Abstract
We have studied the pathogenesis of influenza virus infection in mice that are unable to respond to type I or II interferons due to a targeted disruption of the STAT1 gene. STAT1−/− animals are 100-fold more sensitive to lethal infection with influenza A/WSN/33 virus than are their wild-type (WT) counterparts. Virus replicated only in the lungs of WT animals following intranasal (i.n.) virus inoculation, while STAT1−/− mice developed a fulminant systemic influenza virus infection following either i.n. or intraperitoneal inoculation. We investigated the mechanism underlying this altered virus tropism by comparing levels of virus replication in fibroblast cell lines and murine embryonic fibroblasts derived from WT mice, STAT−/− mice, and mice lacking gamma interferon (IFNγ−/− mice) or the IFN-α receptor (IFNαR−/− mice). Influenza A/WSN/33 virus replicates to high titers in STAT1−/− or IFNαR−/− fibroblasts, while cells derived from WT or IFNγ−/− animals are resistant to influenza virus infection. Immunofluorescence studies using WT fibroblast cell lines demonstrated that only a small subpopulation of WT cells can be infected and that in the few infected WT cells, virus replication is aborted at an early, nuclear phase. In all organs examined except the lung, influenza A WSN/33 virus infection is apparently prevented by an intact type I interferon response. Our results demonstrate that type I interferon plays an important role in determining the pathogenicity and tissue restriction of influenza A/WSN/33 virus in vivo and in vitro.
Influenza virus infections in humans are acquired by inhalation of aerosolized virus in the form of droplet nuclei and are limited to the epithelial lining of the respiratory tract (22). In normal adults, the disease caused by influenza virus is usually a tracheobronchitis but may become a viral pneumonia. The illness has no viremic phase. Inhaled virus replicates in the respiratory epithelium, and new virions bud into the airway lumen. The tissue restriction is thought to be determined at least in part by the viral hemagglutinin (HA) molecule (17). The influenza virus HA mediates attachment to sialic acid-containing receptors on the host cell surface, as well as fusion of the virus envelope with the cellular membrane. Infectivity requires proteolytic processing of the HA precursor protein, HA0, into the disulfide-linked cleavage products HA1 and HA2 (16, 18, 28). The limited occurrence of HA-activating proteases is believed to be responsible for the localization of the viral infection to the pulmonary epithelium (17). Not all strains of influenza virus, however, share this limitation. Animal influenza A virus strains belonging to two HA subtypes, H5 and H7, have HA molecules that can be cleaved by furin and/or other ubiquitous proteases (34). For example, fowl plague virus (FPV) of subtype H7 is extremely virulent, causing a systemic, rapidly fatal disease in afflicted birds (21). The HA molecules of these viruses are characterized by multiple basic residues at the cleavage site. This feature correlates with the recognition of the cleavage site by ubiquitous proteases and with virus pathogenicity (11). In contrast, mammalian and nonpathogenic avian influenza viruses have HA molecules that contain a single arginine at the cleavage site, and this difference is thought to explain their limited tropism (7, 8). Tissue tropism of influenza viruses generally correlates with virus growth behavior in cultured cells. Nonpathogenic avian influenza viruses and mammalian influenza viruses require the addition of an exogenous protease to cleave their HA molecules for multicycle growth in cell culture. The highly pathogenic H5 and H7 avian influenza viruses can replicate in immortalized cell lines in the absence of exogenous protease.
Influenza A/WSN/33 virus (WSN), a mouse-adapted H1N1 human strain, can undergo multiple replication cycles in several cell lines, including Madin-Darby bovine kidney (MDBK) and Madin-Darby canine kidney (MDCK) cells, in the absence of exogenous trypsin. The HA of this virus appears to be cleaved during viral entry by an endosomal protease that is present in MDCK and MDBK cells (3). Although infection with WSN virus in mice is mainly restricted to the respiratory epithelium after intranasal (i.n.) inoculation, WSN virus is unusual in that it is also able to replicate in brains of adult mice when injected intrathecally (41). Surprisingly, the growth properties of WSN virus in MDBK cells and its neurotropism segregate with its neuraminidase (NA) gene (30, 37) rather than with its HA gene, which encodes the canonical single arginine at the cleavage site (10). The NA mutation responsible for the altered tissue tropism of WSN results in the loss of a glycosylation site (19), but the mechanism by which this alteration affects the cleavage of HA is not well understood.
The altered replication characteristics of WSN virus in cultured cells are not associated with significant changes in the spread of virus in mice that acquire the infection by the respiratory route. The failure of WSN virus to spread beyond the pulmonary surface, despite its ability to replicate in cultured cells without exogenous protease, led us to question whether host responses to infection might also play a role in viral targeting.
We compared the infectivity and tropism of two strains of human influenza virus, WSN and A/PR/8/34 (H1N1) (PR8), in wild-type (WT) mice and mice homozygous for a targeted disruption of the signal transducer and activator of transcription 1 (STAT1) gene (STAT−/− mice). STAT1, a cytoplasmic protein in its latent state, is activated by phosphorylation on tyrosine when it associates with ligand-bound type I or type II interferon (IFN) receptor. Once activated, STAT1 translocates to the nucleus, where it acts as part of a protein complex to activate transcription from IFN-regulated genes (29). In the absence of STAT1, IFN-mediated gene induction does not occur. Mice lacking a functional STAT1 gene show extreme sensitivity to many viruses, including vesicular stomatitis virus, and to bacteria of the genus Listeria, by virtue of their inability to respond to IFN (5, 20).
We have found that the administration of WSN virus by different routes gives rise to a fulminant systemic infection in STAT1−/− mice, similar to FPV in birds, whereas infection is limited to the respiratory tract in WT animals. In the STAT1−/− mice, PR8 virus infection remains restricted to the respiratory tract, although mutant animals are much more susceptible than controls to fatal pneumonia. WSN virus also gives rise to systemic infection in mice lacking the IFN-α receptor (IFNαR−/− mice) but not in mice lacking IFN-γ. In addition, fibroblast cell lines derived from either STAT1−/− or IFNαR−/−, but not from WT or IFNγ−/− animals, are permissive for WSN virus replication. This work demonstrates by genetic techniques that although the HA molecule of the mammalian influenza WSN virus can be cleaved by ubiquitous proteases in vivo (4), the replication of this virus in mice infected i.n. is inhibited, except in the respiratory tract, by the type I IFN system. This inhibition is independent of the IFN-induced Mx protein, since all mouse laboratory strains used in our experiments were deficient in the Mx gene (32, 33).
MATERIALS AND METHODS
Mice.
