Abstract
The function of membrane proteins relies on a defined orientation of protein relative to lipid. In apparent correlation to protein anchoring, tryptophan residues are enriched in the lipid headgroup region. To characterize the thermodynamic and structural basis of this relationship in α-helical membrane proteins, we examined the role of three conserved tryptophans in the folding of the heterodimeric integrin αIIbβ3 transmembrane (TM) complex in phospholipid bicelles and mammalian membranes. In the homogenous lipid environment of bicelles, tryptophan was replaceable by residues of distinct polarities. The appropriate polarity was guided by the electrostatic potential of the tryptophan surrounding, suggesting that tryptophan can complement diverse environments by adjusting the orientation of its anisotropic sidechain to achieve site-specific anchoring. As a sole membrane anchor, tryptophan made a contribution of 0.4 kcal/mol to TM complex stability in bicelles. In membranes, it proved more difficult to replace tryptophan even by tyrosine, indicating a superior capacity to interact with heterogeneous lipids of biological membranes. Interestingly, at intracellular TM helix ends where integrin activation is initiated, sequence motifs that interact with lipids via opposing polarity patterns were found to restrict TM helix orientations beyond tryptophan anchoring. In contrast to bicelles, phenylalanine became the least accepted substitute in membranes, demonstrating an increased role of the hydrophobic effect. Altogether, our study implicates a wide amphiphilic range of tryptophan, membrane complexity, and the hydrophobic effect to be important factors in tryptophan membrane anchoring.
INTRODUCTION
Membrane proteins fold, function and interact in two-dimensional lipid bilayers that are solvated by water. Whereas lipid hydrocarbon tails and bulk water represent relatively uniform environments, the interface between these two environments, which contains diverse lipid headgroups, is chemically complex.1,2 The composition of these three different regions influences the prevalence of particular amino acids.3–6 For example, Trp and Tyr are overrepresented in the headgroup region. Trp has generated considerable interest as it has the strongest preference for the headgroup interface7 and contributes to the stability, folding and function of a range of integral membrane proteins.8–11 In β-barrel membrane proteins, the contribution of Trp to protein stability has been studied experimentally.12,13 Such proteins fold from an aqueous, unfolded state to form β-barrel structures of closed networks of interstrand hydrogen bonds. In contrast, α-helical membrane proteins fold within the membrane by assembling individually formed transmembrane (TM) helices into bundles.14,15 Therein Trp may influence both the unfolded and folded state of α-helical membrane proteins; whereas in β-barrel membrane proteins the influence of Trp on the free energy of the unfolded, random coil state appears modest relative to the other residues. This circumstance renders the folding pathways of α-helical and β-sheet membrane proteins distinct. Considering α-helical membrane proteins form most membrane proteins of eukaryotic origin and represent drug targets of primary importance to human health, it is essential to define Trp anchoring in this class of proteins.
Complementary to the amphiphilic environment of lipid headgroups (Figure 1A), the character of the indole sidechain of Trp is amphiphilic with a polar N-H bond, permanent dipole moment and hydrophobic ring of anisotropic electron distribution (Figure 1B). The interaction of (the) indole (sidechain) with lipid bilayers encompasses electrostatic, dipolar, hydrophobic and steric effects.16–20 Indole N1 proton hydrogen bonding with lipid carbonyl and phosphate moieties is detected.18,21,22 The π electron cloud of the indole ring facilitates interactions with lipid cations, most prominently the choline group (Figure 1A).18,22 The distinct electrostatic gradient across the lipid bilayer may contribute to the localization of the molecular dipole of indole in the headgroup region.17,21 The hydrophobic effect favors the exclusion of indole from the aqueous milieu, while the immersion of the large, flat indole ring structure into the hydrophobic core may be entropically unfavorable.6,16,17,23 Despite the many studies on Trp, there is a lack of a consensus view on the relative importance of the outlined contributions. This uncertainty is compounded by the frequent study of isolated indole, outside its protein helical environment, and the use of model lipid environments that simplify the lipid heterogeneity of native membranes.
Figure 1.
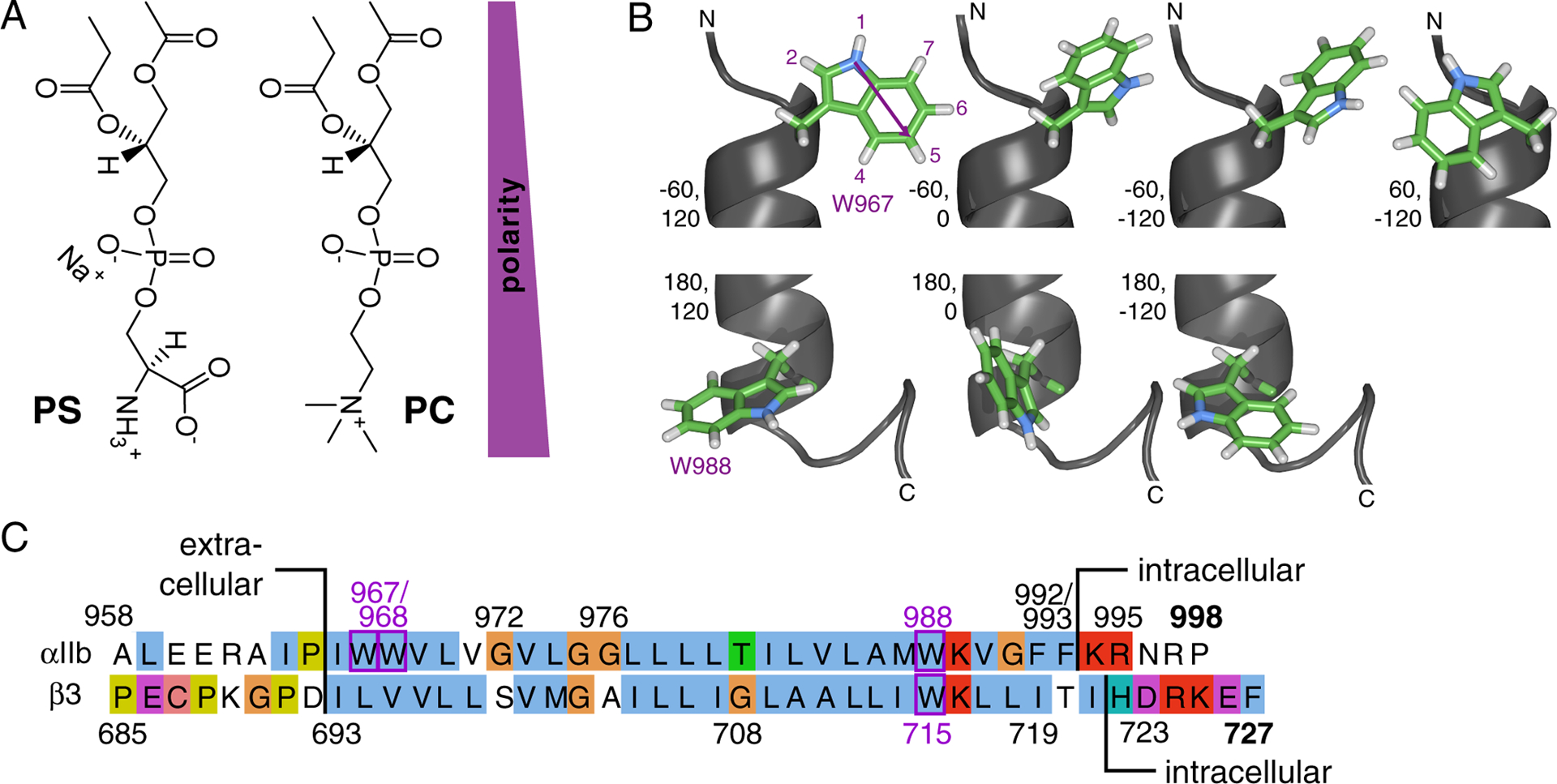
Overview of experimental system: lipids, tryptophan and integrin TM sequences. (A) Illustration of headgroup structures of phosphatidylserine (PS) and -choline (PC)-based lipids in extended conformations. (B) Structure of the Trp sidechain and rotamers populated.54 The indole ring has a permanent dipole moment of 2.1 Debye pointing from N1 in the five-membered ring to C5 in the six-membered ring.55 The N1 proton is the most acidic proton on indole and a hydrogen bond donor. Depicted are N-terminal W967 (χ1 ≈ −60°) and C-terminal W988 (χ1 ≈ 180°) of the integrin αIIb TM helix (PDB ID 2k1a)26 with χ2 ≈ −120°, 0°, 120°. For the benefit of completeness, W967 is also depicted with χ1 ≈ 60°, χ2 ≈ −120°. Rotamers were built using chimera.56 (C) Amino acid sequence of human integrin αIIb and β3 TM segments and flanking regions.
Here, we examine the folding of the integrin αIIbβ3 TM complex from its constituent αIIb and β3 TM helices with respect to three of its four Trp residues in phospholipid bicelles and mammalian membranes. αIIb(W967) and αIIb(W968) populate the extracellular headgroup region, whereas αIIb(W988) and β3(W715) reside in the intracellular membrane interface (Figure 1C).24–26 In addition, αIIb(W968) and β3(W715) participate in intersubunit contacts. Among the 18 and 8 human integrin α and β subunits, respectively, Trp at position 715 is conserved in 75% of β subunits, at positions 967 and 988 in 61% of α subunits, and only αIIb exhibits Trp at position 968 (Figure S1). Accordingly, W967 and W988 can serve as paradigms of extra- and intracellular Trp anchors and W715 as prototypical anchor involved in interhelical packing. Physiologically, the assembled αβ TM complex maintains the resting, non-adhesive state of integrin cell adhesion receptors,27,28 which additionally brings forth insight into the function and design principles of this widespread receptor family.29 Specifically, the present study defines the contribution of Trp to α-helical membrane protein stability, provides insight into the structural basis underlying its anchoring ability, and unveils functional implications of anchoring in integrins.
