Eosinophilia is defined as an elevation of the peripheral blood (PB) eosinophil count above 0.5 × 109/L [1]. Among these, hypereosinophilia has been historically defined as a persistent eosinophil count of at least 1.5×109/L [2]. This is conventionally divided into four main categories: primary (clonal/neoplastic), secondary (reactive), hereditary (familial) and idiopathic/undetermined significance [1,2]. Primary hypereosinophilia is a clonal hematologic disorder in which the eosinophils form a part of the neoplastic clone. It is seen in the context of a myeloid neoplasm, and is most frequently associated with chronic myeloid neoplasms, including myeloproliferative neoplasms (MPN) or myelodysplastic/myeloproliferative neoplasms (MDS/MPN), and more rarely with acute myeloid leukemia (AML) and B- or T-lymphoblastic leukemia/lymphoma [2]. In addition, primary hypereosinophilia generated by dysregulated fusion tyrosine kinase genes is recognized, and is classified as myeloid/lymphoid neoplasms with eosinophilia and gene rearrangement of PDGFRA, PDGFRB or FGFR1, or with PCM1-JAK2 [3]. Secondary reactive/non-clonal hypereosinophilia may be driven by a wide range of underlying conditions including allergic disorders, autoimmunity, infectious diseases, lymphoproliferative disorders, solid tumors, drug reactions and other conditions through overproduction of eosinophilopoietic cytokines such as interleukin 3 (IL-3), IL-5, or granulocyte-macrophage colony-stimulating factor (GM-CSF) [4]. Among these, reactive eosinophilia resulting from excess production of eosinophilopoietic cytokines secreted by immunophenotypically aberrant clones of T-cells is recognized and is called lymphocyte-variant hypereosinophilia [5].
Recently, STAT5B p.N642H, activating mutation of STAT5B, was reported as a recurrent mutation in patients with myeloid neoplasms with eosinophilia, identified in 1.6% of patients with myeloid neoplasms with eosinophilia by Cross et al [6]. Few sporadic cases were also reported showing myeloid neoplasm with eosinophilia and STAT5B p.N642H, and no other STAT5B mutations were reported in relation with eosinophilia in myeloid neoplasms to date [6–8]. STAT5B is an essential factor in the signal transduction pathways for several cytokines and a critical downstream mediator of transformation by oncogenic tyrosine kinase such as BCR-ABL1, JAK2 V617F, FLT3-ITD and ZNF198-FGFR1, and its mutations have been reported in various T-cell disorders including T-cell large granular lymphocytic leukemia, pediatric T-lymphoblastic leukemia/lymphoma, T-cell prolymphocytic leukemia and hepatosplenic T-cell lymphoma[6,9,10]. Although STAT5B mutations have been well studied in T-cell lymphomas, clinical presentations and detailed genetic profiles of myeloid neoplasms with STAT5B mutations are unclear. To investigate these, we performed a systemic search to identify myeloid neoplasms with STAT5B mutations to evaluate the clinical and pathological features of these patients. We also evaluated the difference of genomic findings between STAT5B mutated myeloid neoplasm with eosinophilia and STAT5B mutated lymphocyte-variant hypereosinophilia using flow cytometry sorting and next generation sequencing (NGS).
Biospecimens were retrieved from the pathology archives between March 2014 and March 2022 at Memorial Sloan Kettering Cancer Center under the declaration of Helsinki and the approval by institutional review board. Diagnosis was rendered based on criteria specified in the WHO 2016 classification by morphological assessment in conjunction with standard immunohistochemistry panel, flow cytometry and molecular analyses [11]. NGS was performed on bone marrow aspirate using Memorial Sloan Kettering (MSK) laboratory-developed hybrid-capture-based 400 gene panels (MSK-IMPACT). DNA isolation, sequencing library preparation, and paired-end sequencing on an illumina HiSeq 2500 instrument were performed as described previously. Mutations were called based on a paired analysis using the patient normal sample (if available) and a pooled unmatched normal, and only somatic variants were reported [12].
