Abstract
Redox signaling, a mode of signal transduction that involves the transfer of electrons from a nucleophilic to electrophilic molecule, has emerged as an essential regulator of inflammatory macrophages. Redox reactions are driven by reactive oxygen/nitrogen species (ROS and RNS) and redox-sensitive metabolites such as fumarate and itaconate, which can post-translationally modify specific cysteine residues in target proteins. In the past decade our understanding of how ROS, RNS, and redox-sensitive metabolites control macrophage function has expanded dramatically. In this review, we discuss the latest evidence of how ROS, RNS, and metabolites regulate macrophage function and how this is dysregulated with disease. We highlight the key tools to assess redox signaling and important questions that remain.
1. Introduction
Redox (reduction-oxidation) signaling is a mode of signal transduction that involves the transfer of electrons from a nucleophilic to electrophilic molecule [1] and is an essential regulator of many biological processes across all cell types. These reactions are driven by reactive oxygen/nitrogen species (ROS and RNS) and redox-sensitive metabolites. Specific modifications of cysteine residues by these species is a core element of redox signaling [1].
ROS are reactive molecules, or oxidants, that originate from molecular oxygen. They play a pivotal role in a multitude of physiological and pathological processes. The most prominent ROS are superoxide (O2.-) and hydrogen peroxide (H2O2). Superoxide is the precursor of most ROS and H2O2, formed by the dismutation of O2.-, is the most abundant ROS [2]. A primary mode of ROS signaling is oxidation of redox-sensitive cysteine residues. Oxidation of cysteine residues can be reversed by the antioxidant defense system, and serves as a crucial signal for regulating cell function [[3], [4], [5], [6], [7]] and a myriad of biological processes [[8], [9], [10], [11], [12]]. However, excessive ROS production can lead to irreversible cysteine oxidation, resulting in protein dysfunction and disease [[13], [14], [15], [16]].
Macrophages are innate immune cells that serve as the first line of defense against invading pathogens. Macrophages have diverse and essential functions including phagocytosis of invading pathogens, inflammatory cytokine production, chemokine release to recruit other immune cells, antigen presentation, and tissue repair [17]. In addition to their critical roles in immunity, tissue resident macrophages (TRMs) have essential, highly specialized, and diverse homeostatic functions across tissues. Such functions are critical to our physiology. These include control of insulin sensitivity (adipose) [18], neuronal homeostasis (brain) [19], microbial clearance (liver, and other) [20], immune surveillance (lung) [21], and iron metabolism (spleen) [22]. However, macrophages also contribute to the pathology of a range of diseases, particularly those characterized by metabolic dysfunction and chronic inflammation. These include obesity and type 2 diabetes (adipose) [23], neurodegeneration (brain) [24], non-alcoholic steatohepatitis (liver) [25], alveolar proteinosis (lung) [21], and sepsis (spleen, and other) [26]. To perform these diverse functions, macrophages must sense and respond to their microenvironment. Macrophages can sense structures broadly shared by invading microbes, termed pathogen-associated molecular patterns (PAMPs), or mediators induced by cell damage, so-called damage- or danger-associated molecular patterns (DAMPs) [27]. PAMPs and DAMPs are recognized by pattern-recognition receptors (PRRs). Macrophages are highly plastic cells that detect PAMPs and DAMPs and alter their phenotype to mount an appropriate response. Once the insult is cleared macrophages are also a critical component of the subsequent pro-resolution or tissue repair process that follows [17].
ROS produced by macrophages act as critical antimicrobial agents to kill invading microorganisms [28,29]. However, ROS can also protect against infection in several indirect ways through redox signaling, for example by regulating the secretion of pro-inflammatory cytokines, activation of inflammasomes, and orchestration of the subsequent adaptive immune response via antigen presentation [[30], [31], [32]]. Redox signaling also regulates signal transduction, gene expression, metabolic reprogramming, differentiation, and polarization in macrophages [[33], [34], [35], [36], [37], [38], [39]].
