Abstract
The herpes simplex virus 1 infected cell protein 22 (ICP22), the product of the α22 gene, is a nucleotidylylated and phosphorylated nuclear protein with properties of a transcriptional factor required for the expression of a subset of viral genes. Here, we report the following. (i) ICP22 interacts with a previously unknown cellular factor designated p78 in the yeast two-hybrid system. The p78 cDNA encodes a polypeptide with a distribution of leucines reminiscent of a leucine zipper. (ii) In uninfected and infected cells, antibody to p78 reacts with two major bands with an apparent Mr of 78,000 and two minor bands with apparent Mrs of 62,000 and 55,000. (ii) p78 also interacts with ICP22 in vitro. (iii) In uninfected cells, p78 was dispersed largely in the nucleoplasm in HeLa cells and in the nucleoplasm and cytoplasm in HEp-2 cells. After infection, p78 formed large dense bodies which did not colocalize with the viral regulatory protein ICP0. (iv) Accumulation of p78 was cell cycle dependent, being highest very early in S phase. (v) The accumulation of ICP22 in synchronized cells was highest in early S phase, in contrast to the accumulation of another protein, ICP27, which was relatively independent of the cell cycle. (vi) In the course of the cell cycle, ICP22 was transiently modified in an aberrant fashion, and this modification coincided with expression of p78. The results suggest that ICP22 interacts with and may be stabilized by cell cycle-dependent proteins.
The herpes simplex virus 1 (HSV-1) genome encodes at least 84 different proteins whose synthesis is coordinately regulated and sequentially ordered in a cascade fashion (15, 16). The first group of genes to be expressed are the α genes. The expression of α genes does not require de novo protein synthesis, whereas the expression of β and γ genes expressed later in infection requires functional α proteins. With one exception, that of the α47 gene, the α proteins regulate viral gene expression (for reviews, see references 34 and 35). The product of the α47 gene has been reported to interfere with the presentation of antigenic peptides (40). The focus of this report is on the α22 gene. The domain of the α22 gene encodes two transcripts. The transcript traversing the entire open reading frame (ORF) encodes a 420-amino-acid protein designated infected cell protein 22 (ICP22) (14–16). A smaller transcript originating in the domain of the gene, designated US1.5, directs the synthesis of a protein containing the carboxyl-terminal 273 amino acids of ICP22 (8). Although in this report we shall trace the expression of ICP22, we are not able to separate the function of ICP22 from that of the US1.5 protein.
The α22 gene is dispensable for viral replication in some cells but not others. In primary human cells and in continuous cell lines of rodent derivation, a recombinant virus (R325) lacking the carboxyl-terminal amino acids replicates poorly and is relatively avirulent in experimental animal systems (24, 29, 38). Subsequent studies from this laboratory have suggested that ICP22 may be a transcriptional factor. Thus, Purves et al. have shown that in cells requiring the α22 gene for efficient viral replication, there was a decrease in steady-state levels of mRNA of the α0 gene and of a subset of late genes (e.g., US11) and, concurrently, a decrease in the accumulation of proteins encoded by these genes (31). In still more recent studies, ICP22 was shown to be required for the utilization of one of the four splice acceptor sites of intron 1 of the α0 gene (9). The role of ICP22 in late gene expression became apparent from studies showing that concurrent with the onset of viral DNA synthesis, ICP22 and ICP4, the major regulatory protein, aggregate in nuclear structures with nascent viral DNA, RNA polymerase II, and other proteins and that this aggregation is essential for late gene expression (22). ICP22 has also been reported to be responsible for the aberrant phosphorylation of the large-subunit carboxyl-terminal domain of RNA polymerase II upon HSV-1 infection (32, 33).
ICP22 undergoes extensive posttranslational modifications. These include phosphorylation by viral protein kinases encoded by US3 and UL13, and nucleotidylylation by casein kinase II (5, 25, 26, 30, 31). A virus deleted in the UL13 gene is phenotypically similar to R325, suggesting that phosphorylation is necessary for at least some functions of ICP22 (31). Indeed, in cells infected with mutants from which UL13 had been deleted, ICP22 fails to aggregate in the nuclear structures containing nascent DNA, ICP4, RNA polymerase II, and other factors (22).
In this report, we show that (i) ICP22 interacts with a hitherto unidentified cellular protein designated p78, (ii) p78 synthesis is cell cycle dependent, since the protein is detected only early in the S phase, (iii) the accumulation of ICP22, but not of ICP27, in cells infected with wild-type virus strongly depends on the phase of the cell cycle at the time of infection, with the highest expression of ICP22 occurring during S phase, and (iv) ICP22 made in early S phase is transiently modified in a fashion different from that of the corresponding protein accumulating later in the cell cycle or in asynchronized cells.
These results suggest that p78 may be a regulator of ICP22 expression and that ICP22 is capable of interacting with cell cycle-regulated proteins.
