Abstract
We have examined the viral selection that may occur during transmission by studying the env gene sequences from four cases of mother-to-child transmission of human immunodeficiency virus type 1. The V3 region sequences were directly amplified from both plasma viral RNA and peripheral blood mononuclear cells containing proviral DNA from mothers at delivery and at the time of diagnosis for children. Transmission occurred perinatally in three cases. The similarity of the viral sequences in each infant sample contrasted with the heterogeneous viral populations in the mothers. Phylogenetic analysis indicated the transmission of one or a few closely related maternal minor virus variants. In contrast, the child virus population in the fourth case was as heterogeneous as that of his mother, and phylogenetic analysis strongly suggested the transmission of multiple maternal variants. This case of multiple transmission was confirmed by analyzing sequences obtained at three times after delivery. Strains with sequences corresponding to the syncytium-inducing phenotype were also transmitted in this fourth case, and this was associated with the rapid development of disease in the child. There was no evidence for transmission of particular viral variants from mother to infant. We have thus described a particular case of vertical human immunodeficiency virus type 1 transmission with the transmission of multiple maternal variants to the infant and a rapid, fatal outcome in the child.
The transmission of human immunodeficiency virus type 1 (HIV-1) from mother to child is the main cause of pediatric infection. Transmission rates are about 14% in industrialized countries but can be over 35% in developing countries (4, 24). The use of zidovudine plus avoiding breast feeding has greatly reduced HIV transmission to less than 5% (18). But this prevention cannot mask our lack of knowledge of how the vertical transmission of HIV-1 occurs. Many questions still have no clear answers, despite their importance for optimizing prevention strategies and understanding the pathophysiology of HIV-1 infection in children.
Transmission from mother to child may occur in utero, intrapartum, or postnatally by breast feeding. The development of an HIV-based clinical disease in children seems to be correlated with the timing of the vertical transmission (35). The disease develops slowly in about two-thirds of children, and they are believed to have been infected at the very end of pregnancy or at delivery. The remaining one-third progress rapidly to AIDS, with increased indices of viral replication (8); these children appear to have been infected during pregnancy. Molecular variability studies have shown that infected children with slow progression to AIDS have a higher viral diversity than do children who progress rapidly (11, 31), as reported for adults (9). It has been proposed that the detection of no virus in the child at birth indicates that contamination took place at or shortly before delivery (5). But detection of the virus at birth indicates in utero contamination. Virus is usually detected by HIV coculture or sensitive PCR analyses of cell-associated proviral DNA and/or plasma RNA.
There can be genetic variations in HIV-1, especially in the V3 region of the envelope glycoprotein gene, within infected individuals (13). Virus variants arise in the course of infection and form quasispecies. These variants are generated by errors of reverse transcription at an estimated rate of 3.4 × 10−5 (17) and by recombination during viral replication. The rate at which variants appear is enhanced by the high viral population turnover, with reported production of more than 109 HIV-1 virions per day (12). Once generated, each variant undergoes selective pressure from the host environment, with cellular tropism, the host immune response, and antiretroviral therapy. The best adapted variants survive, although the survival of variants with similar replicative capacities in a given individual may also be influenced by chance (3). Since the V3 loop is an important determinant of cellular tropism and virus neutralization, it could affect vertical transmission and the subsequent development of an HIV-1 infection within children (11). The viral populations of most children are usually more homogeneous than the HIV-1 populations in their mothers (1, 7, 19–21, 36), and the HIV-1 strains isolated soon after contamination usually have a macrophage-tropic phenotype (22, 26, 33). The mechanisms involved in variant selection remain unclear. One of the main problems is to determine whether selection occurs when the virus passes to the infant or some time after transmission. Selection at transmission should result in only viruses having particular, appropriate characteristics infecting the child. The loss of glycosylation sites on the V3 env region (36) and some other sequence features (21, 34) may facilitate vertical transmission. If there is selection after transmission, viruses are transmitted by chance and only those well adapted to this new host (the child) persist. This precludes the transmission of multiple maternal viral variants. This issue is controversial because the majority of reported cases suggest that one or few closely related variants are transmitted and also because the samples may have been contaminated in the few reported cases of multiple viral variant transmission (16).
We have therefore examined the molecular mechanisms involved in the vertical transmission of HIV-1 by comparing the C2V3 sequences from four cases of vertical transmission. Both the circulating virus (viral RNA in plasma) and the virus in infected cells (proviral DNA in peripheral blood mononuclear cells [PBMCs]) were studied in each of the four mother-child pairs to identify the mother’s population responsible for transmission.
MATERIALS AND METHODS
Patients.
Four HIV-1-infected mother-infant pairs were studied. The mothers’ peripheral blood samples, CD4+ T cell counts, and clinical stage according to the Centers for Disease Control (CDC) criteria were provided by A. Berrebi, Department of Gynecology and Obstetrics, La Grave Hospital, Toulouse, France. The peripheral blood samples from the children were provided by J. Tricoire, Pediatrics Department, Purpan Hospital, Toulouse, France. The viral populations of the mothers were studied with samples collected as close as possible to the time of delivery. Child FR was born at term by cesarean section for cervical dystocia, after 11 h of membrane rupture, and had a normal weight. This child rapidly developed AIDS and died shortly after he was 1 year old. The other three children (AZ, BO, and RO) were full-term infants born by spontaneous vaginal delivery and had not developed AIDS by 1 year of age. Proviral DNA analyses were done on PBMC samples by PCR with the Amplicor HIV-1 kit (Roche Diagnostic Systems, Neuilly, France). Viral RNA was detected and quantified with the Amplicor HIV-1 monitor kit version 1.5 (Roche), allowing PCR amplification from HIV-1 B and non-B-subtype RNA. The children’s samples that were negative for proviral DNA were also tested for plasma RNA by an ultrasensitive protocol (detection limit of 20 HIV-1 RNA copies per ml). Infectious HIV-1 was detected by coculture (27). The clinical characteristics and times of sampling for each mother-child pair are shown in Table 1.
TABLE 1.
