Abstract
Rolling-circle replication is initiated by a replicon-encoded endonuclease which introduces a single-strand nick into specific origin sequences, becoming covalently attached to the 5′ end of the DNA at the nick and providing a 3′ hydroxyl to prime unidirectional, leading-strand synthesis. Parvoviruses, such as minute virus of mice (MVM), have adapted this mechanism to amplify their linear single-stranded genomes by using hairpin telomeres which sequentially unfold and refold to shuttle the replication fork back and forth along the genome, creating a continuous, multimeric DNA strand. The viral initiator protein, NS1, then excises individual genomes from this continuum by nicking and reinitiating synthesis at specific origins present within the hairpin sequences. Using in vitro assays to study ATP-dependent initiation within the right-hand (5′) MVM hairpin, we have characterized a HeLa cell factor which is absolutely required to allow NS1 to nick this origin. Unlike parvovirus initiation factor (PIF), the cellular complex which activates NS1 endonuclease activity at the left-hand (3′) viral origin, the host factor which activates the right-hand hairpin elutes from phosphocellulose in high salt, has a molecular mass of around 25 kDa, and appears to bind preferentially to structured DNA, suggesting that it might be a member of the high-mobility group 1/2 (HMG1/2) protein family. This prediction was confirmed by showing that purified calf thymus HMG1 and recombinant human HMG1 or murine HMG2 could each substitute for the HeLa factor, activating the NS1 endonuclease in an origin-specific nicking reaction.
Although originally thought to be confined to prokaryotic replicons, such as single-stranded coliphages and conjugative plasmids, closely related rolling-circle DNA replication (RCR) mechanisms have since been observed in several small eukaryotic viruses (14, 22, 24, 26), all of which rely heavily on the synthetic machinery of their host cells. Parvoviruses are an unusual member of this group because their single-stranded genomes are linear, and they have adapted the RCR system to amplify themselves by a unique “rolling hairpin” process, mediated by the palindromic viral telomeres. In virion DNA, these telomeres fold back on themselves to form imperfect hairpin duplexes which sequentially unfold, to allow the terminus to be copied once, and then refold to shuttle the unidirectional cellular replication fork back along the linear genome, creating covalently continuous hairpin termini. Since this process can be repeated indefinitely at each end of the genome, multimeric duplex intermediates containing a single continuous DNA strand are generated, rather than the more classical RCR circles. Unit-length genomes are then excised from this continuum, and their termini are duplicated, by the introduction of a single-strand nick into specific origin sequences generated within the viral telomeres, which provides a base-paired 3′ hydroxyl group to prime de novo DNA synthesis. As in all RCR systems, these site-specific nicks are generated by a trans-esterification reaction mediated by the virally coded initiator endonuclease, termed Rep or NS1, which becomes covalently attached to the 5′ side of the nick.
Minute virus of mice (MVM) encapsidates a single, negative-sense DNA strand containing a 4.8-kb single-stranded coding region bracketed by imperfect hairpin duplexes of 120 and 248 nucleotides at its 3′ (left) and 5′ (right) termini, respectively. Its initiator endonuclease, NS1, is an abundant and long-lived nuclear phosphoprotein of 83 kDa with helicase and ATPase activities (14, 25, 30). NS1 contains two critical sequence motifs encoding a putative metal coordination site and an active-site tyrosine, thought to comprise the catalytic site of the nickase, which have been recognized in prokaryotic RCR initiators (20, 25). As would be expected, NS1 is also a site-specific DNA-binding protein; it recognizes the sequence (ACCA)1–3, present both in the viral origins and reiterated at multiple sites throughout the viral genome (9). Since most MVM sequences of 100 bp or more contain at least one copy of this recognition sequence, and some regions contain multiple tandem and inverted reiterations, NS1 can bind throughout MVM replicative-form DNA (11a). This finding not only suggests a potential role for NS1 in viral chromatin structure and progeny strand packaging but also implies that its endonuclease function must be strictly regulated to confine this otherwise disruptive activity to the viral replication origins.
Although the two telomeres of MVM differ markedly from each other in size, primary sequence, and secondary structure, they both contain sequence elements which ultimately become rearranged to create NS1-dependent origins. Sequences from the left-end telomere form a functional origin only on one side of a fully base-paired, duplex copy of the palindrome which bridges adjacent genomes in a dimer duplex intermediate (11, 13, 23). In contrast, the right-end origin is active in the covalently closed hairpin configuration (2, 11a). We have previously focused on defining the minimal left-end origin (11, 13), which we have shown contains an NS1-binding site (9) and a critically spaced binding site for a novel, sequence-specific cellular DNA-binding factor, dubbed PIF, for parvovirus initiation factor (6, 7). PIF is a heterodimer of two related polypeptide chains, p96 and p79, which functions as an essential cofactor in replication initiation at the left-end origin, binding to a repeated ACGT motif located immediately next to the NS1-binding site and activating the NS1 endonuclease function in some as yet undefined way (4a, 7). In this study, we examined whether the MVM right-hand origin is activated by the initiator protein alone, as observed for the hairpin origins of the helper-dependent parvovirus adeno-associated virus type 2 (AAV2) (17), or whether it is also dependent on a cellular cofactor.
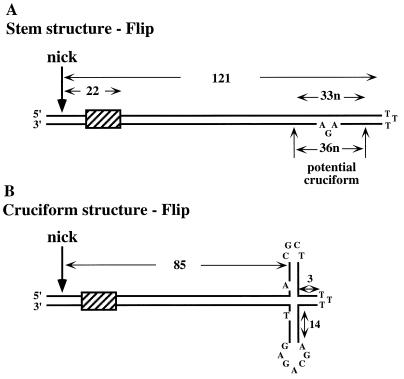
To this end, we prepared substrates which mimic the active MVM right-hand origin, in its covalently closed hairpin form (Fig. 1). Two distinguishable forms of the right-end hairpin, dubbed flip and flop, are generated in equimolar amounts in vivo (1). These two forms, which both function as viral origins, are the inverted complements of each other and can be depicted as simple, base-paired stem structures with three unpaired nucleotides at the tip of the stem and another three unpaired nucleotides forming a small asymmetric bubble near the distal end of one strand (Fig. 1A). As indicated, these stem structures harbor a small internal palindrome surrounding the three-nucleotide bubble, so that an alternative structure, in which the stem assumes an asymmetric cruciform configuration, is also thermodynamically probable (1). Using an in vitro nicking assay with 3′-end-labeled hairpin substrates that mimic this origin, we show that a cellular DNA-binding protein is absolutely required to allow NS1 to nick this hairpin and that this protein is a member of the high-mobility group 1/2 (HMG1/2) family of chromatin-associated polypeptides.