Mice homozygous for a targeted deletion of STAT1 were generated as previously described (4). IFNγ−/− mice on a C57B6 background were purchased from Jackson Laboratories. IFNαR−/− mice on a 129 background have been described previously (13). Specific-pathogen-free C57B6, 129/J, and CD1 mice were purchased from Taconic Farms or from Jackson Laboratories. Mice were used at 6 to 12 weeks of age.
Viruses and infectivity titrations.
WSN virus and transfectant influenza virus CAT2A/NAmodII were grown in MDBK cells in reinforced minimal essential medium. The transfectant CAT2A/NAmodII virus, which is a genetically engineered WSN virus expressing chloramphenicol acetyltransferase (CAT), has been described previously (24). PR8 virus was grown in the allantoic cavities of 10-day-old embryonated eggs.
For quantitation of WSN virus, tissue samples were homogenized in phosphate-buffered saline (PBS), and dilutions of clarified homogenates were adsorbed for 1 h at 37°C onto monolayers of MDCK cells. Infected monolayers were then overlaid with a solution of minimal essential medium containing 0.1% bovine serum albumin (BSA), 0.01% DEAE-dextran, 0.1% NaHCO3, and 1% agar. Plates were incubated 2 to 3 days until plaques could be visualized. Tissue culture infectious dose (TCID) assays to titrate virus from PR8-infected samples were carried out as follows. Confluent monolayers of MDCK cells in 96-well plates were incubated with log dilutions of clarified tissue homogenates in media. Two to three days after inoculation, 0.05-ml aliquots from each well were assessed for viral growth by hemagglutination assay (HA assay).
The HA assay was carried out in V-bottom 96-well plates. Serial twofold dilutions of each sample in PBS were incubated for 1 h on ice with an equal volume of a 0.5% suspension of chicken erythrocytes in PBS. Positive wells contained an adherent, homogeneous layer of erythrocytes; negative wells contained a nonadherent pellet.
Infections.
Inoculations by the i.n. route were performed in anesthetized mice, using 0.1-ml aliquots of virus stock diluted into PBS. For intraperitoneal (i.p.) infections, 0.2-ml aliquots of virus in PBS were injected into the peritoneal cavity. Animals were monitored daily, and animals observed to be in extremis were sacrificed. All procedures were in accord with National Institutes of Health guidelines on care and use of laboratory animals.
Cells.
Mouse embryo fibroblasts (MEFs) derived from 14- to 16-day mouse embryos were maintained in DMEM (Dulbecco’s minimal essential medium) with 10% newborn calf serum (NCS). Immortalized fibroblast cell lines were derived from MEFs by continuous passage (39). Primary cultures were replated every 3 days at a density of up to 3 × 105 cells per 6-cm-diameter plate until 3T3-like continuous cell lines were established. These were cultured in DMEM with 10% NCS. Subclones of fibroblast cell lines were obtained by limiting dilution.
Histology and immunohistochemistry.
Samples for microscopic examination were fixed in 10% buffered formalin and embedded in paraffin. Five-micrometer sections were stained with hematoxylin-eosin or deparaffinized for immunohistochemistry. Deparaffinized sections were incubated with a 1:1,000 dilution of polyclonal rabbit serum raised against whole WSN virus. The secondary, biotin-conjugated, goat anti-rabbit antibody was purchased from Vector. Immunostaining was developed by incubation with peroxidase-labeled streptavidin (Vector) and aminoethyl carbazole.
Immunoprecipitation of viral proteins.
MEF cell monolayers in 35-mm-diameter dishes were infected at a multiplicity of infection (MOI) of 2 with WSN virus in DMEM with 0.1% BSA. At intervals postinfection (p.i.), cells were labeled with l-[35S]cysteine and l-[35S]methionine for the indicated times. For immunoprecipitation, labeled cells were lysed in 10 mM Tris-HCl (pH 7.4) containing 150 mM NaCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10% glycerol, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate (SDS). Proteins were immunoprecipitated by overnight incubation at 4°C with rabbit polyclonal anti-WSN serum followed by 1 h of incubation at room temperature with protein A-Sepharose. Immunoprecipitated proteins were analyzed by SDS–12% polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography following fluorography using Amplify (Amersham).
CAT assay.
Confluent cell monolayers in 35-mm-diameter dishes were infected with transfectant CAT2A/NAmodII virus at an MOI of 2. Cells were harvested at different times p.i. into 100 μl of 0.25 M Tris-HCl (pH 7.5) and lysed by three rounds of freeze-thawing. Cell extracts were clarified by microcentrifugation for 2 min at 3,000 rpm. CAT enzyme activities in cell extracts were determined as previously described by thin-layer chromatography of the resulting products of enzymatic reactions, using [14C]chloramphenicol as substrate (24). CAT activity was calculated as the percentage of chloramphenicol conversion into its acetylated forms and normalized to the total protein concentration in the extracts as measured in the Bradford assay (Bio-Rad). One unit is defined as the amount of enzyme required to acetylate 1 nmol of [14C]chloramphenicol in 1 min under the reaction conditions (24).
Extraction of vRNA and primer extension assay.
Confluent cell monolayers in 35-mm-diameter dishes were infected with WSN virus at an MOI of 5. Cells were harvested at the indicated times and lysed with guanidinium isothiocyanate. Total RNA was purified by cesium chloride ultracentrifugation, and the amount of viral RNA (vRNA) corresponding to the NA and NS segments of WSN virus was measured by primer extension as previously described (42). Primers 5′-GGAACAATTAGGTCAGAAGT-3′, complementary to the region between nucleotides 695 and 715 of the NS-specific vRNA, and 5′-GTGGCAATAACTAATCGGTCA-3′, complementary to the region between nucleotides 1151 and 1171 of the NA-specific vRNA, were used in these studies. Briefly, 5 μg of total RNA was reverse transcribed in the presence of 3 × 105 cpm of the end-labeled NS- and NA-specific primers, and the extended products were analyzed on a 6% polyacrylamide gel containing 7 M urea. The amount of product was determined with a phosphorimaging system (Molecular Dynamics).
Immunofluorescence studies.
STAT1+/+ or STAT1−/− 3T3-like cells grown on coverslips were infected at an MOI of 2 with WSN virus. At 14 h p.i., cells were fixed for 15 min at −20°C with acetone-methanol (1:1), washed with PBS, and incubated for 1 h at room temperature with a 1:500 dilution in PBS–3% BSA of the NP-specific monoclonal antibody HT103 (23). After three washes with PBS, cells were incubated for 1 h at room temperature with a 1:750 dilution in PBS–3% BSA of a fluorescein isothiocyanate-labeled rabbit anti-mouse antibody (Boehringer Mannheim). Samples were then analyzed by confocal microscopy.
Virus adsorption assay.