Clinical status, laboratory parameters, and time of sampling for mother-infant pairsa
| Pair | Mother
|
Child
|
||||||
|---|---|---|---|---|---|---|---|---|
| Time of sampling | CD4 cells/mm3 at delivery | Clinical stage | Plasmid RNA (copies/ml) at delivery | Time of sampling | Cell-associated proviral DNA | Plasma RNA | Coculture | |
| AZ | D + 6 mo | 185 | A3 | 127 | D + 2 days | − | <12 | − |
| D + 9 days | + | NT | + | |||||
| BO | D − 45 days | 577 | A1 | 19,000 | D + 5 days | − | NT | − |
| D + 40 days | + | NT | + | |||||
| RO | D − 5 mo | 410 | A2 | <84 | D + 5 days | − | <17 | − |
| D + 3 mo | + | NT | + | |||||
| FR | D + 2 days | 110 | A3 | 850,000 | D + 2 | − | 6,600 | − |
| D + 1 yr | D + 23 days | + | NT | + | ||||
| D + 1 yr | + | NT | NT | |||||
D corresponds to the day of delivery in each mother-infant pair. NT, not tested.
Nucleic acids extraction and cDNA synthesis.
Peripheral blood samples collected in citrate anticoagulant were centrifuged over Lymphocyte Separation Medium (Organon Teknika, Malvern, Pa.) density gradients. The PBMC samples were washed, pelleted, and stored at −80°C. Plasma was prepared by centrifugation at 600 × g for 10 min and clarified by centrifugation for 15 min at 3,000 × g to ensure cell-free specimens; it was stored at −80°C. Viral RNA was extracted with 300 μl of TRIzol (Life Technologies, Inc., Gaithersburg, Md.) per 100 μl of plasma, followed by two phenol-chloroform extractions and ethanol precipitation in the presence of 1 μg of glycogen. The RNA pellet was suspended in 30 μl of RNase-free water. The products of three extractions were pooled for cDNA synthesis. The extracted RNA was immediately reverse transcribed into cDNA by using the primer E2 (2). The reverse transcription mixture (20 μl) contained a final concentration of 50 mM Tris-HCl (pH 8.3), 40 mM KCl, 1 mM dithiothreitol, 6 mM MgCl2, 1 mM concentrations of each deoxynucleoside triphosphate, 60 pmol of primer E2, 20 U of Moloney murine leukemia virus reverse transcriptase (Life Technologies), and 20 U of RNase inhibitor (Boehringer Mannheim GmbH). Reverse transcription was performed for 1 h at 42°C. PBMCs were recovered from the Ficoll gradient, washed twice with phosphate-buffered saline, and counted. Five million PBMCs were pelleted, dried, and stored at −80°C. Each PBMC pellet was lysed for 2 h at 56°C in a mixture containing 10 mM Tris-HCl (pH 8.5), 50 mM KCl, 2.5 mM MgCl2, 0.45% Nonidet P-40, 0.45% Tween 20, and 80 μg of proteinase K per ml. The proteinase K was then inactivated by heating the mixture for 2 min at 96°C. Semiquantitative PCR were performed on the PBMCs to ensure the presence of at least 100 copies of proviral genomes as a template for amplification.
Molecular cloning and sequencing.
A region of 313 nucleotides (positions 6615 to 6928 in the HIV-LAI genome) encoding the gp120 V3 loop was amplified as previously described (2). Samples were processed one at a time to avoid cross-contamination. Negative controls and blanks were included in each PCR run; they were samples from healthy blood donors and lysis buffer. DNA (10 μl) or cDNA (20 μl) was amplified by nested PCR. Each cDNA underwent the PCR amplification in parallel with its corresponding RNA as a negative DNA contamination control. The outer primers E1 and E2 and the two inner primers E3 and E4 have been described previously (2). The PCR product amplified by the inner primers was purified with the QIAquick PCR purification kit (Qiagen GmbH) and cloned by using the pGEM-T TA cloning Vector system (Promega Corp.). Ligated vector was used to transform DH5α competent cells (Life Technologies). Multiple recombinant plasmids from each sample were sequenced with dye-labeled universal and reverse M13 primers (ABI PRISM Dye Primer Cycle Sequencing Ready Reaction Kit; Applied Biosystems) on an ABI 377 automated sequencer. The rate of misincorporation generated by the above protocol was evaluated by sequencing 20 clones from two different PCR experiments on LAV-8E5 cells containing one copy of HIV-1 provirus per cell. The misincorporation rate was 0.047% (1/2,086), corresponding to three point mutations.
Analysis of sequence data.
Multiple alignments were done with Sequence Navigator (Perkin-Elmer Applied Biosystems) and CLUSTALW version 1.7 (32) programs. The alignment was adjusted by hand before phylogenetic analysis with version 3.572c of the Phylogeny Inference Package (PHYLIP). Phylogenetic distances of sequences within each isolate and among all isolate sequences were calculated with the two-parameter Kimura algorithm (DNADIST from PHYLIP). Dendograms were created by the neighbor-joining and maximum likelihood methods with the CLUSTALW, PHYLIP, and MEGA programs. Tree diagrams were plotted with the TREEVIEW version 1.4 program. Bootstrapping was performed on the neighbor-joining tree with the CLUSTALW and MEGA programs. Loop charge calculations were derived from the peptide sequence of the V3 loop region by using DNAid 1.8 software (Frederic Dardel, Palaiseau, France).
Statistical analysis.
Student’s t test was used to evaluate the significance of differences in nucleotide distances within an individual and between individuals.
Nucleotide sequence accession numbers.
The sequences have been submitted to EMBL with accession no. AJ008670 to AJ009112.
RESULTS
Patient characteristics.