FIG. 1.
Structure of the right-hand hairpin. (A) The 5′ (right) hairpin of MVM displayed as a simple duplex stem structure, in the flip configuration. Flip comprises 121 potential bp, with three unpaired thymidine residues forming the turnaround at the top of the stem. Twenty base pairs in from this end, three unpaired residues, AGA, form a small asymmetric bubble. This region of the stem harbors a small internal palindrome centered on the three-nucleotide (n) mismatch, and thus an alternative structure in which the duplex assumes an asymmetric cruciform configuration is possible. This cruciform is created by the concertina-like rearrangement of the palindrome, so that its size and position on the stem can vary slightly. At its most extended position (B), the four-way junction at its core is 85 bp from the nick site. Two nucleotides which would be unpaired in the cruciform structure are also indicated. The flop sequence is simply the inverted complement of flip. The cross-hatched box indicates the sequence 5′-AACCAACTGAACCA-3′, which forms the binding site for an NS1 molecule whose footprint would project over the nick site.
MATERIALS AND METHODS
Plasmids.
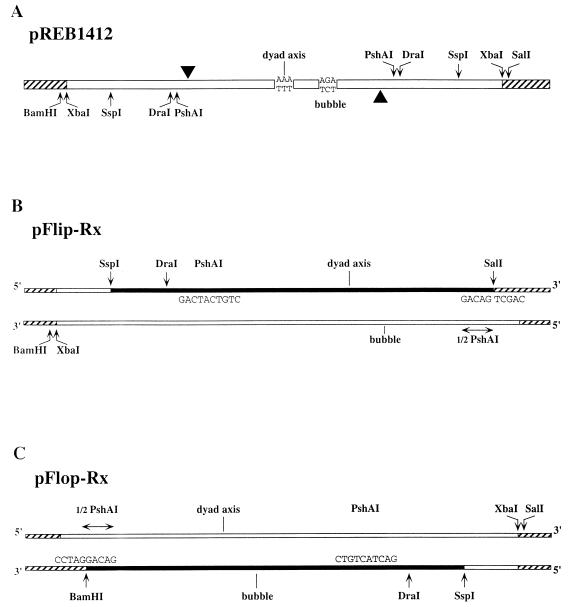
The pUC19-based plasmid pREB1412 (Fig. 2A) has been described previously (12). It contains the 1,412-bp palindromic fragment which lies between XbaI sites at MVM nucleotide 4342 in the right-end/right-end junction of tetrameric MVM replicative-form DNA. Derivatives pFlip-Rx and pFlop-Rx were constructed by partial deletion and religation of the pREB1412 insert, between an internal PshAI site and SalI or BamHI sites, respectively, in the polylinker, as described in the legend to Fig. 2. In each of these plasmids, the religated PshAI site was destroyed, leaving a single PshAI site in the construct, while the polylinker site was recreated so that it could be used to release the viral insert precisely from the vector sequences.
FIG. 2.
Derivation and structures of plasmids used for generating the hairpin substrates. (A) pREB1412, which contains a single duplex copy of the right-hand hairpin sequence as present in the junction between two dimers in a tetrameric replication intermediate. The axis of dyad symmetry is at MVM nucleotide 5048, flanked on each side by a palindromic arrangement of downstream sequence extending to the viral XbaI site at nucleotide 4342. These flanking sequences each contain a single PshAI site, located just 7 bp inboard of the nick site at nucleotide 4916. One or other arm of the flanking sequence was deleted by digesting pREB1412 at a single PshAI site, cutting in the polylinker sequence with either SalI or BamHI, and religating the product. This gave one construct, designated pFlop-Rx (C), which lacked the flanking sequence between PshAI site A and the polylinker BamHI site, and a second construct, pFlip-Rx (B), which lacked the sequence between PshAI site B and the SalI site. A hatched line indicates plasmid sequence; a filled line indicates the strand, released by denaturation of the excised insert, which contains a recessed 3′ end and serves as a primer-template for Sequenase to produce the end-labeled hairpin substrate for nicking reactions, as described in Materials and Methods.
Substrates.
Flip or flop substrates were prepared by excision from the appropriate plasmid, pFlip-Rx or pFlop-Rx, using SalI or BamHI, respectively, in combination with SspI or DraI, as indicated in Fig. 2. Gel-purified fragments were melted by heating to 95°C for 5 min and then rapidly cooled, allowing individual strands to reanneal, forming complete right-end hairpin structures with single-stranded tails. One strand from each insert (filled lines in Fig. 2) reannealed so that its 3′ end was located at the PshAI site, base paired to the other viral arm, thus forming a template-primer substrate. This was filled in, using Sequenase (U.S. Biochemical, Cleveland, Ohio) and a mixture of unlabeled and 32P-labeled nucleotides, to generate a 3′-end-labeled template for the nicking assay. Prior to use, substrates were purified by electrophoresis through nondenaturing polyacrylamide gels.
Cell extracts and fractions.
S100 extracts were prepared from HeLa cells and chromatographed on phosphocellulose as previously described (6). Eluted fractions (referred to as P-cell 1, P-cell 2, etc.) were dialyzed into buffer A (25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.01% Nonidet P-40 [NP-40], 1 mM dithiothreitol, 25 mM NaCl, 10% glycerol) containing 20% (wt/vol) sucrose, flash-frozen, and stored at −80°C. Mono Q fast protein liquid chromatography (FPLC) fractions containing PIF were obtained from fraction P-cell 1 as described previously (6). Fraction P-cell 3 was analyzed by FPLC gel filtration on Superose 12 (HR10/30) equilibrated in buffer A adjusted to 50 mM NaCl. Dextran blue, carbonic anhydrase, and lysozyme were used as molecular weight markers, and fractions were flash-frozen and stored at −80°C. Calf thymus HMG1 was the kind gift of Michael Bustin (National Institutes of Health).
Recombinant proteins.
MVM NS1 with an amino-terminal histidine tag was expressed from a recombinant baculovirus and purified as described previously (5). Plasmids expressing inducible, histidine-tagged, recombinant human HMG1 (pET-11 HMG1-6His [15]) and murine HMG2 (pMFpKA HMG-2 [32]) were the gifts of Robert Roeder (Rockefeller University, New York, N.Y.) and Thomas Wirth (Wuerzburg, Germany), respectively. Transformed into Escherichia coli BL21(DE3)pLys5, these constructs were induced for the expression of T7 polymerase with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), which then transcribed the HMG gene. Two hours after induction cells were harvested, washed, and lysed by freeze-thawing in buffer A containing 50 mM NaCl and 0.2 mg of lysozyme per ml. Supernatants were adjusted to 200 mM NaCl, applied to phosphocellulose, washed with buffer A containing 400 mM NaCl, and eluted with buffer A containing 800 mM NaCl (fraction P-cell 3). This fraction was then diluted fourfold with buffer A alone and applied to Ni2+-agarose, and the column was eluted with buffer A containing 50 mM NaCl and 100 mM imidazole.