MATERIALS and METHODS
Protein Expression and Purification.
Human integrin αIIb(A958-P998) and β3(P685-F727) TM peptides incorporating β3(C687S) were produced as described.25,26 The examined Trp substitutions were introduced by QuikChange mutagenesis, and the preparation of the corresponding peptides followed the protocols for wild-type peptides. Peptide concentrations were measured by UV spectroscopy using ε(αIIb)280nm= 16,500 M−1cm−1 and ε(β3)280nm= 5,500 M−1cm−1 for wild-type peptides and adjusted by −4,010 and −5,500 M−1cm−1 for tyrosine and all other substitutions, respectively. For β3 variants with ε280nm= 0 M−1cm−1, the absorbance at 205 nm was used instead with ε205nm calculated following ref.30. Purified, freeze-dried peptides were dissolved in 67% CH3CN/33% H2O for concentration measurements and again freeze-dried.
Fluorescence Spectroscopy.
Measurements were carried out at room temperature using a Varian Cary Eclipse fluorescence spectrophotometer with an excitation wavelength of 290 nm and excitation and emission slit widths of 10 and 2.5 nm, respectively. The intrinsic fluorescence of single Trp variants of αIIbβ3 were measured at the peptide concentrations tabulated in Figure S2. Freeze-dried peptides were dissolved in 43 mM DHPC (1,2-dihexanoly-sn-glycero-3-phosphocholine), 17 mM POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), 25 mM NaH2PO4/Na2HPO4 (pH 7.4) solution. Three samples were prepared independently for each data point. Dimer samples were mixed in 67% CH3CN/33% H2O before freeze-drying. Peak emission maxima, λmax, and widths, Γ, were obtained by fits to a log-normal distribution.31,32
Isothermal Titration Calorimetry (ITC).
Measurements were carried on a Microcal VP-ITC calorimeter at 28 °C in 43 mM DHPC, 17 mM POPC, 25 mM NaH2PO4/Na2HPO4 (pH 7.4) solution. 10 μM of β3 peptide in the 1.425 ml sample cell was titrated with αIIb peptide by injecting 9 μl aliquots over a period of 10 s until binding saturation was reached (Figure S3). Prior to data analysis, the measurements were corrected for the heat of dilutions of the αIIb and β3 peptides. The reaction enthalpy (ΔH°) and the αIIb + β3 ⇆ αIIbβ3 equilibrium constant on the mole fraction scale (KXY) were calculated from the measured heat changes, δHi, with the αIIbβ3 complex stoichiometry fixed at 1:1 as described previously.33 The free energy change ΔG° is obtained as −RT ln KXY where R denotes the gas constant and T the absolute temperature. The entropy change, ΔS°, is obtained as (ΔH° − ΔG°) / T. The previous assessed reproducibility and accuracy of individual titrations were found to lie within approximately 0.05 kcal/mol.33,34 For that reason, only one titration experiment was conducted for most of the studied substitutions.
Integrin αIIbβ3 TM Complex Displacement Assay.
The previously introduced mini integrin αIIb and β3 constructs were used in which the FLAG sequence is fused to the N-terminus of TM-cytosolic tail sequence of αIIb and the extracellular domain of Tac to the corresponding β3 sequence.35 Chinese hamster ovary (CHO) cells stably expressing integrin αIIbβ3 were transfected with the desired mini integrin. To avoid sequestration of endogenously expressed talin by β3 constructs,36 the Y747A substitution, which disrupts talin binding,37 was introduced in all β3 mini integrins. As negative controls for the competitive effects of αIIb and β3 mini integrins on integrin αIIbβ3 activation, a non-interacting, neutral membrane protein was studied; the protein Tie2 was fused to the FLAG tag and the extracellular domain of Tac. Cells were detached at 24 hr post-transfection, and approximately 5×105 cells were incubated with anti-FLAG or anti-Tac antibody and integrin αIIbβ3 ligand-mimetic antibody PAC1.38 After washing, cells were further incubated with fluorophore-conjugated secondary reagents and analyzed by a FACSCalibur (BD Biosciences). The mean fluorescence intensities (MFIs) of PAC1 binding to cells expressing different, moderate levels of mini integrins were calculated, as described,39 and the MFIs of PAC1 binding to negative control-transfected cells were subtracted. The data were fit to a one-site competitive binding curve, MFI(PAC1) = A2 + (A1-A2) / (1 + 10^(log(MFI(FLAG/Tac))-log(A0))), to extract PAC1max= A2-A1 (Figure S4). For each Trp substitution, six independent experiments were conducted and averaged.
RESULTS and DISCUSSION
Integrin Tryptophans Primarily Reflect the Electrostatic Potential of Surrounding Amino Acid Residues.
To obtain a quantitative indicator of Trp structural environments, we determined the fluorescence emission maxima, λmax, of individual Trp residues in monomeric and dimeric αIIbβ3 TM states. Specifically, to detect only one Trp, we substituted all others with amino acids that did not significantly affect TM complex stability (see below). Invariant TM complex stabilities suggest invariant TM helix orientations between mutants and wild type. Phospholipid bicelles consisting of 1,2-dihexanoly-sn-glycero-3-phosphocholine (DHPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) with an effective q-factor, qeff=[POPC]/[DHPC]bound, of 0.5 served as membrane mimic for integrins and was validated previously.25,27,40,41 For the monomeric TM domains, λmax values were almost evenly distributed between 328 and 342 nm (Figure 2A and Table 1), which identified different structural environments for each Trp that range from purely hydrophobic to partially water exposed milieus.42,43 The wide range of λmax value is noteworthy for a membrane protein;9 it reveals a wide variety of Trp membrane environments and shows that integrin αIIbβ3 is a good model system for representing diverse interfacial Trp.
Figure 2.
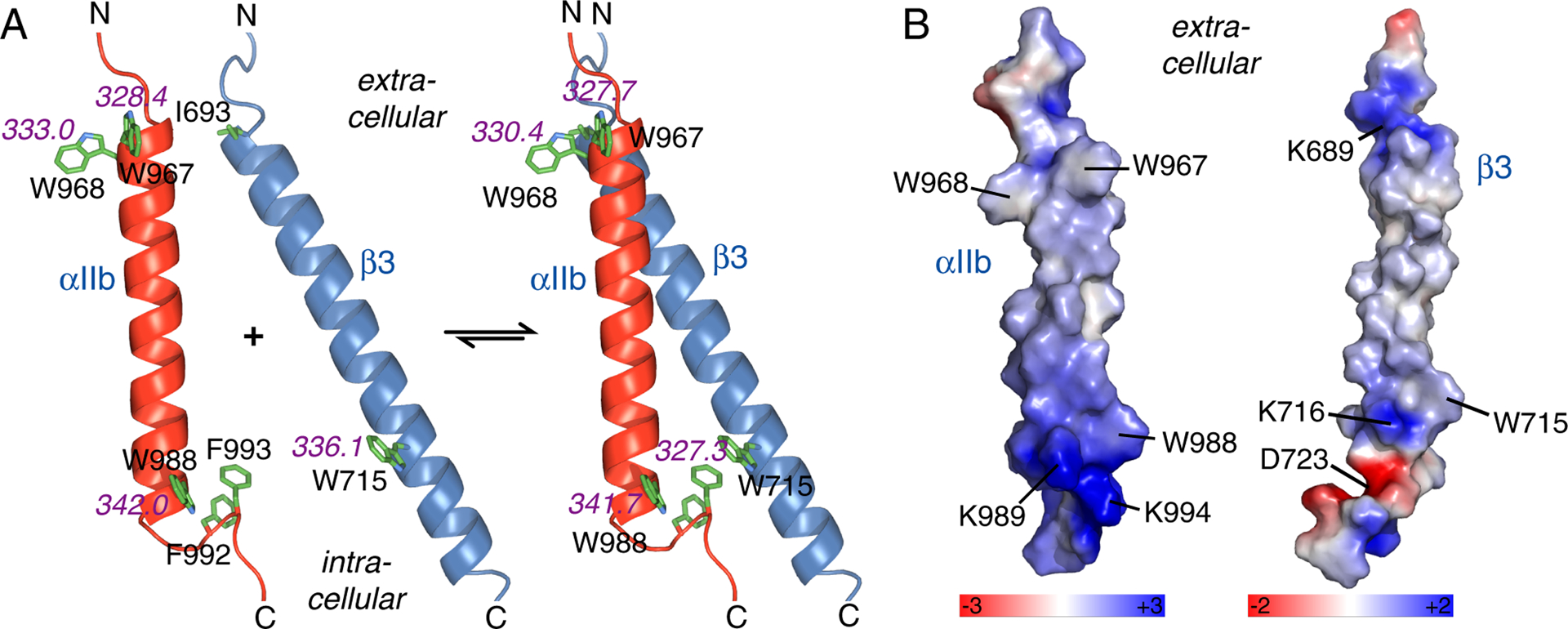
Properties of tryptophan residues in integrin αIIbβ3 TM domains. (A) The measured fluorescence emission maxima (λmax) of the four TM tryptophans in monomeric and dimeric states25–27 are shown in magenta (Table 1 and Figure S2). For structural context, αIIb(Phe992-Phe993) and β3(I963) are also depicted. (B) The calculated electrostatic environment of the integrin tryptophans in monomeric TM helix states is illustrated. Specifically, the molecular surfaces of αIIb and β3 TM helices were color-coded by electrostatic potential as depicted. The potential was calculated with the PyMOL APBS tool.
