Abstract
The Epstein-Barr virus transactivator Zta triggers lytic gene expression and is essential for replication of the lytic origin, oriLyt. Previous analysis indicated that the Zta activation domain contributed a replication-specific function. We now show that the Zta activation domain interacts with components of the EBV helicase-primase complex. The three helicase-primase proteins BBLF4 (helicase), BSLF1 (primase), and BBLF2/3 (primase-associated factor) were expressed fused to the Myc epitope. When expression plasmids for BBLF4 or BBLF2/3 plus BSLF1 (primase subcomplex) were separately transfected, the proteins localized to the cytoplasm. Interaction between Zta and the components of the helicase-primase complex was tested by examining the ability of Zta to alter the intracellular localization of these proteins. Cotransfection of Zta with Myc-BBLF4 resulted in nuclear translocation of Myc-BBLF4; similarly, cotransfection of Zta with the primase subcomplex led to nuclear translocation of the Myc-BSLF1 and Myc-BBLF2/3 proteins. This relocalization provides evidence for an interaction between Zta and the helicase and Zta and the primase subcomplex. An affinity assay using glutathione S-transferase–Zta fusion proteins demonstrated that Myc-BBLF4 and Myc-BBLF2/3 plus BSLF1 bound to the Zta activation domain (amino acids 1 to 133). In the nuclear relocalization assay, the amino-terminal 25 amino acids of Zta were required for efficient interaction with the primase subcomplex but not for interaction with BBLF4. Evidence for interaction between oriLyt bound Zta and the helicase-primase complex was obtained in a superactivation assay using an oriLyt-chloramphenicol acetyltransferase (CAT) reporter. Zta activated expression from a CAT reporter containing the complete oriLyt region and regulated by the oriLyt BHLF1 promoter. Cotransfection of the helicase-primase proteins, one of which was fused to a heterologous activation domain, led to Zta-dependent superactivation of CAT expression. This assay also provided evidence for an interaction between the single-stranded DNA binding protein, BALF2, and the Zta-tethered helicase-primase complex. The helicase-primase interaction is consistent with a role for Zta in stabilizing the formation of an origin-bound replication complex.
Expression of the Zta (BZLF1, ZEBRA) transactivator induces lytic cycle reactivation in latently Epstein-Barr virus (EBV)-infected lymphoblastoid cell lines (15, 56, 65). Zta is a bZip transcriptional transactivator that has a nonconsensus dimerization domain (10, 24) and binds as a homodimer to AP-1 sites and to related sequences called Zta response elements (ZREs) (7, 10, 20, 41, 66). Zta stabilizes the formation of a DNA-bound complex containing the basal transcription factors TFIID and TFIIA (14, 39), and it is most active on promoters containing noncanonical TATA boxes that have reduced affinity for TFIID (38, 42). Both DNA binding and activation of endogenous viral promoters are modified by phosphorylation of Zta (25, 34). In addition to regulating EBV lytic promoters, Zta modifies cellular gene expression (8, 9, 23) and has been found to interact with a variety of cellular proteins, including the retinoic acid receptor, NF-κB/p65, and p53 (29, 54, 64, 73).
Zta has a second role in lytic cycle reactivation, serving as an essential regulatory protein for replication of the lytic origin, oriLyt (1, 21, 58, 61). EBV oriLyt is duplicated with one copy located within the BamHI H fragment and a second copy located in the region that is deleted in the sequenced B95-8 isolate (30, 35). The origin comprises two essential elements and one auxiliary domain (30, 60, 62). The essential BHLF1 promoter and leader region and the auxiliary upstream enhancer domain contain seven ZREs. The promoter ZREs are essential for replication, while the enhancer ZREs are dispensable (60). The enhancer domain also contains two binding sites for the Rta transactivator (12, 27, 31). Rta is not essential for oriLyt replication but has a significant effect on replication efficiency (21). In addition to Zta and Rta, oriLyt replication requires six EBV-encoded replication proteins that were originally defined in a Challberg cotransfection replication assay (21, 22). The six core replication proteins have homologs in the other herpesviruses (17, 48, 53, 59, 71). Their functions are sufficiently conserved that the six core herpes simplex virus (HSV) proteins plus Zta and Rta can replicate EBV oriLyt (21); similarly, the six core EBV proteins plus the cytomegalovirus ancillary proteins can replicate cytomegalovirus oriLyt (59). The core EBV proteins are the DNA polymerase (BALF5), the polymerase accessory protein (BMRF1), the single-stranded DNA binding protein (BALF2), the helicase (BBLF4), the primase (BSLF1), and the primase-associated protein (BBLF2/3). Relatively few studies characterizing the core replication proteins have been performed. The polymerase and polymerase accessory protein (BMRF1) interact and together mediate processive DNA replication with strand displacement in model replication systems (33, 36, 43, 68, 70). BMRF1 binds DNA nonspecifically (32). Further, BMRF1 has been found to interact with the bZip domain of Zta (75) and to function independently as a transcriptional activator in transient expression assays (52, 75). Although the mechanism of this activation has not been established, a region within the second essential domain of oriLyt that is required for BMRF1 transactivation of the BHLF1(oriLyt) promoter has been mapped (74). BALF2 has been shown to have single-stranded DNA binding properties, and a role in melting hairpin structures to facilitate processive DNA replication has been postulated (18, 69). The EBV helicase-primase proteins have not been subjected to functional analyses.
Attempts to determine the role of Zta in oriLyt replication have focused largely on the contribution of the activation domain, and different studies have reached different conclusions. The Zta activation domain is encoded within the first exon, which comprises amino acids (aa) 1 to 167 (55). In transcription assays, the activation domain is both modular and redundant in that loss of individual subdomains can be compensated for by multimerization of the remaining domains and by multimerization of Zta binding sites in the target promoter (7, 13). Three studies have used EBV-positive cell lines to evaluate the requirement for Zta activation domain sequences in oriLyt replication. Schepers et al. (61) found that the activation domain could not be substituted by the activation domain from HSV VP16 and, using chimeric Zta-E2 and Gal4-Zta constructions and a modified oriLyt reporter in which the ZREs were converted to either E2 or Gal4 binding sites, that Zta aa 28 to 103 were necessary for replication (60). Using EBV-positive cells and an unmodified oriLyt reporter, Askovic and Baumann (1) observed oriLyt replication when the Zta activation domain was exchanged for a heterologous activation domain; in this system, deletions of individual regions of the activation domain did not identify any region that was specifically required for replication.
We have previously used a cotransfection replication assay to examine the contribution of Zta (21, 58). This assay system is strictly defined in that EBV-negative cells are transfected with an oriLyt-containing plasmid, and replication is totally dependent on the cotransfected EBV replication genes. In this system, deletion of the Zta activation domain between aa 2 and 10, aa 25 and 86, or aa 93 and 141 did not affect replication. However deletion of aa 2 to 25, and more specifically aa 13 to 19, severely impaired replication efficiency, implicating a region between aa 11 and 25 as serving a replication function. We have further pursued the requirement for Zta activation domain sequences by examining interactions between Zta and the core replication proteins. Evidence of an interaction with the helicase-primase complex is presented. The finding that the amino terminus of the Zta activation domain is involved in the helicase-primase interaction strongly suggests that this interaction is functionally relevant and that Zta contributes to oriLyt replication, in part, by stabilizing formation of a tethered replication complex.
MATERIALS AND METHODS
Plasmids.