Maternal blood samples were collected before delivery from mothers BO and RO and after delivery from mothers FR and AZ (Table 1). The phylogenetic analysis of sequences from the four mother-child pairs compared to reference sequences from the 10 env clades of HIV-1 disclosed that the sequences from pairs AZ, BO, and FR belonged to clade B and those from RO belonged to clade A. This was confirmed by a heteroduplex mobility assay (data not shown). The four children were tested for HIV-1 infection by assaying for proviral DNA and culturing PBMC samples taken 2 to 5 days after birth. All were negative. The HIV-1 RNA PCR was positive 2 days after birth for child FR. The second samples from the children were positive for proviral DNA as determined by PCR and cocultures of samples collected 9 days (AZ), 23 days (FR), 40 days (BO), and 3 months (RO) after delivery.
Global analysis of sequences from mother-child pairs.
Pairwise genetic distances were calculated between sequence sets (DNA and RNA) from each mother-child pair. Interpair distances were 9.54 to 29.9% of the mean pairwise nucleotide genetic distance, a level significantly greater (P < 0.01) than that for the intrapair distances (1.45 to 4.44%) (Table 2). The minimal genetic distance between pairs was 4.7% for FR and BO. The greatest interpair distances were between the RO sequences and all other sequences. The RO sequences belonged to a different HIV-1 subtype than the sequences from the other three pairs.
TABLE 2.
Intrapair and interpair mean pairwise nucleotide genetic distancesa
| Pair | Mean nucleotide genetic distances (range) for pair:
|
|||
|---|---|---|---|---|
| AZ (n = 80) | BO (n = 85) | RO (n = 88) | FR (n = 86) | |
| AZ | 4.44 (0–11.8) | |||
| BO | 16.37 (12.8–19.7) | 1.45 (0–3.5) | ||
| RO | 29.99 (25.3–35.0) | 26.11 (22.6–29.8) | 2.66 (0–6.6) | |
| FR | 17.15 (12.7–21.3) | 9.54 (4.7–15.1) | 29.06 (24.6–33.9) | 3.80 (0–8.2) |
Sequence set for each pair includes DNA and RNA sequences from mother and child.
Sequences (DNA plus RNA) from the AZ, BO, and RO infants indicated a more uniform virus population (0.35, 0.10, and 0.37% intrasample mean nucleotide distances) than the maternal samples (5.1, 1.71, and 3.02%) (Table 3). In contrast, the sample from infant FR (at delivery and at 23 days) was as heterogeneous (3.7%) as that of his mother (3.6%). The experimental misincorporation rate was estimated to be 0.047%, a value much lower than the sequence variations that might be considered to be true variations.
TABLE 3.
Mean of pairwise nucleotide genetic distances of all sequences (DNA and RNA) from mothers and children
| Sample | Mean genetic distance (%) |
|---|---|
| AZ | |
| Mother | 5.1 |
| Child | 0.35 |
| BO | |
| Mother | 1.71 |
| Child | 0.10 |
| RO | |
| Mother | 3.02 |
| Child | 0.37 |
| FR | |
| Mother | 3.6 |
| Child | 3.7 |
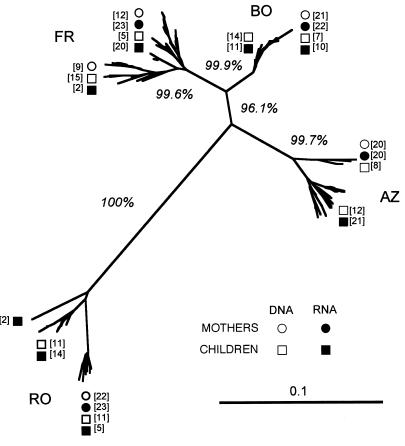
A phylogenetic tree was reconstructed by using the neighbor-joining method for the 340 env sequences (Fig. 1). Sequences from each mother-child pair clustered together and were clearly separated from those of the other mother-child pairs. This was emphasized by high bootstrap values (over 99%) obtained for the four mother-child pair sequence sets. The RO sequence subtree, belonging to subtype A, was the most divergent from the other three subtrees, whose sequences belonged to subtype B.
FIG. 1.
Unrooted neighbor-joining tree for the four mother-child pairs. The scale bar corresponds to 1% of nucleotide sequence divergence. Open symbols indicate DNA sequences, and closed symbols indicate RNA sequences; squares denote maternal sequences (RNA, ■; DNA, □), and circles denote child sequences (RNA, •; DNA, ○). Numbers in parentheses indicate the numbers of identical sequences at each position. Bootstrap values are expressed as percentages for each branch and represent the percent occurrence of that branch per 1,000 bootstrap replicates.
This first phylogenetic analysis and the mean genetic distances for the mother and child sequences suggested that the children of three mother-child pairs (AZ, BO, and RO) were infected by the transmission of one maternal variant. In contrast, the FR child seems to have received multiple maternal variants.
Analysis of sequences from mother-child pairs AZ, BO, and RO.
The three cases that may have been due to transmission of one or a few closely related maternal variants were AZ, BO, and RO. Pairwise genetic nucleotide distances were calculated for each sample (DNA or RNA sequences) and between samples within each mother-child pair (Table 4). Genetic distances on viral RNA sequences were generally more uniform than the proviral DNA sequences. The mean nucleotide distance between each mother-child pair of samples was 1.64 to 9.15%. The greatest intrapair mean distance between samples was for the AZ pair, perhaps because of the time lapse between obtaining the mother and child samples.
TABLE 4.
Intra- and intersample means of pairwise nucleotide genetic distances within each mother-child paira
| Pair | Motherb
|
Childc
|
||
|---|---|---|---|---|
| RNA | DNA | RNA | DNA | |
| AZ | ||||
| Mother RNA | 5.2 | |||
| Mother DNA | 5.10 | 3.4 | ||
| Child RNA | 7.37 | 9.15 | 0.43 | |
| Child DNA | 7.25 | 8.69 | 0.35 | 0.28 |
| BO | ||||
| Mother RNA | 0.83 | |||
| Mother DNA | 2.19 | 1.59 | ||
| Child RNA | 2.42 | 1.64 | 0.17 | |
| Child DNA | 2.35 | 1.59 | 0.10 | 0.03 |
| RO | ||||
| Mother RNA | 2.63 | |||
| Mother DNA | 3.10 | 3.21 | ||
| Child RNA | 3.53 | 3.66 | 0.29 | |
| Child DNA | 3.64 | 3.75 | 0.37 | 0.46 |
| FR | ||||
| Mother RNA | 2.70 | |||
| Mother DNA | 4.14 | 3.70 | ||
| Child RNA | 2.65 | 4.75 | 1.29 | |
| Child DNA | 4.53 | 3.87 | 5.20 | 3.01 |
Each mother-child pair includes four samples: RNA and DNA sequences from the child and RNA and DNA sequences from the mother.