More recently, metabolites have emerged as key players in redox regulation of macrophages. Certain metabolites can directly modify redox sensitive cysteine residues through a diverse set of post-translational modifications (PTMs). These include succination by fumarate [40] and 2,3-dicarboxypropylation, or alkylation, by itaconate [41].
In this review, we provide a brief overview of ROS (primarily focusing on O2.- and H2O2) production in macrophages. We summarize the diverse roles of ROS in both direct and indirect regulation of the antimicrobial response in macrophages. We discuss the crucial redox signals that regulate macrophage function with a focus on PTM of redox sensitive cysteines. We also review the latest tools and methods to detect and examine redox-sensitive cysteines. We discuss the impact of dysfunctional redox signaling in macrophages on inflammatory diseases and cancer, and finally, we highlight key questions that remain to be resolved.
2. Sources of ROS and RNS
Two of the major endogenous sources of O2.- and H2O2 in macrophages are cytosolic NADPH oxidases (NOX) and mitochondria [[42], [43], [44], [45], [46]]. NOX are a family of transmembrane enzymes that are dedicated to the production of cytosolic O2.- via the transfer of an electron from NADPH to molecular oxygen [47,48]. In phagocytes such as neutrophils and macrophages, O2.- is rapidly and robustly produced by NOX, the so-called oxidative burst, in response to bacterial or viral infections. The oxidative burst is an essential mechanism for the elimination of invading microorganisms upon phagocytosis [[49], [50], [51], [52]]. This is discussed in detail below.
Another major site of O2.- production, which is then converted to H2O2 and other oxidants in macrophages, is the mitochondrial electron transport chain (ETC) [45,53]. Complex I of the ETC is the major contributor of mitochondrial ROS (mtROS) production in macrophages, however complex II and III also produce mtROS [[54], [55], [56]]. During oxidative phosphorylation, electrons travel from complexes I to IV of the ETC to form a proton gradient that drives the activity of ATP synthase to catalyze the synthesis of ATP from ADP. Electrons from complex I and complex III cause a partial reduction of oxygen that leads to the generation of O2.-, which is released into the mitochondrial matrix. Additionally, O2.- can be released into the intermembrane space via complex III. O2.- is quickly converted into H2O2 by superoxide dismutases (SOD), SOD2 and SOD1 in the matrix and intermembrane space respectively [42,46,47]. ROS released into different mitochondrial compartments can interact with, and modify, different redox-sensitive proteins residing at various subcellular locations [57].
In addition to NOX enzymes and ETC complexes in the mitochondria, other enzymes including xanthine oxidase (XOR), cyclooxygenases, and cytochrome P450 all generate O2.- during their respective biochemical processes [[58], [59], [60]]. Similarly, non-mitochondrial organelles such as peroxisomes and the endoplasmic reticulum (ER) are important sites of ROS production [[61], [62], [63]].
RNS are derived from nitric oxide (NO) whose production from arginine is catalyzed by NO synthase (NOS) enzymes including neuronal NOS (NOS1 or nNOS), inducible NOS (NOS2 or iNOS), and endothelial NOS (NOS3 or eNOS) [64]. iNOS is highly expressed in macrophages [64,65] and its transcription is potently induced by proinflammatory signals such as lipopolysaccharide (LPS) and interferon gamma [[66], [67], [68]]. Like ROS, RNS have important roles in microbial killing and inflammatory macrophage function [64,69,70]. Interestingly, RNS production is species-dependent with reports suggesting that human macrophages produce less RNS than mouse macrophages in response to inflammatory stimuli [64,71,72].