The SG5 vector (Stratagene), which uses the simian virus 40 early promoter to drive expression, was modified by introduction into the vector BamHI site of a sequence encoding aa 425 to 434 of the human c-myc gene to create pJH363. The double-stranded insert was generated by annealing the oligonucleotides 5′-GATCTAAGATGGCGGAACAAAAGCTTATTTCTGAAGAAGACTTGG and 5′-GATCCCAAGTCTTCTTCAGAAATAAGCTTTTGTTCCGCCATCTTA. The five replication genes for which immunological reagents were not available were introduced into this vector. The previously described expression vectors (59) for BSLF1, BALF5, BBLF4, and BALF2 were modified by conversion of an upstream vector XbaI site into either a Bgl II site (BSLF1, BALF5, and BBLF4) or a BamHI site (BALF2), and DNA fragments containing the open reading frames were isolated by cleavage with either BglII or BamHI and ligated into BglII-cleaved pJH363. The open reading frame for BBLF2/3 was isolated as a BamHI fragment from pEF75A (21).
Another modified SG5 vector, pJH209, contains the sequence encoding the nuclear localization signal and transcriptional activation domain (aa 424 to 487) from EBV EBNA2 introduced into the BglII site of SG5. The EBNA2 sequences were amplified by using PCR techniques and the primers 5′-GCTAGGATCCCCAATACATGAACCGGAG and 5′-GCTAAGATCTCTGGATGGAGGGGCGAGG. The six replication gene open reading frames were introduced into the BglII site of pJH209 as either BamHI or BglII fragments as described above. The individual replication gene expression clones are summarized in Table 1. Expression of the appropriately sized proteins from each construction was confirmed by Western blotting.
TABLE 1.
EBV replication genes and expression plasmids
| Viral gene | Function | Clone | c-Myca | E2TANLSb |
|---|---|---|---|---|
| BMRF1 | Polymerase accessory factor | RTS14 | DH295 | |
| BALF5 | DNA polymerase | RTS13 | DH312 | DH294 |
| BALF2 | Single-stranded DNA binding protein | RTS12 | DH316 | JF3 |
| BSLF1 | Primase | RTS11 | DH310 | JF5 |
| BBLF4 | Helicase | RTS28 | PG14 | JF7 |
| BBLF2/3 | Primase-associated factor | RTS25 | DH318 | JF6 |
Protein is expressed as a fusion with an epitope (aa 425 to 434) from c-Myc.
Protein is expressed as a fusion with the nuclear localization signal and transcriptional activation domain (aa 424 to 487) from EBV EBNA2.
The reporter for glutathione S-transferase (GST) fused to Zta aa 1 to 133 [Zta(1-133)], pDH245, was generated by cleaving pDH237 with SmaI and EcoRI to remove the DNA binding and dimerization domains; a BglII linker was added to the blunted ends, and the DNA was cleaved with BglII and religated. The oriLyt-chloramphenicol acetyltransferase (CAT) reporter, pDH123, has been described elsewhere (40), as have the expression plasmids for Zta (pRTS21), Zta(Δ2-25) (pRTS68), and Zta(Δ13-19) (pDH285) (58).
Immunofluorescence assays.
Vero cells were seeded at 8 × 104 cells per well in two-well slide chambers. Cells were transfected with a maximum of 3 μg of DNA by the calcium phosphate procedure. After transfection, cells were incubated in Dulbecco modified Eagle medium plus 10% fetal bovine serum for 16 h at 35°C in 3% CO2 and, after a medium change, for a further 24 h. Cells were washed in 1× Tris-saline (100 mM Tris-HCl [pH 7.5]), fixed with 1% paraformaldehyde in phosphate-buffered saline (0.144 g of KH2PO4, 9.0 g of NaCl, and 0.795 g of Na2HPO4 · 7H2O per liter) for 10 min at room temperature and permeabilized for 20 min on ice in 0.2% Triton X-100 in phosphate-buffered saline. Cells were incubated with primary antibody for 60 min at 37°C and with secondary antibody at 37°C for 30 min. Antibodies used were anti-Myc mouse monoclonal (1:200) and rabbit polyclonal (1:300) (Santa Cruz Biotechnology Inc., Santa Cruz, Calif.), anti-BMRF1 monoclonal (DAKO Corp., Carpinteria, Calif.), anti-BZLF1 monoclonal (1:1,000; DAKO) and polyclonal (1:800; gift of Marie Hardwick, Johns Hopkins School of Hygiene and Public Health) (31) antibodies; fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG; 1:100; Cappel Organon Teknika, Durham, N.C.); and FITC-conjugated donkey anti-rabbit (1:100) and rhodamine-conjugated donkey anti-mouse (1:100) and anti-rabbit (1:100) IgG (Chemicon, Temecula, Calif.).
CAT assay.
Vero cells were plated in six-well cluster dishes at 2 × 105 cells per well 16 h before transfection with a medium change 4 h before transfection. Cells were transfected by calcium phosphate precipitation with the oriLyt-CAT reporter pDH123 (1 μg), Zta reporter pRTS21, pRTS68, or pDH285 (0.2 μg), and individual replication genes (0.8 μg). Vector SG5 DNA was used to equalize the amount of DNA in each transfection to 4.8 μg. Cells were harvested 40 h after transfection, and CAT activity was assayed (44). CAT activity was quantitated with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
GST affinity assay.
GST and GST-Zta fusion proteins were induced by growth in medium containing 5 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 30°C. Pelleted bacteria were resuspended in binding buffer (20 mM HEPES [pH 7.9], 20 mM KCl, 1 mM MgCl2, 2 mM dithiothreitol, 17% glycerol) and sonicated. Cell debris was removed by centrifugation at 10,000 × g for 10 min. The supernatant was incubated with glutathione-agarose beads (Sigma, St. Louis, Mo.) at 4°C overnight and then washed three times in binding buffer. The amount of protein bound to the beads was determined by Coomassie blue staining of protein separated on a sodium dodecyl sulfate (SDS)-polyacrylamide gel. Equal amounts of each GST protein were used in the affinity assays.
293T cells in 100-mm-diameter dishes were transfected with a maximum of 15 μg of DNA per dish, and cells were harvested 40 h after transfection. Cells were lysed in 500 μl of isotonic buffer (142.5 mM KCl, 5 mM MgCl2 10 mM HEPES [pH 7.2], 1 mM EGTA [pH 8.0], 0.2% Nonidet P-40). The cell extract was incubated with the GST fusion proteins overnight at 4°C, after which the complex was washed five times in binding buffer. The complex was dissociated from the beads by boiling for 5 min in 2× SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer (2% SDS, 10% glycerol, 100 mM dithiothreitol, 60 mM Tris [pH 6.8], 0.02% bromophenol blue), and the proteins were separated by SDS-PAGE on a 10% gel. Proteins were transferred to a nitrocellulose membrane (Bio-Rad, Hercules, Calif.), and the replication proteins were detected by incubation with anti-Myc antibody (1:1,000) followed by visualization by the enhanced chemiluminescence reaction (Amersham Life Science, Buckinghamshire, England).
RESULTS
Intracellular localization of the EBV helicase-primase proteins.
