Abstract
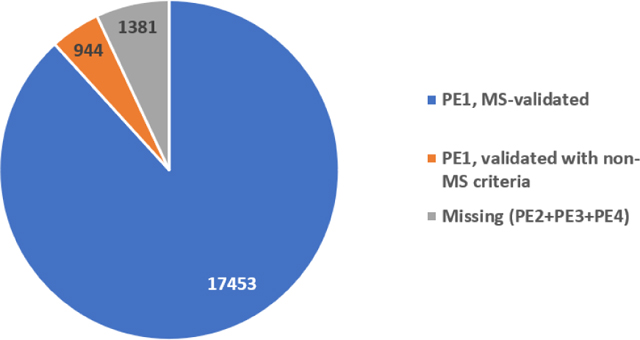
Since 2010, the Human Proteome Project (HPP), the flagship initiative of the Human Proteome Organization (HUPO), has pursued two goals: (1) to credibly identify the protein parts list and (2) to make proteomics an integral part of multi-omics studies of human health and disease. The HPP relies on international collaboration, data sharing, standardized reanalysis of MS data sets by PeptideAtlas and MassIVE-KB using HPP Guidelines for quality assurance, integration and curation of MS and non-MS protein data by neXtProt, plus extensive use of antibody profiling carried out by the Human Protein Atlas. According to the neXtProt release 2023–04-18, protein expression has now been credibly detected (PE1) for 18,397 of the 19,778 neXtProt predicted proteins coded in the human genome (93%). Of these PE1 proteins, 17,453 were detected with mass spectrometry (MS) in accordance with HPP Guidelines and 944 by a variety of non-MS methods. The number of neXtProt PE2, PE3, and PE4 missing proteins now stands at 1381. Achieving the unambiguous identification of 93% of predicted proteins encoded from across all chromosomes represents remarkable experimental progress on the Human Proteome parts list. Meanwhile, there are several categories of predicted proteins that have proved resistant to detection regardless of protein-based methods used. Additionally there are some PE1–4 proteins that probably should be reclassified to PE5, specifically 21 LINC entries and ~30 HERV entries; these are being addressed in the present year. Applying proteomics in a wide array of biological and clinical studies ensures integration with other omics platforms as reported by the Biology and Disease-driven HPP teams and the antibody and pathology resource pillars. Current progress has positioned the HPP to transition to its Grand Challenge Project focused on determining the primary function(s) of every protein itself and in networks and pathways within the context of human health and disease.
Keywords: Human Proteome Organization (HUPO), Human Proteome Project (HPP), neXtProt protein existence (PE) metrics, missing proteins (MP), non-MS PE1 proteins, uncharacterized protein existence 1 (uPE1), Chromosome-centric HPP (C-HPP), Biology and Disease-HPP (B/D-HPP), PeptideAtlas, Mass Spectrometry Interactive Virtual Environment Knowledge Base (MassIVE-KB), Human Protein Atlas, Grand Challenge Project
Graphical Abstract
Progress on the Human Proteome Parts List
As the flagship initiative of the Human Proteome Organization1 (HUPO), the Human Proteome Project2 (HPP) has pursued two goals: (1) to credibly identify the protein “parts list”, primarily but not entirely by mass spectrometry (MS); and (2) to make proteomics an integral part of multi-omics studies of human health and disease.3–5 The HPP has made consistent progress for over ten years through engagement of the global community, data sharing, standardized reanalysis of datasets by PeptideAtlas6,7 and MassIVE-KB,8 curation and integration of data by neXtProt,9 and collaboration with the Human Protein Atlas10 for antibody-based protein expression and intracellular localization. The HPP organization includes 25 teams by nuclear and mitochondrial chromosomes, 16 teams based on biological and clinical categories, and four resource pillars5. The work is supported by advances in data formats and standards by the HUPO Proteome Standards Initiative11,12 (HUPO-PSI), ProteomeXchange Consortium data repositories, and the concomitant growth of UniProtKB,13 Swiss-Prot, and neXtProt.
As the primary HPP knowledge base, neXtProt plays a central role in the HPP documentation and dissemination of information about the human proteins confidently identified with protein-level evidence (PE1). Table 1 shows the progress in identifying proteins from predicted protein-coding genes, with 13,975 PE1 proteins in neXtProt release 2012–02 to 18,397 PE1 proteins in neXtProt release 2023–04-18, representing 93.0% of the predicted proteins, and an increase of 1.7% in canonical proteins in PeptideAtlas from 2022. neXtProt also tabulates known sequence variants, splice variants, and post-translational modifications (PTMs) for these protein entries. Table 1 further documents the decline in the number of “missing proteins” (PE2,3,4). PE2 entries are thought to be translated but, thus far, have only transcript evidence with no or insufficient protein supportive data. PE3 entries have evidence only by homology in non-human species. PE4 entries are computationally predicted to be translated, but do not have translation, transcription, or homology evidence. PE5 entries are considered dubious by UniProtKB, are often pseudogenes, but may have some weak evidence, and may rarely be upgraded to PE1 based on new reports.
Table 1.
Numbers of proteins by neXtProt protein existence (PE) evidence levels from 2012–02 to 2023–04, showing progress in identifying PE2,3,4 missing proteins to become PE1 proteins and PeptideAtlas canonical proteins by mass spectrometry.a,b,c
| PE Level | 2012–02 | 2013–09 | 2014–10 | 2016–01 | 2017–01 | 2019–01 | 2020–01 | 2021–02 | 2022–02 | 2023–04 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1:Evidence at protein level | 13,975 | 15,646 | 16,491 | 16,518 | 17,008 | 17,694 | 17,874 | 18,357 | 18,407 | 18,397 |
| 2:Evidence at transcript level | 5205 | 3570 | 2647 | 2290 | 1939 | 1548 | 1596 | 1265 | 1135 | 1151 |
| 3:Inferred from homology | 218 | 187 | 214 | 565 | 563 | 510 | 253 | 147 | 195 | 215 |
| 4:Predicted | 88 | 87 | 87 | 94 | 77 | 71 | 50 | 9 | 13 | 15 |
| MP = PE2 + PE3 + PE4 | 5511 | 3844 | 2948 | 2949 | 2579 | 2129 | 1899 | 1421 | 1343 | 1381 |
| Human PeptideAtlas canonical proteins | 12,509 | 13,377 | 14,928 | 14,569 | 15,173 | 16,293 | 16,655 | 16,702 | 16,957 | 17,245 |
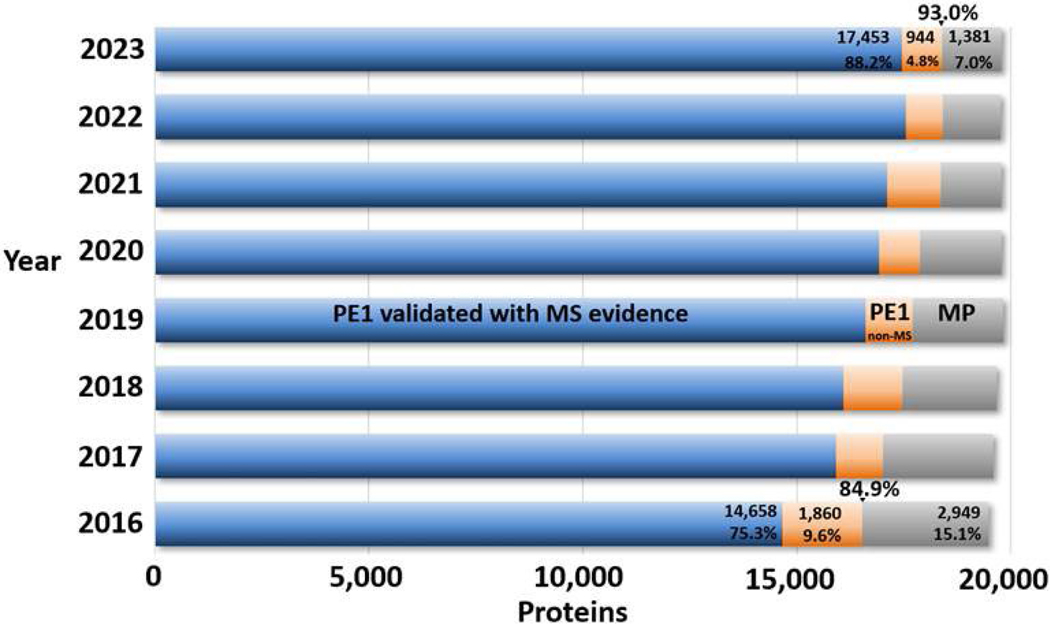
Figure 1 shows the progress in the eight years since 2016, when stringency criteria were substantially tightened with the introduction of the HPP Mass Spectrometry Data Interpretation Guidelines v2.1.14 The total length of the bars shows the sum of PE1–4 proteins, which we note has changed over the years as the reference proteomes are continually refined by UniProtKB/Swiss-Prot. The blue portions represent the PE1 entries that have been validated via mass spectrometry evidence that meets the HPP guidelines. Substantial progress has been made every year since 2016, except from 2022 to 2023, as will be discussed below. The orange portions represent entries that have PE1 status without MS detection, but, rather, via other methods such as protein-protein interactions, Edman degradation, and/or 3D structure information. The gray portions represent the missing proteins (PE2–4), which are predicted to be translated but remain without sufficient direct evidence.
Figure 1.
This bar chart documents progress in reducing PE2,3,4 missing proteins (gray) and increasing PE1 proteins (blue + orange). Within the PE1 proteins, there is a shift from non-MS PE1 proteins (orange) to MS-based PE1 proteins (blue). The base year is 2016, after the introduction of the much more stringent HPP Mass Spectrometry Data Interpretation Guidelines v2.1.
The current version 3.0 of the HPP Mass Spectrometry Data Interpretation Guidelines was released on 2019–10-15. It is presented as a checklist available at https://hupo.org/HPP-Data-Interpretation-Guidelines and is described in detail by Deutsch et al.15 It has three major groups of guidelines, the first of which describes data deposition requirements to a ProteomeXchange repository and use of a recent reference proteome for analysis. The second group of guidelines describes how false discovery rate (FDR) control should be carefully described, including handling for dataset merging. The final group of guidelines describes specific requirements for claims of detection of missing proteins or proposed additions to the reference proteome in neXtProt. These include a baseline requirement of at least two uniquely mapping peptides of length nine or more residues, where one peptide may not be fully nested within the other, and together they must cover at least 18 amino acids of the protein. For these two peptides, there is a described requirement for the presentation of manually scrutinized, high signal-to-noise ratio, high mass accuracy annotated spectra along with Universal Spectrum Identifiers (USIs), comparison with matching synthetic peptide peak intensities, and consideration for potential alternative peptide-to-protein mappings, taking variants into account. Substantial additional nuances and clarifications are presented within the guidelines document.
In the top bar for 2023, the 944 non-MS-based PE1 proteins received PE1 status almost entirely from UniProtKB: 54 are based primarily on Edman degradation, 39 on 3D structures in Protein Data Bank, 472 on protein-protein interactions (including 8 by neXtProt), 33 on antibody studies (one by neXtProt), 89 on PTMs and processing, 85 on genetic mutations, and 172 from biochemical studies (4 by neXtProt). Many are supported by multiple types of studies (see below); some entries have some MS data, but the evidence for these does not meet the HPP MS Guidelines.
In order to increase the transparency in the promotion of a missing protein from PE2,3,4 to PE1 in neXtProt based on MS-based proteomics data, entries must have at least two non-overlapping proteotypic peptides (peptides uniquely matched to a single protein ID) of nine amino acids or more which have been detected in the same specimen and confirmed upon reanalysis by PeptideAtlas or MassIVE-KB. These proteins now can be identified or queried with the keyword KW-1267 Proteomics Identification in the neXtProt interface. These bars can be easily generated using saved SPARQL queries with identifiers at neXtProt: PE1 with MS query NXQ_00301, PE1 with non-MS NXQ_00302, and PE2–4 missing proteins with NXQ_00204. These queries will always produce the most up-to-date lists as new data are curated in neXtProt. For example, the latest list of missing proteins is available via https://www.nextprot.org/proteins/search?mode=advanced&queryId=NXQ_00204.
It is important to note that the reference human proteome is continually changing as the community learns more about the human genome and proteome and the literature is curated by UniProtKB/Swiss-Prot.16 Figure 2 shows the changes in the reference proteome in UniProtKB/Swiss-Prot and PE levels in neXtProt between 2022 and 2023. The total number of PE1–5 entries increased from 20359 to 20389, but this did not result from only a simple addition of entries. Rather, on the left side are shown the number of PE1, MP, and PE5 entries that were removed from the reference proteome for a variety of reasons. On the right margin are shown the number of additions to the reference proteome: 20 new PE1 entries, 33 new MP entries, and four new PE5 entries. Notably, the 20 new PE1 and 33 new PE2,3,4 proteins include 12 TATA-box-binding protein associated factor 11-like proteins 3–14 (2 PE1, 10 PE2) (per neXtProt weblink in the Figure). Some of the deletions and additions represent merging and splitting of entries as more is learned about the proteins.
Figure 2.
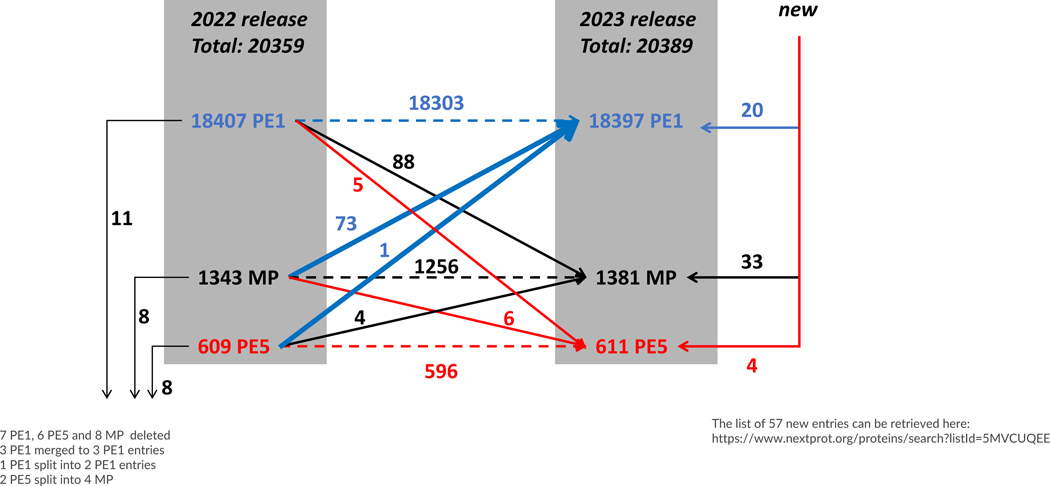
Flow diagram of the changes in numbers of PE1, PE2, PE3, PE4 (and PE5) classes of predicted proteins from neXtProt 2022–02 to 2023–04, reflecting newly reported findings and changes by curators, including the inclusion of previously unrecognized proteins (left and right margins).