Table 1.
Integrin αIIbβ3 TM tryptophan fluorescence emission peak maxima (λmax) and widths at half-height (Γ)a
| Tryptophanb | Monomerc λmax [nm] |
Monomerc Γ [nm] |
Dimerc λmax [nm] |
Dimerc Γ [nm] |
|---|---|---|---|---|
| αllb(W967) | 328.4 ± 0.1 | 51.8 ± 0.1 | 327.7 ± 0.2 | 50.0 ± 0.6 |
| αllb(W968) | 333.0 ± 0.1 | 53.8 ± 0.1 | 330.4 ± 0.1 | 51.6 ± 0.2 |
| αllb(W988) | 342.0 ± 0.2 | 59.9 ± 0.6 | 341.7 ± 0.4 | 58.0 ± 2.3 |
| β3(W715) | 336.1 ± 0.2 | 56.5 ± 0.6 | 327.3 ± 0.2 | 49.2 ± 0.4 |
Measurements were performed in 43 mM DHPC, 17 mM POPC, 25 mM NaH2PO4/Na2HPO4 (pH 7.4) solution at room temperature (Figure S1). The presence of 9 mM free, non bicellar DHPC57 and resulting effective q-factor (qeff) of 0.5 is noted.
The fluorescence of αllb(W988), αllb(W968) and αllb(W967) was determined using αllb(W967V/W968V), αllb(W988H/W967V) and αllb(W988H/W968V) peptides, respectively, in the absence and presence of saturating amounts of β3(W715I) peptide. The fluorescence of β3(W715) was determined using β3 peptide in the absence and presence of saturating amounts of αllb(W967V/W968V/W988H) peptide.
Mean and SEM determined from three independently prepared samples.
Notably, the Trp residues on the extracellular side experienced a significantly more hydrophobic environment compared to their intracellular counterparts. In terms of membrane immersion, the αIIb TM helix, which encompasses I966-K989, is centered in the bicelle44 positioning W967 and W988 at similar immersion depths. Nonetheless, αIIb(W967) and αIIb(W988) resided in the most apolar and polar environments, respectively (Figure 2A). The electrostatic potential surface of the TM helices shows that W967 and W968 are surrounded by hydrophobic sidechains whereas W715 and W988 reside in proximity to K716 and K989, respectively (Figure 2B). In addition, a second Lys at position 994 is a structural neighbor of W988. Upon dimerization, λmax of αIIb(W967) and αIIb(W988) remained unchanged confirming their absence from the helix-helix interface. In contrast, β3(W715) and, to a lesser extent, αIIb(W968) experienced blue shifts (Figure 2A). Upon TM complex formation αIIb(F993) inserts between β3(W715) and β3(K716) shielding Trp from Lys. The apolar environment created by αIIb(F993) and a relatively deep membrane immersion of β3(W715) can explain its large shift of −8.8 nm upon dimerization (Table 1), which reflects a fully apolar environment. The −2.6 nm shift for αIIb(W968) upon dimerization can be explained by a reduction of polar interactions, which likely originated from lipid headgroups, by the proximity of β3(I693) (Figure 2A). The association of αIIb and β3 TM helices was further accompanied by a reduction in fluorescence peak widths (Figure S2). Peak widths are sensitive to the heterogeneity of tryptophan environments,31,42 thus indicating a reduction in conformational heterogeneity upon TM complex assembly. In sum, it appears that interfacial Trp primarily and versatilely responded to the local electrostatic potentials created by the TM helices with the bicelle lipids adjusting to this environment rather than altering it.
A Single Tryptophan Anchors the αIIb TM Helix to Extracellular Lipids.
A complementary approach of probing the nature of the Trp structural environment is to assess the consequences of Trp substitutions on the free energy of αIIbβ3 TM complex formation, termed ΔG°. The change of ΔG° upon substituting Trp with amino acid X, termed ΔΔG°WX, gives not only insight into the nature of protein-lipid interactions but also into the contribution of Trp to protein stability and anchoring, and, if localized in the dimer interface, the strength of local helix-helix contacts. Moreover, ΔΔG°WX is sensitive to differences between wild type and mutant at the level of individual, dissociated TM helices. This dependence relates to the fact that ΔG° will be most favorable if TM helix orientations match between dissociated and associated TM helices (Figure 3). Therefore encompassed in the definition of TM helix orientation are TM helix tilt and rotation angles, TM helix immersion depths, and the amplitudes of fluctuation around mean parameters. ΔΔG°WX was measured by isothermal titration calorimetry in DHPC/POPC bicelles (qeff=0.5) (Figure S3). In this environment, the TM helices are predominantly surrounded by long-chain lipids44 and experience contributions from helix-helix preorientation33,45 while ideal solvent behavior is maintained.33,46 Moreover, kinetic parameters indicate the reversibility of αIIbβ3 TM association33,40 and ΔG° measurements at different protein-lipid ratios indicate a measurement accuracy of approximately 0.05 kcal/mol,34 which enables the differentiation of even subtle amino acid substitutions. In the present study, we substituted the conserved αIIb(W967), αIIb(W988) and β3(W715) residues with aromatic and select aliphatic residues including Ala.
Figure 3.
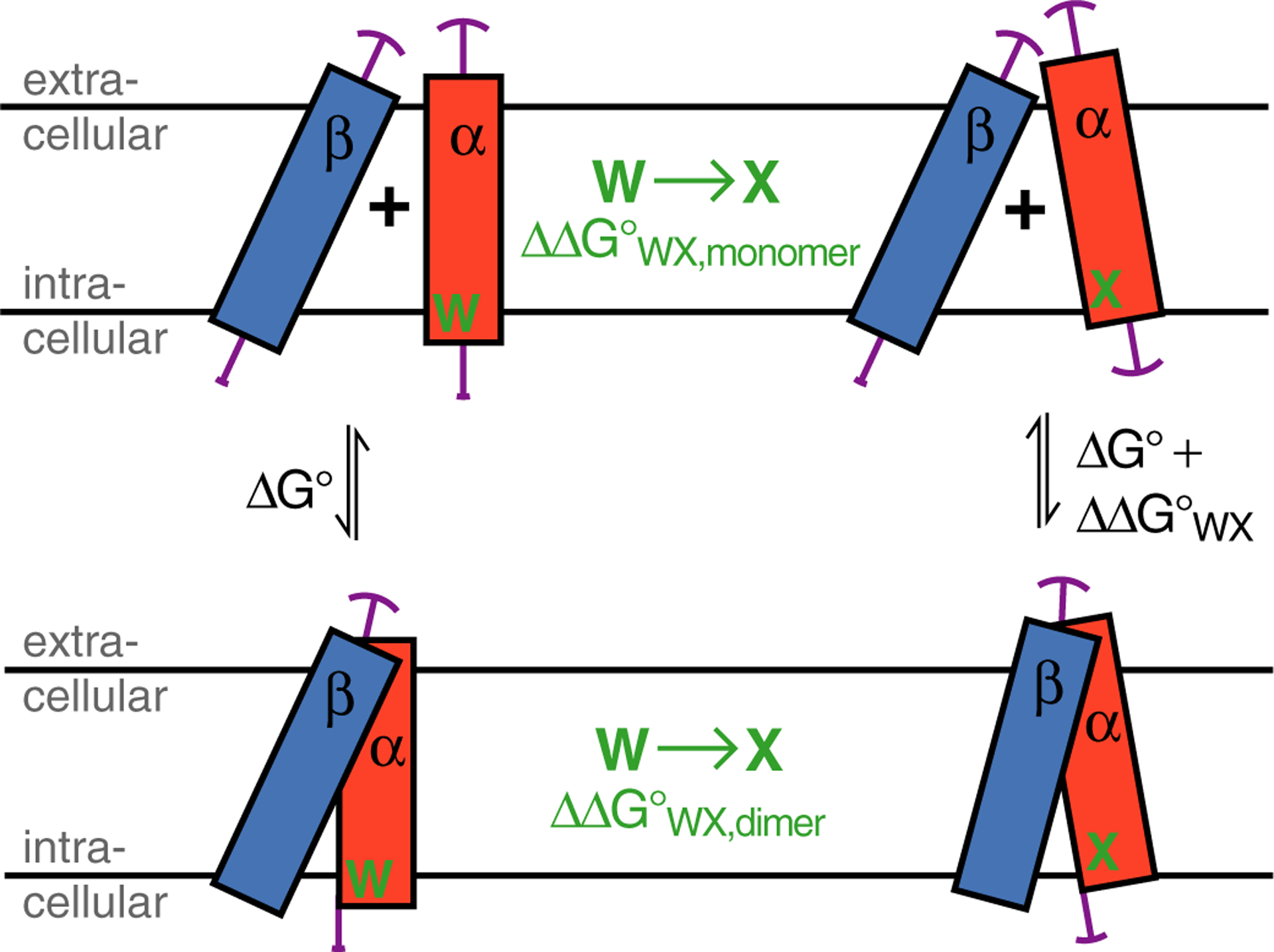
Illustration of the possible effects of Trp substitution on the folding of the integrin TM complex. A Trp substitution in integrin subunit α to residue X, termed WX, may alter its TM helix tilt and rotation, immersion depth and amplitudes of fluctuation around mean positions (illustrated by magenta arcs). The subsequent association with unperturbed subunit β is a compromise between ideal αβ helix-helix association and any required change to β TM helix orientation in order to reach the dimeric state. The αβ TM complex stability, termed ΔG°, changes upon WX substitution by ΔΔG°WX. Moreover, the effects of WX can also be seen in isolation for the monomeric and dimeric state, respectively, i.e., ΔΔG°WA,monomer underlies the transition from α to α(WX) orientations and ΔΔG°WX,dimer describes the change from αβ to α(WX)β orientations. Altogether, ΔG° + ΔΔG°WX,dimer = ΔΔG°WX,monomer + (ΔG° + ΔΔG°WX), which simplifies to ΔΔG°WX,dimer = ΔΔG°WX,monomer + ΔΔG°WX.