We identified 2266 myeloid neoplasm patients with comprehensive genomic data by NGS. Among them, 5 patients (0.22%) had STAT5B mutation(s). Diagnosis by WHO 2016 classification and clinical and pathological findings of these 5 patients are summarized in Table 1 and Figure 1. Among these, eosinophilia was observed in case #4 and #5. Other 3 patients (case #1, #2 and #3) diagnosed with AML did not show eosinophilia. Abnormal T-cell population was not detected by flow cytometry in all tested cases including case #4 and #5. In these 5 patients, 4 patients had STAT5B p.N642H activating mutation. One patient (case #4) had two STAT5B activating mutations, p.T628S and p.V712E, neither of which has been previously reported in myeloid neoplasms with eosinophilia [13]. All 5 cases had co-mutations. TET2 was the most frequent co-mutation, observed in 3 patients, followed by NPM1, IDH2 and SRSF2, which were observed in 2 patients (Suppl Table 1). Examination of the variant allele frequencies (VAF) revealed that the STAT5B mutations were identified with high VAF in 40% (2/5) of cases (Case #1, #5), and were subclonal in 60% (3/5) of cases (Case #2, #3, #4) (Table 1, Suppl Table 1).
Table 1.
Myeloid neoplasms with STAT5B mutations
| Case # | Age | Gender | Diagnosis by WHO classification | WBC (/mcl) | Eosinophilia (Y/N) | PB eosinophils (%) | PB absolute eosinophil count (/mcl) | BM Blast (%) | Abnormal T-cells by flow cytometry | Karyotype | STAT5B mutation(s) (VAF%) | Therapy | Follow up (months) | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 77 | F | AML with with mutated NPM1 | 40300 | N | 1 | 400 | 66 | Not detected | 46,XX[20] | p.N642H (38.9%) | Decitabine, Enasidenib | 12 | DOD |
| 2 | 70 | F | AML, therapy-related* | 2200 | N | 4 | 100 | 34 | ND | 46,XX[20] | p.N642H (10.1%) | Cytarabine, daunorubicin, HIDAC | 24 | AWD |
| 3 | 71 | F | AML, NOS** | 8000 | N | 3 | 200 | 8 | Not detected | 46,X,t(X;9)(p22.1;q34.3)[15]/46,XX[5] | p.N642H (12.2%) | CLAG-M, Enasidenib | 30 | DOD |
| 4 | 88 | M | MDS-RS-SLD | 7100 | Y | 24 | 1700 | 4 | Not detected | 47,XY,+8[16]/46,idem,-Y[8]/46,XY[1] | p.T628S (5.4%); p.V712E (1.8%) | Decitabine, EPO, G-CSF | 60 | AWD |