MATERIALS AND METHODS
Cell lines and viruses.
HeLa and HEp-2 cell lines were obtained from the American Type Culture Collection. The human 143TK− cell line was obtained from Carlo Croce. A strain of human foreskin fibroblasts (HFF) was the gift of George Kemble (Aviron Inc., Mountainview Calif.). HSV-1(F) is the prototype HSV-1 strain used in this laboratory (11). Recombinant virus R7353 is deleted for the UL13 and US3 genes and has been described previously (31).
Plasmids.
pRB4965 was constructed by ligating the 1.5-kb HeLa cDNA encoding the carboxyl-terminal 370 amino acids of p78 as an EcoRI/XhoI fragment in frame with the glutathione S-transferase (GST) gene in pGEX4T-3 (Pharmacia). The GST-ORF P recombinant pRB4966 has been described elsewhere (7). pRB5113 contains the entire coding sequence of the 22/US1.5 α genes fused in frame with the DNA-binding domain of GAL4 in pGBT9 (Clontech).
Yeast two-hybrid system and isolation of cDNA clones.
pRB5113 was transformed into yeast strain HF7c (Clontech). Transformants were grown to large scale and transformed with a HeLa cDNA library cloned in pGADGH (Clontech) or a cDNA library derived from an Epstein-Barr virus-immortalized human peripheral blood B-lymphocyte cell line (Clontech). Positive clones encoding interacting polypeptides were selected and isolated as described previously (7). cDNA clones relevant to these studies were isolated by hybridization of a HeLa S3 cDNA library cloned into λgt11 (human HeLa S3 5′-STRETCH PLUS cDNA library; Clontech) with the 1.5-kb HeLa cDNA isolated from the two-hybrid screen as a probe. Seven positive clones were isolated and shown to contain inserts ranging from 0.5 to 2.6 kbp. The largest insert was subcloned into pGEM3Zf(−) (Promega, Madison, Wis.) and sequenced on both strands.
Preparation of cell extracts.
HeLa cell extract for GST affinity precipitation was prepared as follows. Subconfluent HeLa cells grown in a 150-cm2 flask were infected with 10 PFU of HSV-1(F) per cell. After incubation for 18 h at 37°C, the infected cells were scraped into phosphate-buffered saline (PBS), washed once with PBS, and resuspended in 600 μl of PBS* (1% deoxycholate–1% Nonidet P-40–10 M tolylsulfonyl phenylalanyl chloromethyl ketone–10 M α-tosyl-l-lysine chloromethyl ketone in PBS). Binding reactions were done with 150 to 200 μl of cell extract. For electrophoretic separation of proteins in denaturing gels, cells from 25 and 150-cm2 flasks were scraped into PBS, rinsed with PBS, and resuspended in 200 and 500 μl, respectively, of 2× disruption buffer (100 mM Tris-HCl [pH 6.8], 200 mM dithiothreitol, 4% sodium dodecyl sulfate, 0.2% bromphenol blue, 20% glycerol).
Antibodies.
Monoclonal antibodies to ICP0 and ICP27, purchased from the Goodwin Institute, Plantation, Fla., and the polyclonal antiserum R77 to ICP22 have been described elsewhere (1, 2). Monoclonal antibody to coilin protein was obtained from Maria Carmo-Fonseca, University of Lisbon, Lisbon, Portugal. The polyclonal serum to p78 was generated as follows. Escherichia coli BL21 was transformed with pRB4965, and the fusion protein encoded by the plasmid was purified from a large-scale culture as recommended by the manufacturer (Pharmacia). Two rabbits were injected at Josman Laboratories (Napa, Calif.) subcutaneously with 0.3 to 0.5 mg of fusion protein each time at 14-day intervals. The serum used in the studies reported here was collected 1 week after the fourth immunization.
Immunoblots.
Cell extracts were separated in sodium dodecyl sulfate–7% denaturing polyacrylamide gels as described by Sambrook et al. (36). Proteins were then electrically transferred to nitrocellulose sheets (Bio-Rad), blocked for 1 h in 5% milk (in PBS) at room temperature, and then reacted for 2 h at room temperature with the primary antibody diluted in 1% bovine serum albumin (BSA) (in PBS). Monoclonal antibodies to ICP27 and polyclonal antibody to ICP22 were diluted 1:500, whereas the p78 antiserum was diluted 1:1,500. The immunoblots were rinsed four times with 5% milk, reacted for 1 h at room temperature with goat anti-rabbit or goat anti-mouse antibody conjugated to alkaline phosphatase diluted 1:3,000 in 5% milk, and rinsed once with 5% milk and four times with PBS. Colorimetric reactions were done as recommended by the manufacturer (Bio-Rad).
Immunofluorescence.
