Abstract
Background & Aims
Epithelial disruption in eosinophilic esophagitis (EoE) encompasses both impaired differentiation and diminished barrier integrity. We have shown that lysyl oxidase (LOX), a collagen cross-linking enzyme, is up-regulated in the esophageal epithelium in EoE. However, the functional roles of LOX in the esophageal epithelium remains unknown.
Methods
We investigated roles for LOX in the human esophageal epithelium using 3-dimensional organoid and air–liquid interface cultures stimulated with interleukin (IL)13 to recapitulate the EoE inflammatory milieu, followed by single-cell RNA sequencing, quantitative reverse-transcription polymerase chain reaction, Western blot, histology, and functional analyses of barrier integrity.
Results
Single-cell RNA sequencing analysis on patient-derived organoids revealed that LOX was induced by IL13 in differentiated cells. LOX-overexpressing organoids showed suppressed basal and up-regulated differentiation markers. In addition, LOX overexpression enhanced junctional protein genes and transepithelial electrical resistance. LOX overexpression restored the impaired differentiation and barrier function, including in the setting of IL13 stimulation. Transcriptome analyses on LOX-overexpressing organoids identified an enriched bone morphogenetic protein (BMP) signaling pathway compared with wild-type organoids. In particular, LOX overexpression increased BMP2 and decreased the BMP antagonist follistatin. Finally, we found that BMP2 treatment restored the balance of basal and differentiated cells.
Conclusions
Our data support a model whereby LOX exhibits noncanonical roles as a signaling molecule important for epithelial homeostasis in the setting of inflammation via activation of the BMP pathway in the esophagus. The LOX/BMP axis may be integral in esophageal epithelial differentiation and a promising target for future therapies.
Keywords: Lysyl Oxidase, Organoid, BMP, Eosinophilic Esophagitis
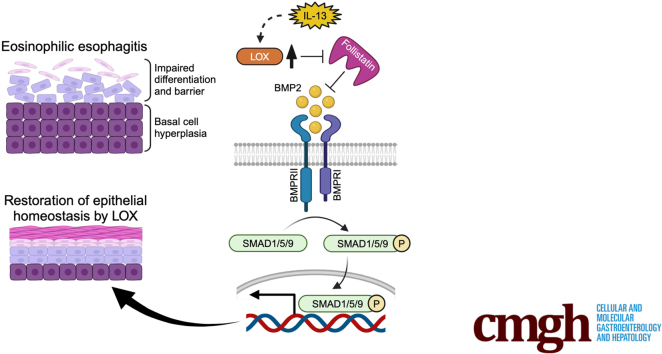
Graphical abstract
Summary.
Lysyl oxidase re-established impaired epithelial homeostasis via activation of the bone morphogenetic protein pathway in esophagus. The lysyl oxidase/bone morphogenetic protein axis may be a promising approach for eosinophilic esophagitis.
The stratified squamous epithelium of the esophagus is the first line of protection against luminal contents including food, bacteria, and other pathogens. Epithelial barrier disruption as well as impaired epithelial differentiation are universal histologic findings in eosinophilic esophagitis (EoE), a chronic allergic disease that affects children and adults.1 Patients with EoE have chronic swallowing issues, vomiting, weight loss, and, over time, develop esophageal fibrosis and strictures. We previously have shown there is ongoing epithelial disruption and symptomatology in EoE patients despite achieving histologic remission.2 Furthermore, a recent study using single-cell RNA sequencing (scRNA-seq) showed that epithelium of inactive EoE patients fails to normalize at the cellular and molecular levels.3 These data underscore the need to better understand the perturbations that occur in the epithelial barrier of the esophagus. Such knowledge may inform strategies to prevent progression of symptomatology and lifelong esophageal dysfunction.
In our previous publication, we found that lysyl oxidase (LOX), a collagen cross-linking enzyme, was increased in the esophageal epithelium of patients with EoE, and to a larger degree in patients with fibrostenosis.4 LOX catalyzes extracellular collagen to form intermolecular and intramolecular cross-links, thus forming collagen fibers.5 LOX is a requisite for normal tissue structure and integrity, with global murine deletion causing perinatal fatality resulting from aortic aneurysms and pulmonary abnormalities.6,7 In the setting of inflammation, enhanced cross-linking within tissue has been shown to promote tissue stiffness in the context of liver fibrosis, cardiovascular disease, and breast cancer.8, 9, 10 Although its role in perpetuating fibroblast activation and tissue stiffness has been described, little is known about the functional role of LOX outside of extracellular matrix remodeling and tissue stiffness.
LOX has been shown to have noncollagen cross-linking functions in bone, skin, muscle, and blood through effects on chemotaxis, gene regulation, and differentiation.11, 12, 13 It has been shown to be both protumorigenic and antitumorigenic, making the organ and the context particularly important in its evaluation.14 In the skin, LOX expression has been shown specifically in differentiated keratinocytes. LOX silencing inhibits keratinocyte differentiation in vitro and causes decreased expression of terminal differentiation markers filaggrin (FLG) and keratin 10.14, 15, 16 Although it seems to have a role in squamous differentiation, the function of LOX in the esophageal epithelium is unknown. LOX is up-regulated in the EoE epithelium, however, its role in the esophagus beyond collagen cross-linking is not well understood.
Herein, we sought to determine the role of LOX in the esophageal epithelium in the context of EoE inflammation using 3-dimensional (3D) organoid and air–liquid interface (ALI) cultures.17,18 We evaluate the effect of LOX on epithelial differentiation and barrier integrity and describe a novel cytoprotective role for LOX within the inflamed esophageal epithelium.
Results
Interleukin 13 Induces LOX in the Differentiated Epithelium in Human Esophagus
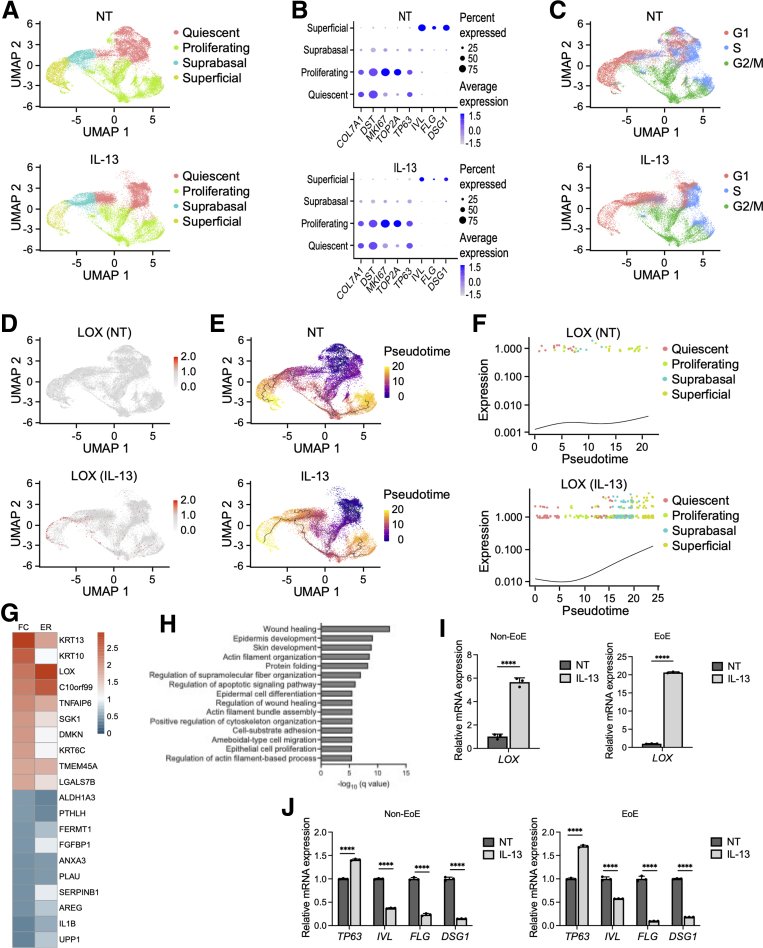
To determine the characteristics of LOX in human esophageal epithelium, we performed scRNA-seq on patient-derived organoids (PDOs) (Table 1) treated with interleukin (IL)13. 3D esophageal epithelial organoids allow for evaluation of the esophageal epithelial dynamics in vitro.19 Our prior work showed that stimulation with IL13, the major effector cytokine in EoE, recapitulates the epithelial reactive changes (such as basal cell hyperplasia) seen in EoE.17,18,20 PDOs were derived from 3 non-EoE subjects20 and we performed scRNA-seq on each line in the presence and absence of IL13. The integrated analysis identified 9 esophageal cell populations in the uniform manifold approximation and projection (UMAP). The 9 clusters then were categorized into 4 groups: quiescent basal, proliferating basal, suprabasal, and superficial, based on the expression of known epithelial makers: COL7A1, dystonin DST, marker of proliferation Ki-67 MKI67, TOP2A, tumor protein TP63, IVL, FLG, and DSG119,20 (Figure 1A). High TP63 expression and low IVL, FLG, and DSG1 expression were observed in the basal clusters. We detected COL7A1 and DST transcripts in the quiescent basal cluster, and MKI67 and TOP2A transcripts in the proliferating basal cluster, respectively. In contrast, IVL, FLG, and DSG1 were highly expressed in the differentiated (superficial) cluster (Figure 1B). We further constructed progression mapping of cell-cycle phases. In agreement with expression profiling, the majority of cells from proliferating basal cluster and differentiated clusters were located in the G2/M and G1 cell-cycle phase, respectively (Figure 1C).
