Abstract
The Epstein-Barr virus (EBV) DNA polymerase (pol) mRNA, which contains a noncanonical polyadenylation signal, UAUAAA, is cleaved and polyadenylated inefficiently (S. C. S. Key and J. S. Pagano, Virology 234:147–159, 1997). We postulated that the EBV early proteins SM and M, which appear to act posttranscriptionally and are homologs of herpes simplex virus (HSV) ICP27, might compensate for the inefficient processing of pol pre-mRNA. Here we show that the SM and M proteins interact with each other in vitro. In addition, glutathione S-transferase–SM/M fusion proteins precipitate the heterogeneous ribonucleoprotein (hnRNP) C1 splicing protein. Further, the SM protein is coimmunoprecipitated from SM-expressing cell extracts with an antibody to the hnRNP A1/A2 proteins, which are splicing and nuclear shuttling proteins. Finally, the amount of processed EBV DNA polymerase mRNA was increased three- to fourfold in a HeLa cell line expressing SM; this increase was not due to enhanced transcription. Thus, inefficient processing of EBV pol RNA by cellular cleavage and polyadenylation factors appears to be compensated for and may be regulated by the early EBV protein, SM, perhaps via RNA 3′-end formation.
At least two of the Epstein-Barr virus (EBV)-associated diseases, infectious mononucleosis and oral hairy leukoplakia, are the result of primary infection and the cytolytic phase of replication of the virus. The primary gene whose product is needed for viral replication is the EBV DNA polymerase gene (pol), which is one of six early viral genes identified as being necessary and sufficient for transient in vitro EBV DNA replication, closely following the requirements for herpes simplex virus (HSV) replication (6, 7). In addition, replication is dependent on the products of three cytolytic-cycle genes: the BZLF1 and BRLF1 open reading frames (ORFs), which are immediate-early (IE) genes, and BSLF2/BMLF1 (SM/M), which are early genes. Although the SM/M-encoding genes are not among the six genes essential for viral DNA replication, there is evidence that the SM/M proteins may indirectly facilitate replication by in some way upregulating the expression of one or more of the replication factors (7). However, a specific function for SM/M has not been identified.
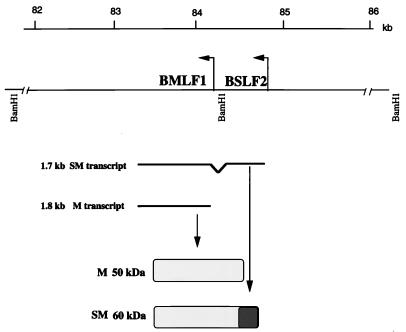
The 1.7- and 1.8-kb poly(A) transcripts from the BSLF2 and BMLF1 ORFs express the SM (60-kDa) and M (50-kDa) proteins, respectively (4, 38, 48), SM results from splicing of the BSLF2 ORF immediately upstream of BMLF1 into the M reading frame (Fig. 1). Various sizes for the SM/M proteins have been reported, possibly due to the different EBV cell lines used for viral induction or to posttranslational modifications (4).
FIG. 1.
EBV BSLF2/BMLF1 (SM/M) genes, transcripts, and protein products. The portion of the B95-8 genome that encodes the BSLF2/BMLF1 proteins is shown below a size scale. Transcription occurs in the leftward direction, generating two transcripts of 1.8 (SM) and 1.7 (M) kb in length. The phosphoproteins migrate at approximately 50 and 60 kDa in SDS-PAGE. The shaded portions of SM and M are identical; the hatched region of SM represents the extra N-terminal 41 amino acids encoded by BSLF2.
At first thought to be promiscuous transcriptional activators of human immunodeficiency virus (HIV), adenovirus, and EBV promoters (21, 48), SM/M proteins were later shown to affect gene expression through a posttranscriptional mechanism (2, 4, 16, 27). However, the endogenous genes that SM/M proteins targeted were not identified. It was surmised that the SM/M proteins might be involved in coordinating viral DNA replication (9, 10, 32). SM/M proteins have homologs in the alpha- and betaherpesviruses, one of which is HSV ICP27 (IE63). This protein is multifunctional, participating in poly(A) site selection and splicing as well as viral DNA replication (28, 33, 35, 41). Homology to SM/M is in the carboxyl terminus of ICP27, which is required for many of its functions (1). Additionally, the amino-terminal portions of SM/M and ICP27 proteins are arginine rich, and this region is also important in ICP27 function (35, 41).
The EBV DNA polymerase mRNA is apparently unique among the herpesviruses in that it has a noncanonical poly(A) signal which results in inefficient cleavage and polyadenylation (10, 18). We therefore postulated that SM/M proteins might enhance processing of the EBV DNA polymerase pre-mRNA and regulate the supply of functional mRNA of this critical replication gene. Possible mechanisms included interaction of SM/M proteins with cellular proteins involved in pre-mRNA processing, including the heterogeneous nuclear ribonucleoprotein (hnRNP) family (47).
We report here that SM/M proteins interact with the hnRNP complex, probably through the hnRNP C1/C2 and hnRNP A1/A2 proteins. Using an SM-expressing HeLa cell line, we show that in RNase protection assays, SM increases levels of pol mRNA, presumably through enhancement of its processing.
MATERIALS AND METHODS
Plasmids and constructs.
The glutathione S-transferase (GST)-SM and GST-M constructs were generated with the use of reverse transcription-PCR to amplify the appropriate region of EBV B95-8 cDNA and subcloned into pBS+ (Stratagene) and pGEX2 (Promega). The first strand was generated by using the oligo(dT) primer 5′-GGACTGAGTGACATCGA(T)17-3′, containing restrict-ion sites for SalI, XhoI, and ClaI. Amplification was by Vent polymerase (New England Biolabs) and 5′ and 3′ primers directed to amplify the EBV DNA from positions 84318 (BSLF2) or 82746 (BMLF1) to 82086 with HindIII sites engineered into 5′ primers for directional cloning. Constructs were sequenced with a Sequenase version 2.0 kit (United States Biochemical/Amersham). The SM KpnI fragment from pCEP-SM (kind gift of Paul Farrell [4]) was blunt-end ligated into the EcoRV site of pcDNA3 (Invitrogen) to generate pcSM.
