ABSTRACT
Chlamydia trachomatis is an intracellular bacterial pathogen that undergoes a biphasic developmental cycle, consisting of intracellular reticulate bodies and extracellular infectious elementary bodies. A conserved bacterial protease, HtrA, was shown previously to be essential for Chlamydia during the reticulate body phase, using a novel inhibitor (JO146). In this study, isolates selected for the survival of JO146 treatment were found to have polymorphisms in the acyl-acyl carrier protein synthetase gene (aasC). AasC encodes the enzyme responsible for activating fatty acids from the host cell or synthesis to be incorporated into lipid bilayers. The isolates had distinct lipidomes with varied fatty acid compositions. A reduction in the lipid compositions that HtrA prefers to bind to was detected, yet HtrA and MOMP (a key outer membrane protein) were present at higher levels in the variants. Reduced progeny production and an earlier cellular exit were observed. Transcriptome analysis identified that multiple genes were downregulated in the variants especially stress and DNA processing factors. Here, we have shown that the fatty acid composition of chlamydial lipids, HtrA, and membrane proteins interplay and, when disrupted, impact chlamydial stress response that could trigger early cellular exit.
IMPORTANCE
Chlamydia trachomatis is an important obligate intracellular pathogen that has a unique biphasic developmental cycle. HtrA is an essential stress or virulence protease in many bacteria, with many different functions. Previously, we demonstrated that HtrA is critical for Chlamydia using a novel inhibitor. In the present study, we characterized genetic variants of Chlamydia trachomatis with reduced susceptibility to the HtrA inhibitor. The variants were changed in membrane fatty acid composition, outer membrane proteins, and transcription of stress genes. Earlier and more synchronous cellular exit was observed. Combined, this links stress response to fatty acids, membrane proteins, and HtrA interplay with the outcome of disrupted timing of chlamydial cellular exit.
KEYWORDS: intracellular, pathogen, protease
INTRODUCTION
Chlamydia (C.) trachomatis is a Gram-negative obligate intracellular bacterial pathogen and is a prevalent sexually transmitted infection. Infections can result in serious sequelae including pelvic inflammatory disease, tubal factor infertility, and ectopic pregnancies in women (1). All Chlamydia spp. have a biphasic developmental cycle with an extracellular infectious phase [elementary body (EB)] and an intracellular replicative phase [reticulate body (RB)] that is located inside a vacuole called the inclusion vacuole (2). As it has co-evolved with its host, Chlamydia spp. have undergone reductive evolution and considerable gene loss (3). Nonetheless, C. trachomatis has a near complete set of genes required for phospholipid synthesis (3, 4). C. trachomatis can synthesize the glycerophospholipids commonly found in Gram-negative bacterial membranes including phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin. However, it relies on the host to obtain the required precursors isoleucine, serine, and glucose (5). C. trachomatis can only synthesize saturated, but not unsaturated, fatty acids de novo (3, 4, 6). The PE phospholipid class and branched chain 15:0 fatty acid species are the most abundant C. trachomatis lipid species (4, 7). To incorporate unsaturated fatty acids and perhaps preserve resources, Chlamydia activates host cell-derived fatty acids using an acyl-acyl carrier protein synthetase (AasC). These activated fatty acids then enter the de novo biosynthetic pathways or type II fatty acid synthesis pathway for elongation (3, 4, 6).
A chemical biology approach using an inhibitor, JO146, identified the protein HtrA to be essential for the survival of C. trachomatis during the mid-replicative phase (8, 9). C. trachomatis HtrA is hypothesized to have a role in outer membrane protein stability, like its Escherichia (E.) coli ortholog DegP (10–12). While genetic manipulation strategies have advanced [reviewed in reference (13)] and it is now possible to implement most genetic approaches against chlamydia, high-throughput genetic methods remain limited. Hence, we implemented a random mutation and selection protocol, to further characterize the function of HtrA in Chlamydia. We hypothesized that mutants with resistance to the HtrA inhibitor, JO146, would identify factors that are functionally involved in HtrA’s essential role in the chlamydial replicative phase. We report the selection and characterization of three independently isolated genetic variants of C. trachomatis with reduced susceptibility to the previously described HtrA inhibitor JO146 (8); all three isolates had single nucleotide variants (SNVs) in aasC (ct_776). The variants had an impacted fatty acid composition, validating the SNV impact on function. Considerable phenotypic impacts were observed in the isolates along with transcriptional changes, implicating a stress response process that is likely linked to the early exit phenotype observed.
RESULTS
Chlamydial isolates with reduced susceptibility to the HtrA inhibitor JO146 all have single nucleotide variants in loci related to fatty acids
A selection experiment was conducted using the repeated passage of Ethyl methanesulfonate (EMS) mutated and non-mutated pools of Chlamydia in the presence of JO146. The purpose was to select C. trachomatis isolates with resistance or reduced susceptibility to JO146 (see the supplementary results for full details). Sequence analysis of the pools of isolates that survived the selection conditions identified that four genetic hotspots were selected for through the experiment [Table S1; CT776 (aasC), CT206 (putative esterase), CT544 (uhpC), and CT587 (enolase)]. Three isolates, subsequently referred to as 1A3, 2A3, and 1B3, were cultured from independent selection pools, plaque-purified (Fig. S1), and tested to confirm reduced susceptibility to JO146 (Fig. 1a). 1A3 was the most susceptible isolate with similar infectious yields to the wild type (WT). 1B3 was less susceptible to JO146 than 1A3, while 2A3 was the least susceptible isolate. The isolates were characterized by whole genome sequencing, identifying that all three isolates had a distinct SNV in CT776, the gene encoding for aasC (Table S2). In the case of isolate 1B3, this was the only genetic variation detected on the entire genome of the isolate characterized, meaning CT776 is solely responsible for any changes observed. In the case of 1A3, one other variation, a G to A transition in a non-coding region was detected, which may indicate that the CT776 change is the only functionally relevant change. There were a number of mutations in 2A3 that may be relevant to the phenotype including a putative esterase CT206 that was a hotspot for selection in the overall experiment (see Table S2). CT776 is the only common gene with a SNV in all purified isolates with reduced susceptibility to JO146 and the only/major change in two of the isolates. In silico bioinformatics and structural modeling of the sequence variants indicated a likely impact on AasC function with all three SNVs located around what appears to be a pocket in the predicted structure with impacts on hydrogen bonding (Fig. S2).
Fig 1.
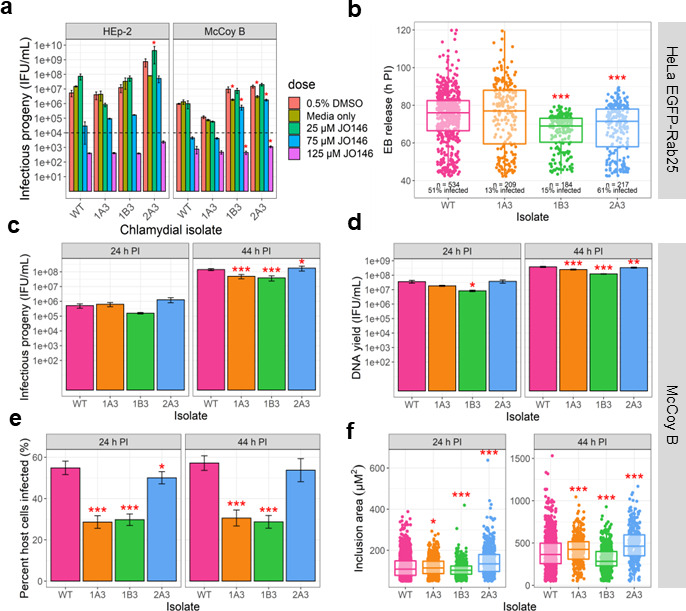
Analysis of phenotypes of variants isolated with reduced susceptibility to JO146. (a) Recovery of infectious progeny following JO146 treatment of chlamydial variants cultured in HEp-2 and McCoy B cell lines that were treated at 16 h post-infection (PI) with JO146 doses and harvested at 44 h PI, prior to re-infection to determine inclusion forming unit (IFU)/milliliter yields indicated in this figure. Error bars represent the standard error of the mean (SEM) of three independent experiments (n = 3). The dashed line indicates the threshold of accuracy (1 × 104 IFU/mL) for this enumeration method. *P < 0.05 compared to dimethylsulfoxide (DMSO) control as measured by two-way ANOVA with Dunnett’s multiple comparisons test. The % reductions for each isolate at 75 µM JO146 treatment relative to the solvent are HEp-2: WT: 99.46%, 1A3: 97.73%, 1B3: 98.68%, and 2A3: 92.87% and McCoy B: WT: 99.53%, 1A3: 94.39%, 1B3: 94.23%, and 2A3: 88.31%. (b) EB release from cells for each isolate. Real-time microscopy analysis of EB release from HeLa EGFP-Rab25 host cells is shown in hours post-infection (h PI) (y-axis) and variants (x-axis). The number of inclusions monitored and % infectivity are indicated under the data set for each variant. The microscopy was conducted from 42 to 120 h PI with data collected every 30 min; every visible inclusion in each field of view was monitored until no longer visible in the cell or the cell was also no longer visible, and this was recorded as the exit point. (c) Infectious progeny yields of isolates at 24 and 44 h PI. Three biological replicates were enumerated in duplicate for each isolate at each timepoint. (d) Yield of chromosomal DNA at 24 and 44 h PI, determined by quantitative polymerase chain reaction (qPCR). Three biological replicates were quantified in duplicate for each isolate at each timepoint. (e) The percent of McCoy B host cells infected by each isolate at 24 and 44 h PI. (f) Inclusion vacuole size at 24 and 44 h PI. The inclusion vacuole size was measured as a two-dimensional area (µM2). Triplicates of each isolate at each timepoint were visualized by microscopy with multiple fields of view or samples analyzed. Error bars represent SEM from multiple experiments. *P-value ≤0.05, **P-value ≤0.001, and ***P-value ≤0.0001, as measured by Student’s t-test with Holm–Sidak’s test for multiple comparisons.
