Abstract
The adenovirus E1B 55-kDa and E4 34-kDa oncoproteins bind and inactivate the p53 tumor suppressor gene product, resulting in cell transformation. A recently discovered cellular protein, p73, shows extensive similarities to p53 in structure and function. Here we show that the simultaneous transient expression of E1B 55-kDa and E4 34-kDa proteins is sufficient to drastically shorten the intracellular half-life of p53, leading to strongly reduced steady-state p53 levels. Concomitantly, the E1B 55-kDa and E4 34-kDa proteins act synergistically to inactivate the transcriptional activity of p53. Mutational analysis suggests that physical interactions between the E1B 55-kDa protein and p53 and between the E1B 55-kDa and E4 34-kDa proteins are both required for p53 degradation. In contrast, the ability of p53 to interact with the cellular mdm2 oncoprotein or with its cognate DNA element appears to be dispensable for its destabilization by adenovirus gene products. The adenovirus E1B 55-kDa protein did not detectably interact with p73 and failed to inhibit p73-mediated transcription; also, the E1B 55-kDa and E4 34-kDa proteins did not promote p73 degradation. When five amino acids near the amino termini were exchanged at corresponding positions between p53 and p73, this rendered p53 resistant and p73 susceptible to complex formation and inactivation by the E1B 55-kDa protein. Our results suggest that while p53 inactivation is a central step in virus-induced tumor development, efficient transformation can occur without targeting p73.
The development of malignant tumors commonly includes mechanisms to inactivate the p53 tumor suppressor gene product. Viral oncoproteins bind and inactivate p53. Two adenovirus proteins, the E1B 55-kDa and E4 34-kDa proteins, form a complex with a dual function. First, these proteins modulate the nuclear export of mRNA during virus infection (1, 10, 24) and undergo nucleocytoplasmic shuttling (7). On the other hand, both proteins were reported to bind p53 and antagonize p53-mediated transcription (8, 25, 30). In cell transformation assays, the combination of the E1B 55-kDa and E4 34-kDa proteins promotes the formation of colonies more strongly than does the E1B 55-kDa protein alone (20, 21), raising the possibility that the two proteins act synergistically to inactivate p53.
Some p53 antagonists are known to promote the intracellular degradation of p53. This destabilization of p53 is an activity common to oncoproteins of human papillomaviruses (HPVs) (32), and the cellular mdm2 protein (11, 16, 27). Intriguingly, the half-life of p53 was shown to be reduced during adenovirus infection (25, 33), depending on the presence of the E1B 55-kDa and E4 34-kDa proteins. Furthermore, the steady-state level of p53 is downregulated after transformation with the E1B 55-kDa and E4 34-kDa proteins (20, 21), leading to the hypothesis that the E1B 55-kDa and E4 34-kDa proteins might be sufficient to accelerate the degradation of p53 even without the context of virus infection.
A recently discovered cellular protein, p73, shows many homologies to p53 (14). The sequence homologous between p53 and p73 covers the N-terminal domain of p53, which is known to interact with the adenovirus E1B 55-kDa protein (15), raising the question whether p73 might also interact with this protein.
The homology of p53 and p73 is particularly extensive within the DNA binding region and includes all amino acids known to form contact sites between p53 and DNA. Both proteins activate transcription from p53-responsive promoters and were reported to induce apoptosis (13). To date, the only known functional difference between p53 and p73 consists of the upregulation of p53 but not p73 levels in response to DNA damage. The fact that at least some p53-responsive promoters can also be activated by p73, along with the structural similarities between p53 and p73, initially suggested that p53 antagonists might also inactivate p73 to achieve complete transcriptional inhibition. Therefore, we analyzed the potential of adenovirus oncoproteins to inactivate p73 in addition to p53.
We show that the simultaneous transient expression of the adenovirus E1B 55-kDa and E4 34-kDa proteins is sufficient to strongly promote the intracellular degradation of p53. In contrast, the adenovirus proteins did not inhibit p73-mediated transcription, nor did they destabilize p73. The E1B 55-kDa protein selectively binds p53 but not p73, due to a 5-amino-acid difference between the primary structures of p53 and p73. Thus, despite the similar transcriptional activities of p53 and p73, p73 does not represent a target of the adenovirus p53 antagonists.
MATERIALS AND METHODS
Cell culture and plasmid construction.
Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum. The expression plasmids for p53 and mutants (18), HA-tagged E1B 55-kDa protein (termed pCGNE1B) (7), and E4 34-kDa protein (8) have been described. An expression plasmid for nontagged E1B 55-kDa protein was obtained by cloning the E1B 55-kDa protein coding region from pCGNE1B into the pCG vector (34) with BamHI. To obtain expression constructs for nontagged p73β, the corresponding human cDNA was amplified with the primers GCGGGATCCGCGGCCGCCACCATGGCCCAGTCCACCGCCACCTCC and GCGTCTAGAGGTCACGGTCCCCAAGTTCTGACGAGGC and the PCR product was cloned into pcDNA3 (Invitrogen) with BamHI and XbaI. To obtain an expression plasmid for nontagged p73α, the procedure was carried out with the second primer replaced with GCGTCTAGAGGTCAGTGGATCTCGGCCTCCGTGAAC. An expression construct for C-terminally tagged p73β was obtained by performing the same procedure but replacing the second primer with the oligonucleotide GCGTCTAGAGGTCAGCTTGCGTAATCCGGTACATCGTAAGGGTACGGTCCCCAAGTTCTGACGAGGC. Expression plasmids for mutant p53 and p73 were obtained by site-directed mutagenesis (QuikChange; Stratagene). The constructs were confirmed by sequencing.