The number of samples for RNA and DNA were 21 and 20, (AZ), 21 and 21 (BO), 21 and 22 (RO), and 22 and 20 (FR), respectively.
The number of samples for RNA and DNA were 20 and 20 (AZ), 21 and 22 (BO), 23 and 22 (RO), and 23 and 21 (FR), respectively.
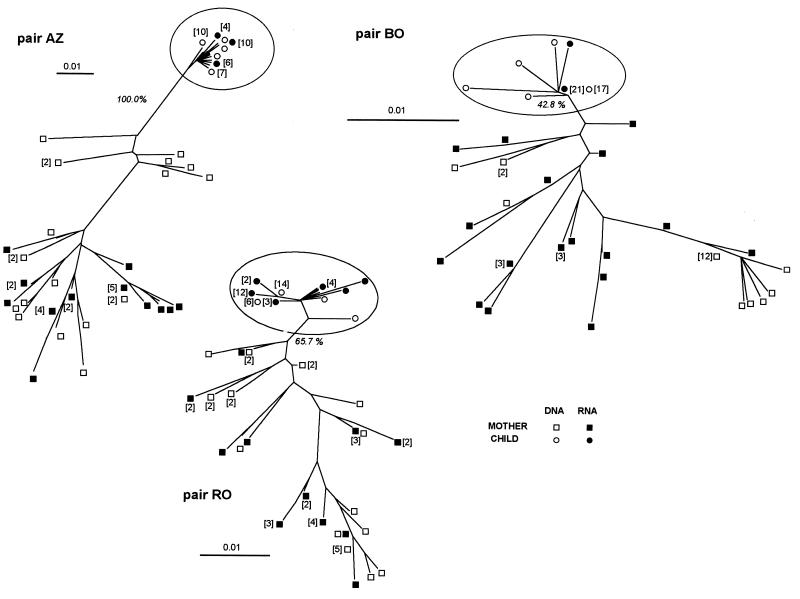
We further investigated the relationship between the maternal and infant sequences by phylogenetic analysis of each mother-child nucleotide sequence set (Fig. 2). The resulting phylogenetic trees showed that the maternal sequences were in different branches, suggesting a multiple maternal lineage. The infant sequences were more uniform than those of their mothers and lay within a single branch of the phylogenetic tree. Viral variants of the child were clustered distinctly from the sequences of the mother for pair AZ, as supported by high bootstrap values (100%). For pairs RO and BO the confidence level for the clustering of children viral sequences decreased, with bootstrap values of 65.7% (RO) and 42.8% (BO).
FIG. 2.
Unrooted neighbor-joining trees for each of the four mother-child pairs. The scale bar corresponds to 1% of nucleotide sequence divergence. Open symbols indicate DNA sequences, and closed symbols indicate RNA sequences; squares denote maternal sequences (RNA, ■; DNA, □), and circles denote child sequences (RNA, •; DNA, ○). The subtree clusters of child sequences in each tree are circled. Numbers in parentheses indicate the numbers of identical sequences at each position.
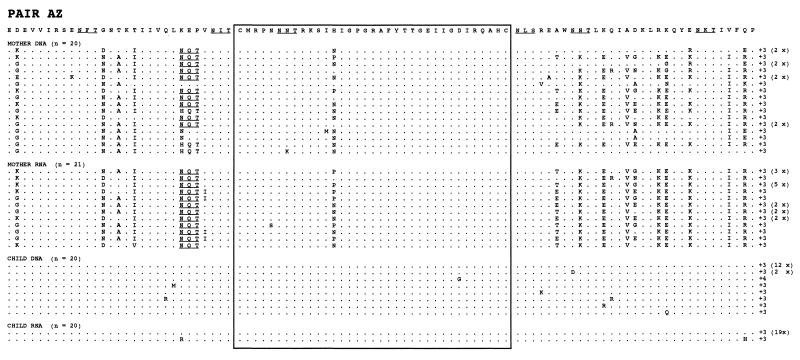
We next attempted to trace the origin of the children’s sequences by comparing sequences from the mothers’ proviral DNA in PBMC samples and in plasma virus RNA. Phylogenetic trees showed that the sequences from child AZ were more similar to the maternal proviral DNA-derived sequences, whereas the sequences of child BO were more similar to the maternal RNA sequences (Fig. 2). The sequences from child RO were equidistant from the maternal DNA and RNA sequences. The deduced amino acid sequences of the V3 loop and flanking regions are shown in Fig. 3. The coding open reading frame was present in all of the sequences, and the two cysteines flanking the V3 loop were present in all of sequences. The samples from the three children contained a small number of variants, all from a single predominant sequence. This major sequence was found both as a proviral DNA and as a viral RNA sequence in each child. These major child sequences represented 77.5 to 93.0% of all of the sequences in a child. There was only one type of V3 loop central motif in each of these three mother-child pairs, and there was no systematic loss or acquisition of N-glycosylation sites during transmission. The AZ and RO mother-child pairs had a potential N-X-T glycosylation site located six amino acids upstream from the first cysteine of the V3 loop in the majority of sequences from the maternal variants (AZ, 93%; RO, 51%), which was absent from the child sequence set. The BO mother-child pair had two glycosylation sites downstream from the V3 loop that were absent from some maternal variants and from all of the child sequences. The RO sequences showed the deletion of one amino acid (position 90) in most maternal sequences but not from the child sequences. The net charge in the V3 loop region for mothers was +3 for AZ, +3 to +5 for BO, and +3 to +4 for RO (Fig. 3). No amino acid distribution associated with the syncytium-inducing (SI) phenotype (6, 10) was found in the deduced amino acid sequences.
