Abstract
Purpose:
TPST-1120 is a first-in-class oral inhibitor of peroxisome proliferator-activated receptor α (PPARα), a fatty acid ligand-activated transcription factor that regulates genes involved in fatty acid oxidation, angiogenesis, and inflammation, and is a novel target for cancer therapy. TPST-1120 displayed antitumor activity in xenograft models and synergistic tumor reduction in syngeneic tumor models when combined with anti-PD-1 agents.
Experimental Design:
This phase I, open-label, dose-escalation study (NCT03829436) evaluated TPST-1120 as monotherapy in patients with advanced solid tumors and in combination with nivolumab in patients with renal cell carcinoma (RCC), cholangiocarcinoma (CCA), or hepatocellular carcinoma. Objectives included evaluation of safety, pharmacokinetics, pharmacodynamics, and preliminary antitumor activity (RECIST v1.1).
Results:
A total of 39 patients enrolled with 38 treated (20 monotherapy, 18 combination; median 3 prior lines of therapy). The most common treatment-related adverse events (TRAE) were grade 1–2 nausea, fatigue, and diarrhea. No grade 4–5 TRAEs or dose-limiting toxicities were reported. In the monotherapy group, 53% (10/19) of evaluable patients had a best objective response of stable disease. In the combination group, 3 patients had partial responses, for an objective response rate of 20% (3/15) across all doses and 30% (3/10) at TPST-1120 ≥400 mg twice daily. Responses occurred in 2 patients with RCC, both of whom had previously progressed on anti-PD-1 therapy, and 1 patient with late-line CCA.
Conclusions:
TPST-1120 was well tolerated as monotherapy and in combination with nivolumab and the combination showed preliminary evidence of clinical activity in PD-1 inhibitor refractory and immune compromised cancers.
Significance:
TPST-1120 is a first-in-class oral inhibitor of PPARα, whose roles in metabolic and immune regulation are implicated in tumor proliferation/survival and inhibition of anticancer immunity. This first-in-human study of TPST-1120 alone and in combination with nivolumab supports proof-of-concept of PPARα inhibition as a target of therapeutic intervention in solid tumors.
Introduction
Metabolic reprogramming and evasion of immune destruction are hallmarks of cancer (1). While the most widely recognized metabolic adaptation in cancer is an increase in aerobic glycolysis (the Warburg effect), cancer cells can utilize other metabolic pathways such as increased fatty acid oxidation (FAO) to support tumorigenesis. FAO also promotes stemness, drug resistance, and metastasis (2–4), and modulates immune cell function within the tumor microenvironment, which enables tumors to evade antitumor immune responses (5–7). Enhanced FAO is described in multiple cancers and has been reported to correlate with poor patient outcomes (8).
Peroxisome proliferator-activated receptor α (PPARα) is a fatty acid ligand-activated transcription factor that controls the expression of over 100 genes involved in FAO, angiogenesis, and inflammation (9–11). PPARα is critical for maintaining physiologic metabolic homeostasis under conditions when fatty acids are the predominant source of energy, such as during fasting or diet-induced lipid overload. In addition to upregulating genes involved in FAO, activated PPARα dampens Th1-promoting inflammatory responses to metabolic disturbances by directly enhancing transcription of anti-inflammatory proteins, such as IκBα, and by antagonizing the activity of proinflammatory transcription factors, such as NFκB and AP-1, through transrepression, a mechanism involving direct protein–protein interactions (12, 13).
Because of its critical roles in metabolic regulation and immune function, PPARα has emerged as a target of interest for cancer therapy. The dual effect of PPARα both promoting tumor cell growth and inhibiting anticancer immunity is supported by preclinical experiments in which Ppara deficiency in either implanted tumor cells or recipient host reduced tumor growth in vivo, while Ppara deficiency in both implanted tumor cells and recipient host resulted in significantly greater tumor inhibition than in either compartment alone (14). The critical antitumor role of PPARα inhibition specifically in immune cells was further demonstrated in chimeric animals where bone marrow from Ppara knockout (KO) mouse transplanted into a Ppara wild-type animal, but not the reverse, profoundly inhibited tumor growth (14).
TPST-1120 is a first-in-class, oral, small molecule, competitive antagonist of PPARα, with nanomolar potency (IC50 0.04 µmol/L) for human PPARα and high specificity (>250-fold) for PPARα over the other PPAR isoforms (PPAR β/δ and γ; ref. 15). In xenograft and syngeneic tumor models, TPST-1120 inhibited tumor growth in vivo as monotherapy, and the combination of TPST-1120 plus anti-PD-1 therapy resulted in synergistic tumor reduction and durable antitumor immunity in multiple syngeneic mouse models (16). Adoptive transfer of splenocytes from syngeneic mice bearing MC38 colon tumors treated and cured with TPST-1120 plus anti-PD-1 conferred resistance to tumor challenge in naïve mice, similar to the results observed in Ppara KO studies (16). This first-in-human study was conducted to evaluate TPST-1120 as monotherapy and in combination with the PD-1 inhibitor nivolumab in patients with select advanced solid tumors.
Materials and Methods
Study Design
This was a first-in-human, phase I, open-label, 3+3 design, dose-escalation study (ClinicalTrials.gov identifier: NCT03829436). The primary study objectives were to investigate the safety and tolerability, and to determine the MTD or optimal biological dose (OBD), of TPST-1120 as monotherapy and in combination with nivolumab. Key additional objectives included evaluation of pharmacokinetics, pharmacodynamics, and preliminary assessment of anticancer activity. Exploratory objectives included investigation of immunomodulatory effects of treatment in peripheral blood.
The trial was designed by employees of the study Sponsor, Tempest Therapeutics, in collaboration with the study investigators.
Patient Selection
Key inclusion criteria included diagnosis of advanced/metastatic solid tumor previously treated with standard systemic therapy for the disease; Eastern Cooperative Oncology Group (ECOG) performance status of 0–1 with estimated life expectancy of at least 12 weeks; adequate organ function; and measurable disease according to the RECIST version 1.1. Tumor types eligible for monotherapy dose escalation [cholangiocarcinoma (CCA), colorectal cancer, metastatic castration-resistant prostate cancer, gastroesophageal cancer, hepatocellular carcinoma (HCC), non–small cell lung cancer, pancreatic cancer, renal cell carcinoma (RCC), sarcoma, head and neck squamous cell carcinoma, triple-negative breast cancer, and urothelial bladder cancer] were selected based upon elevated expression of PPARA and associated genes in The Cancer Genome Atlas in these indications. In the nivolumab combination portion of the study, eligible tumor types were limited to those displaying the highest expression of PPARA and associated genes: clear cell RCC, CCA, and HCC (16). Key exclusion criteria included history of intolerable or unresolved immune-related adverse event (AE) resulting from prior immune checkpoint inhibitor (ICI) therapy; ongoing use of immunosuppressive medications or active autoimmune disease; untreated/active central nervous system metastases; or use of fibrates within 28 days of enrollment.
Treatment Plan
Monotherapy TPST-1120 was administered orally in 21-day cycles until disease progression or unacceptable toxicity with a starting dose of 100 mg twice daily (200 mg/day). In a standard 3+3 design, patients were sequentially enrolled at progressively higher dose levels of TPST-1120, evaluating 100, 200, 300, 400, and 600 mg twice daily.
Dose escalation of TPST-1120 in combination with nivolumab was initiated after the 300 mg twice daily (600 mg/day) monotherapy cohort successfully cleared the dose-limiting toxicity (DLT) evaluation period. TPST-1120 was evaluated in 28-day cycles at progressively higher dose levels of 200, 300, 400, and 600 mg orally twice daily in combination with standard dose nivolumab (480 mg intravenous infusion every 4 weeks). Treatment continued until disease progression or unacceptable toxicity.