Other recent additions to the human proteome include a number of small ORFs or microproteins, which are added to the reference set by UniProt expert curators on a case-by-case basis when publications describing the function of these proteins become available in the scientific literature. The canonical amino acid sequence displayed in each entry is regularly reviewed and updated as part of the MANE17 project, in collaboration with curators from Ensembl and RefSeq. Entries may be removed or recategorized if, for example, it is decided the gene is actually a pseudogene and most probably non-coding.
In the central part of Figure 2, we show the transitions in PE level in neXtProt between 2022 and 2023. neXtProt incorporates reanalyzed MS data both from PeptideAtlas and, since 2019, MassIVE-KB. There are 88 PE1 proteins demoted to PE2,3,4 MP status (black line) and only 73 MPs elevated to PE1 (blue line).
There were five formerly PE1 entries that were demoted to PE5 by UniProt curators (red line). O95411 and Q86SI9 have no peptides in PeptideAtlas at all; therefore, their demotion to PE5 is consistent on the basis of available MS evidence. However, Q8N319 has several uniquely mapping peptides confirmed by manual inspection that are detected in HLA immunopeptidome datasets. The gene (LINC03040) that encodes this protein is designated as non-coding by Ensembl, RefSeq, and HGNC; therefore, UniProt curators made the decision to add a “Caution” comment to this effect, which then designates this protein as PE5. However, in response to the proteomics data, the protein status is currently being re-reviewed. Q5XLA6 has several peptides that meet HPP guidelines criteria and pass manual inspection in CPTAC cancer datasets. Q92637 has several uniquely mapping peptides in PeptideAtlas, but is highly similar to PE1 protein P12314. In summary, three of the five cases of PE1 demotions to PE5 from 2022 to 2023 are questionable. This observation is why UniProt continues to maintain proteins designated PE5 in the proteome, to enable such observations to be considered by MS proteomics researchers.
Because neXtProt serves as the primary knowledge base for the HPP, its periodic updates begin with the list of ~20,000 human entries in UniProtKB/Swiss-Prot with information added from various other sources. One of the additions is the upgrade of PE2,3,4 entries to PE1 based on uniquely mapping peptides that meet HPP guidelines from PeptideAtlas and/or MassIVE-KB. Both PeptideAtlas and MassIVE-KB collect publicly available MS proteomics data sets from ProteomeXchange Consortium repositories, reprocess those data sets with a uniform pipeline, and produce high-stringency compendia of detected peptides and proteins with full provenance back to the original spectra. Both resources then send their list of peptides to neXtProt, which remaps the peptides to its reference proteome and upgrades UniProtKB/Swiss-Prot PE2,3,4 entries to PE1 if peptides from either PeptideAtlas or MassIVE-KB (or both) meet HPP guidelines. Entries are not upgraded if only the merging of two different peptide sets produces the two uniquely mapping peptides required by the HPP Guidelines.
The implications of this process are that there are some differences between UniProtKB/Swiss-Prot and neXtProt. neXtProt uses the same entries as UniProtKB/Swiss-Prot, with the exception that releases of neXtProt follow after UniProtKB/Swiss-Prot releases by a few months, and neXtProt prefixes their identifiers with “NX_”. Thus, the changes in the left and right margins of Figure 2 are first made in UniProtKB/Swiss-Prot and then replicated to neXtProt for the next release. Furthermore, the PE level classifications differ somewhat between UniProtKB/Swiss-Prot and neXtProt due to the addition of reanalyzed proteomics MS evidence from PeptideAtlas and MassIVE-KB. The current differences in PE level are summarized in Table 2. The difference of 24 entries in the total row in Table 2 reflects the lag in releases. The difference of 1760 entries in the PE1 row of Table 2 is very largely due to the upgrade to PE1 status by neXtProt due to MS evidence. neXtProt has upgraded 13 of those protein entries via non-MS evidence (see above).
Table 2.
Comparison of proteins by PE level in UniProtKB/Swiss-Prot and neXtProt
| Protein existence level | UniProtKB/ Swiss-Prot 2023-07-23 | neXtProt 2023-03-25 release | Delta |
|---|---|---|---|
| PE1 | 16637 | 18397 | 1760 |
| PE2 | 2249 | 1151 | −1098 |
| PE3 | 776 | 215 | −561 |
| PE4 | 140 | 15 | −125 |
| Total PE1-4 | 19802 | 19778 | −24 |
The numbers of PE1–4 neXtProt entries (out of 19778 neXtProt entries) that satisfy the HPP guidelines criteria of at least two non-nested uniquely mapping peptides of at least nine amino acids in length and covering at least 18 amino acids in extent for PeptideAtlas and for MassIVE-KB in 2022 and 2023 are shown in Table 3. While the number of PeptideAtlas entries supporting PE1 status continued to grow (by 264) from 2022 to 2023 in neXtProt, there was a very substantial reduction in PE1 proteins supported by MassIVE-KB (by 619 from 17,033 to 16,414). Correspondingly, there was an increase of 731 from 405 to 1136 in the number of PE1 entries documented only by PeptideAtlas while the number only from MassIVE-KB dropped by 253 from 365 to 112.
Table 3.
Change in the number of neXtProt entries between 2022 and 2023 in PeptideAtlas and MassIVE-KB that meet the HPP Guidelines v3.0
| Set | 2022 | 2023 | Delta |
|---|---|---|---|
| PeptideAtlas | 17174 | 17438 | 264 |
| MassIVE-KB | 17033 | 16414 | −619 |
| Both | 16668 | 16302 | −366 |
| PeptideAtlas only | 405 | 1136 | 731 |
| MassIVE-KB only | 365 | 112 | −253 |
The reason for this dramatic change was a substantial increase in size and stringency in the MassIVE-KB pipeline between 2022 and 2023. The number of distinct peptides detected by MassIVE-KB increased by over 50% to ~2 million peptides. In order to maintain a low false discovery rate, the build process was updated to use all decoys from all the searches even if those decoys do not make it into MassIVE-KB. This change was made because MassIVE-KB introduced in 2023 a protein-level 0.1% FDR threshold to determine which precursors to keep. A post-hoc evaluation of the 1% protein-level FDR threshold used by MassIVE-KB in 2022 resulted in five olfactory receptor entries meeting the guidelines, even though manual inspection clearly revealed these as false positives. Thus, the 0.1% protein-level FDR was retained by MassIVE.
A substantial part of the advance in the number of canonical proteins in PeptideAtlas came from data sets derived from HLA immunopeptidome samples.18–21 These samples are enriched in processed peptides that are presented on cell surfaces as part of the HLA antigen presentation system, and thus provide a new opportunity to detect proteins that would otherwise be too low in abundance to detect in ordinary protein mixtures. Figure 3 shows the data sets that contributed five or more new canonical proteins to PeptideAtlas. The top contributor by far was the HLA Ligand Atlas18 data from 227 benign tissue samples via PXD019643 with 32 new canonical proteins. Three additional data sets19–21 in the top seven were also derived from HLA immunopeptidome samples. The largest non-immunopeptidome contributor used the TAILS N-terminomics approach to enrich for protein N-terminal peptides.22 Data from PXD02038923 and PXD02864724 explored neuroblastoma and aneuploidy cell lines, respectively, to provide detection of five new canonical proteins each. Dozens of additional datasets, reflecting in part the activity of the B/D Immunopeptidomics community, provide a few additional canonical proteins. PeptideAtlas focused on adding immunopeptidome data sets as it was suspected that this might make a substantial contribution to new canonical proteins.
Figure 3.
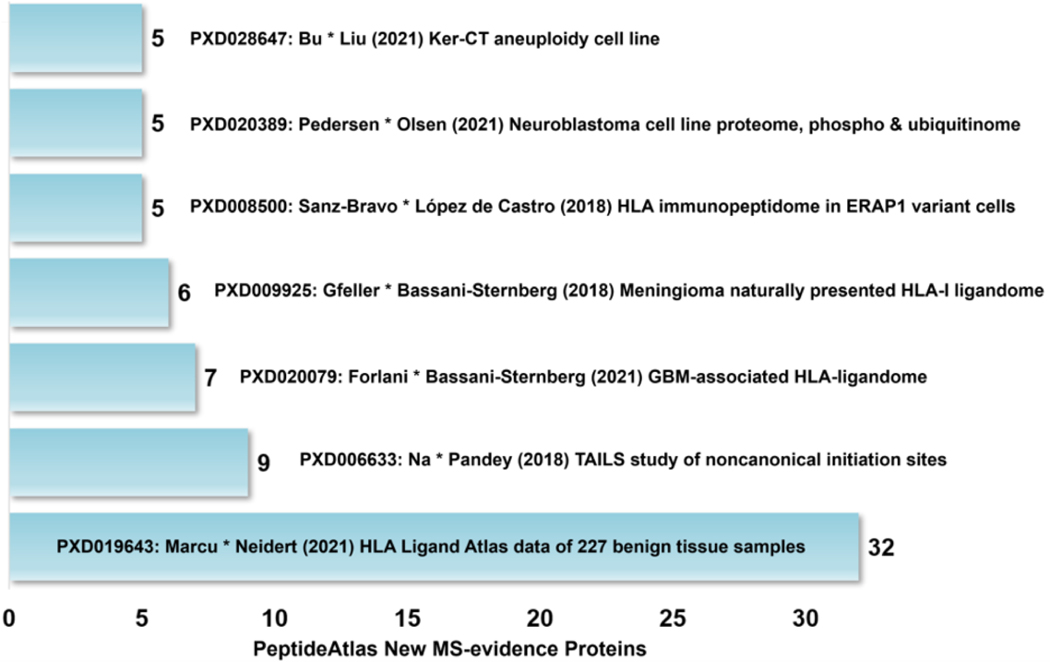
Data sets that each contributed five or more new canonical proteins in the 2023–01 PeptideAtlas build.
An open question under active discussion is whether all 1381 current missing protein entries really ought to be on the HPP target list, or whether some should be demoted to PE5. One clear group of entries is those labeled with a gene symbol beginning with “LINC” (long intergenic non-coding RNA). It would seem that the most likely hypothesis is that these genes do not code for proteins, and have already been annotated as such. There are currently 90 entries with a gene symbol beginning with “LINC”. Of these, 68 are already PE5 and not on the HPP missing protein target list. Of the remaining 22, only one is currently PE1, LINC02914 (Q52M58) with one apparent detection in sperm cells.25 The other 21 PE2–4 entries have no detections in PeptideAtlas that come near HPP guidelines. We propose that these 21 entries be demoted to PE5 until there are credible detections at the protein level that warrant promotion to PE1. This would reduce the list of MPs to 1360.
Another important group of proteins is the olfactory receptors. There are 427 PE1–5 entries that have a description that includes “olfactory receptor”, of which 25 are PE1 and another 383 are MPs (PE2–4). There is no doubt that many of these are translated since they are required for successful human olfaction, but they are very difficult to detect by MS due to their many transmembrane regions and highly specific localization, which has rarely been sampled. Thus, olfactory receptors should remain on the HPP target list. Most of the current 25 PE1 olfactory receptors have been detected by means other than MS. In fact, only one olfactory receptor meets the lowest level of HPP guidelines in PeptideAtlas, OR51E2 (Q9H255). A detailed review of the evidence is provided in the Supplementary Material. The conservative conclusion is that these PSMs do not provide the sort of conclusive evidence that would be expected to claim detection of an olfactory receptor by MS. The postulate that all purported MS detections of olfactory receptors are false positives seems safe for now.
One final group discussed here is the human endogenous retrovirus (HERV) proteins. There are 66 neXtProt entries that contain “endogenous retrovir” in their description. Excluded from these are a few famous examples of originally HERV proteins that have been put to use in the human system, notably Syncytin-1 (ERVW-1) (Q9UQF0), Syncytin-2 (ERVFRD-1) (P60508), and Suppressyn (ERVH48–1) (M5A8F1). Of the 66, only one is PE5, whereas 35 are listed as PE1. A few have very clear evidence in PeptideAtlas, with ERV3–1 (Q14264) being the best example, having 19 distinct peptides from 25 different experiments, of which nearly all are uniquely mapping. However, many entries are very highly similar to each other and more than half have no MS evidence whatsoever. A careful review of these is warranted and likely many (~30) should be demoted to PE5. It would be appropriate for the HPP community to consider whether these are useful to keep on the HPP target list, especially if the few that have wisps of detections in cancers or other samples may well be non-canonical translations yielding non-functional products. The major sequence resources UniProt, Ensembl, RefSeq, HGNA, and InterPro are already discussing how to address this situation.
Human proteins in UniProt are mapped to the equivalent sequence in Ensembl (requiring 100% identity over 100% of the length of the two sequences) and are made available to search and download as a human proteome set (Homo sapiens proteome ID:UP000005640). At the time of this writing the T2T-CHM13v2.0 assembly and annotation is currently only available through Ensembl Rapid Release (https://rapid.ensembl.org/Homo_sapiens_GCA_009914755.4/Info/Index) and is not integrated into the UniProt import pipeline but discussions are ongoing with the Ensembl, RefSeq, Human Pangenome, and HGNC teams to enable as near concurrence between genome, transcriptome, and proteome as possible.
The Chromosome-centric HPP (C-HPP) component enables a natural division of effort by chromosome, undertaken by 25 different teams from many different countries (www.hupo.org/C-HPP). Although the initial focus has been on the detection of missing proteins, substantial effort has shifted to trying to understand the function of those proteins lacking any known function. Table 4 shows the current state of proteins by PE level for each chromosome. For most chromosomes, the percentage of missing proteins is in the single digits with a median of 5%. However, several chromosomes stand out with double-digit percentages of missing proteins, notably 7, 11, 14, 21, and Y. Most striking is that chromosome 11 had 175 missing olfactory receptor proteins in the summary by Adhikari et. al.3
Table 4.