3. ROS and RNS as signaling molecules
For many years, the term “ROS” was inadvertently equated with damaging molecules and byproducts of cellular stress. More recently, many studies have unveiled the essential, but often overlooked, role of ROS as signaling molecules that regulate a myriad of biological processes [47,73]. Some of the early evidence for this was in the context of cell growth, where intracellular ROS, more specifically H2O2, were found to be required for tyrosine phosphorylation of proteins that promote cell growth via epidermal growth factor (EGF) and platelet-derived growth factor (PDGF) signaling pathways [3,5]. Other studies soon after detailed a mechanism linking ROS to hypoxia-inducible factor 1 alpha (HIF-1α)-dependent cellular processes. HIF-1α is a master transcription factor regulating many biological processes, most notably the response to hypoxia, and was the first transcription factor identified to act via environmental sensing [74]. It was shown that during hypoxia mtROS stabilize HIF-1α by inhibiting its molecular degraders, prolyl hydroxylases (PHDs). This allows HIF-1α to localize to the nucleus and drive the transcription of target genes controlling many biological processes including erythropoiesis, metabolism, angiogenesis, and autophagy [8,[75], [76], [77], [78], [79]]. These studies are fundamental to our understanding of ROS as important endogenous signaling molecules.
RNS can also act as important signaling molecules. NO regulates many heme-containing proteins such as soluble guanylate cyclase (sCG) [80], cytochrome P450 [81], complex IV [82,83], and NOS enzymes [84,85] by reacting with the heme Fe group which is essential for the activity of hemeproteins [86]. The formation of the heme-NO complex can inhibit enzyme activity by blocking heme incorporation into hemeproteins, as is the case for cytochrome P450 and NOS enzymes, or by competitively inhibiting oxygen binding [82,83]. In some cases like sCG, heme-NO formation can boost enzyme activity by invoking structural changes [80]. RNS also regulate the activity of metalloenzymes containing catalytic iron-sulfur (Fe–S) clusters such as complex I and II of the ETC [87,88], and mitochondrial aconitase [89]. Like heme, Fe–S clusters are protein prosthetic groups residing in the active sites of enzymes that are essential for electron transfer [90]. The Fe–S cluster in aconitase mediates the Lewis acid reaction that converts citrate to aconitate, while Fe–S clusters in the ETC complex I, II, and III facilitate electron flow within the ETC [90]. NO can inhibit the activity of Fe–S-containing enzymes such as aconitase and complex II [88,91,92] by oxidizing and removing an Fe atom from the Fe–S cluster (see reviews [65,93] for further discussion). NO can also inhibit complex I activity via PTM of cysteine (Cys)39 on the ND3 subunit [87].
3.1. Post-translational modification of cysteine residues in redox signaling
A key mechanism by which endogenous ROS act as signaling molecules is via oxidation of cysteine residues in target proteins. At physiological pH, the thiol (-SH) group on cysteine residues is deprotonated to form a charged thiolate (-S-). While both thiol and thiolates can act as nucleophiles, the negative charge of thiolates makes them more reactive than thiols [94]. As such, the reactivity of protein cysteine residues varies with pH and the distinct protein environment which affects deprotonation. Susceptibility to oxidation is similarly pKa-dependent i.e. the pH at which thiol- and thiolate-bearing cysteines are in equilibrium. Redox-sensitive cysteines can undergo rapid oxidation by a reactive oxidant. Oxidation of cysteine residues can alter protein function by adding/removing disulfide bonds or changing the electrostatic state in important active sites, resulting in alterations in protein stability, activity, localization, and protein-protein interactions [42]. It should also be noted that the initial oxidation, or sulfenylation, of a cysteine residue is reversible and can be reduced back to a free thiol by endogenous antioxidants such as glutathione (GSH) [42,95] however hyperoxidation of sulfenylated thiols to sulfinic or sulfonic acid is irreversible.
Oxidation, or sulfenylation, is only one of many reversible redox modifications. Others include disulfide bond formation, glutathionylation, nitrosylation, and palmitoylation. Disulfide bond formation occurs when an unstable -sulfenylated cysteine reacts with a proximal cysteine residue to form an inter- or intramolecular bond (RSSR').