In all dose cohorts, TPST-1120 was taken with food and with water. Patients were required to fast for a minimum of 8 hours prior to protocol-specified laboratory assessments on cycle 1 day 1, cycle 1 day 8, cycle 2 day 1, and cycle 3 day 1.
Efficacy and Safety Assessments
Safety was assessed on the basis of incidence of AEs, laboratory results, and physical examinations on day 1 and day 8 of the first cycle, then on day 1 of each subsequent cycle. AEs were recorded by Common Toxicity Criteria for Adverse Events v5.0 from the first dose of study therapy through 28 days after the last dose for monotherapy and 90 days after the last treatment dose for combination therapy.
Disease assessments using CT scans or MRI were performed at baseline and on day 1 of cycle 3, day 1 of cycle 5, and every 9 weeks thereafter for monotherapy, and day 1 of every odd cycle thereafter for combination therapy. Antitumor response was assessed using RECIST v1.1.
Pharmacokinetics and Pharmacodynamics
Blood for pharmacokinetic and research assessments was collected at screening, cycle 1 days 1, 2, and 8, cycle 2 day 1, cycle 3 day 1, cycle 4 day 1 (combination only), and cycle 5 day 1 (monotherapy only). Tumor biopsies were not required.
Plasma TPST-1120 concentrations were quantified using a validated tandem mass spectrometry assay. Noncompartmental analysis was performed on observed data following the first dose and at steady state on day 8. To assess pharmacodynamic gene expression changes, RNA was extracted from whole blood samples collected in PAXgene Blood RNA tubes on cycle 1 days 1 and 8 and cycle 3 day 1 and assayed on the nCounter instrument (NanoString, Inc.) using the nCounter PanCancer Immune Profiling panel supplemented with an additional 30 PPARα-associated genes (Supplementary Table S1) according to manufacturer's protocols. Associations between gene expression changes and TPST-1120 exposure levels were assessed by linear regression analysis between AUC0–24 and baseline-normalized values on day 8, adjusting for the FDR using the method of Benjamini and Hochberg (ref. 17; FDR P < 0.05) and demonstrating an effect size greater than 0.5. Similar exposure-related changes in expression had to be observed on cycle 3 day 1 for a gene to be denoted as a pharmacodynamic biomarker. Differences in gene expression change magnitudes between exposure tertiles were assessed using Wilcoxon pairwise method (α = 0.05). To identify associations of gene changes with clinical response, patients were stratified on the basis of best overall response (BOR), and linear discriminant analysis (LDA) was performed to identify gene expression changes on day 8 associated with each response category. Change magnitudes of identified genes were compared between partial responders and stable disease (SD) or progressive disease (PD) patients using Mann–Whitney U tests (α = 0.05).
Lipidomics analysis was performed by the UCLA Lipidomics Lab using a LC/MS technique, as described previously (18). Data were analyzed using the method of Su and colleagues (19).
DLT Definition and OBD Determination
Dose escalation followed a standard 3+3 design with a minimum of 3 patients assigned per dose level. If 0 of 3 or 1 of 6 patients experienced a DLT, dose-escalation continued to the next higher dose level cohort until the MTD was identified or evaluation of the top protocol-defined dose level was completed. Six patients were to be enrolled in the highest dose level cohort that did not exceed the MTD. The MTD was exceeded if >1 of 3 or ≥2 of 6 patients experienced a DLT.
DLTs were evaluated during the first treatment cycle (21 days for monotherapy and 28 days for TPST-1120 plus nivolumab). Patients were evaluable for DLTs if they received at least 85% of planned TPST-1120 doses (and all planned nivolumab doses for combination patients) in the first treatment cycle unless this exposure threshold was not met due to a DLT. Patients who did not meet the DLT evaluability criteria were replaced. AEs occurring during the first treatment cycle and assessed as related to study treatment were considered DLTs following criteria established in the study protocol. OBD determination was based on emerging pharmacokinetics, any pharmacokinetic/pharmacodynamic relationships which could be established, and overall safety and tolerability.
Statistical Methods
Analysis was performed using descriptive statistics. All patients who received at least one dose of TPST-1120 were included in the safety analysis.
Overall response rate (ORR) was defined as the proportion of patients with complete response or partial response (PR). Disease control rate (DCR) was defined as ORR + SD of at least one scan. The efficacy population (EP) comprised all safety-evaluable patients who had at least one postbaseline tumor assessment, as well as patients who discontinued from study treatment due to PD without undergoing a follow-up radiographic assessment.
Study Approval
This study was reviewed and approved by the individual site Institutional Review Boards and/or ethics committees where the study was opened and by the FDA. The study was conducted in accordance with the provisions of the Declaration of Helsinki and the International Conference on Harmonization Guidelines for Good Clinical Practice. All patients provided written informed consent.
Data Availability
The data generated in this study are available upon request from the corresponding author.
Results
Patient Characteristics
Between May 13, 2019 and September 07, 2022, a total of 39 patients enrolled at 11 centers in the United States: 21 in the monotherapy cohort and 18 in the combination therapy cohort. One of the 21 patients enrolled in the monotherapy cohort withdrew consent prior to treatment initiation and was not included in the analysis. The baseline characteristics of the study population are presented in Table 1. The mean age was 61.7 years in the monotherapy cohort (range, 41–78 years) and 63.4 years in the combination therapy cohort (range, 43–84 years). In the monotherapy cohort, the primary malignancies were pancreatic cancer [8 (40.0%) patients], CCA [5 (25.0%) patients], and colorectal cancer [4 (20.0%) patients]. In the combination therapy cohort, 9 patients (50.0%) had a primary diagnosis of CCA, while the rest had diagnoses of RCC [5 (27.8%) patients] or HCC [4 (22.2%) patients]. The median number of prior systemic therapies was 3 (range, 2–9) for monotherapy and 3 (range, 1–6) for combination patients. In the combination cohort, all patients with HCC or RCC had received at least one prior anti-PD-(L)1 therapy as part of standard of care and had discontinued the most recent anti-PD-(L)1 therapy for disease progression.
TABLE 1.
Demographics and patient characteristics of the monotherapy and combination cohorts
| Baseline characteristics | TPST-1120 Monotherapy (n = 20) |
TPST-1120 + Nivolumab (n = 18) |
|
|---|---|---|---|
| Age, mean, years (range) | 61.7 (41–78) | 63.4 (43–84) | |
| Female, n (%) | 10 (50) | 9 (50) | |
| TPST-1120 dose, n (%) | 100 mg BID | 3 (15) | — |
| 200 mg BID | 4 (20) | 3 (17) | |
| 300 mg BID | 3 (15) | 3 (17) | |
| 400 mg BID | 4 (20) | 3 (17) | |
| 600 mg BID | 6 (30) | 9 (50) | |
| Primary cancer type, n (%) | Castration-resistant prostate cancer | 1 (5) | — |
| Cholangiocarcinoma | 5 (25) | 9 (50) | |
| Colorectal cancer | 4 (20) | — | |
| Hepatocellular carcinoma | 1 (5) | 4 (22) | |
| Non–small cell lung cancer | 1 (5) | — | |
| Pancreatic cancer | 8 (40) | — | |
| Renal cell carcinoma | — | 5 (28) | |
| Prior systemic regimens | Median (range) | 3 (2–9) | 3 (1–6) |
| Prior α-PD-1/α-PD-L1, n (%) | 6 (30) | 10 (56) | |
| ECOG PS, n (%) | 0 | 5 (25) | 8 (44) |
| 1 | 15 (75) | 10 (56) |
Abbreviations: BID, twice daily; ECOG PS, Eastern Cooperative Oncology Group performance status.