The passage of the αIIb TM helix through extracellular lipid headgroups is devoid of polar residues and appears to rely solely on Trp anchoring by W967-W968 (Figure 1C). Based on its low λmax value, αIIb(W967) was expected to tolerate hydrophobic substitutions, however, in the presence of αIIb(W968), it was insensitive to any of the implemented substitutions (Figure 4A and Table 2). Small TM complex stabilizations were even observed for all examined except His substitutions. The polar nature of His implied that local lipid associations have changed; however, perhaps in relation to the continued presence of adjacent W968, TM helix orientation was apparently little affected. We tested this assumption by examining αIIb(W967) substitutions with non-conserved W968 mutated to Val and obtained pronounced TM complex destabilizations (Figure 4A). This behavior highlights that W968 indeed compensated for the loss of W967 and shows that one Trp is already a good membrane anchor, consistent with the presence of a single Trp (W967) at the extracellular border of most integrin α subunits (Figure S1). Among the tested substitutions (Figure 4A and Table 2), Tyr provided the best compensation for W967 within the W968V background, highlighting their common amphiphilic nature.
Figure 4.
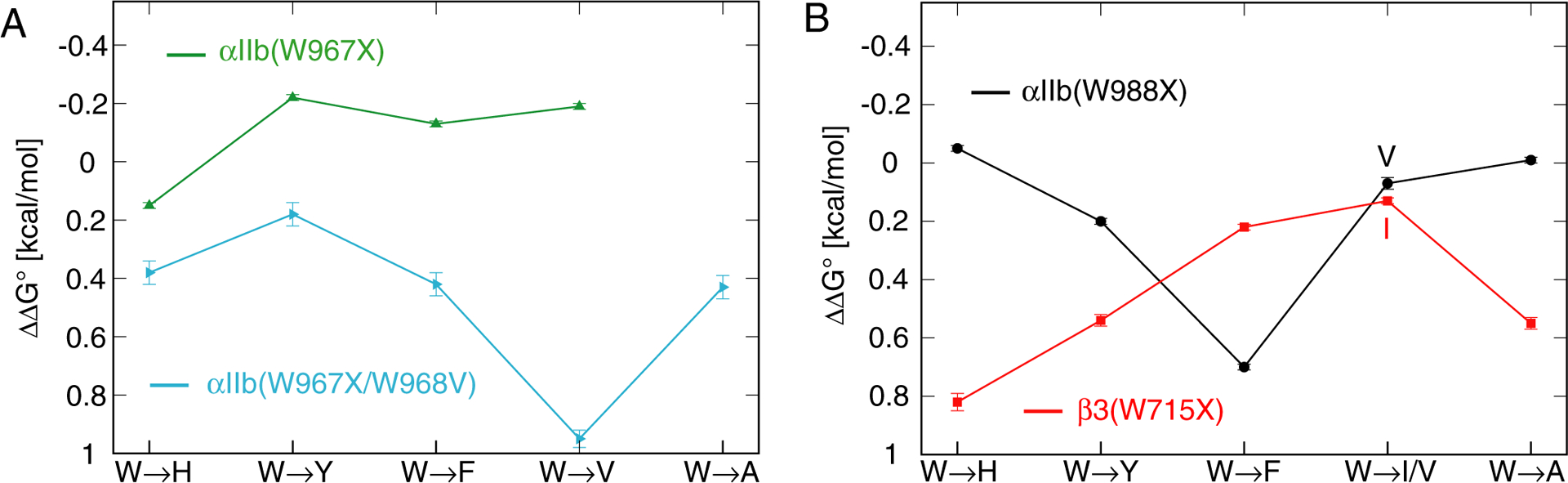
Integrin αIIbβ3 TM complex stability in bicelles as a function of tryptophan substitution. (A-B) For the depicted substitutions, the change of TM complex stability, ΔΔG° = ΔG°mutant − ΔG°wildtype, was measured by ITC in qeff=0.5 bicelles (Table 2).
Table 2.
Integrin αIIbβ3 TM complex stability as a function of tryptophan substitutionsa
| Peptides | KXYa | ΔH° [kcal/mol] |
TΔS° [kcal/mol] |
ΔG° [kcal/mol] |
ΔΔG° [kcal/mol]b |
|---|---|---|---|---|---|
| αIIb + β3 | 3250 ± 60 | −16.0 ± 0.1 | −11.1 ± 0.1 | −4.84 ± 0.01 | – |
| αIIb(W988H) + β3 | 3540 ± 40 | −16.4 ± 0.1 | −11.5 ± 0.1 | −4.89 ± 0.01 | −0.05 ± 0.01 |
| αIIb(W988Y) + β3 | 2320 ± 50 | −19.2 ± 0.2 | −14.6 ± 0.2 | −4.64 ± 0.01 | 0.20 ± 0.01 |
| αIIb(W988F) + β3 | 1000 ± 20 | −16.4 ± 0.2 | −12.3 ± 0.2 | −4.14 ± 0.01 | 0.70 ± 0.01 |
| αIIb(W988V) + β3 | 2900 ± 100 | −13.7 ± 0.2 | −8.9 ± 0.2 | −4.77 ± 0.02 | 0.07 ± 0.02 |
| αIIb(W988A) + β3 | 3320 ± 70 | −17.3 ± 0.1 | −12.4 ± 0.1 | −4.85 ± 0.01 | −0.01 ± 0.01 |
| αIIb + β3(W715H) | 820 ± 40 | −8.7 ± 0.2 | −4.6 ± 0.3 | −4.02 ± 0.03 | 0.82 ± 0.03 |
| αIIb + β3(W715Y) | 1320 ± 40 | −14.2 ± 0.2 | −9.9 ± 0.2 | −4.30 ± 0.02 | 0.54 ± 0.02 |
| αIIb + β3(W715F) | 2230 ± 50 | −17.2 ± 0.2 | −12.6 ± 0.2 | −4.62 ± 0.01 | 0.22 ± 0.01 |
| αIIb + β3(W715I) | 2600 ± 70 | −9.6 ± 0.1 | −4.9 ± 0.1 | −4.71 ± 0.01 | 0.13 ± 0.01 |
| αIIb + β3(W715A) | 1300 ± 40 | −15.5 ± 0.3 | −11.2 ± 0.3 | −4.29 ± 0.02 | 0.55 ± 0.02 |
| αIIb(W967H) + β3 | 2530 ± 50 | −15.4 ± 0.1 | −10.7 ± 0.1 | −4.69 ± 0.01 | 0.15 ± 0.01 |
| αIIb(W967F) + β3 | 4030 ± 90 | −12.6 ± 0.1 | −7.6 ± 0.1 | −4.97 ± 0.01 | −0.13 ± 0.01 |
| αIIb(W967Y) + β3 | 4700 ± 100 | −13.7 ± 0.2 | −8.6 ± 0.2 | −5.06 ± 0.01 | −0.22 ± 0.01 |
| αIIb(W967V) + β3 | 4450 ± 80 | −13.1 ± 0.1 | −8.0 ± 0.1 | −5.03 ± 0.01 | −0.19 ± 0.01 |
| αIIb(W968V) + β3 | 13200 ± 600 | −14.3 ± 0.2 | −8.6 ± 0.2 | −5.68 ± 0.03 | – |
| αIIb(W967H/W968V) + β3 | 7000 ± 200 | −14.9 ± 0.2 | −9.6 ± 0.2 | −5.30 ± 0.02 | 0.38 ± 0.04 |
| αIIb(W967F/W968V) + β3 | 6500 ± 200 | −15.7 ± 0.2 | −10.5 ± 0.2 | −5.26 ± 0.02 | 0.42 ± 0.04 |
| αIIb(W967Y/W968V) + β3 | 9800 ± 400 | −14.0 ± 0.2 | −8.5 ± 0.2 | −5.50 ± 0.02 | 0.18 ± 0.04 |
| αIIb(W967V/W968V) + β3 | 2680 ± 60 | −17.8 ± 0.2 | −13.0 ± 0.2 | −4.73 ± 0.01 | 0.95 ± 0.03 |
| αIIb(W967A/W968V) + β3 | 6400 ± 200 | −18.4 ± 0.2 | −13.1 ± 0.2 | −5.25 ± 0.02 | 0.43 ± 0.04 |
Measurements in 43 mM DHPC, 17 mM POPC, 25 mM NaH2PO4/Na2HPO4 (pH 7.4) at 28 °C. The presence of 9 mM free, non bicellar DHPC57 and resulting effective q-factor (qeff) of 0.5 is noted.
ΔΔG° = ΔG°mutant – ΔG°wildtype and, for αIIb(W967/W968V) substitutions, ΔG°mutant – ΔG°αIIb(W968V)β3.