Table 1.
Patient Demographics
| Sex | Age, y | Race | Ethnicity | Indication for esophogastroduodenoscopy | Histologic findings | |
|---|---|---|---|---|---|---|
| Single-cell RNA sequencing | ||||||
| PDO1 | Male | 16.4 | White | Non-Hispanic/Latino | Abdominal pain | Normal |
| PDO2 | Male | 11.3 | Other | Non-Hispanic/Latino | Nausea | Mild esophagitis |
| PDO3 | Female | 3.2 | Other | Hispanic/Latino | Vomiting | Normal |
| PCR/IHC | ||||||
| Non-EoE 1 | Male | 17.1 | White | Non-Hispanic/Latino | Abdominal pain | Normal |
| Non-EoE 2 | Male | 15.1 | White | Non-Hispanic/Latino | Abdominal pain | Normal |
| Non-EoE 3 | Female | 8.0 | White | Non-Hispanic/Latino | Abdominal pain | Mild esophagitis |
| Non-EoE 4 | Male | 9.7 | White | Non-Hispanic/Latino | Abdominal pain, vomiting | Normal |
| EoE 1 | Male | 15.1 | White | Non-Hispanic/Latino | Eosinophilic esophagitis | Severely inflamed squamous mucosa with >40 eos/hpf, marked basal cell hyperplasia, and fibrosis of the lamina propria |
| EoE 2 | Male | 12.7 | Other | Hispanic/Latino | Eosinophilic esophagitis | Esophagitis with focal intraepithelial eosinophils (up to 35 eos/hpf) |
eos/hpf, eosinophils per high power field; IHC, immunohistochemistry.
Figure 1.
scRNA-seq from PDOs reveals specific distribution of LOX in esophageal epithelium. (A–H) scRNA-seq analyses based on PDOs from 3 non-EoE patients. PDOs were cultured with or without IL13 (10 μg/mL) from days 7 to 11 and then harvested on day 11. (A) UMAP plots displaying 4 distinct cell types (quiescent basal, proliferating basal, suprabasal, and superficial) in nontreated (NT) and IL13-stimulated PDOs. (B) Dot plots showing the expression of COL7A1, DST, MKI67, TOP2A, TP63, IVL, FLG, and DSG1 in NT and IL13-stimulated PDOs. (C) UMAP plots showing cell-cycle phases in NT and IL13-stimulated PDOs. (D) UMAP plots showing the expression of LOX in NT and IL13-stimulated PDOs. (E) Inferred cell fate trajectories in NT and IL13-stimulated PDOs. Cells are colored by pseudotime value. Dark purple and bright yellow imply the earliest cells and the latest cells, respectively. (F) Expression kinetics for LOX across the pseudotime axis in NT and IL13-stimulated PDOs. (G) Heatmap representation of fold change (FC) and expression ratio (ER) of the average expression across all cells of the top 10 differentially expressed genes in LOX-expressing cells (LOX+) compared with LOX nonexpressing (LOX-) cells in the superficial cluster of IL13-stimulated PDOs. (H) Gene Ontology analysis of DEG profiles of LOX+ cells compared with LOX- cells in the superficial cluster of IL13-stimulated PDOs. (I) Relative expression, via qRT-PCR, of LOX gene in EoE or non-EoE PDOs, cultured with or without IL13 (representative results are shown, total n = 2/2 EoE/non-EoE lines). (J) Relative expression, via qRT-PCR, of TP63, IVL, FLG, and DSG1 genes in EoE or non-EoE PDOs, cultured with or without IL13 (representative results are shown, total n = 2/1 EoE/non-EoE lines).. ∗∗∗∗P < .0001
Although LOX expression was low in untreated samples, it was up-regulated markedly by IL13 treatment. Interestingly, the UMAP cells expressing LOX (LOX+) mainly emerged in the differentiated clusters (Figure 1D). Pseudotime analysis was performed to determine developmental relationships between the epithelial populations in human esophagus. Inferred trajectories identified 2 unique cell fates for the quiescent cell cluster in response to IL13 treatment, as follows: (1) toward the proliferating basal cluster and (2) toward the terminally differentiated cluster (Figure 1E). Relative expression values of LOX were increased at the late pseudotime (Figure 1F), suggesting that IL13 up-regulates LOX specifically in differentiated populations within human esophageal epithelium. We further focused on the heterogeneity of LOX+ cells in the differentiated clusters. Only 10% of the superficial cells were expressing LOX in IL13-stimulated PDOs (Figure 1D). To reveal the features of LOX+ cells, we performed differentially expressed gene (DEG) analysis between LOX+ cells and LOX nonexpressing (LOX-) cells in the IL13-treated superficial population. Expression patterns of the top 10 up-regulated and down-regulated DEGs are shown in Figure 1G. The most highly expressed DEGs in the LOX+ cells compared with the LOX- cells was KRT13, followed by KRT10, which are known as differentiation markers in stratified epithelium. Gene Ontology analysis using the DEGs showed that LOX+ cells were enriched for regulation of cell migration, squamous cell differentiation, and cytoskeleton in IL13-stimulated PDOs (Figure 1H). We next validated that LOX expression is increased in the setting of IL13 in PDOs from both non-EoE and EoE subjects (Figure 1I). Furthermore, we validated the findings that IL13 disrupts barrier integrity and differentiation in PDOs from both EoE and non-EoE subjects, consistent with previous reports showing the effects of IL13 on the esophageal epithelium in EoE.
LOX Promotes Cell Differentiation in Esophageal Epithelium
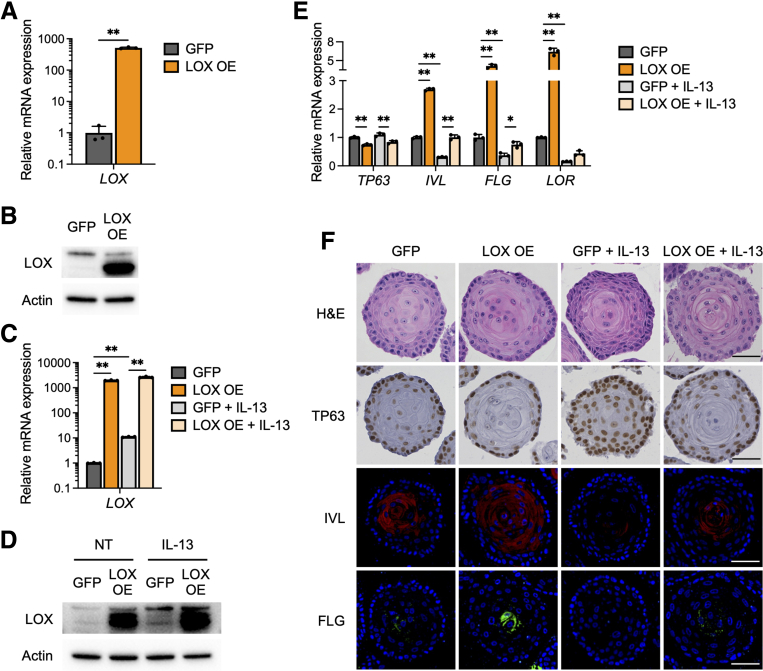
To investigate the impact of induced LOX in the esophageal epithelium, we overexpressed LOX (LOX OE) in the immortalized nontransformed normal human esophageal epithelial cell line (EPC2-hTERT).4,18,20,21 Green fluorescent protein-transduced (GFP) cells were used as a control. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and immunoblotting confirmed increased expression in LOX mRNA and protein in OE EPC2-hTERT cells compared with GFP cells in monolayer culture (Figure 2A and B), at levels higher than LOX induced by IL13 (Figure 2C and D).
Figure 2.