FIG. 3.
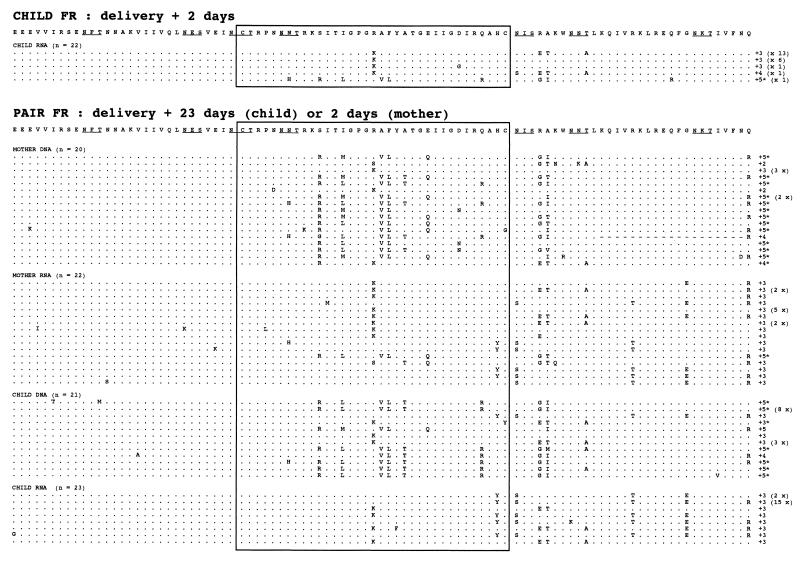
Multiple alignment of the amino acid sequences of the C2V3 region. The C2V3 amino acid sequences were aligned by comparison with the consensus sequence within each mother-child pair. Dots indicate the identity with the consensus sequence for each pair, dashes are amino acid deletions, and “x” indicates a stop codon. The charges of the V3 loop amino acids and frequencies of clones with identical amino acid sequences are indicated at the end of each sequence. Sequences containing amino acid distributions compatible with the SI phenotype are indicated by an asterisk. Potential N-linked glycosylation sites are indicated by underlined letters.
Analysis of sequences from mother-child pair FR.
In contrast to the mother-child pairs described above, there may have been transmission of multiple maternal variants to the child in the FR pair. The mean genetic distances of sequences of the mother-child pair FR were similar for the first mother sample and the first DNA and RNA positive child sample. Mean pairwise genetic distances were 1.29% for child sample viral RNA and 3.01% for the proviral DNA, whereas the mean genetic distances were 2.70% for the mother’s viral RNA and 3.70% for her proviral DNA (Table 4).
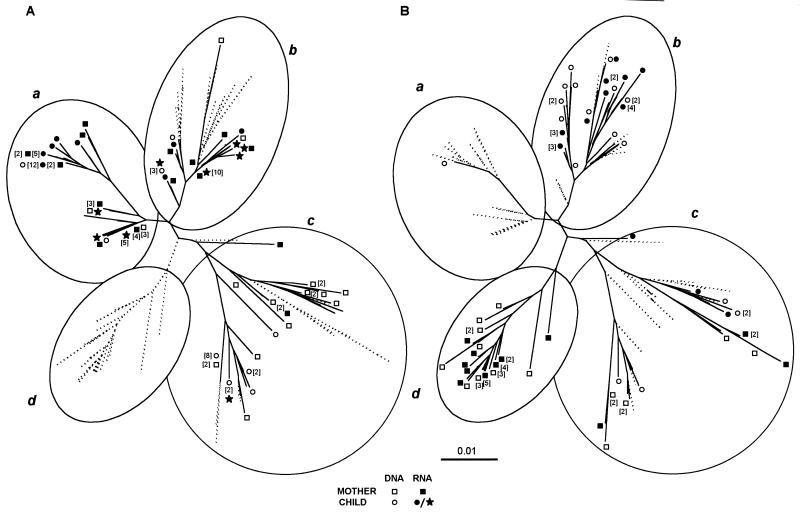
The changes in the sequences between samples within the FR mother-child pair were assessed by phylogenetic analysis of all of the sequences obtained (Fig. 4). Two identical trees were drawn with different legends to discriminate between the sequences from samples taken near delivery (Fig. 4A) and the sequences from samples collected 1 year after delivery (Fig. 4B). Maternal and infant sequence were divided in four subtrees (a, b, c, and d). The sequences in samples from the mother and child taken close to delivery were intermingled in subtrees a, b, and c. The distributions of the infant and mother sequences were similar, supporting the transmission of multiple maternal variants to the child. The viral RNA sequences from the child sample collected 2 days after birth were distributed in the three previously described subtrees (a, b, and c).
FIG. 4.
Unrooted neighbor-joining tree of sequences obtained from all FR mother-child pair sequences in samples taken at birth and 1 year later. The scale bar corresponds to 1% of nucleotide sequence divergence. Open symbols indicate DNA sequences, and closed symbols indicate RNA sequences; squares denote maternal sequences, and circles denote child sequences. Numbers in parentheses indicate the numbers of identical sequences at each position. Each tree is divided into four circled subtrees—a, b, c, and d—to facilitate analysis. Numbers inside brackets refer to the numbering of the clonal sequences if more than one clone was obtained. (A) Sequences obtained from the child (RNA, ★) and the mother (RNA, ■; DNA, □) 2 days after delivery and from the child (RNA, •; DNA, ○) 23 days after delivery are shown. The shadowed branches correspond to sequences observed 1 year later. (B) Sequences obtained from the child (RNA, •, DNA, ○) and the mother (RNA, ■; DNA, □) 1 year after delivery. The shadowed branches correspond to extinct branches.