At the Intracellular Membrane Border, αIIb(W988) is Part of a Multi-Pronged Anchoring Motif.
On the intracellular side, W988 and K989 terminate the TM helix. Furthermore, the succeeding, universally conserved GFF motif of integrins (Figure 1C and Figure S1) positions the aromatic Phe rings back into the membrane core (Figure 2A).26 For that reason, anchoring to intracellular lipids and the ensuing TM helix orientation may depend on lipid interactions beyond Trp. Consistent with λmax = 342 nm of αIIb(W988) in both monomeric and dimeric states, His fully compensated Trp at this position (Figure 4B). αIIb(W988) was remarkably sensitive to substitution by Phe (ΔΔG°= 0.70 ± 0.01 kcal/mol; Table 2), while the Tyr mutation was intermediate. Judging from the structure of αIIb, W988F would enhance the lipid immersion of Phe992-Phe993 (Figure 2A). The strong effect of W988F on ΔG° was contrasted by the minor effects of W988V and W988A (Figure 4B). We hypothesize that the membrane immersion tendency of Phe is stronger than that of Val or Ala and apparently above a threshold value required to change TM orientation. In other words, the αIIb TM orientation at the intracellular membrane border changed in a stepwise rather than linear manner. The invariance of Val and Ala mutations in case of αIIb(W988) appears to relate to a multi-pronged anchoring of αIIb at the intracellular leaflet. Phe992-Phe993 have strong tendencies to be membrane immersed, whereas K989 and W988 act as polar counterbalances in a multi-pronged anchoring motif (Figure 5). In conclusion, a more narrowly defined αIIb TM helix orientation can ensue at the C-terminal as compared to the N-terminal helix end (Figure 3) that, in bicelles, can do without W988 anchoring.
Figure 5.
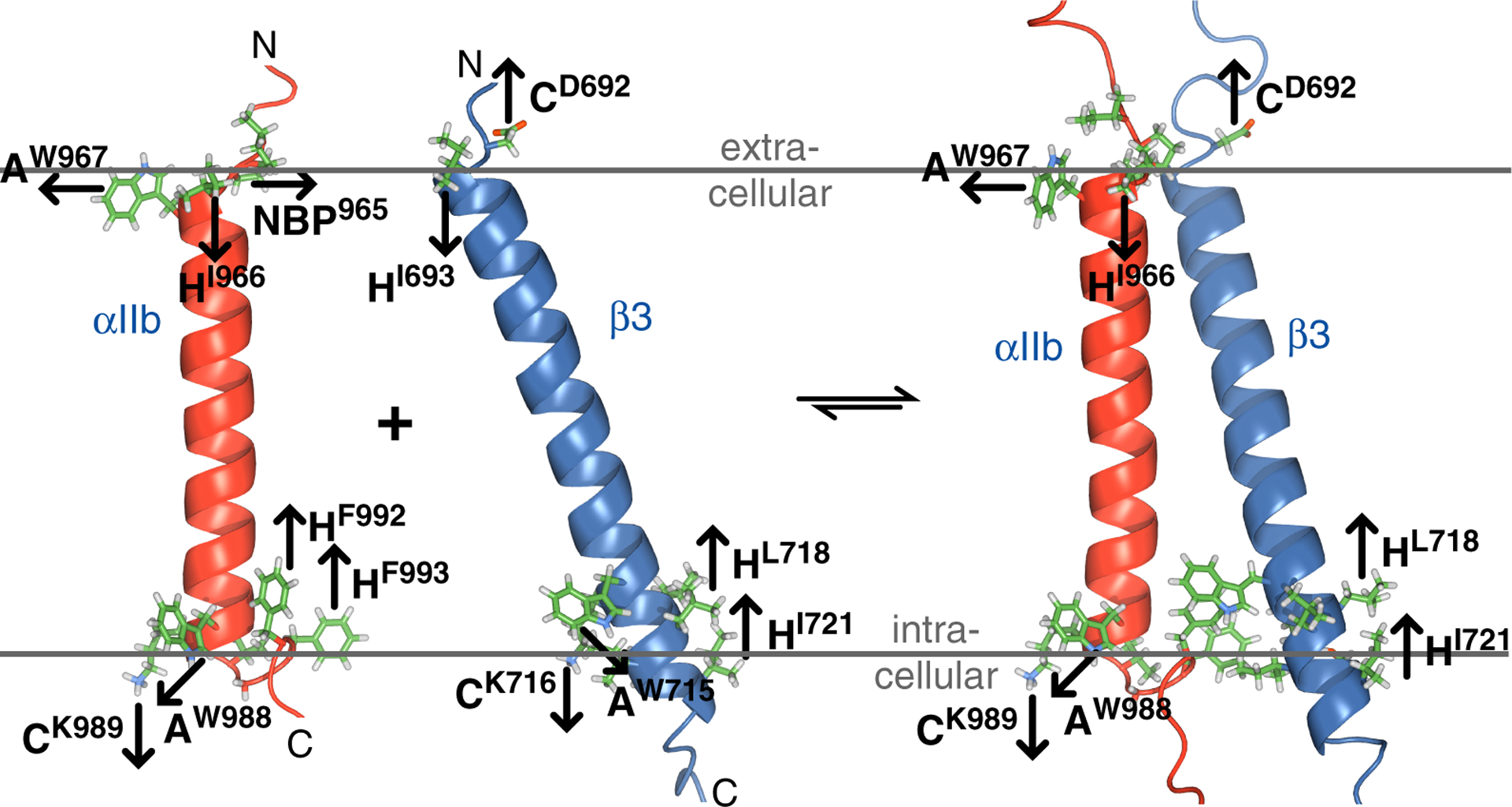
Membrane anchoring of integrin αIIbβ3 TM domains. Proposed anchoring tendencies of αIIb and β3 residues at membrane borders in monomeric and dimeric states are indicated by arrows that depict tendencies to reach the membrane core, the lipid headgroup region or the aqueous solution. Trp is shown with site-specific anchoring tendencies. Amphiphilic, charged and hydrophobic residues are abbreviated A, C and H, respectively. NBP denotes an N-terminal border proline.49 For visual clarity, not all H residues of the AαIIb(W988)CαIIb(K989)HαIIb(G991-F993) and Aβ3(W715)Cβ3(K716)Hβ3(L717-I721) anchoring motifs are shown.
The Structural Environment of β3(W715) Highlights Its Superiority over Tyrosine.
In homology to αIIb, W715 and K716 are found at the intracellular membrane interface of the β3 TM helix (Figure 1C). However, the succeeding hydrophobic β3(LLITI) sequence continues the TM helix in contrast to αIIb(GFF) (Figure 1C and 2A). Although structurally different from αIIb, an Amphiphilic-Charged-Hydrophobic (ACH) motif is again formed where Aβ3(W715) and Cβ3(K716) constitute a counterbalance to Hβ3(L717-I721). Notably, upon dimerization, Aβ3(W715), Cβ3(K716) and HαIIb(G991-F993) become part of the helix-helix interface, and a new, combined ACH motif, AαIIb(W988)CαIIb(K989)Hβ3(L717-I721), develops instead (Figure 5). This structural context differentiates β3(W715) from αIIb(W988).
Based on the hydrophobic environment of β3(W715) in the dimeric state (λmax= 327.3 ± 0.2 nm; Figure 2A), W715 should not be replaceable with His like αIIb(W988). Its substitution pattern validated this view: β3(W715) did not tolerate His and, remarkably, likewise Tyr was poorly accepted (Figure 4B). In contrast, Phe was accepted with little penalty and W715I tolerated best. The tolerance of Ile and Phe only appears possible because Cβ3(K716) maintains a polar counterbalance to Hβ3(L717-I721). The tolerance of the loss of the W715 aromatic rings (Figure 1B) implies that αIIb(Phe993)-β3(W715) interactions (Figure 2A) were mostly steric in nature. This view was supported by the loss of TM complex stability for the Ala when compared to Ile mutation (Figure 4B).
Despite the good replacement of β3(W715) with Ile in bicelles, we consider this site to constitute the most challenging Trp environment. Trp has to provide anchoring interactions of polar nature in the monomer (λmax= 336.1 ± 0.2 nm) and hydrophobic intersubunit contacts in the dimer (λmax= 327.3 ± 0.2 nm). In bicelles, it was sufficient to replicate the hydrophobic characteristic but membranes may not forgive the absence of polar properties. The substitution pattern of β3(W715) identifies a relatively poor performance by Tyr. While Tyr also has amphiphilic properties, as shown here by αIIb(W967Y/W968V) anchoring (Figure 4A and Table 2), it appears unable to match the range of polarities that Trp can display. In relation to the larger and more anisotropic sidechain of Trp compared to Tyr, the different Trp sidechain rotamers may confer site-specific polarities (Figure 1B).
Contribution of a Lone Tryptophan to Integrin αIIbβ3 TM Complex Stability.
Within the αIIb(W968V) background, the N-terminal membrane anchoring of αIIb primarily depends on W967 (Figure 1C and 4A) and, thus, offers a suitable setting to assess the free energy contribution of a lone Trp to α-helical membrane protein stability. Its Ala substitution led to a loss of 0.4 kcal/mol in bicelles (Table 2). Because W967 is not in the dimerization interface (Figure 2A), this loss will arise from weakened TM helix-helix interactions that originated from changes in αIIb TM helix orientation in monomer and/or dimer (Figure 3). In the context of other membrane proteins, this value will depend on the sensitivity of interhelical interactions on TM helix orientation (Figure 3). More destabilizing substitutions of the studied Trp residues lost approximately 0.8 kcal/mol (Figure 4), which may approximate an upper limit for the contribution of Trp anchoring in α-helical membrane proteins.