pCMV-W91 was created by PCR amplification of the EBV BamHI A pol reading frame, and the XbaI-BamHI product was ligated into the pBS+ vector containing the cytomegalovirus (CMV) enhancer and the BamHI-KpnI fragment of BamHI-I of the EBV genome. Mutations were generated by use of an oligonucleotide-mediated method (Sculptor kit; Amersham). The construct for riboprobe analysis, pBS-313RPA, was made through PCR amplification of a 300-bp fragment encompassing the 3′ cleavage site and directionally cloned into the EcoRI and HindIII sites of the pBS vector. pCMV-W91 (22) was created through PCR amplification of the BALF5-encoding portion of the BamHI A fragment and subcloning the amplimer into the pBS+ vector (Stratagene). The BamHI-KpnI fragment of BamHI-I was then subcloned downstream of the BamHI A fragment. The CMV IE promoter/enhancer was placed upstream of BALF5 DNA in a PstI fragment. Linker-scanner mutations were created essentially as described previously (18, 22). Sequenase version 2.0 (Amersham/Life Sciences) data confirmed all constructs.
In vitro translation of SM/M proteins, generation of GST fusion proteins, and radioactive labeling of cellular proteins.
SM/M proteins were transcribed and translated in the presence of [35S]methionine with a coupled method (Promega). pBS+SM/M and pcSM constructs were linearized with HindIII and XbaI, respectively, and transcribed with T7 RNA polymerase.
One liter of Escherichia coli BL21(pLysS) cells expressing the GST, GST-Z, GST-M, or GST-SM fusion proteins was grown to a density of about 0.5 (A600)/ml and then induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 2 h at 37°C. Cultures were pelleted, resuspended in 10 ml of cold phosphate-buffered saline solution (PBS), and stored at −70°C. Lysates were made by freezing and thawing, and fusion proteins were then conjugated to glutathione-S Sepharose 4B (Pharmacia), washed four times with PBS, and used to precipitate labeled cellular and in vitro-translated proteins. Akata cells (5 × 107) were grown in the presence of [35S]methionine (1 mCi) for 24 h, and whole-cell lysates were prepared in PBS.
GST precipitation, immunoprecipitation, and Western assays.
GST precipitation was performed as described previously (45) except that ELB-2 buffer (0.45 M NaCl, 0.1% Nonidet P-40, 0.05 M HEPES [pH 7.3], 0.5 mM EDTA) was used. Immunoprecipitations with monoclonal antibodies to the hnRNP C1/C2 proteins, 4F4 and 1B12, and against hnRNP A1/A2 proteins, 1A1, were performed as reported previously (3, 47). A polyclonal SM antibody, αSM53 (a generous gift from Paul Farrell), was used for Western blot analysis as described elsewhere (4).
Cell lines.
The Akata cell line (46) was maintained in RPMI 1640 in 10% fetal calf serum. The SM-HeLa and pcDNA3-HeLa cell lines were generated as follows. Plasmids pcSM (described above) and pcDNA3 were transfected into HeLa S3 cells with Lipofectamine (Gibco-BRL). The transfectants were selected in Dulbecco’s modified Eagle’s medium H (DMEM-H) containing G418 (500 μg/ml) after cultivation for 2 weeks. A single SM protein-expressing clone was subcloned. Cells were maintained in DMEM-H with 10% serum and G418, the concentration of which was gradually increased to 700 μg/ml over 4 weeks (addition of 50 μg/ml/week) to retain SM expression. SM-Akata was cultivated in the same way as the Akata cell line (4, 46). CdCl2 (10 mmol/ml of medium) was used to induce expression of SM by exposure of the cells for 6 h.
mRNA harvesting and RNase protection assays.
Cells were lysed, and mRNA was collected with the use of the Oligotex direct system protocol (Qiagen). mRNA (0.5 to 1 μg) was hybridized overnight with DNA pol 3′ untranslated region (UTR) antisense probe and glutaraldehyde phosphate dehydrogenase (GAPDH) probe (Gibco-BRL). RNase protection assays were performed as specified by the manufacturer (Gibco-BRL). The 313-nucleotide (nt) probe to the pol transcript was generated by linearizing pBS-313RPA with HindIII and transcribing with T7 RNA polymerase in the presence of [32P]UTP. Products were separated through a 5% polyacrylamide gel containing 7 M urea and analyzed by phosphorimagery or autoradiography.
β-Gal enzyme assay.
HeLa cells (5 × 105) were transiently transfected with Lipofectamine and 6 μg of total DNA for 4 h in DMEM-H without serum. Then cells were incubated in DMEM-H with 10% serum as indicated in the supplier’s protocols (Lipofectamine; Gibco-BRL). β-Galactosidase (β-Gal) enzyme activity was examined by two methods. Two experimental sets were assayed with the Luminescent β-gal Genetic Reporter System II kit (Clonetech) and analyzed on a luminometer (AutoLumat LB-953; Bertholf GmbH & Co.). One experimental set was assayed according to specifications for the Promega β-Gal enzyme assay kit and analyzed with a spectrophotometer.
RESULTS
SM/M protein-protein interactions.
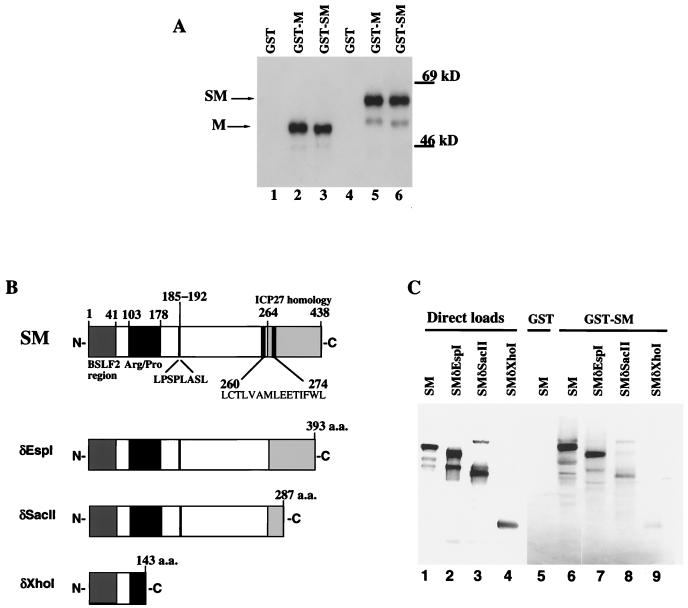
SM and M are produced by differential splicing (Fig. 1). Therefore, potential interactions of the SM and M proteins were tested with the use of GST-SM and GST-M fusion proteins and in vitro-translated proteins from pBS-SM and pBS-M constructs. Both of the fusion proteins precipitated in vitro-translated M (50-kDa) and SM (60-kDa) proteins (Fig. 2A, lanes 2, 3, 5, and 6), whereas GST alone did not (lanes 1 and 4). These data suggest that the SM and M interact with each other and may be capable of other protein-protein interactions.