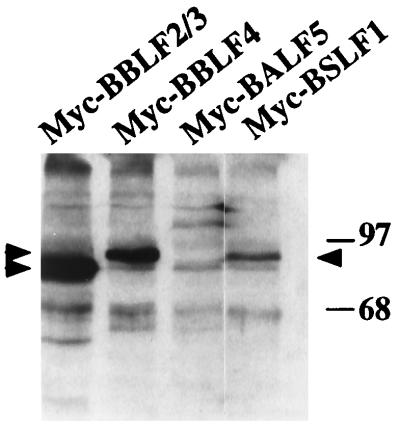
BALF2, BALF5, and BMRF1 are known to individually localize to the nucleus (33, 69). However, the three EBV helicase-primase proteins have not been extensively characterized, and their intracellular localization has not been examined. As an initial step in evaluating potential interactions between Zta and the core replication proteins, we generated vectors expressing Myc-tagged replication proteins. These reagents were used to examine the localization of the individual core replication proteins in transfected cells (Fig. 1). As previously described, the BMRF1, BALF5, and BALF2 proteins localized to the nucleus (exemplified by Myc-BALF2 in Fig. 1). When individually transfected, Myc-BBLF2/3 showed mixed nuclear and cytoplasmic staining, Myc-BSLF1 was perinuclear, and Myc-BBLF4 localized to the cytoplasm. The vectors for these three latter Myc-proteins express proteins in the appropriate size range, as shown in Fig. 2. The open reading frames for BBLF2/3, BSLF1, and BBLF4 are predicted to encode 765, 874, and 811 aa, respectively, while the control DNA polymerase protein is predicted to contain 1,014 aa. The effect of secondary modifications on the relative mobility of these proteins is not known.
FIG. 1.
Immunofluorescence assay showing the intracellular localization of the proteins of the helicase-primase complex. Vero cells were transfected with Myc-tagged BSLF1, BBLF4, and BBLF2/3 and a control, BALF2. Transfected proteins were visualized with anti-Myc antibody and FITC-conjugated anti-mouse IgG secondary antibody.
FIG. 2.
Expression of the Myc-tagged helicase-primase proteins. Expression vectors for Myc-BBLF2/3, Myc-BBLF4, and Myc-BSLF1 and for a control replication protein, Myc-BALF5, were individually transfected into Cos cells, and protein expression was analyzed by Western blotting using anti-Myc antibody and visualization by chemiluminescence. Positions of the helicase-primase protein bands are indicated with arrowheads; positions of the 97- and 68-kDa molecular size markers are indicated on the right.
The three helicase-primase proteins encoded by HSV have been shown to form a tripartite complex which influences intracellular localization (6, 16). Evidence that the EBV homologs also interact came from an examination of the intracellular localization of the helicase-primase proteins in triply transfected cells. Cotransfection of Myc-BSLF1 in the presence of BBLF2/3 and BBLF4 resulted in discrete nuclear localization of Myc-BSLF1. Similarly, cotransfection of Myc-BBLF2/3 with BSLF1 plus BBLF4 resulted in localization of Myc-BBLF2/3 to the nucleus (Fig. 3). The nuclear localization of BBLF2/3 and BSLF1 required the concurrent presence of all three members of the complex. Myc-BBLF2/3 in the presence of BSLF1 gave a diffuse cytoplasmic pattern, and Myc-BSLF1 in the presence of BBLF2/3 was also cytoplasmic (Fig. 3). Similarly, Myc-BBLF4 in the presence of either BSLF1 or BBLF2/3 remained cytoplasmic.
FIG. 3.
Immunofluorescence assay showing the ability of BBLF4, BSLF1, and BBLF2/3 to form a tripartite complex that localizes to the nucleus in cotransfected Vero cells. Cotransfection of any two of the helicase-primase proteins did not confer nuclear localization. Transfected proteins were visualized with anti-Myc antibody and FITC-conjugated anti-mouse IgG secondary antibody.
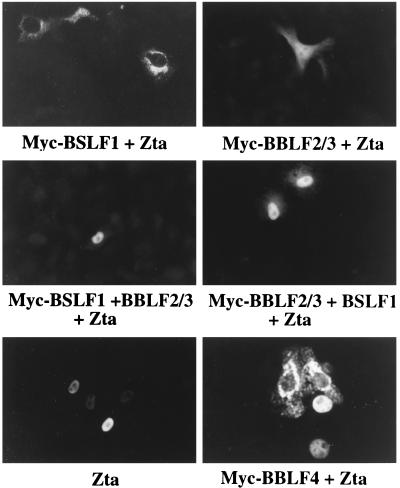
Zta relocates both the helicase and the primase subcomplex into the nucleus.
In contrast to the cytoplasmic localization of the individual members of the helicase-primase complex, Zta is a nuclear protein (Fig. 4). The differential localization of Zta and the helicase-primase proteins provided an assay for potential interactions between Zta and these proteins. Cotransfection of Zta with either Myc-BBLF2/3 or Myc-BSLF1 did not change the localization of these proteins from that observed for the individually transfected proteins shown in Fig. 1. Myc-BBLF2/3 showed the mixed nuclear and cytoplasmic pattern, and Myc-BSLF1 remained perinuclear (Fig. 4). However, cotransfection of Zta with the combination of BSLF1 and BBLF2/3 converted the localization of these proteins from the distinctly cytoplasmic pattern observed in doubly transfected cells to a dominant nuclear pattern (Fig. 4). Cotransfection of Zta with Myc-BBLF4 converted the strictly cytoplasmic BBLF4 pattern to a strictly nuclear pattern (Fig. 4) indicative of a separate interaction between Zta and BBLF4. In the experiment shown in Fig. 4, the cells were transfected with Zta and BBLF4 at a ratio of 2:1. At this ratio, some cells with cytoplasmic staining are also visible.
FIG. 4.
Immunofluorescence assay demonstrating relocalization of Myc-BBLF4, Myc-BBLF2/3 plus BSLF1, and Myc-BSLF1 plus BBLF2/3 in the presence of cotransfected Zta. Myc-tagged proteins were visualized with anti-Myc antibody and FITC-conjugated anti-mouse IgG secondary antibody. Zta was detected with mouse monoclonal antibody. The ratio of cotransfected Zta to Myc-tagged expression vector was 2:1. Both nuclear and cytoplasmic Myc-BBLF4 were detected at this ratio.
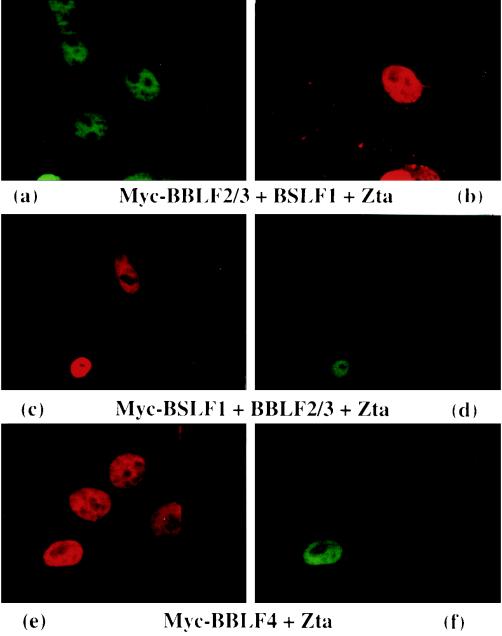
Not all of the cotransfected cells expressed both proteins. At the 2:1 ratio for the Zta-Myc fusion protein expression plasmids, cells expressing Zta but not the Myc-tagged proteins were more common than those expressing only the Myc-tagged proteins. As shown by double staining (Fig. 5), all cells in which Myc-BBLF4 and the Myc-BBLF2/3-plus-BSLF1 subcomplex were present in the nucleus also expressed Zta. Different combinations of antibody and fluorescence tag were used to ensure the specificity of the results. For example, Zta was detected with polyclonal anti-Zta rabbit antiserum and FITC-tagged anti-rabbit antibody in the cotransfection with Myc-BBLF2/3 plus BSLF1, polyclonal anti-Zta rabbit antiserum and rhodamine-tagged anti-rabbit antiserum in the cotransfection with Myc-BSLF1 plus BBLF2/3, and monoclonal mouse antiserum in the cotransfection with Myc-BBLF4.