Chromosome-by-Chromosome Status of Predicted Proteins in neXtProt 2023–03, Showing Missing Proteins (MP) and Unannotated Proteins (uPE1)a
| Chromosome | PE1 | PE2 | PE3 | PE4 | PE1–4 | MP | %MP/ PE1–4 | uPE1 | %uPE1/PE1 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1864 | 117 | 35 | 2 | 2018 | 154 | 7.63% | 141 | 7.56 |
| 2 | 1229 | 45 | 3 | 0 | 1277 | 48 | 3.76% | 71 | 5.78 |
| 3 | 1006 | 45 | 6 | 0 | 1057 | 51 | 4.82% | 56 | 5.57 |
| 4 | 707 | 27 | 12 | 0 | 746 | 39 | 5.23% | 45 | 6.36 |
| 5 | 838 | 34 | 2 | 0 | 874 | 36 | 4.12% | 48 | 5.73 |
| 6 | 938 | 52 | 4 | 1 | 995 | 57 | 5.73% | 56 | 5.97 |
| 7 | 878 | 90 | 7 | 3 | 978 | 100 | 10.22% | 48 | 5.47 |
| 8 | 627 | 32 | 10 | 1 | 670 | 43 | 6.42% | 34 | 5.42 |
| 9 | 703 | 51 | 8 | 2 | 764 | 61 | 7.98% | 54 | 7.68 |
| 10 | 690 | 38 | 1 | 1 | 730 | 40 | 5.48% | 42 | 6.09 |
| 11 | 1072 | 151 | 69 | 0 | 1292 | 220 | 17.03% | 61 | 5.69 |
| 12 | 962 | 40 | 8 | 0 | 1010 | 48 | 4.75% | 45 | 4.68 |
| 13 | 310 | 10 | 2 | 0 | 322 | 12 | 3.73% | 27 | 8.71 |
| 14 | 648 | 58 | 15 | 2 | 723 | 75 | 10.37% | 32 | 4.94 |
| 15 | 534 | 41 | 3 | 0 | 578 | 44 | 7.61% | 33 | 6.18 |
| 16 | 775 | 40 | 2 | 1 | 818 | 43 | 5.26% | 42 | 5.42 |
| 17 | 1080 | 60 | 4 | 0 | 1144 | 64 | 5.59% | 58 | 5.37 |
| 18 | 255 | 5 | 0 | 0 | 260 | 5 | 1.92% | 10 | 3.92 |
| 19 | 1318 | 77 | 12 | 1 | 1408 | 90 | 6.39% | 68 | 5.16 |
| 20 | 509 | 24 | 1 | 0 | 534 | 25 | 4.68% | 32 | 6.29 |
| 21 | 198 | 22 | 4 | 0 | 224 | 26 | 11.61% | 11 | 5.56 |
| 22 | 454 | 24 | 4 | 1 | 483 | 29 | 6.00% | 34 | 7.49 |
| X | 758 | 57 | 2 | 0 | 817 | 59 | 7.22% | 85 | 11.21 |
| Y | 33 | 7 | 1 | 0 | 41 | 8 | 19.51% | 1 | 3.03 |
| MT | 15 | 0 | 0 | 0 | 15 | 0 | 0.00% | 0 | 0.00 |
| ? | 2 | 4 | 0 | 0 | 6 | 4 | 66.67% | 0 | 0.00 |
| All | 18,403 | 1,151 | 215 | 15 | 19,784 | 1,381 | 1,134 |
Note: The total from neXtProt (18,403) differs slightly from the sum of all PE1 proteins (18,397), which includes six whose genes are duplicated on a second chromosome.
Table 4 also lists the PE1 proteins with unknown function (designated uPE1). A protein is classified as having unknown function if there are no associated Gene Ontology terms, or the only associated terms are so vague as not to be considered useful functional information (e.g. ATP binding, protein binding, nucleic acid binding) as defined in the neXtProt SPARQL query NXQ_00022 (https://www.nextprot.org/proteins/search?mode=advanced&queryId=NXQ_00022). Efforts to determine the function of more of these uPE1 proteins are discussed below in the Grand Challenge section.
There has recently been substantial progress in characterizing the translation of non-canonical open reading frames (ncORFs). An initial set of 7264 ncORFs with high-quality evidence of ribosome activity from Ribo-Seq assays by Mudge et. al.26 has led to a concerted effort by PeptideAtlas to search for these ORFs. The analysis is still ongoing but preliminary evidence in the 2023–01 PeptideAtlas build (https://peptideatlas.org/builds/human/) indicates that translation evidence from ncORFs is very sparse in ordinary tryptic digest data sets, with under a dozen detections in over 2 billion spectra searched. As mentioned earlier, HLA immunopeptidome data sets are highly enriched in peptides presented on the surface of cells by the HLA system; interestingly, already over 15% of these 7264 ncORFs appear to be detectable by MS. ncORFs are generally upstream, downstream, or internal but out-of-frame to the coding regions of the primary proteins of the human proteome. These detections raise the prospect that these might also be proteins in their own right and dramatically increase the size of the human proteome. Discussions in the community are still ongoing, but prevailing thoughts seem to be that these ncORFs may best be classified in a separate category and not be included in the primary pantheon of human proteins.27
Progress from other Components of the HPP
The HPP is organized into 25 teams by nuclear and mitochondrial chromosomes, 16 teams by biological and disease categories, and four resource pillars for antibody-based protein localization, MS, knowledge bases, and pathology. HUPO is developing a set of technology groups that may serve HPP as well as other initiatives. Here we report highlights from these HPP groups.
Biology and Disease-driven HPP
The Biology and Disease-driven Human Proteome Project (B/D-HPP) focuses on the strategies and the methods to determine and interpret the biological functions of human proteins within disease contexts. A common aim is to discover biomarkers that could guide and enable preventive and therapeutic interventions. As such, it is crucial to the new HPP Grand Challenge of assigning function(s) to every protein. The B/D-HPP has stimulated an exciting burst of activity from the international teams of scientists in 13 areas of biological and clinical relevance (see https://hupo.org/B/D-HPP) with new chairs, co-chairs, and committee members, two new initiatives, and three others under review (Table 5 and Figure 4). Moving forward, the B/D-HPP teams aim to collaborate and to draw upon the technology groups evolving in HUPO. Here we highlight recent accomplishments of several B/D-HPP teams.
Table 5.
Current active B/D HPP initiatives
| Status | Initiative |
|---|---|
| Updated | Cancer HPP, Human Glycoproteomics Initiative (HGI), Human Immuno-Peptidome Project (HIPP), Infectious Disease, Model Organism Proteomes (iMOP), Plasma (HPPP), Human Brain Proteome Project (HBPP), Rheumatoid Arthritis Disorders, Cardiovascular Initiative (CVI), Food & Nutrition (FaN) Proteomics, EyeOME |
| New | Single Cell (SCP), Metaproteomics |
| Under Review | Urine Proteome, Liber (HLPP), Proteomics and Large AI Models |
Figure 4.
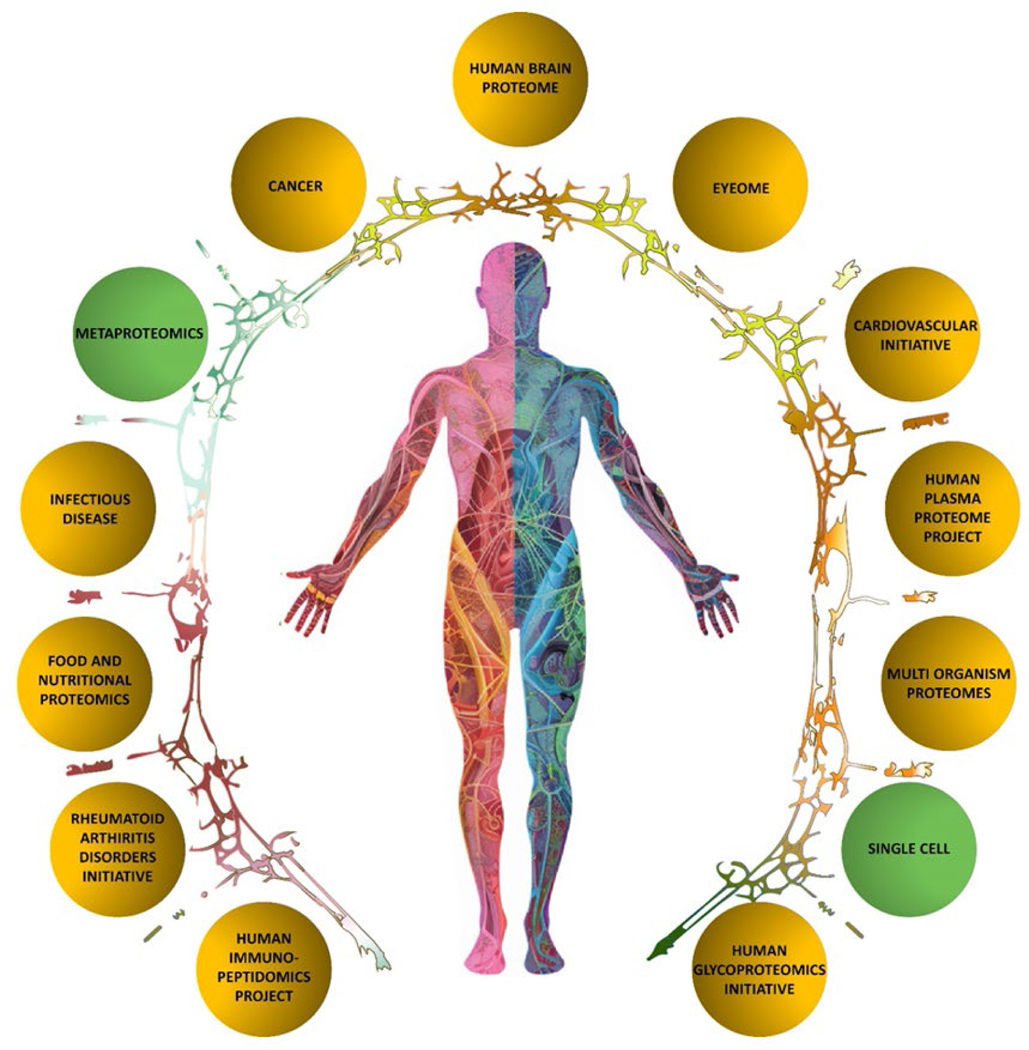
The B/D HPP Initiatives in 2023. New initiatives on single-cell proteomics and on meta-proteomics are shown in green. For details of leadership and progress, see the HPP website https://hupo.org/B/D-HPP.
The B/D HPP tradition of hosted webinars has continued in 2023 as part of the B/D-HPP strategy to disseminate and distribute knowledge about the B/D-HPP fields of activity. Two stimulating webinars held in 2023 drew 161 attendees for the Single Cell Webinar (May 2) and 41 attendees for the Aging & Disease Webinar (July 20). Each had a stellar group of speakers and lively discussions, which are available here: https://www.hupo.org/Webinars-and-Virtual-Presentations. These recordings had subsequently been viewed 5700 and 1100 times, respectively, as of July 2023.
Single Cell Proteomics (SCP).
Single Cell Proteomics (SCP) aims to better understand and differentiate the diversity of proteins expressed within individual cells, which is technically and bioinformatically challenging. In traditional proteomics, data from tissues or cells are averaged over many cells. Isolation of single cells traditionally by flow cytometers is now augmented by other devices that use different printing or aliquoting approaches, such as the HP100 printer and the CellenIon robotic piezo-pipettor combined with TMT isobaric labeling with carrier channels. For examples, up to 360 cells were quantified with TMT18 plex and 384 well plates in 24 hours.28 Several label-free approaches are now in use29 in combination with Bruker TimsTOF spectrometers or WISH-DIA-MS using the Thermo-Fisher Astral mass analyzer, as examples. The goal of the SCP initiative is to bring together the international community to develop transferable, transparent laboratory-based methods that can be coupled with new data analytic, statistical, and bioinformatic methods to provide molecular characterization of cellular heterogeneity at the single cell level and explain the heterogeneity of responses to drugs using clouds of isolated cells.
Metaproteomics.
The community-driven Metaproteomics Initiative, which joined B/D in June 2023, promotes dissemination of fundamentals, advances, and applications through collaborative networking in microbiome research. Its aim is to be the central information hub and open meeting place where newcomers and experts interact to communicate, standardize, and accelerate experimental and bioinformatic methodologies in this field.
This Initiative has launched a series of Critical Assessment of MetaProteome Investigations (CAMPI). The first was published in December 2021.30 CAMPI-2 investigates sample handling and stabilization as key challenges. Two different microbiota samples, human feces and soil, were shared across 10 laboratories, allowing five different protocols for protein identification to be compared. The results are expected to generate standard procedures for sample handling, reduce sampling bias, enhance sample stability from collection to protein extraction, and facilitate metaproteomic analyses of samples collected away from laboratory facilities. CAMPI-3 has focused on bioinformatics to identify the strengths and limitations of the workflows for taxonomic and functional profiles. Defined synthetic communities were produced with different levels of complexity and challenges for bioinformatic analyses. Raw MS files have been shared with many research groups to test their favorite bioinformatic pipelines. The results with these synthetic mixes should pave the way for international projects of broader scope and help establish workflow recommendations and enhance data transparency.
Human Glycoproteomics Initiative (HGI).
The glycosylation of proteins affects the biological functions and diseases of many tissues and organs. Analysis of these PTMs has always been difficult due to the complexity of the structures, the MS data obtained, and the quality of the software available to interpret the data. Therefore, to make glycopeptide analysis available to more proteomics researchers, the major focus of this initiative has been on providing a community-endorsed bioinformatic pipeline and data reporting criteria to support the interpretation of glycoproteomics results within the larger motivation of the B/D-HPP. This year, the HGI is comparing current software, both commercial and academic, available for the interpretation of serum glycopeptide mass spectrometric data.31 Following the successful first community evaluation of glycoproteomics informatics solutions, for serum glycopeptide analysis of the same MS data, the HGI’s second community challenge focuses on comparing and improving the bioinformatic tools for N- and O-glycopeptide identification and quantitation obtained on the same sample by different analytical workflows. Headed by Stacy Malaker (Yale) and Nick Riley (University of Washington), this study involves 20 software developer teams from around the world, with completion due in 2024. Additionally, the HGI community has teamed up with the Beilstein Institute in Germany to draft a set of glycoproteomics guidelines that dictate the minimum information required for a glycoproteomics experiment (MIRAGE) to standardize the reporting of glycoproteomics data. Final guidelines are expected early 2024.
EyeOME.
The EyeOME initiative aims to provide proteomics knowledge that may translate into direct applications for ophthalmologic diseases in high need of treatments and prevention, starting with blepharitis, central retinal vein occlusion (CRVO), and hereditary retinal disease.
Blepharitis is an inflammatory condition of the eyelid, often the first step in the development of severe dry eye disease or corneal infection. EyeOME members studied the tear film proteome of blepharitis and reported high levels of plakin proteins, indicating significant organizational changes to the cytoskeleton. These findings may open and guide experimental interventional approaches focused on this important family of proteins.32CRVO is caused by an impaired outflow from the major outflow vessel of the eye33 with visual loss related to ischemia and macular edema, an accumulation of fluid in the retinal area of high-resolution vision. Analyzing the ocular fluid from treatment-naïve patients with CRVO, EyeOME researchers uncovered the proteome of central retinal vein occlusion (CRVO).34 Inflammation is a significant driving force in the loss of visual function in CRVO. Upon treatment of an experimental porcine model of CRVO35 with a dexamethasone implant in the posterior ocular cavity, proteomic analyses identified a number of corticosteroid-sensitive proteins thought to modulate the inflammatory response.36Proteomic advances continue to bring insights for hereditary retinal diseases. Uncovering the retinal proteome of X-linked juvenile retinoschisis in a murine knockout model, Ambrosio et al. identified downregulation of phototransduction proteins as an under-recognized role of photoreceptor function.37
Initiative for Model Organism Proteomics (iMOP).
Through the use of various relevant biological models, iMOP aims to advance knowledge of proteins important for human health and disease, and more generally for the environment, as part of the “One-Health” concept and is in line with the expanded HUPO mission statement. Various projects carried out by iMOP members illustrate this broad spectrum of research. For example, the combination of metabolomics and discovery proteomics carried out on the black garden ant demonstrated that age and social environment both shape the molecular phenotype of animals.38 Targeted proteomics, in which 3001 dynamic MRM transitions were monitored, revealed the precise molecular effects of Cd, Ag, and Zn metal exposures on the caecal proteome of Gammarus fossarum.39 A new methodology has been proposed to identify the initiating molecular events when model cell lines are exposed to toxic substances,40 starting with the demonstration of altered solubility of the integral proteome of HepG2 cells exposed to tetrachlorinated dioxins. Proteomics has also identified a circadian clock in cyanobacteria, a key biological mechanism in new prokaryotic models.41 Proteogenomics can identify and characterize the structure of new coding sequences to better define their function through short open reading frames and their possible PTMs.42 New methodologies may improve the functional characterization of protein-protein interactomes.43 Overall, iMOP aims to federate expertise to improve structural and functional knowledge of proteins to answer fundamental biological questions using the most relevant biological model.