Pharmacokinetics
Data from 31 patients were available for pharmacokinetic analysis on day 8. TPST-1120 steady-state exposure levels increased in a linear, dose-dependent manner and were not affected by nivolumab (Supplementary Fig. S1). Key pharmacokinetic parameters of patients receiving TPST-1120 at 600 mg following single dose and at steady state are listed in Supplementary Table S2.
Safety
No DLTs occurred during dose escalation. Two monotherapy patients (1 at 200 mg twice daily and 1 at 400 mg twice daily) were not evaluable for DLTs due to missing more than 15% of the required TPST-1120 doses during the evaluation period due to AEs unrelated to treatment and were replaced. An MTD was not established for TPST-1120 as monotherapy or in combination with nivolumab.
In the monotherapy cohort, 10 patients (50%) experienced an AE that was deemed as related to TPST-1120 (Table 2). The most frequently reported treatment-related AEs (TRAE) were nausea [4 (20%) patients], fatigue [3 (15%) patients], and diarrhea [2 (10%) patients], all grade 1–2. One patient experienced grade 3 hypertension at the 600 mg twice daily dose of TPST-1120 that was assessed as treatment-related. There were no grade 4 or grade 5 TRAEs, and no patient discontinued study treatment due to a TRAE.
TABLE 2.
Treatment-related treatment-emergent AEs by preferred term in ≥1 patient, any grade and grade 3
| AE, n (%) | Grades 1–3a | Grade 3 |
|---|---|---|
| TPST-1120 Monotherapy (n = 20) | ||
| Patients with ≥1 TRAE | 10 (50.0) | 1 (5.0) |
| Nausea | 4 (20.0) | — |
| Fatigue | 3 (15.0) | — |
| Diarrhea | 2 (10.0) | — |
| Hypertension | 1 (5.0) | 1 (5.0) |
| TPST-1120 + Nivolumab (n = 18) | ||
| Patients with ≥1 TRAEb | 14 (77.8) | 3 (16.7) |
| Fatigue | 6 (33.3) | — |
| Diarrhea | 4 (22.2) | — |
| Nausea | 3 (16.7) | — |
| Abdominal pain | 2 (11.1) | — |
| Arthralgia | 1 (5.6) | 1 (5.6) |
| Hepatic enzyme increased | 1 (5.6) | 1 (5.6) |
| Muscle spasms | 1 (5.6) | 1 (5.6) |
aNo grade 4 or 5 TRAEs.
bRelated to either TPST-1120 or nivolumab.
In the combination therapy cohort, 14 patients (77.8%) experienced TRAEs related to either TPST-1120 or nivolumab (Table 2). The most frequent TRAEs were fatigue [6 (33.3%) patients], diarrhea [4 (22%) patients], and nausea [3 (17%) patients], all grade 1–2. Three patients experienced grade 3 TRAEs: 1 each of arthralgia (TPST-1120 400 mg twice daily), hepatic enzymes increased (TPST-1120 600 mg twice daily), and muscle spasms (TPST-1120 600 mg twice daily). Two of these TRAEs (grade 3 arthralgia and grade 3 hepatic enzymes increased) were considered immune-related and treated with systemic steroids. There were no grade 4 or grade 5 TRAEs. The patient with grade 3 hepatic enzymes increased was the sole patient treated with combination therapy (TPST-1120 600 mg orally twice daily) to discontinue treatment due to a TRAE.
Efficacy
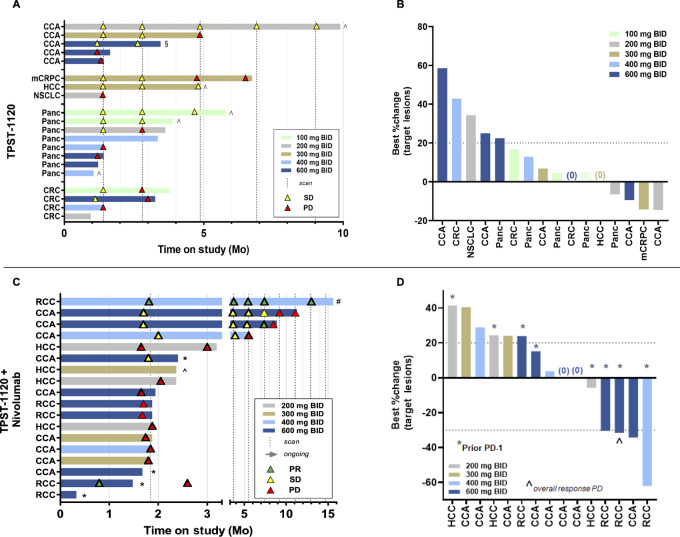
Clinical efficacy for TPST-1120 monotherapy and in combination with nivolumab is summarized in Fig. 1. Four treated patients were not included in the EP due to discontinuation of treatment for either symptomatic deterioration without objective evidence of PD (1 monotherapy, 1 combination) or due to unrelated AE (2 combination patients) prior to obtaining a postbaseline tumor assessment. The monotherapy efficacy evaluable population included 3 patients who did not have a postbaseline RECIST assessment but discontinued treatment for investigator-assessed disease progression not confirmed by RECIST. Among the 19 efficacy evaluable patients in the monotherapy cohort, the BOR was SD for 10 (53.0%) patients and PD for 6 (32.0%) patients, for a DCR of 53%. Tumor shrinkage of target lesions was observed in 4 patients (21%) with no target lesion growth as the best relative change from baseline in 3 additional patients. Of the 10 patients who had SD, 5 (50%) were on treatment >20 weeks. Among 5 patients with CCA, 3 had at least two assessments of SD, including 1 patient who was on treatment for almost 10 months before discontinuing due to symptomatic deterioration while still meeting RECIST for SD (Fig. 1A; Supplementary Fig. S2).
FIGURE 1.
Clinical efficacy of TPST-1120 monotherapy (A, B) and in combination with nivolumab (C, D). In swimmer plots shown in A and C, study treatment discontinuations for other than disease progression are shown as *Adverse event, ^Symptomatic deterioration, #Investigator decision, or §Consent withdrawn. For monotherapy, scans were every 6 weeks until week 12, followed by an increase to 9-week intervals between scans. For combination therapy, scans occurred every 8 weeks. The RCC responder in the 600 mg BID + nivolumab dose group had an unscheduled scan at day 24 during hospitalization for treatment-unrelated AEs. This patient discontinued treatment one day prior to the scan but was unable to restart and had disease progression during the posttreatment follow-up period. BID, twice daily; CCA, cholangiocarcinoma; CRC, colorectal carcinoma; HCC, hepatocellular carcinoma; mCRPC, metastatic castration-resistant prostate cancer; NSCLC, non–small cell lung cancer; Panc, pancreatic cancer; PD, progressive disease; PR, partial response; RCC, renal cell carcinoma; SD, stable disease.