| 5 | 74 | M | CMML | 17000 | Y | 14.7 | 2500 | 3 | Not detected | 46,XY[20] | p.N642H (44.9%) | Observation | 10 | DOC |
AML; acute myeloid leukemia. NOS; not otherwise specified. MDS-RS-SLD; myelodysplasic syndrome with ring sideroblasts and single lineage dysplasia. CMML; chronic myelomonocytic leukemia. PB; peripheral blood. BM; bone marrow.
ND; not done. HIDAC; high-dose cytarabine. CLAG-M; Cladribine, Cytarabine, G-CSF, Mitoxantrone. DOD; died of disease. AWD; alive with disease. DOC; died of complications
History of marginal zone lymphona, status post chemotherapy
Persistent AML, NOS, showing 8% blast in the bone marrow after induction therapy
Figure 1:
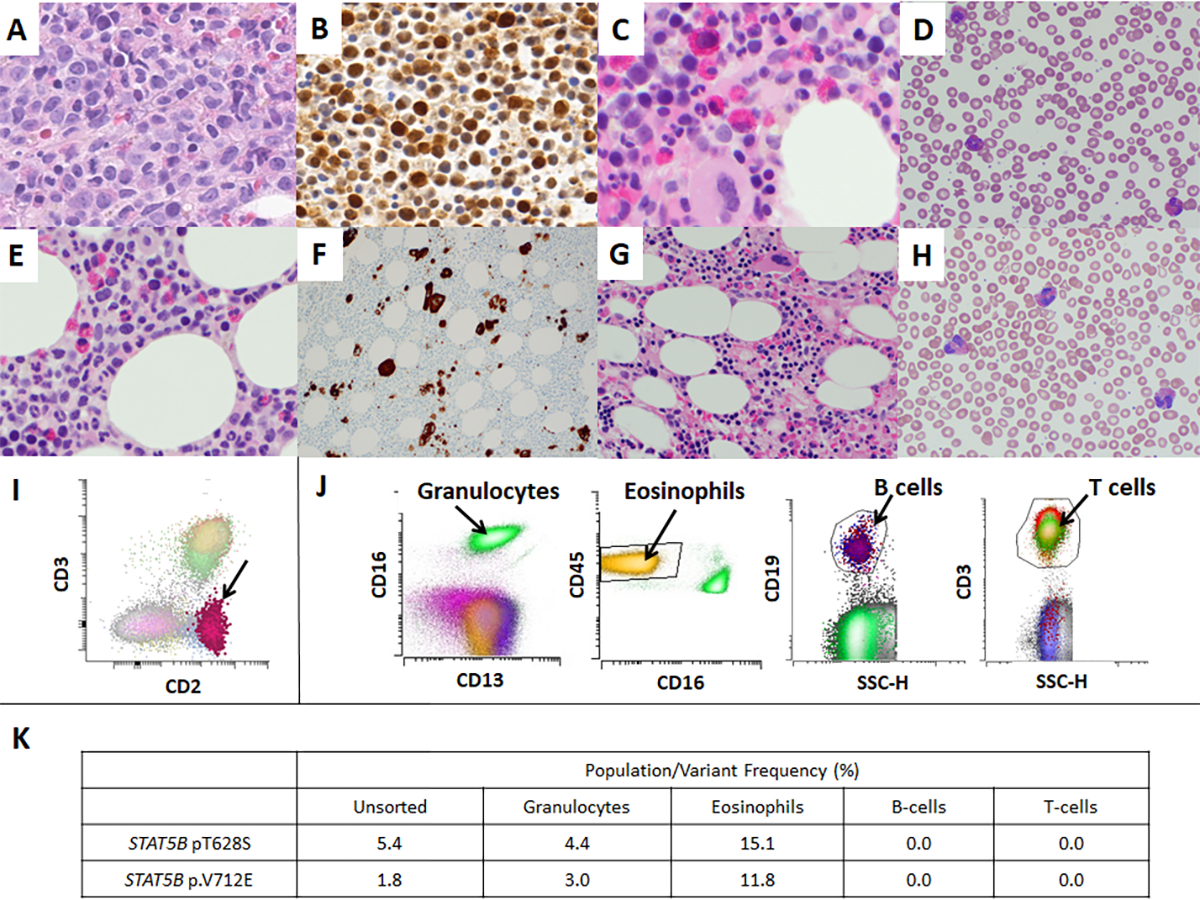
Microscopic pictures of myeloid neoplasms with STAT5B mutations (A-F) and bone marrow of a patient with peripheral T-cell lymphoma (PTCL)-NOS with STAT5B mutation (G-H). A: Marrow shows sheets of myeloblasts (Case #1, Bone marrow biopsy, HE x400), B: pSTAT5 immunohistochemistry shows immunoreactivity in myeloblasts (Case #1, Bone marrow biopsy, pSTAT5 ×400), C: Eosinophils and precursors are increased (Case #4, Bone marrow biopsy, HE x400), D: Peripheral blood shows increase in eosinophils (Case #4, Peripheral blood, Giemsa x500), E: Eosinophils and precursors are increased (Case #5, Bone marrow biopsy, HE x400), F: CD61 highlights numerous dysplastic megakaryocytes (Case #5, Bone marrow biopsy, CD61 ×200), G: Bone marrow shows increased eosinophils and precursors (PTCL-NOS, Bone marrow biopsy, HE x200), H: Peripheral blood shows increase in eosinophils (PTCL-NOS, Peripheral blood, Giemsa x500), I: Flow cytometry performed with bone marrow aspirate sample of PTCL-NOS patient shows small abnormal T-cell population with loss of surface CD3 (0.26% of total WBC), consistent with minimal bone marrow involvement by PTCL-NOS. J: Flow sorting was performed with a peripheral blood sample of Case #4 to isolate granulocytes, eosinophils, B-cells and T-cells to further perform NGS in each population, K: The results of subsequent next generation sequencing in each population of Case #4. STAT5B mutations were identified in granulocytes and eosinophils, but not in B-cells or T-cells.