Transfections and reporter assays.
Saos-2 cells (5 × 105 per assay) were transfected with a cationic lipid preparation (Fugene 6; Boehringer Mannheim). Luciferase activities were determined with a premanufactured assay system (Promega).
Western blotting.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide), transferred to nitrocellulose, incubated with antibodies in phosphate-buffered saline (PBS) containing 5% milk powder and 0.05% Tween, and subjected to chemiluminescent detection (Pierce) of peroxidase-coupled secondary antibody (Jackson). Antibody Pab1801 to p53 was from Calbiochem; antibody HA.11 against the HA epitope was from Babco.
Pulse-chase analysis.
Cells (5 × 105 per lane) were transfected and starved for 30 min in starving medium (DMEM lacking methionine and cysteine) and then subjected to incubation with 35S-labelled amino acids (Promix; Amersham) diluted 1:30 in starving medium. After 10 min, the medium was changed to DMEM containing 10% fetal bovine serum. After various chase times, the cells were lysed and p53 was immunoprecipitated as described previously (26).
Immunofluorescence.
Transfected cells were fixed for 15 min with 4% paraformaldehyde in PBS, permeabilized for 25 min with 0.2% Triton X-100 in PBS, and incubated with antibody as described previously (6). To stain the HA tag, a rabbit polyclonal antibody (Santa Cruz) was used, followed by a Texas red-labeled secondary antibody (Jackson). To detect the E1B 55-kDa protein, the murine monoclonal antibody 2A6 (31) was used, followed by a fluorescein isothiocyanate-conjugated secondary antibody (Jackson).
Immunoprecipitation.
In each experiment, 2 × 106 293 cells constitutively expressing the E1B 55-kDa protein were lysed, incubated with in vitro-translated p53 or p73 proteins, and immunoprecipitated with antibody 2A6 (31) against the E1B 55-kDa protein by a previously described procedure (36).
RESULTS
p53 degradation mediated by transiently expressed E1B 55-kDa and E4 34-kDa proteins.
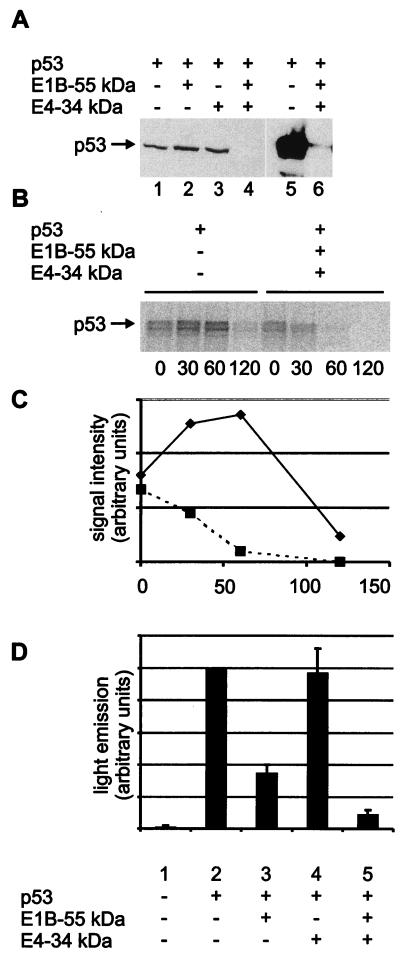
To determine whether the adenovirus type 5 E1B 55-kDa and E4 34-kDa proteins are sufficient to promote p53 degradation, we transiently expressed p53 in Saos-2 cells, an osteosarcoma cell line lacking p53, and subjected the cells to immunological detection of p53. The level of p53 was not detectably affected by the presence of either the E1B 55-kDa protein or the E4 34-kDa protein separately (Fig. 1A, lanes 1 to 3). However, when both viral proteins were coexpressed with p53, the amount of p53 was reduced more than 100-fold (lanes 4 to 6). This effect was considerably stronger than the p53 reduction achievable with coexpressed mdm2 or HPV E6 proteins (data not shown). The proteasome inhibitor MG132 elevated to some extent the amount of detectable p53 in the presence of the adenovirus oncoproteins (data not shown) but only when it was used at unusually high concentration (400 μM). It is therefore uncertain whether proteasome-mediated degradation might be contributing to the observed drop in p53 levels. Calpain inhibitors did not change the amount of detected p53 (data not shown). To further address the possibility that p53 is destabilized in the presence of the E1B 55-kDa and E4 34-kDa proteins, the transfected cells were pulse-labeled with [35S]methionine and the decay of p53 was monitored over time (Fig. 1B). Quantitation of the nondegraded protein (Fig. 1C) revealed that the biological half-life of p53 was drastically shortened by the simultaneous expression of the E1B 55-kDa and E4 34-kDa proteins. We conclude that the E1B 55-kDa and E4 34-kDa proteins act in concert to trigger the destabilization of intracellular p53 and may thereby promote cell transformation.