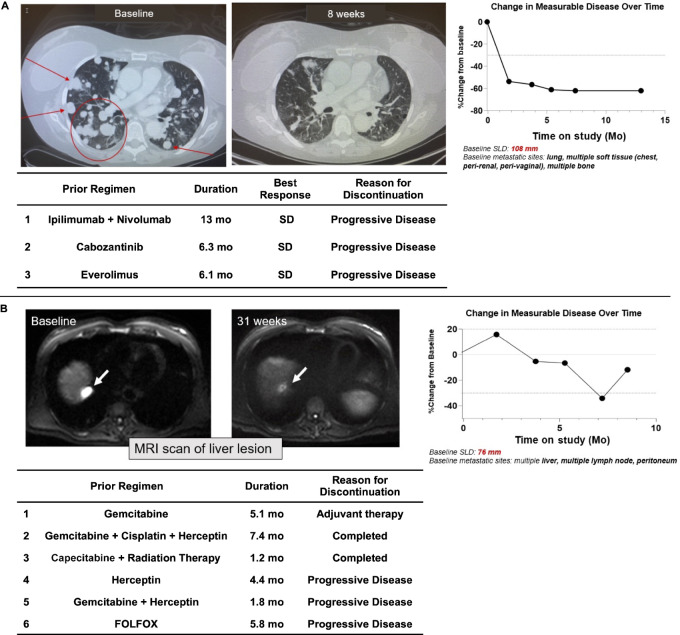
Of the 15 response-evaluable patients in the combination therapy cohort, 3 patients (20.0%) achieved a PR (1 confirmed), while a best response of SD occurred in 3 patients (20%), and 9 patients (60%) had PD. All responders were treated at the two highest doses of TPST-1120 (≥400 mg twice daily) for an ORR of 30% (3/10) at the 400 and 600 mg doses. In addition, of 4 evaluable patients with RCC, 2 achieved a PR for an ORR of 50% in this indication (Fig. 2A for one of the responders) and a third demonstrated a target lesion regression of −31% but also a new lesion in a mixed response (Fig. 1D). One additional PR was achieved in a patient with heavily pretreated CCA that was PD-L1 negative, mismatch repair proficient, with a tumor mutational burden of 10 mut/Mb; he had received six prior lines of systemic therapy before study enrollment (Fig. 2B).
FIGURE 2.
A, PR in a 54-year-old female with RCC who had disease progression on prior anti-PD-1 therapy. Prior treatment included ipilimumab + nivolumab (first line), cabozantinib (second line), and everolimus (third line) with no better than SD on any regimen. She achieved a PR (−54%) after 8 weeks of treatment with TPST-1120 + nivolumab that deepened to −62% and was durable for more than 1 year. Changes in pulmonary lesions are shown in CT chest scans taken during screening and on-treatment. SD, stable disease. B, PR in an 84-year-old male with heavily pretreated extrahepatic cholangiocarcinoma. He had an initial increase in tumor burden followed by serial shrinking tumor scans before achieving a nadir of −34% and an overall response of PR at scan 4. SLD: sum of longest diameters.
Notably, both patients with RCC who responded to the TPST-1120 combination regimen (and the patient with RCC with a −31% mixed response) were previously treated with at least one anti-PD-1 containing regimen with a best response of SD and discontinued the most recent anti-PD-1 therapy for disease progression. The RCC responder who achieved a −54% RECIST PR at the first tumor assessment (8 weeks of treatment) had previously received first-line ipilimumab + nivolumab followed by cabozantinib and everolimus, discontinuing each regimen for PD after a best response of SD. The RCC responder who achieved a −30% RECIST PR had previously received first-line pembrolizumab + axitinib followed by cabozantinib and discontinued both regimens for disease progression after a best response of SD.
Biomarker Exploration
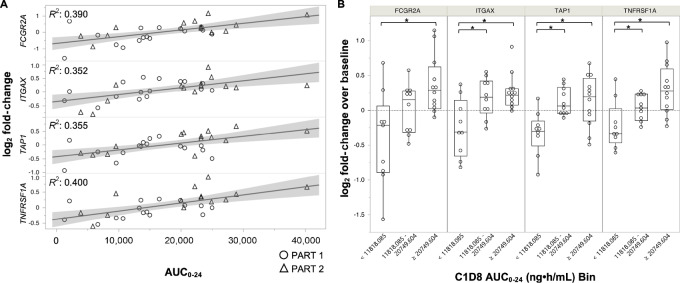
Analysis of differential expression levels of 780 genes in whole blood specimens revealed that four were increased as a function of TPST-1120 AUC0–24 on day 8 (FDR P < 0.05, effect size > 0.5, Fig. 3A). Similar associations between expression levels of these genes and TPST-1120 exposure were also observed on cycle 3 day 1 (Supplementary Fig. S3). Modulated genes included FCGRIIA (CD32; ref. 20), ITGAX (CD11c; ref. 21), TAP1 (22, 23), and TNFRSF1A (CD120a; ref. 24), all of which are gene targets of transcription factors inhibited by PPARα through transrepression (Table 3). Analysis of gene expression changes by exposure tertile demonstrated that median expression levels of three of the four target genes at the middle exposure tertile, 11,818–20,749 ng⋅hour/mL, were statistically elevated above that observed in the lowest tertile (P < 0.05 by Wilcoxon pairwise comparison, Fig. 3B). This corresponds to 60% of patients with TPST-1120 steady-state exposures of at least 11,819 ng⋅hour/mL who demonstrated target gene elevations above baseline levels and identifies a minimum exposure for induction of pharmacodynamic activity.
FIGURE 3.
Genes differentially expressed on treatment day 8 versus treatment baseline, as a function of TPST-1120 exposure. A, Linear associations of day 8 log2 fold change in expression levels of indicated genes and TPST-1120 AUC0–24. All genes exhibited FDR P < 0.05 and effect size > 0.5. Data are shown for both TPST-1120 monotherapy (part 1) and TPST-1120 + nivolumab combination therapy (part 2). An AUC0–24 value of 66,180 ng⋅hour/mL was excluded as an outlier. B, Comparison of differential gene expression on day 8 as a function of AUC0–24 tertile. Median elevation magnitudes in highest tertile were statistically increased above baseline values (*, P < 0.05 by Wilcoxon pair-wise comparison).
TABLE 3.
Summary of genes associated with TPST-1120 exposure levels
| Gene | Name | Function in immune cells | FDR P-value, effect size | Transcription factor associated with PPARα transrepression |
|---|---|---|---|---|
| FCGR2A | Fc-γ RIIa, CD32 | Enhances antibody-dependent cytotoxicity of tumor cells, increased phagocytosis and cytokine release by myeloid cells (25) | 0.047, 0.732 | STAT1 (20) |
| ITGAX | Integrin α-X, CD11c | Marker of conventional dendritic cells that cross-present tumor antigens, enhances phagocytosis of tumor cells by macrophages (26) | 0.049, 0.566 | Fos, Jun, C/EBP (21) |
| TAP1 | Transporter associated with antigen processing-1 | Increases endogenous antigen processing and presentation by MHC-I molecules | 0.049, 0.550 | STAT1 (22), NFκB (23) |
| TNFRSF1A | TNFα R1, CD120a | Enhances responsiveness to TNFα, activates NFκB-responsive genes (27) | 0.047, 0.648 | STAT3 (24) |
Abbreviation: FDR, false discovery rate.
LDA of day 8 gene expression levels among BOR categories revealed clear distinctions in expression patterns between PD or SD patients and PR patients (Supplementary Fig. S4A). PR patients were observed to express statistically decreased levels of CFB and PVR (CD155) and increased levels of APOE, MAGEA12, RORC, and SYT17 (P < 0.05 by Mann–Whitney U test; Supplementary Table S3; Supplementary Fig. S4B).
Lipid analysis revealed on-treatment elevation in circulating free fatty acids (FFA) from baseline to day 57 and day 85 in patients demonstrating PRs that were not detected in patients with a best response of PD/SD (Supplementary Fig. S5).