Cells grown in wells on 1- by 3-in. slides were infected with 106 PFU of HSV-1(F). After incubation at 37°C, the cells were fixed in methanol for 20 min at −20°C, reacted for 30 to 60 min in PBS containing 20% normal human serum and 1% BSA at room temperature, rinsed once with PBS, and then reacted for 18 to 24 h at 4°C with the primary antibodies diluted in PBS containing 10% normal human serum and 1% BSA. Dilutions were 1:300 for the p78 antiserum and 1:200 for the ICP0 antibody. The cells were then washed three times with PBS, reacted for 1 h at room temperature with goat anti-rabbit immunoglobulin G (IgG) conjugated to fluorescein isothiocyanate (FITC; Sigma) and goat anti-mouse IgG conjugated to Texas red (Molecular Probes), rinsed again three times with PBS, and mounted in PBS containing 90% glycerol and 1 mg of p-phenylenediamine per ml. The slides were examined with a Zeiss confocal fluorescence microscope, and digitized images were acquired with software provided with the microscope and printed by a Tektronix 440 phaser printer. Single-color images were acquired by excitation using an argon-krypton laser at 488 nm (FITC) or 568 nm (Texas red). Double-stained images were acquired by a split image of both fluorochromes filtered by a 515- to 540-nm band pass (FITC) and 590 nm long-pass (Texas red) filters and subsequent overlay of the two-color images.
Synchronization of HeLa cells.
HeLa cells were blocked in early S phase as described by Johnson et al. (18). HeLa cells (106 cells per 25-cm2 flask) were incubated in medium supplemented with 2 mM thymidine for 24 to 25 h at 37°C, washed twice in regular medium, and incubated for 12 to 14 h in regular medium. Cells were then incubated again in medium containing 2 mM thymidine for another 24 h and released from the block by rinsing the cells twice in regular medium.
RESULTS
ICP22 interacts with a previously unidentified cellular factor.
In this series of experiments, we screened a HeLa cDNA library cloned in the two-hybrid system vector pGADGH for cellular gene products interacting with ICP22, using the entire α22 gene as bait. Approximately 2 × 106 yeast colonies were screened, and three clones were shown to specifically interact with ICP22 in the yeast two-hybrid systems when assayed in a β-galactosidase assay. The three clones contained the same 1.5-kb insert, as shown by restriction enzyme digestion and partial DNA sequencing (data not shown). A clone containing the same sequence was found in a screen using a cDNA library derived from an Epstein-Barr virus-immortalized human peripheral blood B-lymphocyte cell line (Clontech). One insert was sequenced completely on both strands of the DNA and found to contain a 370-codon ORF. A HeLa cDNA library was screened with the original 1.5-kb insert as a probe, and several clones were isolated. The largest clone contained an insert of approximately 2.6 kb. Sequencing of this clone revealed an ORF of 534 codons preceded by a 500-bp 5′ untranslated sequence (Fig. 1). The ORF in the 1.5-kb insert corresponds to the 3′ domain of the 534 codon ORF in the 2.6-kb cDNA. We conclude that ICP22 interacts with the carboxyl-terminal 370 amino acids of the protein encoded by the 2.6-kb DNA.
FIG. 1.
Nucleotide and amino acid sequences of p78. The amino acid sequence is shown below the DNA sequence. The underlined sequence refers to the putative leucine zipper region. Hydrophobic residues occurring every seven amino acids are circled. The boxed amino acid sequences are potential nuclear localization signals. Numbers on the right refer to the DNA sequence.
No significant homologies to any known DNA or amino acid sequence were detected by either the FastA or BLAST computer program (3, 27). The protein was designated p78 since an antiserum raised against a bacterial fusion protein containing the original 370 amino acids reacted with two major polypeptide bands with an apparent Mr of 78,000 in an immunoblot (see Materials and Methods and below). The predicted amino acid sequence of p78 (Fig. 1) contained a stretch of hydrophobic residues (leucines and methionines) between amino acid residues 152 and 201, reminiscent of a leucine zipper (4, 17).
GST-p78 chimeric protein binds ICP22 from infected cell lysates.
To verify the results of the two-hybrid-system assays, we constructed a fusion protein consisting of GST fused in frame with the 370 codons contained in the original 1.5-kbp cDNA insert and shown to react with ICP22 in the two-hybrid system. The chimeric protein GST-p78 was induced and purified from bacterial cultures and then reacted with HSV-infected HeLa cell extract as described in Materials and Methods. As shown in Fig. 2, the GST-p78 chimeric protein bound native ICP22, whereas GST alone or an irrelevant GST fusion protein did not. The GST-p78 chimeric protein did not bind any of the other α proteins, ICP0, ICP4, and ICP27 (data not shown). Interestingly, of the four ICP22 bands prominent in infected HeLa cell extract, the second-fastest-migrating band was the most intense in the GST pull-down experiment even though it was not the most abundant ICP22 isoform in the infected cell extract (compare lanes 2 and 4 in Fig. 2). These results suggest that p78 has a higher affinity for this isoform of ICP22 than for the other ones. This band most likely represents an ICP22 posttranslationally modified differently from the ICP22 isoforms forming the other bands (2, 30).