Human Immuno-Peptidome Project (HIPP).
Over the past two years, the HUPO-HIPP has successfully fostered and strengthened the immunopeptidomics community within HUPO. cThe HIPP has organized several high-profile webinars. The second HUPO-HIPP Summer School in Oxford, UK, in September 2022 addressed HLA-antibody specificities and their impact on sample quality, optimized workflows for improved identification of PTMs,44 advantages in automating sample preparation now emerging in specialized laboratories,45–47 improved HLA-peptide acquisition technology,48,49 and the importance of accurate and improved spectral annotation and validation strategies,50,51 specifically in the context of tumor-specific, non-canonical peptide discovery.52,53 The 3rd HUPO-HIPP Summer School will take place in Montreal in August 2024. The HIPP organized a pre-HUPO Congress workshop in Korea on the non-canonical proteome – a novel class of clinically targetable T cell antigens in conjunction with genome annotation and Ribo-Seq experts. The aim is to identify optimal strategies for identifying unannotated translation products within the human proteome from non-canonical open reading frames.
Cancers.
Proteomics continues to play an important role in understanding the underlying biology for multiple cancers, including renal cell carcinomas, colorectal cancers, triple-negative breast cancers, intrahepatic cholangiocarcinomas, and leukemias. There has been continued emphasis on serum and/or plasma proteomics, frequently assisted by advances in technology and bioinformatics. Multiomics-based protocols [genomics, transcriptomics, proteomics, phosphoproteomics, metabolomics] are established in the Clinical Proteomics Tumor Analysis Consortium (CPTAC) and the International Clinical Proteomics Consortium (ICPC). Human blood proteoform atlases from the Human Protein Atlas are identifying biomarkers for development of personalized/precision medicine. PTM analyses are a significant interest, with phosphorylation and glycosylation most widely studied,54,55 as well as acetylation, ubiquitinylation, and persulfidation.56 Top-down proteomics also has been used. Protein-protein interaction studies provide insight into normal and disease-related biological pathways. An integrated model of the formation and architecture of a large signalosome, the TNF-receptor signaling complex (TNF-RSC), linked the modular proteome with cellular function.57 Reactive oxygen species cause metabolic disturbances and signaling aberrations which promote malignant progression and provide a rationale for targeting oxidative stress for cancer treatment.58 Metabolomics has revealed biomarkers for diagnosis and monitoring of therapeutic response.59,60
The role of the microbiome in a range of pathologies, including cancers, is of interest61 as is single cell proteomics driven by improvements in instrument design, sample preparation, separation techniques, and use of AI.62 These topics are now new initiatives in the B/D HPP.
Drug repurposing is increasingly becoming an attractive proposition since it may lower overall development costs and shorten development timelines, especially if drug safety has already been established. Proteomics can aid such studies.63,64 Mass cytometry characterized the single-cell signaling profiles of 62 breast cancer cell lines and five healthy controls and accurately predicted drug sensitivity.65
Human Brain Proteome Project (HBPP).
The HBPP (https://hupo.org/brain) comprises a diverse group of scientists promoting the connections of neuroproteomics and actively encouraging the inclusion of young investigators. Annual HBPP workshops around the world have congregated and connected the international neuroproteomics community. In 2019 and 2021 the HBPP workshops were online, while the 2020 workshop was canceled due to the pandemic. The 32nd HUPO Human Brain Proteome Project Workshop took place in person in May 2023 in São Sebastião, Brazil. Participants from four continents addressed clinical, animal, and cellular findings for neurodegeneration and neuropsychiatric conditions.. Researchers unraveled the mechanisms involved in SARS-CoV-2 brain infection during COVID-19.66 Synaptic deficits in brains from people with schizophrenias were elegantly shown with synaptosome proteomics.67 Large-scale protein arrays identified serum protein patterns in bipolar disorder patients.68 For scientists in Latin America, hosting an international scientific event marked another stride towards enhancing equity in science. The 33rd HUPO HBPP workshop will be held in Dublin, Ireland, in May 2024.
Cardiovascular Initiative.
The Cardiovascular Initiative (CVI, hupo.org/CVI) continues to develop improved methodologies that enhance our understanding of heart and vascular physiology and disease. Highlights include: (1) a high-throughput standardized workflow for detecting biomarkers from naïve plasma, depleted plasma, and dried blood;69 (2) an analysis of the platelet proteome during myocardial infarction, discovering that protein S100A8/A9 is released from neutrophils and taken up by platelets, making neutrophils potential targets for prevention of thrombotic complications in cardiovascular disease;70 (3) using the novel CellSurfer platform to generate the first experimental map of cell surface glycoproteins on human cardiac cells, revealing cell type- and region-restricted proteins,71 and to compare the surfaceome among pluripotent stem cell derivatives and their primary cardiac counterparts, with implications for drug screening and disease modeling; (4) accurately measuring protein half-life in vivo in different mouse tissues while addressing complications from label kinetics,72 to understand how regulation of mRNA translation drives cardiac hypertrophy; and (5) refinement of top-down mass spectrometry-based cardioproteomics to assess full-length proteoforms in cardiac sarcomeres in diseases.73
Human Plasma Proteome Project (HPPP).
The Human Plasma Proteome Project74 released two new PeptideAtlas builds in 2023 based on an ensemble reanalysis with the Trans-Proteomics Pipeline75,76 of many data-dependent acquisition mass spectrometry experiments deposited to ProteomeXchange77,78 repositories. The first comprises 113 ProteomeXchange data sets (PXDs) derived from plasma and serum samples, yielding 108 million peptide-spectrum matches (PSMs) and 4608 canonical proteins (https://peptideatlas.org/builds/human/plasma/). The second contains 33 PXDs derived from circulating extracellular vesicle-enriched samples, yielding 10 million peptide-spectrum matches (PSMs) and 4985 canonical proteins (https://peptideatlas.org/builds/human/plasma_ev/). Manuscripts describing the results of these builds as well as our current understanding and prospects for learning more about the circulating proteome are in preparation. In addition, major efforts by the Human Pharma Plasma Proteomics Project (HPPPP) using antibody detection systems (Olink) have now processed over 54,000 samples from the UK Biobank plasma repository paving the way for population based studies of protein associations and variants and their impact on disease.79
Infectious Diseases.
The Infectious Disease Initiative has continued to promote technological advances needed for applying proteomics to pathogen infection studies, as well as to characterize the complex interactions between pathogens and hosts to discover drivers of infection-induced pathologies. A focus is understanding the array of responses elicited by host cells to prevent the spread of an infection, including immune signaling. In this vein, the use of targeted proteomics defined the absolute numbers of protein molecules per cell in the mouse macrophage pathogen recognition receptor and chemotaxis pathways, providing the means for pathway modeling and simulation.80 Combinatorial phosphorylation, protein interaction, and microscopy studies have demonstrated that the immune factor IFI16, upon recognition and binding to viral DNA, undergoes phosphorylation-controlled liquid-liquid phase separation that regulates innate immune signaling.81 To counteract such host responses, viruses have acquired a range of mechanisms for immune evasion. Demonstrating the value of understanding N-terminal protein processing, viral 3C proteases from poliovirus and coxsackievirus B3 were shown to target and cleave 14–3-3ε to inhibit its functions and prevent the activation of host antiviral RIG-I signaling.82 Crosslinking mass spectrometry and quantitative proteomics spatially resolved a protein interactome map of human cytomegalovirus virions, demonstrating the layer-specific incorporation of host proteins.83
This year marked the first proteomic investigation of a virus microenvironment formed via cell-to-cell communication following a viral infection. A herpesvirus infection primes the cell cycle state of neighboring cells to facilitate subsequent rounds of viral infection with the same virus or other nuclear-replicating viruses, while cells farther away from the site of infection displayed increased levels of immune factors.84 From continued work on the COVID-19 pandemic, databases were constructed for the MS identification of mutated peptides from SARS-CoV-2 variants across different geographical regions.85 A MALDI-ToF-MS method was optimized for the rapid detection of the SARS-CoV-2 nucleoprotein at a level of 8 amol/μl.86 Demonstrating the significance of the clinical application of proteomics, analysis of diverse clinical specimens from thousands of patients identified proteomic signatures that can distinguish between severe and mild disease outcomes.87 Tissue-specific alterations in host proteomes were discovered, pointing to opportunities for organ-specific therapeutic interventions.88
Antibody Pillar
The Antibody Resource Pillar works in close collaboration with the Human Protein Atlas (HPA) project, www.proteinatlas.org, one of the world’s largest biological resources, that maps the human proteome based on transcriptomics and spatial antibody-based imaging. The open-access data are updated on a yearly basis; in HPA version 23 that builds upon the Ensembl version 109 genome release for annotation of all protein-coding genes, the proteome analysis is based on 27,520 antibodies covering 17,288 unique proteins. The HPA now contains 12 main sections, comprehensively summarizing different aspects of the human proteome and transcriptome.
Since last year, three completely new sections have been added. The Disease section allows exploration of protein levels and proteome signatures in blood in patients with different diseases. In the first release of this new section, plasma profiles of 1,463 proteins from 1,400 cancer patients representing 12 major cancer types were measured in blood plasma using proximity extension assay (Olink) and targeted proteomics.89 Differential expression profiling and machine learning-based disease prediction highlighted proteins associated with each of the analyzed cancer types, constituting panels of proteins suitable for future diagnostic studies. The second newly added section of the HPA focuses on 3D protein structures for a majority of the human proteins. The data are based on predicted 3D structures from the AlphaFold Protein Structure database, together with experimentally determined structures from the Protein Data Bank (PDB). The interactive visualization allows for exploring antigens corresponding to many of the antibodies analyzed in the HPA, and also includes clinical and population variants displayed on the structures. Finally, the HPA now has a section on interactions, displaying protein-protein interaction data from the EMBL-EBI IntAct database for 11,351 proteins, together with metabolic maps for 2,912 proteins.90 All networks are interactive and can be integrated with other data in the HPA, allowing for exploration of interactions in relation to, e.g., subcellular localization or tissue specificity.
In addition to the three newly added sections, major updates have been introduced in the Tissue section, focusing on cell type specific expression levels of proteins with a spatial resolution covering all major human tissues. With help from single cell RNA sequencing data,91 subsets of cells that are challenging to distinguish by the human eye have been identified, and multiplex antibody profiles mapping these different subsets have been created. By using such multiplex panels together with proteins that lack previous data on function or cell type specificity, their exact location can be mapped. Since some proteins are expressed only in a specific cell state within a short period of time, in some cases linked to certain cell cycle events such as mitosis or meiosis, mapping the exact spatial localization to these structures adds important insights on potential protein function. This newly generated data will likely be of major importance for the HPP Grand Challenge, linking the detailed spatial localization to quantitative protein data produced by other methods. At present, 742 proteins have been analyzed in 16 cell types in testis and five cell types in kidney using the novel multiplex protein profiling workflow; more organs and protein targets will be included in future versions of the HPA.
Several ongoing projects related to the Tissue section focus on temporal changes in protein expression, e.g., in relation to life-span or the menstrual cycle. With the addition of more specialized samples, proteins that could not be mapped using the previous tissue collection have been successfully analyzed. One such example is extended analysis of ovarian samples from women of fertile age, that allowed for localizing multiple proteins defined as “missing proteins” to various structures of follicle cells.92 As a follow-up to this study, four new ovary RNA samples have been sequenced, including samples from prepubertal women and young adults, from whom oocytes were captured and analyzed, as well as granulosa, stromal, and follicular cells.93 This paper is an outstanding example of biology-driven studies yielding several previously missing proteins. Paraffin blocks from the same individuals are available for antibody-based profiling, opening up unique possibilities to identify and validate protein candidates expressed in these rare structures that are challenging to target with other methods.
Pathology Pillar
Pathology is the core medical discipline to bring proteomics to patients via a new class of “postgenomic” protein-directed assays for early disease detection, risk prediction, choice of therapy and combination therapies, and surveillance. Combined with improvements in instrument design,94 orthogonal95,96 and multiomic approaches,97–100, rapid progress in advanced computational capabilities including AI and machine learning76,101–105 can now facilitate the development of personalized/precision medicine106 for the improvement of human health.101,106,107
The U.S. National Cancer Institute’s Clinical Proteomics Tumor Analysis Consortium (CPTAC) and International Cancer Proteomics Consortium (ICPC) have set standards, established procedures, provided reagents, and populated massive datasets on more than a dozen major types of cancers (see Cancer B-D/HPP report). Proteomics has also advanced insights into COVID-19,85,108,109 cardiovascular76,77 and neurological disorders, and diseases of the eye. New workflows are being developed for working with low-level44,49,110–112 and low-volume samples.29,113 Network analysis continues to inform studies on disease-related pathways.114–117
The microbiome, now considered as an “organ” in its own right,118 plays key roles in multiple pathologies. Understanding cellular heterogeneity in cancers and other diseases can have important clinical applications, including the development of improved diagnostic tools and targeted treatments.119 Best practices, quality controls, and data-reporting recommendations assist in the broad adoption of reliable quantitative workflows for single-cell proteomics.120
HUPO Human Proteome Project Grand Challenge
The HPP Grand Challenge to determine a function or multiple functions for every human protein continues to gather momentum since its introduction in 2021 as a recommendation of the HPP Scientific Advisory Board, chaired by Ruedi Aebersold.5 Several countries have now taken on projects to focus on the HPP Grand Challenge, including China, France, and other interested consortia. Charles Pineau, HPP Chair (2022–2024), presented the Human Proteome Project and the HPP Grand Challenge to the French National Research Agency (ANR) to stimulate interest in funding applications to address the Grand Challenge goals. Several French teams from various biological and clinical fields submitted projects for funding from the ANR. In July 2023, one project was funded and initiated. Hopefully the next few years will see multiple projects funded by the French ANR in connection with the HPP Grand Challenge. The Grand Challenge also was highlighted during the annual proteomics congress organized by the French, Spanish, and Portuguese societies at ProteoAix 2023.
In 2023, an initiative was launched between the HPP and ChemBioFrance, one of the French national research infrastructures. ChemBioFrance draws upon four different resources and networks: the national chemical library, a network of screening platforms, a distributed chemoinformatics platform, and a network of ADME platforms. A first pilot call entitled “A Protein, a Ligand, a Function” was launched. Four projects were selected, with a second round of funding planned for the fall of 2023. Selected projects aim at using a biologically active small molecule from the national chemical library to alter the proteome of a specific cell line and study at the global level the variation of protein abundances in response to the compound using a differential proteomics strategy. Several concentrations will be explored in order to estimate the specific effects of the compound over a 100-fold concentration range. Proteomic and data analyses are carried out by one of the French core facilities involved in the HPP. The first results were presented at HUPO 2023 in Busan, Korea; protocols and data repository will be considered as a general framework for HPP Grand Challenge results. Productive discussions at HUPO 2022 in Cancun also included the incorporation of new findings using AlphaFold2 and I-TASSER/COFACTOR algorithms to predict functions and provide guidance for experimentalists.