LOX overexpression promotes cell differentiation in esophageal epithelium. (A) qRT-PCR for LOX of monolayer-cultured EPC2-hTERT cells overexpressing GFP or LOX OE (n = 3). (B) Representative immunoblot for LOX of the monolayer-cultured GFP and LOX OE cells. (C–E) IL13 treatment induces aberrant LOX expression in EPC2-hTERT organoids. GFP and LOX OE organoids were cultured with or without IL13 (10 μg/mL) from days 7 to 11 and then harvested on day 11. (C) qRT-PCR and (D) immunoblot for LOX in GFP and LOX OE cells, monolayer-cultured with or without IL13. (E) qRT-PCR for SOX2, KRT14, TP63, IVL, FLG, and LOR of the GFP and LOX OE organoids (n = 3). (F) Representative images of H&E staining, immunohistochemistry for TP63, and immunofluorescence staining for IVL (red), FLG (green), and 4′,6-diamidino-2-phenylindole (blue), in the GFP and LOX OE organoids. Scale bar: 50 μm. Data are representative of 3 independent experiments and expressed as means ± SDs. (A) A 2-tailed Student t test and (C and E) 1-way analysis of variance were performed for statistical analyses. ∗P < .05, ∗∗P < .01.
We next evaluated 3D organoid cultures16,17 stimulated with IL13.22,23 Ectopic LOX expression resulted in decreased expression of basal cell marker genes SOX2, KRT14, and TP63, and increased expression of differentiation marker genes IVL, FLG, and LOR, in both nontreated and IL13-treated organoids (Figure 2E). We also assessed organoid morphology. H&E staining revealed that LOX OE organoids had advanced inner core hyperkeratosis compared with GFP organoids (Figure 2F). IL13-treated GFP organoids had expansion of the basal cell population, as seen in EoE,1,2 with thickening of the outer basaloid layer. However, this effect was attenuated in IL13-treated LOX OE organoids (Figure 2F). Immunohistochemistry and immunofluorescence staining showed an increase of TP63 and depression of IVL and FLG levels in response to IL13 in GFP organoids. On the other hand, LOX OE organoids showed reduced expression of TP63 and enhanced expression of IVL and FLG in both untreated and IL13 conditions, compared with GFP organoids (Figure 2F). These findings support the conclusion that LOX partially mitigates the disrupted cellular gradient caused by IL13 stimulus.
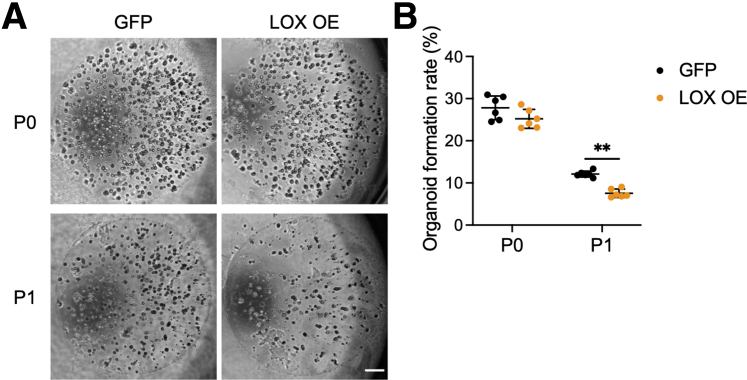
Because only basaloid cells (and not terminally differentiated cells) are capable of forming organoids, we assessed the organoid formation rate (OFR) to confirm whether LOX OE cells have reduced OFR owing to enhanced differentiation. Although OFR did not change significantly after seeding from 2-dimensional basaloid monolayers into organoids passage 0 (P0), it was decreased significantly in LOX OE organoids after passage (P1), suggesting that LOX OE organoids comprise more differentiated cells than GFP organoids (Figure 3A and B). Taken together, these results confirm that overexpressing LOX promotes epithelial differentiation.
Figure 3.
LOX overexpression attenuates organoid formation capacity. (A) Representative phase contrast images of EPC2-hTERT organoids overexpressing GFP or LOX OE on day 11. The OFR was assessed on day 11 (P0), followed by passage. OFR was assessed again on day 11 (P1). Scale bar: 50 μm. (B) OFR was calculated as the number of organoids (≥50 μm) divided by the number of total seeded cells. Data are representative of 3 independent experiments and expressed as means ± SDs (n = 6). A 2-tailed Student t test was performed for statistical analyses. ∗∗P < .01.
LOX Improves Barrier Integrity in Esophageal Epithelium
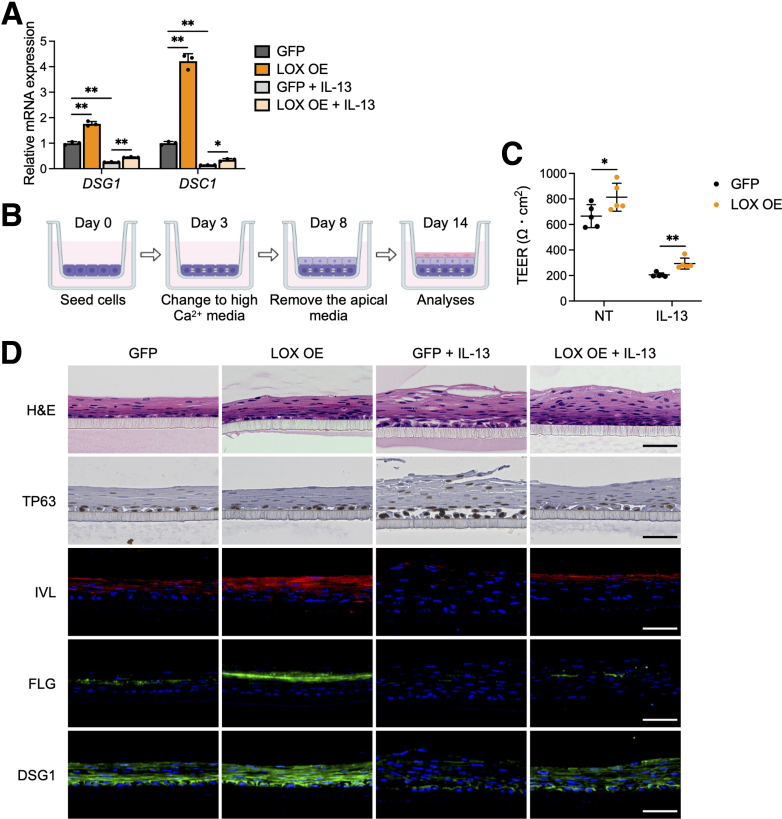
Disturbed squamous cell differentiation alters epithelial barrier integrity.24,25 This disruption is crucial in the pathogenesis of EoE.22, 23, 24 Thus, we sought to elucidate if LOX is implicated in barrier regulation. DSG1 and desmocollin-1 (DSC1), members of the desmosomal cadherin family, are required for adhesive intercellular junctions and maintenance of epithelial homeostasis.25 Interestingly, although IL13 treatment decreased transcript levels of DSG1 and DSC1 in organoids, LOX overexpression increased their expression at baseline and partially rescued the effect of IL13 (Figure 4A).
Figure 4.
LOX overexpression improves epithelial barrier integrity. (A) qRT-PCR for DSG1 and DSC1 in EPC2-hTERT organoids overexpressing GFP or LOX OE. GFP and LOX OE organoids were cultured with or without IL13 (10 μg/mL) from days 7 to 11 and then harvested on day 11 (n = 3). Data are representative of 3 independent experiments. (B) Schematic of ALI model. GFP and LOX OE EPC2-hTERT cells were cultured in low-calcium (0.09 mmol/L Ca2+) media for 3 days, followed by high-calcium media (1.8 mmol/L Ca2+) for 5 days, and then brought to ALI on day 8. ALI-cultured cells were stimulated with IL13 (10 μg/mL) from days 9 to 14. (C) TEER (Ω ∗ cm2) of the GFP and LOX OE EPC2-hTERT ALI cultures (n = 5). (D) Representative images of H&E staining, immunohistochemistry for TP63, and immunofluorescence staining for IVL (red), FLG (green), and DSG1 (green) of the GFP and LOX OE EPC2-hTERT ALI cultures. 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar: 50 μm. Data are representative of 2 independent experiments and expressed as means ± SDs. (A) One-way analysis of variance and (C) 2-tailed Student t test were performed for statistical analyses. ∗P < .05, ∗∗P < .01. NT, nontreated.
To directly assess the effect of LOX on barrier function, we used the ALI system, which mimics an in vivo epithelial environment, and measured transepithelial electrical resistance (TEER)26 (Figure 4B). IL13 treatment increased barrier permeability in the epithelium as measured by TEER. Conversely, LOX overexpression improved the IL13-induced barrier deficiency by 1.4-fold compared with GFP cells (Figure 4C), albeit not to the levels without IL13 stimulation. H&E staining of GFP cultures also showed impaired squamous stratification in response to IL13. However, LOX overexpression counteracted the disruption caused by IL13. We performed staining for TP63, IVL, FLG, and DSG1 in ALI cultures. As seen in organoids, although IL13 stimulation prevented differentiation and barrier-related protein expression in the epithelium, LOX overexpression enhanced IVL, FLG, and DSG1 in both untreated and IL13-stimulated cultures. Collectively, these data show that LOX promotes normal cell differentiation and supports epithelial integrity, including in the setting of IL13 stimulation.