Confluent cell monolayers in 35-mm-diameter dishes were incubated with WSN virus at a ratio of 10 PFU per cell for 1 h at 4°C. Cells were then washed three times with ice-cold PBS and disrupted by freeze-thawing in DMEM containing 0.1% BSA. Samples were incubated for 45 min at 37°C with Clostridium perfringens neuraminidase (5 U/ml; Sigma) to liberate adsorbed virus, which was titrated by plaque assay in MDBK cells.
RESULTS
Altered influenza virus sensitivity and clearance in STAT1−/− mice.
We compared the sensitivities of WT and STAT1−/− animals to infection by two strains of H1N1 influenza A virus. Tenfold dilutions of PR8 or WSN virus were administered i.n. to three to four anesthetized animals of each genotype, and survival was scored. In each case, the STAT1−/− animals were more sensitive than control animals, with 50% mortality at doses of virus 1 to 2 logs lower than for their WT counterparts. In WT animals, the 50% lethal doses (LD50s) were 1,000 PFU and 10 TCID for WSN and PR8 viruses, respectively; in STAT1−/− mice, the corresponding LD50s were 10 PFU and 1 TCID.
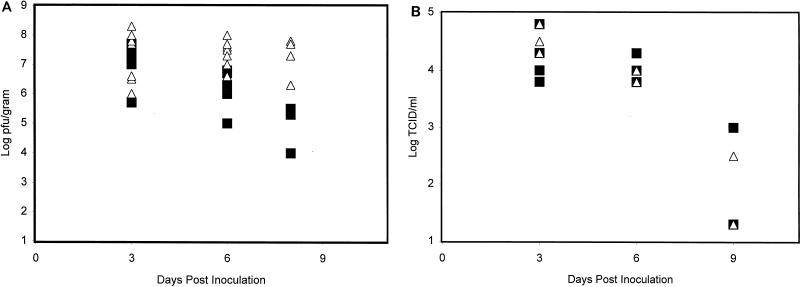
Mice of both genotypes (WT and STAT1−/−) were infected i.n. with a dose of PR8 or WSN virus equivalent to 1 LD50 for a WT animal. This was 1,000 PFU for the experiment carried out with the WSN strain or 10 TCID for the experiment carried out with the PR8 strain. Four to five animals of each genotype were sacrificed 3, 6, and 8 or 9 days after infection. Lung viral titers were determined for each animal (Fig. 1). Virus titers rose with the same kinetics for both control and mutant animals, with peak lung titers occurring 3 days after i.n. inoculation. Peak viral titers were similar in WT and STAT1−/− animals for both virus strains. Virus clearance was complete for all PR8-infected animals by 9 days postinoculation. While WSN clearance was well under way in WT animals by day 8 postinoculation, in STAT1−/− mice WSN titers remained maximal.
FIG. 1.
Determination of lung virus titers over a 9-day period for WT (■) and STAT1−/− (▵) CD1 mice inoculated i.n. with WSN and PR8 viruses. (A) Animals of each genotype were inoculated i.n. with 1,000 PFU (1 LD50 for a WT animal) of WSN virus in 50 μl of PBS. Four to five WT and STAT1−/− animals were sacrificed at each time point. Results from two experiments are included. Viral titers in lung homogenates from each animal (PFU/gram of tissue) were determined by plaque assay in MDCK cells. (B) Fifteen animals of each genotype were inoculated i.n. with 10 TCID (1 LD50 for a WT animal) of PR8 virus in 100 μl of PBS. Five WT and five STAT1−/− animals were sacrificed at days 3, 6, and 9 p.i. Viral titers in lung homogenates from each animal were determined as TCID/milliliter.
Fatal, disseminated influenza virus infection following i.p. inoculation of STAT1−/− mice.
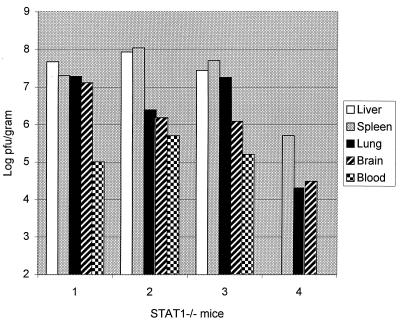
We wished to examine whether the failure of STAT1−/− mice to clear the WSN strain of influenza virus might be related to an altered virus tropism in these animals. To test this hypothesis, four STAT1−/− mice, and three control animals were each given 107 PFU of WSN virus by i.p. injection. Mice were observed until mutant animals began to show signs of illness, at 4 days postinoculation. WT animals appeared unaffected. Tissues were harvested from all animals at day 4 for determination of viral titers and histological examination. No virus could be detected in any tissue from any WT animal. All STAT1−/− animals had a significant virus load in liver, spleen, lung, brain, and blood, ranging from 104 to 108 PFU/g of tissue (Fig. 2). Two repeat experiments yielded similar results.
FIG. 2.
Virus titers following i.p. WSN virus inoculation of STAT1−/− mice. Four STAT1−/− and three WT mice were injected i.p. with 107 PFU of WSN virus. Animals were sacrificed at day 4 postinoculation when the mutant animals showed signs of illness. Viral titers of tissue homogenates were determined by plaque assay in MDCK cells and are expressed as PFU/gram of tissue. Virus titers for each tissue are shown for each of the STAT1−/− animals. No virus could be detected in samples obtained from three WT animals.