In order to assess if the loci were isolated as an indirect impact related to JO146 and HtrA function rather than being a direct “off-target” protein that is bound by JO146, we first investigated if JO146 binds to either of the two proteins. We were not able to detect any evidence of binding of JO146 to either AasC or CT206. This was ruled out by AasC enzyme activity that showed no change when JO146 was added (Fig. S3). CT206 was analyzed as it was initially identified in the selection pools associated with the variant 1B3 and is confirmed to be mutated in 2A3. CT206 did not bind to JO146 (cy5) using recombinant purified protein (Fig. S3); full methods were outlined in the Supplementary Information.
Variants have differences in infectivity, inclusion size, progeny production, and exit time frames
In order to determine the impact of the variants or mutations on Chlamydia, a series of characterizations were conducted. Phenotypic analysis was conducted on cultures in McCoy B cells in comparison to the wild-type strain (referred to as CtDpp or WT), with infectious yield, chromosome counts, infectivity, and inclusion size assessed. This experiment was conducted in McCoy B cells as C. trachomatis has been observed to produce uniformly distributed growth in McCoy B cells versus patchy growth in epithelial cell lines (14). Variant 1B3 had significant reductions compared to WT in IFU production, DNA copy number, infectivity, and inclusion size (Fig. 1). Variant 1A3 also had reduced productivity, inclusion size, and infectivity compared to WT, although not to the same extent as observed for 1B3. A live microscopy experiment was used to assess chlamydial exit from the host cell, accounting for both inclusion vacuole extrusion and/or cell lysis. WT and 1A3 isolates had the same range and mean time of EB release (Fig. 1). EBs from both 1B3 and 2A3 variants were released earlier than both WT and 1A3 isolates (P-value <0.0001; mean time of EB release: WT = 74.0 h PI; 1A3 = 74.6 h PI; 1B3 = 65.9 h PI; 2A3 = 67.6 h PI). The overall range of EB release was the smallest (more synchronous and early) for the 1B3 variant, with all detected inclusion vacuoles completing lysis or extrusion by 80 h PI. An examination of the gross morphology of the chlamydial inclusion vacuoles, using confocal microscopy, throughout the culture phases revealed no apparent differences between the variants and WT (Fig. S3).
Variants have distinct lipid compositions throughout the developmental cycle
We conducted a lipidomic analysis at 24 and 44 h PI as the dominant role for fatty acids in bacterial cells is as constituents of the lipid bilayer. PE and PG lipid classes were analyzed as they are the major glycerophospholipid classes autonomously synthesized by C. trachomatis (5). A total of 116 lipids, including 75 PE species and 41 PG species, were detected with robust reproducibility. In some cases, in the analysis used here, more than one lipid (typically isomers) was detected at the same point in the chromatogram in the workflow used for this analysis; in order to ensure the accuracy of assignments or provide clarity when the assignment is ambiguous, dual assignment is listed in the figure/table. A comparison of WT samples and uninfected HEp-2 cells was initially performed to select lipids significantly increased in abundance during infection. HEp-2 cells were used as the overall yields, for all variants were greater in this cell line. As the WT samples (but not variant samples) were matched with uninfected host cells as controls, the exclusion of lipids not significantly associated with WT infection ensured that any significant differences between WT and variants were attributed to the bacteria. Prior to proceeding, it was confirmed that all the lipid species identified in the variants were also detected in the WT. A total of 30 lipid species, 21 PE and 9 PG, were significantly associated with C. trachomatis infection (Fig. 2). Of the 30 lipids associated with C. trachomatis infection, 24 contain the 15:0 fatty acid which is known to be abundant in C. trachomatis (4). A range of unsaturated fatty acids were identified to be significantly associated with infection (Table S3).
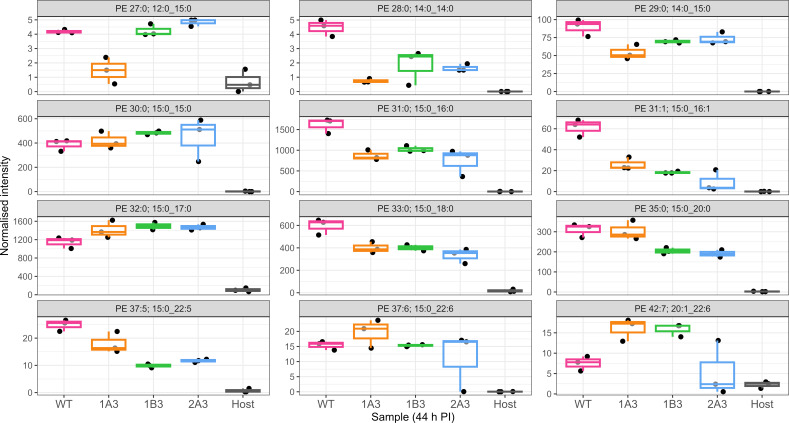
Fig 2.
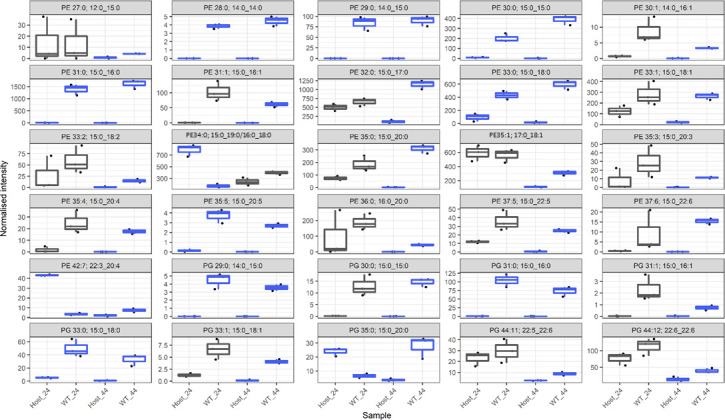
Normalized intensity of 30 lipid species with significantly different abundance in WT relative to uninfected host cells. This figure identifies the lipids that are significantly associated with infection. In order to focus further analysis of the differences in the variants and WT, subsequent analysis (Fig. 4 and 5) focused only on these lipids that are significantly associated with infection. The data are presented as box plots, with the x-axis indicating the sample as indicated in the bottom row. The normalized intensity is indicated by the y-axis. The y-axis is intentionally different for each box as significant differences for less abundant lipids may still be biologically relevant. Gray boxes indicate no significant difference at that timepoint (P > 0.05), and blue boxes indicate a significant difference (P < 0.05). Significance was measured via a t-test (n = 3) performed in MetaboAnalyst v5.0 as outlined in Materials and Methods. In some cases, dual assignments to the same MS2 feature occurred in different specimens; this is indicated by the multiple assignments at the top of the figure for that species, PE 34:0; 15:0_19:0/16:0_18:0, and PE 35:0; 15:0_20:0/17:0_18:0.
Across the lipids significantly associated with infection, we observe a substantial difference in the lipid profile of the variants compared to the WT control, with 25 significant differences identified at 24 h PI (Fig. 3; Fig. S5). In broad terms, there was an increase in most of the significantly different PE and PG species at 24 h PI with the increasing species all containing 15:0 except for PE 36:0 (PE 18:0_18:0). For example, the most abundant PE lipids associated with C. trachomatis infection, PE 32:0, 35:1, 34:0, 31:0, 36:0, and 33:1, were generally elevated in the variants compared to WT, and except for PE 31:0, all of these lipids were significantly elevated in either 1B3 or 1B3 and 2A3. The 1A3 variant appears to be following a similar trend although the differences fail to reach significance. The trend is similar in the PG species with three of the four most abundant species, PG 44:12, PG 44:11, and PG 35:0, showing a general increase among the variants (Fig. S5). A summary of the lipidomic data listing the infection-associated PE and PG lipids with significant differences between WT and any variant at either 24 or 44 h PI is shown in Table 1.
Fig 3.
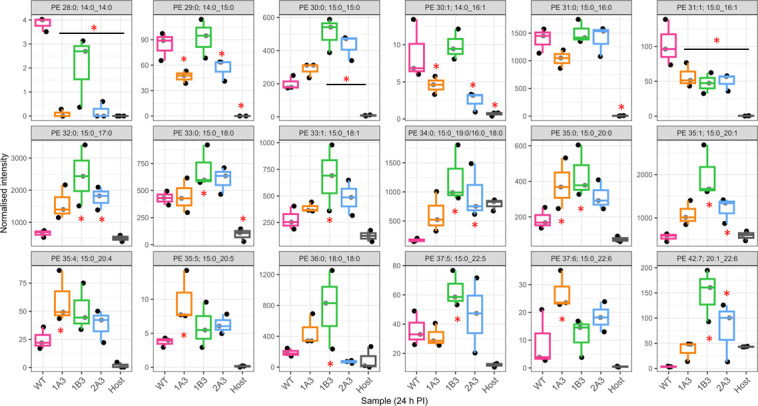
Normalized intensities of PE lipid species at 24 h PI in WT, variants, and host cells. The data are presented as box plots, color-coded for each strain indicated on the x-axis of each box. The normalized intensities are indicated on the y-axis. Different normalized intensities are used to show differences more clearly, as differences in low abundant lipids may still be biologically significant. Significance was measured by one-way ANOVA, performed using MetaboAnalyst as described in Materials and Methods. An asterisk indicates a significant difference (P < 0.05) in normalized intensity compared to WT (n = 3 each). The best match/assignment across the samples is identified in the graphs, noting that assignments can vary based on the methodology used. In some cases, more than one best match/assignment from the MS/MS data has potential; both are listed in the figure (e.g., PE34:0 15:0_19:0/16:0_18:0).