Transcriptome Profiling Reveals Pathways Associated With LOX Expression in Esophageal Epithelium
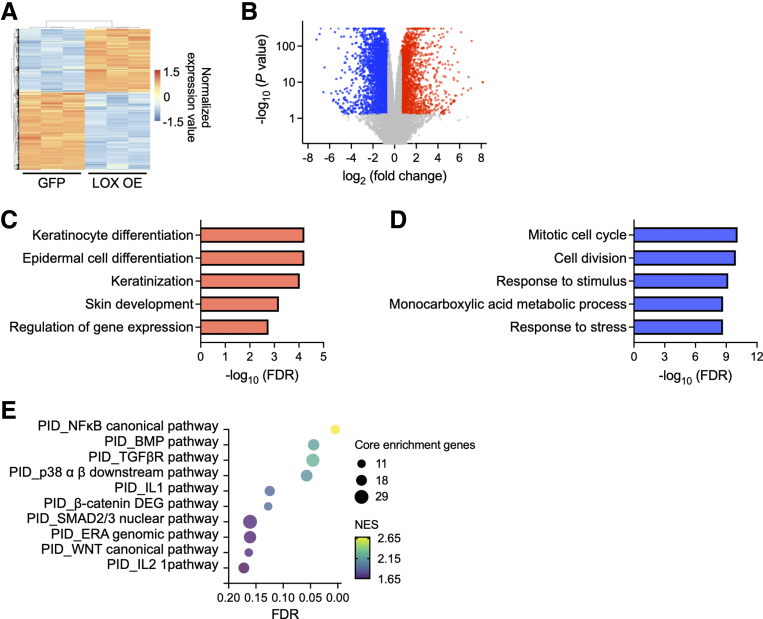
To understand the mechanism by which LOX regulates epithelial integrity, we performed RNA sequencing on LOX OE organoids. DEG analysis using DESeq227 identified 2446 up-regulated (P < .05 and log2(fold change) ≥ 0.585) and 2923 down-regulated (P < .05 and log2(fold change) ≤ -0.585) genes in LOX OE organoids compared with GFP organoids (Figure 5A and B). Gene Ontology analysis of DEGs revealed that cell differentiation and keratinization-related terms were enriched in LOX OE organoids (Figure 5C). By contrast, cell proliferation–related terms were decreased (Figure 5D). Furthermore, Gene Set Enrichment Analysis (GSEA) on the Pathway Interaction Database (PID)28 revealed that gene signatures associated with bone morphogenetic protein (BMP), transforming growth factor β (TGFβ), and WNT pathways were enriched significantly in LOX OE organoids (false discovery rate, <0.25) (Figure 5E). These results are consistent with our previous results showing a role for LOX in modulating esophageal epithelial differentiation.
Figure 5.
Transcriptome analysis identifies the functional roles of LOX in esophageal epithelium. (A) Heatmap and (B) volcano plot of DEGs based on RNA sequencing data from GFP or LOX OE EPC2-hTERT organoids. Up-regulated and down-regulated DEGs in LOX OE organoids are shown with red and blue, respectively. Top 5 (C) enriched and (D) depleted terms in LOX OE organoids based on Gene Ontology analysis. (E) Gene Set Enrichment Analysis based on the PID. The top 10 enriched pathways in LOX OE organoids are shown. Dot size and color represent the number of core enrichment genes and normalized enrichment score (NES) for the pathway, respectively. FDR, false discovery rate.
BMP Signaling Pathway Is Activated in LOX Overexpressing Organoids
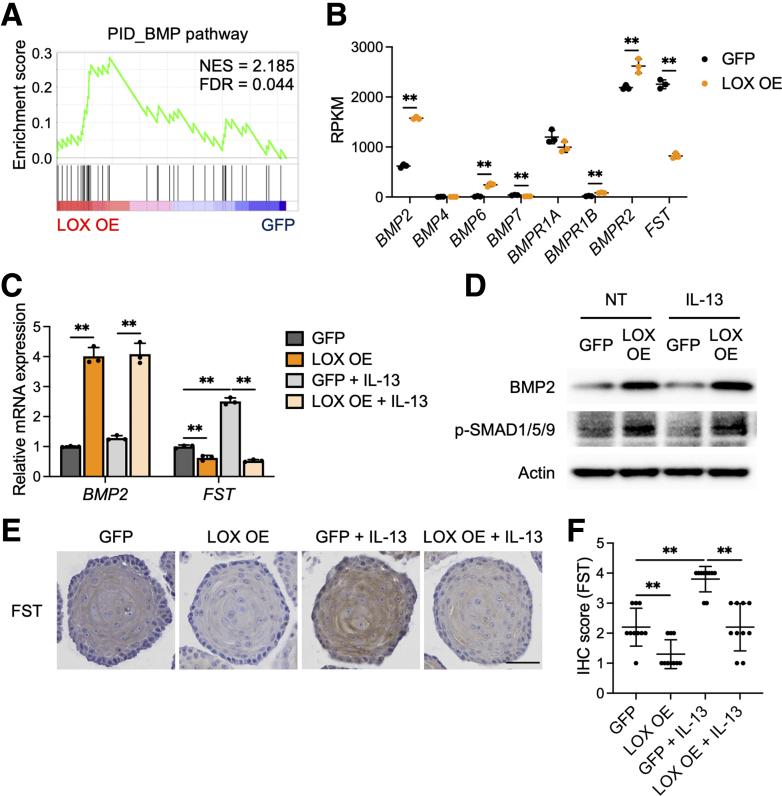
Recent studies have shown that the BMP pathway is essential for esophageal progenitor cell differentiation and disrupted BMP signaling results in basal cell hyperplasia in EoE.29,30 Therefore, we postulated that the BMP signaling pathway could be integral in LOX-supported differentiation (GSEA trace presented in Figure 6A). We investigated BMP ligands and BMP receptors from the PID gene set presented in Figures 5E and 6A. BMP2, BMP6, BMPR1B, and BMPR2 were increased significantly in LOX OE organoids compared with GFP organoids (by 2.5-fold, 18.3-fold, 4.8-fold, and 1.2-fold, respectively). Intriguingly, we found that the BMP antagonist follistatin (FST) was decreased significantly by 0.36-fold in LOX OE organoids (Figure 6B).
Figure 6.
BMP signaling pathway is activated in LOX overexpressing organoids. (A) Gene Set Enrichment Analysis for BMP pathway in the PID based on the differentially expressed genes in LOX OE organoids. (B) Normalized expression of genes relevant to the BMP pathway gene set, plotted as reads per kilobase per million (RPKM) (n = 3). (C–F) Validation of the BMP pathway activation in EPC2-hTERT organoids. GFP and LOX OE organoids were cultured with or without IL13 (10 μg/mL) from days 7 to 11 and then harvested on day 11. (C) qRT-PCR for BMP2 and FST in the GFP and LOX OE organoids (n = 3). (D) Representative immunoblot for BMP2 and phosphorylated SMAD1/5/9 (p-SMAD1/5/9) and (E) immunohistochemistry staining for FST in the GFP and LOX OE organoids. Scale bar: 50 μm. (F) FST protein levels in panel E, quantified (n = 10). Data are representative of 3 independent experiments and expressed as means ± SDs. (B) Two-tailed Student t test and (C and F) 1-way analysis of variance were performed for statistical analyses. ∗∗P < .01. FDR, false discovery rate; IHC, immunohistochemistry; NES, normalized enrichment score; NT, nontreated.
We validated these findings by qRT-PCR, Western blot, and immunohistochemistry in the setting of IL13 stimulation (Figure 6C–F). Transcript levels of BMP2 were increased by 4.0-fold and 3.2-fold in untreated and IL13-stimulated LOX OE organoids compared with GFP organoids, respectively (Figure 6C). IL13 stimulus led to increased expression of FST in GFP organoids, whereas LOX overexpression reduced this effect by 0.62-fold and 0.21-fold in untreated and IL13-stimulated LOX OE organoids, respectively (Figure 6C). Protein levels of BMP2 and phosphorylated forms of the downstream transcription factors Suppressor of Mothers against Decapentaplegic (SMAD) 1/5/9 also were increased in LOX OE organoids (Figure 6D). Follistatin, which was expressed robustly in IL13-stimulated organoids, was decreased in the setting of LOX overexpression (Figure 6E and F). These data support a model in which overexpression of LOX inhibits FST, leading to increased BMP signaling.