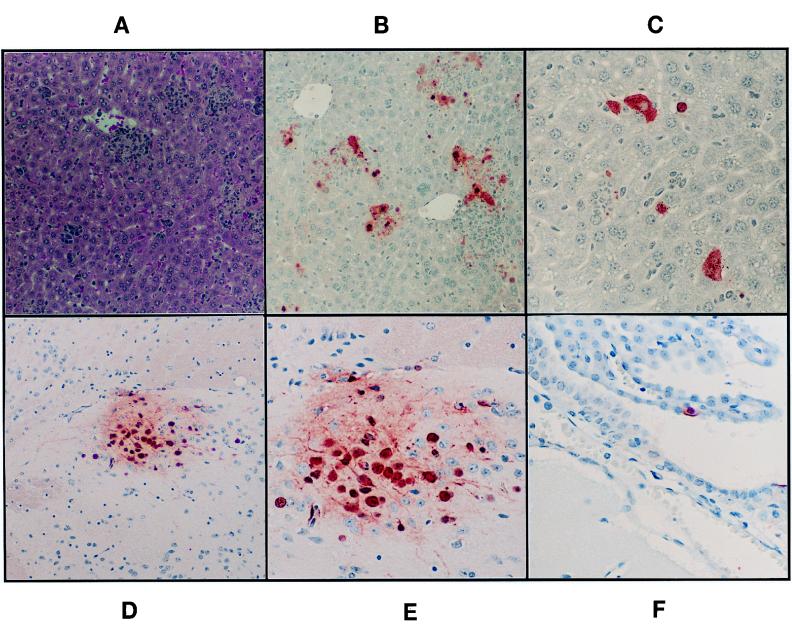
Tissues harvested from i.p.-infected mice were examined histologically. No lesions could be found in any tissue obtained from control animals. In contrast, large necrotic foci were present in spleens of all STAT1−/− mice, as were widespread granulomatous hepatic lesions (Fig. 3). Tissue sections were stained with polyclonal anti-influenza virus antibodies to demonstrate the specificity of these lesions. Splenic lesions in the mutant animals stained intensely with the influenza virus-specific antibodies. In control animals, only rare splenic macrophages were positive for the presence of viral antigen (data not shown). In the livers of STAT1−/− animals, a small fraction of the inflammatory cells making up the hepatic lesions stained with antiviral antibody. Many hepatocytes showed strong staining, demonstrating that virus was actively replicating in this tissue. In the brains of mutant animals, foci of cortical neurons and rare ependymal cells showed evidence of virus replication in the absence of other pathology. Lungs from infected STAT1−/− animals had infiltrates of mononuclear cells within alveolar septa throughout the parenchyma.
FIG. 3.
Pathology of hepatic and central nervous system lesions of STAT1−/− mice following i.p. injection with WSN virus. (A) Hematoxylin-eosin-stained section of liver showing multiple aggregates of inflammatory cells within the hepatic parenchyma. Magnification, ×98. (B [magnification, ×197] and C [magnification, ×394]) Sections reacted with polyclonal antibody against influenza A virus. Infected cells are bright red. Only a fraction of inflammatory cells constituting the liver lesions show positive staining for influenza virus antigens. Multiple hepatocytes stain strongly, some showing only nuclear staining, indicating an early phase of virus replication. In other hepatocytes, viral proteins are also present in the cytoplasm. (D) Section of brain with a focus staining positively for the presence of viral antigen (magnification, ×98). In panel E (magnification, ×394), it can be appreciated that many of the red-staining cells are neurons. Glial cells within the focus also stain positively. (F) Single infected ependymal cell (magnification, ×394).
Intraperitoneal inoculation of five WT and five STAT1−/− mice with the PR8 strain of influenza virus (also 107 PFU/animal) was performed as described for WSN virus. No illness was apparent in animals of either genotype by 4 days p.i. When mice were sacrificed and tissue homogenates were assayed for live virus, none could be detected (data not shown).
Disseminated influenza virus infection following i.n. inoculation in STAT1−/− and IFNαR−/− mice.
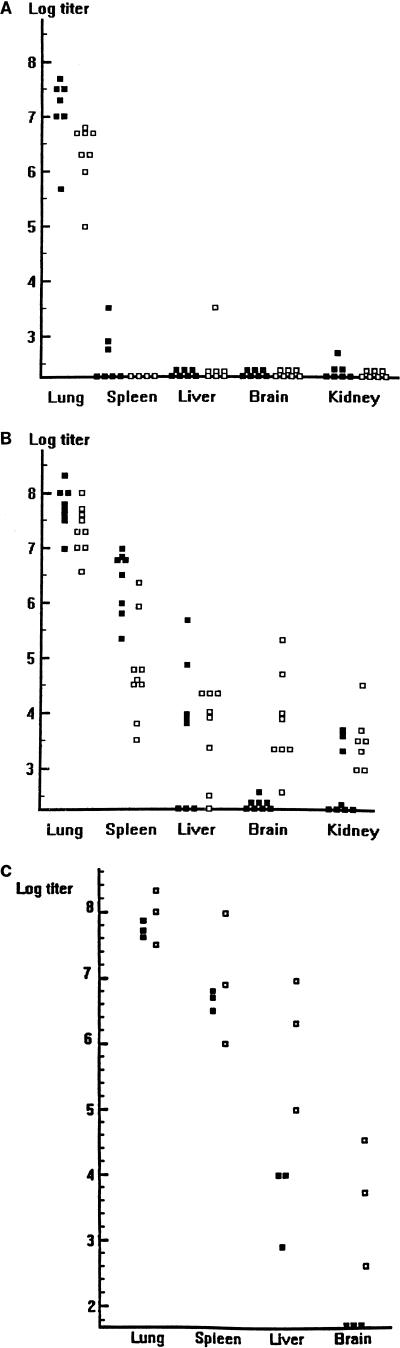
We wished to determine whether influenza virus infection could become disseminated following the more physiologic i.n. inoculation route. To answer this question, each of 15 WT and 15 STAT1−/− animals was infected with 103 PFU of WSN virus. Four to five animals of each genotype were sacrificed 3 and 6 days postinoculation, and tissue extracts were analyzed by plaque assay. Data from two such determinations are included in Fig. 4. In WT animals, virus replication was largely limited to the lungs on day 3, although low levels of virus were detected in spleen and kidney extracts of some animals. By day 6, lung titers decreased by approximately 1 log. Low levels of virus were again found in the spleen, liver, and kidney extracts from some animals (Fig. 4A).
FIG. 4.
Comparison of tissue virus titers in WT (A), STAT1−/− (B), and IFNαR−/− (C) mice at different days after i.n. inoculation with WSN virus. (A and B) Eight to 10 WT (A) or STAT1−/− (B) mice were infected i.n. with 1,000 PFU of WSN virus, and viral titers in the indicated tissues at days 3 (■) and 6 (□) p.i. were determined by plaque assay in MDCK cells. Results from two experiments are expressed as PFU/gram of tissue. (C) Six IFNαR−/− mice were infected i.n. with 1,000 PFU of WSN virus, and viral titers at days 4 (■) and 8 (□) p.i. were determined by plaque assay in MDCK cells. Results are expressed as PFU/gram of tissue.
In STAT1−/− mice, spleen and lung titers were very high at day 3 p.i., with significant virus also present in the liver and kidney extracts of most animals. At day 6 p.i., WSN virus was found in all organs assayed from all mutant mice (Fig. 4B). When this experiment was repeated with IFNαR−/− mice, the results were similar: 8 days after i.n. inoculation with WSN virus, disseminated infection was present in all of three IFNαR−/− mice (Fig. 4C). Although the STAT1−/− and IFNαR−/− mice were of different genetic backgrounds, there were no significant differences in the ability of WSN virus to replicate in the tissues assayed.
STAT1−/− and IFNαR−/− fibroblasts are permissive for influenza virus replication.
