google Scholar]
- 7. Cui J, Kim G, Kim S, Kwon JE, Park SY. 2020. Ultra‐pH‐sensitive small molecule probe showing a ratiometric fluorescence color change. ChemPhotoChem 4:393–397. doi: 10.1002/cptc.202000023 [DOI] [Google Scholar]
- 8. Han J, Burgess K. 2010. Fluorescent indicators for intracellular pH. Chem Rev 110:2709–2728. doi: 10.1021/cr900249z [DOI] [PubMed] [Google Scholar]
- 9. Miesenböck G, De Angelis DA, Rothman JE. 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394:192–195. doi: 10.1038/28190 [DOI] [PubMed] [Google Scholar]
- 10. Poburko D, Santo-Domingo J, Demaurex N. 2011. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J Biol Chem 286:11672–11684. doi: 10.1074/jbc.M110.159962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chakraborty S, Kenney LJ. 2018. A new role of OmpR in acid and osmotic stress in Salmonella and E. coli. Front. Microbiol 9:2656. doi: 10.3389/fmicb.2018.02656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stincone A, Daudi N, Rahman AS, Antczak P, Henderson I, Cole J, Johnson MD, Lund P, Falciani F. 2011. A systems biology approach sheds new light on Escherichia coli acid resistance. Nucleic Acids Res 39:7512–7528. doi: 10.1093/nar/gkr338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kenney LJ. 2019. The role of acid stress in Salmonella pathogenesis. Curr Opin Microbiol 47:45–51. doi: 10.1016/j.mib.2018.11.006 [DOI] [PubMed] [Google Scholar]
- 14. Liew ATF, Foo YH, Gao Y, Zangoui P, Singh MK, Gulvady R, Kenney LJ. 2019. Single cell, super-resolution imaging reveals an acid pH-dependent conformational switch in SsrB regulates SPI-2. Elife 8:e45311. doi: 10.7554/eLife.45311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheeseman GC, Fuller R. 1968. Changes in the pH activity profile of the lysine decarboxylase during incubation of Escherichia coli. J Appl Bacteriol 31:253–258. doi: 10.1111/j.1365-2672.1968.tb00365.x [DOI] [PubMed] [Google Scholar]
- 16. Bearson BL, Wilson L, Foster JW. 1998. A low pH-inducible, phoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J Bacteriol 180:2409–2417. doi: 10.1128/JB.180.9.2409-2417.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Choi J, Groisman EA. 2016. Acidic pH sensing in the bacterial cytoplasm is required for Salmonella virulence. Mol Microbiol 101:1024–1038. doi: 10.1111/mmi.13439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang LC, Morgan LK, Godakumbura P, Kenney LJ, Anand GS. 2012. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J 31:2648–2659. doi: 10.1038/emboj.2012.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol Microbiol 49:1309–1320. doi: 10.1046/j.1365-2958.2003.03633.x [DOI] [PubMed] [Google Scholar]
- 20. Castanié-Cornet M-P, Cam K, Bastiat B, Cros A, Bordes P, Gutierrez C. 2010. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nuc Acids Res 38:3546–3554. doi: 10.1093/nar/gkq097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Biase D, Lund PA. 2015. The Escherichia coli acid stress response and its significance for pathogenesis. Adv Appl Microbiol 92:49–88. doi: 10.1016/bs.aambs.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 22. Itou J, Eguchi Y, Utsumi R. 2009. Molecular mechanism of transcriptional cascade initiated by the EvgS/EvgA system in Escherichia coli K-12. Biosci Biotechnol Biochem 73:870–878. doi: 10.1271/bbb.80795 [DOI] [PubMed] [Google Scholar]
- 23. Masuda N, Church GM. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol Microbiol 48:699–712. doi: 10.1046/j.1365-2958.2003.03477.x [DOI] [PubMed] [Google Scholar]
- 24. Eguchi Y, Ishii E, Hata K, Utsumi R. 2011. Regulation of acid resistance by connectors of two-component signal transduction systems in Escherichia coli. J Bacteriol 193:1222–1228. doi: 10.1128/JB.01124-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eguchi Y, Itou J, Yamane M, Demizu R, Yamato F, Okada A, Mori H, Kato A, Utsumi R. 2007. B1500, a small membrane protein, connects the two-component systems EvgS/EvgA and phoQ/phoP in Escherichia coli. Proc Natl Acad Sci U S A 104:18712–18717. doi: 10.1073/pnas.0705768104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gorelik MG, Yakhnin H, Pannuri A, Walker AC, Pourciau C, Czyz D, Romeo T, Babitzke P. 2024. Multitier regulation of the E. coli extreme acid stress response by CsrA. J Bacteriol 206:e00354-23. doi: 10.1128/jb.00354-23 [DOI] [PMC free article] [PubMed] [Google Scholar]