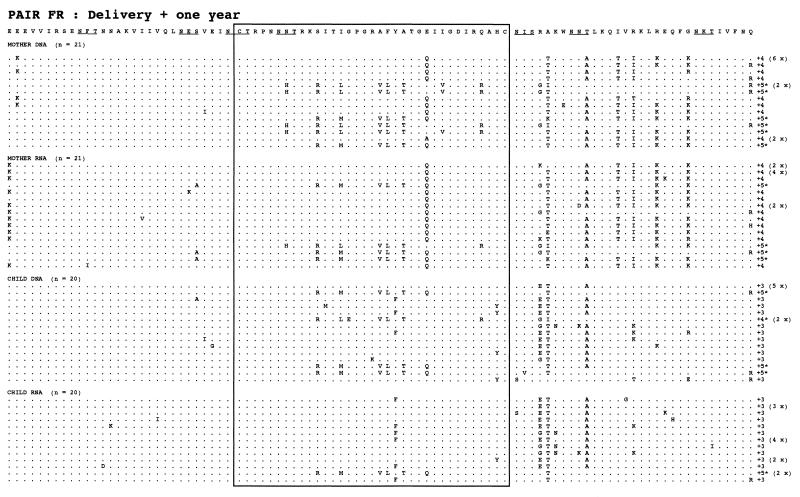
Samples from the FR mother-child pair were also analyzed 1 year later (Fig. 4). The child sample contained viral variants from all the subtrees just after birth (a, b, and c), with particular development of one branch (b), which initially contained sequences from both mother and child. This subtree (b) contained 80% of the child sequences, while the remaining 20% were distributed in the rest of the tree (a and c). The maternal sequences formed two major populations; one was not present at delivery (d), and the other (c) developed from a population present 1 year earlier.
The deduced amino acid sequences from the mother FR had three types of V3 loop central motif—GPGR (50%), GPGK (45%), and GPGS (5%)—while the sequences from the FR child had only two: GPGR (80%) and GPGK (20%). The FR mother-child pair had four potential N-X-T or N-X-S glycosylation sites randomly distributed through all of the mother and child sequence sets. The net charge in the V3 loop was +2 to +5 for the FR mother-child pair. Some amino acid sequences from the FR mother and child samples had a polar amino acid distribution in the V3 loop that is associated with the SI phenotype (6, 10).
DISCUSSION
We have compared the sequences from the V3 loop and flanking regions of env genes found in four mother-child pairs with vertical transmission of HIV-1. We identified a case in which multiple maternal variants were transmitted to the child. The phylogenetic trees strongly suggest that there was no ex vivo interpair contamination because the four mother-child pair sequence sets were clustered in four subtrees with high bootstrap values. These results are in agreement with what the findings would be in epidemiologically unrelated individuals.
None of the children had proviral DNA detectable by PCR or infectious HIV by coculture just after birth, and they all became positive after they were 1 week old. The three children AZ, BO, and RO, who also had no detectable RNA at birth, fulfilled the criteria for contamination at the time of delivery, and their disease progressed slowly. The viral populations in the four mothers were very heterogeneous for viral RNA and for proviral DNA V3 sequences. This variability fits well with those reported for mothers in other studies (1, 2, 15, 19, 21, 28, 29). In contrast to the maternal samples, the three children AZ, BO, and RO had uniform sequences with very few variations. This homogeneity was unlikely to be due to sequence selection occurring during nucleic acid extraction or amplification since the minimal quantity of proviral DNA used as template was checked and the DNA and RNA sequence results were concordant. These three mother-child pairs had the sequence and phylogenetic characteristics expected from the transmission of one or a few closely related maternal variants to the child. Phylogenetic analysis showed a homogeneous population in the child samples, suggesting the transmission of one maternal variant. The viral population in the children located outside (AZ) or on the periphery of the mother’s subtree suggested the transmission of a minor maternal variant. This type of mother-child phylogeny seems to be the most common (1, 19, 28, 29, 36). Transmission of a major variant seems to be less frequent (28).
In contrast, child FR had a positive viral RNA as determined by PCR 2 days after delivery and then did not strictly fit the criteria for contamination at delivery. Nevertheless, this child was born at term and had a normal birth weight, no detectable proviral DNA, and a negative HIV coculture in a sample collected 2 days after delivery. This case has three novel features. First, plasma viral RNA was detected in the first child sample (day 2) but not cell-associated proviral DNA. It has been reported that plasma HIV RNA may be detectable earlier than proviral DNA (30). There is likely to be no maternal virus in the child’s blood at this time, since the half-life of virions is believed to be 6 h (12). Contamination at birth may have been followed by sequestration of infected maternal cells into lymphoid organs (e.g., the spleen or thymus). Alternatively, the detection of proviral DNA may have been less sensitive. Second, child FR was born after 11 h of membrane rupture and was infected by a heterogeneous viral population. Delivery after more than 4 h of membrane rupture greatly increases the risk of contamination (14). This child may then have been infected by an ascending route during labor. The diversity of the child’s viral population could thus have been due to the transmission of multiple maternal viral variants, as was strongly suggested by the phylogenetic analysis. Contamination with the mother’s sample or between the child’s samples is unlikely, since those samples were taken at different times and were treated separately; the negative and blank controls were always negative. Sequence analysis strongly supports the transmission of multiple maternal virus variants. Third, in contrast to the other three children, child FR rapidly developed AIDS and died. Sequences from pair FR had amino acid distributions that suggest the presence of SI, highly replicative strains. Since this sequence was also found in the child sequences at birth and 1 year later, they could have contributed to his rapid deterioration. This unusual case of vertical HIV-1 transmission combines the timing of transmission compatible with ascendant contamination, the transmission of multiple maternal variants (including possible SI phenotype strains), and rapid disease development.