FIG. 5.
Immunofluorescence assay showing that cotransfected cells in which Myc-BBLF4 or Myc-BBLF2/3 plus BSLF1 localize to the nucleus express Zta. Vero cells were cotransfected with Zta plus Myc-BBLF4 or with Zta plus Myc-BBLF2/3 and BSLF1 and stained with anti-Zta (a, c, and e) and anti-Myc (b, d, and f) primary antibodies. Secondary antibodies: (a and f) FITC-conjugated donkey anti-rabbit; (b and e) rhodamine-conjugated goat anti-mouse; (c) rhodamine-conjugated donkey anti-rabbit; (d) FITC-conjugated goat anti-mouse. Not all cells express both cotransfected proteins, but cells expressing nuclear Myc-tagged proteins always also express Zta.
BBLF4 and the BBLF2/3-plus-BSLF1 primase subcomplex interact with the activation domain of Zta.
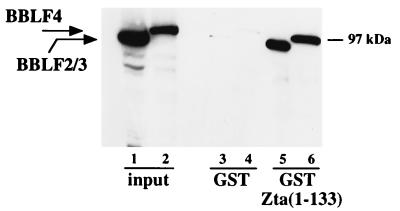
We had previously presented evidence that the activation domain of Zta also contributed a replication-specific function. To determine whether the activation domain mediated the interaction between Zta and BBLF4 and between Zta and the BSLF1-plus-BBLF2/3 primase subcomplex, a GST-Zta affinity assay was performed with GST fusion proteins containing the activation domain of Zta, GST-Zta(1-133), and control GST protein. Extracts of 293T cells cotransfected with either Myc-BBLF4 or Myc-BBLF2/3 plus BSLF1 were incubated with equal amounts of the GST proteins bound to beads. After washing, the bound proteins were solubilized by boiling in SDS, separated on a denaturing polyacrylamide gel, and transferred to a nitrocellulose membrane. Reactive proteins were visualized using anti-Myc antibody and a chemiluminescence detection system (Fig. 6). Myc-BBLF4 and Myc-BBLF2/3 each bound to GST-Zta(1-133). Neither protein bound significantly to the control GST protein. Thus, the interaction between Zta and the helicase-primase complex is mediated by the Zta activation domain.
FIG. 6.
BBLF4 and BBLF2/3 plus BSLF1 interact with the Zta activation domain, as determined by GST affinity assay in which extracts of 293T cells transfected with Myc-BBLF4 (lanes 2, 4, and 6) or Myc-BBLF2/3 plus BSLF1 (lanes 1, 3, and 5) were incubated with GST-Zta(1-133) and control GST proteins. Bound protein was separated by SDS-PAGE, transferred to a nitrocellulose membrane, and incubated with anti-Myc antibody. Reactive proteins were visualized by chemiluminescence. The input lanes were loaded with 1/15 of the amount of extract incubated with the GST beads. The position of the 97-kDa molecular size marker is indicated on the right.
Effect of N-terminal deletions of the Zta activation domain.
We then used an immunofluorescence assay and anti-Myc antibody to evaluate the effect of Zta N-terminal activation domain deletions in cotransfected cells. Zta(Δ2-25) and Zta(Δ13-19) have been previously characterized (58). The ability of Zta(Δ2-25) and Zta(Δ13-19) to relocalize cotransfected Myc-BBLF2/3 plus BSLF1 was examined in cells transfected with Zta and Myc-BBLF2/3 plus BSLF1 at a ratio of 5:1. As summarized in Table 2, Myc-BBLF2/3 plus BSLF1 gave a completely cytoplasmic signal. Cotransfection with wild-type Zta resulted in approximately 90% of the fluorescent cells showing either nuclear or mixed nuclear plus cytoplasmic staining. Cells scored as nuclear staining had immunofluorescence signal only in the nucleus. Cells scored as mixed nuclear plus cytoplasmic showed nuclear staining that was equal to or stronger than that in the cytoplasm. The mixed staining pattern was not seen in the absence of Zta and presumably reflects incomplete relocalization of the Myc-tagged protein. In contrast to the effect of wild-type Zta, between 88 and 97% of the stained cells cotransfected with Myc-BBLF2/3 and either Zta(Δ2-25) or Zta(Δ13-19) showed cytoplasmic fluorescence. Wild-type Zta, Zta(Δ2-25), and Zta(Δ13-19) each give a discrete nuclear pattern in the transfected cells (reference 58 and data not shown). Thus, in cotransfected cells, deletion of amino-terminal sequences of Zta severely impaired interaction with the primase subcomplex. Deletion of Zta N-terminal sequences had less effect on the interaction of Zta with Myc-BBLF4. The results obtained with Zta and Myc-BBLF4 cotransfected at a 5:1 ratio are also listed in Table 2. At this ratio, wild-type Zta converted the Myc-BBLF4 staining pattern from cytoplasmic to predominantly nuclear, with a small proportion of the cells showing mixed nuclear plus cytoplasmic staining. Zta(Δ2-25) behaved similarly to wild-type Zta. Zta(Δ13-19) was less efficient at nuclear relocation than Zta(Δ2-25), but a significant proportion (61%) of the cells positive for Myc-BBLF4 expression had nuclear staining in the presence of this mutant. The Δ13-19 deletion may have a more severe effect on protein conformation than the Δ2-25 deletion. The total number of stained cells was less in cells cotransfected with Myc-BBLF4 plus Zta or the Zta mutants than in cells transfected with Myc-BBLF4 alone, and the interaction between Zta and Myc-BBLF4 may render the Myc tag less available for antibody recognition.
TABLE 2.
Effect of Zta activation domain deletions on localization of BBLF4 and BBLF2/3 plus BSLF1
| Localization (% of positive cells)a
|
|||
|---|---|---|---|
| Nucleus | Nucleus + cytoplasm | Cytoplasm | |
| Myc-BBLF2/3 + BSLF1 | 0 | 0 | 100 |
| +wt Zta | 61 ± 2 | 28 ± 1 | 11 ± 2 |
| +Zta(Δ2-25) | 7 ± 4 | 5 ± 5 | 88 ± 2 |
| +Zta(Δ13-19) | 2 ± 2 | 1 ± 1 | 97 ± 3 |
| Myc-BBLF4 | 0 | 0 | 100 |
| +wt Zta | 91 ± 5 | 9 ± 5 | 0 |
| +Zta(Δ2-25) | 91 ± 1 | 9 ± 1 | 0 |
| +Zta(Δ13-19) | 61 ± 1 | 30 ± 1 | 9 ± 1 |
Myc-tagged proteins were detected in transfected Vero cells by indirect immunofluorescence using anti-Myc antibody and FITC-conjugated anti-mouse Ig. Values are averages of two experiments.