TABLE 1.
Summary of PE and PG lipids identified to have significant differences between WT and any variant at either 24 or 44 h PI
| Lipid species | 24 h log2 FC variant vs WTa | 44 h log2 FC variant vs WTa | ||||
|---|---|---|---|---|---|---|
| 1A3 | 1B3 | 2A3 | 1A3 | 1B3 | 2A3 | |
| PE | ||||||
| PE 28:0 (14:0_14:0) | −5.35 | −0.90 | −4.26 | −2.59 | −1.28 | −1.45 |
| PE 29:0 (14:0_15:0) | −0.85 | 0.14 | −0.58 | −0.74 | −0.37 | −0.29 |
| PE 30:0 (15:0_15:0) | 0.51 | 1.33 | 1.09 | 0.11 | 0.32 | 0.21 |
| PE 30:1 (14:0_16:1) | −0.94 | 0.17 | −1.82 | −1.57 | −1.45 | −0.2 |
| PE 31:0 (15:0_16:0) | −0.42 | 0.12 | 0.01 | −0.89 | −0.65 | −1.13 |
| PE 31:1 (15:0_16:1) | −0.85 | −1.12 | −1.04 | −1.23 | −1.75 | −2.77 |
| PE 32:0 (15:0_17:0) | 1.26 | 1.90 | 1.43 | 0.31 | 0.39 | 0.36 |
| PE 33:0 (15:0_18:0) | 0.05 | 0.70 | 0.49 | −0.58 | −0.57 | −0.84 |
| PE 33:1 (15:0_18:1) | 0.46 | 1.26 | 0.78 | 0.23 | −0.39 | 1.19 |
| PE 34:0 (15:0_19:0/16:0_18:0) | 1.85 | 2.85 | 2.48 | 0.33 | 0.3 | 0.78 |
| PE 35:0 (15:0_20:0) | 1.04 | 1.23 | 0.76 | −0.03 | −0.60 | −0.70 |
| PE 35:1 (15:0_20:1) | 0.96 | 1.83 | 1.12 | 0.44 | 0.1 | 0.97 |
| PE 35:4 (15:0_20:4) | 1.26 | 1.03 | 0.61 | 0.89 | −0.03 | 4.05 |
| PE 35:5 (15:0_20:5) | 1.40 | 0.68 | 0.75 | 0.97 | 0.09 | 4.04 |
| PE 36:0 (18:0_18:0) | 1.28 | 2.04 | −1.44 | 1.53 | 0.18 | 2.61 |
| PE 37:5 (15:0_22:5) | −0.19 | 0.80 | 0.37 | −0.47 | −1.33 | −1.09 |
| PE 37:6 (15:0_22:6) | 1.56 | 0.35 | 1.00 | 0.35 | −0.01 | −0.46 |
| PE 42:7 (20:1_22:6) | 3.34 | 5.36 | 4.46 | 1.09 | 1.07 | −0.49 |
| PG | ||||||
| PG 29:0 (14:0_15:0) | −0.98 | −0.06 | −0.44 | −0.74 | −0.36 | −0.66 |
| PG 30:0 (15:0_15:0) | 0.02 | 0.60 | 0.64 | 0.30 | 0.25 | 1.02 |
| PG 31:1 (15:0_16:1) | −5.47 | −3.36 | −4.60 | −3.55 | −3.88 | 5.57 |
| PG 33:0 (15:0_18:0) | −0.15 | 0.32 | −0.36 | −0.83 | −0.79 | −0.96 |
| PG 33:1 (15:0_18:1) | −1.13 | −0.28 | −0.49 | −1.28 | −1.99 | −0.74 |
| PG 35:0 (15:0_20:0) | 2.79 | 4.40 | 3.98 | −0.11 | −0.54 | −0.35 |
| PG 44:11 (22:5_22:6) | 0.83 | 2.26 | 0.90 | 0.79 | 0.32 | 1.26 |
| PG 44:12 (22:6_22:6) | 1.37 | 1.89 | 1.06 | 0.98 | 0.59 | 1.92 |
Bolded values indicate P < 0.05 (based on one-way ANOVA); non-bolded values indicate non-significant P-values.
In contrast to the general increased trend among the infection-associated PE and PG species, PE 28:0, 29:0, 30:1, and 31:1 as well as PG 29:0 and 31:1 all showed evidence of a decrease relative to WT. These lipids comprise all of the lipids associated with infection, which contain 14:0, and with the exception of PE/PG 31:1, they all contain this fatty acid. Similarly, PE/PG 31:1 and PE 30:1 were all decreased and comprise the set of 16:1 containing the lipids examined here. Taken together, this suggests that an underlying decrease in 14:0 and 16:1 incorporation in the bacterial membrane is associated with a reduced susceptibility to JO146.
Analysis of lipid intensities 44 h PI demonstrated significant differences between strains (adjusted P < 0.05) in 17 lipids, with 13 determined to be non-significant (Fig. 4; Fig. S6). PE 28:0 (14:0_14:0) was significantly lower in abundance in the variants compared to the WT result (at 24 h PI) (Fig. S6). PE 29:0 (14:0_15:0) was lower in abundance at 24 h PI in 1A3 and 2A3, and by 44 h PI, this lipid was also significantly decreased in 1B3. Conversely, at 24 h PI, no significant differences in PE 30:0 (15:0_15:0) were observed between isolates. However, PE 31:0 (15:0_16:0) was found to be significantly decreased in variants compared to WT, although no differences were seen at 24 h PI. There was significantly less PE 33:0 (15:0_18:0) and PE 37:5 (15:0_22:5) in all variants compared to WT. PE 32:0 (15:0_17:0) was increased in 1B3 and 2A3 at 24 h PI but was found to be significantly increased in all three variants at 44 h PI. It was the only lipid found to have an increased abundance in variants compared to WT at this timepoint. One PG species had a significantly smaller normalized intensity in all three variants compared to WT, PG 33:0 (15:0_18:0). 1A3 had significantly less PG 29:0 (14:0_15:0), which was also observed at 24 h PI. 1A3 and 1B3 had significantly less PG 33:1 (15:0_18:1) than WT, with a large variation in this lipid for 2A3.
Fig 4.
Normalized intensities of PE lipids at 44 h PI in WT, variants, and host cells. The data are presented as box plots, color-coded for each strain indicated on the x-axis of each box. The normalized intensities are indicated on the y-axis. Different normalized intensities are used to more clearly show differences, as differences in low abundant lipids may still be biologically significant. Significance was measured by one-way ANOVA, performed using MetaboAnalyst as described in Materials and Methods. Each species is indicated in the gray bar at the top; WT, variants, and host cell only are indicated on the x-axis. An asterisk indicates a significant difference (P < 0.05) in normalized intensity compared to WT (n = 3 each). The best match/assignment across the samples is identified in the graph.
Changes in lipid composition changes do not affect cellular access of JO146 but likely impact the HtrA–membrane interaction and the protein composition of the membranes
The reduced susceptibility to JO146 observed could be a consequence of the reduced cellular access of JO146 mediated by the distinct lipid compositions of the variants. A JO146–JO146-Cy5 cell permeability and competitive binding assay was performed at the replicative phase of growth to assess cellular access. Cultures were treated with JO146 (an irreversible binding mechanism), and then, competitive binding of JO146-Cy5 on lysates was subsequently conducted to determine if there were differences in access to the JO146 relative to the total HtrA levels. There was no significant difference in JO146 access and binding detected once the normalized levels of HtrA were analyzed (Fig. 5). These data, while semi-quantitative, suggest that the reduced susceptibility to JO146 does not relate to reduced cellular access of the compound.
Fig 5.