Nicking assays.
Nicking assay mixtures (15 μl) contained 30 mM HEPES-KOH (pH 7.8), 75 mM sodium acetate, 7 mM MgCl2, 5 mM dithiothreitol, 2.5 mM ATP, and 0.1% NP-40. Fifty nano-grams of purified baculovirus NS1 was preincubated in sample buffer for 15 min on ice with total HeLa S100 extracts, with fraction P-cell 3, or with various column fractions, as indicated below, in the presence of 0.25 μg of a blunt-ended, nonspecific, duplex 29-mer oligonucleotide competitor (scramble oligonucleotide; 5′-GAT CTA GAG AGT CGA TGT ATC TGC AGA TC-3′) and 0.25 μg of blunt-ended, nonspecific, duplex DNA fragments (100 to 800 bp in length) derived from plasmid pCRII (Invitrogen, San Diego, Calif.) by digestion with RsaI and NciI. These nonspecific duplex competitor DNAs served to sequester nonspecific DNA-binding proteins that might otherwise mask the substrate. Poly(dI-dC) and sonicated salmon sperm DNA were not used as nonspecific competitors because they both inhibited nicking, presumably due to the presence of heterogeneous structured DNA in these preparations. Where required, specific oligonucleotide competitors (0.25 μg, approximately equivalent to a 1,000-fold molar excess for a 25-mer duplex oligonucleotide in a reaction mixture containing 4 ng of a 400-bp substrate) were also added in the preincubation step. 32P-labeled substrate DNA (1 to 4 ng) was then added, and the reaction mixture was incubated at 37°C for 45 min. The duplex competitors used for Fig. 5 were oligonucleotide 1 (5′-CAT TAG TAT TAC TAT GTT TT-3′ [MVM nucleotides 5150 to 5131]); oligonucleotide 2 (5′-TAG GGT GGG AGG GTG GGA-3′ [MVM nucleotides 4963 to 4980]); oligonucleotide 3 (5′-GAT ACA TGT GTT CGC TAT GAG CG-3′ [MVM nucleotides 4981 to 4903]); oligonucleotide 4 (a duplex formed from 5′-CTG GTT GGT TGC GCT CAA CCA ACC-3′ [MVM nucleotides 5081 to 5058] annealed to 5′-CTG GTT GGT TGA GCA GAG CAA CCA ACC-3′ [nucleotides 5039 to 5013]); and TR56 (5′-GAT CGA TCT GTC AGA GCA CCT CGC GAG CGT ACG TGC CTC AGG AAG TGA CGC ACA GC-3′ [an unrelated control sequence derived from the promoter of the transferrin receptor gene]).
FIG. 5.
Duplex DNA sequences from the origin do not compete for the HeLa factor, but long complex single-stranded DNA will inhibit nicking. (A) The right-hand hairpin of MVM, displayed as a simple duplex stem structure in the flip configuration, showing the positions of oligonucleotides (oligo) 1 through 4 (see Materials and Methods for sequences). (B) 32P-labeled hairpin probe in the flip orientation was incubated with NS1 and partially purified HeLa extract under standard reaction conditions (lane 1) or in the presence of 0.25 μg of competing pCRII DNA fragments (lane 2) or duplex oligonucleotide 1 (lane 3), 2 (lane 4), 3 (lane 5), or 4 (lane 6). (C) 32P-labeled flip hairpin probe was incubated with NS1 and partially purified HeLa extract under standard reaction conditions (lane 1) or in the presence of 0.25 μg of competing duplex (lane 2) and single-stranded (lane 3) pCRII DNA fragments. In lane 4, the competing DNA was single-stranded phage M13 DNA; in lanes 5 and 6, the competitor was a 29-mer oligonucleotide in duplex and single-stranded forms, respectively; in lanes 7 and 8, the competitor was a 56-mer oligonucleotide, also in duplex and single-stranded forms, respectively. pCRII DNA comprised duplex fragments of 100 to 800 bp derived from pCRII vector DNA as described in Materials and Methods. The sequences of the 29-mer and 56-mer oligonucleotides are also given in Materials and Methods. Single-stranded forms of the competitors were prepared by heating to 95°C for 5 min immediately prior to use in the reaction. C, nicked complex; P, free probe.
Unless otherwise specified, the substrate was a hairpin DNA fragment containing 418 bp (and 6 unpaired nucleotides) of viral sequence, in the flip configuration, extending from an SspI site at MVM nucleotide 4626 to the tip of the right-end hairpin. Reactions were stopped by the addition of 10 μl of 10 mM Tris-HCl (pH 7.5)–10 mM EDTA–1% sodium dodecyl sulfate (SDS), and samples were incubated at 60°C for 15 min. Aliquots then received a half-volume of sample buffer (40% glycerol, 50 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol, 0.5% SDS) prior to electrophoresis for 2.5 h at 200 V through a 4.5% nondenaturing polyacrylamide gel (39:1 acrylamide:bisacrylamide) which had been preelectrophoresed for 1 h at 200 V in electrophoresis buffer containing 0.2% SDS. For immunoprecipitation, aliquots were diluted into immunoprecipitation buffer (150 mM NaCl, 20 mM Tris-HCl [pH 8.0], 2 mM EDTA, 1% NP-40) containing 1% SDS and incubated at 60°C for 30 min. They were then diluted with immunoprecipitation buffer alone to a final SDS concentration of 0.25% and precipitated as previously described (10). Where indicated, aliquots of the reaction products or immunoprecipitates were incubated with proteinase K 50 μg/ml at 60°C for 30 min prior to addition of the sample buffer or were melted by heating to 95°C for 5 min immediately prior to electrophoresis.
RESULTS
A host protein is absolutely required to allow NS1 to nick the right-hand origin.