FIG. 1.
Degradation of p53 by the adenovirus type 5 E1B 55-kDa and E4 34-kDa proteins. (A) Expression plasmids for the p53 (0.5 μg), E1B 55-kDa (0.5 μg), and E4 34-kDa (1.0 μg) proteins or “empty” vector constructs were transfected into Saos-2 cells as indicated. After 24 h, the cells were lysed and subjected to SDS-PAGE and Western blot analysis. p53 was detected with monoclonal antibody Pab1801 (lanes 1 to 6). In a second experiment (lanes 5 and 6), the film was overexposed to allow detection of residual p53 in the presence of the E1B 34-kDa and E4 34-kDa proteins. (B) Expression plasmids for the p53 (1 μg), E1B 55-kDa (330 ng), and E4 34-kDa (660 ng) proteins or “empty” vector constructs were transfected into Saos-2 cells as indicated. After 24 h, the cells were labeled with [35S]methionine and [35S]cysteine for 10 min and then incubated in nonradioactive medium (chase). After the time points indicated (minutes), the cells were harvested and subjected to immunoprecipitation with monoclonal antibody Pab421 directed against p53, followed by SDS-PAGE and autoradiography. Note that the signal intensities obtained with p53 alone initially increase, possibly reflecting the incorporation of radioactively labeled amino acids that were internalized into the cells but not yet assembled into protein at the start of the chase. (C) The signal intensities obtained in the same experiment with p53 in the absence (diamonds) or presence (squares) of the E1B 55-kDa and E4 34-kDa proteins were quantified with a Bio Imaging Analyzer (Fuji) and plotted against the time after removal of the radioactive medium. (D) Expression plasmids for the p53 (50 ng), E1B 55-kDa (1.0 μg), and E4 34-kDa (0.5 μg) proteins were transfected as indicated into Saos-2 cells along with a reporter plasmid containing a p53-responsive promoter driving luciferase expression (pBP100luc [27], 0.5 μg). After 24 h, the cells were lysed and subjected to a luciferase assay. Luciferase activity is indicated in relative units, and the value obtained with p53 in the absence of antagonists was set to 100%. Error bars reflect the standard deviation of at least three independent experiments.
We next asked if accelerated degradation by these viral oncoproteins results in decreased transcriptional activity of p53. Transiently expressed p53 activated expression from a cotransfected reporter plasmid containing a p53-responsive promoter (Fig. 1D, lane 2). Transactivation was reduced in the presence of the E1B 55-kDa protein (lane 3). While the E4 34-kDa protein alone had no apparent effect on p53-mediated transcription (lane 4), the combination of the E1B 55-kDa and E4 34-kDa proteins reduced the transcriptional activity of p53 more profoundly than did the E1B 55-kDa protein alone (lane 5).
Mutational analysis of p53 degradation by the E1B 55-kDa and E4 34-kDa proteins.
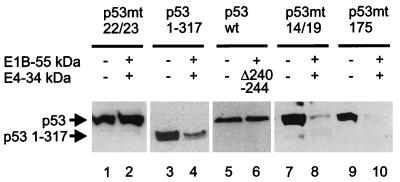
When two amino acids within the N terminus (positions 22 and 23) of p53 are mutated, the interaction of p53 with the E1B 55-kDa protein is abolished (18). The same mutation completely protected p53 from intracellular degradation by the E1B 55-kDa and E4 34-kDa proteins (Fig. 2, lanes 1 and 2), suggesting that the interaction between p53 and the E1B 55-kDa protein is a prerequisite for oncogene-mediated p53 degradation. In contrast, p53 levels were still reduced by the E1B 55-kDa and E4 34-kDa proteins, albeit less strongly, when the C-terminal domain of p53 was removed (leaving residues 1 to 317) (lanes 3 and 4). Since the C-terminal domain of p53 was previously mapped to interact with the E4 34-kDa protein (8), this argues that direct interactions between p53 and the E4 34-kDa protein might contribute to but are not necessary for the degradation of p53. Finally, a mutant form of the E4 34-kDa protein (deletion of amino acids 240 to 244) that lacks the ability to relocate the E1B 55-kDa protein from the cytoplasm to the nucleus (35a) did not reduce p53 levels when coexpressed with the E1B 55-kDa protein (lanes 5 and 6), suggesting that the interaction between the E1B 55-kDa and E4 34-kDa proteins is a requirement for p53 degradation. Mutation of p53 amino acids 14 and 19 is known to abolish complex formation between p53 and the mdm2 protein while preserving the ability of p53 to associate with the E1B 55-kDa protein (18). The abundance of this p53 mutant was downregulated by the adenovirus oncoproteins (lanes 7 and 8), arguing that mdm2 is not involved in adenovirus-mediated p53 degradation. Finally, a tumor-derived mutation of p53 (R175H) that abolishes promoter-binding activity did not stabilize the protein in the presence of the E1B 55-kDa and E4 34-kDa proteins (lanes 9 and 10), suggesting that adenovirus-mediated degradation of p53 occurs regardless of the specific DNA binding activity of p53.