We subsequently performed flow sorting and NGS to evaluate the difference of genomic findings between STAT5B mutated myeloid neoplasm with eosinophilia and STAT5B mutated lymphocyte-variant hypereosinophilia. We identified a case of peripheral T-cell lymphoma, not otherwise specified (PTCL-NOS) with STAT5B p.T628S, diagnosed with retroperitoneal lymph node biopsy in our pathology archives. This patient presented with hypereosinophilia (3300/mcL), and bone marrow biopsy performed for disease staging showed small abnormal T-cell population by flow cytometry (0.26% of total WBC), consistent with minimal bone marrow involvement by PTCL-NOS (Figure 1). We performed flow cytometry cell sorting of peripheral blood sample of patient #4 and bone marrow aspirate of PTCL-NOS patient to isolate granulocytes, eosinophils, B-cells, and T-cell population, and performed NGS in each fraction. Cell sorting was performed on BD FACS Aria Fusion (BD Biosciences, San Jose, CA) by four-way sorting using an 85-μm nozzle. The T-cell population and the B-cell population were sorted using CD3+ gate and CD19+ gate, respectively, into RPMI-1640 (Corning Inc., Corning, NY) with 20% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA) in 5-mL flow tube and chilled at 4 °C via water recirculator. Sorting for the granulocytes and eosinophil population were performed with the same methodology. The results of NGS combined with flow cytometry sorting were summarized in Figure 1 and Suppl Table 2. In case #4, STAT5B p.T628S and p.V712E variants were identified in the granulocytes and eosinophils, but not in B-cells or T-cells. In lymphocyte-variant hypereosinophilia case, STAT5B p.T628S was identified in T-cells but not in eosinophils.
STAT5B N642H mutation is located in the center of the STAT5B SH2 domain, the phosphotyrosine-binding domain that is essential for the formation of parallel STAT5 dimers and efficient nuclear translocation and has been described as a gain-of-function mutation that causes enhanced and prolonged tyrosine phosphorylation [10]. In our study, not all myeloid neoplasm with STAT5B activating mutation showed hypereosinophilia. 3 cases of AML did not show eosinophilia despite the presence of STAT5B p.H642H with relatively high allele frequency. Case series of Cross et al. also do not have AML among 27 cases, whereas this study includes 7 cases of idiopathic hypereosinophilic syndrome (HES) and 2 cases of chronic eosinophilic leukemia, not otherwise specified (CEL-NOS). These are likely related to the status of co-mutations/additional genomic aberrations either leading or not leading to eosinophilic differentiation and expansion of clonal eosinophils. In our study, we confirmed the absence of STAT5B mutations in T-cells by next generation sequencing with flow cytometry sorted samples in case #4. This finding supports that the eosinophilia seen in myeloid neoplasm with STAT5B mutations is primary eosinophilia which the eosinophils form a part of the neoplastic clone. The findings of Cross et al, demonstrating the absence of p.N642H variant in cultured T-cells from 4 cases in their cohort also support our conclusion [6].
Abnormal T-cell population in lymphocyte-variant hypereosinophilia is typically evaluated with flow cytometry; therefore, genetic profile of abnormal T-cell population in lymphocyte-variant hypereosinophilia has not been fully studied to date. It is unclear if specific mutations in T-cells are associated with lymphocyte-variant hypereosinophilia, or mutation profile is similar to those seen in T-cell lymphoma general. Our case had STAT5B p.T628S in abnormal T-cells, confirmed by next generation sequencing performed with flow-sorted T-cells, but not in eosinophils (Suppl Table 2). Two pediatric cases of somatic STAT5B p.N642H mutation presenting nonclonal eosinophilia, urticaria and dermatitis were reported, and this report also shows the presence of STAT5B p.N642H in T-cells, confirmed by cell sorting and digital droplet PCR [8]. Of note, one of the 2 patients in this report presented with marked eosinophilia peaking as high as 77000/mcl and STAT5B p.N642H was detected in sorted eosinophils. Therefore, the diagnosis of primary hypereosinophilia would be rendered for this case rather than reactive eosinophilia as reported.