After extensive discussion with the Grand Challenge Project team and collaborators across the community, Tiannan Guo (Westlake University, China) developed a new project called ProtTalks. They cultured 15 triple-negative breast cancer (TNBC) cell lines and two non-TNBC cell lines, and perturbed them by in vitro exposure for 6, 24, and 48 hours with each of 63 clinically-approved drugs for patients with such tumors. They applied microflow short-gradient DIA-MS to acquire the proteome before and after perturbation, with biological triplicates. Also included were breast cancer cells perturbed with drug combinations and with CRISPR/Cas9 technology. A QC system has been established and a total of over 16,000 high-quality perturbed proteomes have been acquired. Artificial intelligence (AI) methods applied to this data resource will help reveal the functions of targeted proteins and pathways, and their sophisticated interactions. Guo et al have called this research domain “perturbation proteomics”, in line with recommendations of the HPP SAB Report that stimulated this Grand Challenge on Function.5 The aim is to perturb biological systems with chemical or biological agents or genetic technologies, and record the proteome before and after the perturbation for understanding the complex systems biology outcomes in interactions and consequences of these perturbations. This work was presented in the HUPO 2023 World Congress in Busan, Korea.
The π-HuB consortium led by Fuchu He from the Beijing Proteome Research Center achieved quite significant progress during the past year with establishment of the pilot center in Guangzhou housing 17 mass spectrometers and over 100 staff. A permanent 300,000 sq/ft purpose-built center will be completed in 2024 on Bio-Island, Guangzhou as the central home of π-HuB for both national and international proteomics research. A collaboration with over 100 proteomics scientists, clinicians, and computer scientists in China and outside China produced a white paper v1.0 which describes the rationale and goals of this 30-year program. Briefly, the ultimate goals of the π-HuB Consortium are 1) charting a ‘reference-space’ of the human proteome, 2) defining diverse ‘state-spaces’ for human proteome dynamics, and 3) building the π-HuB navigator with the ultimate goal of building knowledge towards proteomics driven Phronesis Medicine. The success of this project will depend on human biospecimens, technology innovations, Big-Science infrastructure, open resources, and participating researchers from related fields. The first ten years will focus on building cell-type resolved proteome atlases, life-oriented adaptive proteome atlases, organ-centric disease proteome atlases, and the π-HuB navigator version 1.0. The success of the project will rely on patient advocacy. Active discussions of the organization and implementation of these goals are ongoing. It is very likely that this enormous project will fuel major progress in clinical proteomics and in precision medicine.
Conclusion
The HUPO Human Proteome Project continues to demonstrate progress throughout the global proteomics community in credibly identifying expression of predicted proteins not previously observed, using a widening range of specimens. Its Biology and Disease-driven HPP teams have been refreshed and have expanded their research and outreach. The HPP Grand Challenge to find function(s) for every human protein is gaining traction toward making a significant contribution to precision medicine.
Supplementary Material
Acknowledgements
We appreciate the guidance from the HPP Executive Committee and the participation of all HPP investigators. We thank the UniProt groups at SIB, EBI, and PIR for providing high-quality annotations for the human proteins in UniProtKB/Swiss-Prot. The neXtProt server is hosted at SIB Swiss Institute of Bioinformatics in Switzerland, ProteomeXchange and PRIDE at the European Bioinformatics Institute in Cambridge, UK, PeptideAtlas at the Institute for Systems Biology in Seattle, and MassIVE-KB at the University of California San Diego. G.S.O. acknowledges support from National Institutes of Health Grants P30ES017885–11-S1 and U24CA271037; E.W.D. and R.L.M. from National Institutes of Health Grants R01GM087221, R24GM127667, U19AG023122, S10OD026936, and from National Science Foundation Grant DBI-1933311; C.M.O. by Canadian Institutes of Health Research Foundation Grant 148408 and a Canada Research Chair in Protease Proteomics and Systems Biology; N.B. from NIH grant R01LM013115, and NSF grant ABI1759980; C.L. by the Knut and Alice Wallenberg Foundation for the Human Protein Atlas; and M.H.R by National Institutes of Health Grants R21CA263262, U01CA253217, R21CA251992, P30CA008748 (MSKCC CCSG, Pathology Component), NIH-Leidos CPTAC contract 17X173, and Farmer Family Foundation. M.H.A.R. acknowledges grants from the NIH/NCI (R21CA251992, R21CA263262, and U01CA263986), a Cycle for Survival Equinox Innovation Grant, and an Investigator Grant from the Neuroendocrine Tumor Research Foundation (NETRF).
Footnotes
Author Information
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
T.G. is the founder of Westlake Omics Inc. The other authors declare no competing financial interest.
Supporting Information
Supplementary Material: Olfactory receptor OR51E2 (Q9H255) in PeptideAtlas
References
- (1).Hanash S; Celis JE The Human Proteome Organization: A Mission to Advance Proteome Knowledge. Mol. Cell. Proteomics MCP 2002, 1 (6), 413–414. [DOI] [PubMed] [Google Scholar]
- (2).Legrain P; Aebersold R; Archakov A; Bairoch A; Bala K; Beretta L; Bergeron J; Borchers CH; Corthals GL; Costello CE; Deutsch EW; Domon B; Hancock W; He F; Hochstrasser D; Marko-Varga G; Salekdeh GH; Sechi S; Snyder M; Srivastava S; Uhlén M; Wu CH; Yamamoto T; Paik Y-K; Omenn GS The Human Proteome Project: Current State and Future Direction. Mol. Cell. Proteomics 2011, 10 (7), M111.009993. 10.1074/mcp.M111.009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Adhikari S; Nice EC; Deutsch EW; Lane L; Omenn GS; Pennington SR; Paik Y-K; Overall CM; Corrales FJ; Cristea IM; Van Eyk JE; Uhlén M; Lindskog C; Chan DW; Bairoch A; Waddington JC; Justice JL; LaBaer J; Rodriguez H; He F; Kostrzewa M; Ping P; Gundry RL; Stewart P; Srivastava S; Srivastava S; Nogueira FCS; Domont GB; Vandenbrouck Y; Lam MPY; Wennersten S; Vizcaino JA; Wilkins M; Schwenk JM; Lundberg E; Bandeira N; Marko-Varga G; Weintraub ST; Pineau C; Kusebauch U; Moritz RL; Ahn SB; Palmblad M; Snyder MP; Aebersold R; Baker MS A High-Stringency Blueprint of the Human Proteome. Nat. Commun. 2020, 11 (1), 5301. 10.1038/s41467-020-19045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Omenn GS Reflections on the HUPO Human Proteome Project, the Flagship Project of the Human Proteome Organization, at 10 Years. Mol. Cell. Proteomics 2021, 20, 100062. 10.1016/j.mcpro.2021.100062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Omenn GS; Lane L; Overall CM; Pineau C; Packer NH; Cristea IM; Lindskog C; Weintraub ST; Orchard S; Roehrl MHA; Nice E; Liu S; Bandeira N; Chen Y-J; Guo T; Aebersold R; Moritz RL; Deutsch EW The 2022 Report on the Human Proteome from the HUPO Human Proteome Project. J. Proteome Res. 2023, 22 (4), 1024–1042. 10.1021/acs.jproteome.2c00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Desiere F; Deutsch EW; Nesvizhskii AI; Mallick P; King NL; Eng JK; Aderem A; Boyle R; Brunner E; Donohoe S; Fausto N; Hafen E; Hood L; Katze MG; Kennedy KA; Kregenow F; Lee H; Lin B; Martin D; Ranish JA; Rawlings DJ; Samelson LE; Shiio Y; Watts JD; Wollscheid B; Wright ME; Yan W; Yang L; Yi EC; Zhang H; Aebersold R. Integration with the Human Genome of Peptide Sequences Obtained by High-Throughput Mass Spectrometry. Genome Biol. 2005, 6 (1), R9. 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Desiere F; Deutsch EW; King NL; Nesvizhskii AI; Mallick P; Eng J; Chen S; Eddes J; Loevenich SN; Aebersold R. The PeptideAtlas Project. Nucleic Acids Res. 2006, 34 (Database issue), D655–658. 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Pullman BS; Wertz J; Carver J; Bandeira N. ProteinExplorer: A Repository-Scale Resource for Exploration of Protein Detection in Public Mass Spectrometry Data Sets. J. Proteome Res. 2018, 17 (12), 4227–4234. 10.1021/acs.jproteome.8b00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zahn-Zabal M; Michel P-A; Gateau A; Nikitin F; Schaeffer M; Audot E; Gaudet P; Duek PD; Teixeira D; Rech de Laval V; Samarasinghe K; Bairoch A; Lane L. The neXtProt Knowledgebase in 2020: Data, Tools and Usability Improvements. Nucleic Acids Res. 2020, 48 (D1), D328–D334. 10.1093/nar/gkz995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Uhlén M; Fagerberg L; Hallström BM; Lindskog C; Oksvold P; Mardinoglu A; Sivertsson Å; Kampf C; Sjöstedt E; Asplund A; Olsson I; Edlund K; Lundberg E; Navani S; Szigyarto CA-K; Odeberg J; Djureinovic D; Takanen JO; Hober S; Alm T; Edqvist P-H; Berling H; Tegel H; Mulder J; Rockberg J; Nilsson P; Schwenk JM; Hamsten M; von Feilitzen K; Forsberg M; Persson L; Johansson F; Zwahlen M; von Heijne G; Nielsen J; Pontén F. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347 (6220), 1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- (11).Orchard S; Hermjakob H; Apweiler R. The Proteomics Standards Initiative. Proteomics 2003, 3 (7), 1374–1376. 10.1002/pmic.200300496. [DOI] [PubMed] [Google Scholar]
- (12).Deutsch EW; Vizcaíno JA; Jones AR; Binz P-A; Lam H; Klein J; Bittremieux W; Perez-Riverol Y; Tabb DL; Walzer M; Ricard-Blum S; Hermjakob H; Neumann S; Mak TD; Kawano S; Mendoza L; Van Den Bossche T; Gabriels R; Bandeira N; Carver J; Pullman B; Sun Z; Hoffmann N; Shofstahl J; Zhu Y; Licata L; Quaglia F; Tosatto SCE; Orchard SE Proteomics Standards Initiative at Twenty Years: Current Activities and Future Work. J. Proteome Res. 2023, 22 (2), 287–301. 10.1021/acs.jproteome.2c00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Consortium UniProt. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51 (D1), D523–D531. 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Deutsch EW; Overall CM; Van Eyk JE; Baker MS; Paik Y-K; Weintraub ST; Lane L; Martens L; Vandenbrouck Y; Kusebauch U; Hancock WS; Hermjakob H; Aebersold R; Moritz RL; Omenn GS Human Proteome Project Mass Spectrometry Data Interpretation Guidelines 2.1. J. Proteome Res. 2016, 15 (11), 3961–3970. 10.1021/acs.jproteome.6b00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Deutsch EW; Lane L; Overall CM; Bandeira N; Baker MS; Pineau C; Moritz RL; Corrales F; Orchard S; Van Eyk JE; Paik Y-K; Weintraub ST; Vandenbrouck Y; Omenn GS Human Proteome Project Mass Spectrometry Data Interpretation Guidelines 3.0. J. Proteome Res. 2019, 18 (12), 4108–4116. 10.1021/acs.jproteome.9b00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bowler-Barnett EH; Fan J; Luo J; Magrane M; Martin MJ; Orchard S; UniProt Consortium. UniProt and Mass Spectrometry-Based Proteomics-A 2-Way Working Relationship. Mol. Cell. Proteomics MCP 2023, 22 (8), 100591. 10.1016/j.mcpro.2023.100591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Morales J; Pujar S; Loveland JE; Astashyn A; Bennett R; Berry A; Cox E; Davidson C; Ermolaeva O; Farrell CM; Fatima R; Gil L; Goldfarb T; Gonzalez JM; Haddad D; Hardy M; Hunt T; Jackson J; Joardar VS; Kay M; Kodali VK; McGarvey KM; McMahon A; Mudge JM; Murphy DN; Murphy MR; Rajput B; Rangwala SH; Riddick LD; Thibaud-Nissen F; Threadgold G; Vatsan AR; Wallin C; Webb D; Flicek P; Birney E; Pruitt KD; Frankish A; Cunningham F; Murphy TD A Joint NCBI and EMBL-EBI Transcript Set for Clinical Genomics and Research. Nature 2022, 604 (7905), 310–315. 10.1038/s41586-022-04558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Marcu A; Bichmann L; Kuchenbecker L; Kowalewski DJ; Freudenmann LK; Backert L; Mühlenbruch L; Szolek A; Lübke M; Wagner P; Engler T; Matovina S; Wang J; Hauri-Hohl M; Martin R; Kapolou K; Walz JS; Velz J; Moch H; Regli L; Silginer M; Weller M; Löffler MW; Erhard F; Schlosser A; Kohlbacher O; Stevanović S; Rammensee H-G; Neidert MC HLA Ligand Atlas: A Benign Reference of HLA-Presented Peptides to Improve T-Cell-Based Cancer Immunotherapy. J. Immunother. Cancer 2021, 9 (4), e002071. 10.1136/jitc-2020-002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Forlani G; Michaux J; Pak H; Huber F; Marie Joseph EL; Ramia E; Stevenson BJ; Linnebacher M; Accolla RS; Bassani-Sternberg M. CIITA-Transduced Glioblastoma Cells Uncover a Rich Repertoire of Clinically Relevant Tumor-Associated HLA-II Antigens. Mol. Cell. Proteomics MCP 2021, 20, 100032. 10.1074/mcp.RA120.002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Gfeller D; Guillaume P; Michaux J; Pak H-S; Daniel RT; Racle J; Coukos G; Bassani-Sternberg M. The Length Distribution and Multiple Specificity of Naturally Presented HLA-I Ligands. J. Immunol. Baltim. Md 1950 2018, 201 (12), 3705–3716. 10.4049/jimmunol.1800914. [DOI] [PubMed] [Google Scholar]