FIG. 2.
Mutational analysis of p53 degradation. Wild-type or mutant versions of p53 and the E4 34-kDa protein were transiently expressed as indicated, along with the E1B 55-kDa protein as in Fig. 1A, and subjected to Western blot detection of p53.
Selective inactivation of p53 but not p73 by adenovirus oncoproteins.
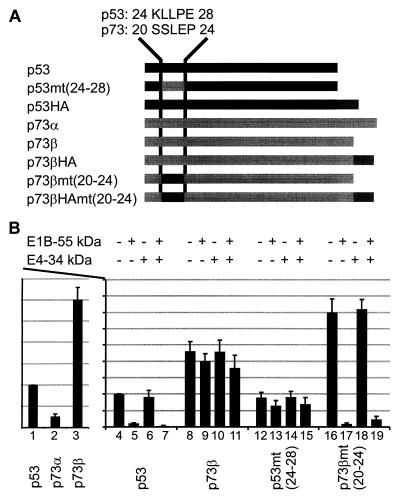
Given the structural and functional homology between the p53 and p73 proteins, it has been proposed that both proteins might be regulated by the same antagonists (14). To test this, the transcriptional activities of p53 and the α and β forms of p73 (p73α and p73β) were assessed by using a luciferase reporter. p73α was found to be a considerably weaker transcriptional activator than p53 or p73β (Fig. 3B, lanes 1 to 3), possibly due to its reported failure to form oligomers (14). Therefore, p73β was chosen for further analysis. The proportion of transactivation was roughly maintained among p53, p73α, and p73β when several different p53-responsive promoters were used (data not shown), suggesting that p53 and p73 have similar or identical target sequences.
FIG. 3.
Inactivation of p53 but not p73 by the E1B 55-kDa and E4 34-kDa proteins. (A) p53, p73, and chimeras were constructed as outlined. Amino acid residues 24 to 28 in p53 correspond to residues 20 to 24 in p73, according to homology-based alignment of the primary structures (14). These residues were exchanged between p53 and p73β to create p53mt(24–28) and p73βmt(20–24), respectively. The proteins were expressed with or without a carboxy-terminally fused HA tag for immunodetection. (B) Expression plasmids for p53, p73α, and p73β (25 ng each) were transfected along with the luciferase reporter plasmid pBP100luc (500 ng) and then subjected to a luciferase assay (lanes 1 to 3). Then expression constructs for p53 (25 ng) or p73β (10 ng) were transfected together with the reporter (500 ng) and expression plasmids for the E1B 55-kDa (500 ng) and E4 34-kDa (1 μg) proteins, as indicated above the diagram (lanes 4 to 19), and subjected to a luciferase assay.
Next, the E1B 55-kDa and/or E4 34-kDa protein was coexpressed with p53 or p73β, and transcription was quantified by measuring the luciferase activity (Fig. 3B, lanes 4 to 11). The transcriptional activity of p53 was reduced by the E1B 55-kDa protein alone (lane 5) and even more strongly by the two proteins together (lane 7), but p73β remained unaffected by the adenovirus oncoproteins (lanes 8 to 11).
Based on previously reported mutational analysis (18), we suspected that five residues near the amino terminus of p53 (amino acids 24 to 28) might be critical for E1B binding and hence for transcriptional inactivation. The amino acids at the corresponding positions are not conserved between p53 and p73 (Fig. 3A, compare residues 24 to 28 in p53 with amino acids 20 to 24 in p73). We hypothesized that this difference in primary structure might constitute the differential response of p53 and p73 activity to adenovirus oncoproteins. To test this hypothesis, chimeric proteins were designed with these five residues exchanged between p53 and p73 (Fig. 3A); they are termed p53mt(24–28) and p73βmt(20–24), respectively. This replacement resulted in complete resistance of p53 to E1B- and/or E4-mediated inhibition (Fig. 3B, lanes 12 to 15). In turn, p73βmt(20–24) was fully susceptible to E1B-mediated inactivation (lane 17), even though the E4 34-kDa protein did not further enhance this effect (lane 19).
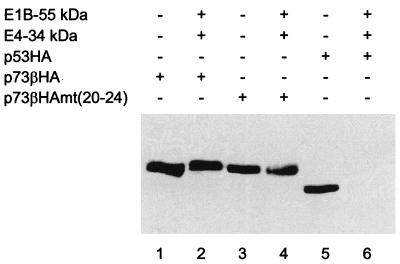
The abundance of p53 but not p73 is reduced in the presence of the E1B 55-kDa and E4 34-kDa proteins.