To explore right-hand hairpin processing in vitro, we engineered plasmids which would allow us to recover and label hairpin substrates. Plasmid pREB1412 contains a palindromic viral insert derived from a right-end/right-end junction fragment which spans adjacent genomes in a tetrameric, duplex replication intermediate. As illustrated in Fig. 2A, this sequence is a duplex copy of the entire right-end hairpin sandwiched between inverted copies of the adjacent coding sequences, extending to an XbaI site at MVM nucleotide 4342, cloned into the XbaI site of pUC19. In the presence of a HeLa cell extract and recombinant NS1, this junction can be resolved and replicated in vitro in a process which generates two extended-form viral telomeres containing the characteristic flip and flop sequence inversions observed in vivo (10). pREB1412 was modified, by partial deletion as described in Fig. 2, to produce the two plasmids, pFlip-Rx and pFlop-Rx. When the inserts from these plasmids are excised with SspI and either SalI or BamHI, respectively, denatured, and allowed to reanneal intramolecularly, they yield hairpin strands which are perfectly base paired at their SalI- or BamHI-cut ends, within the internal PshAI site. One strand in each released fragment, represented in Fig. 2B and C by filled lines, is a primer-template for DNA polymerase, which can complete the recessed 3′ strand, allowing controlled labeling of the resulting hairpin. 32P-labeled hairpin substrates prepared in this way from pFlip-Rx exactly mimic the origin sequence present at the right end of covalently closed viral intermediates in the flip configuration, while those similarly prepared from pFlop-Rx mimic origins in the flop configuration.
These substrates were incubated in the presence or absence of recombinant NS1 and ATP, and the products were analyzed by electrophoresis through nondenaturing polyacrylamide gels in the presence of SDS, included to eliminate noncovalent protein-DNA interactions. In salt concentrations of less than 75 mM, NS1 will bind to all DNA in a non-sequence-specific manner (9). Thus, in order to leave the initiator free to bind its cognate site, and therefore increase the efficiency and specificity of the nicking reaction, 75 mM salt was included in all assays. Unlike the PIF-NS1 complex formed on the left-hand origin, the hairpin nickase complex would not tolerate high Cl− levels but would tolerate equivalent acetate concentrations, which were found to be equally effective at inhibiting nonspecific binding to both duplex and single-stranded DNA in the standard DNA-binding assay (data not shown).
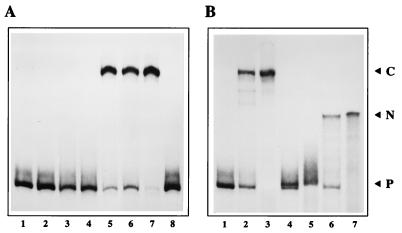
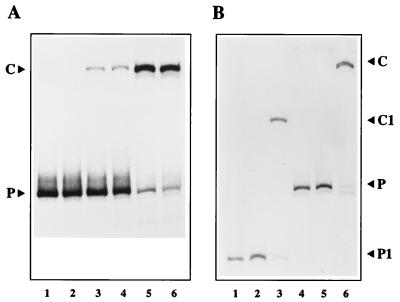
Although NS1 bound efficiently to the hairpin origin under these salt conditions when analyzed by coimmunoprecipitation (data not shown), by itself it was unable to nick the substrate (Fig. 3A, lane 2). Similarly, HeLa cell S100 extracts failed to modify the substrate (lane 3), but when NS1 and HeLa extracts were combined, a new form of the origin which showed reduced electrophoretic mobility was generated (lane 5). This product was not generated if ATP was omitted from the reaction (lane 4) and was generated with an efficiency which varied with the amount of NS1 added (lanes 6 and 7). Significantly, it was not generated in the presence of NS1, ATP, and HeLa cell PIF (7), the DNA-binding factor which activates the nickase function of NS1 at the MVM left-hand origin (lane 8).
FIG. 3.
(A) NS1 can nick the right-hand origin only in the presence of a host protein. A 32P 3′-end-labeled right-hand hairpin origin probe in the flip orientation (lane 1) was incubated with 50 ng of NS1 alone (lane 2) or ∼2 μg of HeLa S100 extract alone (lane 3) in the presence of ATP. ATP was included in all reactions except that shown in lane 4, where probe, NS1, and HeLa extract were coincubated in its absence. HeLa extract was incubated with 50 ng of NS1 (lane 5), with 25 ng of NS1 (lane 6), or with 100 ng of NS1 (lane 7), and NS1 (50 ng) was incubated with partially purified PIF (lane 8). Reactions were terminated by heating to 60°C in 0.5% SDS, and the products were electrophoresed through 4.5% native acrylamide gels. (B) Structure of the nicked product. The 32P-labeled right-hand hairpin probe in the flip orientation (lane 1) was incubated with 50 ng of NS1 and ∼2 μg of HeLa S100 extract in the presence of ATP (lane 2). Reaction products equivalent to those shown in lane 2 were heated to 60°C in SDS and immunoprecipitated with a polyclonal antibody directed against the amino terminus of NS1 (lane 3). Total reaction products and anti-NS1 immunoprecipitates equivalent to those shown in lanes 2 and 3 were incubated with proteinase K (lanes 4 and 5, respectively) or incubated with proteinase K and then melted prior to electrophoresis (lanes 6 and 7, respectively). P, free probe; C, the nicked complex in which NS1 has become covalently attached to the probe at the nick site; N, the 32P-labeled, partially single-stranded product remaining after removal of both NS1 and the unlabeled duplex strand from the nicked hairpin complex.
The nicked product is covalently attached to NS1.
To initiate DNA replication, NS1 must nick one strand of the duplex at a specific site (Fig. 1). Cleavage appears to occur via a trans-esterification reaction in which the phosphodiester bond is attacked by the hydroxyl of NS1 tyrosine 210 and results in the formation of a DNA–(5′-phosphotyrosyl)–NS1 complex (25). To determine whether the retarded form of the origin observed in Fig. 3A was modified in this way, total reaction products were first incubated in SDS at 60°C for 30 min and then immunoprecipitated with antiserum directed against NS1. As seen in Fig. 3B, lane 3, only the retarded form of the origin was precipitated under these conditions, indicating that it had become covalently-associated with NS1. When either the total reaction product or an anti-NS1 immunoprecipitate was digested with proteinase K (lanes 4 or 5, respectively), the modified origin band was lost and replaced by a deproteinized band which comigrated with the input probe, indicating that the modified complex retained all of the DNA sequences present in the original substrate. If this deproteinized reaction product contained the expected single-strand nick, melting the duplex immediately prior to electrophoresis would release a long unlabeled fragment and leave a 3′-end-labeled molecule with a single-stranded tail. Since regions of single-stranded DNA severely decrease fragment mobility in this gel system, such a molecule would be expected to migrate more slowly than the input probe. When proteinase K-treated reaction products and anti-NS1 immunoprecipitates were melted by heating to 95°C prior to electrophoresis, a new labeled species with the predicted lower mobility was generated (lanes 6 and 7, respectively). Since proteinase K treatment alone failed to reveal any molecules with comparable mobility among the unmelted reaction products (lanes 4 and 5), NS1 does not exhibit helicase activity under the reaction conditions used here. Deproteinized reaction products were also analyzed on sequencing gels, and the approximate position of the nick site was verified by comparison with sequencing ladders (data not shown).