The tripartite helicase-primase complex interacts with oriLyt-bound Zta.
The ability of Zta to alter the intracellular localization of components of the helicase-primase complex indicates an interaction between Zta and these proteins. However, the tripartite helicase-primase complex is also capable of localizing to the nucleus in the absence of Zta, suggesting that the interaction with Zta probably serves another function in the infected cell. Zta binds to multiple ZRE sites within the promoter and enhancer elements of oriLyt, and interactions between Zta and the replication proteins could potentiate formation of a stable replication initiation complex. An assay was designed to test for an interaction between oriLyt-bound Zta and the helicase-primase proteins. The three components of the helicase-primase complex and the single-stranded DNA binding protein were recloned such that they would be expressed as fusions with the transactivation domain and nuclear localization signal of EBNA2 (E2TANLS). Cells individually transfected with BBLF4-E2TANLS and BBLF2/3-E2TANLS showed nuclear localization of these proteins, but the addition of the E2TANLS sequences was insufficient to alter the localization of BSLF1-E2TANLS, which remained predominantly perinuclear in its distribution (Fig. 7). The previously nuclear BALF2 remained nuclear as the BALF2-E2TANLS form (Fig. 7).
FIG. 7.
Immunofluorescence assay showing the intracellular localization in transfected Vero cells of the three helicase-primase proteins and BALF2 expressed as fusions with E2TANLS. Transfected proteins were visualized with anti-EBNA2 antibody and FITC-conjugated anti-mouse IgG secondary antibody. Addition of the tag sequences resulted in nuclear localization of BBLF4 and BBLF2/3 but was insufficient to relocate individually transfected BSLF1. The localization of the nuclear BALF2 protein was not affected by the addition of the tag.
We established a transactivation assay in which an oriLyt-CAT reporter was cotransfected in Vero cells with low levels (200 ng) of a Zta expression vector. The oriLyt reporter contains the complete 1,000-bp oriLyt region, including the oriLyt enhancer. The oriLyt promoter (BHLF1p) drives CAT expression, and there are seven Zta binding sites present, four in the promoter region and three in the oriLyt enhancer. Binding of Zta to the ZREs in oriLyt results in transactivation of CAT activity (Fig. 8). The use of 200 ng of transfected Zta was designed to give low-level transactivation such that if a second activation domain were tethered to the reporter, then the effects of this second activator would also be detectable. Cotransfections were performed in which the cells received oriLyt-CAT and the E2TANLS-tagged proteins, either individually or in groups and in the presence or absence of Zta. As shown in a representative assay (Fig. 8), cotransfection of BALF2-E2TANLS had no effect on Zta activation of the oriLyt-CAT reporter. Cotransfection of BBLF4-E2TANLS, BSLF1-E2TANLS, and BBLF2/3-E2TANLS plasmids individually gave limited activation over that seen with Zta alone, while cotransfection of the three helicase-primase expression plasmids with either BBLF2/3 or BSLF1 carrying the activation domain tag consistently resulted in a fourfold activation of the reporter above that seen with Zta alone. This activation was Zta specific and was not seen in the absence of Zta.
FIG. 8.
Transient expression assay using Zta-dependent superactivation of expression from an oriLyt-CAT reporter as a measure of Zta-helicase-primase interaction. The results shown are representative of results obtained in three different experiments. Vero cells were cotransfected with oriLyt-CAT plus or minus Zta, and the indicated individual replication genes were expressed as E2TANLS fusions (indicated by an asterisk) or groups of replication proteins, one of which was expressed as an E2TANLS fusion. Cotransfection of the three helicase-primase proteins with one member tagged with the E2TANLS consistently resulted in Zta-dependent superactivation of CAT expression.
Interestingly, BALF2-E2TANLS, which had no effect on Zta activation individually, produced significant superactivation in the presence of the untagged BBLF4, BSLF1, and BBLF2/3 proteins, suggesting that BALF2 may interact with the helicase-primase complex.
DISCUSSION
Studies on Zta transcriptional activation have provided evidence for Zta-mediated stabilization of a DNA-bound TFIIA-TFIID complex. In the stepwise addition model of transcription complex assembly, recruitment of TFIID and recruitment of TFIIA represent the initial steps in the assembly of the core initiation complex. The role of Zta in oriLyt replication has been less clear. Zta binds to multiple sites in oriLyt, including the four oriLyt (BHLF1) promoter ZREs that are essential for oriLyt replication (41, 60, 61). A transcriptional contribution to replication by Zta seems likely. Removal of the Zta transcriptional activation domain abolishes replication activity (1, 58, 60), and fusion of an additional heterologous activation domain increases reactivation efficiency (2). As a transcription factor, Zta may contribute either by disrupting nucleosome formation and increasing the accessibility of the origin to the replication complex or by introducing topological changes in the DNA that facilitate replication initiation (28, 37). However, in the replication assay performed with EBV-negative cells where all of the EBV genes are introduced individually on expression plasmids, there was not a direct correlation between the ability of Zta to function as a transcriptional activator and its ability to support oriLyt replication (58). As has been reported for the papillomavirus E2 transcriptional activator (57), mutations in the Zta activation domain revealed a separable DNA replication function. Zta activation domain mutants such as Zta(Δ2-25) and Zta(Δ13-19) were able to activate expression from the endogenous BMRF1 promoter but were unable to provide replication function, suggesting that the amino terminus of the activation domain might specify an additional replication-specific activity. We have now presented experimental data indicating that amino-terminal Zta activation domain sequences are required for efficient interaction with the core replication proteins of the primase subcomplex.
When individually transfected, the Myc-tagged helicase (BBLF4) and primase (BSLF1) proteins localized to the cytoplasm, while the primase-associated protein (BBLF2/3) showed a mixed nuclear and cytoplasmic distribution. These three proteins were originally designated on the basis of their amino acid homology with HSV replication proteins, and the EBV proteins themselves have not been functionally characterized. The immunofluorescence assays provided evidence that, like their HSV counterparts, BSLF1 and BBLF2/3 interact to form a primase subcomplex. Interaction with HSV UL8 (the BBLF2/3 homolog) leads to more efficient primer synthesis by the primase (63, 67). When individually transfected, Myc-BBLF2/3 showed mixed nuclear and cytoplasmic staining, but when it was cotransfected with BSLF1, the pattern changed to that of BSLF1 and was strictly cytoplasmic. Further, the presence of Zta did not affect the localization of either BSLF1 or BBLF2/3 when they were individually cotransfected with Zta. Nuclear relocalization by Zta required the concurrent presence of both BSLF1 and BBLF2/3, suggesting that interaction between BSLF1 and BBLF2/3 produces a conformational change in one or other protein. Triple transfection of BBLF4, BBLF2/3, and BSLF1 also resulted in nuclear localization of these proteins, indicating that they form a helicase-primase complex. This behavior is identical to that observed with the three HSV helicase-primase proteins (6).