OBD Selection
The 600 mg twice daily dose of TPST-1120 was determined as the OBD for both monotherapy and combination therapy regimens. Analysis of pharmacokinetics across doses and for both monotherapy and combination showed a linear relationship between TPST-1120 dose and plasma exposure, with no saturation at 600 mg twice daily, the highest dose level tested. Pharmacodynamic analysis of the association between TPST-1120 steady-state exposures and gene expression changes in peripheral blood demonstrated exposure-dependent increases in expression of a subset of genes known to be regulated by transcription factors transrepressed by PPARα, and the identified minimum exposure for induction of pharmacodynamic activity was achieved in all patients receiving TPST-1120 at 400 or 600 mg twice daily (Supplementary Fig. S1). Review of safety data showed no evidence of dose-dependent toxicity through the highest dose tested of 600 mg twice daily, either for single agent TPST-1120 or when administered with nivolumab. Finally, although limited by small numbers, RECIST responses in the combination cohort all occurred at the two highest TPST-1120 dose levels tested, consistent with dose-responsive antitumor activity.
Discussion
TPST-1120 is a novel investigational agent designed to therapeutically target cancer cells and enhance anticancer immunity by inhibiting the fatty acid ligand-activated transcription factor PPARα. In this phase I first-in-human study, which is the first to report clinical data on PPARα modulation in solid tumors, we tested the hypothesis that inhibiting PPARα with TPST-1120 would be tolerable in patients with advanced cancer and would have anticancer activity. TPST-1120 was well tolerated both as monotherapy and in combination with the PD-1 inhibitor, nivolumab, including no DLTs during dose escalation and predominantly grade 1–2 and manageable TRAEs. In the 18 patients who received TPST-1120 in combination with nivolumab, there was no evidence of synergistic or unexpected toxicity, and the AEs were consistent with the profiles of the two drugs.
TPST-1120 also showed evidence of clinical activity. In the monotherapy group, disease control including target lesion shrinkage was demonstrated in a subset of patients with highly refractory pancreatic, CCA, and late-line colorectal cancers. The combination of TPST-1120 and nivolumab demonstrated an ORR of 20% across all doses and 30% at the two highest doses of TPST-1120. Responses were seen in patients with RCC previously refractory to ICI therapy and in a patient with late-line, PD-L1–negative and microsatellite stable CCA—a tumor type poorly responsive to ICI monotherapy (e.g., pembrolizumab ORR of 2.9% in patients with PD-L1 combined positive score ≤1 treated in the KEYNOTE-158 trial; refs. 28–31). It is notable that among 4 evaluable patients with RCC, 2 (50%) achieved a PR with the TPST-1120 + nivolumab combination, including a deep and very durable response in a patient who had already progressed on nivolumab + ipilimumab after achieving SD as best response. In a recent prospective randomized phase III study, the addition of atezolizumab to cabozantinib provided no additional benefit compared with cabozantinib alone in second-line treatment of patients with RCC who had received ICI treatment in first line (32). In retrospective reports, the ORR of patients with RCC treated with nivolumab monotherapy after previous ICI therapy was 16% (33) and 23% (34). Acknowledging the limitation of small patient numbers, these response data in immunotherapy-refractory patients are consistent with the preclinical results showing that TPST-1120 modulates the immune phenotype away from suppressor populations and combines synergistically with anti-PD-1 therapy (16), and consistent with the literature that genetic KO of PPARα increases inflammation and decreases tumor growth in mouse models (14).
Further supporting the immune mechanism, an exploratory analysis of gene expression changes in whole blood of patients treated in this study demonstrated TPST-1120 exposure-dependent increases in the expression of genes transcriptionally regulated by Th1-promoting proinflammatory transcription factors that are subject to PPARα transrepressive activities, including NFκB and STAT1. This transcriptional dose–response allows for establishment of a minimum exposure threshold for pharmacodynamic activity corresponding to a TPST-1120 dose of at least 400 mg twice daily. Consistent with the exposure threshold, all patients who achieved a PR were dosed with TPST-1120 at ≥400 mg twice daily. Further exploratory analyses of associations between BOR and changes in both gene expression and lipids in study patients showed increases in circulating FFA levels, indicating a reduction in lipid catabolism, and increases in RORC, encoding RORγt, the master transcriptional regulator of Th17 cells, consistent with literature reports that Th17 cells are increased in PPARα-deficient animals (35, 36).
The increased expression of genes downstream of proinflammatory transcription factor targets of PPARα transrepression suggests that immune cell function is enhanced with TPST-1120 treatment. These results are consistent with literature indicating PPARα deficiency increases inflammation, one such example being the promotion of M1 macrophage polarization by Ppara KO in myeloid cells—shown as increased IL1 and TNFα mRNA and decreased arginase mRNA expression upon lipopolysaccharide stimulation of bone marrow–derived macrophages (37). Lack of PPARα, as well as treatment with the PPARα antagonist IS001, resulted in increased IFNγ production by CD4+ and CD8+ T lymphocytes and natural killer T cells (38). Correspondingly, augmented or sustained responses to inflammatory stimuli across multiple models of inflammation were observed when Ppara was deleted and support a role for PPARα in immune regulation (39–41).
In characterizing the antitumor activity of TPST-1120, it is of interest whether these dual PPARα functions—transrepression associated with immunomodulatory activity and PPAR response element (PPRE)-mediated transcription controlling lipid metabolism—are linked, as increased utilization of FAO is a well-studied metabolic characteristic of suppressive immune cells. Experiments conducted using a mutant PPARα protein unable to activate transcription of PPRE-dependent genes and consequently devoid of its lipid-regulating activity, showed that mutant PPARα retained the ability to attenuate inflammation in a mouse model of liver fibrosis, demonstrating that these two functions can be uncoupled and are distinct (42). Future preclinical work will focus on dissecting the contribution of each function to the antitumor activity displayed by TPST-1120.
Limitations of this study include small numbers of patients treated at different TPST-1120 dose levels. In addition, imbalanced enrollment of pancreaticobiliary cancers in the monotherapy cohort reduced the assessment of the PPARα inhibition mechanism in other solid tumor types. Evaluation of TPST-1120 pharmacodynamic activity was restricted to the challenging and complex setting of peripheral blood cells and circulating lipids by the lack of fresh tumor biopsies.
Further clinical development of TPST-1120 in RCC and CCA is planned based upon these study results. A separate study is ongoing in HCC, a tumor type not adequately represented in this phase I study, but of particular interest due to the high expression of PPARα. A global randomized phase Ib/II study is evaluating the combination of atezolizumab plus bevacizumab, with or without TPST-1120, in patients with first-line advanced HCC (NCT04524871). Initial efficacy results from this randomized study are encouraging and have been publicly reported (43). The identification of novel biomarkers that correlate with sensitivity to TPST-1120 may further improve patient selection for future trials and inform the development of novel therapeutic combinations.
Supplementary Material
Additional PPAR-α Associated Genes Monitored
Summary of pharmacokinetics of 600 mg TPST-1120 twice daily as a single agent and in combination with nivolumab after single dose or at steady state (cycle 1 day 8)
Summary of Differentially Expressed Genes in PR Patients (p-value < 0.05 by Mann-Whitney U-test vs. PR/SD patients) on Cycle 1 Day 8
Relationship between dose (mg BID) and AUC0-24 of TPST-1120 at steady state on Cycle 1 Day 8. Monotherapy and nivolumab combination data points offset slightly for clarity.