The ability of WSN virus to replicate in fibroblast cell lines derived from STAT1−/− and STAT1+/+ mice was determined. Cell monolayers were infected with an MOI of 0.001. Two days later, virus was harvested and assayed by plaque assay on MDBK cells. WSN virus failed to grow or grew poorly in four independent continuous cell lines derived from WT or STAT1+/− embryos. Viral titers of between 106 and 8 × 106 PFU/ml were obtained from all of four independently derived STAT1−/− cell lines (Table 1).
TABLE 1.
Virus replication in STAT1+/+ and STAT1−/− cells
| Clonea | WSN (PFU/ml)b |
|---|---|
| CD1 +/+ A | <10 |
| CD1 +/+ B | <10 |
| CD1 +/+ I | 7 × 103 |
| B6 +/− 13 | 6 × 104 |
| CD1 −/− 7 | 8 × 106 |
| CD1 −/− 3 | 1 × 106 |
| CD1 −/− I | 4 × 106 |
| B6 −/− 6 | 1 × 106 |
Independent cell lines were derived from primary MEFs from CD1 STAT1+/+ mice (clones CD1 +/+ A, CD1 +/+ B, and CD1 +/+ I), C57B6 STAT1+/− mice (clone B6 +/− 13), CD1 STAT1−/− mice (clones CD1 −/− 7, CD1 −/− 3, and CD1 −/− I), and C57B6 STAT1−/− mice (clone B6−/− 6).
Cell monolayers were infected with WSN virus at an MOI of 0.001 and incubated at 37°C; 2 days p.i., virus in supernatants was quantitated by plaque assay in MDBK cells.
Growth of WSN virus was then assayed on MEFs derived from STAT1−/−, IFNγ−/−, IFNαR−/−, and WT mice. Confluent monolayers were again infected with an MOI of 0.001. Samples were harvested every 12 h and titrated by HA assay. Virus could be detected in the media of STAT1−/− and IFNαR−/− cells by 48 h. No virus was detected by HA assay in WT or IFNγ−/− MEFs (Table 2).
TABLE 2.
WSN virus growth determined by HA assay
| Fibroblast culture | Virus titera
|
|||
|---|---|---|---|---|
| 48 h | 60 h | 72 h | 84 h | |
| WT cell line | <2 | <2 | <2 | <2 |
| WT MEFs | <2 | <2 | <2 | <2 |
| STAT1−/− cell line | <2 | 8 | 16 | 32 |
| STAT1−/− MEFs | 16 | 32 | 64 | ND |
| IFNγ−/− MEFs | <2 | <2 | <2 | <2 |
| IFNαR−/− MEFs | 32 | 64 | 64 | 64 |
Cell monolayers were infected with WSN virus at an MOI of 0.001 and incubated at 37°C. At the indicated time points, the amount of virus in the supernatant was determined by HA assay. At 12, 24, and 36 h p.i., titers in all cultures were <2. ND, not done.
Mechanism of STAT1-mediated resistance to influenza virus.
A number of 3T3-like continuous cell lines were derived by continuous passage from WT and STAT1−/− embryonic fibroblasts. Two of these immortalized cell lines, one WT (designated A) and one mutant (numbered 7), were used in the following experiments. These two clones were chosen because they showed the greatest differences in the ability to support WSN virus replication (Table 1). To investigate whether the poor influenza virus growth in the WT fibroblasts was due to a decrease in virus binding, we compared STAT1+/+ fibroblasts, STAT1−/− fibroblasts, and MDBK cells by viral adsorption assay. STAT1+/+ and STAT1−/− fibroblasts adsorbed virus with equal efficiencies (106 PFU/35-mm-diameter plate); MDBK cells adsorbed 3 × 106 PFU/35-mm-diameter plate.
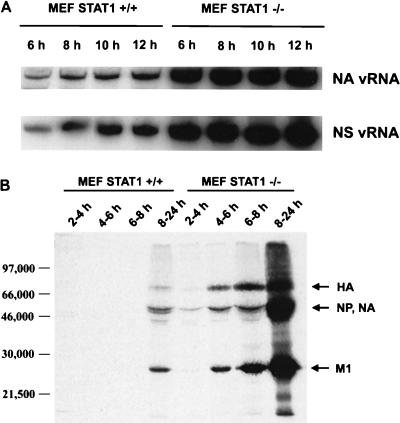
Levels of vRNA and protein were then compared for the STAT1+/+ and STAT1−/− cells infected with WSN virus. For vRNA measurement, cells were infected with an MOI of 5 and total RNA was harvested at 6, 8, 10, and 12 p.i. Viral genomic RNA was measured by primer extension using NA- and NS-specific primers (Fig. 5A). While NA and NS vRNAs were produced by both cell types, there was a marked (10- to 20-fold) reduction in synthesis by STAT1+/+ fibroblasts. This correlates well with our measurement of viral protein expression in these cells. Viral protein levels were measured in STAT1+/+ or STAT1−/− cells labeled with l-[35S]cysteine and l-[35S]methionine at the indicated time points after WSN virus infection. After labeling, viral proteins were immunoprecipitated from cell extracts and separated by SDS-PAGE (Fig. 5B). Quantitation of the 35S signal indicates that 20- to 30-fold less viral protein was synthesized by infected STAT1+/+ cells.
FIG. 5.
Viral RNA and protein expression levels in MEFs derived from WT (STAT+/+) and STAT−/− CD1 mice. (A) STAT+/+ and STAT−/− MEFs were infected with WSN virus at an MOI of 5, and vRNA levels specific for the NA and NS genes were determined by PAGE analysis of primer extension products at different times p.i. (B) STAT+/+ and STAT−/− MEFs were infected with WSN virus at an MOI of 2 and 35S labeled at the indicated time points, and total amount of viral proteins was immunoprecipitated with a polyclonal antiserum against WSN virus. Immunoprecipitated products were analyzed by SDS-PAGE.
We also compared levels of viral protein expression in the STAT1+/+ and STAT1−/− fibroblast cell lines by using a transfectant influenza virus, CAT2A/NAmodII, which directs CAT expression. Cells were infected with the transfectant virus at an MOI of 2, and CAT assays were performed at different times p.i. CAT activity was detected in extracts from infected STAT1−/− cells beginning at 4 h p.i. but was undetectable in WT extracts until 10 h p.i. After 12 h of infection, CAT activity was 20-fold higher in STAT1−/− cells than in WT cells (data not shown).