BMP2 Promotes Cell Differentiation in Esophageal Epithelium
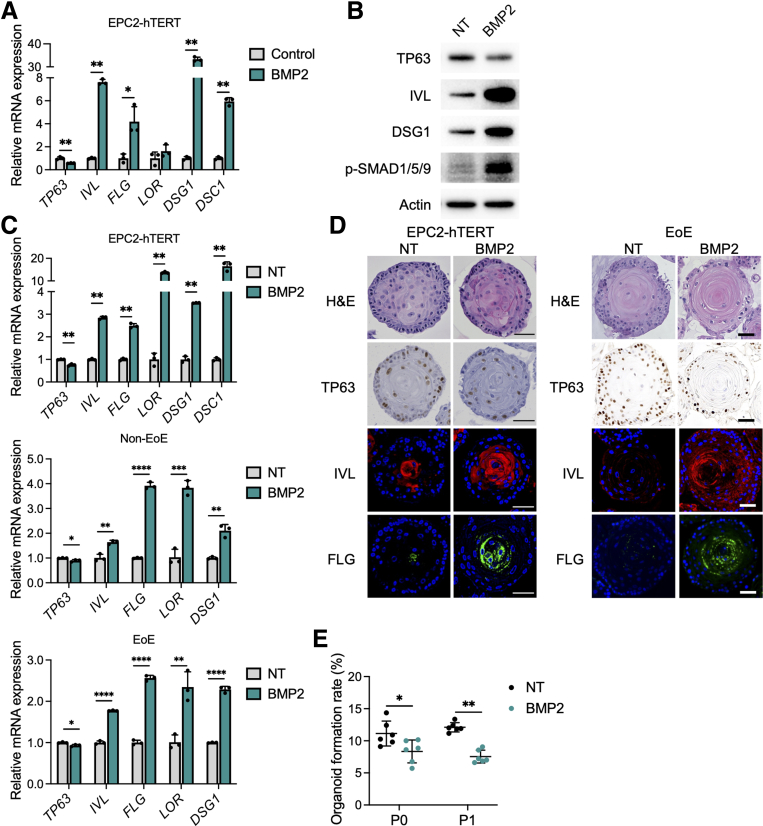
Finally, we investigated whether BMP2 is involved in esophageal epithelial differentiation and barrier integrity. Treatment with recombinant BMP2 led to decreased TP63 mRNA expression and increased IVL, FLG, DSG1, and DSC1 mRNA expression in monolayer-cultured EPC2-hTERT cells (Figure 7A). Immunoblotting showed similar results with increased phosphorylated SMAD1/5/9, IVL, and DSG1 in BMP2-treated cells (Figure 7B). Consistent with the results in monolayer culture, BMP2-treated organoids and PDOs from EoE or non-EoE control patients showed decreased expression of basal genes (SOX2 and TP63) and increased expression of differentiation and junctional protein genes (IVL, FLG, LOR, DSG1, and DSC1) (Figure 7C and D). BMP2 treatment also reduced organoid formation capacity, further supporting the notion that BMP2 enhances differentiation and reduces the basal population (Figure 7E). In summary, our findings suggest that LOX has a protective role in the esophageal epithelium in response to type 2 inflammation, in which it acts to restore homeostasis in EoE via activation of BMP signaling.
Figure 7.
BMP2 treatment induces cell differentiation in esophageal epithelium. (A and B) BMP2 treatment in monolayer culture of EPC2-hTERT cells. EPC2-hTERT cells were treated with recombinant BMP2 protein (10 μg/mL) for 72 hours in high-calcium (1.8 mmol/L Ca2+) media. (A) qRT-PCR for TP63, IVL, FLG, LOR, DSG1, and DSC1 in the EPC2-hTERT cells (n = 3). (B) Representative immunoblot for TP63, IVL, DSG1, and phosphorylated SMAD1/5/9 (p-SMAD1/5/9) in the EPC2-hTERT cells. (C–E) EPC2-hTERT organoids were treated with recombinant BMP2 protein (10 μg/mL) from days 7 to 11 and then harvested on day 11. (C) qRT-PCR for SOX2, KRT14, TP63, IVL, FLG, LOR, DSG1, and DSC1 in the EPC2-hTERT organoids or PDOs (representative from 5 PDOs: 2 EoE and 3 non-EoE cultures). (D) Representative images of H&E staining, immunohistochemistry for TP63, and immunofluorescence staining for IVL (red), FLG (green), and 4′,6-diamidino-2-phenylindole (blue), in the EPC2-hTERT organoids or PDOs. Scale bar: 50 μm. (E) OFR was assessed on day 11 (P0), followed by passage. OFR was assessed again on day 11 (P1). OFR was calculated as the number of organoids (≥50 μm) divided by the number of total seeded cells (n = 6). Data are representative of 3 independent experiments and expressed as means ± SDs. (A, C, and E) Two-tailed Student t test was performed for statistical analyses. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001. NT, nontreated.
Discussion
LOX is an extracellular matrix remodeling enzyme that acts to cross-link collagen, thereby enhancing tissue stiffness. LOX is expressed in the epithelium of the prostate, retina, and skin, as well as other locations,15,31 however, its role in the esophageal epithelium in homeostasis and disease is unknown. Herein, we describe a noncanonical role for LOX in the esophageal epithelium and a potential protective role in epithelial differentiation and barrier integrity. Building on our previous work showing that LOX was up-regulated in the EoE epithelium, we now show that LOX is expressed in the differentiated esophageal epithelium where it has noncollagen cross-linking roles. LOX overexpression models show enhanced differentiation and barrier integrity, even in the setting of IL13 stimulation in the esophageal epithelium. Using unbiased transcriptomic approaches, we found that the BMP pathway was enriched in LOX OE cells and that LOX OE cells have increased expression of BMP2, whereas the BMP antagonist follistatin is decreased. This effect is maintained despite IL13 stimulation. These results help to elucidate the contribution of LOX-supported differentiation and barrier integrity both in homeostasis and in the context of allergic insult.
Epithelial changes in EoE disrupt the mucosal barrier, which normally provides protection from acid and food particles during normal swallows. Current treatment strategies are aimed at decreasing the invasion of inflammatory cells into the esophagus. However, we have found that epithelial changes persist even in patients in remission with low eosinophil counts.2 Thus, determining mechanisms to restore homeostasis to the esophageal epithelium represents an unexplored avenue of research with therapeutic potential. We now highlight a novel role for LOX in restoration of epithelial homeostasis. LOX silencing has been shown to impair keratinocyte differentiation of the skin.15 In fact, in vitro skin models have shown that LOX expression is increased in early differentiation and knockdown inhibits terminal differentiation. Taken together, these results suggest that increased LOX in the setting of inflammation may serve to re-establish differentiation and barrier integrity during injury in the squamous mucosa. However, it is important to note that although our in vitro findings suggest that LOX overexpression acts to enhance differentiation, its up-regulation in the setting of EoE disease is not enough to heal the EoE epithelium and there is an ongoing differentiation defect despite increased LOX. This may be because there is a complex inflammatory infiltrate at play in EoE with multiple cell types secreting multiple cytokines, whereas our in vitro model focuses on IL13 alone.
Fibrosis is a major complication in EoE, and because LOX is a collagen cross-linker, it would be tempting to use LOX inhibitors to prevent fibrosis. However, our data showing the protective roles of LOX in the esophageal epithelium in the context of T-helper type 2 (Th2) inflammation would suggest that global LOX inhibition would have detrimental effects on the epithelial barrier. It therefore would be advantageous to target the cross-linker activity of LOX without affecting its non–cross-linking functions or vice versa. In that regard, our identification of the beneficial role of the LOX/BMP axis in the esophageal epithelium may represent a new approach to mitigating EoE, which could be used via a topical approach. Topical steroid preparations often are used in EoE to spare the negative consequences of systemic treatment with steroids.32,33 Furthermore, recombinant human BMP2 currently is approved by the Food and Drug Administration to promote bone healing in orthopedics.34, 35, 36 Building off of this and our in vitro findings of the protective role of BMP2 in the setting of IL13 stimulation (Figure 7), it may be possible to use preparations of BMP2 agonists to enhance esophageal epithelial barrier integrity.
BMP2, a member of the TGFβ superfamily, is essential for embryogenesis and development of the gut, and the BMP pathway affects morphogenesis of esophageal epithelial progenitor cells.30,37, 38, 39 We revealed that overexpression of LOX results in increased expression of BMP2 and increased squamous differentiation in esophageal organoids. Interestingly, down-regulated BMP2 expression by fibroblast growth factor 9, which is up-regulated in patients with EoE, has been proposed as the mechanism of hyperplasia in EoE.40 Furthermore, basal cell hyperplasia in the EoE epithelium, specifically in the setting of IL13 stimulation, is associated with inhibition of the BMP pathway in human disease and murine models.34 Herein, we demonstrate the role of LOX specifically in differentiated cells to maintain barrier integrity and restore homeostasis. Future work will interrogate the mechanism by which LOX activates the BMP pathway to exert these protective roles. One possibility is down-regulation of the BMP antagonist follistatin41 by LOX. We showed decreased follistatin levels in LOX OE organoids, even in the setting of IL13 stimulation (Figure 6). Interestingly, the IL13/signal transducer and activator of transcription (STAT)6 pathway directly up-regulates follistatin, and its increased levels have been reported in EoE patients and murine models.29 Correspondingly, knockdown of follistatin accelerates epithelial differentiation in esophageal cells. To date, although reactive oxidative stress has been shown to mediate differentiation by the activated BMP in EoE,29 the specific mechanism remains to be elucidated.