Monotherapy tumor control in late-line cholangiocarcinoma. Change in measurable tumor burden over time is shown for two patients with late-line cholangiocarcinoma demonstrating prolonged disease control achieved with monotherapy TPST-1120, including patient B who achieved multiple stable disease scans with serial shrinkage of tumor burden to a nadir of -13% by RECIST over a duration of 9.5 months on treatment. Prior systemic treatment for patient A included cisplatin/gemcitabine, an investigational multi-kinase inhibitor, and an investigational anti-PD-1, while patient B received carboplatin/taxol, gemcitabine, oxaliplatin/capecitabine, and an investigational anti-PD-1/indoleamine 2,3-dioxygenase 1 inhibitor combination. Both patients discontinued the most recent therapy regimen received prior to TPST-1120 treatment for progressive disease.
Genes differentially expressed as a function of BOR on cycle 3 day 1. Genes differentially expressed on C3D1 versus treatment baseline, as a function of TPST-1120 exposure. Linear associations of C3D1 Log2 fold change in expression levels of indicated genes and TPST-1120 AUC0-24. Data are shown for both TPST-1120 monotherapy (part 1) and TPST-1120 + nivolumab combination therapy (part 2).
Genes differentially expressed as a function of BOR on day 8. (A) Linear discriminant analysis of Log2 fold change in expression levels of 780 genes in patients stratified based on BOR. PR: partial response; SD: stable disease; PD: progressive disease. (B) Genes that maximally discriminate between PR and PD patients enrolled in combination therapy arm. * p<0.05 by Mann-Whitney U-test.
Elevated circulating free fatty acids (FFA) at days 57 and 85 in PR patients. Log2-fold changes in baseline normalized FFA in patients enrolled in Part 2. Samples were collected prior to TPST-1120 dose administration on day 1 of each 28-day cycle. Blue symbols: PD/SD patients; Red symbols: PR patients.
Acknowledgments
Editorial support was provided by Ingrid Koo, PhD, and funded by Tempest Therapeutics.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Communications Online (https://aacrjournals.org/cancerrescommun/).
Authors’ Disclosures
M. Yarchoan reports grants and personal fees from Genentech; grants from Bristol Myers Squibb and Incyte; personal fees from Exelixis, Eisai, AstraZeneca, Replimune, Hepion, Lantheus; and other from Adventris outside the submitted work. J.D. Powderly reports personal fees and other from Tempest Therapeutics during the conduct of the study; other from Carolina BioOncology Institute, PLLC, BioCytics Inc., Genentech Roche, AstraZeneca Medimmune, AbbVie, Aavocyte, AavoBioCytics, Phanes Therapeutics, BMS, EMD Serono, Macrogenics, Top Alliance BioSciences, Alkermes, Calico, Atreca, Precision for Medicine, Stem Cell, Sequenom, Replimuune, Engineered BioPharmaceuticals, Therapeutic Brainpower, Merck, PIOMA, Xilis, Repertoire Immune Medicines, NextCure, Xilio Therapeutics, Nuvation Bio, Cullinan Oncology, Immune-Onc Therapeutics, Trethera, Fate Therapeutics, Conjupro BioTherapeutics, Ascendis Pharma, PEEL Therapeutics, CUE BioPharma, Pieris Pharmaceuticals, RiboScience, Moderna TX, SK Life Sciences, Harbour BioMed, Allarity Therapeutics, GI Innovation, Aulos BioScience, IgM BioSciences, Aptevo Therapeutics, Medikine, Sairopa, IconOVir Bio, Qurgen, Glenmark, FBD Biologics, MBrace, Astellas, BioNTech, SK Life Sciences, Revolution Medicines, and Multitude; personal fees from Terumo and Boxer Capital outside the submitted work; in addition, J.D. Powderly has a patent to BioCytics is developing personalized cellular immunotherapies pending; and J.D. Powderly is the founder and owner of Carolina BioOncology Institute PLLC (phase 1 clinic) and BioCytics Inc (Human Applications Lab), of which both of these entities are developing intellectual property on the delivery of personalized cellular immunotherapies manufactured at the point of care. Both entities perform sponsored contract professional services for many small biotech/biopharma entities as pre-IND, IND, and CRO/CDMO consulting services. O.V. Crysler reports grants from Tempest Therapeutics during the conduct of the study; grants from Servier Pharmaceuticals LLC, Genentech Inc, ACCRU, Surface Oncology, and AstraZeneca outside the submitted work. P.N. Munster reports grants from Tempest during the conduct of the study; grants from Oric, Arvinas, Actuate, GSK, Revolution Medicine, Merck, Xynomics, Amgen, Bliss Bio, Blueprint, IGM, Inventis BIo, Novartis, Cyteir; other from AtlasMedex, RasCal, Catalent, and Alessa outside the submitted work. M.A. McKean reports grants from Tempest Therapeutics during the conduct of the study; grants from Aadi Biosciences, Alpine Immune Sciences, Arcus Biosciences, Arvinas, Ascentage Pharma Group, ASCO, Astellas, Aulos Bioscience, Bayer, Bicycle Therapeutics, BioMed Valley Discoveries, BioNTech, Bristol Myers Squibb, C4 Therapeutics, Dragonfly Therapeutics, EMD Serono, Epizyme, Erasca, Exelixis, Foghorn Therapeutics, G1 Therapeutics, Genentech/Roche, Gilead Sciences, GlaxoSmithKline, IDEAYA Biosciences, Ikena Oncology, ImmVira Pharma, Infinity Pharmaceuticals, Jacobio Pharmaceuticals, Jazz Pharmaceuticals, Kechow Pharma, Kezar Life Sciences, Kinnate BioPharma, Krystal Biotech, MedImmune, Mereo BioPharma, Metabomed, NBE Therapeutics, Nektar, Novartis, NucMito Pharmaceuticals, OncoC4, Oncorus, OnKure, PACT Pharma, Plexxikon, Poseida, Prelude Therapeutics, Pyramid Biosciences, Regeneron, Remix Therapeutics, Sapience Therapeutics, Scholar Rock, Seattle Genetics, Synthrox, Takeda Pharmaceuticals, Teneobio, Tempest Therapeutics, Tizona Therapeutics, TMUNITY Therapeutics, TopAlliance Biosciences, Xilio; grants and other from Moderna, Pfizer; other from Castle Biosciences, IQVIA, and Merck outside the submitted work. L.A. Emens reports grants from Tempest during the conduct of the study; grants from Ankyra Therapeutics, AstraZeneca, Bolt Therapeutics, Bristol Myers Squibb, Compugen, CytomX, EMD Serono, Genentech, Roche, Immune Onc, Merck, Next Cure, Silverback, and Takeda outside the submitted work; and consulting for AstraZeneca, Bioline Rx, DNAMx, Genentech, Roche, GPCR, Gilead, Immune Onc, Immunitas, Immutep, Lilly, Macrogenics Mersana, Shionogi member SAB Bioline RX member CAB Immutep potential for equity with Ankyra Therapeutics potential for future royalties from Molecuvax. R. Stagg reports personal fees and other from Tempest outside the submitted work; and Clinical consultant for Tempest Therapeutics. S. Smith reports personal fees from Tempest Therapeutics during the conduct of the study. C.C. Whiting reports other from Tempest Therapeutics outside the submitted work; in addition, C.C. Whiting has a patent to us2023/078674 pending. A. Moon reports personal fees, non-financial support, and other from Tempest Therapeutics during the conduct of the study. P. Prasit reports other from Tempest Therapeutics outside the submitted work. N. Standifer reports other from AstraZeneca, Amgen, Tempest Therapeutics, and personal fees from Vincerx outside the submitted work; in addition, N. Standifer has a patent to Triazolone compounds and uses thereof pending. T.W. Dubensky reports personal fees and other from Tempest Therapeutics during the conduct of the study; in addition, T.W. Dubensky has a patent to several TPST-1120 composition, methods of use and biomarker pending and issued. S.H. Whiting reports employed by Tempest Therapeutics. S.V. Ulahannan reports grants from Tempest Therapeutics during the conduct of the study; other from Eisai Pharmaceutical, AstraZeneca, IGM Biosciences; grants from AbbVie, Inc, Adlai Nortye, Artios Pharma, ArQule, Inc, AstraZeneca, Atreca, Boehringer Ingelheim, Bristol Myers Squibb, Celgene Corporation, Ciclomed LLC, Eisai Pharmaceutical, Erasca, Evelo Biosciences, Inc, Exelexis, G1 Therapeutics, Inc, GlaxoSmithKline GSK, IGM Biosciences, Immunitas Therpeutics, Incyte, Isofol, Jazz Pharmaceuticals, Klus Pharma, Inc, Macrogenics, Merck Co. Inc, Mersana Therapeutics, Omega Therapeutics, OncoMed Pharmaceuticals, Inc, Pfizer, Purple Biotech, Rgeneron Pharmaceuticals, Inc, Revolution Medicines, Inc, Synermore Biologics Co, Takeda, Tarveda, Tesaro, Tempest Therapeutics, Theradex, Totos Medicines, Inc, Tvardi Therapeutics, and Vigeo Therapeutics Inc outside the submitted work. No other disclosures were reported.