The low-mobility form of the origin generated in the presence of NS1, HeLa extract, and ATP is therefore a full-length template which has been nicked and is covalently linked to NS1. Together, these assays show that a cellular factor(s) is absolutely required to allow NS1 to nick the MVM right-hand origin. Since PIF-enriched fractions could not induce this reaction, either a third factor is needed in addition to PIF or different cellular factors are required to activate the NS1 endonuclease at each of the two viral origins.
Flip and flop sequences and hairpins with minimal flanking sequences are substrates for the nickase complex.
In vivo, right-end hairpins are generated in two alternate configurations, called flip and flop. Since these both contribute equally to the covalently closed intermediate pool in the cell, they presumably both serve as substrates for the NS1 endonuclease reaction. This was shown to be the case in the assay illustrated in Fig. 4A, where hairpin probes in the two configurations, labeled to the same specific activity, were exposed to NS1 in the presence of increasing concentrations of partially purified HeLa extract and were nicked with equal efficiency.
FIG. 4.
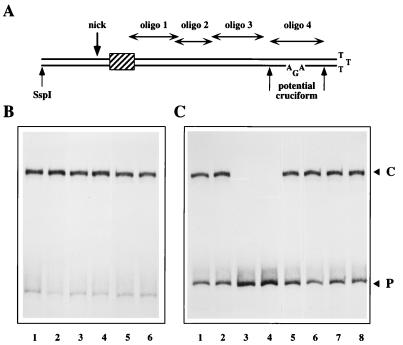
(A) The HeLa protein promotes nicking of both flip and flop forms of the hairpin. 32P-labeled hairpin probes in the flop (lanes 1, 3, and 5) or flip (lanes 2, 4, and 6) orientation were incubated with NS1 alone (lanes 1 and 2) or in the presence of 1 μl (∼0.2 μg) of partially purified HeLa extract (lanes 3 and 4) or with 3 μl (∼0.6 μg) of the same extract (lanes 5 and 6). (B) A hairpin origin with 27 nucleotides of flanking sequence is nicked in the presence of HeLa protein. 32P-labeled hairpin probes in the flip orientation extending 27 bp beyond the nick site to the DraI site at MVM nucleotide 4896 (P1 in lanes 1 to 3) or 297 bp beyond the nick site to an SspI site at MVM nucleotide 4626 (P in lanes 4 to 6) were incubated with NS1 alone (lanes 2 and 5) or in the presence of 3 μl of partially purified HeLa extract (lanes 3 and 6). C and C1 denote the complexes formed with P and P1, respectively.
Since the hairpin substrates used in these assays had extensive MVM flanking sequences extending to an SspI site located 297 bp beyond the nick site, it was possible that the activating cofactor recognized sequences outside the hairpin. To explore this possibility, a second, shorter substrate which only contained 27 bp of flanking sequence was constructed and labeled to approximately the same specific activity as the longer probe. As illustrated in Fig. 4B, both of these substrates are nicked efficiently in a HeLa- and NS1-dependent reaction, indicating that the activator operates either within the hairpin or in its immediate environment.
Duplex DNA sequences from within the hairpin fail to compete for the host factor.
In the small duplex origin derived from the left end of the viral genome, PIF, the activating host factor, binds to a duplex DNA sequence located immediately next to the NS1-binding site, on the side distal to the nick site. Hence, duplex synthetic oligonucleotides which contain this sequence efficiently compete with the labeled origin for the activator (7). To determine whether the cellular activator for the right-end origin binds in a similar way, we asked if duplex oligonucleotides which contained sequences from various parts of the hairpin stem, as indicated in Fig. 5, would inhibit nicking when added in 2,000-fold molar excess over the labeled probe. Since all of these failed to compete for the activator (Fig. 5A), we also tested the possibility that the hypothetical binding site for this molecule might span adjacent oligonucleotides. Longer duplex fragments from the stem region, generated by PCR using juxtaposed pairs of oligonucleotides 1 to 3, again failed to suppress the reaction (data not shown), suggesting that the HeLa cell factor does not recognize a specific duplex sequence within the hairpin stem.
The potential cruciform region (oligonucleotide 4) is more difficult to manipulate for two reasons: first, because each individual oligonucleotide can self-anneal to form a short duplex hairpin with a terminal loop which recreates the structure found in one or other of the cruciform arms; and second, because the palindromic sequences themselves form potential NS1-binding sites. However, a 500 to 2,000-fold molar excess of a self-annealed hairpin oligonucleotide from either strand of sequence 4 failed to compete (data not shown). NS1 binds asymmetrically over its cognate site, protecting approximately 30 nucleotides on the 5′ side of the 5′-ACCA-3′ sequence but only 4 residues on the 3′ side (9), so that two NS1 molecules bound on the sequence 4 duplex would likely project into each other, while one molecule bound at one of the two sites, or to the self-annealed form of either oligonucleotide, would project over the end of the DNA. Since NS1 binds stably only to long duplex DNA fragments (5, 9), it would not be expected to bind stably to these short competitors, a prediction strongly supported by the observation that duplex oligonucleotide 4 does not inhibit nicking.
Long, complex single-stranded DNA can compete for the host factor.
Since the cruciform configuration of the right-end hairpin contains sequences which can be considered both single stranded and structured, we explored the possibility that the activating factor could bind to such elements. Duplex plasmid DNA which had been cut with restriction enzymes into blunt-ended fragments of between 100 and 800 nucleotides was used routinely as a nonspecific competitor in all of these assays. As a control we included an additional 0.25 μg of this duplex competitor, which, as expected, failed to impede nicking (Fig. 5B; cf. lanes 1 and 2). In contrast, if the same DNA was melted prior to addition, it completely inhibited the nicking reaction (lane 3). Similarly, 0.25 μg of single-stranded M13 DNA eliminated nicking (lane 4), but 29-mer and 56-mer synthetic oligonucleotides failed to compete for the HeLa cofactor, as either double- or single-stranded DNA (lanes 5 to 8). Thus, the activating factor does not appear to bind efficiently to single-stranded DNA per se but does recognize long, complex single-stranded DNA, suggesting that it binds preferentially to structured elements.
The cellular factor has the characteristics of an HMG1/2 family member.