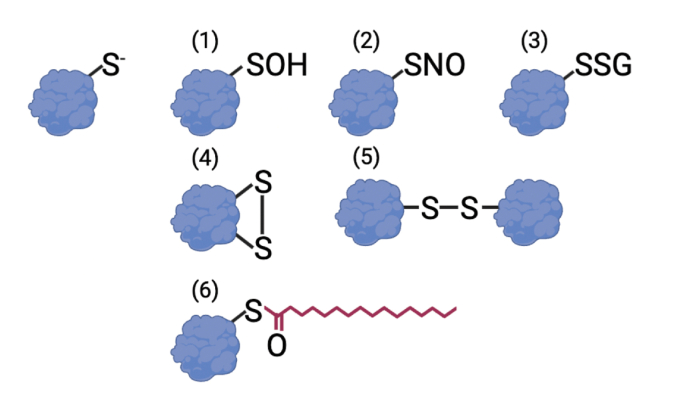
S-nitrosylation or nitrosylation takes place on cysteine residues when NO is incorporated into the thiol group (-SNO) while glutathionylation arises from a disulfide formation between a cysteine residue and the reduced form of GSH [96,97] (Fig. 1). These PTMs are reversible and dynamic and regulate a range of biological processes. As such, cysteine residues are important rheostats that can fine-tune protein function.
Fig. 1.
Reversible post-translational modification (PTM) of cysteine residues.
Free thiolates (top left) can undergo a range of reversible PTMs. These include sulfenylation (1), S-nitrosylation (2), glutathionylation (3), intra- (4), and inter- (5) molecular disulfide bonds, and palmitoylation (6).
Palmitoylation and geranylgeranylation (GGylation) are lipid-mediated PTMs generated by a covalent link between a cysteine thiol and the respective lipid molecules (Fig. 1). Palmitoylation is a unique covalent lipid modification as it is reversible and functionally dynamic. The transmembrane palmitoyltransferase DHHC family is the main catalyst of this modification [98] while depalmitoylating enzymes including several protein thioesterases [[99], [100], [101], [102]] and α/β-hydrolase domain-containing proteins [103] remove this PTM. As a bulky and hydrophobic group, the addition of palmitate to a protein usually leads to a significant alteration in its physical properties, playing an important role in protein trafficking and protein-protein interactions [98].
GGylation is an irreversible lipid modification involving the covalent attachment of geranylgeranyl isoprene to the thiol group of cysteine residues. This PTM is catalyzed by the Rab geranylgeranyltransferase enzyme and is important for protein transport across the secretory and endocytosis pathways [[104], [105], [106], [107]].
These diverse cysteine PTMs can profoundly alter protein and cellular function but our understanding of what endows the specificity of a cysteine to undergo one PTM versus another, and the breadth of these modifications, is poorly understood. It should also be noted that reversible redox modification can also occur at methionine residues. However, its sulfur is in a less reactive thioether form than the thiol/thiolate of cysteine [94].
4. The anti-microbial role of ROS/RNS and the oxidative burst
Macrophages produce large amounts of ROS as a defense mechanism against invading microorganisms. But more recently, our understanding of the role of ROS in macrophages has expanded from its role as an anti-bacterial agent to a role in cell signaling and inflammation [108,109].
A central function of macrophages is to phagocytose or engulf invading microorganisms and destroy them in the phagosome. NOX-derived ROS play a critical role in this process. NOX are membrane-bound protein complexes of which there are 7 members. NOX2 (or gp91phox) is the best characterized catalytic subunit of the NOX complexes for its role in phagocytosis. Upon phagocytic activation, NOX2 forms a heterodimer with cytochrome B light chain (CYBA or p22phox) and translocates to the plasma membrane to produce ROS (for a detailed review see Ref. [110]). The generation of NOX-mediated ROS is regulated by Toll-like receptor (TLR) signaling [111,112]. Ablation of NOX2 significantly impairs cellular ROS production in activated macrophages [30,111]. Peritoneal macrophages from NOX2-deficient mice fail to produce cellular ROS and this impairs ROS-dependent pro-inflammatory cytokine secretion in response to infection [30]. Another NOX enzyme, NOX4, is also implicated in ROS production in human monocyte-derived macrophages [49].