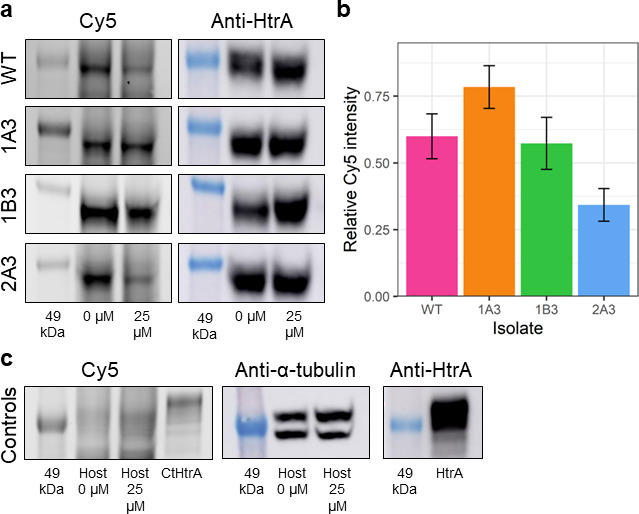
SDS-PAGE gels and western blots of WT and mutant chlamydial lysates pre-incubated with JO146 and competitive binding with JO146-Cy5. RBs from each isolate were pre-incubated with 0 or 25 µM JO146. Cultures were lysed and subsequently incubated with Cy5-JO146 in a competitive binding assay to indicate permeability to JO146 of the RB or replicative phase of the cultures (consistent with the most impactful time of JO146 treatment). (a) Representative SDS-PAGE gels (Cy5 gel) and western blots (anti-HtrA) for each chlamydial isolate, with the 49-kDa molecular weight marker indicated in the leftmost lane of each image. (b) Signal intensity of Cy5 following incubation with 25 µM JO146, relative to 0 µM, and normalized to the relative intensity of anti-HtrA, as described in the Supplementary Methods. (c) Representative SDS-PAGE gels and western blots for host-only uninfected controls and recombinant HtrA. These are controls to demonstrate gel loading consistency. Error bars represent the SEM (n = 3). Statistical differences in the normalized intensity of Cy5 in cells pre-incubated with 25 µM relative to the untreated controls were tested using one-way ANOVA. There was no significant difference between WT and any mutant.
The connection between HtrA inhibition (via the JO146 selection) and changes in the lipid composition identified here led us to test for a direct HtrA interaction with lipid bilayers. Tethered bilayer lipid membranes (tBLMs) in conjunction with electrical impedance spectroscopy were used to monitor changes in membrane conductance and capacitance that would only occur if a protein is binding to the membrane (15, 16). Changes in membrane conduction are due to a membrane disruption event altering the transport of ions in a solution across the membrane. Increases in membrane capacitances are an indication of a decrease in membrane thickness and/or the presence of water in the membrane. Recombinant HtrA was shown to cause an increase in both membrane conductance and capacitance in tBLMs, more favorably with negatively charged palmitoyl-oleoyl-phosphatidylglycerol (POPG) (phosphatidylglycerol head group—red lines) containing tBLMs than zwitterionic 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) compositions (phosphatidylethanolamine head group—black lines) (Fig. 6). These data indicate that, inside the cells, HtrA could be closely associated with the membrane.
Fig 6.
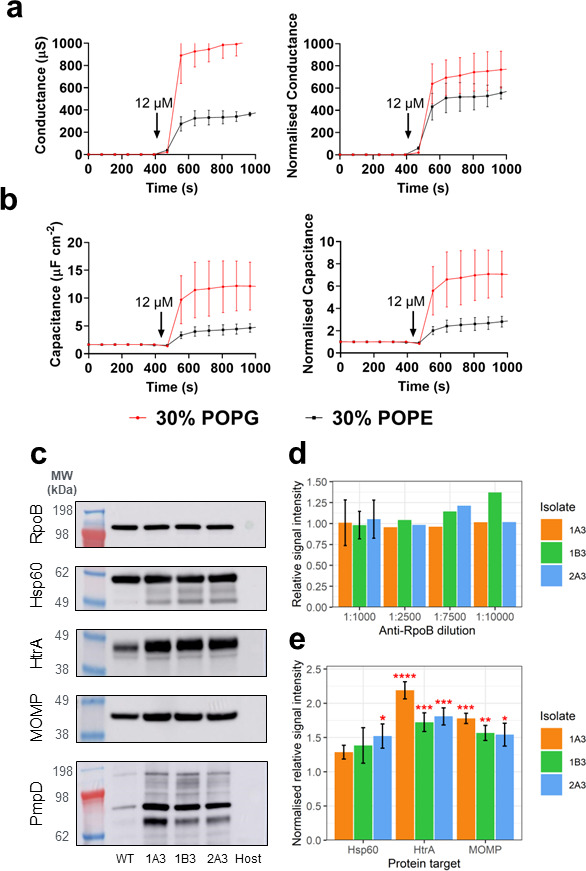
Chlamydial HtrA binding to tethered lipid bilayer and chlamydial protein levels. The figure shows the (a) membrane conductance and (b) capacitance changes (a measure of membrane thickness and/or water content) to lipid bilayers after the addition of HtrA. The data are membrane conductance (top, y-axis) and capacitance (bottom, y-axis) with the two distinct membrane compositions tested (red line 30% POPG, black line 30% POPE). The arrow indicates the timepoint that the recombinant protein was added. (c) Representative images of each western blot. Host, uninfected host-only control. Western blots of select stress response and membrane proteins in WT and variant chlamydial lysates. EBs from each isolate were harvested at 44 h PI from cultures infected with standardized multiplicity of infections (MOIs), and western blots were performed to assess the relative quantities of the proteins RpoB, Hsp60, HtrA, MOMP, and PmpD. (d) Signal intensity of RpoB across a dilution series, relative to WT. (e) Relative signal intensity of Hsp60, HtrA, and MOMP in each mutant, normalized to the mean RpoB relative intensity (PmpD was not analyzed due to the multiple bands). Error bars represent the SEM (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 compared to RpoB as measured by two-way ANOVA with Dunnett’s multiple comparisons test.
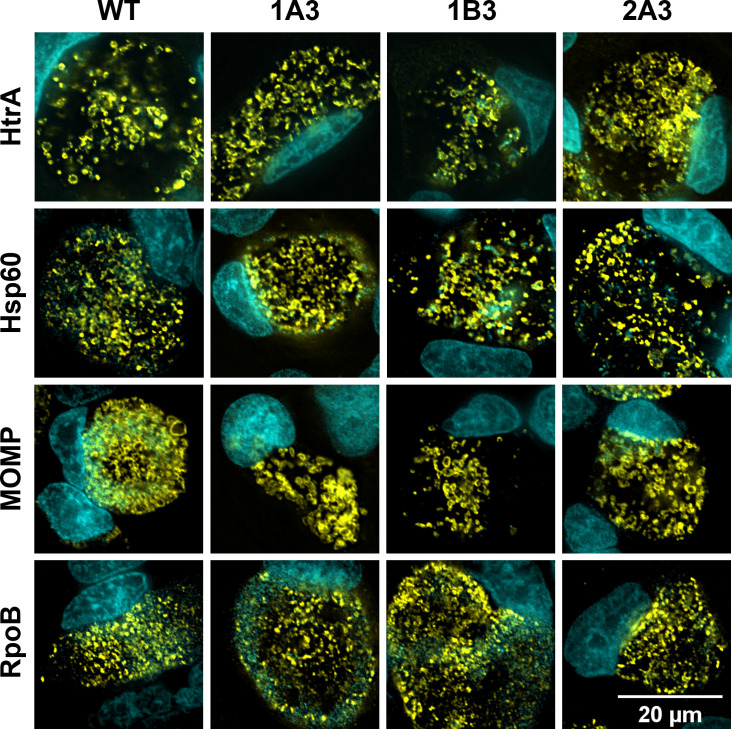
To determine the changed lipid profile impact on the abundance of key proteins, western blot analysis was conducted on selected protein targets on 44 h PI cultures. HtrA and the membrane proteins MOMP and PmpD were found to be higher in protein abundance in the variants than WT (Fig. 6). HtrA and MOMP intensities were measured using densitometry, and both were significantly higher than in WT (1.7–2.2× and 1.5–1.8× higher, respectively). In order to determine if this was mediated by transcriptional changes, we conducted reverse transcriptase quantitative PCR (RT-qPCR) analysis of these targets, aasC, and developmental stage genes. The transcript levels for almost all genes measured demonstrated no significant changes, with log2 fold change (FC) of <1 between WT and variants at 24 and 44 h PI (Fig. S7). The transcripts measured included aasC (CT776); htrA; hsp60, which, like htrA, is involved in the stress response; euo, a transcriptional repressor; two genes encoding outer membrane proteins, omcB and ompA (momp), which were assessed with 16S rRNA as the normalizing gene; and rpoB (an additional housekeeping gene). In 1B3 at 24 h PI, hsp60 was 1.3-fold downregulated (P-value <0.05); however, this result was not replicated at 44 h PI or in any other isolate (Fig. S7). The localization of chlamydial proteins (HtrA, MOMP, Hsp60, and RpoB) in WT and variants was assessed by confocal microscopy at 44 h PI in HEp-2 cell cultures and revealed no detectable differences in protein localization between any isolates at this resolution (Fig. 7).
Fig 7.
Protein localization in WT and variants. Microscopy images of infected cell cultures that were fixed to slides, and HtrA, Hsp60, MOMP, and RpoB proteins were immunocytochemically labeled, visible in yellow. Mammalian and bacterial DNA were labeled with DAPI, visible in cyan. The scale bar denotes 20 µM for all panels.
Variants have altered gene expression profiles for stress response and early and mid-developmental cycle genes
Given the changes in some protein abundance and HSP60 transcript change detected at 24 h PI, we considered that there may be other genes with distinct expression profiles in the variants. RNA-sequencing was conducted at 20, 36, and 44 h, and the transcriptome of the variants was compared to the WT (Table S6). In all variants compared to WT, 46, 5, and 7 genes were significantly downregulated at 20, 36, and 44 h, respectively (fold change <2, P-adjust <0.05) (Table 2). No upregulated genes were common in all variants compared to WT. CT158, a putative phospholipase gene, was significantly downregulated at 36 and 44 h PI. DNA processing-related genes, genes encoding virulence proteins, protein fate factors, and some developmental cycle phasing factors were impacted (including some Inc proteins and Tsp at 20 h PI).