One weakness of this study is that we rely on cell culture techniques because there is no in vivo model of LOX overexpression. LOX knockout mouse models would complement this work, however, these models are embryonic lethal. To contend with these limitations, we use 3D cultures (organoid and ALI culture) to replicate the proliferation and renewal patterns in vitro. Another potential weakness is that although LOX is up-regulated in EoE, our overexpression model induces higher levels of LOX than observed in vivo or with IL13 stimulation. It may be that such high levels of LOX are necessary to demonstrate the protective effect we observe in vitro and that the level of increase observed in the EoE patients is not enough to heal the epithelium. However, the benefit of this model is that it allows for evaluation of the mechanistic effects of LOX without the secondary effects of inflammatory cytokines.
We previously showed that TGFβ and tumor necrosis factor α work synergistically to increase LOX production in the esophageal epithelium. Herein, we begin to examine our previous LOX observation that IL13 also up-regulates LOX production.4 How IL13 stimulates LOX has yet to be determined, however, other investigators have confirmed STAT3 binding in the LOX promoter regulates its expression in both kidney42 and lung.43 Furthermore, murine models of esophageal IL13 overexpression have shown STAT3 activation,44 suggesting the potential for IL13 up-regulation of LOX via STAT3. Future studies should work to evaluate the specific role of Th2 cytokines (IL13 and IL4) in the development and propagation of LOX-mediated fibrosis.
The esophageal epithelial barrier is maintained by an exquisitely regulated proliferation and differentiation gradient. The perturbations lead to symptoms as well as inflammation. Herein, we describe a novel role for LOX involving maintenance of differentiation and barrier integrity independent of its effects in subepithelial matrix remodeling. Epithelial LOX may serve to re-establish homeostasis in the setting of inflammation via BMP activation, underscoring the diverse functions of LOX in the esophagus. Investigation of the LOX-BMP pathway may provide novel insights into the pathogenesis and be a promising therapeutic approach for EoE. Furthermore, elucidating these mechanisms may have implications for epithelial disruption in disorders beyond EoE, including caustic ingestions and gastroesophageal reflux disease.
Methods
Cell Line and Monolayer Culture
EPC2-hTERT or primary patient-derived cells were cultured in keratinocyte–serum-free medium with 0.09 mmol/L Ca2+ (KSFM; Thermo Fisher Scientific, Inc, Waltham, MA) supplemented with bovine pituitary extract (50 μg/mL), human recombinant epidermal growth factor (1 ng/mL), and 1% penicillin-streptomycin. The cells were incubated at 37°C in a 5% humidified CO2 atmosphere.45,46
3D Esophageal Organoid Culture
EPC2-hTERT or PDOs (Table 1) were cultured as described previously.17,18,20 Briefly, EPC2-hTERT cells were dissociated into a single-cell suspension and placed into Matrigel basement membrane matrix (Corning, Inc, Corning, NY) under KSFM modified with 0.6 mmol/L Ca2+. Organoids were grown for 7 days, followed by treatment with 10 ng/mL IL13, 10 ng/mL recombinant BMP2 protein (R&D Systems, Inc, Minneapolis, MN), or vehicle (phosphate-buffered saline for IL13 and hydrochloride for BMP2) for 4 days. We then used the day 11 organoids for further analyses. OFR was defined as the number of organoids ≥50 μm divided by the total seeded cells in each well.
ALI Culture
EPC2-hTERT cells were seeded on Transwell permeable supports with a 0.4-μm pore (3470; Corning) and grown with KSFM (0.09 mmol/L Ca2+) for the initial 3 days to confluency. Cultures then were switched to high-calcium KSFM (1.8 mmol/L Ca2+) for 5 days. The media was removed from the apical compartment on day 8 to induce epithelial differentiation and stratification. A total of 10 μg/mL IL13 (or vehicle) was applied in the basolateral compartment from days 9 to 14. To assess epithelial barrier integrity, TEER was measured with an Epithelial Volt/Ohm Meter (World Precision Instruments, Sarasota, FL). The day 14 ALI-cultured epithelium was used for TEER measurement and histology.
Bioinformatic Analysis of scRNA-seq Data
PDOs were stimulated with vehicle or IL13 (10 μg/mL) from days 7 to 11, and the day 11 PDOs then were dissociated into a single-cell suspension for scRNA-seq.20 The Dead Cell Removal Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) was used to guarantee cell viability. The raw count matrix with barcode and feature information for each sample were imported and transformed to Seurat (Seurat Technologies version 4.2.0) objects for further processing. Genes expressed in 3 or fewer cells were excluded from analysis. To eliminate dead cells or doublets, cells with an expression of less than 700 or more than 6000 genes, respectively, were excluded. In addition, cells with more than 10% of their transcripts consisting of mitochondrial genes were excluded. Seurat integration workflow was used to integrate the top 3000 variable genes, as anchors, across cells for control samples.47 After integration, dimensionality reduction used the genes and values that were preprocessed using the integration workflow. Principal component analysis was used for initial dimensionality reduction and later for clustering, resulting in 20 principal components. The components then were used as input to the UMAP dimensionality reduction procedure using 20 neighbors for local neighborhood approximation and embedding into 2 components for visualization. Clustering was initialized with a Shared Nearest Neighbor graph by first determining 20 nearest neighbors for each cell, and then determined by a modularity optimization algorithm by Waltman and Van Eck.48 Cell type annotations for each cluster are based on the expression of marker genes and DEGs. To better compare the IL13-treated samples with the control data, IL13 data were projected to control data after filtering and integration using Seurat projection/query workflow. For DEG analysis on the IL13-treated PDOs, cells with a LOX expression greater than 0 were defined as LOX+ cells and cells with a LOX expression of 0 were defined as LOX- cells.
Trajectory Analysis
Monocle 3 (version 1.0.0) was used to infer the trajectory analysis based on the scRNA-seq data. Seurat objects from upstream analysis were converted to Monocle objects, and then reversed graph embedding was applied to yield a principal graph that is allowed to branch from the reduced dimension space.49 Pseudotime trajectory are defined and derived by selecting the specific cells as roots based on the prior knowledge of the cell type.
Bulk mRNA Sequencing and Gene Expression Analysis
GFP and LOX OE organoids were grown for 11 days and then harvested for RNA sequencing. Sequencing libraries were constructed from total RNA (1 μg) using a TruSeq Stranded mRNA Library Prep (Illumina, Inc, San Diego, CA). RNA sequencing was performed on an Illumina HiSeq2000 platform. We used kallisto50 and human reference genome hg38 for alignment. Mapped reads were analyzed with DESeq2.28
DEGs were determined as P < .05 and log2(fold change) ≥ 0.585 or ≤ -0.585. Using DEGs, Gene Ontology enrichment analysis was provided by the topGO package.51 The top 5 enriched or depleted terms with the lowest false discovery rate values in LOX OE organoids were selected. The stat output field from DESeq2 then was used as input for GSEA52,53 preranked analysis to identify enriched pathways from the PID.
Lentivirus-Mediated Gene Transfer
Lentiviral vectors pLX304-eGFP and pLX304-LOX were constructed by Gateway LR reaction of entry clones pENTR223-LOX (HsCD00378945, (Plasmid Information Database (PlasmID) Harvard Medical School) and pDONR221-eGFP (Addgene vector 25899) with destination vector pLX304 (Addgene vector 25890). Vesicular stomatitis virus g (VSV-g)–coated lentiviral particles were prepared by transfecting 293T cells with 9 μg psPAX, 0.9 μg pMD2.G, and 9 μg overexpression vector using Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific, Inc). EPC2-hTERT cells were transduced by spinfection and selected with 10 μg/mL blasticidin.
qRT-PCR
RNA extraction and reverse transcription were performed as described.21,51 Real-time qRT-PCR was performed with TaqMan Gene Expression Assays (Thermo Fisher Scientific, Inc) for LOX (Hs00942480_m1), SOX2 (Hs01053049_s1), KRT14 (Hs03044364_m1), TP63 (Hs00978340_m1), IVL (Hs00846307_s1), FLG (Hs00856927_g1), LOR (Hs01894962_s1), BMP2 (Hs00154192_m1), FST (Hs01121165_g1), and GAPDH (Hs02786624_g1) using the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific, Inc). Relative mRNA levels of each gene were normalized to GAPDH levels as a housekeeping control.