Interaction between Zta and the helicase-primase complex was demonstrable in three different assays. The ability of Zta to independently translocate to the nucleus the cotransfected helicase, Myc-BBLF4, and the cotransfected primase subcomplex indicates that Zta contacts at least two of the helicase-primase proteins. The presence of both BSLF1 and BBLF2/3 was required for the primase subcomplex interaction with Zta, and which of these two proteins actually contacts Zta cannot be determined from the experiments described here. The HSV origin binding protein, UL9, has been found to contact UL8, the BBLF2/3 analog (49). Although nuclear localization was initially used as a measure of interaction with Zta, it seems unlikely that this is the primary replication-specific function for Zta in the infected cell since the tripartite helicase-primase complex is itself capable of nuclear localization. Transcription factors may also be involved in directly recruiting core replication proteins or stabilizing the formation of the replisome (11, 19, 50). If Zta were to fulfill such a role, then the interaction with the helicase-primase complex would likely take place with oriLyt-bound Zta. A superactivation assay designed to test this possibility indicated that Zta bound to an oriLyt-CAT reporter interacted with the helicase-primase proteins. Stable interaction was most clearly demonstrable in the presence of the tripartite complex, which consistently gave a fourfold activation when either member of the primase subcomplex was expressed fused to a heterologous transactivation domain. Most models of replication postulate that the initial steps in DNA synthesis involve sequence-specific recognition by an initiator protein followed by localized DNA melting or unwinding (26). A number of other viruses encode origin binding proteins that have helicase or unwinding activity. Examples include simian virus 40 T antigen (4), HSV UL9 (5), and papillomavirus E1 (72). Zta acts as the EBV oriLyt origin binding protein and may ensure localized unwinding through the combination of transcriptional stimulation and direct recruitment of the helicase-primase complex.
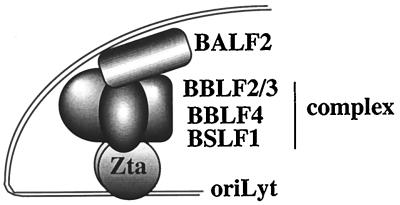
The oriLyt-CAT superactivation assay also raised the possibility of an interaction between the Zta-tethered helicase-primase complex and the single-stranded DNA binding protein BALF2. Studies of HSV replication complex assembly in both virus-infected and DNA-transfected cells have indicated that the helicase primase proteins, UL5, UL8, and UL52, are needed for localization of the HSV single-stranded DNA binding protein, UL29, to prereplicative sites (45, 46, 76). Thus, the proposed HSV assembly model involves the single-stranded DNA binding protein assembling into a complex with the helicase primase proteins, with polymerase and polymerase accessory protein, UL42, being recruited subsequently. The potential exists for multiple contacts within the replication complex. Contacts between the HSV single-stranded DNA binding protein and polymerase and the origin binding protein, UL9, have also been documented (3, 51), as have contacts between UL8 and polymerase (47). The information available for assembly of the EBV replication complex is much more limited, but the present study provides parallels with the HSV assembly model in that initial steps in assembly seem to involve tethering of the helicase-primase complex at the origin by interaction with DNA-bound Zta and entry of the single-stranded DNA binding protein, BALF2, through interaction with the helicase-primase proteins. A pictorial summation of the EBV data is presented in Fig. 9.
FIG. 9.
Representation of the initial stages of core replication protein assembly as deduced from this study. OriLyt bound dimers of Zta interact through the activation domain with both the helicase (BBLF4) and primase subcomplex (BSLF1 plus BBLF2/3) to provide a stabilizing tether for the core replication complex. The single-stranded DNA binding protein BALF2 may in turn contact the helicase-primase complex.
We had previously found that Zta(Δ2-25) and Zta(Δ13-19) were unable to replicate oriLyt in a cotransfection replication assay (58). However, in studies in other laboratories using different assay systems for oriLyt replication, replacement or mutation of the Zta activation domain had not been able to uncover a replication specific contribution for this domain (1, 60). While the reason for the different results cannot be ascribed with certainty, the present study allows speculation. In the nuclear relocation assay, deletion of Zta aa 2 to 25 or 13 to 19 had little effect on the interaction of Zta with BBLF4, although the deletions severely reduced Zta interaction with the primase subcomplex. Since the helicase primase proteins themselves interact to form a tripartite complex, it seems likely that deletion of Zta aa 2 to 25 might destabilize the interaction with the tripartite complex by eliminating the contact point or structural context for the BBLF2/3-plus-BSLF1 interaction but that the complex could still be tethered to Zta at reduced affinity through the contacts made by BBLF4. In the same vein, in the cotransfection replication assay mutation of Zta at codons 12 and 13, Zta(m12/13), did not allow detectable replication in the absence of Rta but supported oriLyt replication quite effectively in the presence of Rta (58). This result was interpreted to imply that Rta may also make some contacts with the replication complex, stabilizing complex formation. Other replication systems rely on EBV-positive cells, and Rta is always present. The combination of multiple contact points by the helicase-primase complex and the stabilizing contribution of Rta may have hindered detection of the helicase-primase interaction in these systems. Another possible complication for Zta mutagenesis studies is the observation that the bZip domain of Zta can interact with the polymerase accessory factor BMRF1 (75). Such an interaction could potentially provide another tethering point for the replication complex. In the same way that the Zta activation domain is functionally redundant in transcriptional activation assays, multiple contacts between Zta and the core replication proteins generate functional redundancy for oriLyt replication.
ACKNOWLEDGMENTS
We thank Michael Delannoy for assistance with the confocal microscopy, Mabel Chiu for technical assistance, and Feng Chang for manuscript preparation.
This work was funded by National Institute of Health grant RO1 CA30356. S.D.H. was supported by American Cancer Society grant FRA429, and A.K. received support from NIH Anti-Cancer Drug Development training grant 5 T32 CA09243.