TABLE 2.
Functional categories of genes downregulated in expression in all variants compared to wild type
| Total # genes downregulated in variants | Hypothetical protein | Deubiquitinase | Amino acids and derivatives | Co-factors, vitamins, prosthetic groups | DNA processing | Energy and precursor metabolites | Fatty acids, lipids, and isoprenoids | Functional category | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Porphyrin metabolism | Protein fate | RNA processing | Stress response, defense, virulence | Transporter/binding proteins | |||||||||
| 20h PI | 46 | 13 | 1 | 3 | 2 | 9 | 1 | 2 | 2 | 2b | 2 | 8a | 1 |
| 36h PI | 5 | 1 | 1 | 3a | |||||||||
| 48h PI | 7 | 4 | 1 | 2a | |||||||||
Including Inc proteins and phospholipase.
including tsp.
DISCUSSION
In this study, the selection of C. trachomatis variants with resistance to JO146, an inhibitor of HtrA, resulted in three isolates carrying SNVs in the aasC gene, encoding the acyl-acyl carrier protein synthetase. AasC is the enzyme responsible for activating and ligating fatty acids scavenged from the host for incorporation into phospholipids (5). The variants had distinct PE and PG lipid species when compared to the wild type. While there were differences between variants, there were consistent findings for all three variants, as described here and in Fig. 8. Figure 8 summarizes the impact of altered aasC observed across the variants and how this implicates HtrA function with membrane composition and chlamydial responses. Notably, there was a substantial increase in PE species containing the 15:0 fatty acid and a decrease in PE and PG species containing either 14:0 or 16:1 fatty acid in the JO146-resistant variants compared to WT. These results suggest that the variants may increase de novo fatty acid synthesis of 15:0 while concurrently reducing the incorporation of myristic (14:0) and palmitoleic (16:1) fatty acids derived from the host cell. CT158, a phospholipase gene that has previously been reported as likely involved in hydrolyzing host phosphatidylcholine for lipid update (17), was also significantly downregulated in gene expression at later stages of the developmental cycle for the variants, as were stress response and DNA processing factors. The variants had an increase in the abundance of HtrA and MOMP proteins, not mediated by transcription. The increased levels of these proteins are likely due to the variation in the physicochemical or biophysical properties of the membrane bilayer impacting protein interactions and stability. Recombinant HtrA showed a lower binding preference for lipids with a PE group compared to PG, which could indicate that HtrA may have less contact with the membrane in the variants that have higher PE abundance. Despite differences in the lipid profiles, the variants were not significantly less permeable to JO146 than WT. The isolate (1B3) with the most notable phenotypic differences, including reduced infectivity, reduced progeny production, and earlier exit, also had the most marked shifts in PE and PG lipid compositions.
Fig 8.
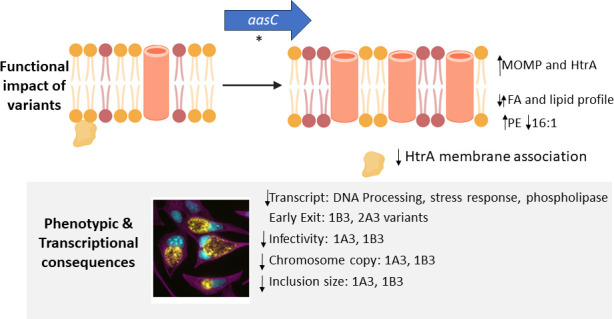
A model summarizing the functional impacts of AasC variants on the membrane and associated proteins and the observed downstream transcriptional and phenotypic impacts. The figure shows a model showing the findings of the role of AasC and how it relates to HtrA function. The figure at the top shows that the composition of the membrane shifted with aasC mutations to have changed lipid composition (purple indicating more PE head groups). The increased level of MOMP (outer membrane beta barrel shown in orange) and likely decreased association of HtrA (orange shape) with the membrane. The phenotypic and transcriptional changes observed in the variants due to these functional changes are shown in the lower gray box.
Given the asynchronous nature of chlamydial growth, it was important to validate the selection protocol. As the selection experiment resulted in the consistent selection of distinct variants in aasC in the three isolates characterized, this supports a stringent selection. All were associated with reduced susceptibility to JO146. A range of phenotypic changes were apparent in the variants; notably, a marked earlier exit and synchronicity in EB exit were apparent in two isolates, as well as reduced inclusion size and reduced infectious progeny and transcriptional differences with stress response and lipid-associated genes.
The consistent selection of SNVs in AasC led us to investigate the functional outcomes of the loci being impacted using a lipidomic approach. The untargeted liquid chromatography tandem mass spectrometry (LC-MS/MS) lipidomic analysis identified all PE and PG species previously reported for Chlamydia, validating that our findings are consistent with previous literature (5, 7, 18). The aasC variants had distinct lipid profiles compared to WT, indicating that the SNVs are impacting AasC selectivity. Several polyunsaturated species were increased in the variants but represented a relatively low overall constituent, while several abundant PE species had higher abundance in the variants, which could indicate a shift to increased overall PE composition of the membranes. These results indicate that the aasC variants have changed selectivity for fatty acids and that the variants are increasing PE species in general. The predicted structure suggests that the site of the mutations is all around a potential pocket that might be a substrate binding site. It is possible that the lipids that were observed to be significantly different between WT and variants at 24 h PI but not 44 h PI may reflect differences in RB vs EB numbers rather than differences in the lipid composition of RBs. For example, PE 30:0 (15:0_15:0) was significantly different in 1B3 and 2A3 compared to WT at 24 h PI but not at 44 h PI, owing to an increase in abundance in WT (log2 FC = 0.9, P = 0.05) and no change in the variants at 44 h PI. Thus, for this lipid, 1B3 and 2A3 already had a composition at 24 h PI that was consistent with the composition of all isolates at 44 h PI. The phenotypic data also indicated that 1B3 and 2A3 had a more “synchronous” or earlier overall release of EBs (several other growth parameters were impacted in 1B3).
There was also an increase in HtrA and MOMP protein abundance (in the absence of increased gene transcripts). However, the mechanism of JO146 resistance in these isolates cannot be explained entirely by the increase in HtrA protein, as the isolate most sensitive to JO146 (1A3) also produced the most HtrA. MOMP, PmpD, and other outer membrane proteins are predicted substrates of HtrA (10–12). As HtrA was significantly increased in the variants, if the increased MOMP is owing to an accumulation of unfolded protein, then this should be readily degraded and cleared by HtrA. Membrane lipid composition is known to affect membrane protein function and stability [reviewed in reference (19)]; thus, the variants may accommodate more MOMP (and possibly other proteins) in their membranes, or the proteins are more stable and accumulate over time to a greater extent. HtrA was found to have a reduced preference for lipid bilayers with more PE head groups, likely implicating a reduced association of HtrA with the membranes inside these variant cells. The differences in the transcriptome suggest an impact on the gene expression of the variants, with a variety of DNA processing, stress response, and virulence factors (e.g., Inc proteins) impacted from 20 to 28 h PI in all three variants.
This work demonstrates an interplay between HtrA, fatty acid metabolism, and membrane proteins in C. trachomatis. Similar biological interactions were previously reported in E. coli; therefore, we suggest that this may be a conserved mechanism found in many bacteria. Certainly, a link to stress response is well understood for many bacterial HtrA. However, the link via membrane compositions is less established; the evidence for this includes DegP (HtrA in E. coli) being upregulated by a lack of cellular PE and PG (20), increased lipoproteins (21), and accumulation of unfolded membrane proteins (22). DegP likely interacts with the inner membrane (23, 24) and forms multimeric structures on liposomes, depending on liposome fluidity (25). The phenotype and impacts here are also consistent with recent chlamydial work demonstrating that developmental cycle phases, such as the RB–EB transition, are based on an intrinsic signal (26).
In summary, the continued JO146 selection over serial passages has resulted in the selection of variants with altered phospholipid composition, increased levels of HtrA and outer membrane proteins, and several phenotypic impacts including earlier cellular exit and less infectious progeny production in the most impacted variant. This implies that HtrA and membrane protein–lipid compositions are part of the chlamydial stress response, which may impact cellular exit.
MATERIALS AND METHODS
Chlamydia culture and phenotypic assays
HEp-2 (human epithelial type 2, ATCC CCL-23) were used in most cell culture experiments and for all maintenance cultures and bulk growth. Chlamydia trachomatis D/UW-3/Cx (ATCC VR-885) was plaque-purified, and this isolate (CtDpp, referred to as WT) was used to generate the variants isolated during this study. Cell cultures were routinely conducted in a 96-well plate at a density of 25,000 cells/well. Chlamydia cultures were conducted after 24 h of fresh cell culture, when Chlamydia were added to the cultures at the stated MOI. The infection was routinely synchronized by centrifugation at 500 × g/28°C for 30 min, and at 4 h PI, infectious media was replaced with fresh supplemented media (where, for bulking or conducting growth curves, 1 µg/mL cycloheximide was added at this point). At 16 h PI, cells were treated with 25, 75, and 125 µM doses of JO146 in addition to media only (0 µM JO146) and 0.5% DMSO controls, and impacts were measured on cultures harvested at 44 h PI (or other timepoints as specified).