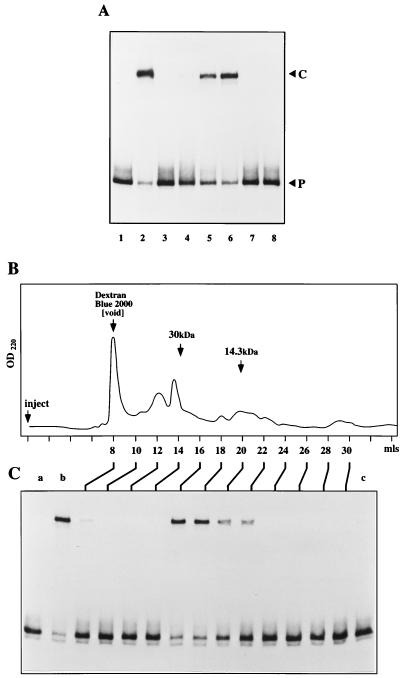
To characterize the host factor further, HeLa S100 extracts were first chromatographed on phosphocellulose, and the fractions were analyzed for their ability to activate the NS1-hairpin complex (Fig. 6A). Fraction P-cell 1 contains components which fail to bind to phosphocellulose in 150 mM salt and includes PIF, the left-end origin activator. Neither this fraction nor fraction P-cell 2, which contains material eluted in 400 mM salt, would support nicking (Fig. 6A, lanes 3 and 4), but the nicking reaction was efficiently activated by addition of fraction P-cell 3, which contains material eluted in 800 mM salt (lanes 5 and 6). Thus, PIF has no role in the activation of the right-end origin, since this is carried out exclusively by factors which exhibit a much higher affinity for phosphocellulose.
FIG. 6.
The HeLa factor elutes from phosphocellulose in high salt and has a molecular mass of around 25 kDa. (A) A 32P-labeled hairpin probe in the flop orientation was incubated under standard reaction conditions with NS1 alone (lane 1) or with NS1 in the presence of 1 μl (∼2 μg) of unfractionated HeLa extract (lane 2), 3 μl of phosphocellulose fractions P-cell 1 (lane 3) and P-cell 2 (lane 4), or 1 and 3 μl of P-cell 3 (lanes 5 and 6, respectively). Fraction P-cell 3 (3 μl) had no effect on the probe when incubated in the absence of NS1 (lane 7). A reaction carried out in the absence of any nonspecific competitor DNA, but otherwise exactly equivalent to that shown in lane 6, is shown in lane 8. (B) The elution profile obtained when concentrated P-cell 3 fractions were chromatographed on an FPLC Superose 12 column, together with the positions of dextran blue and marker proteins carbonic anhydrase (30 kDa) and lysozyme (14.3 kDa) determined by calibration immediately prior to use. OD220, optical density at 220 nm. Fractions eluting from the column 8 to 30 ml after injection were screened for nicking activity in the presence of NS1 (C). The 32P-labeled hairpin probe in the flip orientation used for this assay was incubated with NS1 alone (lane a), with NS1 and the P-cell 3 sample used to load the column (lane b), and with column load alone (lane c).
Nicking assays were routinely carried out in the presence of nonspecific duplex competitor DNA, which served to sequester nonspecific DNA-binding proteins that might otherwise mask the substrate. Since P-cell 3 was particularly rich in such proteins, the assay shown in Fig. 6A was highly dependent on the presence of a nonspecific competitor. This is illustrated in lane 8, where nonspecific DNA was omitted from a P-cell 3/NS1 reaction equivalent to the one shown in lane 6, with the result that all nicking activity was effectively masked. This increased need for nonspecific competitor DNA complicated attempts to quantitate cofactor enrichment during purification. However, estimates of specific activity derived by relating the ability of each fraction to support NS1-mediated nicking with its protein concentration, determined from Coomassie blue-stained gels, suggested that the P-cell 3 fraction was around 12 times more active than the starting S100 extract.
We next subjected the HeLa P-cell 3 fraction to FPLC gel filtration on Superose 12. As shown in Fig. 6B and C, the major peak of activity eluted 16 to 18 ml following injection and corresponded to proteins with molecular masses of approximately 25 kDa. This peak also had a trailing shoulder eluting between 20 and 22 ml, corresponding to proteins of around 14 kDa. When active fractions were titrated individually into the nicking assay, it became clear that this trailing shoulder constituted a second, minor peak (data not shown). Since individual fractions from each of these regions of the profile were active in the nicking assay, a single cellular protein species, of either ∼25 or ∼14 kDa, appears sufficient to activate the origin.
A search of known proteins with these characteristics suggested that the cellular factor might be a member of the high-mobility group of small, nonhistone chromosomal proteins, which bind tightly to phosphocellulose and are the most abundant structure-dependent DNA-binding proteins in the eukaryotic cell (3). In particular, members of the HMG1/2 family have molecular masses of around 25 kDa and would thus migrate with the major peak of activity seen in the gel filtration profile. Gel shift analyses in which the right-end hairpin substrate was incubated with these fractions indicated that they contained a DNA-binding activity which could be supershifted with antisera directed against calf thymus HMG1 (data not shown). To explore this possibility further, highly purified calf thymus HMG1 (obtained from Michael Bustin) was directly titrated into the nicking assay, where it potently activated the NS1 endonuclease (Fig. 7, lanes 3 through 5). We then obtained plasmid pET-11 HMG1-6His, expressing inducible, recombinant, histidine-tagged human HMG1 (15), and plasmid pMFpKA HMG-2, expressing inducible, recombinant, histidine-tagged murine HMG2 (32). When expressed in E. coli and substantially purified by phosphocellulose and nickel agarose chromatography, both of these proteins were able to activate NS1 to nick the right-end hairpin substrate (lanes 7 and 9). We therefore conclude that a member of the HMG1/2 family is the factor present in HeLa P-cell 3 which is necessary and sufficient to activate the NS1 endonuclease within this DNA sequence context and that these proteins are therefore essential for initiation of DNA replication from this viral origin.
FIG. 7.
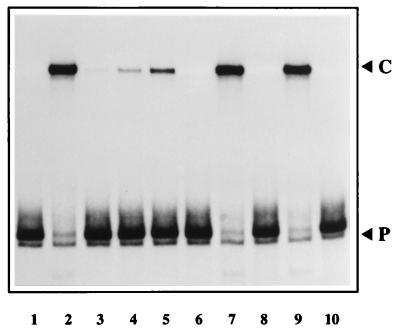
NS1 can nick the origin in the presence of purified HMG1 or HMG2. A 32P-labeled hairpin probe in the flip orientation was incubated with NS1 alone (lane 1) or with NS1 plus 3 μl of fraction P-cell 3 (lane 2), with 25, 50, and 100 ng of purified calf thymus HMG1 (lanes 3, 4, and 5 respectively), or with ∼500 ng of partially purified recombinant human HMG1 (lane 7) or murine HMG2 (lane 9). Calf thymus HMG1 (100 ng) and recombinant HMG1 and HMG2 alone had no effect on the probe (lanes 6, 8, and 10 respectively).