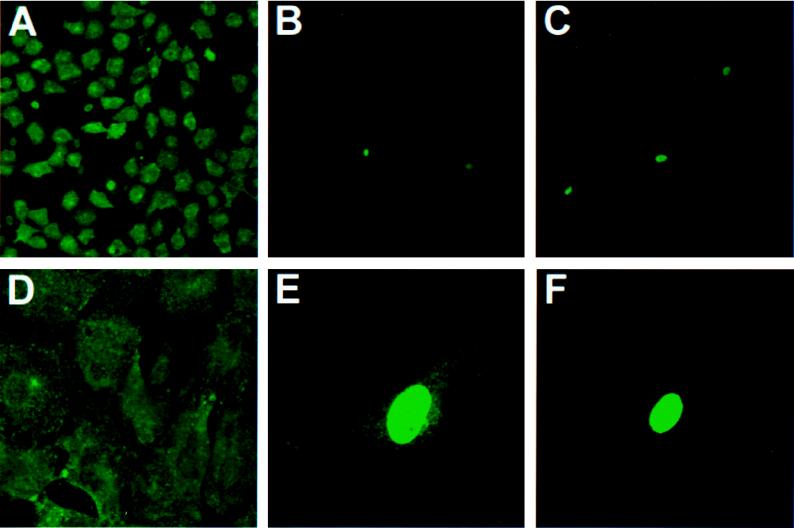
To visualize the cells and cellular compartments where viral protein synthesis was occurring, STAT1+/+ and STAT1−/− fibroblasts were seeded on coverslips and infected with WSN at an MOI of 2. After 14 h of incubation, the cells were fixed with acetone-methanol and stained with a monoclonal antibody against NP. The NP protein of influenza virus is the major structural component of the genomic ribonucleoprotein (RNP) complexes and localizes in the nucleus of the infected cell at early times after infection. During the later stages of infection newly synthesized RNPs exit the nucleus and migrate to the plasma membrane, where viral budding takes place (22). As can be seen in Fig. 6A and D, essentially all STAT1−/− cells showed evidence of infection by 14 h p.i., with the expected nuclear and cytoplasmic NP staining. In contrast, only a small fraction (approximately 5%) of STAT1+/+ cells showed NP staining (Fig. 6B and C), consistent with the low levels of viral RNA and protein synthesis. When the STAT1+/+ (A) cell line was subcloned, all 10 independent subclones exhibited the parental pattern of viral expression in only a minor subpopulation of cells (data not shown). We conclude, therefore, that infection in WT cells is impaired at a very early step, so that about 95% of the cells fail to develop any detectable immunofluorescence at all. In the few NP-stained cells in each STAT1+/+ clone, the staining was intense but remained almost entirely nuclear (Fig. 6E and F), indicating that infection was delayed or aborted at an early, nuclear phase. Taken together, these results suggest the existence of at least two blocks to influenza virus replication in STAT1+/+ cells.
FIG. 6.
Immunofluorescence analysis of NP expression in WT and STAT1−/− MEFs infected with WSN virus. Cells were infected with WSN virus (MOI = 2) and stained with a monoclonal antibody against NP 14 h p.i. (A and D) Different-magnification fields of STAT1−/− cells. The majority of the cells showed a cytoplasmic NP staining, indicative of a late phase of virus replication. (B and C) Low magnification of two different fields of WT-infected cells. Although cell densities were roughly similar between the WT and STAT1−/− samples, only a few WT cells showed positive NP staining. (E and F) Higher magnification of individual positive-stained WT cells. Note that the majority of the NP staining is nuclear, which indicates a delayed or abortive viral replication.
DISCUSSION
Pathogenicity of influenza virus in birds has been associated with a disseminated viral infection affecting multiple tissues and organs. This broad tissue tropism apparently results, at least in part, from the ability of the HA to be activated by ubiquitous proteases (16). Factors determining the severity of human influenza virus disease are probably complex, involving epidemiological considerations as well as inherent viral properties. The catastrophic mortality of the influenza pandemic of 1918, for instance, was undoubtedly the result, at least in part, of a major antigenic shift. Whether the 1918 strain of influenza A virus might also have had unusual virulence determinants is still unknown, as only fragments of its genome have been recovered and sequenced (38). Prediction of the severity of continuously emerging human influenza virus strains remains a high public health priority, but it is limited by our incomplete understanding of the molecular determinants of pathogenicity in this disease.
In this work, we have compared the courses of influenza A virus infection in WT mice and in mice homozygous for a targeted disruption of the STAT1 gene. In the absence of STAT1, IFN-mediated gene induction does not occur, and mice lacking a functional STAT1 gene are IFN unresponsive (5, 20). We have used these animals to enhance our understanding of the role that IFN plays in the host response to influenza virus infection. The LD50 for each of two different strains of influenza virus, WSN and PR8, was determined for STAT1−/− mice. Both strains are known to be pneumotropic in i.n.-inoculated WT mice. The WSN strain was derived from a human influenza virus isolate by passage in suckling mouse brain and is capable of replicating in ependymal cells when inoculation is intrathecal (35). When viruses were inoculated i.n., the LD50s of PR8 and WSN viruses were 10 to 100 times lower in STAT1−/− mice than in WT mice. This increase in sensitivity to killing by influenza virus infection occurred despite the fact that viruses replicated to equal peak titers in the lungs over the same time course in control and mutant animals. One striking dissimilarity between control and STAT1−/− mice was the inability of the STAT1-deficient animals to clear virus from the lungs following infection with the WSN strain. Hypothesizing a tropism shift in the absence of IFN responsiveness, we inoculated WT and STAT1−/− animals i.p. with 107 PFU of WSN or PR8 virus, a dose that would be lethal if delivered i.n. PR8 virus caused no disease in any animal receiving virus i.p. WSN virus administered i.p. was harmless to WT animals but caused fulminant, generalized disease in mice lacking STAT1. Following i.n. inoculation of 103 PFU of WSN virus, infection quickly disseminated in the STAT1−/− animals but not in WT animals. In contrast to our results, Castrucci and Kawaoka (4) observed systemic infection of WT mice after i.n. inoculation of influenza A/WSN virus. The difference is likely due to the much larger dose of virus in their study and/or to mouse strain differences. While the absence of IFN responsiveness in STAT1−/− mice may lead to further disruptions in the development of normal antiviral immunity and thereby contribute to the enhanced lethality of influenza virus infection (6), the results of the present study indicate a critical early role for IFN in determining the tissue tropism of influenza virus.