FIG. 2.
GST-SM and GST-M precipitation of SM and M in vitro-translated proteins. GST proteins were incubated with 5 μl of in vitro-translated proteins. Precipitated proteins were separated by SDS-PAGE (10% gel) and exposed to X-ray film. (A) The pBS-M and pBS-SM constructs were linearized with HindIII and then transcribed and translated in the presence of [35S]methionine. Lanes 1 to 3 are precipitations of the in vitro-made M protein; lanes 4 to 6 are precipitations made with the SM-programmed lysates. (B) Illustration of the in vitro-translated proteins arising from the pcSM deletions at the EspI, SacII, and XhoI restriction sites. The amino acid (a.a.) residue immediately preceding the site of cleavage is indicated at the end of each truncation. The hatched box corresponds to the region encoded by BSLF2; the solid box indicates an arginine/proline-rich region, and the shaded box represents the ICP27 homology region. Residues 185 to 192 represent a potential nuclear export signal, and residues 260 to 274 indicate a leucine-rich region. (C) Lanes 6 to 9, GST-SM precipitation of in vitro-translated SM and truncated SM proteins; lane 5, GST alone; lanes 1 to 4, lighter exposure of 5 μl of programmed lysates loaded directly.
To define the region required for SM interactions, deletions were generated in the pcSM construct by restriction digestion followed by in vitro transcription-translation (Fig. 2B). The larger protein, GST-SM, was used in precipitation assays. Equivalent amounts of in vitro-translated proteins were added to each precipitation reaction and are comparable to the direct loads (Fig. 2C, lanes 1 to 4). All three of the truncated proteins were precipitated by the GST-SM fusion protein (lanes 7 to 9), as was the full-length in vitro-translated SM protein (lane 6). GST-SM precipitation of the in vitro-translated SM proteins appeared significantly reduced when the SacII-truncated protein was included in the assay (compare lanes 6 and 7 with lane 8). The truncated protein resulting from the XhoI deletion, which removes a putative leucine-heptad region (20), also reduced the SM/SM interaction (lane 9).
The SacII truncation data suggest that part of the SM domain homologous to the ICP27 protein (shaded area in Fig. 2B) may be required for SM/SM interaction. Additionally, these data indicate that an SM/SM protein interaction domain may lie in the XhoI-SacII segment of the BMLF1-encoding region. Since none of the C-terminal truncations abolished SM/SM protein interaction, the N terminus may also be involved. Additionally, since the truncations were large, it is possible that conformational changes reduced accessibility to the N terminus, which may be the authentic site of self-interaction. Thus, the data indicate that SM/SM protein interaction may occur at more than one domain but is optimal with the intact protein.
SM/M proteins interact with components of the hnRNP complex.
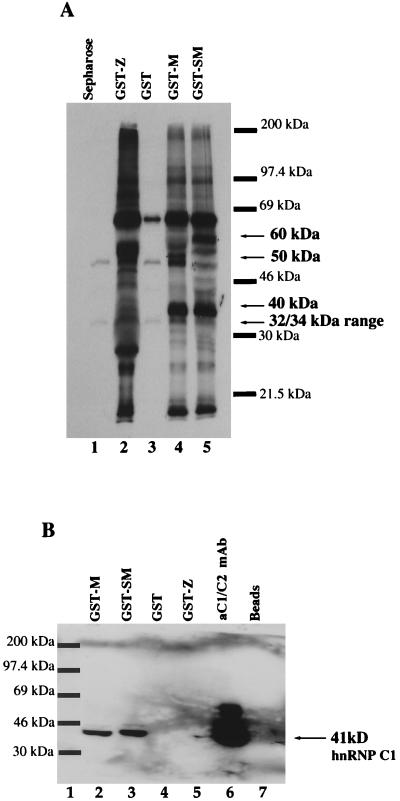
Since SM/M proteins appeared capable of self-interactions, we determined whether SM/M could interact with cellular proteins, specifically, members of the RNA splicing and 3′ processing families. Initially, labeled proteins were precipitated from Akata cell lysates with the use of the GST-M and GST-SM proteins. We detected several proteins that appeared to precipitate specifically with the SM/M fusion proteins (Fig. 3A, lanes 4 and 5) and not with beads or GST alone or with GST-Z, another EBV protein capable of a number of protein interactions (lanes 1 to 3). The approximate sizes of the most prominent proteins are 40 kDa, 50 kDa (upper band, lane 4), and 60 kDa. GST-M and GST-SM proteins precipitated a common protein (40 kDa) as well as unique proteins (50 and 60 kDa) (compare lanes 4 and 5). Additionally, GST-M and GST-SM proteins specifically and reproducibly precipitated other less prominent proteins (compare lane 3 to lanes 4 and 5), including polypeptides in the 32- to 34-kDa range. To examine whether SM might interact with members of the RNA 3′ processing family, we tested antibodies directed to these factors. After GST-M/SM precipitation of Akata cell lysates, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed in Western blot assays. No interactions were detected with antibodies against members of the general 3′ processing complex, including the 50- and 64-kDa polypeptides of the cleavage-stimulatory factor (CSF) complex, and 100- and 160-kDa proteins of the cleavage and polyadenylation specificity factor (CPSF) complex, poly(A) polymerase (PAP), and poly(A) binding protein II (PAB II) (Table 1; for a review of the processing complex, see reference 15). (Antibodies to CSF/CPSF and PAP/PAB II were generous gifts from W. Keller and E. Wahle). Additionally, the small nuclear ribonucleoprotein (snRNP) U1A, which has a role in 3′ processing (25, 26) as well as splicing (24), did not interact with the GST-SM/M proteins (Table 1) (U1A antibody was a generous gift from C. Lutz and J. Alwine). All antibodies used detected the appropriate proteins in lanes loaded with Akata cell extract (data not shown). However, a member of the hnRNP family, hnRNP C1 (a 41-kDa protein), was repeatedly precipitated with the GST-SM/M proteins (Fig. 3B, lanes 2 and 3) and detected by Western blot with an anti-C1/C2 antibody, 4F4 (gift from G. Dreyfuss). The faint upper band represents the 43-kDa hnRNP C2 protein (lanes 2 and 3); on longer exposure, the C2 protein was prominent (data not shown).