DISCUSSION
We have shown here that NS1, the MVM replication initiator protein, can introduce a site-specific nick into an artificial hairpin structure which recapitulates the origin generated at the right end of the viral genome in a covalently continuous, duplex replication intermediate. This ATP-dependent endonuclease reaction, which leaves the NS1 covalently attached to the DNA at the nick site, does not occur unless a member of the HMG1/2 family of non-sequence-specific DNA-binding proteins is present. HMG1/2 proteins are expressed at high copy number in all eukaryotic cells and have traditionally been viewed as structural or architectural elements in the chromatin, associated with regions of less condensed DNA and high metabolic activity (reviewed in reference 3). They bind both single-stranded and double-stranded DNA with low affinity and with little or no specificity for the target DNA sequence. However, they show significantly higher affinity for four-way junctions or otherwise bent DNA, binding exclusively into the minor groove of the helix, distorting the groove and bending the DNA (3).
In prokaryotic rolling circle replication, initiation is a complex process in which the initiator protein binds to a specific duplex sequence in the replication origin and induces an ordered series of conformational changes in the DNA, which both bend the origin and melt the duplex, before the initiator protein is able to nick the exposed single strand at an adjacent specific site (16, 21). Although in some systems these conformational changes appear to be carried out by the endonuclease acting alone, there are also many examples where accessory proteins, encoded by the replicon and/or its host, are absolutely required. Thus, for example, cleavage at oriT during conjugal DNA transfer of bacterial sex factor plasmid R100 requires both the replicon-encoded TraY and TraI proteins and a host-encoded DNA-bending protein, integration host factor (IHF) (18). Whether HMG1/2 proteins could substitute for IHF in this reaction is not known, but in the somewhat analogous situation where IHF is normally required to allow phage lambda site-specific recombination in vitro, other DNA-bending proteins such as HMG1, HMG2, Hu, and the eukaryotic core histone dimer H2A-H2B can substitute for IHF (28). In the latter instance, it is clearly the bending activity of IHF, rather than any direct protein-protein contacts established between IHF and the endonuclease, which is required to activate nicking. Similarly, members of the HMG1/2 family might activate the MVM NS1-origin complex simply by binding directly to the DNA and bending it into a critical configuration.
However, in recent years it has become apparent that HMG1/2 proteins are potent modulators of a broad range of DNA-protein interactions central to transcription, replication, and recombination and that in most instances, this activity is not mediated solely through sequence nonspecific manipulation of DNA structure. Instead, their binding is focused at specific sites in duplex DNA by direct protein-protein interactions established with site-specific cellular effectors molecules. For example, HMG1 has been shown to promote the binding and activity of certain HOX proteins (31), and HMG2 has been shown to promote the binding and activity of octamer transcription factors Oct1 and Oct2 (32). This is effected through a direct interaction between the DNA-binding domain of the activator, the HMG box, and the DNA-binding domain of the transcription factor, the homeodomain, leading to the formation of a tertiary complex on the DNA which has both sequence and structural specificity (31). Thus, if HMG1/2 proteins prove to bind directly to NS1, it is possible that they help to promote nicking at the MVM 5′ origin by a similar type of domain-specific interaction with NS1, linked to coordinate DNA binding near the nick site.
A direct protein-protein interaction between HMG1 and the initiator endonuclease (Rep) of a helper-dependent parvovirus, AAV2, has recently been demonstrated (8). Unlike NS1, Rep is able to nick the smaller AAV hairpin origin efficiently in vitro without the aid of an accessory protein (17). Although Rep does nick substrates in the hairpin configuration more efficiently than their linear counterparts (4, 29), this enhanced activity appears to correlate to a difference in the affinity with which Rep binds the two substrates, rather than any need for a structure-dependent accessory protein. Thus, it recognizes a single cognate Rep-binding site in the linear stem, while in a hairpinned substrate it makes an additional contact with a CTTTG motif in one of the two distal arms (27). While the influence of HMG1 on initiation at an AAV hairpin origin was not monitored, it was shown to promote nicking of a linear substrate under situations where the Rep concentration was severely limiting (8). Although modest relative to the effects of HMG1 on NS1-mediated initiation, the activity reported was undoubtedly mediated by a direct interaction between HMG1 and Rep, leading to the enhanced binding of Rep at its cognate site as a tertiary complex with HMG1. Additionally, HMG1 was shown to stimulate the ATPase activity of Rep in solution, in a situation where DNA binding could not occur, suggesting that it was able to directly modify the activity of the Rep protein itself and not merely up-regulate its DNA-binding activity (8). It is thus tempting to speculate that HMG1 may bind directly to NS1 in an analogous fashion and mediate at least some of its influence through this interaction. However, such a mechanism would suggest that nicking was controlled predominantly in the immediate vicinity of the nick site, which would be incompatible with genetic evidence showing that removal of just a few bases from the outer arm of the cruciform region, some 117 bp away, totally destroys origin function (11a).
A simple combination of these two possible modes of binding, in which HMG1 binds both to the DNA and to the NS1 complex at the nick site, thus seems most likely. A genetically unrelated group of DNA-bending proteins, the high-mobility, nonhistone chromosomal proteins of the HMG-I/Y family, have been shown to mediate just this sort of cooperative interaction. For example, four HMG-1 molecules are required to bind directly to specific sites in the promoter of the gene encoding the interleukin-2 receptor alpha subunit to allow mitogen-induced expression of this gene, but these direct interactions, which appear to bend the DNA, are supplemented by protein-protein interactions established between HMG-I and the essential transcription factors, NF-κB and Elf-1 (19). Whether such a complex interaction occurs in the MVM right-hand origin remains to be determined, but one characteristic of HMG molecules remains invariant: wherever and however they interact with DNA, they bind into the minor groove, distorting the helix and bending the duplex. This finding suggests that HMG1/2 activates the endonuclease function of NS1, at least in part, by modifying the structure of its DNA substrate, a possibility we are now investigating.
ACKNOWLEDGMENTS
We thank Michael Bustin for providing us with purified calf thymus HMG1 and antisera directed against this molecule, Richard Roeder and Thomas Wirth for allowing us to use their prokaryotic HMG expression constructs, and David Schatz and Liz Corbett for facilitating this arrangement. We thank Jessica Bratton for technical support with cell culture, and we are particularly indebted to Jesper Christensen for providing cell extracts and guidance with fractionation procedures.
This work was supported by Public Health Service grants AI26109 and CA29303 from the National Institutes of Health.