Next, we asked if p73 is also resistant to degradation mediated by adenovirus oncoproteins. To address this question, a hemagglutinin epitope was fused to the carboxy-terminal ends of p53 and p73β (Fig. 3A) to allow parallel quantitation. As expected, p73β levels were not reduced when the proteins were coexpressed with the adenovirus oncoproteins (Fig. 4, lanes 1 and 2), while tagged p53 was suppressed below detectability (compare lanes 5 and 6). Surprisingly, the chimeric version of p73 [p73βmt(20–24) (Fig. 3A)] that was inhibitable by the E1B 55-kDa protein was not detectably destabilized by the adenovirus oncoproteins (Fig. 4, lanes 3 and 4). This is consistent with the finding that the E4 34-kDa protein did not further downregulate the transcriptional activity of this mutant on top of the effect of the E1B 55-kDa protein (Fig. 3B, lane 19). Hence, intrinsic properties of p53 other than E1B binding are required for efficient degradation in the presence of adenovirus oncoproteins.
FIG. 4.
Reduction of p53 but not p73 levels by adenovirus proteins. The steady-state levels of HA-tagged p53 and p73 were determined by Western blotting. Transfections were carried out as described in the legend to Fig. 1A and were followed by immunodetection of the HA epitope.
The E1B 55-kDa protein relocalizes p53 but not p73.
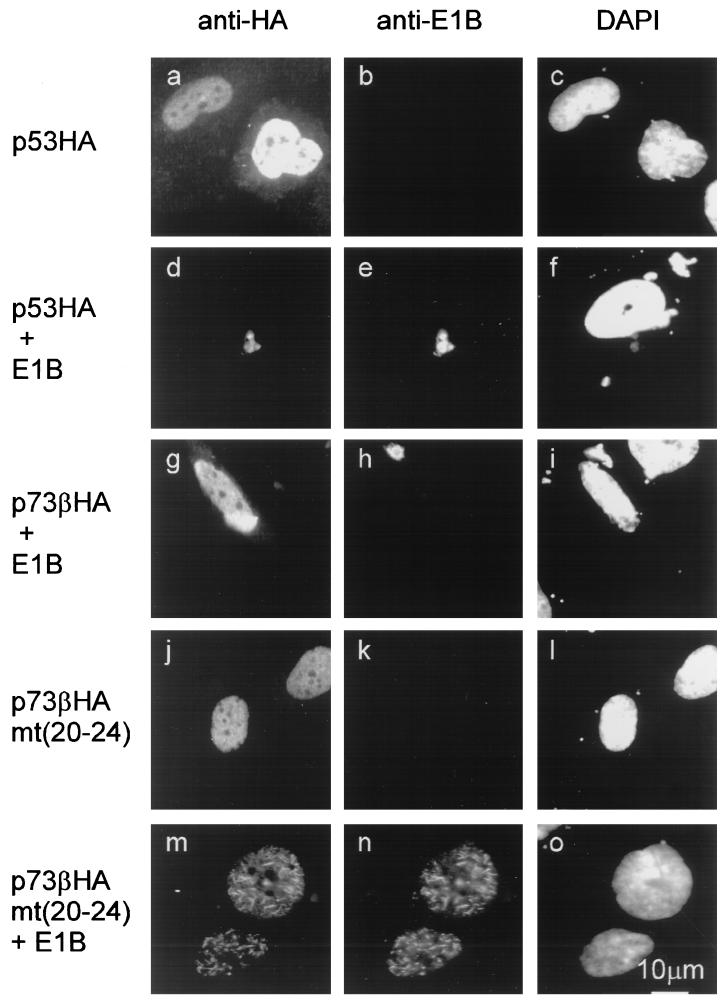
The intracellular complex formation between the E1B 55-kDa protein and p53 or p73 was further analyzed by simultaneous immunofluorescent labeling of coexpressed proteins (Fig. 5). The E1B 55-kDa protein relocalizes p53 into characteristic cytoplasmic clusters (Fig. 5, compare panels a to c with panels d to f) that were described previously (2, 37). In contrast, p73β did not colocalize with the E1B 55-kDa protein (panels g to i). However, when five residues near the amino terminus of p73β were replaced by the analogous amino acids from p53, colocalization with the E1B 55-kDa protein was restored in transfected cells (compare panels j to l with panels m to o). Both proteins were then found in nuclear clusters, possibly reflecting a nuclear localization signal within p73 that maintains its activity despite the association with the E1B 55-kDa protein.
FIG. 5.
Colocalization of the E1B 55-kDa protein with p53 but not p73. The E1B 55-kDa protein and HA-tagged p53 or p73 were examined for their intracellular location by immunofluorescence. Wild-type or chimeric forms of the proteins were transiently expressed in the presence or absence of an expression plasmid for nontagged E1B 55-kDa protein (the latter in threefold excess) as indicated above the panels. HA-tagged proteins and E1B were detected by using antibodies coupled to Texas red or fluorescein isothiocyanate, respectively. The location of the cell nuclei was determined with a fluorescent stain (4′,6-diamidino-2-phenylindole [DAPI]) specific for DNA.