The Hydrophobic Effect is Larger in Membranes than Bicelles.
To our knowledge, thermodynamic aspects of Trp anchoring has only been studied systematically in synthetic membranes.12,13 Native membranes are continuous bilayers that contain a wide range of different lipids in terms of hydrocarbon tail length and headgroup identity.47,48 Intrinsically, their increased complexity relative to bicelles makes them more likely to discriminate between different Trp substitutions. To explore a bona fide membrane environment, we have developed an assay that allows the quantification of the relative stabilities of Trp substitutions in mammalian membranes. Specifically, we set up a displacement assay in CHO cell membranes where the TM complex of full-length integrin αIIbβ3 is exposed to a competing interaction from increasing amounts of an overexpressed integrin TM peptide (Figure 6A). Whenever the overexpressed peptide is able to form a non-receptor TM complex, the inactive ectodomains lose their TM complex-conferred stabilization, permitting receptor activation. Receptor activity levels were monitored by the ligand-mimetic PAC1 antibody (Figure 6A)27,49 and the amount of bound PAC1 at saturating peptide concentrations, termed PAC1max, was quantified by fluorescence spectroscopy (Figure S4). Accordingly, the efficiency of receptor activation by the competing peptide provides quantitative insight into the affinity of the overexpressed peptide for its partnering subunit.
Figure 6.
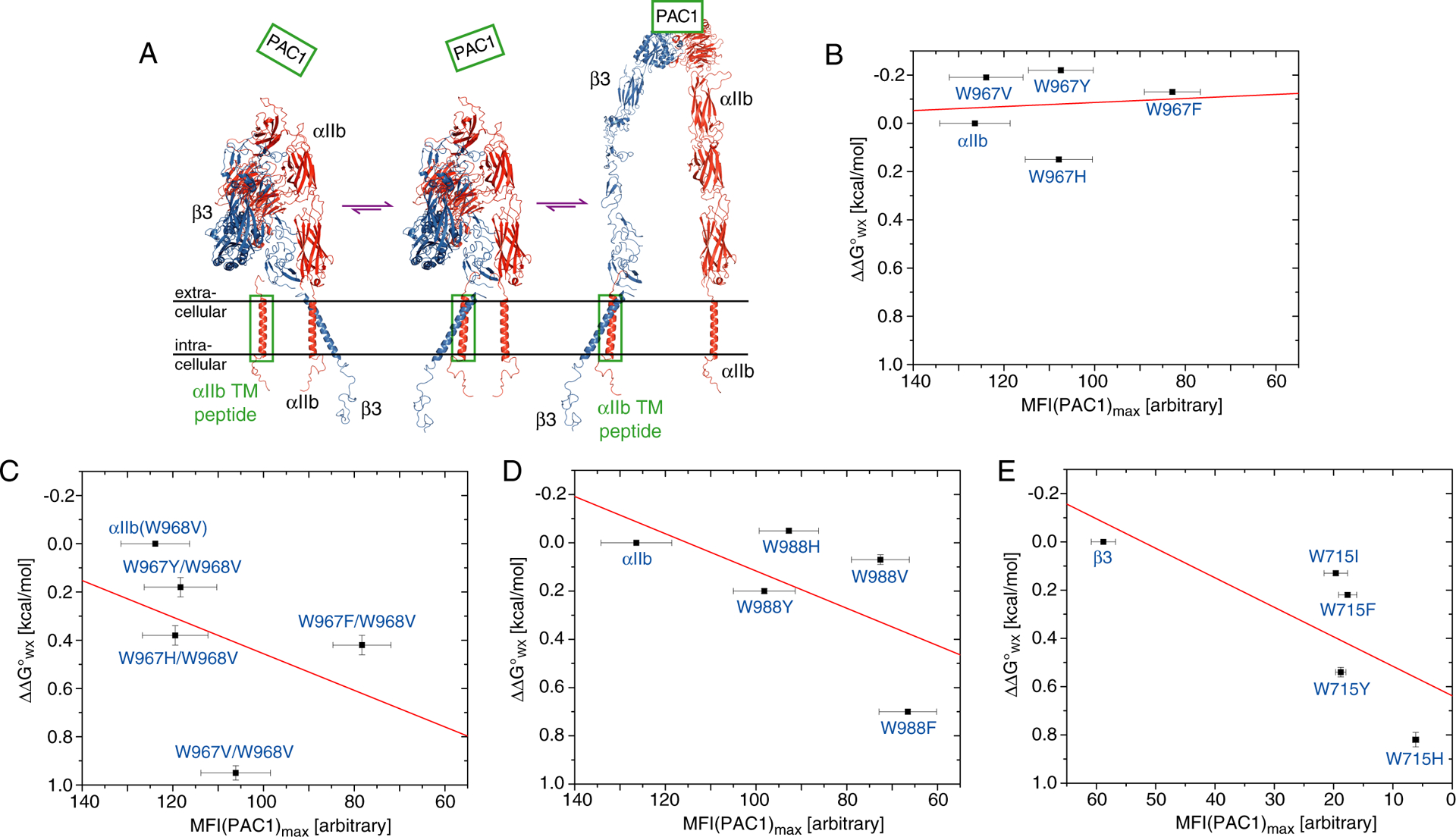
Correlation of αIIbβ3 TM complex stabilities in phospholipid bicelles and mammalian membranes. (A) Displacement assay employed to measure relative TM complex stabilities in membrane. Increasing amount of overexpressed integrin TM peptide, boxed in green, displaces its corresponding TM segment in the native receptor, leading to integrin activation as monitored by the binding of the ligand-mimetic antibody PAC138 (Figure S4). (B-E) For the depicted substitutions, the level of maximal receptor activation, MFI(PAC1max), obtained at saturating concentration of overexpressed TM peptide is correlated with the change of TM complex stability in bicelles, ΔΔG°WX (Figure 4 and Table 2). Linear fits are shown in red mainly to illustrate the absence of any strong correlation.
To assess whether peptide-lipid interactions parallel each other in bicelles and membranes, we compared PAC1max with ΔΔG°WX for substitutions of αIIb(W967), αIIb(W967/W968V), αIIb(W988) and β3(W715). As in bicelles, both αIIb(W967) and αIIb(W967/W968V) were replaceable in membranes within experimental uncertainties (Figure 6B–C). Val was able to replace W967 in the membrane, but, unlike in bicelles, Tyr, His and Phe were now inferior. Evidently, residue selectivity increased and, adjacent to W968, only Val had the adequate hydrophobicity to maintain the proper TM helix orientation. In the W968V background, W967V became destabilizing, indicating a too high hydrophobicity in this context. This view is further supported by the ability of the W967Y and W967H substitutions to maintain TM complex stabilities close to the wild type when paired with W968V (Figure 6C). The biggest difference between membrane and bicelle was the strongly destabilizing nature of W967F and W967F/W968V substitutions in membranes (Figure 6B–C). Phe is perhaps the most hydrophobic residues.50 As such, a larger hydrophobic effect in membranes than in bicelles is likely to pull the N-terminal end of the W967F-substituted αIIb TM helix into the membrane core.
Trp Becomes Superior Over Other Residues in Mammalian Membranes.
Within experimental uncertainties, PAC1max values of αIIb, αIIb(W967V), αIIb(W967Y/W968V) and αIIb(W967H/W968V) overlapped. However, we note a trend to lower PAC1max values for all Trp substitutions (Figure 6B–C). This may indicate somewhat lower stabilities for the Trp-substituted αIIbβ3 TM complexes in accordance with the high conservation of one Trp at the αIIb extracellular face (Figure S1). The superiority of intracellular Trp over other residues was more pronounced; all examined αIIb(W988) and β3(W715) substitutions were destabilizing in membranes (Figure 6D–E). We hypothesize that the balance between the immersion tendencies of A/C and H, in their respective ACH motifs, was lost. Specifically, the loss of this balance made these Trp substitutions more destabilizing than the mere loss of Trp anchoring as observed in αIIb(W967/W968V) (Figure 6C).
In the case of αIIb(W988), W988F followed the bicelle data as the least accepted substitution (Figure 6D) and, as the biggest difference between membrane and bicelle, W988V turned inferior to the more polar W988H and W988Y substitutions. The increased strength of the hydrophobic effect in membranes may be responsible for the changed order of acceptance of Trp substitutions. For β3(W715), W715H was, as in bicelles, again the least accepted substitution (Figure 6E), suggesting that it decisively interfered with helix-helix contacts of hydrophobic nature (λmax= 327.3 ± 0.2 nm; Table 2). Ile, Tyr and Phe were destabilizing to a similar nature either from lacking polar characteristics (Ile, Phe) or from being too polar (Tyr). The β3(W715) site again shows the versatility of Trp to adapt to environments of different polarities and its superiority over other residues, including Tyr, in membranes.
Relevance of Membrane Anchoring for Integrin Activation.