REFERENCES
- 1.Astell C R, Thomson M, Chow M B, Ward D C. Structure and replication of minute virus of mice DNA. Cold Spring Harbor Symp Quant Biol. 1983;47:751–762. doi: 10.1101/sqb.1983.047.01.086. [DOI] [PubMed] [Google Scholar]
- 2.Baldauf A Q, Willwand K, Mumtsidu E, Nuesch J P, Rommelaere J. Specific initiation of replication at the right-end telomere of the closed species of minute virus of mice replicative-form DNA. J Virol. 1997;71:971–980. doi: 10.1128/jvi.71.2.971-980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 4.Chiorini J A, Wiener S M, Owens R A, Kyostio S R, Kotin R M, Safer B. Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J Virol. 1994;68:7448–7457. doi: 10.1128/jvi.68.11.7448-7457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Christensen, J., S. F. Cotmore, and P. Tattersall. Unpublished data.
- 5.Christensen J, Cotmore S F, Tattersall P. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J Virol. 1995;69:5422–5430. doi: 10.1128/jvi.69.9.5422-5430.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen J, Cotmore S F, Tattersall P. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J Virol. 1997;71:1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen J, Cotmore S F, Tattersall P. Parvovirus initiation factor PIF: a novel human DNA-binding factor which coordinately recognizes two ACGT motifs. J Virol. 1997;71:5733–5741. doi: 10.1128/jvi.71.8.5733-5741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costello E, Saudan P, Winocour E, Pizer L, Beard P. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 1997;16:5943–5954. doi: 10.1093/emboj/16.19.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotmore S F, Christensen J, Nuesch J P, Tattersall P. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2–3. J Virol. 1995;69:1652–1660. doi: 10.1128/jvi.69.3.1652-1660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotmore S F, Nuesch J P, Tattersall P. In vitro excision and replication of 5′ telomeres of minute virus of mice DNA from cloned palindromic concatemer junctions. Virology. 1992;190:365–377. doi: 10.1016/0042-6822(92)91223-h. [DOI] [PubMed] [Google Scholar]
- 11.Cotmore S F, Nuesch J P, Tattersall P. Asymmetric resolution of a parvovirus palindrome in vitro. J Virol. 1993;67:1579–1589. doi: 10.1128/jvi.67.3.1579-1589.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Cotmore, S. F., and P. Tattersall. Unpublished data.
- 12.Cotmore S F, Tattersall P. In vivo resolution of circular plasmids containing concatemer junction fragments from minute virus of mice DNA and their subsequent replication as linear molecules. J Virol. 1992;66:420–431. doi: 10.1128/jvi.66.1.420-431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotmore S F, Tattersall P. An asymmetric nucleotide in the parvoviral 3′ hairpin directs segregation of a single active origin of DNA replication. EMBO J. 1994;13:4145–4152. doi: 10.1002/j.1460-2075.1994.tb06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotmore S F, Tattersall P. Parvovirus DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 799–813. [Google Scholar]
- 15.Ge H, Roeder R G. The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J Biol Chem. 1994;269:17136–17140. [PubMed] [Google Scholar]
- 16.Higashitani A, Greenstein D, Hirokawa H, Asano S, Horiuchi K. Multiple DNA conformational changes induced by an initiator protein precede the nicking reaction in a rolling circle replication origin. J Mol Biol. 1994;237:388–400. doi: 10.1006/jmbi.1994.1242. [DOI] [PubMed] [Google Scholar]
- 17.Im D S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 18.Inamoto S, Fukuda H, Abo T, Ohtsubo E. Site- and strand-specific nicking at oriT of plasmid R100 in a purified system: enhancement of the nicking activity of TraI (helicase I) with TraY and IHF. J Biochem. 1994;116:838–844. doi: 10.1093/oxfordjournals.jbchem.a124604. [DOI] [PubMed] [Google Scholar]
- 19.John S, Reeves R B, Lin J X, Child R, Leiden J M, Thompson C B, Leonard W J. Regulation of cell-type-specific interleukin-2 receptor alpha-chain gene expression: potential role of physical interactions between Elf-1, HMG-I(Y), and NF-κB family proteins. Mol Cell Biol. 1995;15:1786–1796. doi: 10.1128/mcb.15.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin E V, Ilyina T V. Computer-assisted dissection of rolling circle DNA replication. Biosystems. 1993;30:241–268. doi: 10.1016/0303-2647(93)90074-m. [DOI] [PubMed] [Google Scholar]
- 21.Kornberg A, Baker T A. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman; 1991. [Google Scholar]
- 22.Laufs J, Jupin I, David C, Schumacher S, Heyraud-Nitschke F, Gronenborn B. Geminivirus replication: genetic and biochemical characterization of Rep protein function, a review. Biochimie. 1995;77:765–773. doi: 10.1016/0300-9084(96)88194-6. [DOI] [PubMed] [Google Scholar]
- 23.Liu Q, Yong C B, Astell C R. In vitro resolution of the dimer bridge of the minute virus of mice (MVM) genome supports the modified rolling hairpin model for MVM replication. Virology. 1994;201:251–262. doi: 10.1006/viro.1994.1290. [DOI] [PubMed] [Google Scholar]
- 24.Meehan B M, Creelan J L, McNulty M S, Todd D. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J Gen Virol. 1997;78:221–227. doi: 10.1099/0022-1317-78-1-221. [DOI] [PubMed] [Google Scholar]
- 25.Nuesch J P, Cotmore S F, Tattersall P. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology. 1995;209:122–135. doi: 10.1006/viro.1995.1236. [DOI] [PubMed] [Google Scholar]
- 26.Orozco B M, Miller A B, Settlage S B, Hanley-Bowdoin L. Functional domains of a geminivirus replication protein. J Biol Chem. 1997;272:9840–9846. doi: 10.1074/jbc.272.15.9840. [DOI] [PubMed] [Google Scholar]
- 27.Ryan J H, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segall A M, Goodman S D, Nash H A. Architectural elements in nucleoprotein complexes: interchangeability of specific and non-specific DNA binding proteins. EMBO J. 1994;13:4536–4548. doi: 10.1002/j.1460-2075.1994.tb06775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snyder R O, Im D S, Ni T, Xiao X, Samulski R J, Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol. 1993;67:6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson G M, Jindal H K, Yeung D E, Chen W, Astell C R. Expression of minute virus of mice major nonstructural protein insect cells: purification and identification of ATPase and helicase activities. Virology. 1991;185:90–98. doi: 10.1016/0042-6822(91)90757-3. [DOI] [PubMed] [Google Scholar]
- 31.Zappavigna V, Falciola L, Citterich M H, Mavilio F, Bianchi M E. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]
- 32.Zwilling S, Konig H, Wirth T. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]