The E1B 55-kDa protein forms a specific complex with p53 but not p73.
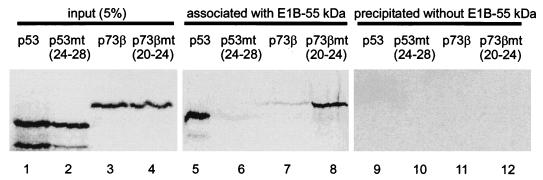
Finally, the interaction of the E1B 55-kDa protein with p53 and/or p73 was tested in vitro based on coimmunoprecipitation. While p53 efficiently associated with the E1B 55-kDa protein (Fig. 6, lane 5), little if any p73β bound this protein (lane 7). Conversely, p53mt(24–28) bound weakly if at all (lane 6) whereas mutant p73β, containing five p53-derived amino acids, was recovered as a complex with the E1B 55-kDa protein (lane 8). In the absence of the E1B 55-kDa protein, the antibody failed to precipitate any detectable p53 or p73 (lanes 9 to 12). We conclude that the adenovirus oncoproteins under study specifically inactivate p53 but not p73 and that this difference can be ascribed to a small sequence element within p53 that allows binding to the E1B 55-kDa protein.
FIG. 6.
Complex formation by the E1B 55-kDa protein with p53 but not p73. To detect association with the E1B 55-kDa protein, p53 and p73 as well as chimeras were translated in vitro (lanes 1 to 4) and then incubated with lysates from 293 cells that contain the E1B 55-kDa protein. After immunoprecipitation with an antibody to the E1B 55-kDa protein, associated p53 or p73 proteins were detected by autoradiography (lanes 5 to 8). In a control experiment (lanes 9 to 12), lysate from HeLa cells (lacking the E1B 55-kDa protein) instead of 293 cells was used to perform the analogous immunoprecipitation.
DISCUSSION
We have shown that p53 is rapidly degraded when coexpressed with the combination of the adenovirus type 5 E1B 55-kDa and E4 34-kDa proteins. p73β activates p53-responsive promoters at least as strongly as p53 itself does, but it is not antagonized or destabilized by the adenovirus oncoproteins. The E1B 55-kDa protein binds and relocalizes p53 but not p73, and this difference was pinpointed to five N-terminal amino acid residues that are not conserved between p53 and p73.
As suggested by mutational analysis, both the interaction between the E1B 55-kDa and E4 34-kDa proteins and the association of the E1B 55-kDa protein with p53 seem to be needed for accelerated p53 degradation (Fig. 2). Possibly, a complex that contains all three proteins is formed. It remains to be determined which mechanism(s) ultimately leads to the degradation of p53 in the presence of adenovirus oncoproteins. So far, proteasome-mediated proteolysis (5, 9) and calpain-mediated proteolysis (19, 23) have been reported to shorten the life span of p53. However, a specific inhibitor for the proteasome only weakly inhibited p53 degradation, and calpain inhibitors completely failed to protect p53 in the presence of the E1B 55-kDa and E4 34-kDa proteins. Therefore, it remains possible that the adenovirus oncoproteins trigger p53 degradation by a third mechanism. The ability of such a novel pathway to destroy p53 and its role in the absence of viral proteins are subjects for further studies.
In our hands, the E1B 55-kDa protein reduces the ability of p53 to activate transcription. In contrast, the E4 34-kDa protein has only an auxiliary effect on p53 inhibition when coexpressed with the E1B 55-kDa protein but does not downregulate the activity of p53 in the absence of the E1B 55-kDa protein (Fig. 1D). This was consistently observed over a wide range of plasmid amounts transfected to express p53 and the E4 34-kDa protein (25a), in contrast to a previous report (8). We therefore assume that direct effects of the E4 34-kDa protein alone on p53 activity may be restricted to special conditions but do not represent a generally observable phenomenon. However, the E1B 55-kDa and E4 34-kDa proteins act synergistically to destabilize and inactivate p53.
The E1B 55-kDa and E4 34-kDa proteins are known to shuttle between the nucleus and cytoplasm (7). It is tempting to speculate that this transport phenomenon might be part of the degradation mechanism. Therefore, we compared the E4 34-kDa protein with a mutant carrying a defective nuclear export signal (NES) and asked if their abilities to degrade p53 in the presence of E1B might be different. Indeed, the mutation within the NES resulted in a decreased ability to destabilize p53 (data not shown). However, the reduction of the p53 levels was still readily observable under these conditions, suggesting that nuclear export is not an absolute requirement to mediate p53 degradation. Furthermore, it cannot be excluded that the NES mutation might reduce the ability of the E4 34-kDa protein to associate with the E1B 55-kDa protein and p53. Therefore, we still consider the role, if any, played by nuclear export of adenovirus oncoproteins in the degradation of p53 to be an open question.