FIG. 3.
Precipitation of cellular proteins by GST-SM/M fusion proteins including hnRNP C1 from Akata cell lysates. (A) Akata cells were incubated in the presence of [35S]methionine-cysteine (Tran35S-label; ICN) for 24 h, harvested, and lysed. Proteins from lysates were precipitated with GST-SM or GST-M fusion protein (lanes 4 and 5) or controls (lanes 1 to 3). Proteins were separated by SDS-PAGE (10% gel) and visualized by autoradiography. (B) Western analysis for the 41- and 43-kDa hnRNP C1/C2 proteins. Unlabeled proteins were precipitated from Akata lysates with GST and GST fusion proteins. Samples were electrophoresed, transferred to Immobilon, and analyzed for the hnRNP C1/C2 proteins with the 4F4 monoclonal antibody (gift from G. Dreyfuss). Lane 6 shows immunoprecipitation of hnRNP C1/C2 by the 4F4 antibody.
TABLE 1.
RNA processing factors that are either precipitated by the EBV GST-SM/M fusion proteins or coimmunoprecipitated with SM/M proteins by antisera to the processing factors
| Protein | GST-SM/M precipitationa | Coimmunoprecipitation of SMb | Known function |
|---|---|---|---|
| PAP | − | NTc | Adenylation |
| PAB II | − | NT | Binds oligo(A); stability |
| CSF | |||
| 50 kDa | − | NT | Cleavage |
| 64 kDa | − | NT | Cleavage; binds U-rich element |
| CSPF | |||
| 160 kDa | NT | − | Cleavage and polyadenylation; binds AAUAAA |
| 100 kDa | − | − | Cleavage and polyadenylation |
| hnRNP C1/C2 | + | − | Splicing; nuclear export; binds U-rich element |
| hnRNP A1/A2 | − | + | Splicing; nuclear export |
| hnRNP B1/B2 | − | − | Splicing; nuclear export |
| snRNP U1A | − | NT | Cleavage; splicing; binds AUUUGURA |
Detection by Western blotting in Akata cell lysates.
Antisera to processing factors used to coimmunoprecipitate SM protein from SM-HeLa cells; detection by Western blotting with SM antibody.
NT, not tested.
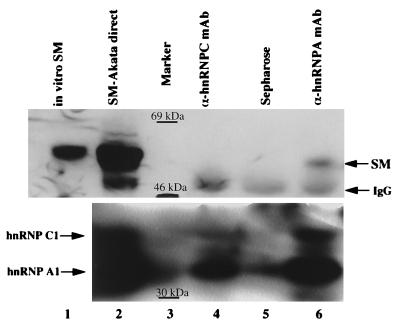
Thereafter, we focused on SM because both SM and M proteins appeared to interact with the hnRNP C1/C2 protein. Also, reports suggest that SM/M proteins function equivalently and that SM protein is more abundant than M during the early viral replicative phase (4, 16, 21, 45). Antibodies directed against members of the hnRNP complex were used to coprecipitate the SM protein from nuclear extracts of the SM-Akata cell line that was induced with CdCl2 (see Materials and Methods). Proteins were separated by SDS-PAGE, transferred, and analyzed with a polyclonal antibody directed against SM (αSM53; gift from Paul Farrell). The hnRNP C1/C2 antibody, 1B12 (gift from J. Wilusz), was unable to coprecipitate SM protein (Fig. 4, lane 4), perhaps because the hnRNP C1/C2 epitope was masked by the SM interaction. However, an antibody directed against the A1/A2 (32/34-kDa) members of the hnRNP complex, 1A1 (gift from J. Wilusz), reproducibly precipitated SM (lane 6). Under the conditions used, the 1A1 monoclonal antibody precipitates C1/C2 as well as A1/A2 proteins (47). The rabbit anti-mouse immunoglobulin G bridging antibody conjugated to protein A-Sepharose did not precipitate SM (lane 5). The antibodies precipitated their respective proteins, as determined with the 4E4 antibody (gift of J. Wilusz) directed against the six major hnRNP polypeptides, A1/A2, B1/B2, and C1/C2 (Fig. 4, bottom; results are summarized in Table 1). Reciprocal coimmunoprecipitation experiments with the SM antibody, αSM53, could not be performed because of the inability of this antibody to immunoprecipitate (data not shown). Thus, SM clearly interacts with the hnRNP complex; candidate proteins for specific interactions are A1/A2 and C1/C2.
FIG. 4.
hnRNP A1/A2 antibody coimmunoprecipitates SM protein. SM expression was induced with 10 mM CdCl2 per ml of medium for 6 h. (Top) Proteins were precipitated from induced SM-Akata nuclear extracts with the 1A1 monoclonal antibody (lane 6) or an antibody against the hnRNP C1/C2 protein, 1B12 (lane 4). The precipitates were separated through an SDS–10% polyacrylamide gel and then transferred to Immobilon. The αSM53 polyclonal antibody was used to detect SM protein. Lane 1, in vitro-translated SM; lane 2, direct load of 30 μg of SM-Akata nuclear extract used in the immunoprecipitations. (Bottom) Western analysis of the same gel with the 4E4 monoclonal antibody, which recognizes all hnRNP A and C proteins. IgG, immunoglobulin G.
The amount of processed EBV DNA polymerase transcript is increased in the presence of SM protein in vivo.