The pattern of disease that we describe for STAT1−/− mice infected with WSN virus resembles that seen in birds infected with highly pathogenic strains of FPV (21). Strains of FPV capable of causing fatal, disseminated influenza disease have been intensively studied and have in common a variant of the HA molecule, H5 or H7, which is readily cleaved by ubiquitous proteases (7, 8, 17). The H5 and H7 serotypes had never been found to be responsible for infections in humans until recently, when several deaths in Hong Kong were attributed to an H5N1 strain of highly pathogenic avian influenza virus (1, 36). The limitation of infection by human influenza A viruses to the respiratory mucosa is thought to be due to the cleavage requirements of the HA molecule. HA cleavage generally requires the presence of a trypsin-like serine protease for infectivity. Such a molecule has been isolated from Clara cells of the lung, and it is assumed that it is this enzyme which allows for multiple cycles of viral replication in rodents (16, 17). This view of influenza A virus tropism is supported by experiments carried out by Vallbracht et al. (40). These investigators were able to produce a generalized infection in mice by using reassortant PR8 viruses containing an HA gene from FPV. In addition, there is clear evidence that the disseminated infection in birds which is associated with pathogenic avian influenza viruses correlates with the cleavability of the viral HA molecule (2, 11, 15, 25, 31).
The work described here adds a new variable to this model of influenza virus pathogenicity and tissue tropism. The WSN strain of influenza A virus is unique among mammalian influenza A viruses for its ability to undergo multiple rounds of replication on MDBK cells in the absence of trypsin as well as for its neurovirulence when inoculated intracranially into adult mice. Its capacity for growth in MDBK cells and mouse brain has been found to depend on its NA gene, which lacks a conserved glycosylation site (3, 19, 41). In some manner, the NA mutation of WSN virus is able to facilitate HA cleavage. Comparison of PR8 and WSN virus infections demonstrates that this alteration is not sufficient to alter the pattern of disease following i.n. inoculation in a WT mouse: infection remains limited primarily to the respiratory tract (although very low virus titers could be detected in the spleen and liver of some animals). In contrast, i.n. instillation of a WSN virus inoculum equivalent to 1 LD50 in a WT animal inevitably led to systemic infection in STAT1−/− and IFNαR−/− mice. These experiments demonstrate that although WSN virus can be cleaved by a ubiquitous protease, WSN replication remains in check in all organs except the respiratory tract in the presence of an intact response to IFN-α.
The experiments described above demonstrate an important role for IFN-α in limiting the spread of influenza virus infection. Using cultured fibroblasts derived from WT, STAT1−/−, IFNγ−/−, and IFNαR−/− mice, we have investigated the nature of the IFN-mediated block. Only a fraction of WT cells could be infected, and in this small subpopulation infection seemed to be delayed or stopped at an early nuclear phase, possibly resulting from blockage of the nuclear export of viral RNPs (23). This resistance to infection was shared by IFNγ−/− MEFs, while STAT1−/− and IFNαR−/− MEFs were susceptible and capable of significant virus production. Thus, IFN-α blocks influenza virus replication in MEFs at a minimum of two steps: at an early step after adsorption and a later step, before nuclear export.
The ability of WSN to cause tracheobronchitis and pneumonia in the presence of a WT innate immune response may be related to the relative IFN insensitivity of lung epithelium. Ronni et al. (27) have demonstrated a poor IFN response to influenza A virus in the human lung epithelial cell line A549 as well as in primary human fetal lung cells. These lung-derived cells are incapable of restricting influenza virus infection in vitro, while other cell types, such as macrophages, respond quickly to influenza virus with vigorous type I IFN production followed by the rapid accumulation of IFN-regulated gene products (22, 26). This differential tissue response may explain lung susceptibility in WT animals. In addition, various levels of IFN production upon viral infection and/or various IFN susceptibilities among cell lines might also explain the ability of some cell lines, like MDCK cells, to support influenza virus replication. We conclude that some strains of influenza A virus, such as PR8, may in fact be limited in their tissue tropism by failure of HA cleavage outside the respiratory tract. In the case of WSN virus, whose HA is more likely processed by a ubiquitous enzyme, infection is still limited to the respiratory tract by the IFN-α response when the virus is administered i.n. No single influenza virus gene determines the pathogenicity of a given virus strain; rather, it is a combination of viral genes and host susceptibility which determines disease outcome. Subtle differences among individuals in IFN responsiveness could play some part in the determination of disease severity. It will be of interest to determine the ability of other strains of influenza virus to replicate outside the respiratory tree in mice defective in the IFN pathway.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Institutes of Health to A.G.-S., D.E.L., and P.P. J.E.D. was supported by a Child Health Research grant from the NICHD.
We thank Sara Mertz for expert technical assistance.
REFERENCES
- 1.Anonymous. Isolation of avian influenza A(H5N1) viruses from humans—Hong Kong, May–December 1997. Morbid Mortal Weekly Rep. 1997;46:1204–1207. [PubMed] [Google Scholar]
- 2.Bosch F X, Garten W, Klenk H-D, Rott R. Proteolytic cleavage of influenza virus hemagglutinins: primary structure of the connecting peptide between HA1 and HA2 determines proteolytic cleavability and pathogenicity of avian influenza viruses. Virology. 1981;113:725–735. doi: 10.1016/0042-6822(81)90201-4. [DOI] [PubMed] [Google Scholar]
- 3.Boycott R, Klenk H-D, Ohuchi M. Cell tropism of influenza virus mediated by hemagglutinin activation at the stage of virus entry. Virology. 1994;203:313–319. doi: 10.1006/viro.1994.1489. [DOI] [PubMed] [Google Scholar]
- 4.Castrucci M R, Kawaoka Y. Biologic importance of the neuraminidase stalk length in influenza A virus. J Virol. 1993;67:759–764. doi: 10.1128/jvi.67.2.759-764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 6.Durbin, J. E., A. Garcia-Sastre, A. Fernandez-Sesma, T. Moran, T. D. Rao, A. B. Frey, and D. E. Levy. Unpublished data.