FIG. 2.
Photographic image of cell proteins bound to GST fusion proteins, electrophoretically separated in denaturing gels and reacted with a serum to ICP22. Recombinant pGEX vectors were grown in E. coli BL21, and fusion proteins were isolated as recommended by the manufacturer (Pharmacia). Fusion protein (2 to 5 μg) was mixed with HSV-1(F)-infected HeLa extract and incubated at 4°C for 2 to 4 h. Beads were collected by centrifugation, washed three times with PBS* (see Materials and Methods), and resuspended in 60 μl of 2× disruption buffer. Proteins were separated in 7% denaturing polyacrylamide gel, transferred to nitrocellulose, blocked, reacted with R77 and then with a goat anti-rabbit antibody conjugated to alkaline phosphatase, and processed as described by the manufacturer (Bio-Rad). Lanes 1 to 3, HeLa cell extract bound to GST, GST-p78, and GST-ORF P, respectively; lane 4, cell extract from infected HeLa cells.
Accumulation of the p78-related protein is cell cycle dependent.
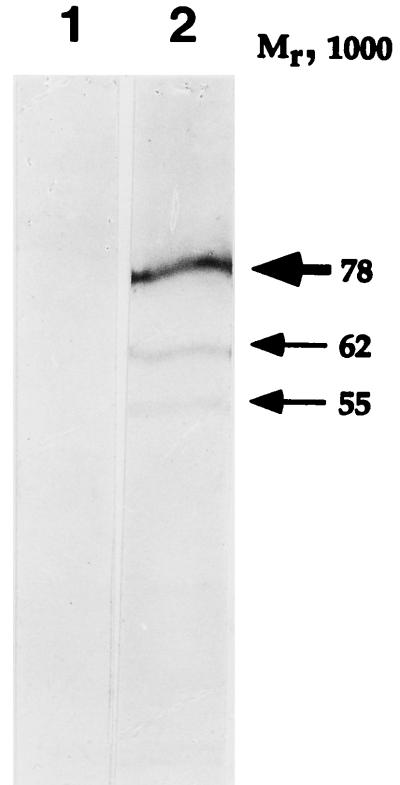
To determine the properties of the native eukaryotic cell protein, a polyclonal rabbit antiserum was raised against the GST-p78 chimeric protein as described in Materials and Methods. In the first series of experiments, uninfected HeLa cell extract was assayed for the presence of p78 by immunoblotting. The p78 antiserum reacted with four protein bands, whereas the preimmune serum did not react at all (Fig. 3). The doublet containing most of the protein had an apparent Mr of approximately 78,000, whereas the two other bands had apparent Mrs of 62,000 and 55,000. At present it is not clear whether the two minor bands represent the native protein, the degradation products of the high-molecular-weight form, or, for example, products of alternatively spliced mRNAs. Intriguingly, an in vitro transcription-translation reaction of the 2.6-kb cDNA described above yielded a polypeptide with an apparent Mr of approximately 55,000 (data not shown), well within the expected range for an ORF encoding a protein of 534 amino acids. These results suggest that p78 represented a highly modified polypeptide. It should be stressed that this pattern was very reproducible from one experiment to another and was also characteristic of lysates of infected HEp-2 cells, 143TK− cells, or HFF (see below and data not shown).
FIG. 3.
Photographic image of HeLa cell proteins, electrophoretically separated in denaturing gels and reacted with a preimmune serum (lane 1) or with a serum to p78 (lane 2). Uninfected HeLa cell extract was separated in a 10% denaturing polyacrylamide gel, blocked, reacted with a rabbit preimmune serum or serum to p78 (see Materials and Methods) and then with a goat anti-rabbit antibody conjugated to alkaline phosphatase, and processed as described by the manufacturer (Bio-Rad). The major p78 band resolves as a doublet in lower-percentage polyacrylamide gels.
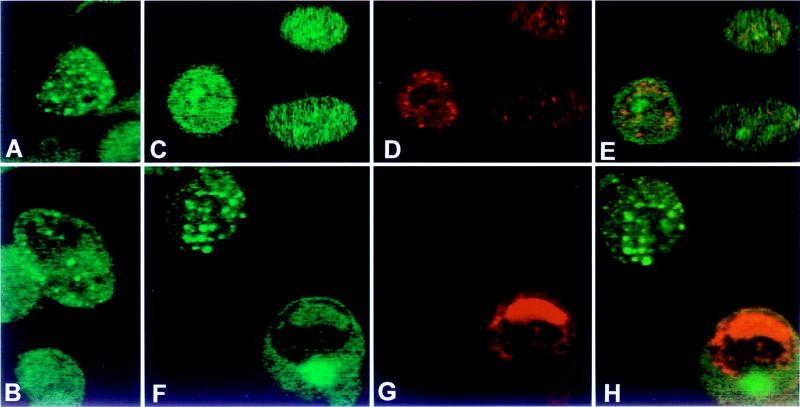
The purpose of the second series of experiments was to localize by immunofluorescence the cellular compartment in which the native p78 protein accumulated in infected and uninfected cells. The results were as follows (Fig. 4). (i) Only a small fraction of total cells reacted with the anti-p78 antibody (Fig. 4). (ii) The localization varied depending on the cell line. The protein localized primarily in the nuclei of HeLa cells and in both the nuclei and cytoplasm of HEp-2 cells (Fig. 4). (iii) In infected HEp-2 cells, p78 was localized in a few dense bodies in the nucleus or at the nuclear membrane. However, these structures did not colocalize with the regulatory protein ICP0 at early or late times after infection (Fig. 4) or with the coilin protein (data not shown).