Previous studies have shown that the EBV pol pre-mRNA is inefficiently cleaved and polyadenylated due to the presence of the variant poly(A) signal, UAUAAA, and flanking elements (18). Since the SM/M proteins are expressed early in the viral replicative cycle and could enhance expression of essential replication factors, we determined whether SM/M proteins could increase posttranscriptional processing of EBV DNA pol mRNA. To test whether the SM protein could increase the levels of the EBV DNA pol transcript in the absence of its promoter, a pol construct driven by the CMV IE promoter/enhancer, pCMV-W91, was generated. Also, the SM-HeLa cell line was created by stably transfecting HeLa cells with a construct, pcSM, in which SM expression was placed under the control of the CMV IE promoter, and selected by gentamicin resistance (see Materials and Methods).
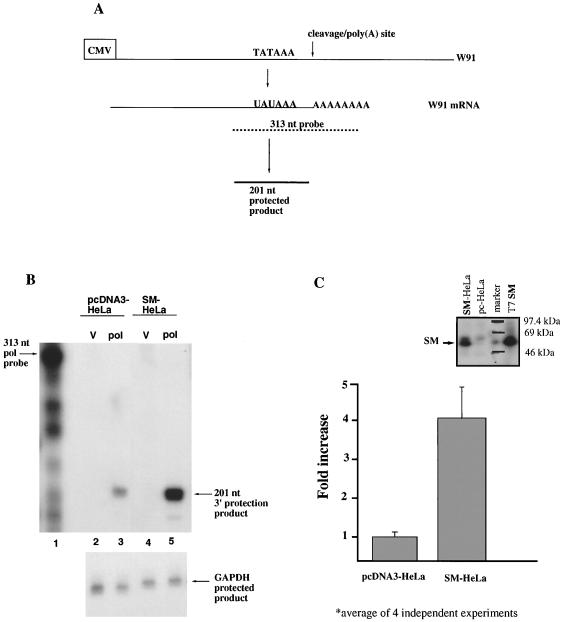
After transient transfections of SM-HeLa and pcDNA3-HeLa cell lines with pCMV-W91 containing the entire EBV DNA pol gene, including its 3′ UTR or with vector DNA, mRNA was selected by using the Oligotex kit protocol (Qiagen) and analyzed for the processed pol transcript. A 313-nt probe was used in a ribonuclease protection assay. This probe is antisense to a region of pol mRNA spanning the poly(A) signal and cleavage/poly(A) site (Fig. 5A). After cleavage, hybridization of the 313-nt probe to the processed pol mRNA should produce a 201-nt protected product (Fig. 5A). Protected RNA of this size was detected with the RNA from pcDNA3-HeLa when pCMV-W91, encoding EBV DNA polymerase, was introduced (Fig. 5B, lane 3), but not with vector alone (Fig. 5B, lane 2). The level of the 201-nt product was specifically and strikingly increased in SM-HeLa mRNA but not in the vector-transfected mRNA sample (Fig. 5B; compare lanes 4 and 5). The amount of endogenous GAPDH transcript remained equivalent in all pcDNA3-HeLa and SM-HeLa samples (Fig. 5B, bottom, lanes 2 to 5). Transfection efficiency, monitored by β-Gal staining, was about 10% in both cell lines. Western blot analysis with the polyclonal antibody against SM protein (gift from P. Farrell) demonstrated that the cell line was expressing SM for each of four independent transfections (inset to Fig. 5C and data not shown). A three- to fourfold enhancement in the amount of processed pol transcript was consistently detected in the SM-HeLa cells (Fig. 5C). Thus, SM protein appears to enhance 3′ RNA processing of the EBV DNA polymerase mRNA, which contains an inefficient poly(A) signal.
FIG. 5.
Comparison of the amounts of processed EBV DNA polymerase transcript detected in the SM-HeLa cell line and the pcDNA3-HeLa cell line by RNase protection assays. SM-HeLa and pcDNA3-HeLa cell lines were transiently transfected with the use of Lipofectamine with either the pCMV-W91 or the pBS+ vector. (A) Diagram illustrating the hybridization of the 313-nt riboprobe generated from pBS-313wtRPA to W91 mRNA. When the RNA-RNA hybrid is treated with RNases T and A1, a 201-nt protected fragment results. (B) RNase protection assay of 1 μg of mRNA from pcDNA3-HeLa (lanes 2 and 3) or SM-HeLa (lanes 4 and 5) cells transfected with vector (V) or pCMV-W91 (pol). GAPDH (Amersham) protected bands are shown at the bottom. This experiment was repeated four times. (C) Average fold increase, calculated from four experiments as the ratio of the counts per minute of the protected mRNA products of pol to GAPDH from SM-HeLa cells divided by the same ratio as detected with pcDNA3-HeLa cell mRNA. The inset is an αSM53 Western blot of pcDNA-HeLa and SM-HeLa cell extracts.
Although it seemed likely that the increased pol mRNA levels were the result of a posttranscriptional mechanism, earlier reports claimed that SM/M activates heterologous viral promoters (17, 21, 48). Later reports concluded that SM/M works through a posttranscriptional mechanism but did not completely exclude the possibility of an effect on transcription (4, 16, 27). Thus, we tested whether SM affected the CMV IE promoter/enhancer to increase pol transcription. CMVβgal and the promoterless BASICβgal constructs (Clontech) were used in transient pcSM cotransfection assays in HeLa or C33 cells, since they are efficiently transfected. The β-Gal constructs contain the simian virus 40 (SV40) poly(A) signal (AAUAAA), which is more efficient than the EBV DNA poly(A) signal (UAUAAA). Expression of BMLF1 was reported not to affect the activity of a β-Gal reporter that contained the SV40 signal (27). Cells were transfected and harvested 48 h later. Lysates were prepared and assayed for β-Gal activity with chemiluminescent reagents and a luminometer (AutoLumat LB-953; Bertholf GmbH & Co.) or by a spectrophotometric method (Promega). The results of three sets of triplicate transfections indicate a slight but insignificant increase in β-Gal activity in the presence of pcSM compared with its vector background, pcDNA3 (Table 2). A fourth set of triplicate β-Gal assays was performed with the stable cell lines SM-HeLa and pcDNA3-HeLa. Again, β-Gal activity was unaffected by the presence of SM (data not shown). Thus, there was little if any effect of SM on the CMV IE promoter/enhancer. The data support the hypothesis that SM increases pol mRNA levels by affecting processing of pre-mRNA.
TABLE 2.