REFERENCES
- 1.Askovic S, Baumann R. Activation domain requirements for disruption of Epstein-Barr virus latency by ZEBRA. J Virol. 1997;71:6547–6554. doi: 10.1128/jvi.71.9.6547-6554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann R, Grogan E, Ptashne M, Miller G. Changing Epstein-Barr viral ZEBRA protein into a more powerful activator enhances its capacity to disrupt latency. Proc Natl Acad Sci USA. 1993;90:4436–4440. doi: 10.1073/pnas.90.10.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehmer P E, Lehman I R. Physical interaction between the herpes simplex virus 1 origin-binding protein and single-stranded DNA-binding protein ICP8. Proc Natl Acad Sci USA. 1993;90:8444–8448. doi: 10.1073/pnas.90.18.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borowiec J A, Dean F B, Bullock P A, Hurwitz J. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell. 1990;60:181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- 5.Bruckner R C, Crute J J, Dodson M S, Lehman I R. The herpes simplex virus 1 origin binding protein: a DNA helicase. J Biol Chem. 1991;266:2669–2674. [PubMed] [Google Scholar]
- 6.Calder J M, Stow E C, Stow N D. On the cellular localization of the components of the herpes simplex virus type 1 helicase-primase complex and the viral origin-binding protein. J Gen Virol. 1992;73:531–538. doi: 10.1099/0022-1317-73-3-531. [DOI] [PubMed] [Google Scholar]
- 7.Carey M, Kolman J, Katz D A, Gradoville L, Barberis L, Miller G. Transcriptional synergy by the Epstein-Barr virus transactivator ZEBRA. J Virol. 1992;66:4803–4813. doi: 10.1128/jvi.66.8.4803-4813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cayrol C, Flemington E K. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 9.Cayrol C, Flemington E K. Identification of cellular target genes of the Epstein-Barr virus transactivator Zta: activation of transforming growth factor βigh3 (TGF-βigh3) and TGF-β 1. J Virol. 1995;69:4206–4212. doi: 10.1128/jvi.69.7.4206-4212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Y-N, Dong D L-Y, Hayward G S, Hayward S D. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J Virol. 1990;64:3358–3369. doi: 10.1128/jvi.64.7.3358-3369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen M, Mermod N, Horwitz M. Protein-protein interactions between adenovirus DNA polymerase and nuclear factor 1 mediate formation of the DNA replication preinitiation complex. J Biol Chem. 1990;265:18634–18642. [PubMed] [Google Scholar]
- 12.Chevallier-Greco A, Gruffat H, Manet A, Calender A, Sergeant A. The Epstein-Barr virus (EBV) DR enhancer contains two functionally different domains: domain A is constitutive and cell specific, domain B is transactivated by the EBV early protein R. J Virol. 1989;63:615–623. doi: 10.1128/jvi.63.2.615-623.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi T, Carey M. The ZEBRA activation domain: modular organization and mechanism of action. Mol Cell Biol. 1993;13:7045–7055. doi: 10.1128/mcb.13.11.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 15.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;81:7632–7636. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crute J J, Tsurumi T, Zhu L, Weller S K, Olivo P D, Challberg M D, Mocarski E S, Lehman I R. Herpes simplex virus 1 helicase-primase: a complex of three herpes-encoded gene products. Proc Natl Acad Sci USA. 1989;86:2186–2189. doi: 10.1073/pnas.86.7.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davison A J, Taylor P. Genetic relations between varicella-zoster virus and Epstein-Barr virus. J Gen Virol. 1987;68:1067–1079. doi: 10.1099/0022-1317-68-4-1067. [DOI] [PubMed] [Google Scholar]
- 18.Decaussin G, Leclerc V, Ooka T. The lytic cycle of Epstein-Barr virus in the nonproducer Raji line can be rescued by the expression of a 135-kilodalton protein encoded by the BALF2 open reading frame. J Virol. 1995;69:7309–7314. doi: 10.1128/jvi.69.11.7309-7314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dornreiter I, Erdile L F, Gilbert I U, von Winkler D, Kelly T J, Fanning E. Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrell P, Rowe D, Rooney C, Kouzarides J. EBV BZLF-1 transactivator specifically binds to consensus AP-1 sites and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fixman E D, Hayward G S, Hayward S D. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol. 1995;69:2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fixman E D, Hayward G S, Hayward S D. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol. 1992;66:5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flemington E, Speck S H. Epstein-Barr virus BZLF1 trans activator induces the promoter of a cellular cognate gene, c-fos. J Virol. 1990;64:4549–4552. doi: 10.1128/jvi.64.9.4549-4552.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flemington E, Speck S H. Evidence for coiled-coil dimer formation by an Epstein-Barr virus transactivator that lacks a heptad repeat of leucine residues. Proc Natl Acad Sci USA. 1990;87:9459–9463. doi: 10.1073/pnas.87.23.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Francis A L, Gradoville L, Miller G. Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J Virol. 1997;71:3054–3061. doi: 10.1128/jvi.71.4.3054-3061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gavin K A, Hidaka M, Stillman B. Conserved initiator proteins in eukaryotes. Science. 1995;270:1667–1671. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- 27.Gruffat H, Manet E, Rigolet A, Sergeant A. The enhancer factor R of Epstein-Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res. 1990;18:6835–6843. doi: 10.1093/nar/18.23.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Z-S, DePamphilis M L. Specific transcription factors stimulate simian virus 40 and polyomavirus origins of DNA replication. Mol Cell Biol. 1992;12:2514–2524. doi: 10.1128/mcb.12.6.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutsch D E, Holley-Guthrie E A, Zhang Q, Stein B, Blanar M A, Baldwin A S, Kenney S C. The bZip transactivator of Epstein-Barr virus, BZLF1, functionally and physically interacts with the p65 subunit of NF-κ B. Mol Cell Biol. 1994;14:1939–1948. doi: 10.1128/mcb.14.3.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 31.Hardwick J M, Lieberman P M, Hayward S D. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol. 1988;62:2274–2284. doi: 10.1128/jvi.62.7.2274-2284.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiehl A, Dorsky D I. Bipartite DNA-binding region of the Epstein-Barr virus BMRF1 product essential for DNA polymerase accessory function. J Virol. 1995;69:1669–1677. doi: 10.1128/jvi.69.3.1669-1677.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiehl A, Dorsky D I. Corperation of EBV DNA polymerase and EA-D(BMFR1) in vitro, and colocalization in nuclei of infected cells. Virology. 1991;184:330–340. doi: 10.1016/0042-6822(91)90849-7. [DOI] [PubMed] [Google Scholar]
- 34.Kolman J L, Taylor N, Marshak D R, Miller G. Serine-173 of the Epstein-Barr virus ZEBRA protein is required for DNA binding and is a target for casein kinase II phosphorylation. Proc Natl Acad Sci USA. 1993;90:10115–10119. doi: 10.1073/pnas.90.21.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laux G, Freese U K, Bornkamm G W. Structure and evolution of two related transcription units of Epstein-Barr virus carrying small tandem repeats. J Virol. 1985;56:987–995. doi: 10.1128/jvi.56.3.987-995.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J-S, Zhou B-S, Dutschman G E, Grill S P, Tan R-S, Cheng Y-C. Association of Epstein-Barr virus early antigen diffuse component and virus-specified DNA polymerase activity. J Virol. 1987;61:2947–2949. doi: 10.1128/jvi.61.9.2947-2949.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li R, Botchan M R. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc Natl Acad Sci USA. 1994;91:7051–7055. doi: 10.1073/pnas.91.15.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lieberman P. Identification of functional targets of the Zta transcriptional activator by formation of stable preinitiation complex intermediates. Mol Cell Biol. 1994;14:8365–8375. doi: 10.1128/mcb.14.12.8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieberman P M, Berk A J. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 40.Lieberman P M, Hardwick J M, Hayward S D. Responsiveness of the Epstein-Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J Virol. 1989;63:3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieberman P M, Hardwick J M, Sample J, Hayward G S, Hayward S D. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J Virol. 