We have attempted to trace the origin of children’s viral sequences by comparing the viral sequences obtained from the plasma and the PBMCs of their mothers. The phylogenetic trees showed that the sequences from child AZ were more similar to the DNA-derived maternal sequences, but that the sequences from the child BO were more similar to the RNA-derived maternal sequences. The maternal sequences most similar to sequences from child RO viruses were from both the DNA and RNA viral genomes. The maternal and infant sequences of the FR pair were closely intermingled, so that it was impossible to discriminate between the roles of maternal RNA and DNA-derived sequences by phylogenetic analysis. These results agree with those obtained in a previous study on 10 mother-child pairs, with four cases of possible RNA sequence transmission and four cases for DNA (29). Viral RNA reflects the sequences from the replicative pool of viruses, although it is only a minor part of the proviral DNA sequence. These proviral sequences can be from productive infected cells, cells with latent infection, or cells with defective viruses. It also reflects different kinds of infected cells that could be in contact with the infant. Infected cells in tissues could also be involved in child infection, since PBMC proviral DNA only reflects the monocyte and part of the lymphocyte populations. Infected genital tract cells in particular are in close contact with the child during delivery. The distributions of maternal RNA and DNA sequences were often similar and overlapping. It is still difficult to determine whether transmission is due to infected cells or free virus. Since the virus populations in the genital tract may be quantitatively and qualitatively different from those in the blood (23, 25, 37), a study of these viral sequences in intrapartum contamination may help to determine the origin of the virus. Analysis of both viral RNA and proviral DNA will probably provide a better evaluation of viral polymorphism than will analysis of DNA or RNA alone.
In conclusion, we have found one case of maternal multiple-variant HIV-1 transmission that probably took place near delivery. Multiple-variant transmission is possible but uncommon, since the three other cases of transmission analyzed in this study and in the large majority of reported cases indicate the transmission of a single variant. Either cell-free virus or cell-associated virus can be transmitted to the child at the time of delivery.
ACKNOWLEDGMENTS
We thank H. Coppin for critical review and Owen Parkes for linguistic advice.
This work was supported by a grant from SIDACTION.
REFERENCES
- 1.Ahmad N, Baroudy B M, Baker R C, Chappey C. Genetic analysis of human immunodeficiency virus type 1 envelope V3 region isolates from mothers and infants after perinatal transmission. J Virol. 1995;69:1001–1012. doi: 10.1128/jvi.69.2.1001-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briant L, Wade C M, Puel J, Brown A J, Guyader M. Analysis of envelope sequence variants suggests multiple mechanisms of mother-to-child transmission of human immunodeficiency virus type 1. J Virol. 1995;69:3778–3788. doi: 10.1128/jvi.69.6.3778-3788.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown A J. Analysis of HIV-1 env gene sequences reveals evidence for a low effective number in the viral population. Proc Natl Acad Sci USA. 1997;94:1862–1865. doi: 10.1073/pnas.94.5.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryson Y J. Perinatal HIV-1 transmission: recent advances and therapeutic interventions. AIDS. 1996;10:S33–S42. [PubMed] [Google Scholar]
- 5.Bryson Y J, Luzuriaga K, Sullivan J L, Wara D W. Proposed definitions for in utero versus intrapartum transmission of HIV-1. N Engl J Med. 1992;327:1246–1247. doi: 10.1056/NEJM199210223271718. [DOI] [PubMed] [Google Scholar]
- 6.Chesebro B, Wehrly K, Nishio J, Perryman S. Mapping of independent V3 envelope determinants of human immunodeficiency virus type 1 macrophage tropism and syncytium formation in lymphocytes. J Virol. 1996;70:9055–9059. doi: 10.1128/jvi.70.12.9055-9059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Contag C H, Ehrnst A, Duda J, Bohlin A-B, Lindgren S, Learn G H, Mullins J I. Mother-to-infant transmission of human immunodeficiency virus type 1 involving five envelope sequence subtypes. J Virol. 1997;71:1292–1300. doi: 10.1128/jvi.71.2.1292-1300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Rossi A, Masiero S, Giaquinto C, Ruga E, Comar M, Giacca M, Chieco-Bianchi L. Dynamics of viral replication in infants with vertically acquired human immunodeficiency virus type 1 infection. J Clin Infect. 1996;2:323–330. doi: 10.1172/JCI118419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delwart E L, Pan H, Sheppard H W, Wolpert D, Neumann A U, Korber B, Mullins J I. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J Virol. 1997;71:7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halapi E, Gigliotti D, Hodara V, Scarlatti G, Tovo P A, DeMaria A, Wigezll H, Rossi P. Detection of CD8 T-cell expansions with restricted T-cell receptor V usage in infants vertically infected by HIV-1. AIDS. 1996;10:1621–1626. doi: 10.1097/00002030-199612000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 13.Holmes E C, Zhang L Q, Simmonds P, Ludlam C A, Brown A J L. Convergent and divergent sequence evolution in the surface envelope glycoprotein of human immunodeficiency virus type 1 within a single infected patient. Proc Natl Acad Sci USA. 1992;89:4835–4839. doi: 10.1073/pnas.89.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn L, Abrams E J, Matheson P B, Thomas P A, Lambert G, Bamji M, Greenberg B, Steketee R W, Thea D M the New York City Perinatal HIV Transmission Collaborative Study Group. Timing of maternal-infant HIV transmission—associations between intrapartum factors and early polymerase chain reaction results. AIDS. 1997;11:429–435. doi: 10.1097/00002030-199704000-00005. [DOI] [PubMed] [Google Scholar]