SM protein does not transactivate the CMV promoter in a β-Gal reporter assay
| Expta | Reporter construct | Fold increasec |
|---|---|---|
| 1 | BASICβgalb | 1.3 ± 0.20 |
| CMVβgal | 1.6 ± 0.65 | |
| 2 | BASICβgal | 1.0 ± 0.14 |
| CMVβgal | 1.7 ± 0.14 | |
| 3 | BASICβgal | 1.0 ± 0.01 |
| CMVβgal | 1.2 ± 0.13 |
Data for experiments 1 and 2 were measured by luminometry; data for experiment were measured by spectrophotometry.
Contains no promoter.
Ratio of results with pcSM over results with vector pcDNA3.
DISCUSSION
Pre-mRNA of the EBV DNA polymerase gene is cleaved and polyadenylated inefficiently in vitro because of a noncanonical poly(A) signal (18). In this report, we show that the abundance of processed pol mRNA is increased in vivo in cells expressing the EBV SM early protein. In the absence of SM, which, like its homolog the HSV ICP27 protein, acts posttranscriptionally, little mRNA can be detected. Thus, SM protein appears to compensate for the deficient processing of EBV pol RNA. Based on previous in vitro results, SM most likely affects the cleavage/polyadenylation step in processing of the RNA (18). However, the possibility that SM enhances nuclear export or increases mRNA stability has not been excluded. Effects of these mechanisms are difficult to distinguish since all result in higher levels of processed mRNA in the cytoplasm and indeed may not be mutually exclusive (12, 19). In any case, what is normally a cellular process, namely, cleavage/polyadenylation and export of mRNA from the nucleus, is clearly facilitated by a viral protein, SM. Although the functions of HSV ICP27 have some resemblance (4, 28, 33), SM’s effects are distinctive in that the EBV protein produces a three- to fourfold enhancement in the level of the early mRNA, pol, for which levels are otherwise low.
At least 11 polypeptides orchestrate the cleavage and polyadenylation of pre-mRNA, including four members of the CPSF complex, three members of the CSF complex, poly(A) PAP, cleavage factors I and II, and PAB II (for a review, see reference 15). So far, interaction of SM has not been detected with any of the six proteins that we tested. Although hnRNPs are not known to participate in cleavage and polyadenylation, hnRNP C1 binds to the same downstream U-rich element recognized by the 64-kDa member of the CSF complex (47). In this work, we implicate hnRNP C1/C2 and hnRNP A1/A2 in the pre-mRNA 3′ processing of the intronless pol transcript indirectly by showing that these proteins interact with SM.
The hnRNP family in eukaryotes includes more than 20 proteins, designated A through U, that are localized to the nucleus (5). The core hnRNP complex is composed of six highly related proteins, hnRNP A1/A2, hnRNP C1/C2, and hnRNP B1/B2 (3). hnRNP family members bind RNA through two copies of an amino-terminal RNP motif, an RNA-binding domain, and a carboxyl-terminal glycine-rich domain (44). Various members of this complex have been implicated in pre-mRNA/mRNA processing events, including splicing and nuclear shuttling (30). In particular, recent studies of A1 indicate a specific role in mRNA nuclear export (31, 34). Interestingly, SM/M proteins contain an extensive leucine-rich domain (SM coordinates 185 to 274), which includes a putative nuclear export signal, LPSPLASLTL, similar to that of Rev/Rex required for rapid nuclear export of retroviral RNAs (reviewed in references 11 and 19). One report suggests that in order for Rev to transport an HIV RNA rapidly, the polyadenylation machinery must interact with that RNA (14). Perhaps SM/M-hnRNP complexes favor rapid export of EBV transcripts. However, the definitive role(s) of each hnRNP member is unclear. Thus, it is difficult to assign a particular role to SM/M-hnRNP interaction complexes other than their involvement in pre-mRNA processing events.
Little is known about pre-mRNA processing of EBV transcripts and the role of SM/M proteins. The SM/M proteins, assuming that they function to enhance 3′ processing of pre-mRNA, may first form self- and non-self-interactions through several domains, including a putative leucine zipper domain. Second, the data obtained so far suggest that SM/M proteins may enhance 3′ processing of pol pre-mRNA by directly interacting with members of the pre-mRNA processing complex, namely, hnRNP C1/C2 and/or hnRNP A1/A2. Recent reports implicate coordination of splicing with 3′ processing, with splicing factors playing an essential role in regulation of RNA 3′-end formation. For example, the snRNP U1A interacts with PAP to inhibit 3′ processing of its own transcript (13) but contacts the 160-kDa member of the CPSF complex to enhance processing of the late SV40 mRNA (26). However, the EBV DNA polymerase pre-mRNA does not contain an intron. It is not spliced and is inefficiently cleaved and polyadenylated (10, 18). Thus, in this case SM/M may interact with splicing factors, sequestering the splicing machinery and perhaps favoring the processing of an intronless viral transcript. Indeed, there is precedence for such a scenario, as SM/M’s HSV counterpart, ICP27, appears to increase the production of viral transcripts that are intronless and/or are inefficiently cleaved and polyadenylated (25, 29, 33, 42). Recent evidence suggests that the M protein may suppress the use of cryptic 5′ splice sites (43a).
ICP27 appears to have multiple functions. HSV DNA replication requires the IE protein ICP27 (35). Interestingly, expression of the HSV ICP27 gene can substitute for M in an in vitro EBV ori-Lyt replication assay (7), suggesting a functional equivalence. Additionally, ICP27 is required for shutoff of host cellular protein synthesis, most probably by redistributing the snRNP and the SC35 splicing factors during infection (40, 41). ICP27 is coimmunoprecipitated with snRNPs by the SM-specific antibody, which recognizes the shared epitope of the snRNP family, suggesting that redistribution of splicing proteins by ICP27 expression is carried out through direct interaction (40). Recently, the HSV protein has been shown to bind directly to several cellular 3′ UTRs, apparently stabilizing labile transcripts (1). Homology of SM/M and ICP27 proteins lies in the C-terminal region and includes the zinc knuckle RNA-binding domain of ICP27 (1). In preliminary experiments, the GST-SM/M proteins bound to the EBV pol transcript but not to an HIV transcript containing the Rev-responsive element (data not shown). ICP27 may also facilitate the preferential nuclear export of HSV mRNAs transcribed from intronless genes (33). Most notably, expression of ICP27 appears to be an essential step in the switch from early to late viral gene expression, and the event seems to involve increased processing of transcripts with weak polyadenylation signals (28, 29, 39, 42).