- (21).Sanz-Bravo A; Alvarez-Navarro C; Martín-Esteban A; Barnea E; Admon A; López de Castro JA Ranking the Contribution of Ankylosing Spondylitis-Associated Endoplasmic Reticulum Aminopeptidase 1 (ERAP1) Polymorphisms to Shaping the HLA-B*27 Peptidome. Mol. Cell. Proteomics MCP 2018, 17 (7), 1308–1323. 10.1074/mcp.RA117.000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Na CH; Barbhuiya MA; Kim M-S; Verbruggen S; Eacker SM; Pletnikova O; Troncoso JC; Halushka MK; Menschaert G; Overall CM; Pandey A. Discovery of Noncanonical Translation Initiation Sites through Mass Spectrometric Analysis of Protein N Termini. Genome Res. 2018, 28 (1), 25–36. 10.1101/gr.226050.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Pedersen A-K; Pfeiffer A; Karemore G; Akimov V; Bekker-Jensen DB; Blagoev B; Francavilla C; Olsen JV Proteomic Investigation of Cbl and Cbl-b in Neuroblastoma Cell Differentiation Highlights Roles for SHP-2 and CDK16. iScience 2021, 24 (4), 102321. 10.1016/j.isci.2021.102321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Bu F; Cheng Q; Zhang Y; Zhang X; Yan K; Liu F; Li Z; Lu X; Ren Y; Liu S. Discovery of Missing Proteins from an Aneuploidy Cell Line Using a Proteogenomic Approach. J. Proteome Res. 2021, 20 (12), 5329–5339. 10.1021/acs.jproteome.1c00772. [DOI] [PubMed] [Google Scholar]
- (25).Carapito C; Duek P; Macron C; Seffals M; Rondel K; Delalande F; Lindskog C; Fréour T; Vandenbrouck Y; Lane L; Pineau C. Validating Missing Proteins in Human Sperm Cells by Targeted Mass-Spectrometry- and Antibody-Based Methods. J. Proteome Res. 2017, 16 (12), 4340–4351. 10.1021/acs.jproteome.7b00374. [DOI] [PubMed] [Google Scholar]
- (26).Mudge JM; Ruiz-Orera J; Prensner JR; Brunet MA; Calvet F; Jungreis I; Gonzalez JM; Magrane M; Martinez TF; Schulz JF; Yang YT; Albà MM; Aspden JL; Baranov PV; Bazzini AA; Bruford E; Martin MJ; Calviello L; Carvunis A-R; Chen J; Couso JP; Deutsch EW; Flicek P; Frankish A; Gerstein M; Hubner N; Ingolia NT; Kellis M; Menschaert G; Moritz RL; Ohler U; Roucou X; Saghatelian A; Weissman JS; van Heesch S. Standardized Annotation of Translated Open Reading Frames. Nat. Biotechnol. 2022, 40 (7), 994–999. 10.1038/s41587-022-01369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Prensner JR; Abelin JG; Kok LW; Clauser KR; Mudge JM; Ruiz-Orera J; Bassani-Sternberg M; Moritz RL; Deutsch EW; van Heesch S. What Can Ribo-Seq, Immunopeptidomics, and Proteomics Tell Us about the Non-Canonical Proteome? Mol. Cell. Proteomics MCP 2023, 100631. 10.1016/j.mcpro.2023.100631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ahmad R; Budnik B. A Review of the Current State of Single-Cell Proteomics and Future Perspective. Anal. Bioanal. Chem. 2023. 10.1007/s00216-023-04759-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Matzinger M; Müller E; Dürnberger G; Pichler P; Mechtler K. Robust and Easy-to-Use One-Pot Workflow for Label-Free Single-Cell Proteomics. Anal. Chem. 2023, 95 (9), 4435–4445. 10.1021/acs.analchem.2c05022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Van Den Bossche T; Arntzen MØ; Becher D; Benndorf D; Eijsink VGH; Henry C; Jagtap PD; Jehmlich N; Juste C; Kunath BJ; Mesuere B; Muth T; Pope PB; Seifert J; Tanca A; Uzzau S; Wilmes P; Hettich RL; Armengaud J. The Metaproteomics Initiative: A Coordinated Approach for Propelling the Functional Characterization of Microbiomes. Microbiome 2021, 9 (1), 243. 10.1186/s40168-021-01176-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Kawahara R; Chernykh A; Alagesan K; Bern M; Cao W; Chalkley RJ; Cheng K; Choo MS; Edwards N; Goldman R; Hoffmann M; Hu Y; Huang Y; Kim JY; Kletter D; Liquet B; Liu M; Mechref Y; Meng B; Neelamegham S; Nguyen-Khuong T; Nilsson J; Pap A; Park GW; Parker BL; Pegg CL; Penninger JM; Phung TK; Pioch M; Rapp E; Sakalli E; Sanda M; Schulz BL; Scott NE; Sofronov G; Stadlmann J; Vakhrushev SY; Woo CM; Wu H-Y; Yang P; Ying W; Zhang H; Zhang Y; Zhao J; Zaia J; Haslam SM; Palmisano G; Yoo JS; Larson G; Khoo K-H; Medzihradszky KF; Kolarich D; Packer NH; Thaysen-Andersen M. Community Evaluation of Glycoproteomics Informatics Solutions Reveals High-Performance Search Strategies for Serum Glycopeptide Analysis. Nat. Methods 2021, 18 (11), 1304–1316. 10.1038/s41592-021-01309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Cehofski LJ; Honoré B; Vorum H. A Review: Proteomics in Retinal Artery Occlusion, Retinal Vein Occlusion, Diabetic Retinopathy and Acquired Macular Disorders. Int. J. Mol. Sci. 2017, 18 (5), 907. 10.3390/ijms18050907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Cehofski LJ; Kojima K; Kusada N; Rasmussen M; Muttuvelu DV; Grauslund J; Vorum H; Honoré B. Macular Edema in Central Retinal Vein Occlusion Correlates With Aqueous Fibrinogen Alpha Chain. Invest. Ophthalmol. Vis. Sci. 2023, 64 (2), 23. 10.1167/iovs.64.2.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Cehofski LJ; Kruse A; Mæng MO; Sejergaard BF; Schlosser A; Sorensen GL; Grauslund J; Honoré B; Vorum H. Dexamethasone Intravitreal Implant Is Active at the Molecular Level Eight Weeks after Implantation in Experimental Central Retinal Vein Occlusion. Mol. Basel Switz. 2022, 27 (17), 5687. 10.3390/molecules27175687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Mæng MO; Roshanth N; Kruse A; Nielsen JE; Kjærgaard B; Honoré B; Vorum H; Cehofski LJ Laser-Induced Porcine Model of Experimental Retinal Vein Occlusion: An Optimized Reproducible Approach. Med. Kaunas Lith. 2023, 59 (2), 243. 10.3390/medicina59020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Muttuvelu DV; Cehofski LJ; Muhammad MGF; Chen X; Utheim TP; Khan AM; Abduljabar AB; Kristensen K; Rasmussen MLR; Vorum H; Heegaard S; Honoré B. Anterior Blepharitis Is Associated with Elevated Plectin Levels Consistent with a Pronounced Intracellular Response. Ocul. Surf. 2023, 29, 444–455. 10.1016/j.jtos.2023.06.010. [DOI] [PubMed] [Google Scholar]
- (37).Ambrosio L; Akula JD; Harman JC; Arellano IA; Fulton AB Do the Retinal Abnormalities in X-Linked Juvenile Retinoschisis Include Impaired Phototransduction? Exp. Eye Res. 2023, 234, 109591. 10.1016/j.exer.2023.109591. [DOI] [PubMed] [Google Scholar]
- (38).Quque M; Brun C; Villette C; Sueur C; Criscuolo F; Heintz D; Bertile F. Both Age and Social Environment Shape the Phenotype of Ant Workers. Sci. Rep. 2023, 13 (1), 186. 10.1038/s41598-022-26515-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Leprêtre M; Chaumot A; Aboud R; Delorme N; Espeyte A; Salvador A; Ayciriex S; Armengaud J; Coquery M; Geffard O; Degli-Esposti D. Dynamic Multiple Reaction Monitoring of Amphipod Gammarus Fossarum Caeca Expands Molecular Information for Understanding the Impact of Contaminants. Sci. Total Environ. 2023, 893, 164875. 10.1016/j.scitotenv.2023.164875. [DOI] [PubMed] [Google Scholar]
- (40).Lizano-Fallas V; Carrasco Del Amor A; Cristobal S. Prediction of Molecular Initiating Events for Adverse Outcome Pathways Using High-Throughput Identification of Chemical Targets. Toxics 2023, 11 (2), 189. 10.3390/toxics11020189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Géron A; Werner J; Wattiez R; Matallana-Surget S. Towards the Discovery of Novel Molecular Clocks in Prokaryotes. Crit. Rev. Microbiol. 2023, 1–13. 10.1080/1040841X.2023.2220789. [DOI] [PubMed] [Google Scholar]
- (42).Cassidy L; Kaulich PT; Tholey A. Proteoforms Expand the World of Microproteins and Short Open Reading Frame-Encoded Peptides. iScience 2023, 26 (2), 106069. 10.1016/j.isci.2023.106069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Garcia-Del Rio DF; Cardon T; Eyckerman S; Fournier I; Bonnefond A; Gevaert K; Salzet M. Employing Non-Targeted Interactomics Approach and Subcellular Fractionation to Increase Our Understanding of the Ghost Proteome. iScience 2023, 26 (2), 105943. 10.1016/j.isci.2023.105943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Abelin JG; Bergstrom EJ; Rivera KD; Taylor HB; Klaeger S; Xu C; Verzani EK; Jackson White C; Woldemichael HB; Virshup M; Olive ME; Maynard M; Vartany SA; Allen JD; Phulphagar K; Harry Kane M; Rachimi S; Mani DR; Gillette MA; Satpathy S; Clauser KR; Udeshi ND; Carr SA Workflow Enabling Deepscale Immunopeptidome, Proteome, Ubiquitylome, Phosphoproteome, and Acetylome Analyses of Sample-Limited Tissues. Nat. Commun. 2023, 14 (1), 1851. 10.1038/s41467-023-37547-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Li X; Pak HS; Huber F; Michaux J; Taillandier-Coindard M; Altimiras ER; Bassani-Sternberg M. A Microfluidics-Enabled Automated Workflow of Sample Preparation for MS-Based Immunopeptidomics. Cell Rep. Methods 2023, 3 (6), 100479. 10.1016/j.crmeth.2023.100479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Lim Kam Sian TCC; Goncalves G; Steele JR; Shamekhi T; Bramberger L; Jin D; Shahbazy M; Purcell AW; Ramarathinam S; Stoychev S; Faridi P. SAPrIm, a Semi-Automated Protocol for Mid-Throughput Immunopeptidomics. Front. Immunol. 2023, 14, 1107576. 10.3389/fimmu.2023.1107576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Bruno PM; Timms RT; Abdelfattah NS; Leng Y; Lelis FJN; Wesemann DR; Yu XG; Elledge SJ High-Throughput, Targeted MHC Class I Immunopeptidomics Using a Functional Genetics Screening Platform. Nat. Biotechnol. 2023, 41 (7), 980–992. 10.1038/s41587-022-01566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Gomez-Zepeda D; Arnold-Schild D; Beyrle J; Kumm E; Distler U; Schild H; Tenzer S. Thunder-DDA-PASEF Enables High-Coverage Immunopeptidomics and Identifies HLA Class-I Presented SarsCov-2 Spike Protein Epitopes; preprint; In Review, 2023. 10.21203/rs.3.rs-2625909/v1. [DOI] [Google Scholar]
- (49).Phulphagar KM; Ctortecka C; Jacome ASV; Klaeger S; Verzani EK; Hernandez GM; Udeshi ND; Clauser KR; Abelin JG; Carr SA Sensitive, High-Throughput HLA-I and HLA-II Immunopeptidomics Using Parallel Accumulation-Serial Fragmentation Mass Spectrometry. Mol. Cell. Proteomics MCP 2023, 22 (6), 100563. 10.1016/j.mcpro.2023.100563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Liao H; Barra C; Peng X; Woodhouse I; Tailor A; Parker R; Carré A; Mallone R; Hogan M; Paes W; Eisenlohr L; Nielsen M; Ternette N. MARS: Improved De Novo Peptide Candidate Selection for Non-Canonical Antigen Target Discovery in Cancer; preprint; In Review, 2022. 10.21203/rs.3.rs-1890352/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Xin L; Qiao R; Chen X; Tran H; Pan S; Rabinoviz S; Bian H; He X; Morse B; Shan B; Li M. A Streamlined Platform for Analyzing Tera-Scale DDA and DIA Mass Spectrometry Data Enables Highly Sensitive Immunopeptidomics. Nat. Commun. 2022, 13 (1), 3108. 10.1038/s41467-022-30867-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Millar DG; Yang SYC; Sayad A; Zhao Q; Nguyen LT; Warner K; Sangster AG; Nakatsugawa M; Murata K; Wang BX; Shaw P; Clarke B; Bernardini MQ; Pugh T; Thibault P; Hirano N; Perreault C; Ohashi PS Identification of Antigenic Epitopes Recognized by Tumor Infiltrating Lymphocytes in High Grade Serous Ovarian Cancer by Multi-Omics Profiling of the Auto-Antigen Repertoire. Cancer Immunol. Immunother. CII 2023, 72 (7), 2375–2392. 10.1007/s00262-023-03413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Merlotti A; Sadacca B; Arribas YA; Ngoma M; Burbage M; Goudot C; Houy A; Rocañín-Arjó A; Lalanne A; Seguin-Givelet A; Lefevre M; Heurtebise-Chrétien S; Baudon B; Oliveira G; Loew D; Carrascal M; Wu CJ; Lantz O; Stern M-H; Girard N; Waterfall JJ; Amigorena S. Noncanonical Splicing Junctions between Exons and Transposable Elements Represent a Source of Immunogenic Recurrent Neo-Antigens in Patients with Lung Cancer. Sci. Immunol. 2023, 8 (80), eabm6359. 10.1126/sciimmunol.abm6359. [DOI] [PubMed] [Google Scholar]
- (54).Cucchi DGJ; Van Alphen C; Zweegman S; Van Kuijk B; Kwidama ZJ; Al Hinai A; Henneman AA; Knol JC; Piersma SR; Pham TV; Jimenez CR; Cloos J; Janssen JJWM. Phosphoproteomic Characterization of Primary AML Samples and Relevance for Response Toward FLT3-Inhibitors. HemaSphere 2021, 5 (7), e606. 10.1097/HS9.0000000000000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Haga Y; Ueda K. Glycosylation in Cancer: Its Application as a Biomarker and Recent Advances of Analytical Techniques. Glycoconj. J. 2022, 39 (2), 303–313. 10.1007/s10719-022-10043-1. [DOI] [PubMed] [Google Scholar]