Authors’ Contributions
M. Yarchoan: Conceptualization, formal analysis, investigation, writing-original draft, writing-review and editing. J.D. Powderly: Investigation, writing-review and editing. B.R. Bastos: Investigation, writing-review and editing. T.B. Karasic: Investigation, writing-review and editing. O.V. Crysler: Investigation, writing-review and editing. P.N. Munster: Investigation, writing-review and editing. M.A. McKean: Investigation, writing-review and editing. L.A. Emens: Investigation, writing-review and editing. Y.M. Saenger: Investigation, writing-review and editing. Y. Ged: Investigation, writing-review and editing. R. Stagg: Investigation, writing-review and editing. S. Smith: Investigation, writing-review and editing. C.C. Whiting: Data curation, supervision, investigation, writing-review and editing. A. Moon: Data curation, formal analysis, supervision, investigation, writing-review and editing. P. Prasit: Data curation, formal analysis, investigation, writing-review and editing. Y. Jenkins: Data curation, formal analysis, supervision, investigation, writing-original draft, project administration, writing-review and editing. N. Standifer: Data curation, formal analysis, investigation, writing-review and editing. T.W. Dubensky: Conceptualization, supervision, funding acquisition, investigation, writing-review and editing. S.H. Whiting: Data curation, formal analysis, supervision, investigation, writing-original draft, project administration, writing-review and editing. S.V. Ulahannan: Supervision, investigation, writing-review and editing.
References
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 2. Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer 2013;13:227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ma Y, Temkin SM, Hawkridge AM, Guo C, Wang W, Wang X-Y, et al. Fatty acid oxidation: an emerging facet of metabolic transformation in cancer. Cancer Lett 2018;435:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koundouros N, Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br J Cancer 2020;122:4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol 2011;186:3299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang SC-C, Everts B, Ivanova Y, O'Sullivan D, Nascimento M, Smith AM, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol 2014;15:846–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, et al. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab 2006;4:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carracedo A, Weiss D, Leliaert AK, Bhasin M, de Boer VCJ, Laurent G, et al. A metabolic prosurvival role for PML in breast cancer. J Clin Investig 2012;122:3088–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bougarne N, Weyers B, Desmet SJ, Deckers J, Ray DW, Staels B, et al. Molecular actions of PPARα in lipid metabolism and inflammation. Endocr Rev 2018;39:760–802. [DOI] [PubMed] [Google Scholar]
- 10. Zeng W, Yin X, Jiang Y, Jin L, Liang W. PPARα at the crossroad of metabolic–immune regulation in cancer. FEBS J 2022;289:7726–39. [DOI] [PubMed] [Google Scholar]
- 11. Szalowska E, Tesfay HA, van Hijum SAFT, Kersten S. Transcriptomic signatures of peroxisome proliferator-activated receptor α (PPARα) in different mouse liver models identify novel aspects of its biology. BMC Genomics 2014;15:1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wahli W, Michalik L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol Metab 2012;23:351–63. [DOI] [PubMed] [Google Scholar]
- 13. Pawlak M, Lefebvre P, Staels B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol 2015;62:720–33. [DOI] [PubMed] [Google Scholar]
- 14. Kaipainen A, Kieran MW, Huang S, Butterfield C, Bielenberg D, Mostoslavsky G, et al. PPARα deficiency in inflammatory cells suppresses tumor growth. PLoS One 2007;2:e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stock NS, Chih-Yu CA, Mostofi BY, Duarte JJ, Melissa BJ, Andrew SB, et al. US Patent No.676,754 B2. Triazolone compounds and uses thereof; 2017.
- 16. Whiting C, Stock N, Messmer D, Olafson T, Metzger D, Enstrom A, et al. Blockade of the PPARα metabolic checkpoint with TPST-1120 suppresses tumor growth and stimulates anti-tumor immunity [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2019; 2019 Mar 29–Apr 3; Atlanta, GA. Philadelphia (PA): AACR; Cancer Res 2019;79(13 Suppl):Abstract nr 3606. [Google Scholar]
- 17. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 1995;57:289–300. [Google Scholar]
- 18. Hsieh W-Y, Williams KJ, Su B, Bensinger SJ. Profiling of mouse macrophage lipidome using direct infusion shotgun mass spectrometry. STAR Protoc 2021;2:100235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Su B, Bettcher LF, Hsieh W-Y, Hornburg D, Pearson MJ, Blomberg N, et al. A DMS shotgun lipidomics workflow application to facilitate high-throughput, comprehensive lipidomics. J Am Soc Mass Spectrom 2021;32:2655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dahlqvist J, Fulco CP, Ray JP, Liechti T, de Boer CG, Lieb DJ, et al. Systematic identification of genomic elements that regulate FCGR2A expression and harbor variants linked with autoimmune disease. Hum Mol Genet 2021;31:1946–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. López-Rodríguez C, Botella L, Corbí AL. CCAAT-Enhancer-binding Proteins (C/EBP) regulate the tissue specific activity of the CD11c integrin gene promoter through functional interactions with Sp1 proteins*. J Biol Chem 1997;272:29120–6. [DOI] [PubMed] [Google Scholar]
- 22. Cramer LA, Nelson SL, Klemsz MJ. Synergistic induction of the tap-1 gene by IFN-γ and lipopolysaccharide in macrophages is regulated by STAT1. J Immunol 2000;165:3190–7. [DOI] [PubMed] [Google Scholar]
- 23. Wright KL, White LC, Kelly A, Beck S, Trowsdale J, Ting JP. Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J Exp Med 1995;181:1459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Egusquiaguirre SP, Yeh JE, Walker SR, Liu S, Frank DA. The STAT3 target gene TNFRSF1A modulates the NF-κB pathway in breast cancer cells. Neoplasia 2018;20:489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anania JC, Chenoweth AM, Wines BD, Hogarth PM. The human FcγRII (CD32) family of leukocyte FcR in health and disease. Front Immunol 2019;10:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tang Z, Davidson D, Li R, Zhong M-C, Qian J, Chen J, et al. Inflammatory macrophages exploit unconventional pro-phagocytic integrins for phagocytosis and anti-tumor immunity. Cell Rep 2021;37:110111. [DOI] [PubMed] [Google Scholar]