Prognostic significance and a role of tyrosine kinase inhibitors in myeloid neoplasm with STAT5B mutation are unclear. The median overall survival of STAT5B mutated idiopathic HES cases was only 30 months, compared with previously reported 87% of 5-year survival in a cohort of 98 patients with idiopathic HES [6,14]. A few reports have shown the efficacy of tyrosine kinase inhibitors in some of these patients, suggesting a potential therapeutic target [15]. Further studies will be needed to determine the prognostic significance and efficacy of target therapy in these patients.
In conclusion, although rare, activating STAT5B mutations are seen in myeloid neoplasms, and often, but not always, associated with eosinophilia. Same STAT5B mutation can lead lymphocyte-variant hypereosinophilia when occurred in T-cell population. STAT5B mutations are potential diagnostic, prognostic and targetable marker and should be screened in patients with eosinophilia.
Supplementary Material
Footnotes
Declaration of interest
AD has received personal consultancy fees from InCycle, EUSA and Loxo, and has received research support from Roche and Takeda. MR participates in a scientific advisory board for and has equity in Auron Therapeutics, Inc., has received research support from Roshe, NGM and Beat AML. MY has received personal consultancy fee from Janssen Research and Development. The remaining authors declare no financial interests.
References
- 1.Butt NM, Lambert J, Ali S, et al. Guideline for the investigation and management of eosinophilia. Br J Haematol 2017;176:553–572. [DOI] [PubMed] [Google Scholar]
- 2.Valent P, Gleich GJ, Reiter A, et al. Pathogenesis and classification of eosinophil disorders: a review of recent developments in the field. Expert Rev Hematol 2012;5:157–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bain BJHHP, Arber DA., Tefferi A., Hasserjian RP. Myeloid/lymphoid neoplasms with eosinophilia and gene rearrangement of rearrangement of PDGFRA, PDGFRB or FGFR1, or with PCM1-JAK2 In: Swerdlow SHCE, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J., editor. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (Revised 4th edition). Lyon: IARC; 2017. p 71–80. [Google Scholar]
- 4.Rothenberg ME. Eosinophilia. N Engl J Med 1998;338:1592–1600. [DOI] [PubMed] [Google Scholar]
- 5.Simon HU, Plotz SG, Dummer R, Blaser K. Abnormal clones of T cells producing interleukin-5 in idiopathic eosinophilia. N Engl J Med 1999;341:1112–1120. [DOI] [PubMed] [Google Scholar]
- 6.Cross NCP, Hoade Y, Tapper WJ, et al. Recurrent activating STAT5B N642H mutation in myeloid neoplasms with eosinophilia. Leukemia 2019;33:415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sreedharanunni S, Jamwal M, Balakrishnan A, et al. Chronic eosinophilic leukemia with recurrent STAT5B N642H mutation-An entity with features of myelodysplastic syndrome/ myeloproliferative neoplasm overlap. Leuk Res 2022;112:106753. [DOI] [PubMed] [Google Scholar]
- 8.Ma CA, Xi L, Cauff B, et al. Somatic STAT5b gain-of-function mutations in early onset nonclonal eosinophilia, urticaria, dermatitis, and diarrhea. Blood 2017;129:650–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson EI, Tanahashi T, Sekiguchi N, et al. High incidence of activating STAT5B mutations in CD4-positive T-cell large granular lymphocyte leukemia. Blood 2016;128:2465–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pham HTT, Maurer B, Prchal-Murphy M, et al. STAT5BN642H is a driver mutation for T cell neoplasia. J Clin Invest 2018;128:387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swerdlow SH CE, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (Revised 4th edition). Lyon: IARC; 2017. [Google Scholar]
- 12.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinney M, Moffitt AB, Gaulard P, et al. The Genetic Basis of Hepatosplenic T-cell Lymphoma. Cancer Discov 2017;7:369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardanani A, Lasho T, Wassie E, et al. Predictors of survival in WHO-defined hypereosinophilic syndrome and idiopathic hypereosinophilia and the role of next-generation sequencing. Leukemia 2016;30:1924–1926. [DOI] [PubMed] [Google Scholar]
- 15.Eisenberg R, Gans MD, Leahy TR, et al. JAK inhibition in early-onset somatic, nonclonal STAT5B gain-of-function disease. J Allergy Clin Immunol Pract 2021;9:1008–1010 e1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