- 7.Garten W, Bosch F X, Linder D, Rott R, Klenk H-D. Proteolytic activation of the influenza virus hemagglutinin: the structure of the cleavage site and the enzymes involved in cleavage. Virology. 1981;115:361–374. doi: 10.1016/0042-6822(81)90117-3. [DOI] [PubMed] [Google Scholar]
- 8.Garten W, Linder D, Rott R, Klenk H-D. The cleavage site of the hemagglutinin of fowl plague virus. Virology. 1982;122:186–190. doi: 10.1016/0042-6822(82)90387-7. [DOI] [PubMed] [Google Scholar]
- 9.Haller O. A mouse hepatotropic variant of influenza virus. Arch Virol. 1975;49:99–116. doi: 10.1007/BF01317530. [DOI] [PubMed] [Google Scholar]
- 10.Hiti A L, Davis A R, Nayak D P. Complete sequence analysis shows that the hemagglutinins of the H0 and H2 subtypes of human influenza virus are closely related. Virology. 1981;111:113–124. doi: 10.1016/0042-6822(81)90658-9. [DOI] [PubMed] [Google Scholar]
- 11.Horimoto T, Kawaoka Y. Reverse genetics provides direct evidence for a correlation of hemagglutinin cleavability and virulence of an avian influenza A virus. J Virol. 1994;68:3120–3128. doi: 10.1128/jvi.68.5.3120-3128.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horisberger M A. Interferons, Mx genes, and resistance to influenza virus. Am J Respir Crit Care Med. 1995;152:S67–S71. doi: 10.1164/ajrccm/152.4_Pt_2.S67. [DOI] [PubMed] [Google Scholar]
- 13.Hwang S Y, Hertzog P J, Holland K A, Sumarsono S H, Tymms M J, Hamilton J A, Whitty G, Bertoncello I, Kola I. A null mutation encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc Natl Acad Sci USA. 1995;92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isaacs A, Lindemann J. Virus interference. I. The interferon. Proc R Soc Lond Ser B. 1957;147:258–267. [PubMed] [Google Scholar]
- 15.Kawaoka Y, Nestorowicz A, Alexander D J, Webster R G. Molecular analyses of the hemagglutinin genes of H5 influenza viruses: origin of a virulent turkey strain. Virology. 1987;158:218–227. doi: 10.1016/0042-6822(87)90256-x. [DOI] [PubMed] [Google Scholar]
- 16.Kido H, Yokogoshi Y, Sakai K, Tashiro M, Kishino Y, Fukutomi A, Katunuma N. Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J Biol Chem. 1992;267:13573–13579. [PubMed] [Google Scholar]
- 17.Klenk H-D, Garten W. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 1994;2:39–43. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 18.Klenk H-D, Rott R. The molecular biology of influenza virus pathogenicity. Adv Virus Res. 1988;34:247–281. doi: 10.1016/S0065-3527(08)60520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Schulman J, Itamura S, Palese P. Glycosylation of neuraminidase determines the neurovirulence of influenza A/WSN/33 virus. J Virol. 1993;67:6667–6673. doi: 10.1128/jvi.67.11.6667-6673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meraz M A, White M J, Sheehan K C F, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, Carver-Moore K, DuBois R N, Clark R, Aguet M, Schreiber R D. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 21.Mo I P, Brugh M, Fletcher O J, Rowland G N, Swayne D E. Comparative pathology of chickens experimentally inoculated with avian influenza viruses of low and high pathogenicity. Avian Dis. 1997;41:125–136. [PubMed] [Google Scholar]
- 22.Murphy B R, Webster R G. Orthomyxoviruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1397–1445. [Google Scholar]
- 23.O’Neill R E, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Percy N, Barclay W S, García-Sastre A, Palese P. Expression of a foreign protein by influenza A virus. J Virol. 1994;68:4486–4492. doi: 10.1128/jvi.68.7.4486-4492.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perdue M L, García M, Senne D, Fraire M. Virulence-associated sequence duplication at the hemagglutinin cleavage site of avian influenza viruses. Virus Res. 1997;49:173–186. doi: 10.1016/s0168-1702(97)01468-8. [DOI] [PubMed] [Google Scholar]
- 26.Ronni T, Sareneva T, Pirhonen J, Julkunen I. Activation of IFN-α, IFN-γ, MxA, and IFN regulatory factor 1 genes in influenza A virus-infected human peripheral blood mononuclear cells. J Immunol. 1995;154:2764–2774. [PubMed] [Google Scholar]
- 27.Ronni T, Matikainen S, Sareneva T, Melen K, Pirhonen J, Keskinen P, Julkunen I. Regulation of IFN α/β, MxA, 2′,5′-oligoadenylate synthase, and HLA gene expression in influenza A-infected human lung epithelial cells. J Immunol. 1997;158:2363–2374. [PubMed] [Google Scholar]
- 28.Rott R, Klenk H-D, Nagai Y, Tashiro M. Influenza viruses, cell enzymes and pathogenicity. Am J Respir Crit Care Med. 1995;152:S16–S19. doi: 10.1164/ajrccm/152.4_Pt_2.S16. [DOI] [PubMed] [Google Scholar]
- 29.Schindler C, Darnell J E. Transcriptional responses to polypeptide ligands: the Jak-Stat pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 30.Schulman J L, Palese P. Virulence factors of influenza A viruses: WSN virus neuraminidase required for plaque production in MDBK cells. J Virol. 1977;24:170–176. doi: 10.1128/jvi.24.1.170-176.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senne D A, Panigraphy B, Kawaoka Y, Pearson J E, Süss J, Lipkind M, Kida H, Webster R G. Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 avian influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential. Avian Dis. 1996;40:425–437. [PubMed] [Google Scholar]
- 32.Staeheli P. Interferon-induced proteins and the antiviral state. Adv Virus Res. 1990;38:147–200. doi: 10.1016/s0065-3527(08)60862-3. [DOI] [PubMed] [Google Scholar]
- 33.Staeheli P, Grob R, Meier E, Sutcliffe J G, Haller O. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol Cell Biol. 1988;8:4518–4523. doi: 10.1128/mcb.8.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stieneke-Gröber A, Vey M, Angliker H, Shaw E, Thomas G, Roberts C, Klenk H-D, Garten W. Influenza virus hemagglutinin with multi-basic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992;11:2407–2414. doi: 10.1002/j.1460-2075.1992.tb05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuart-Harris C. Neurotropic strain of human influenza virus. Lancet. 1939;i:497–499. [Google Scholar]
- 36.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A virus (H5N1) isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 37.Sugiura A, Ueda M. Neurovirulence of influenza virus in mice. I. Neurovirulence of recombinants between virulent and avirulent strains. Virology. 1980;101:440–449. doi: 10.1016/0042-6822(80)90457-2. [DOI] [PubMed] [Google Scholar]
- 38.Taubenberger J K, Reid A H, Krafft A E, Bijaand N E, Fanning T G. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 39.Todaro G J, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established cell lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vallbracht A, Scholtissek C, Fleming B, Gerth H-J. Recombination of influenza A strains with fowl plague virus can change pneumotropism for mice to a generalized infection with involvement of the central nervous system. Virology. 1980;107:452–460. doi: 10.1016/0042-6822(80)90312-8. [DOI] [PubMed] [Google Scholar]
- 41.Ward A C. Neurovirulence of influenza A virus. J Neurovirol. 1996;2:139–151. doi: 10.3109/13550289609146876. [DOI] [PubMed] [Google Scholar]
- 42.Zheng H, Palese P, García-Sastre A. Nonconserved nucleotides at the 3′ and 5′ ends of an influenza A virus RNA play an important role in RNA replication. Virology. 1996;217:242–251. doi: 10.1006/viro.1996.0111. [DOI] [PubMed] [Google Scholar]