- (56).He B; Zhang Z; Huang Z; Duan X; Wang Y; Cao J; Li L; He K; Nice EC; He W; Gao W; Shen Z. Protein Persulfidation: Rewiring the Hydrogen Sulfide Signaling in Cell Stress Response. Biochem. Pharmacol. 2023, 209, 115444. 10.1016/j.bcp.2023.115444. [DOI] [PubMed] [Google Scholar]
- (57).Ciuffa R; Uliana F; Mannion J; Mehnert M; Tenev T; Marulli C; Satanowski A; Keller LML; Rodilla Ramírez PN; Ori A; Gstaiger M; Meier P; Aebersold R. Novel Biochemical, Structural, and Systems Insights into Inflammatory Signaling Revealed by Contextual Interaction Proteomics. Proc. Natl. Acad. Sci. U. S. A. 2022, 119 (40), e2117175119. 10.1073/pnas.2117175119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Luo M; Zhou L; Huang Z; Li B; Nice EC; Xu J; Huang C. Antioxidant Therapy in Cancer: Rationale and Progress. Antioxid. Basel Switz. 2022, 11 (6), 1128. 10.3390/antiox11061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Guerrero L; Paradela A; Corrales FJ Targeted Proteomics for Monitoring One-Carbon Metabolism in Liver Diseases. Metabolites 2022, 12 (9), 779. 10.3390/metabo12090779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Ghantasala S; Bhat A; Agarwal U; Biswas D; Bhattarai P; Epari S; Moiyadi A; Srivastava S. Deep Proteome Investigation of High-Grade Gliomas Reveals Heterogeneity Driving Differential Metabolism of 5-Aminolevulinic Acid. Neuro-Oncol. Adv. 2023, 5 (1), vdad065. 10.1093/noajnl/vdad065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Fekete EE; Figeys D; Zhang X. Microbiota-Directed Biotherapeutics: Considerations for Quality and Functional Assessment. Gut Microbes 2023, 15 (1), 2186671. 10.1080/19490976.2023.2186671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Mund A; Coscia F; Kriston A; Hollandi R; Kovács F; Brunner A-D; Migh E; Schweizer L; Santos A; Bzorek M; Naimy S; Rahbek-Gjerdrum LM; Dyring-Andersen B; Bulkescher J; Lukas C; Eckert MA; Lengyel E; Gnann C; Lundberg E; Horvath P; Mann M. Deep Visual Proteomics Defines Single-Cell Identity and Heterogeneity. Nat. Biotechnol. 2022, 40 (8), 1231–1240. 10.1038/s41587-022-01302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Barpanda A; Biswas D; Verma A; Parihari S; Singh A; Kapoor S; Kantharia C; Srivastava S. Integrative Proteomic and Pharmacological Analysis of Colon Cancer Reveals the Classical Lipogenic Pathway with Prognostic and Therapeutic Opportunities. J. Proteome Res. 2023, 22 (3), 871–884. 10.1021/acs.jproteome.2c00646. [DOI] [PubMed] [Google Scholar]
- (64).Shi J; Xu J; Li Y; Li B; Ming H; Nice EC; Huang C; Li Q; Wang C. Drug Repurposing in Cancer Neuroscience: From the Viewpoint of the Autophagy-Mediated Innervated Niche. Front. Pharmacol. 2022, 13, 990665. 10.3389/fphar.2022.990665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Tognetti M; Gabor A; Yang M; Cappelletti V; Windhager J; Rueda OM; Charmpi K; Esmaeilishirazifard E; Bruna A; de Souza N; Caldas C; Beyer A; Picotti P; Saez-Rodriguez J; Bodenmiller B. Deciphering the Signaling Network of Breast Cancer Improves Drug Sensitivity Prediction. Cell Syst. 2021, 12 (5), 401–418.e12. 10.1016/j.cels.2021.04.002. [DOI] [PubMed] [Google Scholar]
- (66).Crunfli F; Carregari VC; Veras FP; Silva LS; Nogueira MH; Antunes ASLM; Vendramini PH; Valença AGF; Brandão-Teles C; Zuccoli G. da S; Reis-de-Oliveira G; Silva-Costa LC; Saia-Cereda VM; Smith BJ; Codo AC; de Souza GF; Muraro SP; Parise PL; Toledo-Teixeira DA; Santos de Castro ÍM; Melo BM; Almeida GM; Firmino EMS; Paiva IM; Silva BMS; Guimarães RM; Mendes ND; Ludwig RL; Ruiz GP; Knittel TL; Davanzo GG; Gerhardt JA; Rodrigues PB; Forato J; Amorim MR; Brunetti NS; Martini MC; Benatti MN; Batah SS; Siyuan L; João RB; Aventurato ÍK; Rabelo de Brito M; Mendes MJ; da Costa BA; Alvim MKM; da Silva Júnior JR; Damião LL; de Sousa IMP; da Rocha ED; Gonçalves SM; Lopes da Silva LH; Bettini V; Campos BM; Ludwig G; Tavares LA; Pontelli MC; Viana RMM; Martins RB; Vieira AS; Alves-Filho JC; Arruda E; Podolsky-Gondim GG; Santos MV; Neder L; Damasio A; Rehen S; Vinolo MAR; Munhoz CD; Louzada-Junior P; Oliveira RD; Cunha FQ; Nakaya HI; Mauad T; Duarte-Neto AN; Ferraz da Silva LF; Dolhnikoff M; Saldiva PHN; Farias AS; Cendes F; Moraes-Vieira PMM; Fabro AT; Sebollela A; Proença-Modena JL; Yasuda CL; Mori MA; Cunha TM; Martins-de-Souza D. Morphological, Cellular, and Molecular Basis of Brain Infection in COVID-19 Patients. Proc. Natl. Acad. Sci. U. S. A. 2022, 119 (35), e2200960119. 10.1073/pnas.2200960119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).MacDonald ML; Garver M; Newman J; Sun Z; Kannarkat J; Salisbury R; Glausier J; Ding Y; Lewis DA; Yates N; Sweet RA Synaptic Proteome Alterations in the Primary Auditory Cortex of Individuals With Schizophrenia. JAMA Psychiatry 2020, 77 (1), 86–95. 10.1001/jamapsychiatry.2019.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Oraki Kohshour M; Kannaiyan NR; Falk AJ; Papiol S; Heilbronner U; Budde M; Kalman JL; Schulte EC; Rietschel M; Witt S; Forstner AJ; Heilmann-Heimbach S; Nöthen MM; Spitzer C; Malchow B; Müller T; Wiltfang J; Falkai P; Schmitt A; Rossner MJ; Nilsson P; Schulze TG Comparative Serum Proteomic Analysis of a Selected Protein Panel in Individuals with Schizophrenia and Bipolar Disorder and the Impact of Genetic Risk Burden on Serum Proteomic Profiles. Transl. Psychiatry 2022, 12 (1), 471. 10.1038/s41398-022-02228-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Mc Ardle A; Binek A; Moradian A; Chazarin Orgel B; Rivas A; Washington KE; Phebus C; Manalo D-M; Go J; Venkatraman V; Coutelin Johnson CW; Fu Q; Cheng S; Raedschelders K; Fert-Bober J; Pennington SR; Murray CI; Van Eyk JE Standardized Workflow for Precise Mid- and High-Throughput Proteomics of Blood Biofluids. Clin. Chem. 2022, 68 (3), 450–460. 10.1093/clinchem/hvab202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Joshi A; Schmidt LE; Burnap SA; Lu R; Chan MV; Armstrong PC; Baig F; Gutmann C; Willeit P; Santer P; Barwari T; Theofilatos K; Kiechl S; Willeit J; Warner TD; Mathur A; Mayr M. Neutrophil-Derived Protein S100A8/A9 Alters the Platelet Proteome in Acute Myocardial Infarction and Is Associated With Changes in Platelet Reactivity. Arterioscler. Thromb. Vasc. Biol. 2022, 42 (1), 49–62. 10.1161/ATVBAHA.121.317113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Luecke LB; Waas M; Littrell J; Wojtkiewicz M; Castro C; Burkovetskaya M; Schuette EN; Buchberger AR; Churko JM; Chalise U; Waknitz M; Konfrst S; Teuben R; Morrissette-McAlmon J; Mahr C; Anderson DR; Boheler KR; Gundry RL Surfaceome Mapping of Primary Human Heart Cells with CellSurfer Uncovers Cardiomyocyte Surface Protein LSMEM2 and Proteome Dynamics in Failing Hearts. Nat. Cardiovasc. Res. 2023, 2 (1), 76–95. 10.1038/s44161-022-00200-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Hammond DE; Simpson DM; Franco C; Wright Muelas M; Waters J; Ludwig RW; Prescott MC; Hurst JL; Beynon RJ; Lau E. Harmonizing Labeling and Analytical Strategies to Obtain Protein Turnover Rates in Intact Adult Animals. Mol. Cell. Proteomics MCP 2022, 21 (7), 100252. 10.1016/j.mcpro.2022.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Chapman EA; Aballo TJ; Melby JA; Zhou T; Price SJ; Rossler KJ; Lei I; Tang PC; Ge Y. Defining the Sarcomeric Proteoform Landscape in Ischemic Cardiomyopathy by Top-Down Proteomics. J. Proteome Res. 2023, 22 (3), 931–941. 10.1021/acs.jproteome.2c00729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Omenn GS; States DJ; Adamski M; Blackwell TW; Menon R; Hermjakob H; Apweiler R; Haab BB; Simpson RJ; Eddes JS; Kapp EA; Moritz RL; Chan DW; Rai AJ; Admon A; Aebersold R; Eng J; Hancock WS; Hefta SA; Meyer H; Paik Y-K; Yoo J-S; Ping P; Pounds J; Adkins J; Qian X; Wang R; Wasinger V; Wu CY; Zhao X; Zeng R; Archakov A; Tsugita A; Beer I; Pandey A; Pisano M; Andrews P; Tammen H; Speicher DW; Hanash SM Overview of the HUPO Plasma Proteome Project: Results from the Pilot Phase with 35 Collaborating Laboratories and Multiple Analytical Groups, Generating a Core Dataset of 3020 Proteins and a Publicly-Available Database. Proteomics 2005, 5 (13), 3226–3245. 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- (75).Keller A; Eng J; Zhang N; Li X; Aebersold R. A Uniform Proteomics MS/MS Analysis Platform Utilizing Open XML File Formats. Mol. Syst. Biol. 2005, 1, 2005.0017. 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Deutsch EW; Mendoza L; Shteynberg DD; Hoopmann MR; Sun Z; Eng JK; Moritz RL Trans-Proteomic Pipeline: Robust Mass Spectrometry-Based Proteomics Data Analysis Suite. J. Proteome Res. 2023, 22 (2), 615–624. 10.1021/acs.jproteome.2c00624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Vizcaíno JA; Deutsch EW; Wang R; Csordas A; Reisinger F; Ríos D; Dianes JA; Sun Z; Farrah T; Bandeira N; Binz P-A; Xenarios I; Eisenacher M; Mayer G; Gatto L; Campos A; Chalkley RJ; Kraus H-J; Albar JP; Martinez-Bartolomé S; Apweiler R; Omenn GS; Martens L; Jones AR; Hermjakob H. ProteomeXchange Provides Globally Coordinated Proteomics Data Submission and Dissemination. Nat. Biotechnol. 2014, 32 (3), 223–226. 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Deutsch EW; Bandeira N; Perez-Riverol Y; Sharma V; Carver JJ; Mendoza L; Kundu DJ; Wang S; Bandla C; Kamatchinathan S; Hewapathirana S; Pullman BS; Wertz J; Sun Z; Kawano S; Okuda S; Watanabe Y; MacLean B; MacCoss MJ; Zhu Y; Ishihama Y; Vizcaíno JA The ProteomeXchange Consortium at 10 Years: 2023 Update. Nucleic Acids Res. 2022, gkac1040. 10.1093/nar/gkac1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Sun BB; Chiou J; Traylor M; Benner C; Hsu Y-H; Richardson TG; Surendran P; Mahajan A; Robins C; Vasquez-Grinnell SG; Hou L; Kvikstad EM; Burren OS; Davitte J; Ferber KL; Gillies CE; Hedman ÅK; Hu S; Lin T; Mikkilineni R; Pendergrass RK; Pickering C; Prins B; Baird D; Chen C-Y; Ward LD; Deaton AM; Welsh S; Willis CM; Lehner N; Arnold M; Wörheide MA; Suhre K; Kastenmüller G; Sethi A; Cule M; Raj A; Alnylam Human Genetics; AstraZeneca Genomics Initiative; Biogen Biobank Team; Bristol Myers Squibb; Genentech Human Genetics; GlaxoSmithKline Genomic Sciences; Pfizer Integrative Biology; Population Analytics of Janssen Data Sciences; Regeneron Genetics Center; Burkitt-Gray L; Melamud E; Black MH; Fauman EB; Howson JMM; Kang HM; McCarthy MI; Nioi P; Petrovski S; Scott RA; Smith EN; Szalma S; Waterworth DM; Mitnaul LJ; Szustakowski JD; Gibson BW; Miller MR; Whelan CD Plasma Proteomic Associations with Genetics and Health in the UK Biobank. Nature 2023, 622 (7982), 329–338. 10.1038/s41586-023-06592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Manes NP; Calzola JM; Kaplan PR; Fraser IDC; Germain RN; Meier-Schellersheim M; Nita-Lazar A. Absolute Protein Quantitation of the Mouse Macrophage Toll-like Receptor and Chemotaxis Pathways. Sci. Data 2022, 9 (1), 491. 10.1038/s41597-022-01612-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Liu D; Lum KK; Treen N; Núñez CT; Yang J; Howard TR; Levine M; Cristea IM IFI16 Phase Separation via Multi-Phosphorylation Drives Innate Immune Signaling. Nucleic Acids Res. 2023, 51 (13), 6819–6840. 10.1093/nar/gkad449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Andrews DDT; Vlok M; Akbari Bani D; Hay BN; Mohamud Y; Foster LJ; Luo H; Overall CM; Jan E. Cleavage of 14–3-3ε by the Enteroviral 3C Protease Dampens RIG-I-Mediated Antiviral Signaling. J. Virol. 2023, e0060423. 10.1128/jvi.00604-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Bogdanow B; Gruska I; Mühlberg L; Protze J; Hohensee S; Vetter B; Bosse JB; Lehmann M; Sadeghi M; Wiebusch L; Liu F. Spatially Resolved Protein Map of Intact Human Cytomegalovirus Virions. Nat. Microbiol. 2023. 10.1038/s41564-023-01433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Song B; Sheng X; Justice JL; Lum KK; Metzger PJ; Cook KC; Kostas JC; Cristea IM Intercellular Communication within the Virus Microenvironment Affects the Susceptibility of Cells to Secondary Viral Infections. Sci. Adv. 2023, 9 (19), eadg3433. 10.1126/sciadv.adg3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Rajoria S; Halder A; Tarnekar I; Pal P; Bansal P; Srivastava S. Detection of Mutant Peptides of SARS-CoV-2 Variants by LC/MS in the DDA Approach Using an In-House Database. J. Proteome Res. 2023, 22 (6), 1816–1827. 10.1021/acs.jproteome.2c00819. [DOI] [PubMed] [Google Scholar]
- (86).Kollhoff L; Kipping M; Rauh M; Ceglarek U; Barka G; Barka F; Sinz A. Development of a Rapid and Specific MALDI-TOF Mass Spectrometric Assay for SARS-CoV-2 Detection. Clin. Proteomics 2023, 20 (1), 26. 10.1186/s12014-023-09415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Zhang F; Luna A; Tan T; Chen Y; Sander C; Guo T. COVIDpro: Database for Mining Protein Dysregulation in Patients with COVID-19. J. Proteome Res. 2023. 10.1021/acs.jproteome.3c00092. [DOI] [PubMed] [Google Scholar]