- 27. Wajant H, Siegmund D. TNFR1 and TNFR2 in the control of the life and death balance of macrophages. Front Cell Dev Biol 2019;7:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Piha-Paul SA, Oh D, Ueno M, Malka D, Chung HC, Nagrial A, et al. Efficacy and safety of pembrolizumab for the treatment of advanced biliary cancer: results from the KEYNOTE-158 and KEYNOTE-028 studies. Int J Cancer 2020;147:2190–8. [DOI] [PubMed] [Google Scholar]
- 29. Kim RD, Chung V, Alese OB, El-Rayes BF, Li D, Al-Toubah TE, et al. A phase 2 multi-institutional study of nivolumab for patients with advanced refractory biliary tract cancer. JAMA Oncol 2020;6:888–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doki Y, Ueno M, Hsu C, Oh D, Park K, Yamamoto N, et al. Tolerability and efficacy of durvalumab, either as monotherapy or in combination with tremelimumab, in patients from Asia with advanced biliary tract, esophageal, or head-and-neck cancer. Cancer Med 2022;11:2550–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yarchoan M, Cope L, Ruggieri AN, Anders RA, Noonan AM, Goff LW, et al. Multicenter randomized phase II trial of atezolizumab with or without cobimetinib in biliary tract cancers. J Clin Invest 2021;131:e152670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pal SK, Albiges L, Tomczak P, Suárez C, Voss MH, de Velasco G, et al. Atezolizumab plus cabozantinib versus cabozantinib monotherapy for patients with renal cell carcinoma after progression with previous immune checkpoint inhibitor treatment (CONTACT-03): a multicentre, randomised, open-label, phase 3 trial. Lancet 2023;402:185–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vauchier C, Auclin E, Barthélémy P, Carril-Ajuria L, Ryckewaert T, Borchiellini D, et al. REchallenge of NIVOlumab (RENIVO) or nivolumab-ipilimumab in metastatic renal cell carcinoma: an ambispective multicenter study. J Oncol 2022;2022:3449660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ravi P, Mantia C, Su C, Sorenson K, Elhag D, Rathi N, et al. Evaluation of the safety and efficacy of immunotherapy rechallenge in patients with renal cell carcinoma. JAMA Oncol 2020;6:1606–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang H, Zhao F, Xie X, Liao Y, Song Y, Liu C, et al. PPARα suppresses Th17 cell differentiation through IL-6/STAT3/RORγt pathway in experimental autoimmune myocarditis. Exp Cell Res 2019;375:22–30. [DOI] [PubMed] [Google Scholar]
- 36. Wei P, Kou W, Fu J, Chen Z, Pan F. Pparα knockout in mice increases the Th17 development by facilitating the IKKα/RORγt and IKKα/Foxp3 complexes. Commun Biol 2023;6:721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xie G, Song Y, Li N, Zhang Z, Wang X, Liu Y, et al. Myeloid peroxisome proliferator-activated receptor α deficiency accelerates liver regeneration via IL-6/STAT3 pathway after 2/3 partial hepatectomy in mice. Hepatobiliary Surg Nutr 2022;11:199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang MA, Ahn JJ, Zhao FL, Selvanantham T, Mallevaey T, Stock N, et al. Antagonizing peroxisome proliferator–activated receptor α activity selectively enhances Th1 immunity in male mice. J Immunol 2015;195:5189–202. [DOI] [PubMed] [Google Scholar]
- 39. Devchand PR, Keller H, Peters JM, Vazquez M, Gonzalez FJ, Wahli W. The PPARα–leukotriene B4 pathway to inflammation control. Nature 1996;384:39–43. [DOI] [PubMed] [Google Scholar]
- 40. Cuzzocrea S, Mazzon E, Paola RD, Peli A, Bonato A, Britti D, et al. The role of the peroxisome proliferator-activated receptor-α (PPAR-α) in the regulation of acute inflammation. J Leukoc Biol 2006;79:999–1010. [DOI] [PubMed] [Google Scholar]
- 41. Gervois P, Mansouri RM. PPARα as a therapeutic target in inflammation-associated diseases. Expert Opin Ther Targets 2012;16:1113–25. [DOI] [PubMed] [Google Scholar]
- 42. Pawlak M, Baugé E, Bourguet W, Bosscher K, Lalloyer F, Tailleux A, et al. The transrepressive activity of peroxisome proliferator-activated receptor alpha is necessary and sufficient to prevent liver fibrosis in mice. Hepatology 2014;60:1593–606. [DOI] [PubMed] [Google Scholar]
- 43. Tempest Therapeutics Inc. Tempest releases new data demonstrating superiority of TPST-1120 arm across multiple study endpoints in randomized first-line HCC study; 2023. Available from: https://ir.tempesttx.com/news-releases/news-release-details/tempest-releases-new-data-demonstrating-superiority-tpst-1120.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional PPAR-α Associated Genes Monitored
Summary of pharmacokinetics of 600 mg TPST-1120 twice daily as a single agent and in combination with nivolumab after single dose or at steady state (cycle 1 day 8)
Summary of Differentially Expressed Genes in PR Patients (p-value < 0.05 by Mann-Whitney U-test vs. PR/SD patients) on Cycle 1 Day 8
Relationship between dose (mg BID) and AUC0-24 of TPST-1120 at steady state on Cycle 1 Day 8. Monotherapy and nivolumab combination data points offset slightly for clarity.
Monotherapy tumor control in late-line cholangiocarcinoma. Change in measurable tumor burden over time is shown for two patients with late-line cholangiocarcinoma demonstrating prolonged disease control achieved with monotherapy TPST-1120, including patient B who achieved multiple stable disease scans with serial shrinkage of tumor burden to a nadir of -13% by RECIST over a duration of 9.5 months on treatment. Prior systemic treatment for patient A included cisplatin/gemcitabine, an investigational multi-kinase inhibitor, and an investigational anti-PD-1, while patient B received carboplatin/taxol, gemcitabine, oxaliplatin/capecitabine, and an investigational anti-PD-1/indoleamine 2,3-dioxygenase 1 inhibitor combination. Both patients discontinued the most recent therapy regimen received prior to TPST-1120 treatment for progressive disease.
Genes differentially expressed as a function of BOR on cycle 3 day 1. Genes differentially expressed on C3D1 versus treatment baseline, as a function of TPST-1120 exposure. Linear associations of C3D1 Log2 fold change in expression levels of indicated genes and TPST-1120 AUC0-24. Data are shown for both TPST-1120 monotherapy (part 1) and TPST-1120 + nivolumab combination therapy (part 2).
Genes differentially expressed as a function of BOR on day 8. (A) Linear discriminant analysis of Log2 fold change in expression levels of 780 genes in patients stratified based on BOR. PR: partial response; SD: stable disease; PD: progressive disease. (B) Genes that maximally discriminate between PR and PD patients enrolled in combination therapy arm. * p<0.05 by Mann-Whitney U-test.
Elevated circulating free fatty acids (FFA) at days 57 and 85 in PR patients. Log2-fold changes in baseline normalized FFA in patients enrolled in Part 2. Samples were collected prior to TPST-1120 dose administration on day 1 of each 28-day cycle. Blue symbols: PD/SD patients; Red symbols: PR patients.
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.