Western Blot
Whole-cell lysates from cells in monolayer culture and 3D organoids were prepared as described previously.17 Equivalent amounts (20–40 μg) of protein were loaded into a NuPAGE 4% to 12% Bis-Tris gel. After electrophoresis, transfer to a polyvinylidene difluoride membrane, and blocking with 5% bovine serum albumin or nonfat milk, membranes were incubated with primary antibodies at 4°C overnight. The primary antibodies used were as follows: anti-LOX (1:500, NB100-2527; Novus Biologicals, Centennial, CO), anti-TP63 (1:1000, ab124762; Abcam, Cambridge, UK), anti-IVL (1:1000, I9018; Millipore Sigma, Burlington, MA), anti-DSG1 (1:1000, sc-137164; Santa Cruz Biotechnology, Dallas, TX), anti-BMP2 (1:1000, ab214821; Abcam), anti–phosphorylated-SMAD1/5/9 (1:1000, 13820S; Cell Signaling Technology, Danvers, MA), and anti–β-actin (1:5000, A5316; Millipore Sigma). Immunoblots were detected with an appropriate horseradish peroxidase–conjugated secondary antibody (1:2000, NA934 or NA 931; Amersham BioSciences, Buckinghamshire, UK) by ECL detection (Bio-Rad Laboratories, Hercules, CA). β-actin served as a loading control.
Immunohistochemistry and Immunofluorescence
Organoids were fixed and embedded as described previously,4,20 and subjected to H&E staining, immunohistochemistry, and immunofluorescence.
For immunohistochemistry, after deparaffinization and rehydration, antigens were retrieved by high-pressure cooking. After peroxidase quenching and blocking with an appropriate serum (Jackson ImmunoResearch, West Grove, PA), sections were incubated with primary anti-TP63 monoclonal antibody (1:1000, ab124762; Abcam) and antifollistatin monoclonal antibody (1:50, MAB669; R&D Systems, Inc) at 4°C overnight and then with an appropriate biotinylated secondary antibody (1:200; Vector Laboratories, Burlingame, CA). The signal was detected with the VECTASTAIN Elite ABC–Horseradish Peroxidase Kit (PK-6100; Vector Laboratories). The (3,3' Diaminobenzidine) DAB Substrate Kit (SK-4100; Vector Laboratories) was used for color reaction. For follistatin staining, slides were quantified with the following scored: 1, negative to weak staining; 2, weak to moderate staining; 3, moderate to strong staining; and 4, strong staining. Ten organoids per condition were used for the evaluation.
For immunofluorescence, sections were stained with primary anti-IVL monoclonal antibody (1:100, I9018; Millipore Sigma), anti-FLG monoclonal antibody (1:100, MA5-13440; Thermo Fisher Scientific, Inc), and anti-DSG1 monoclonal antibody (1:100, sc-137164; Santa Cruz Biotechnology) at 4°C overnight and then with Cy2 or Cy5-AffiniPure Donkey Anti-Mouse IgG (H+L) secondary antibody (1:500, 715-225-150 or 715-175-150; Jackson ImmunoResearch) at room temperature for 1 hour. Nuclei were stained with 4′,6-diamidino-2-phenylindole (17985-50; Electron Microscopy Sciences, Hatfield, PA). Images were taken with an All-in-One Fluorescence Microscope BZ-X710 (KEYENCE Corp, Osaka, Japan). The images were evaluated in 5 different locations in a high-power field per condition, and representatives are shown.
Statistical Analysis
Data are presented as means ± SDs. Continuous variables were analyzed by a 2-tailed Student t test for 2 independent groups or analysis of variance for multiple groups. All statistical analyses were conducted with GraphPad Prism (GraphPad Software, San Diego, CA). A P value less than .05 was considered statistically significant.
All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
We thank to the Molecular Pathology and Imaging Core (Kate Bennett, Rebecca Ly, and Jonathan P. Katz) for technical support. Schematics were created with BioRender.com.
CRediT Authorship Contributions
Amanda Muir, MD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Lead; Supervision: Lead)
Masaru Sasaki (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Methodology: Lead)
Takeo Hara (Conceptualization: Supporting; Data curation: Supporting)
Joshua X. Wang (Data curation: Equal; Supervision: Equal)
Yusen Zhou (Formal analysis: Lead)
Kanak V. Kennedy (Data curation: Equal; Formal analysis: Supporting)
Nicole N. Umeweni (Investigation: Supporting; Methodology: Supporting)
Maiya A. Alston (Investigation: Equal; Project administration: Equal)
Zachary C. Spergel (Data curation: Supporting)
Ritsu Nakagawa (Data curation: Supporting)
Emily A. Mcmillan (Data curation: Supporting; Formal analysis: Supporting)
Kelly A. Whelan (Conceptualization: Supporting; Methodology: Supporting)
Tatiana A. Karakasheva (Conceptualization: Equal; Data curation: Supporting; Formal analysis: Supporting)
Kathryn E. Hamilton (Data curation: Supporting; Investigation: Supporting)
Melanie A. Ruffner (Conceptualization: Supporting; Investigation: Supporting)
Satoshi Ishikawa (Data curation: Supporting; Formal analysis: Supporting)
Ryugo Teranishi (Data curation: Supporting)
Footnotes
Conflicts of interest This author discloses the following: Amanda B. Muir has served on the medical advisory boards for Nexstone Immunology, Bristol Meyers Squib, and Regeneron. The remaining authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grants R01DK121159 (K.A.W.), K08AI148456 (M.A.R.), and R03DK118310 and R01DK124266-01 (A.B.M.); University of Pennsylvania Transdisciplinary Awards Program in Translational Medicine and Therapeutics (A.B.M.); and the Children’s Hospital of Philadelphia Gastrointestinal Epithelial Modeling Program (T.A.K. and A.B.M.). This work also was supported by P30DK050306: Center for Molecular Studies in Digestive and Liver Diseases.
Data Availability The data and materials described in this study will be made available upon request.
References
- 1.Muir A., Falk G.W. Eosinophilic esophagitis: a review. JAMA. 2021;326:1310–1318. doi: 10.1001/jama.2021.14920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whelan K.A., Godwin B.C., Wilkins B., et al. Persistent basal cell hyperplasia is associated with clinical and endoscopic findings in patients with histologically inactive eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2020;18:1475–1482.e1. doi: 10.1016/j.cgh.2019.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rochman M., Wen T., Kotliar M., et al. Single-cell RNA-seq of human esophageal epithelium in homeostasis and allergic inflammation. JCI Insight. 2022;7 doi: 10.1172/jci.insight.159093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasagi Y., Dods K., Wang J.X., et al. Fibrostenotic eosinophilic esophagitis might reflect epithelial lysyl oxidase induction by fibroblast-derived TNF-α. J Allergy Clin Immunol. 2019;144:171–182. doi: 10.1016/j.jaci.2018.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W., Yang A., Jia J., et al. Lysyl oxidase (LOX) family members: rationale and their potential as therapeutic targets for liver fibrosis. Nat Rev Gastroenterol Hepatol. 2020;72:729–741. doi: 10.1002/hep.31236. [DOI] [PubMed] [Google Scholar]
- 6.Mäki J.M., Räsänen J., Tikkanen H., et al. Inactivation of the lysyl oxidase gene Lox leads to aortic aneurysms, cardiovascular dysfunction, and perinatal death in mice. Circulation. 2002;106:2503–2509. doi: 10.1161/01.cir.0000038109.84500.1e. [DOI] [PubMed] [Google Scholar]
- 7.Maki J., Sormunem R., Lippo S., et al. Lysyl oxidase is essential for normal development and function of the respiratory system and for the integrity of elastic and collagen fibers in various tissues. Am J Pathol. 2005;167:927–936. doi: 10.1016/S0002-9440(10)61183-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maller O., Drain A.P., Barrett A.S., et al. Tumour-associated macrophages drive stromal cell-dependent collagen crosslinking and stiffening to promote breast cancer aggression. Nat Mater. 2021;20:548–559. doi: 10.1038/s41563-020-00849-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Kleeck R., Roberts E., Castagnino P., et al. Arterial stiffness and cardiac dysfunction in Hutchinson-Gilford Progeria syndrome corrected by inhibition of lysyl oxidase. Life Sci Alliance. 2021;4 doi: 10.26508/lsa.202000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perepelyuk M., Terajima M., Wang A.Y., et al. Hepatic stellate cells and portal fibroblasts are the major cellular sources of collagens and lysyl oxidases in normal liver and early after injury. Am J Physiol Gastrointest Liver Physiol. 2013;304:605–614. doi: 10.1152/ajpgi.00222.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trackman P.C. Enzymatic and non-enzymatic functions of the lysyl oxidase family in bone. Matrix Biol. 2016;52–54:7–18. doi: 10.1016/j.matbio.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucero H.A., Ravid K., Grimsby J.L., et al. Lysyl oxidase oxidizes cell membrane proteins and enhances the chemotactic response of vascular smooth muscle cells. J Biol Chem. 2008;283:24103–24117. doi: 10.1074/jbc.M709897200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laczko R., Csiszar K. Lysyl oxidase (Lox): functional contributions to signaling pathways. Biomolecules. 2020;10:1093. doi: 10.3390/biom10081093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouez C., Reynaud C., Noblesse E., et al. The lysyl oxidase LOX is absent in basal and squamous cell carcinomas and its knockdown induces an invading phenotype in a skin equivalent model. Clin Cancer Res. 2006;12:1463–1469. doi: 10.1158/1078-0432.CCR-05-1456. [DOI] [PubMed] [Google Scholar]