FIG. 4.
Photomicrographs of uninfected (A and B) and HSV-1(F)-infected (C to E and F to H) HeLa cells (B) and Hep-2 cells (A, C to E, and F to H) reacted with an antibodies to p78 and ICP0 (see Materials and Methods). Infected cells were fixed at 3 h after infection (C to E) or at 18 h after infection (F to H). Immunofluorescence was done as described in Materials and Methods. (A, B, C, and F) FITC-labeled anti-rabbit IgG antibody to p78 antiserum; (D and G) Texas red-labeled anti-mouse IgG to the ICP0 antibody; (E and H) overlays of panels C and D and panels F and G, respectively.
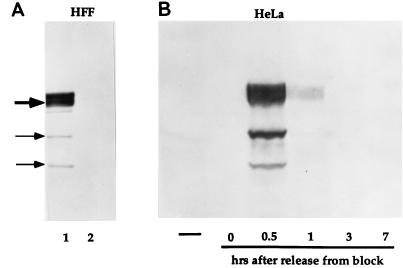
The observation that only a fraction of the cells contained detectable amounts of p78 related, native protein raised the possibility that the expression of the p78 protein was cell cycle dependent. To test this hypothesis, two sets of experiments were done. In the first, HFF were harvested at a cell confluency of approximately 50%, or after they were fully confluent and contact inhibited, and the presence of p78 was then assayed by immunoblotting. As shown in Fig. 5A, p78 was present in dividing HFF cells (lane 1), but could not be detected in non-dividing cells (lane 2).
FIG. 5.
Photographic images of uninfected HFF and HeLa cell extracts, electrophoretically separated in denaturing gels and reacted with a serum to p78. (A) HFF extracts were prepared at 50% cell confluency (lane 1) and at 100% cell confluency (lane 2), separated in 7% denaturing gels, transferred to nitrocellulose, blocked, and reacted with a serum to p78 and then with a goat anti-rabbit antibody conjugated to alkaline phosphatase. Gel loading was monitored by staining for actin (not shown). (B) HeLa cell extracts were prepared from unsynchronized cells (leftmost lane) and from synchronized cells at 0, 0.5, 1, 3, and 7 h after release from block (see Materials and Methods) and processed as described above.
In a second set of experiments, HeLa cells were blocked at the G1/S interphase as described in Materials and Methods, harvested at different time intervals after release from the block, solubilized, subjected to electrophoresis in denaturing polyacrylamide gels, transferred to a nitrocellulose sheet, and probed with the anti-p78 antibody. As shown in Fig. 5B, p78 accumulation was highest at 0.5 h after release from the block. Thus, p78 levels were highest very early in S phase but undetectable in G2, i.e., at 7 h after release from the block.
ICP22 accumulates preferentially in cells infected during early S phase.
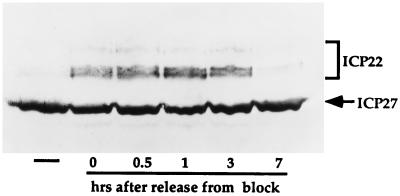
The results described above suggested that the functions of ICP22 may be cell cycle dependent. To determine whether expression of ICP22 was affected by the phase of the cell cycle in which viral gene expression was initiated, HeLa cells were blocked at the G1/S boundary as described above and infected with wild-type HSV-1 at 0, 0.5, 1, 4, or 8 h after release from the block. Cells were harvested 24 h after infection, solubilized, subjected to electrophoresis in denaturing gels, and reacted with antibody to ICP22 and ICP27. The results were as follows. ICP22 levels were highest in cells infected at 0.5 or 1 h after release from the block, but the protein was almost undetectable in cells infected at 7 h (Fig. 6). In contrast, accumulation of ICP27 was not at all or only slightly affected by the phase of the cell cycle at the time of infection. Inasmuch as the patterns of expression of ICP22 and p78 almost overlap (compare Fig. 5 and 6), these results suggest that the accumulation of ICP22 and of p78 may be determined by similar cell cycle-regulated factors.
FIG. 6.