EBV DNA pol contains a functional but inefficient poly(A) signal, UAUAAA (10, 18). Two other viruses, figwort mosaic virus and hepatitis B virus, also contain the UAUAAA poly(A) signal. The figwort mosaic virus P6 protein and the hepatitis B virus precore protein are in some way involved in regulating whether replication or pre-mRNA processing is favored (19, 36, 37, 43). Recent evidence suggests that SM suppresses levels of polyadenylated mRNA from intron-containing genes but increases mRNA levels from intronless genes (37a). Whether SM/M enhances the amount of EBV DNA pol mRNA through a mechanism of pre-mRNA processing, nuclear exportation, RNA stabilization, or a combination of these events remains to be elucidated. This report provides evidence that the level of at least one EBV transcript increases strikingly in the presence of SM/M proteins.
What is the functional significance of SM’s effect on processing of the EBV DNA polymerase mRNA? The products of the BZLF1 (Z), BRLF1 (R), and BMLF1/BSLF2 (SM/M) genes are used in the cytolytic cycle for viral replication (7). Z and R are IE gene products that activate early EBV genes (8, 23), whereas SM/M is an early gene product with a quite different level of action (4, 16, 27). In addition to regulating expression of viral genes through activating transcription, EBV may be able to regulate gene expression at a later stage of the process. The virus may also coordinate the timely expression of its DNA polymerase by enlisting SM/M proteins to enhance the otherwise inefficient processing of the mRNA of this key gene.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
This work was supported in part by the National Cancer Institute (NIH grant P01 CA 19014) and by a grant-in-aid from the Ministry of Education, Science and Culture of Japan (T.Y.).
We thank Gideon Dreyfuss, Clinton MacDonald, Jeffrey Wilusz, Carol Lutz, James Alwine, Elmar Wahle, Andreas Jenny, and Walter Keller for generously providing antibodies directed against pre-mRNA processing proteins. We are grateful to Paul Farrell for providing the αSM53 antibody and constructs. We thank William Marzluff and Nancy Raab-Traub for advice. We thank Maureen Caldwell and Cyd Johnson for help in preparing the manuscript and Chunnan Liu and Luwen Zhang for assistance.
REFERENCES
- 1.Brown C R, Nakamura M S, Mosca J D, Hayward G S, Straus S E, Perera L P. Herpes simplex virus trans-regulatory protein ICP27 stabilizes and binds to 3′ ends of labile mRNA. J Virol. 1995;69:7187–7195. doi: 10.1128/jvi.69.11.7187-7195.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buisson M, Manet E, Trescol-Biemont M-C, Gruffat H, Durand B, Sergeant A. The Epstein-Barr virus (EBV) early protein EB2 is a posttranscriptional activator expressed under the control of EBV transcription factors EB1 and R. J Virol. 1989;63:5276–5284. doi: 10.1128/jvi.63.12.5276-5284.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burd C G, Swanson M S, Gorlach M, Dreyfuss G. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: a diversity of RNA binding proteins is generated by small peptide inserts. Proc Natl Acad Sci USA. 1989;86:9788–9792. doi: 10.1073/pnas.86.24.9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook I D, Shanahan F, Farrell P J. Epstein-Barr virus SM protein. Virology. 1994;205:217–227. doi: 10.1006/viro.1994.1637. [DOI] [PubMed] [Google Scholar]
- 5.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 6.Fixman E D, Hayward G S, Hayward S D. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol. 1992;66:5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fixman E D, Hayward G S, Hayward S D. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol. 1995;69:2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furnari F B, Adams M D, Pagano J S. Regulation of the Epstein-Barr virus DNA polymerase gene. J Virol. 1992;66:2837–2845. doi: 10.1128/jvi.66.5.2837-2845.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furnari F B, Adams M D, Pagano J S. The 3′ ends of EBV DNA polymerase mRNAs are processed without canonical polyadenylation signals. In: Tursz T, et al., editors. The Epstein-Barr virus and associated diseases. Montrouge, France: Colloque INSERM/John Libbey Eurotext Ltd.; 1993. pp. 175–180. [Google Scholar]
- 10.Furnari F B, Adams M D, Pagano J S. Unconventional processing of the 3′ termini of the Epstein-Barr virus DNA polymerase mRNA. Proc Natl Acad Sci USA. 1993;90:378–382. doi: 10.1073/pnas.90.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerace L. Nuclear export signals and the fast track to the cytoplasm. Cell. 1995;82:340–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 12.Green M R. Pre-mRNA processing and mRNA nuclear export. Curr Opin Cell Biol. 1989;1:519–525. doi: 10.1016/0955-0674(89)90014-8. [DOI] [PubMed] [Google Scholar]
- 13.Gunderson S I, Beyer K, Martin G, Keller W, Boelens W C, Mattaj I W. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994;76:531–541. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, Carmichael G G. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller W. No end yet to messenger RNA 3′ processing! Cell. 1995;81:829–832. doi: 10.1016/0092-8674(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 16.Kenney S, Kamine J, Holley G E, Mar E C, Lin J C, Markovitz D, Pagano J. The Epstein-Barr virus immediate-early gene product, BMLF1, acts in trans by a posttranscriptional mechanism which is reporter gene dependent. J Virol. 1989;63:3870–3877. doi: 10.1128/jvi.63.9.3870-3877.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenney S, Kamine J, Markovitz D, Fenrick R, Pagano J. An Epstein-Barr virus immediate-early gene product trans-activates gene expression from the human immunodeficiency virus long terminal repeat. Proc Natl Acad Sci USA. 1988;85:1652–1656. doi: 10.1073/pnas.85.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Key S C S, Pagano J S. A noncanonical poly(A) signal, UAUAAA, and flanking elements in Epstein-Barr virus DNA polymerase mRNA function in cleavage and polyadenylation assays. Virology. 1997;234:147–159. doi: 10.1006/viro.1997.8647. [DOI] [PubMed] [Google Scholar]