- 15.Lamers S L, Sleasman J W, She J X, Barrie K A, Pomeroy S, Barrett D J, Goodenow M M. Persistence of multiple maternal genotypes of human immunodeficiency virus type 1 in infants infected by vertical transmission. J Clin Invest. 1994;93:380–390. doi: 10.1172/JCI116970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Learn G H, Korber B T M, Foley B, Hahn B H, Wolinsky S M, Mullins J I. Maintaining the integrity of human immunodeficiency virus sequence databases. J Virol. 1996;70:5720–5730. doi: 10.1128/jvi.70.8.5720-5730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansky L M, Temin H M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayaux M J, Teglas J, Mandelbrot L, Berrebi A, Jallais H, Matheron S, Ciraru-Vigneron N, Parnet-Mathieu F, Bongain A, Rouzioux C, Delfraissy J, Blanche S. Acceptability and impact of zidovudine prevention on the mother-to-child HIV-1 transmission in France. J Pediatr. 1997;131:857–962. doi: 10.1016/s0022-3476(97)70033-7. [DOI] [PubMed] [Google Scholar]
- 19.Mulder-Kampinga A, Simonon A, Kuiken C L, Dekker J, Scherpbier H J, van de Perre P, Boer K, Goudsmit J. Similarity in env and gag genes between genomic RNAs of human immunodeficiency virus type 1 (HIV-1) from mother and infant is unrelated to time of HIV-1 RNA positivity in the child. J Virol. 1995;69:2285–2296. doi: 10.1128/jvi.69.4.2285-2296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulder-Kampinga G A, Kuiken C, Dekker J, Scherpbier H J, Boer K, Goudsmit J. Genomic human immunodeficiency virus type 1 RNA variation in mother and child following intra-uterine virus transmission. J Gen Virol. 1993;74:1747–1756. doi: 10.1099/0022-1317-74-9-1747. [DOI] [PubMed] [Google Scholar]
- 21.Narwa R, Roques P, Courpotin C, Parnet M F, Boussin F, Roane A, Marce D, Lasfargues G, Dormont D. Characterization of human immunodeficiency virus type 1 p17 matrix protein motifs associated with mother-to-child transmission. J Virol. 1996;70:4474–4483. doi: 10.1128/jvi.70.7.4474-4483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ometto L, Zanotto C, Maccabruni A, Caselli D, Truscia D, Giaquinto C, Ruga E, Chieco B L, De Rossi A. Viral phenotype and host-cell susceptibility to HIV-1 infection as risk factors for mother-to-child HIV-1 transmission. AIDS. 1995;9:427–434. [PubMed] [Google Scholar]
- 23.Overbaugh J, Anderson R J, Ndinya-Achola J O, Kreiss J K. Distinct but related human immunodeficiency virus type 1 variant populations in genital secretions and blood. AIDS Res Hum Retroviruses. 1996;12:107–115. doi: 10.1089/aid.1996.12.107. [DOI] [PubMed] [Google Scholar]
- 24.Peckham C, Gibb D. Mother-to-child transmission of the human immunodeficiency virus. N Engl J Med. 1995;333:298–302. doi: 10.1056/NEJM199508033330507. [DOI] [PubMed] [Google Scholar]
- 25.Poss M, Martin H L, Kreiss J K, Granville L, Chochan B, Nyange P, Mandaliya K, Overbaugh J. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J Virol. 1995;69:8118–8122. doi: 10.1128/jvi.69.12.8118-8122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinhardt P P, Reinhardt B, Lathey J L, Spector S A. Human cord blood mononuclear cells are preferentially infected by non-syncytium-inducing, macrophage-tropic human immunodeficiency virus type 1 isolates. J Clin Microbiol. 1995;33:292–297. doi: 10.1128/jcm.33.2.292-297.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouzioux C, Puel J, Agut H, Brun-Vézinet F, Ferchal F, Tamalet C, Descamps D, Fleury H. Comparative assessment of quantitative HIV viraemia assays. AIDS. 1992;6:373–377. doi: 10.1097/00002030-199204000-00003. [DOI] [PubMed] [Google Scholar]
- 28.Salvatori F, Masiero S, Giaquinto C, Wade C M, Brown A, Chiecobianchi L, Derossi A. Evolution of human immunodeficiency virus type 1 in perinatally infected infants with rapid and slow progression to disease. J Virol. 1997;71:4694–4706. doi: 10.1128/jvi.71.6.4694-4706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scarlatti G, Leitner T, Halapi E, Wahlberg J, Marchisio P, Clerici-Schoeller M A, Wigzell H, Fenyo E M, Albert J, Uhlen M, Rossi P. Comparison of variable region 3 sequences of human immunodeficiency virus type 1 from infected children with the RNA and DNA sequences of the virus populations of their mothers. Proc Natl Acad Sci USA. 1993;90:1721–1725. doi: 10.1073/pnas.90.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steketee R W, Abrams E J, Thea D M, Brown T M, Lambert G, Orloff S, Weedon J, Bamji M, Schoenbaum E E, Rapier J, Kalish M L the New York City Perinatal HIV Transmission Collaborative Study Group. Early detection of perinatal human immunodeficiency virus (HIV) type 1 infection using HIV RNA amplification and detection. J Infect Dis. 1997;175:707–711. doi: 10.1093/infdis/175.3.707. [DOI] [PubMed] [Google Scholar]
- 31.Strunnikova N, Ray S C, Livingston R A, Rubalcaba E, Viscidi R P. Convergent evolution within the V3 loop domain of human immunodeficiency virus type 1 in association with disease progression. J Virol. 1995;69:7548–7558. doi: 10.1128/jvi.69.12.7548-7558.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1996;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van’t Wout A B, Kootstra N A, Mulder-Kampinga G A, Albrecht-van Lent N, Scherpbier H J, Veenstra J, Boer K, Countinho R A, Miedema F, Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang B, Ge Y C, Palasanthiran P, Ziegler J, Bolton W, Xiang S-H, Dwyer D E, Cunningham A L, Saksena N K. HIV type 1 V3 loop sequences derived from peripheral blood of transmitting mothers, their infants, and nontransmitting mothers differ in their own octapeptide motifs. AIDS Res Hum Retroviruses. 1997;13:275–279. doi: 10.1089/aid.1997.13.275. [DOI] [PubMed] [Google Scholar]
- 35.Wilfert C M, Wilson C, Luzuriaga K, Epstein L. Pathogenesis of pediatric human immunodeficiency virus type 1 infection. J Infect Dis. 1994;170:286–292. doi: 10.1093/infdis/170.2.286. [DOI] [PubMed] [Google Scholar]
- 36.Wolinsky S M, Wike C M, Korber B T M, Hutto C, Parks W P, Rosenblum L L, Kunstman K J, Furtado M R, Munoz J L. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science. 1992;255:1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 37.Zhu T, Wang N, Carr A, Nam D S, Moor-Jankowski R, Cooper D A, Ho D D. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]