- (88).Schweizer L; Schaller T; Zwiebel M; Karayel Ö; Müller-Reif JB; Zeng W-F; Dintner S; Nordmann TM; Hirschbühl K; Märkl B; Claus R; Mann M. Quantitative Multiorgan Proteomics of Fatal COVID-19 Uncovers Tissue-Specific Effects beyond Inflammation. EMBO Mol. Med. 2023, e17459. 10.15252/emmm.202317459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Álvez MB; Edfors F; von Feilitzen K; Zwahlen M; Mardinoglu A; Edqvist P-H; Sjöblom T; Lundin E; Rameika N; Enblad G; Lindman H; Höglund M; Hesselager G; Stålberg K; Enblad M; Simonson OE; Häggman M; Axelsson T; Åberg M; Nordlund J; Zhong W; Karlsson M; Gyllensten U; Ponten F; Fagerberg L; Uhlén M. Next Generation Pan-Cancer Blood Proteome Profiling Using Proximity Extension Assay. Nat. Commun. 2023, 14 (1), 4308. 10.1038/s41467-023-39765-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Robinson JL; Kocabaş P; Wang H; Cholley P-E; Cook D; Nilsson A; Anton M; Ferreira R; Domenzain I; Billa V; Limeta A; Hedin A; Gustafsson J; Kerkhoven EJ; Svensson LT; Palsson BO; Mardinoglu A; Hansson L; Uhlén M; Nielsen J. An Atlas of Human Metabolism. Sci. Signal. 2020, 13 (624), eaaz1482. 10.1126/scisignal.aaz1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Karlsson M; Zhang C; Méar L; Zhong W; Digre A; Katona B; Sjöstedt E; Butler L; Odeberg J; Dusart P; Edfors F; Oksvold P; von Feilitzen K; Zwahlen M; Arif M; Altay O; Li X; Ozcan M; Mardinoglu A; Fagerberg L; Mulder J; Luo Y; Ponten F; Uhlén M; Lindskog C. A Single-Cell Type Transcriptomics Map of Human Tissues. Sci. Adv. 2021, 7 (31), eabh2169. 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Méar L; Sutantiwanichkul T; Östman J; Damdimopoulou P; Lindskog C. Spatial Proteomics for Further Exploration of Missing Proteins: A Case Study of the Ovary. J. Proteome Res. 2023, 22 (4), 1071–1079. 10.1021/acs.jproteome.2c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Méar Loren; Hao Xia; Hikmet Feria; Damdimopoulou Pauliina; Rodriguez-Wallberg Kenny; Lindskog Cecilia. TRANSCRIPTOMICS AND SPATIAL PROTEOMICS FOR DISCOVERY AND VALIDATION OF MISSING PROTEINS IN HUMAN OVARY. JPR this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Mathew A; Giskes F; Lekkas A; Greisch J-F; Eijkel GB; Anthony IGM; Fort K; Heck AJR; Papanastasiou D; Makarov AA; Ellis SR; Heeren RMA An Orbitrap/Time-of-Flight Mass Spectrometer for Photofragment Ion Imaging and High-Resolution Mass Analysis of Native Macromolecular Assemblies. J. Am. Soc. Mass Spectrom. 2023, 34 (7), 1359–1371. 10.1021/jasms.3c00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).den Boer MA; Lai S-H; Xue X; van Kampen MD; Bleijlevens B; Heck AJR Comparative Analysis of Antibodies and Heavily Glycosylated Macromolecular Immune Complexes by Size-Exclusion Chromatography Multi-Angle Light Scattering, Native Charge Detection Mass Spectrometry, and Mass Photometry. Anal. Chem. 2022, 94 (2), 892–900. 10.1021/acs.analchem.1c03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Johansson C; Hunt H; Signorelli M; Edfors F; Hober A; Svensson A-S; Tegel H; Forstström B; Aartsma-Rus A; Niks E; Spitali P; Uhlén M; Szigyarto CA-K Orthogonal Proteomics Methods Warrant the Development of Duchenne Muscular Dystrophy Biomarkers. Clin. Proteomics 2023, 20 (1), 23. 10.1186/s12014-023-09412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Shalon D; Culver RN; Grembi JA; Folz J; Treit PV; Shi H; Rosenberger FA; Dethlefsen L; Meng X; Yaffe E; Aranda-Díaz A; Geyer PE; Mueller-Reif JB; Spencer S; Patterson AD; Triadafilopoulos G; Holmes SP; Mann M; Fiehn O; Relman DA; Huang KC Profiling the Human Intestinal Environment under Physiological Conditions. Nature 2023, 617 (7961), 581–591. 10.1038/s41586-023-05989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Mani DR; Krug K; Zhang B; Satpathy S; Clauser KR; Ding L; Ellis M; Gillette MA; Carr SA Cancer Proteogenomics: Current Impact and Future Prospects. Nat. Rev. Cancer 2022, 22 (5), 298–313. 10.1038/s41568-022-00446-5. [DOI] [PubMed] [Google Scholar]
- (99).Boyer S; Lee H-J; Steele N; Zhang L; Sajjakulnukit P; Andren A; Ward MH; Singh R; Basrur V; Zhang Y; Nesvizhskii AI; Pasca di Magliano M; Halbrook CJ; Lyssiotis CA Multiomic Characterization of Pancreatic Cancer-Associated Macrophage Polarization Reveals Deregulated Metabolic Programs Driven by the GM-CSF-PI3K Pathway. eLife 2022, 11, e73796. 10.7554/eLife.73796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Zhang Y-H; Cho MH; Morrow JD; Castaldi PJ; Hersh CP; Midha MK; Hoopmann MR; Lutz SM; Moritz RL; Silverman EK Integrating Genetics, Transcriptomics, and Proteomics in Lung Tissue to Investigate Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cell Mol. Biol. 2023, 68 (6), 651–663. 10.1165/rcmb.2022-0302OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Babu M; Snyder M. Multi-Omics Profiling for Health. Mol. Cell. Proteomics MCP 2023, 22 (6), 100561. 10.1016/j.mcpro.2023.100561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Mohammed Y; Goodlett D; Borchers CH Bioinformatics Tools and Knowledgebases to Assist Generating Targeted Assays for Plasma Proteomics. Methods Mol. Biol. Clifton NJ 2023, 2628, 557–577. 10.1007/978-1-0716-2978-9_32. [DOI] [PubMed] [Google Scholar]
- (103).Tran NK; Kretsch C; LaValley C; Rashidi HH Machine Learning and Artificial Intelligence for the Diagnosis of Infectious Diseases in Immunocompromised Patients. Curr. Opin. Infect. Dis. 2023, 36 (4), 235–242. 10.1097/QCO.0000000000000935. [DOI] [PubMed] [Google Scholar]
- (104).Patel MA; Knauer MJ; Nicholson M; Daley M; Van Nynatten LR; Cepinskas G; Fraser DD Organ and Cell-Specific Biomarkers of Long-COVID Identified with Targeted Proteomics and Machine Learning. Mol. Med. Camb. Mass 2023, 29 (1), 26. 10.1186/s10020-023-00610-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Lam S; Arif M; Song X; Uhlén M; Mardinoglu A. Machine Learning Analysis Reveals Biomarkers for the Detection of Neurological Diseases. Front. Mol. Neurosci. 2022, 15, 889728. 10.3389/fnmol.2022.889728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).He B; Huang Z; Huang C; Nice EC Clinical Applications of Plasma Proteomics and Peptidomics: Towards Precision Medicine. Proteomics Clin. Appl. 2022, 16 (6), e2100097. 10.1002/prca.202100097. [DOI] [PubMed] [Google Scholar]
- (107).Hällqvist J; Lane D; Shapanis A; Davis K; Heywood WE; Doykov I; Śpiewak J; Ghansah N; Keevil B; Gupta P; Jukes-Jones R; Singh R; Foley D; Vissers JPC; Pattison R; Ferries S; Wardle R; Bartlett A; Calton LJ; Anderson L; Razavi M; Pearson T; Pope M; Yip R; Ng LL; Nicholas BI; Bailey A; Noel D; Dalton RN; Heales S; Hopley C; Pitt AR; Barran P; Jones DJL; Mills K; Skipp P; Carling RS Operation Moonshot: Rapid Translation of a SARS-CoV-2 Targeted Peptide Immunoaffinity Liquid Chromatography-Tandem Mass Spectrometry Test from Research into Routine Clinical Use. Clin. Chem. Lab. Med. 2023, 61 (2), 302–310. 10.1515/cclm-2022-1000. [DOI] [PubMed] [Google Scholar]
- (108).Wang JY; Zhang W; Roehrl MW; Roehrl VB; Roehrl MH An Autoantigen Profile from Jurkat T-Lymphoblasts Provides a Molecular Guide for Investigating Autoimmune Sequelae of COVID-19. BioRxiv Prepr. Serv. Biol. 2021, 2021.07.05.451199. 10.1101/2021.07.05.451199. [DOI] [Google Scholar]
- (109).Wang JY; Zhang W; Roehrl VB; Roehrl MW; Roehrl MH An Autoantigen-Ome from HS-Sultan B-Lymphoblasts Offers a Molecular Map for Investigating Autoimmune Sequelae of COVID-19. BioRxiv Prepr. Serv. Biol. 2021, 2021.04.05.438500. 10.1101/2021.04.05.438500. [DOI] [Google Scholar]
- (110).Weingarten-Gabbay S; Klaeger S; Sarkizova S; Pearlman LR; Chen D-Y; Gallagher KME; Bauer MR; Taylor HB; Dunn WA; Tarr C; Sidney J; Rachimi S; Conway HL; Katsis K; Wang Y; Leistritz-Edwards D; Durkin MR; Tomkins-Tinch CH; Finkel Y; Nachshon A; Gentili M; Rivera KD; Carulli IP; Chea VA; Chandrashekar A; Bozkus CC; Carrington M; Bhardwaj N; Barouch DH; Sette A; Maus MV; Rice CM; Clauser KR; Keskin DB; Pregibon DC; Hacohen N; Carr SA; Abelin JG; Saeed M; Sabeti PC Profiling SARS-CoV-2 HLA-I Peptidome Reveals T Cell Epitopes from out-of-Frame ORFs. Cell 2021, 184 (15), 3962–3980.e17. 10.1016/j.cell.2021.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Demichev V; Tober-Lau P; Lemke O; Nazarenko T; Thibeault C; Whitwell H; Röhl A; Freiwald A; Szyrwiel L; Ludwig D; Correia-Melo C; Aulakh SK; Helbig ET; Stubbemann P; Lippert LJ; Grüning N-M; Blyuss O; Vernardis S; White M; Messner CB; Joannidis M; Sonnweber T; Klein SJ; Pizzini A; Wohlfarter Y; Sahanic S; Hilbe R; Schaefer B; Wagner S; Mittermaier M; Machleidt F; Garcia C; Ruwwe-Glösenkamp C; Lingscheid T; Bosquillon de Jarcy L; Stegemann MS; Pfeiffer M; Jürgens L; Denker S; Zickler D; Enghard P; Zelezniak A; Campbell A; Hayward C; Porteous DJ; Marioni RE; Uhrig A; Müller-Redetzky H; Zoller H; Löffler-Ragg J; Keller MA; Tancevski I; Timms JF; Zaikin A; Hippenstiel S; Ramharter M; Witzenrath M; Suttorp N; Lilley K; Mülleder M; Sander LE; Ralser M; Kurth F; Kleinschmidt M; Heim KM; Millet B; Meyer-Arndt L; Hübner RH; Andermann T; Doehn JM; Opitz B; Sawitzki B; Grund D; Radünzel P; Schürmann M; Zoller T; Alius F; Knape P; Breitbart A; Li Y; Bremer F; Pergantis P; Schürmann D; Temmesfeld-Wollbrück B; Wendisch D; Brumhard S; Haenel SS; Conrad C; Georg P; Eckardt K-U; Lehner L; Kruse JM; Ferse C; Körner R; Spies C; Edel A; Weber-Carstens S; Krannich A; Zvorc S; Li L; Behrens U; Schmidt S; Rönnefarth M; Dang-Heine C; Röhle R; Lieker E; Kretzler L; Wirsching I; Wollboldt C; Wu Y; Schwanitz G; Hillus D; Kasper S; Olk N; Horn A; Briesemeister D; Treue D; Hummel M; Corman VM; Drosten C; von Kalle C. A Time-Resolved Proteomic and Prognostic Map of COVID-19. Cell Syst. 2021, S2405471221001605. 10.1016/j.cels.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Mund A; Brunner A-D; Mann M. Unbiased Spatial Proteomics with Single-Cell Resolution in Tissues. Mol. Cell 2022, 82 (12), 2335–2349. 10.1016/j.molcel.2022.05.022. [DOI] [PubMed] [Google Scholar]
- (113).Sobsey CA; Froehlich B; Batist G; Borchers CH Immuno-MALDI-MS for Accurate Quantitation of Targeted Peptides from Volume-Restricted Samples. Methods Mol. Biol. Clifton NJ 2022, 2515, 203–225. 10.1007/978-1-0716-2409-8_13. [DOI] [PubMed] [Google Scholar]
- (114).Cervia LD; Shibue T; Borah AA; Gaeta B; He L; Leung L; Li N; Moyer SM; Shim BH; Dumont N; Gonzalez A; Bick NR; Kazachkova M; Dempster JM; Krill-Burger JM; Piccioni F; Udeshi ND; Olive ME; Carr SA; Root DE; McFarland JM; Vazquez F; Hahn WC A Ubiquitination Cascade Regulating the Integrated Stress Response and Survival in Carcinomas. Cancer Discov. 2023, 13 (3), 766–795. 10.1158/2159-8290.CD-22-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Gentili M; Liu B; Papanastasiou M; Dele-Oni D; Schwartz MA; Carlson RJ; Al’Khafaji AM; Krug K; Brown A; Doench JG; Carr SA; Hacohen N. ESCRT-Dependent STING Degradation Inhibits Steady-State and cGAMP-Induced Signalling. Nat. Commun. 2023, 14 (1), 611. 10.1038/s41467-023-36132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Li X; Yue X; Sepulveda H; Burt RA; Scott DA; A Carr S; A Myers S; Rao A. OGT Controls Mammalian Cell Viability by Regulating the Proteasome/mTOR/ Mitochondrial Axis. Proc. Natl. Acad. Sci. U. S. A. 2023, 120 (3), e2218332120. 10.1073/pnas.2218332120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Hevler JF; Albanese P; Cabrera-Orefice A; Potter A; Jankevics A; Misic J; Scheltema RA; Brandt U; Arnold S; Heck AJ R. MRPS36 Provides a Structural Link in the Eukaryotic 2-Oxoglutarate Dehydrogenase Complex. Open Biol. 2023, 13 (3), 220363. 10.1098/rsob.220363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Baquero F; Nombela C. The Microbiome as a Human Organ. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18 Suppl 4, 2–4. 10.1111/j.1469-0691.2012.03916.x. [DOI] [PubMed] [Google Scholar]
- (119).Polychronidou M; Hou J; Babu MM; Liberali P; Amit I; Deplancke B; Lahav G; Itzkovitz S; Mann M; Saez-Rodriguez J; Theis F; Eils R. Single-Cell Biology: What Does the Future Hold? Mol. Syst. Biol. 2023, 19 (7), e11799. 10.15252/msb.202311799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Gatto L; Aebersold R; Cox J; Demichev V; Derks J; Emmott E; Franks AM; Ivanov AR; Kelly RT; Khoury L; Leduc A; MacCoss MJ; Nemes P; Perlman DH; Petelski AA; Rose CM; Schoof EM; Van Eyk J; Vanderaa C; Yates JR; Slavov N. Initial Recommendations for Performing, Benchmarking and Reporting Single-Cell Proteomics Experiments. Nat. Methods 2023, 20 (3), 375–386. 10.1038/s41592-023-01785-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.