- 19.Kiss-Laszlo Z, Hohn T. Pararetro- and retrovirus RNA: splicing and the control of nuclear export. Trends Microbiol. 1996;4:480–485. doi: 10.1016/s0966-842x(97)82909-5. [DOI] [PubMed] [Google Scholar]
- 20.Kouzarides T, Packham G, Cook A, Farrell P J. The BZLF1 protein of EBV has a coiled coil dimerization domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene. 1991;6:195–204. [PubMed] [Google Scholar]
- 21.Lieberman P M, O’Hare P, Hayward G S, Hayward S D. Promiscuous trans-activation of gene expression by an Epstein-Barr virus-encoded early nuclear protein. J Virol. 1986;60:140–148. doi: 10.1128/jvi.60.1.140-148.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C. Ph.D. thesis. Chapel Hill, N.C: University of North Carolina at Chapel Hill; 1996. [Google Scholar]
- 23.Liu C, Sista N D, Pagano J S. Activation of the Epstein-Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J Virol. 1996;70:2545–2555. doi: 10.1128/jvi.70.4.2545-2555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luhrmann R, Kastner B, Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim Biophys Acta. 1990;1087:265–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- 25.Lutz C S, Alwine J C. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 26.Lutz C S, Murthy K G, Schek N, O’Connor J P, Manley J L, Alwine J C. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 27.Markovitz D M, Kenney S, Kamine J, Smith M S, Davis M, Huang E S, Rosen C, Pagano J S. Disparate effects of two herpesviruses immediate-early gene trans-activators on the HIV-1 LTR. Virology. 1989;173:750–754. doi: 10.1016/0042-6822(89)90591-6. [DOI] [PubMed] [Google Scholar]
- 28.McGregor F, Phelan A, Dunlop J, Clements J B. Regulation of herpes simplex virus poly(A) site usage and the action of immediate-early protein IE63 in the early-late switch. J Virol. 1996;70:1931–1940. doi: 10.1128/jvi.70.3.1931-1940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLauchlan J, Simpson S, Clements J B. Herpes simplex virus induces a processing factor that stimulates poly(A) site usage. Cell. 1989;59:1093–1105. doi: 10.1016/0092-8674(89)90765-4. [DOI] [PubMed] [Google Scholar]
- 30.Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakielny S, Siomi M C, Siomi H, Michael W M, Pollard V, Dreyfuss G. Transportin: nuclear transport receptor of a novel nuclear protein import pathway. Exp Cell Res. 1996;229:261–266. doi: 10.1006/excr.1996.0369. [DOI] [PubMed] [Google Scholar]
- 32.Pagano J S, Sista N D, Furnari F B, Lin J-L. Expression and regulation of the Epstein-Barr virus DNA polymerase. In: Verno R, Nishizuda Y, editors. Biotechnology of cell regulation. Serono Symposia Series, Advances in Experimental Medicine. New York, N.Y: Raven Press; 1991. pp. 153–168. [Google Scholar]
- 33.Phelan A, Dunlop J, Clements J B. Herpes simplex virus type 1 protein IE63 affects the nuclear export of virus intron-containing transcripts. J Virol. 1996;70:5255–5265. doi: 10.1128/jvi.70.8.5255-5265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 35.Rice S A, Lam V, Knipe D M. The acidic amino-terminal region of herpes simplex virus type 1 alpha protein ICP27 is required for an essential lytic function. J Virol. 1993;67:1778–1787. doi: 10.1128/jvi.67.4.1778-1787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russnak R, Ganem D. Sequences 5′ to the polyadenylation signal mediate differential poly(A) site use in hepatitis B viruses. Genes Dev. 1990;4:764–776. doi: 10.1101/gad.4.5.764. [DOI] [PubMed] [Google Scholar]
- 37.Russnak R H. Regulation of polyadenylation in hepatitis B viruses: stimulation by the upstream activating signal PSI is orientation-dependent, distance-independent, and additive. Nucleic Acids Res. 1991;19:6449–6456. doi: 10.1093/nar/19.23.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Ruvolo V, Boyle S, Swaminathan S. The EBV nuclear protein SM is both a post-transcriptional inhibitor and activator of gene expression. Proc Natl Acad Sci USA. 1998;95:8852–8857. doi: 10.1073/pnas.95.15.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sample J, Lancz G, Nonoyama M. Mapping of genes in BamHI fragment M of Epstein-Barr virus DNA that may determine the fate of viral infection. J Virol. 1986;57:145–154. doi: 10.1128/jvi.57.1.145-154.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandri-Goldin R M. Analysis of the regulatory activities of the HSV-1 α protein ICP27. In: Wagner E K, editor. Herpesvirus transcription and its regulation. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 77–103. [Google Scholar]
- 40.Sandri-Goldin R M, Hibbard M K. The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum, and the C terminus appears to be required for this interaction. J Virol. 1996;70:108–118. doi: 10.1128/jvi.70.1.108-118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandri-Goldin R M, Hibbard M K, Hardwicke M A. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandri-Goldin R M, Mendoza G E. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 43.Sanfacon H. Analysis of figwort mosaic virus (plant pararetrovirus) polyadenylation signal. Virology. 1994;198:39–49. doi: 10.1006/viro.1994.1006. [DOI] [PubMed] [Google Scholar]
- 43a.Sergeant, A. Personal communication.
- 44.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith D B. Purification of glutathione-S-transferase fusion proteins. Methods Mol Cell Biol. 1993;4:220–229. [Google Scholar]
- 46.Takada K, Horinouchi K, Ono Y, Aya T, Osato T, Takahashi M, Hayasaka S. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Genes. 1991;5:147–156. doi: 10.1007/BF00571929. [DOI] [PubMed] [Google Scholar]
- 47.Wilusz J, Shenk T. A uridylate tract mediates efficient heterogeneous nuclear ribonucleoprotein C protein-RNA cross-linking and functionally substitutes for the downstream element of the polyadenylation signal. Mol Cell Biol. 1990;10:6397–6407. doi: 10.1128/mcb.10.12.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong K, Levine A J. Characterization of proteins encoded by the Epstein-Barr virus transactivator gene BMLF1. Virology. 1989;168:101–111. doi: 10.1016/0042-6822(89)90408-x. [DOI] [PubMed] [Google Scholar]