The different sensitivities of integrin activation in regards to extra- and intracellular Trp substitutions (Figure 6B–E) suggest different degrees of local TM helix anchoring as depicted in Figure 3. Most integrins share the anchoring patterns of the αIIbβ3 extra- and intracellular membrane transitions. Intracellular ACH motifs are found in all but one α and β subunit (Figure S1). Simpler transitions are present at the extracellular membrane border such as the emergence of a polar or charged residue or a bordering proline following hydrophobic or amphiphilic residues (Figure 5 and Figure S1). We hypothesize the nature of these transitions to relate to the activation mechanism of integrins. Binding of the cytosolic protein talin to the tail of β3 subunit activates the receptor.51 Talin breaks the αIIbβ3 TM complex by perturbing the membrane-proximal αIIb(R995)-β3(D723) salt bridge and by reorienting the β3 TM helix relative to the membrane surface.52,53 Both aspects will be most effective if intracellular TM orientations are narrowly defined and ACH anchors have evolved in apparent correlation to this activating signal. In contrast, on the extracellular face, the activating signal transmits via the loss of αβ TM complex-mediated ectodomain stabilization.27,49 Standard membrane transitions are sufficient for this mechanism, which appears to be tuned decisively by the dynamic properties of ectodomain-TM domain linkers.49 Thus, the homologous membrane anchoring of integrin αβ subunits supports a common activation mechanism for nearly all integrins.
Role of Monomeric TM Helix Orientations for Membrane Protein Stability.
It is perhaps intuitive to regard anchoring interactions formed in the folded state of a membrane protein to be most important. However, as mentioned earlier, in α-helical membrane proteins, the stability of the folded state will additionally depend on the anchoring characteristics of individual TM helices. In the β-barrel membrane protein OmpLA, Trp was found to contribute 3.5 kcal/mol to the folding free energy in liposomes.12 In contrast to the assembly of α-helical membrane proteins from individual helices, β-barrel proteins fold from an aqueous random coil state, which prevents a direct comparison of both classes of membrane proteins. Nonetheless, it is insightful to examine the free energy changes along their folding pathways. ΔΔG°WA,dimer and ΔΔG°WA,monomer shall express the change in free energy of dimer and monomer, respectively, upon WA substitution (Figure 3). As further depicted in Figure 3, ΔΔG°WA,dimer equals ΔΔG°WA,monomer + ΔΔG°WA. Assuming that ΔΔG°WA,monomer is small for unfolded states of β-sheet membrane proteins and that ΔΔG°WA,dimer is similar between α-helical and β-sheet membrane proteins, it follows that ΔΔG°WA,monomer,α-helix ≈ ΔΔG°WA,β-sheet − ΔΔG°WA,α-helix = 3.1 kcal/mol. This large putative contribution illustrates that not only the disturbance of helix-lipid and helix-helix interactions in the folded protein is important for α-helical membrane protein stability but also the response of the monomeric α-helix to the WA substitution. Evidently, the further the orientation of the monomeric TM helix deviates from the folded state, the more destabilizing is WA overall. For example, in the folded αIIbβ3 state, β3(W715) invariably is less important as membrane anchor but more important for direct helix-helix contacts (Figure 2A). With Ile fulfilling the structural role of W715 in the αIIbβ3 dimerization interface (Figure 4B), it may be assumed that the strong bias against this residue in membranes (Figure 6E) arose mainly from compromising the TM helix orientation of monomeric β3.
Insight into the Structural Basis of Tryptophan Membrane Anchoring.
As outlined in Introduction, Trp may engage in a number of different interactions with lipids. The Trp substitution patterns provide insight into the nature of such interactions. Firstly, as discussed above, Phe was the least accepted residue in membranes except for the dual-use β3(W715) location (Figure 6B–E). This observation and the use of opposing polarity pattern in the ACH motifs point to a distinct role of the hydrophobic effect in membrane anchoring. Secondly, within either bicelle or membrane, the order of acceptance of Trp substitutions varied especially between β3(W715) and the other sites (Figure 6B–E), signifying that Trp appeared as adopting different, site-specific polarities. The sidechain of Trp is anisotropic; adjusting its orientation may match its functional groups to the immersion depth-dependent chemical environment of membranes (Figure 1A–B), which is also the basis for its snorkeling behavior.54 In addition, the local environment may affect the polarization of indole itself.19 Thirdly, differences between bicelle and membrane beyond the strength of the hydrophobic effect are apparent in the difference in order of acceptance of Trp substitutions (Figure 6B–E). For example, in bicelles a suitable amino acid was always able to replace Trp in contrast to the more unique role of Trp in biological membranes. The lipid heterogeneity and bilayer continuity of membranes may make this environment more discriminating. For example, Trp may be able to interact with a larger number of lipid types than any other residue. In turn, the inability of other residues to successfully contact a subset of lipids would lead to a free energy penalty in membranes. In sum, the anchoring of α-helical membrane proteins correlates with the ability of tryptophan to thrive in complex membranes and a versatility to expose sidechain regions of different polarities in the background of the hydrophobic effect.
CONCLUSIONS
Studying the role of three conserved Trp residues in the folding of the heterodimeric integrin αIIbβ3 TM complex produced a systematic and quantitative study of Trp anchoring in α-helical membrane proteins. Experiments in phospholipid bicelles and mammalian membranes revealed that the “amphiphilic range” of Trp, i.e., its ability to populate environments ranging from hydrophobic to polar sites, is unsurpassed. This wide range may allow Trp to cope with the heterogeneity of lipids found in mammalian membranes. By itself, Trp anchoring is estimated to contribute 0.4 kcal/mol to membrane protein stability. As shown by the incorporation of Trp in the ACH anchoring motifs of integrin αIIb and β3, Trp anchoring has intrinsic limits. The anchoring of additional TM helices may stabilize the folded protein state, but it should not be overlooked that protein stability also depends on the anchoring characteristics of the monomeric TM helices. In integrins, the narrowing of TM helix orientations beyond mere Trp anchoring correlates with the mode of talin-mediated receptor activation. As such, the degree of TM helix anchoring may not only relate to the folding and stability of membrane proteins but also to important interactions between soluble and membrane proteins.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by American Heart Association grant 15GRNT23200010 (T.S.U.).
Footnotes
Supporting Information
Integrin sequence alignment, tryptophan fluorescence spectra, ITC data, and integrin activation data.
The authors declare no competing financial interest.
REFERENCES
- (1).Nagle JF; Tristram-Nagle S Structure of Lipid Bilayers. Biochim. Biophys. Acta-Rev. Biomembr 2000, 1469, 159–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wiener MC; White SH Structure of a Fluid Dioleoylphosphatidylcholine Bilayer Determined by Joint Refinement of X-Ray and Neutron-Diffraction Data.3. Complete Structure. Biophys. J 1992, 61, 434–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Landoltmarticorena C; Williams KA; Deber CM; Reithmeier RAF Nonrandom Distribution of Amino-Acids in the Transmembrane Segments of Human Type-I Single Span Membrane-Proteins. J. Mol. Biol 1993, 229, 602–608. [DOI] [PubMed] [Google Scholar]
- (4).Ulmschneider MB; Sansom MS P. Amino Acid Distributions in Integral Membrane Protein Structures. Biochim. Biophys. Acta-Biomembr 2001, 1512, 1–14. [DOI] [PubMed] [Google Scholar]
- (5).Adamian L; Nanda V; DeGrado WF; Liang J Empirical Lipid Propensities of Amino Acid Residues in Multispan Alpha Helical Membrane Proteins. Proteins-Structure Function And Bioinformatics 2005, 59, 496–509. [DOI] [PubMed] [Google Scholar]
- (6).MacCallum JL; Bennett WFD; Tieleman DP Distribution of Amino Acids in a Lipid Bilayer from Computer Simulations. Biophys. J 2008, 94, 3393–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wimley WC; White SH Experimentally Determined Hydrophobicity Scale for Proteins at Membrane Interfaces. Nat. Struct. Biol 1996, 3, 842–848. [DOI] [PubMed] [Google Scholar]
- (8).Oconnell AM; Koeppe RE; Andersen OS Kinetics of Gramicidin Channel Formation in Lipid Bilayers - Transmembrane Monomer Association. Science 1990, 250, 1256–1259. [DOI] [PubMed] [Google Scholar]
- (9).Clark EH; East JM; Lee AG The Role of Tryptophan Residues in an Integral Membrane Protein: Diacylglycerol Kinase. Biochemistry 2003, 42, 11065–11073. [DOI] [PubMed] [Google Scholar]
- (10).Garcia JC; Strube M; Leingang K; Keller K; Mueckler MM Amino-Acid Substitutions at Tryptophan-388 and Tryptophan-412 of the Hepg2 (Glut1) Glucose Transporter Inhibit Transport Activity and Targeting to the Plasma-Membrane in Xenopus Oocytes. J. Biol. Chem 1992, 267, 7770–7776. [PubMed] [Google Scholar]