The transcriptional activities of p53 and p73 seem virtually indistinguishable, at least when using the promoters that we have examined so far (data not shown). These included the promoters of hdm2, p21 (waf1), and a synthetic p53-responsive plasmid (3). The ratio between the activities of p53, p73α, and p73β consistently remained the same, with p73β being far more active than p73α. Possibly, the ability of p73β to form homo-oligomers (14) enhances its ability to bind the specific DNA element cooperatively, similar to p53. Given the striking difference in transcriptional activation by the splice variants p73α and p73β, it is conceivable that alternative splicing might be a mechanism to regulate p73 activity. Future studies are aimed at determining if the ratio between p73α and p73β varies between cell types.
Since p73 can activate at least a large subset of the p53-responsive promoters, and given its structural similarities to p53, it was initially assumed that oncoproteins might have evolved to bind and inactivate both p53 and p73 (14). However, at least the inhibitors studied here failed to affect p73. The interaction with oncoproteins is only the second functional difference identified between p53 and p73 (the first difference was the increased amount of p53 but not p73 found in cells after treatment with DNA-damaging agents [14]). It remains to be determined whether other p53 antagonists, e.g., the HPV E6 proteins or the cellular mdm2 protein, also inactivate and degrade p53 but not p73. Our recently obtained results suggest that the simian virus 40 T antigen also binds and inactivates p53 but not p73 (26a).
Why is p73 “neglected” by the oncoproteins studied here while p53 is efficiently inactivated and degraded? One explanation could be that the p73 proteins are controlled by a subset of viral and cellular factors distinct from the p53 antagonists. However, in the case of virus-induced tumor formation, the E1B 55-kDa and E4 34-kDa proteins, along with the adenovirus E1A 13S protein, were shown to be sufficient to strongly promote cell transformation (20, 21) but do not inactivate p73. Thus, it seems that some form of p53 inactivation—preferably destabilization—is a prerequisite for virus-induced tumor development in most cases, while tumors do arise even when p73 is not affected.
A second possibility is that p73 expression and activity is restricted to certain tissues or cell types. However, detectable amounts of p73 were found in most tissues, and several tumor-derived cell lines were shown to express wild-type p73 proteins in considerable amounts while the p53 transcript was mutated (14). Nonetheless, it remains possible that p73 quantities and activities vary between tissues. In this case, p73 may represent a differentiation factor in certain tissues rather than a general “guardian of the genome,” as suggested for p53 (17).
Based on the differential interaction with viral oncoproteins, we propose that p53 has activities that cannot be performed by p73 and that are essential for tumor suppression in at least a subset of cells whereas p73 inactivation seems dispensable for oncogene-mediated tumor induction. What could be the mechanistic nature of such an activity that is unique to p53? One possibility is that some p53-responsive promoters cannot be activated by p73. However, we and others have not found such a differential responsiveness in the promoters tested so far. Alternatively, the activity residing in the proline-rich region of the protein (28, 29, 35), or growth-suppressing functions of p53 that might be unrelated to transcriptional activation (12), may not be maintained in the p73 proteins. Such activities may thus represent critical functions by which the p53 protein plays its role as a protector from tumor development.
To date, it cannot be regarded as certain that p73 fulfils all the criteria of a tumor suppressor gene product (4, 22) or whether it acts so in all cell types. However, should p73 not turn out to play a role as central as p53 in tumorigenesis, its differences from p53 might serve as guidelines to identify as yet unknown p53 functions that are crucial for tumor suppression.
ACKNOWLEDGMENTS
We thank H.-D. Klenk and R. Arnold for their generous support, and we thank M. Kaghad, D. Caput, and W. G. Kaelin for plasmids.
This work was supported by the German Research Foundation, the P. E. Kempkes Foundation, the Fazit Foundation (scholarship to C. König), the Hoechst Scholarship Foundation (to S. Wienzek), and the German Cancer Research Center (Stipendium für Infektionsbiologie to M.D.).
ADDENDUM IN PROOF
While this article was under review, it was reported that the E6 protein of an oncogenic human papillomavirus mediates intracellular degradation of p53 but not p73β (N. S. Prabhu, K. Somasundaram, K. Satyamoorty, M. Iterlyn, and W. S. El-Deiry, Int. J. Oncol. 13:5–9, 1998).