Chlamydia morphological analysis and related infectivity experiments were conducted in McCoy B (mouse fibroblasts, ATCC CRL-1696). A human epithelial HeLa (ATCC CCL-2) cell line stably expressing EGFP-Rab25 (27) was used for the live microscopy experiment. IFU enumeration was performed as previously described (28), imaged using the IN Cell Analyzer 2200 (Cytiva Life Sciences). Cultures were conducted with known multiplicity of infection, and in quantifications, these were either normalized to achieve consistent multiplicity of infection by adding different amounts of culture or normalizing to controls in data analysis depending on the experiment.
The generation of C. trachomatis variants using EMS treatment (0.2 mg/mL) and the determination of the rate of mutations were performed following a previously published procedure (29, 30), outlined in detail in the Supplementary Information (in HEp-2 cells). JO146 [Boc-Val-Pro-ValP(OPh)2] was synthesized by Dr. Joel Tyndall, School of Pharmacy, University of Otago, New Zealand, or sourced by commercial synthesis from GL Biochem (Shanghai, China). Cy5-JO146 ([Sulfo-Cyanine5]-Val-Pro-ValP(OPh)2) was purchased from Cambridge Research Biochemicals, UK, at >95% purity. All cell lines were confirmed every 3 months as Mycoplasma-free [MycoAlert Mycoplasma Detection Kit (Lonza, USA) as per the manufacturer’s instructions]. Chlamydia DNA content was determined via qPCR targeting the C. trachomatis tarP gene (Applied Biosystems, USA, assay ID Ba04646249_s1), in accordance with the manufacturer’s instructions.
Morphology was analyzed on cell cultures that were cultured, fixed, and immunolabeled essentially as previously described (using DAPI, anti-HtrA, and anti-α-tubulin) (28). Cells were imaged on the DeltaVision Elite microscope (Cytiva Life Sciences). To assess infectivity and inclusion size, five FOVs were imaged per slide using the 20× lens objective. To further evaluate morphology and infectivity, 100 fields of view (FOVs) per slide were imaged using the 60× oil objective. All images were deconvolved using the softWoRx software (Cytiva Life Sciences). The percent of host cells infected and inclusion size were measured manually in Fiji (31, 32).
The EB release assay was conducted in the HeLa EGFP-Rab25 cells cultured in six-well plates at a density of 285,000 cells/well. After 24 h, host cells were infected with C. trachomatis WT and isolates (1A3, 1B3, and 2A3) at a median MOI of 5. Uninfected host cells were also cultured. At 2 h PI, infectious media was removed and replaced with fresh supplemented Dulbecco’s modified eagle medium (DMEM) containing 1 µg/mL cycloheximide. Live cell imaging of cultures was started at 42 h PI, near the end of the developmental cycle but before lysis begins. Prior to imaging, culture media was replaced with DMEM without phenol red (product number 21063029, Gibco, USA) and 25 mM HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]. Cultures were imaged using the IN Cell Analyzer 2200 (Cytiva Life Sciences) with brightfield and fluorescein isothiocyanate (FITC channels), at the 20× objective. Images were captured every 30 min, from 42 to 120 h PI, and five FOVs were imaged per well. Images were analyzed using Fiji software (31, 32), where EB release and percent host cells infected were manually measured. The time of EB release was counted as the timepoint at which an inclusion was no longer visible, including both lysis and extrusion. Statistical analysis was performed using GraphPad Prism 7.
Permeability assay
HEp-2 cells were cultured onto six-well plates at a density of 300,000 cells/well. After 24 h, host cells were infected with variants and WT at a median MOI of 18. Cells were centrifuged at 500 × g/28°C for 30 min to synchronize, and media was replaced with fresh supplemented DMEM containing 1 µg/mL cycloheximide at 2 h PI. At 20 h PI, duplicates of each infection were treated with 0 or 25 µM JO146 (0.5% vol/vol DMSO). At 22 h PI, cells were washed with DMEM twice, then harvested via scraping into SPG buffer (250 mM sucrose, 10 mM sodium phosphate and 5 mM L-glutamic acid, pH 7.2). Cells were concentrated by centrifugation initially at 500 × g for 5 min, then at 18,000 × g/1°C for 30 min. Cells were resuspended in a minimal amount of Dulbecco’s Phosphate-Buffered Saline (dPBS), then probe-sonicated at 50% amplitude for 30 seconds to lyse host cells and RBs. Cells were treated with 12.5 µM of Cy5-JO146 and incubated at 37°C for 30 min to allow binding. Approximately 0.6 µg of recombinant CtHtrA was also treated with 1 µM Cy5-JO146 at 37°C for 30 min as a positive control. PAGE analysis of the extracts was conducted followed by imaging of the gels at 635 nm using Typhoon FLA 9500 (Cytiva Life Sciences). Western blots were performed as previously described (28). Densitometry analysis of western blots was performed with Image Studio Lite (LI-COR Biosciences, USA). Local background subtraction was performed, where the median intensity value of a 3-px-wide perimeter around each defined sample area was subtracted from the total pixel intensity for each sample. The intensity of Cy5 following pre-incubation with 25-µM JO146 (relative to 0 µM controls) was normalized to the intensity of anti-HtrA at 25 µM relative to 0 µM controls.
Immunoblot
Pooled EBs from C. trachomatis culture biological quadruplicates of each isolate (WT, 1A3, 1B3, and 2A3) and an uninfected host control lysate were prepared and analyzed for immunoblots. Relative protein densities were measured for the normalization of sample loading. Normalized loading between isolates was first confirmed via western blot using RpoB as a normalizing housekeeping protein, probed with anti-RpoB in a dilution series ranging from 1:1,000 to 1:10,000. Western blots were performed with other primary antibodies as follows: anti-Hsp60 diluted 1:5,000; anti-HtrA diluted 1:1,000; anti-MOMP diluted 1:1,000; and anti-PmpD diluted 1:1,000. Each blot was performed in triplicate, except for PmpD, which was performed in duplicate. Densitometry analysis of western blots with n = 3 was performed with Image Studio Lite (LI-COR Biosciences, USA).
Sequencing and analysis
The wild-type strain (CtDpp, or referred to as WT), cultured isolates from the experiments, and the original six pools from the selection experiment after three rounds of outgrowth without JO146 (passage 29) were whole genome-sequenced. DNA was extracted with the DNeasy mini kit (Qiagen) according to the manufacturer’s instructions. DNA libraries were prepared using the Nextera XT library preparation kit and sequenced on the Illumina MiSeq (2 × 300 bp). Trimmomatic (v 0.39) and FastQC (v 0.11.9) were used to trim and assess the read quality, respectively (33). After trimming, reads were mapped to the reference genome (C. trachomatis D/UW-3/Cx ASM872v1) using BWA mem (v 0.7.17) with default settings. Any unmapped reads were then mapped to the plasmid sequence (pCTDEC1 CP002053.1). SNV calling was performed using Bcftools mpileup (v 1.15.1) and filtered using the following criteria: base quality ≥20, number of reads supporting the SNV ≥20, and proportion of mapped reads supporting the SNV ≥70% (34). Due to the low coverage in some mutant pool samples, SNVs with <20 read supports were manually verified using IGV (v 2.15.4) to ensure the call is supported (35). For RNA-sequencing, McCoy cells were infected with WT and variants in biological triplicates and RNA-harvested at 20, 36, and 44 h post-infection using the RNeasy plus mini kit (Qiagen). Total RNA libraries were prepared using the Illumina stranded total RNA library kit with Ribo-Zero plus to deplete rRNA. The libraries were then sequenced on the NovaSeq 6000 S4 2 × 150 bp flow cell at the UNSW Ramaciotti Centre for Genomics. FastQC was used to assess the read quality, and the reads were then mapped to the C. trachomatis D/UW-3/Cx ASM872v1 reference using Salmon v1.9. DESeq2 (v1.38.3) was used to identify significantly differentially expressed genes (fold change >2 and P-adjust <0.05).
PCR, RT-PCR, and Sanger sequencing
Primers were manually designed to produce an amplicon of the appropriate regions of the genes CT206, CT390, CT664, and CT776, to evaluate the conservation of the polymorphisms selected in the original screen. RT-qPCR and primer design were conducted using standard methods [primer sequences and conditions are provided in Table S5 (36–38)]. The comparative Ct method (39) was used to calculate log2 FC, and a log2 FC ≥ 1 was set as the threshold of significance.