Photographic image of wild-type virus-infected HeLa cell extracts, separated in denaturing gels and reacted with sera to viral α proteins. Unsynchronized HeLa cells (leftmost lane) and synchronized HeLa cells were infected with HSV-1(F) at the time points shown after release from block (see Materials and Methods) for 24 h. Cell extracts were separated in 7% denaturing polyacrylamide gels, transferred to nitrocellulose sheets, blocked, reacted with sera to ICP22 and ICP27 and then with a goat anti-mouse (ICP27) or a goat anti-rabbit (for ICP22) antibody conjugated to alkaline phosphatase, and processed as recommended by the manufacturer.
ICP22 made early in S phase is posttranslationally modified differently from that made in asynchronously dividing HeLa cells.
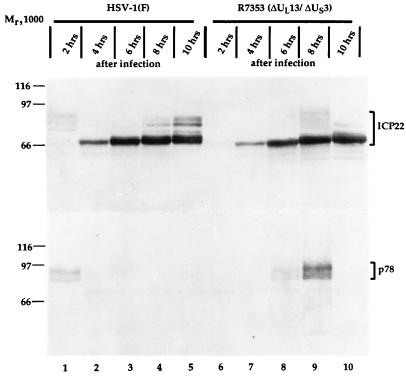
The results described above indicate that the accumulation of ICP22 depended on the phase of the cell cycle at the time of infection and that the highest accumulation occurred in cells infected very early in the S phase. To further investigate this observation, HeLa cells were synchronized as described above and infected with HSV-1(F) 0.5 h after release from the block. The cells were harvested at 2-h intervals after infection, and extracts were prepared and processed for immunoblotting as described in Materials and Methods. As shown in Fig. 7, lane 1, infected synchronized HeLa cells yielded at 2 h after infection novel, hitherto undetected forms of ICP22 in addition to posttranslationally processed forms common to productive infection (lanes 2 to 5) and described earlier (30, 31). This novel modification of ICP22 coincided with expression of p78 (lane 1). This modification was not dependent on the expression of the viral protein kinases encoded by UL13 and US3, respectively, inasmuch as infection of synchronized HeLa cells with the recombinant virus R7353 lacking both UL13 and US3 yielded identical novel forms of ICP22, although at later times after infection than in cells infected with wild-type virus (lanes 6 to 10). As in the case of cells infected with the wild-type virus HSV-1(F), however, the accumulation of this novel form of ICP22 coincided with expression of p78 (lanes 8 and 9).
FIG. 7.
Photographic image of cell extracts infected with wild-type virus or with recombinant R7353, separated in denaturing gels and reacted with sera to ICP22 and p78. Synchronized HeLa cells were infected at 0.5 h after release from block with either HSV-1(F) (lanes 1 to 5) or recombinant R7353 (lanes 6 to 10), and extracts were prepared at the times indicated after infection. Cell extracts were separated in duplicate 7% denaturing polyacrylamide gels, transferred to nitrocellulose sheets, and reacted with sera to ICP22 (upper panel) and p78 (lower panel).
DISCUSSION
The salient features of this report are as follows. (i) We have identified p78, a previously unknown cellular protein that interacts with ICP22 in the yeast two-hybrid system and in in vitro assays. Although p78 does not have significant homology to other known proteins or nucleotide sequences, it contains a leucine zipper-like region. The protein sequence of p78 also contains several stretches of basic residues which could act as nuclear localization signals (Fig. 1; see reference 10 for a review). These features of p78 are characteristic of regulatory proteins (17). The hypothesis that p78 is a regulatory protein which interacts with ICP22 is tantalizing inasmuch as ICP22 has obvious regulatory functions but lacks features characteristic of regulatory proteins such as DNA-binding domains, Zn-binding domains, or ability to transactivate gene expression. This situation is reminiscent of the one described for TIF (VP16) and for the Epstein-Barr virus transactivator EBNA-2 (23). In the latter case, it has been shown that EBNA-2 is targeted to the DNA by a cellular enhancer-binding protein (23). Interestingly, HSV-1 ICP22 and homologs of other alphaherpeviruses share conserved amino acid sequences in their carboxyl-terminal domains that are rich in acidic residues (39). This region could potentially act as a transactivating domain.
(ii) Both p78 and ICP22 are regulated in cell cycle-dependent manner. Evidence in support of this conclusion stems from three observations. First, ICP22 accumulates optimally in cells during the first several hours after release from the G1/S interphase. Surprisingly, ICP27 but not ICP22 was detected in cells infected 7 h after the release and harvested 24 h later. Inasmuch as most asynchronous cells infected at appropriate multiplicity contain ICP22 as detected by immunofluorescence, we hypothesize that ICP22 was made and degraded faster in cells infected in G2 than in cells infected during G1 or S phase. This was the most surprising and unexpected observation made in this study, and it suggests that ICP22 is stabilized by a factor available during G1 or S phase but not later.