REFERENCES
- 1.Babiss L E, Ginsberg H S, Darnell J E., Jr Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol Cell Biol. 1985;5:2552–2558. doi: 10.1128/mcb.5.10.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blair Zajdel M E, Blair G E. The intracellular distribution of the transformation-associated protein p53 in adenovirus-transformed rodent cells. Oncogene. 1988;2:579–584. [PubMed] [Google Scholar]
- 3.Buckbinder L, Talbott R, Seizinger B R, Kley N. Gene regulation by temperature-sensitive p53 mutants: identification of p53 response genes. Proc Natl Acad Sci USA. 1994;91:10640–10644. doi: 10.1073/pnas.91.22.10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dickman S. First p53 relative may be a new tumor suppressor. Science. 1997;277:1605–1606. doi: 10.1126/science.277.5332.1605. [DOI] [PubMed] [Google Scholar]
- 5.Dietrich C, Bartsch T, Schanz F, Oesch F, Wieser R J. p53-dependent cell cycle arrest induced by N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal in platelet-derived growth factor-stimulated human fibroblasts. Proc Natl Acad Sci USA. 1996;93:10815–10819. doi: 10.1073/pnas.93.20.10815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobbelstein M, Arthur A K, Dehde S, van Zee K, Dickmanns A, Fanning E. Intracistronic complementation reveals a new function of SV40 T antigen that co-operates with Rb and p53 binding to stimulate DNA synthesis in quiescent cells. Oncogene. 1992;7:837–847. [PubMed] [Google Scholar]
- 7.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 9.Gonen H, Shkedy D, Barnoy S, Kosower N S, Ciechanover A. On the involvement of calpains in the degradation of the tumor suppressor protein p53. FEBS Lett. 1997;406:17–22. doi: 10.1016/s0014-5793(97)00225-1. [DOI] [PubMed] [Google Scholar]
- 10.Halbert D N, Cutt J R, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 12.Haupt, Y., S. Rowan, E. Shaulian, A. Kazaz, K. Vousden, and M. Oren. 1997. p53 mediated apoptosis in HeLa cells: transcription dependent and independent mechanisms. Leukemia 11(Suppl. 3):337–339. [PubMed]
- 13.Jost C A, Marin M C, Kaelin W G., Jr p73 is a human p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 14.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J C, Valent A, Minty A, Chalon P, Lelias J M, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 15.Kao C C, Yew P R, Berk A J. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology. 1990;179:806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 16.Kubbutat M H, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 17.Lane D P. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 18.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 19.Maki C G, Huibregtse J M, Howley P M. In vivo ubiquitination and proteasome-mediated degradation of p53(1) Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 20.Moore M, Horikoshi N, Shenk T. Oncogenic potential of the adenovirus E4orf6 protein. Proc Natl Acad Sci USA. 1996;93:11295–11301. doi: 10.1073/pnas.93.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc Natl Acad Sci USA. 1997;94:1206–1211. doi: 10.1073/pnas.94.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oren M. Lonely no more: p53 finds its kin in a tumor suppressor haven. Cell. 1997;90:829–832. doi: 10.1016/s0092-8674(00)80347-5. [DOI] [PubMed] [Google Scholar]
- 23.Pariat M, Carillo S, Molinari M, Salvat C, Debussche L, Bracco L, Milner J, Piechaczyk M. Proteolysis by calpains: a possible contribution to degradation of p53. Mol Cell Biol. 1997;17:2806–2815. doi: 10.1128/mcb.17.5.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilder S, Moore M, Logan J, Shenk T. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol Cell Biol. 1986;6:470–476. doi: 10.1128/mcb.6.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Querido E, Marcellus R C, Lai A, Charbonneau R, Teodoro J G, Ketner G, Branton P E. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997;71:3788–3798. doi: 10.1128/jvi.71.5.3788-3798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Roth, J. Unpublished observations.
- 26.Roth J, Dittmer D, Rea D, Tartaglia J, Paoletti E, Levine A J. p53 as a target for cancer vaccines: recombinant canarypox virus vectors expressing p53 protect mice against lethal tumor cell challenge. Proc Natl Acad Sci USA. 1996;93:4781–4786. doi: 10.1073/pnas.93.10.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Roth, J., and M. Dobbelstein. Unpublished observations.
- 27.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruaro E M, Collavin L, Del Sal G, Haffner R, Oren M, Levine A J, Schneider C. A proline-rich motif in p53 is required for transactivation-independent growth arrest as induced by Gas1. Proc Natl Acad Sci USA. 1997;94:4675–4680. doi: 10.1073/pnas.94.9.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakamuro D, Sabbatini P, White E, Prendergast G C. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene. 1997;15:887–898. doi: 10.1038/sj.onc.1201263. [DOI] [PubMed] [Google Scholar]
- 30.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 31.Sarnow P, Sullivan C A, Levine A J. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology. 1982;120:510–517. doi: 10.1016/0042-6822(82)90054-x. [DOI] [PubMed] [Google Scholar]
- 32.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 33.Steegenga W T, Riteco N, Jochemsen A G, Fallaux F J, Bos J L. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene. 1998;16:349–357. doi: 10.1038/sj.onc.1201540. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 35.Walker K K, Levine A J. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci USA. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Wienzek, S. Unpublished data.
- 36.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 37.Zantema A, Fransen J A, Davis-Olivier A, Ramaekers F C, Vooijs G P, DeLeys B, Van der Eb A J. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology. 1985;142:44–58. doi: 10.1016/0042-6822(85)90421-0. [DOI] [PubMed] [Google Scholar]