- 15.Provost G.S.L., Debret R., Cenizo V., et al. Lysyl oxidase silencing impairs keratinocyte differentiation in a reconstructed-epidermis model. Exp Dermatol. 2010;19:1080–1087. doi: 10.1111/j.1600-0625.2010.01135.x. [DOI] [PubMed] [Google Scholar]
- 16.Noblesse E., Cenizo Ã.V.R., Bouez C., et al. Lysyl oxidase-like and lysyl oxidase are present in the dermis and epidermis of a skin equivalent and in human skin and are associated to elastic fibers. J Invest Dermatol. 2004;122:621–630. doi: 10.1111/j.0022-202X.2004.22330.x. [DOI] [PubMed] [Google Scholar]
- 17.Nakagawa H., Kasagi Y., Karakasheva T.A., et al. Modeling epithelial homeostasis and reactive epithelial changes in human and murine three-dimensional esophageal organoids. Curr Protoc Stem Cell Biol. 2020;52:e106. doi: 10.1002/cpsc.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasagi Y., Chandramouleeswaran P.M., Whelan K.A., et al. The esophageal organoid system reveals functional interplay between notch and cytokines in reactive epithelial changes. Cell Mol Gastroenterol Hepatol. 2018;5:333–352. doi: 10.1016/j.jcmgh.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whelan K.A., Muir A.B., Nakagawa H. Esophageal 3D culture systems as modeling tools in esophageal epithelial pathobiology and personalized medicine. Cell Mol Gastroenterol Hepatol. 2018;5:461–478. doi: 10.1016/j.jcmgh.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hara T., Kasagi Y., Wang J., et al. CD73 + epithelial progenitor cells which contribute to homeostasis and renewal are depleted in eosinophilic esophagitis. Cell Mol Gastroenterol Hepatol. 2022;13:1449–1467. doi: 10.1016/j.jcmgh.2022.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muir A.B., Lim D.M., Benitez A.J., et al. Esophageal epithelial and mesenchymal cross-talk leads to features of epithelial to mesenchymal transition in vitro. Exp Cell Res. 2013;319:850–859. doi: 10.1016/j.yexcr.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherrill J.D., Kc K., Wu D., et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 2014;7:718–729. doi: 10.1038/mi.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katzka D.A., Tadi R., Smyrk T.C., et al. Effects of topical steroids on tight junction proteins and spongiosis in esophageal epithelia of patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2014;12:1824–1829.e1. doi: 10.1016/j.cgh.2014.02.039. [DOI] [PubMed] [Google Scholar]
- 24.Katzka D.A., Ravi K., Geno D.M., et al. Endoscopic mucosal impedance measurements correlate with eosinophilia and dilation of intercellular spaces in patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2015;13:1242–1248.e1. doi: 10.1016/j.cgh.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 25.Simpson C.L., Patel D.M., Green K.J. Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nat Rev Mol Cell Biol. 2011;12:565–580. doi: 10.1038/nrm3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruffner M.A., Song L., Maurer K., et al. Toll-like receptor 2 stimulation augments esophageal barrier integrity. Allergy. 2019;74:2449–2460. doi: 10.1111/all.13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaefer C.F., Anthony K., Krupa S., et al. PID: the pathway interaction database. Nucleic Acids Res. 2009;37(Database issue):D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang M., Ku W.Y., Zhou Z., et al. BMP-driven NRF2 activation in esophageal basal cell differentiation and eosinophilic esophagitis. J Clin Invest. 2015;125:1557–1568. doi: 10.1172/JCI78850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Yang Y., Jiang M., et al. 3D modeling of esophageal development using human PSC-derived basal progenitors reveals a critical role for Notch signaling. Cell Stem Cell. 2018;23:516–529.e5. doi: 10.1016/j.stem.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Omori K., Fujiseki Y., Omori K., et al. Regulation of the expression of lysyl oxidase mRNA in cultured rabbit retinal pigment epithelium cells. Matrix Biol. 2002;21:337–348. doi: 10.1016/s0945-053x(02)00013-6. [DOI] [PubMed] [Google Scholar]
- 32.Gupta S.K., Vitanza J.M., Collins M.H. Efficacy and safety of oral budesonide suspension in pediatric patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2015;13:66–76.e3. doi: 10.1016/j.cgh.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 33.Straumann A., Conus S., Degen L., et al. Long-term budesonide maintenance treatment is partially effective for patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2011;9:400–409.e1. doi: 10.1016/j.cgh.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 34.Wang R.N., Green J., Wang Z., et al. Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis. 2014;1:87–105. doi: 10.1016/j.gendis.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali I.H.A., Brazil D.P. Bone morphogenetic proteins and their antagonists: current and emerging clinical uses. Br J Pharmacol. 2014;171:3620–3632. doi: 10.1111/bph.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salazar V.S., Gamer L.W., Rosen V. BMP signalling in skeletal development, disease and repair. Nat Rev Endocrinol. 2016;12:203–221. doi: 10.1038/nrendo.2016.12. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez P., Silva S.D., Oxburgh L., et al. BMP signaling in the development of the mouse esophagus and forestomach. Development. 2010;137:4171–4176. doi: 10.1242/dev.056077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardwick J.C.H., Van Den Brink G.R., Bleuming S.A., et al. Bone morphogenetic protein 2 is expressed by, and acts upon, mature epithelial cells in the colon. Gastroenterology. 2004;126:111–121. doi: 10.1053/j.gastro.2003.10.067. [DOI] [PubMed] [Google Scholar]
- 39.Torihashi S., Hattori T., Hasegawa H., et al. The expression and crucial roles of BMP signaling in development of smooth muscle progenitor cells in the mouse embryonic gut. Differentiation. 2009;77:277–289. doi: 10.1016/j.diff.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Mulder D.J., Pacheco I., Hurlbut D.J., et al. FGF9-induced proliferative response to eosinophilic inflammation in oesophagitis. Gut. 2009;58:166–173. doi: 10.1136/gut.2008.157628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iemura S.I., Yamamoto T.S., Takagi C., et al. Direct binding of follistatin to a complex of bone-morphogenetic protein and its receptor inhibits ventral and epidermal cell fates in early Xenopus embryo. Proc Natl Acad Sci U S A. 1998;95:9337. doi: 10.1073/pnas.95.16.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X., Zhou W., Niu Y., et al. Lysyl oxidase promotes renal fibrosis via accelerating collagen cross-link driving by β-arrestin/ERK/STAT3 pathway. FASEB J. 2022;36 doi: 10.1096/fj.202200573R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ha A.W., Bai T., Ebenezer D.L., et al. Sphingosine kinase 1 regulates lysyl oxidase through STAT3 in hyperoxia-mediated neonatal lung injury. Thorax. 2022;77:47–57. doi: 10.1136/thoraxjnl-2020-216469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marella S., Sharma A., Ganesan V., et al. IL-13–induced STAT3-dependent signaling networks regulate esophageal epithelial proliferation in eosinophilic esophagitis. J Allergy Clin Immunol. 2023;152:1550–1568. doi: 10.1016/j.jaci.2023.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandramouleeswaran P.M., Shen D., Lee A.J., et al. Preferential secretion of thymic stromal lymphopoietin (TSLP) by terminally differentiated esophageal epithelial cells: relevance to eosinophilic esophagitis (EoE) PLoS One. 2016;11 doi: 10.1371/journal.pone.0150968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muir A.B., Dods K., Henry S.J., et al. Eosinophilic esophagitis-associated chemical and mechanical microenvironment shapes esophageal fibroblast behavior. J Pediatr Gastroenterol Nutr. 2016;63:200–209. doi: 10.1097/MPG.0000000000001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stuart T., Butler A., Hoffman P., et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waltman L., Van Eck N.J. A smart local moving algorithm for large-scale modularity-based community detection. Eur Phys J B. 2013;86:478. [Google Scholar]
- 49.Cao J., Spielmann M., Qiu X., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 51.Alexa A., Rahnenführer J., Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 52.Mootha V.K., Lindgren C.M., Eriksson K.-F., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 53.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]