Lipidomics for comparative relative lipid profiles
The HEp-2 cell cultures of the isolates harvested at 24 and 44 h PI were used for lipidomic analysis. The amount of Chlamydia in each sample was normalized to an equivalent of 7 × 106 IFU for all infected samples. Host-only controls were matched to the average volume of WT used for each timepoint. To account for loss and variance during sample processing, 2.5 µg of SPLASH LIPIDOMIX Mass Spec Standard (Avanti Polar Lipids, USA) was added to each normalized sample as an internal standard. Lipid analysis was performed online by LC-MS/MS. Samples were injected onto a Q Exactive HF-X Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, USA) using a Vanquish Horizon Ultra-High Performance Liquid Chromatograph system (Thermo Fisher Scientific, USA) coupled to a 100 mm × 2.1 mm Accucore Vanquish C18 column (Thermo Fisher Scientific, USA). Sample loading, scan conditions, and coefficient of variation (CV) analysis are described in full in the Supplementary Materials. The obtained CV values are in line with those previously reported and demonstrate good technical reproducibility (40, 41). Lipids were identified by searching the MS/MS spectra of selected model samples (WT 24 h PI replicate #2, WT 44 h PI replicate #2, and PQC replicate #1) against LipidBlast (v10 Hiroshi Tsugawa fork) (42) and modified to include the labeled SPLASH standards using MSPepSearch (National Institute of Standards and Technology, USA). The extracted ion chromatograms corresponding to these putatively identified lipids were then extracted using MZmine 2.32 (43). Assignments corresponding to PE and PG species were then manually reviewed based on their retention times relative to other species of the same lipid class. Only PE and PG species were analyzed based on previous literature demonstrating these to be the main glycerophospholipid classes synthesized by C. trachomatis (4). The raw files were converted to mzXML files using MSConvert (44) with centroiding, which were then processed with MZmine 2.26 (43) using the targeted feature extraction module to extract ion chromatograms corresponding to the putatively identified PG and PE species or multiple possible species (full methodological details are provided in the Supplementary Materials). The lipids here were annotated at the molecular species level, meaning the constituent fatty acids are identified, but their sn-position or additional details such as double bond are not (45). The ambiguity with regard to sn-position is reflected in the use of an underscore between fatty acids in the shorthand nomenclature. This level of annotation is possible because of the formation of product ions in negative-mode PE and PG species characteristic of the constituent fatty acids but does not typically reflect their sn-position (46, 47). Data normalization and missing value imputation were performed using MetaboAnalyst v5.0 (48).
AasC assay
AasC was purified as described (4). The assays contained 100 mM Tris pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM ATP, 2 mM DTT, 100 µM S. aureus ACP, 150 µM [14C]palmitic acid, and AasC in a final volume of 50 µL. AasC was added last to start the reaction. Reactions were incubated at 37°C for 15 min; then, 40 µL was spotted on Whatman 3MM paper and dried to stop the reaction. The papers were washed twice for 20 min each in chloroform:methanol:acetic acid (3:6:1, vol/vol) and dried, and [14C]acyl-ACP formation was determined using a scintillation counter. Compound J0146 was dissolved in DMSO, and twofold serial dilutions were made. The final DMSO concentration in all assays was 4%.
Fatty acid extraction and analysis
Fatty acids were extracted from Chlamydia cultures 44 h PI and were analyzed as previously described using gas chromatography-mass spectrometry (49), compared to an external standard (Bacterial Acid Methyl Esters; 47,080 U, Sigma Aldrich).
tBLMs to assess HtrA binding to lipids
tBLMs were prepared using a “T10” architecture consisting of 10% benzyl-disulfide (tetra-ethyleneglycol) C20-phytanyl “tethering” molecules interspersed with 90% benzyl-disulfide-tetra-ethyleneglycol-OH “spacer” molecules analyzed for lipid binding, as previously described (15). These molecules were coordinated onto 2.1 mm2 gold tethering electrodes (SDx Tethered Membranes Pty Ltd, Australia) (16). Two different mobile lipid phases were investigated: either 70% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) with 30% POPG (mol/mol) or 70% POPC with 30% POPE (mol/mol) (Avanti Lipids, USA). Changes in membrane conduction and capacitance resulting from the addition of the recombinant HtrA protein [prepared as previously described (50)] were measured using AC electrical impedance spectrometry. This utilized a 50-mV peak-to-peak AC excitation spanning the frequency range of 0.1–2,000 Hz, with four steps per decade. The measurements were recorded using a TethaPod electrical impedance spectrometer operated with TethaQuick software (SDx Tethered Membranes Pty Ltd, Australia).
Statistical and data analysis
All experiment data (other than lipidomics) were analyzed, graphically displayed, and statistically analyzed using GraphPad Prism 7 and R (v4.2.2).
ACKNOWLEDGMENTS
The authors thank Professor Harlan Caldwell for the supply of PmpD antibody, Lazlo Kari (exchange and discussion on chlamydial genetics), and Joel Tyndall for the preparation and supply of JO146. The lipidomic part of this research was facilitated by access to Sydney Mass Spectrometry, a core research facility at the University of Sydney. The authors acknowledge the use of equipment and software in the Microbial Imaging Facility in the Faculty of Science at the University of Technology Sydney.
N.S. was supported by a UTS Faculty of Science HDR Scholarship. L.L. was supported by a UTS Chancellor’s Research Fellowship. V.O. was supported by a QUT HDR Tuition Fee Award and Supervisor Scholarship. This work was supported by the National Institutes of Health grant GM034496 (C.O.R.) and ALSAC, St. Jude Children’s Research Hospital. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
N.S. contributed to the design of the study, analysis and interpretation of the data, and drafting of the manuscript. L.L. contributed to the design of the study, analysis and interpretation of the data, and drafting of the manuscript. V.O. contributed to the design of the study, analysis and interpretation of the data relating to the initial selection experiment, and drafting of the manuscript. B.A.W. contributed to the analysis and interpretation of the data from the genomics and drafting of the manuscript. M.J.A.P. contributed to the analysis and interpretation of the data related to HtrA lipid binding and drafting of the manuscript. L.M. contributed to the analysis and interpretation of the data related to protein binding by JO146 and drafting of the manuscript. J.R.S. contributed to the analysis and interpretation of the lipidomic and culture data and drafting of the manuscript. C.K.B. contributed to the analysis and interpretation of the lipidomic data and drafting of the manuscript. C.G.C. contributed to the generation and interpretation of the HtrA lipid binding data and drafting of the manuscript. G.M. contributed to the analysis and interpretation of the data related to Chlamydia culture and drafting of the manuscript. R.M. contributed to the analysis and interpretation of the data related to chlamydial culture and drafting of the manuscript. C.R. contributed to the analysis and interpretation of the data related to AasC and drafting of the manuscript. P.T. contributed to the design of the study, analysis, and interpretation of the data and drafting of the manuscript. W.M.H. contributed to the design of the study, analysis and interpretation of the data, and drafting of the manuscript.
Contributor Information
Wilhelmina M. Huston, Email: Wilhelmina.Huston@uts.edu.au.
Michael Y. Galperin, NCBI, NLM, National Institutes of Health, Bethesda, Maryland, USA
DATA AVAILABILITY
Sequence and expression data is available at EBA Project Accession number PRJEB12312 and the corresponding project in the Genbank's Sequence Read Archive.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jb.00371-23.
Supplementary results, methods, tables, and figures.
RNAseq results.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Menon S, Timms P, Allan JA, Alexander K, Rombauts L, Horner P, Keltz M, Hocking J, Huston WM. 2015. Human and pathogen factors associated with Chlamydia trachomatis-related infertility in women. Clin Microbiol Rev 28:969–985. doi: 10.1128/CMR.00035-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. 2004. Chlamydial persistence: beyond the biphasic paradigm. Infect Immun 72:1843–1855. doi: 10.1128/IAI.72.4.1843-1855.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. doi: 10.1126/science.282.5389.754 [DOI] [PubMed] [Google Scholar]
- 4. Yao J, Dodson VJ, Frank MW, Rock CO. 2015. Chlamydia trachomatis scavenges host fatty acids for phospholipid synthesis via an Acyl-Acyl carrier protein synthetase. J Biol Chem 290:22163–22173. doi: 10.1074/jbc.M115.671008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yao J, Cherian PT, Frank MW, Rock CO. 2015. Chlamydia trachomatis relies on autonomous phospholipid synthesis for membrane biogenesis. J Biol Chem 290:18874–18888. doi: 10.1074/jbc.M115.657148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yao J, Abdelrahman YM, Robertson RM, Cox JV, Belland RJ, White SW, Rock CO. 2014. Type II fatty acid synthesis is essential for the replication of Chlamydia trachomatis. J Biol Chem 289:22365–22376. doi: 10.1074/jbc.M114.584185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wylie JL, Hatch GM, McClarty G. 1997. Host cell phospholipids are trafficked to and then modified by Chlamydia trachomatis. J Bacteriol 179:7233–7242. doi: 10.1128/jb.179.23.7233-7242.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gloeckl S, Ong VA, Patel P, Tyndall JDA, Timms P, Beagley KW, Allan JA, Armitage CW, Turnbull L, Whitchurch CB, Merdanovic M, Ehrmann M, Powers JC, Oleksyszyn J, Verdoes M, Bogyo M, Huston WM. 2013. Identification of a serine protease inhibitor which causes inclusion vacuole reduction and is lethal to Chlamydia trachomatis. Mol Microbiol 89:676–689. doi: 10.1111/mmi.12306 [DOI] [PubMed] [Google Scholar]