1990;64:1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieberman P M, Ozer J, Gursel D B. Requirement for transcription factor IIA (TDFIIA)-TFIID recruitment by an activator depends on promoter structure and template competition. Mol Cell Biol. 1997;17:6624–6632. doi: 10.1128/mcb.17.11.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin J-C, Sista N D, Besencon F, Kamine J, Pagano J S. Identification and functional characterization of Epstein-Barr virus DNA polymerase by in vitro transcription-translation of a cloned gene. J Virol. 1991;65:2728–2731. doi: 10.1128/jvi.65.5.2728-2731.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling P D, Ryon J J, Hayward S D. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J Virol. 1993;67:2990–3003. doi: 10.1128/jvi.67.6.2990-3003.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liptak L M, Uprichard S L, Knipe D M. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J Virol. 1996;70:1759–1767. doi: 10.1128/jvi.70.3.1759-1767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukonis C J, Burkham J, Weller S K. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J Virol. 1997;71:4771–4781. doi: 10.1128/jvi.71.6.4771-4781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marsden H S, McLean G W, Barnard E C, Francis G J, MacEachran K, Murphy M, McVey G, Cross A, Abbotts A P, Stow N D. The catalytic subunit of the DNA polymerase of herpes simplex virus type 1 interacts specifically with the C terminus of the UL8 component of the viral helicase-primase complex. J Virol. 1997;71:6390–6397. doi: 10.1128/jvi.71.9.6390-6397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGeoch D J, Dalrymple M A, Dolan A, McNab D, Perry L J, Taylor P, Challberg M D. Structures of herpes virus type 1 genes required for replication of virus DNA. J Virol. 1988;62:444–453. doi: 10.1128/jvi.62.2.444-453.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLean G W, Abbotts A P, Parry M E, Marsden H S, Stow N D. The herpes simplex virus type 1 origin-binding protein interacts specifically with the viral UL8 protein. J Gen Virol. 1994;75:2699–2706. doi: 10.1099/0022-1317-75-10-2699. [DOI] [PubMed] [Google Scholar]
- 50.Mohr I J, Clark R, Sun S, Androphy E J, MacPherson P, Botchan M R. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science. 1991;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 51.O’Donnell M E, Elias P, Funnell B E, Lehman I R. Interaction between the DNA polymerase and single-stranded DNA-binding protein (infected cell protein 8) of herpes simplex virus. J Biol Chem. 1987;262:4260–4266. [PubMed] [Google Scholar]
- 52.Oguro M O, Shimizu N, Ono Y, Takada K. Both the rightward and the leftward open reading frames within the BamHI M DNA fragment of Epstein-Barr virus act as trans-activators of gene expression. J Virol. 1987;61:3310–3313. doi: 10.1128/jvi.61.10.3310-3313.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pari G S, Anders D G. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfitzner E, Becker P, Rolke A, Schule R. Functional anatagonism between the retinoic acid receptor and the viral transactivator BZLF1 is mediated by protein-protein interactions. Proc Natl Acad Sci USA. 1995;92:12265–12269. doi: 10.1073/pnas.92.26.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rooney C M, Rowe D T, Ragot T, Farrell P J. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J Virol. 1989;63:3109–3116. doi: 10.1128/jvi.63.7.3109-3116.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rooney C, Taylor N, Countryman J, Jenson H, Kolman J, Miller G. Genome rearrangements activate the Epstein-Barr virus gene whose product disrupts latency. Proc Natl Acad Sci USA. 1988;85:9801–9805. doi: 10.1073/pnas.85.24.9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakai H, Yasugi T, Benson J D, Dowhanick J J, Howley P M. Targeted mutagenesis of the human papillomavirus type 16 E2 transactivation domain reveals separable transcriptional activation and DNA replication functions. J Virol. 1996;70:1602–1611. doi: 10.1128/jvi.70.3.1602-1611.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarisky R T, Gao Z, Lieberman P M, Fixman E D, Hayward G S, Hayward S D. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J Virol. 1996;70:8340–8347. doi: 10.1128/jvi.70.12.8340-8347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarisky R T, Hayward G S. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J Virol. 1996;70:7398–7413. doi: 10.1128/jvi.70.11.7398-7413.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schepers A, Pich D, Hammerschmidt W. Activation of oriLyt, the lytic origin of DNA replication of Epstein-Barr virus, by BZLF1. Virology. 1996;220:367–376. doi: 10.1006/viro.1996.0325. [DOI] [PubMed] [Google Scholar]
- 61.Schepers A, Pich D, Hammerschmidt W. A transcription factor with homology to the AP-1 family links RNA transcription and DNA replication in the lytic cycle of Epstein-Barr virus. EMBO J. 1993;12:3921–3929. doi: 10.1002/j.1460-2075.1993.tb06070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schepers A, Pich D, Mankertz J, Hammerschmidt W. cis-acting elements in the lytic origin of DNA replication of Epstein-Barr virus. J Virol. 1993;67:4237–4245. doi: 10.1128/jvi.67.7.4237-4245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sherman G, Gottlieb J, Challberg M D. The UL8 subunit of the herpes simplex virus helicase-primase complex is required for efficient primer utilization. J Virol. 1992;66:4884–4892. doi: 10.1128/jvi.66.8.4884-4892.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sista N D, Barry C, Sampson K, Pagano J. Physical and functional interaction of the Epstein-Barr virus BZLF1 transactivator with the retinoic acid receptors RAR alpha and RXR alpha. Nucleic Acids Res. 1995;23:1729–1736. doi: 10.1093/nar/23.10.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takada K, Shimizu N, Sakuma S, Ono Y. trans-activation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J Virol. 1986;57:1016–1022. doi: 10.1128/jvi.57.3.1016-1022.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor N, Flemington E, Kolman J L, Baumann R P, Speck S H, Miller G. ZEBRA and a Fos-GCN4 chimeric protein differ in their DNA-binding specificities for sites in the Epstein-Barr virus BZLF1 promoter. J Virol. 1991;65:4033–4041. doi: 10.1128/jvi.65.8.4033-4041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tenney D J, Hurlburt W W, Micheletti P A, Bifano M, Hamatake R K. The UL8 component of the herpes simplex virus helicase-primase complex stimulates primer synthesis by a subassembly of the UL5 and UL52 components. J Biol Chem. 1994;269:5030–5035. [PubMed] [Google Scholar]
- 68.Tsurumi T, Daikoku T, Kurachi R, Nishiyama Y. Functional interaction between Epstein-Barr virus DNA polymerase catalytic subunit and its accessory subunit in vitro. J Virol. 1993;67:7648–7653. doi: 10.1128/jvi.67.12.7648-7653.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsurumi T, Kobayashi A, Tamai K, Yamada H, Daikoku T, Yamashita Y, Nishiyama Y. Epstein-Barr virus single-stranded DNA-binding protein: purification, characterization, and action on DNA synthesis by the viral DNA polymerase. Virology. 1996;222:352–364. doi: 10.1006/viro.1996.0432. [DOI] [PubMed] [Google Scholar]
- 70.Tsurumi T, Yamada H, Daikoku T, Yamashita Y, Nishiyama Y. Strand displacement associated DNA synthesis catalyzed by the Epstein-Barr virus DNA polymerase. Biochem Biophys Res Commun. 1997;238:33–38. doi: 10.1006/bbrc.1997.7234. [DOI] [PubMed] [Google Scholar]
- 71.Wu C A, Nelson N J, McGeoch D J, Challberg M D. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang L, Mohr I, Fouts E, Lim D A, Nohaile M, Botchan M. The E1 protein of the papillomavirus BPV-1 is an ATP dependent DNA helicase. Proc Natl Acad Sci USA. 1993;90:5086–5090. doi: 10.1073/pnas.90.11.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Q, Gutsch D, Kenney S. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol Cell Biol. 1994;14:1929–1938. doi: 10.1128/mcb.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Q, Holley-Guthrie E, Ge J Q, Dorsky D, Kenney S. The Epstein-Barr virus (EBV) DNA polymerase accessory protein, BMRF1, activates the essential downstream component of the EBV oriLyt. Virology. 1997;230:22–34. doi: 10.1006/viro.1997.8470. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Q, Hong Y, Dorsky D, Holley-Guthrie E, Zalani S, Elshiekh N A, Kiehl A, Le T, Kenney S. Functional and physical interactions between the Epstein-Barr virus (EBV) proteins BZLF1 and BMRF1: effects on EBV transcription and lytic replication. J Virol. 1996;70:5131–5142. doi: 10.1128/jvi.70.8.5131-5142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong L, Hayward G S. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J Virol. 1997;71:3146–3160. doi: 10.1128/jvi.71.4.3146-3160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]