Second, ICP22 made during the first few hours after release from block, and infection with wild-type virus was posttranslationally processed differently from that made in the majority of infected cells dividing asynchronously and infected at stages other than at the G1/S interphase. This form disappears later in infection and is therefore either reprocessed or degraded.
Third, p78 accumulated and was detectable only in lysates of cells in early S phase. The interdependence of p78 and ICP22 on the cell cycle phase was particularly apparent in the experiment with ΔUL13/ΔUS3 recombinant virus. In that experiment, the accumulation of both ICP22 and of p78 was delayed by several hours. Both the modified form of ICP22 and p78 accumulated at the same time after release from block and infection. Although the observation that in cells infected with this recombinant both proteins appeared after several hours of delay was reproducible, it may not be significant since we have observed a delay in the maximal accumulation of both proteins in one experiment involving infection of cells released from block with the wild-type, parental virus. Except for the delay in accumulation of ICP22 and of p78, the results do not support a role for the viral protein kinases UL13 and US3 in the novel processing of ICP22 observed in this study.
A central question is the role of the interaction of ICP22 and p78. The evidence presented in Fig. 2 is that p78 binds all forms of ICP22 but appears to prefer one form, and therefore the two proteins could be expected to interact in the infected cells. The function of the interaction is not known. It should be noted, however, that there is increasing evidence that herpesvirus proteins interact with cellular proteins to either block cellular proteins or stabilize their function. For example, among α proteins, it has been reported that α47 binds to TAP1/TAP2 to block the transport of antigenic peptides to the surface of the infected cell (40), ICP22 has been reported to modify the phosphorylation of RNA polymerase in some infected cells (32, 33), and ICP27 binds and sequesters components of spliceosomes (28, 37), whereas ICP0 binds a ubiquitin-specific protease, binds to and stabilizes cyclin D3, and binds the translation elongation factor 1 (12, 19, 20). Among proteins made later in infection, UL13 protein kinase hyperphosphorylates elongation factor 1, γ134.5 protein binds to protein phosphatase 1α to dephosphorylate the α subunit of eukaryotic translation initiation factor 2, and the US9 protein is ubiquitinated and is bound to proteosomes but appears to be stable (6, 13, 21). The involvement of herpesviruses with cell cycle progression is evident not only from the stabilization of cyclin D3 by ICP0 but also by the observation that some herpesviruses encode a cyclin D3 homolog. It is conceivable that other viral proteins including ICP22 have evolved interactions with cellular proteins to control specific aspects of the cell cycle progression.
Nucleotide sequence accession number. The accession number for p78 is AF068007.
ACKNOWLEDGMENTS
We thank Patricia L. Ward for assistance with confocal microscopy.
This study was aided by PHS grant CA47451 from the National Cancer Institute.
REFERENCES
- 1.Ackermann M, Braun D K, Pereira L, Roizman B. Characterization of herpes simplex virus 1 proteins 0, 4, and 27 with monoclonal antibodies. J Virol. 1984;52:108–118. doi: 10.1128/jvi.52.1.108-118.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann M, Sarmiento M, Roizman B. Application of antibody to synthetic peptides for characterization of the intact and truncated α22 protein specified by herpes simplex 1 and the R325 α22− deletion mutant. J Virol. 1985;56:207–215. doi: 10.1128/jvi.56.1.207-215.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Baxevanis A D, Vinson C R. Interactions of coiled coils in transcription factors: where is the specificity? Curr Opin Genet Dev. 1993;3:278–285. doi: 10.1016/0959-437x(93)90035-n. [DOI] [PubMed] [Google Scholar]
- 5.Blaho J A, Mitchell C, Roizman B. Guanylylation and adenylylation of the α regulatory proteins of the herpes simplex virus require a viral β or γ function. J Virol. 1993;67:3891–3900. doi: 10.1128/jvi.67.7.3891-3900.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandimarti R, Roizman B. US9, a stable lysine-less herpes simplex virus 1 protein, is ubiquitinated before packaging into virions and associates with proteasomes. Proc Natl Acad Sci USA. 1997;94:13973–13978. doi: 10.1073/pnas.94.25.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruni R, Roizman B. Open reading frame P—a herpes simplex virus gene repressed during productive infection encodes a protein that binds a splicing factor and reduces synthesis of viral proteins made from spliced mRNA. Proc Natl Acad Sci USA. 1996;93:10423–10427. doi: 10.1073/pnas.93.19.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter K I, Roizman B. The promoter and transcriptional unit of a novel herpes simplex virus 1 α gene are contained in, and encode a protein in frame with, the open reading frame of the α22 gene. J Virol. 1996;70:172–178. doi: 10.1128/jvi.70.1.172-178.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter K L, Roizman B. Alternatively spliced mRNAs predicted to yield frame-shift proteins and stable intron 1 RNAs of the herpes simplex virus 1 regulatory gene α0 accumulate in the cytoplasm of infected cells. Proc Natl Acad Sci USA. 1996;93:12535–12540. doi: 10.1073/pnas.93.22.