- (11).Hassan KA; Souhani T; Skurray RA; Brown MH Analysis of Tryptophan Residues in the Staphylococcal Multidrug Transporter Qaca Reveals Long-Distance Functional Associations of Residues on Opposite Sides of the Membrane. J. Bacteriol 2008, 190, 2441–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).McDonald SK; Fleming KG Aromatic Side Chain Water-to-Lipid Transfer Free Energies Show a Depth Dependence across the Membrane Normal. J. Am. Chem. Soc 2016, 138, 7946–7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hong HD; Park S; Jimenez RHF; Rinehart D; Tamm LK Role of Aromatic Side Chains in the Folding and Thermodynamic Stability of Integral Membrane Proteins. J. Am. Chem. Soc 2007, 129, 8320–8327. [DOI] [PubMed] [Google Scholar]
- (14).Popot JL; Engelman DM Membrane-Protein Folding and Oligomerization - the 2-Stage Model. Biochemistry 1990, 29, 4031–4037. [DOI] [PubMed] [Google Scholar]
- (15).Li E; Wimley WC; Hristova K Transmembrane Helix Dimerization: Beyond the Search for Sequence Motifs. Biochim. Biophys. Acta-Biomembr 2012, 1818, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Yau WM; Wimley WC; Gawrisch K; White SH The Preference of Tryptophan for Membrane Interfaces. Biochemistry 1998, 37, 14713–14718. [DOI] [PubMed] [Google Scholar]
- (17).Norman KE; Nymeyer H Indole Localization in Lipid Membranes Revealed by Molecular Simulation. Biophys. J 2006, 91, 2046–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Gaede HC; Yau WM; Gawrisch K Electrostatic Contributions to Indole-Lipid Interactions. J. Phys. Chem. B 2005, 109, 13014–13023. [DOI] [PubMed] [Google Scholar]
- (19).Blaser G; Sanderson JM; Wilson MR Free-Energy Relationships for the Interactions of Tryptophan with Phosphocholines. Org. Biomol. Chem 2009, 7, 5119–5128. [DOI] [PubMed] [Google Scholar]
- (20).Esbjorner EK; Caesar CEB; Albinsson B; Lincoln P; Norden B Tryptophan Orientation in Model Lipid Membranes. Biochem. Biophys. Res. Commun 2007, 361, 645–650. [DOI] [PubMed] [Google Scholar]
- (21).Sun H; Greathouse DV; Andersen OS; Koeppe RE The Preference of Tryptophan for Membrane Interfaces - Insights from N-Methylation of Tryptophans in Gramicidin Channels. J. Biol. Chem 2008, 283, 22233–22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Petersen FNR; Jensen MO; Nielsen CH Interfacial Tryptophan Residues: A Role for the Cation-Pi Effect? Biophys. J 2005, 89, 3985–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Persson S; Killian JA; Lindblom G Molecular Ordering of Interfacially Localized Tryptophan Analogs in Ester- and Ether-Lipid Bilayers Studied by H-2-Nmr. Biophys. J 1998, 75, 1365–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Stefansson A; Armulik A; Nilsson IM; von Heijne G; Johansson S Determination of N- and C-Terminal Borders of the Transmembrane Domain of Integrin Subunits. J. Biol. Chem 2004, 279, 21200–21205. [DOI] [PubMed] [Google Scholar]
- (25).Lau T-L; Partridge AP; Ginsberg MH; Ulmer TS Structure of the Integrin Beta3 Transmembrane Segment in Phospholipid Bicelles and Detergent Micelles. Biochemistry 2008, 47, 4008–4016. [DOI] [PubMed] [Google Scholar]
- (26).Lau T-L; Dua V; Ulmer TS Structure of the Integrin Alphaiib Transmembrane Segment. J. Biol. Chem 2008, 283, 16162–16168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lau T-L; Kim C; Ginsberg MH; Ulmer TS The Structure of the Integrin Alphaiibbeta3 Transmembrane Complex Explains Integrin Transmembrane Signalling. EMBO J. 2009, 28, 1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Hughes PE; DiazGonzalez F; Leong L; Wu CY; McDonald JA; Shattil SJ; Ginsberg MH Breaking the Integrin Hinge - a Defined Structural Constraint Regulates Integrin Signaling. J. Biol. Chem 1996, 271, 6571–6574. [DOI] [PubMed] [Google Scholar]
- (29).Hynes RO Integrins: Bidirectional, Allosteric Signaling Machines. Cell 2002, 110, 673–687. [DOI] [PubMed] [Google Scholar]
- (30).Anthis NJ; Clore GM Sequence-Specific Determination of Protein and Peptide Concentrations by Absorbance at 205 Nm. Protein Sci. 2013, 22, 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ladokhin AS; Jayasinghe S; White SH How to Measure and Analyze Tryptophan Fluorescence in Membranes Properly, and Why Bother? Anal. Biochem 2000, 285, 235–245. [DOI] [PubMed] [Google Scholar]
- (32).Burstein EA; Emelyanenko VI Log-Normal Description of Fluorescence Spectra of Organic Fluorophores. Photochem. Photobiol 1996, 64, 316–320. [Google Scholar]
- (33).Situ AJ; Schmidt T; Mazumder P; Ulmer TS Characterization of Membrane Protein Interactions by Isothermal Titration Calorimetry. J. Mol. Biol 2014, 426, 3670–3680. [DOI] [PubMed] [Google Scholar]
- (34).Schmidt T; Situ AJ; Ulmer TS Structural and Thermodynamic Basis of Proline-Induced Transmembrane Complex Stabilization. Sci Rep 2016, 6, 29809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kim C; Lau T-L; Ulmer TS; Ginsberg MH Interactions of Platelet Integrin Alphaiib and Beta3 Transmembrane Domains in Mammalian Cell Membranes and Their Role in Integrin Activation. Blood 2009, 113, 4747–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Calderwood DA; Tai V; Di Paolo G; De Camilli P; Ginsberg MH Competition for Talin Results in Trans-Dominant Inhibition of Integrin Activation. J. Biol. Chem 2004, 279, 28889–28895. [DOI] [PubMed] [Google Scholar]
- (37).Garcia-Alvarez B; de Pereda JM; Calderwood DA; Ulmer TS; Critchley D; Campbell ID; Ginsberg MH; Liddington RC Structural Determinants of Integrin Recognition by Talin. Mol. Cell 2003, 11, 49–58. [DOI] [PubMed] [Google Scholar]
- (38).Shattil SJ; Cunningham M; Hoxie JA Detection of Activated Platelets in Whole-Blood Using Activation-Dependent Monoclonal-Antibodies and Flow-Cytometry. Blood 1987, 70, 307–315. [PubMed] [Google Scholar]
- (39).Ye F; Kim SJ; Kim C Intermolecular Transmembrane Domain Interactions Activate Integrin Alpha Iib Beta 3. J. Biol. Chem 2014, 289, 18507–18513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Suk JE; Situ AJ; Ulmer TS Construction of Covalent Membrane Protein Complexes and High-Throughput Selection of Membrane Mimics. J. Am. Chem. Soc 2012, 134, 9030–9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Surya W; Li Y; Millet O; Diercks T; Torres J Transmembrane and Juxtamembrane Structure of Alpha L Integrin in Bicelles. PLoS One 2013, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Burstein EA; Vedenkina NS; Ivkova MN Fluorescence and Location of Tryptophan Residues in Protein Molecules. Photochem. Photobiol 1973, 18, 263–279. [DOI] [PubMed] [Google Scholar]
- (43).Vivian JT; Callis PR Mechanisms of Tryptophan Fluorescence Shifts in Proteins. Biophys. J 2001, 80, 2093–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Schmidt T; Situ AJ; Ulmer TS Direct Evaluation of Protein-Lipid Contacts Reveals Protein Membrane Immersion and Isotropic Bicelle Structure. J. Phys. Chem. Lett 2016, 7, 4420–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Grasberger B; Minton AP; Delisi C; Metzger H Interaction between Proteins Localized in Membranes. Proc. Natl. Acad. Sci. U. S. A 1986, 83, 6258–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Schmidt T; Suk JE; Ye F; Situ AJ; Mazumder P; Ginsberg MH; Ulmer TS Annular Anionic Lipids Stabilize the Integrin Alpha Iib Beta 3 Transmembrane Complex. J. Biol. Chem 2015, 290, 8283–8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).GarciaGuerra R; GarciaDominguez JA; GonzalezRodriguez J A New Look at the Lipid Composition of the Plasma Membrane of Human Blood Platelets Relative to the Gpiib/Iiia (Integrin Alpha Iib Beta 3) Content. Platelets 1996, 7, 195–205.21043688 [Google Scholar]
- (48).van Meer G Cellular Lipidomics. EMBO J. 2005, 24, 3159–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Schmidt T; Ye F; Situ AJ; An W; Ginsberg MH; Ulmer TS A Conserved Ectodomain-Transmembrane Domain Linker Motif Tunes the Allosteric Regulation of Cell Surface Receptors. J. Biol. Chem 2016, 291, 17536–17546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Moon CP; Fleming KG Side-Chain Hydrophobicity Scale Derived from Transmembrane Protein Folding into Lipid Bilayers. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 10174–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Ye F; Hu GQ; Taylor D; Ratnikov B; Bobkov AA; McLean MA; Sligar SG; Taylor KA; Ginsberg MH Recreation of the Terminal Events in Physiological Integrin Activation. J. Cell Biol 2010, 188, 157–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Kim CH; Ye F; Hu XH; Ginsberg MH Talin Activates Integrins by Altering the Topology of the Beta Transmembrane Domain. J. Cell Biol 2012, 197, 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Anthis NJ; Wegener KL; Ye F; Kim C; Goult BT; Lowe ED; Vakonakis I; Bate N; Critchley DR; Ginsberg MH, et al. The Structure of an Integrin/Talin Complex Reveals the Basis of inside-out Signal Transduction. EMBO J. 2009, 28, 3623–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Chamberlain AK; Bowie JU Analysis of Side-Chain Rotamers in Transmembrane Proteins. Biophys. J 2004, 87, 3460–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).McCellan AL In Tables of Experimental Dipole Moments, San Francisco, W. H. Freeman: San Francisco, 1963; pp 280–325. [Google Scholar]
- (56).Pettersen EF; Goddard TD; Huang CC; Couch GS; Greenblatt DM; Meng EC; Ferrin TE Ucsf Chimera - a Visualization System for Exploratory Research and Analysis. J. Comput. Chem 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- (57).Chou JJ; Baber JL; Bax A Characterization of Phospholipid Mixed Micelles by Translational Diffusion. J. Biomol. NMR 2004, 29, 299–308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