- 9. Ong VA, Marsh JW, Lawrence A, Allan JA, Timms P, Huston WM. 2013. The protease inhibitor JO146 demonstrates a critical role for CtHtrA for Chlamydia trachomatis reversion from penicillin persistence. Front Cell Infect Microbiol 3:100. doi: 10.3389/fcimb.2013.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huston WM, Tyndall JDA, Lott WB, Stansfield SH, Timms P. 2011. Unique residues involved in activation of the multitasking protease/chaperone HtrA from Chlamydia trachomatis. PLoS One 6:e24547. doi: 10.1371/journal.pone.0024547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marsh JW, Lott WB, Tyndall JDA, Huston WWM. 2013. Proteolytic activation of Chlamydia trachomatis HTRA is mediated by PDZ1 domain interactions with protease domain loops L3 and LC and beta strand beta5. Cell Mol Biol Lett 18:522–537. doi: 10.2478/s11658-013-0103-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marsh JW, Ong VA, Lott WB, Timms P, Tyndall JD, Huston WM. 2017. CtHtrA: the lynchpin of the chlamydial surface and a promising therapeutic target. Future Microbiol 12:817–829. doi: 10.2217/fmb-2017-0017 [DOI] [PubMed] [Google Scholar]
- 13. Wan W, Li D, Li D, Jiao J. 2023. Advances in genetic manipulation of Chlamydia trachomatis. Front Immunol 14:1209879. doi: 10.3389/fimmu.2023.1209879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guseva NV, Dessus-Babus S, Moore CG, Whittimore JD, Wyrick PB. 2007. Differences in Chlamydia trachomatis serovar E growth rate in polarized endometrial and endocervical epithelial cells grown in three-dimensional culture. Infect Immun 75:553–564. doi: 10.1128/IAI.01517-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berry T, Dutta D, Chen R, Leong A, Wang H, Donald WA, Parviz M, Cornell B, Willcox M, Kumar N, Cranfield CG. 2018. Lipid membrane interactions of the cationic antimicrobial peptide chimeras melimine and cys-melimine. Langmuir 34:11586–11592. doi: 10.1021/acs.langmuir.8b01701 [DOI] [PubMed] [Google Scholar]
- 16. Cranfield CG, Cornell BA, Grage SL, Duckworth P, Carne S, Ulrich AS, Martinac B. 2014. Transient potential gradients and impedance measures of tethered bilayer lipid membranes: pore-forming peptide insertion and the effect of electroporation. Biophys J 106:182–189. doi: 10.1016/j.bpj.2013.11.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nelson DE, Crane DD, Taylor LD, Dorward DW, Goheen MM, Caldwell HD. 2006. Inhibition of chlamydiae by primary alcohols correlates with the strain-specific complement of plasticity zone phospholipase D genes. Infect Immun 74:73–80. doi: 10.1128/IAI.74.1.73-80.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bidawid S, Chou S, Ng CW, Perry E, Kasatiya S. 1989. Fatty acid profiles of Chlamydia using capillary gas chromatography. Antonie Van Leeuwenhoek 55:123–131. doi: 10.1007/BF00404752 [DOI] [PubMed] [Google Scholar]
- 19. Zhang YM, Rock CO. 2008. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol 6:222–233. doi: 10.1038/nrmicro1839 [DOI] [PubMed] [Google Scholar]
- 20. Rowlett VW, Mallampalli VKPS, Karlstaedt A, Dowhan W, Taegtmeyer H, Margolin W, Vitrac H. 2017. Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J Bacteriol 199:e00849-16. doi: 10.1128/JB.00849-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miyadai H, Tanaka-Masuda K, Matsuyama S, Tokuda H. 2004. Effects of lipoprotein overproduction on the induction of DegP (HtrA) involved in quality control in the Escherichia coli periplasm. J Biol Chem 279:39807–39813. doi: 10.1074/jbc.M406390200 [DOI] [PubMed] [Google Scholar]
- 22. Meltzer M, Hasenbein S, Mamant N, Merdanovic M, Poepsel S, Hauske P, Kaiser M, Huber R, Krojer T, Clausen T, Ehrmann M. 2009. Structure, function and regulation of the conserved serine proteases DegP and DegS of Escherichia coli. Res Microbiol 160:660–666. doi: 10.1016/j.resmic.2009.07.012 [DOI] [PubMed] [Google Scholar]
- 23. Skórko-Glonek J, Lipińska B, Krzewski K, Zolese G, Bertoli E, Tanfani F. 1997. HtrA heat shock protease interacts with phospholipid membranes and undergoes conformational changes. J Biol Chem 272:8974–8982. doi: 10.1074/jbc.272.14.8974 [DOI] [PubMed] [Google Scholar]
- 24. Krojer T, Pangerl K, Kurt J, Sawa J, Stingl C, Mechtler K, Huber R, Ehrmann M, Clausen T. 2008. Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc Natl Acad Sci U S A 105:7702–7707. doi: 10.1073/pnas.0803392105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shen QT, Bai XC, Chang LF, Wu Y, Wang HW, Sui SF. 2009. Bowl-shaped oligomeric structures on membranes as DegP's new functional forms in protein quality control. Proc Natl Acad Sci U S A 106:4858–4863. doi: 10.1073/pnas.0811780106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chiarelli TJ, Grieshaber NA, Omsland A, Remien CH, Grieshaber SS. 2020. Single-inclusion kinetics of Chlamydia trachomatis development. mSystems 5:e00689-20. doi: 10.1128/mSystems.00689-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kerr MC, Gomez GA, Ferguson C, Tanzer MC, Murphy JM, Yap AS, Parton RG, Huston WM, Teasdale RD. 2017. Laser-mediated rupture of chlamydial inclusions triggers pathogen egress and host cell necrosis. Nat Commun 8:14729. doi: 10.1038/ncomms14729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huston WM, Theodoropoulos C, Mathews SA, Timms P. 2008. Chlamydia trachomatis responds to heat shock, penicillin induced persistence, and IFN-gamma persistence by altering levels of the extracytoplasmic stress response protease HtrA. BMC Microbiol 8:190. doi: 10.1186/1471-2180-8-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, Virok D, Rajaram K, Endresz V, McClarty G, Nelson DE, Caldwell HD. 2011. Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci U S A 108:7189–7193. doi: 10.1073/pnas.1102229108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Binet R, Maurelli AT. 2005. Frequency of spontaneous mutations that confer antibiotic resistance in Chlamydia spp. Antimicrob Agents Chemother 49:2865–2873. doi: 10.1128/AAC.49.7.2865-2873.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data PS . 2009. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Bioinformatics 25:2078–2079. doi: 10.6084/M9.FIGSHARE.963153.V1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bashmakov YK, Zigangirova NA, Pashko YP, Kapotina LN, Petyaev IM. 2010. Chlamydia trachomatis growth inhibition and restoration of LDL-receptor level in HepG2 cells treated with mevastatin. Comp Hepatol 9:3. doi: 10.1186/1476-5926-9-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huston WM, Lawrence A, Wee BA, Thomas M, Timms P, Vodstrcil LA, McNulty A, McIvor R, Worthington K, Donovan B, Phillips S, Chen MY, Fairley CK, Hocking JS. 2022. Repeat infections with chlamydia in women may be more transcriptionally active with lower responses from some immune genes. Front Public Health 10:1012835. doi: 10.3389/fpubh.2022.1012835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nunes A, Gomes JP, Mead S, Florindo C, Correia H, Borrego MJ, Dean D. 2007. Comparative expression profiling of the Chlamydia trachomatis pmp gene family for clinical and reference strains. PLoS One 2:e878. doi: 10.1371/journal.pone.0000878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 40. Drotleff B, Lämmerhofer M. 2019. Guidelines for selection of internal standard-based normalization strategies in untargeted lipidomic profiling by LC-HR-MS/MS. Anal Chem 91:9836–9843. doi: 10.1021/acs.analchem.9b01505 [DOI] [PubMed] [Google Scholar]
- 41. Katajamaa M, Oresic M. 2005. Processing methods for differential analysis of LC/MS profile data. BMC Bioinformatics 6:179. doi: 10.1186/1471-2105-6-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kind T, Liu K-H, Lee DY, DeFelice B, Meissen JK, Fiehn O. 2013. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods 10:755–758. doi: 10.1038/nmeth.2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pluskal T, Castillo S, Villar-Briones A, Oresic M. 2010. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11:395. doi: 10.1186/1471-2105-11-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Holman JD, Tabb DL, Mallick P. 2014. Employing ProteoWizard to convert raw mass spectrometry data. Curr Protoc Bioinformatics 46:13. doi: 10.1002/0471250953.bi1324s46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liebisch G, Vizcaíno JA, Köfeler H, Trötzmüller M, Griffiths WJ, Schmitz G, Spener F, Wakelam MJO. 2013. Shorthand notation for lipid structures derived from mass spectrometry. J Lipid Res 54:1523–1530. doi: 10.1194/jlr.M033506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cao W, Cheng S, Yang J, Feng J, Zhang W, Li Z, Chen Q, Xia Y, Ouyang Z, Ma X. 2020. Large-scale lipid analysis with C=C location and sn-position isomer resolving power. Nat Commun 11:375. doi: 10.1038/s41467-019-14180-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee H-C, Yokomizo T. 2018. Applications of mass spectrometry-based targeted and non-targeted lipidomics. Biochem Biophys Res Commun 504:576–581. doi: 10.1016/j.bbrc.2018.03.081 [DOI] [PubMed] [Google Scholar]
- 48. Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, Gauthier C, Jacques P-É, Li S, Xia J. 2021. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 49:W388–W396. doi: 10.1093/nar/gkab382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sasser M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. In MIDI technical NOTE 101. MIDI inc, Newark, DE. [Google Scholar]
- 50. Huston WM, Swedberg JE, Harris JM, Walsh TP, Mathews SA, Timms P. 2007. The temperature activated HtrA protease from pathogen Chlamydia trachomatis acts as both a chaperone and protease at 37 degrees C. FEBS Lett 581:3382–3386. doi: 10.1016/j.febslet.2007.06.039 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary results, methods, tables, and figures.
RNAseq results.
Data Availability Statement
Sequence and expression data is available at EBA Project Accession number PRJEB12312 and the corresponding project in the Genbank's Sequence Read Archive.