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 11.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 12.Everett R D, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:566–577. doi: 10.1093/emboj/16.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He B, Gross M, Roizman B. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honess R W, Roizman B. Proteins specified by herpes simplex virus. XI. Identification and relative molar rates of synthesis of structural and nonstructural herpes virus polypeptides in the infected cell. J Virol. 1973;12:1347–1365. doi: 10.1128/jvi.12.6.1347-1365.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honess R W, Roizman B. Regulation of herpes simplex virus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honess R W, Roizman B. Regulation of herpes simplex virus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci USA. 1975;72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurst H C. Sequences of bZIP proteins. Prot Prof. 1994;1:125–134. [Google Scholar]
- 18.Johnson R T, Downes C S, Meyn R E. The synchronization of mammalian cells. In: Fantes P, Brooks R, editors. The cell cycle. A practical approach. Oxford, England: IRL Press; 1993. pp. 1–24. [Google Scholar]
- 19.Kawaguchi Y, Bruni R, Roizman B. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1Δ:ICP0 affects translational machinery. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi Y, Van Sant C, Roizman B. Eukaryotic elongation factor 1Δ is hyperphosphorylated by the protein kinase encoded by the UL13 gene of herpes simplex virus 1. J Virol. 1998;72:1731–1736. doi: 10.1128/jvi.72.3.1731-1736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leopardi R, Ward P L, Ogle W O, Roizman B. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the UL13 protein kinase. J Virol. 1997;71:1133–1139. doi: 10.1128/jvi.71.2.1133-1139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meignier B, Longnecker R, Mavromara-Nazos P, Sears A E, Roizman B. Virulence of and establishment of latency by genetically engineered mutants of herpes simplex virus 1. Virology. 1988;162:251–254. doi: 10.1016/0042-6822(88)90417-5. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell C, Blaho J A, Roizman B. Casein kinase II specifically nucleotidylylates in vitro the amino acid sequence of the protein encoded by the 22 gene of herpes simplex virus 1. Proc Natl Acad Sci USA. 1994;91:11864–11868. doi: 10.1073/pnas.91.25.11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell C, Blaho J A, McCormick A L, Roizman B. The nucleotidylylation of herpes simplex virus 1 regulatory protein 22 by human casein kinase II. J Biol Chem. 1997;272:25394–25400. doi: 10.1074/jbc.272.40.25394. [DOI] [PubMed] [Google Scholar]
- 27.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phelan A, Carmo-Fonseca M, McLauchlan J, Lamond A I, Clements J B. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc Natl Acad Sci USA. 1993;90:9056–9060. doi: 10.1073/pnas.90.19.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Post L E, Roizman B. A generalized technique for deletion of specific genes in large genomes: α gene 22 of herpes simplex virus is not essential for growth. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 30.Purves F C, Roizman B. The UL13 gene of the herpes simplex 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein 22. Proc Natl Acad Sci USA. 1992;89:7310–7314. doi: 10.1073/pnas.89.16.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purves F C, Ogle W O, Roizman B. Processing of the herpes simplex virus regulatory protein 22 mediated by the UL13 protein kinase determines the accumulation of a subset of and γ mRNAs and proteins in infected cells. Proc Natl Acad Sci USA. 1993;90:6701–6705. doi: 10.1073/pnas.90.14.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice S A, Long M C, Lam V, Spencer C A. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J Virol. 1994;68:988–1001. doi: 10.1128/jvi.68.2.988-1001.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice S A, Long M C, Lam V, Schaffer P A, Spencer C A. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J Virol. 1995;69:5550–5559. doi: 10.1128/jvi.69.9.5550-5559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roizman B. The function of herpes simplex virus genes: a primer for genetic engineering of novel vectors. Proc Natl Acad Sci USA. 1996;93:11307–11312. doi: 10.1073/pnas.93.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 2231–2295. [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sandri-Goldin R M, Hibbard M K. The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum, and the C terminus appears to be required for this interaction. J Virol. 1996;70:108–118. doi: 10.1128/jvi.70.1.108-118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sears A E, Halliburton I W, Meignier B, Silver S, Roizman B. Herpes simplex virus 1 mutant deleted in the α22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J Virol. 1985;55:338–346. doi: 10.1128/jvi.55.2.338-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwyzer M, Wirth U V, Vogt B, Fraefel C. BICP22 of bovine herpesvirus 1 is encoded by a spliced 1.7 kB RNA which exhibits immediate early and late transcription kinetics. J Gen Virol. 1994;75:1703–1711. doi: 10.1099/0022-1317-75-7-1703. [DOI] [PubMed] [Google Scholar]
- 40.York I A, Roop C, Andrews D W, Riddell S R, Graham F L, Johnson D C. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]