Beyond its role in the oxidative burst, NOX-mediated ROS affect the antimicrobial activity of macrophages in additional ways. For instance, ROS production by NOX2 in RAW264.7 macrophages in response to TLR or Fc-gamma receptor (FcγR) stimulation is essential for the recruitment of the autophagy protein LC3 to phagosomes, thereby promoting phagosomal maturation and microbe killing [113,114]. Another study demonstrated that NOX2-dependent ROS trigger a non-autophagic antimicrobial response called LC3-associated phagocytosis during Listeria infection in bone marrow-derived macrophages (BMDMs) [115]. Furthermore, NOX2-derived ROS has been shown to regulate FcγR cross-linking with its ligands to trigger a kinase signaling cascade, ultimately leading to nuclear factor-kappa B (NF-κB)-mediated IL-6 production [50]. Additionally, NOX2 can control antigen presentation in macrophages as NOX2-deficient BMDMs are unable to process certain peptides by cysteine cathepsins, which are proteases with redox-sensitive cysteine catalytic sites [32]. However, it is unclear whether NOX2 depletion in macrophages directly affects the function of cysteine cathepsins.
RNS are also an important constituent of the antimicrobial response in macrophages. iNOS-induced NO production is essential for microbe killing in macrophages and for host survival [69,[116], [117], [118]]. Chemical inhibition of NO production by methylarginine has been shown to inhibit the microbicidal activity of human macrophages [116,118] and iNOS knockout in mice leads to impaired bactericidal function in macrophages and reduced survival during Salmonella infection [69,117]. Interestingly, during Salmonella infection, macrophages from iNOS-knockout mice exert a greater anti-bacterial response than macrophages from NOX2-knockout mice, suggesting a greater contribution of ROS than RNS to antibacterial activity in this context [69]. This may be a result of a compensatory increase in ROS production in iNOS-deficient macrophages.
More recently, it was discovered that mtROS contribute significantly to the antimicrobial ROS pool. This process was first described in response to stimulation of TLR1, 2, and 4 which leads to recruitment of mitochondria to macrophage phagosomes in a tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6)-dependent manner and an increase of bactericidal mtROS (see review [108] for further details). It was later elucidated that TRAF6 forms a complex with ECSIT (evolutionarily conserved signaling intermediate in Toll pathways) to promote mtROS production [119]. Moreover, bacterial sensing via TLR signaling has been reported to increase the activity of complex II of the ETC and glycerol-3-phosphate dehydrogenase (G3PDH), a mitochondrial-bound enzyme that shuttles electrons from cytoplasmic NADH to coenzyme Q [36]. These ETC adaptations enhance the phagosomal activity of macrophages [36]. Furthermore, mtROS were recently found to promote the bactericidal response of macrophages by activating NF-κB-mediated cytokine production [30] (this is discussed in further detail below). These findings demonstrate the often-overlooked role of mtROS as a significant contributor to the anti-microbial activity of macrophages.
5. PTM of cysteines residues as a mode of redox signaling in macrophages
While ROS production in macrophages has a critical role in anti-microbial defense, ROS also regulate macrophage function by acting as important signaling molecules. Beyond their role in phagocytosis, macrophages have a wide range of functions in immunity including inflammatory cytokine production, chemokine release to recruit other immune cells, antigen presentation, tissue repair, and maintenance of tissue homeostasis (see review [17] for further details). To perform such diverse functions, macrophages rewire their metabolism in response to stimuli, and alterations in metabolism are intrinsically linked with changes in redox homeostasis. One of the earliest examples of this metabolic rewiring was the observation that upon stimulation with the TLR4 agonist LPS macrophages switch their metabolism from oxidative phosphorylation (OXPHOS) to glycolysis [120,121]. It was later identified that upon stimulation with LPS macrophages accumulate high levels of the tricarboxylic (TCA) intermediate succinate and exhibit mitochondrial hyperpolarization due to decreased ATP synthase activity [122]. Together, these phenomena trigger high levels of mtROS production at complex I of the ETC. As such, the metabolic rewiring from OXPHOS to glycolysis supports ATP production while the mitochondria are repurposed for mtROS production. mtROS inhibit PHD function to stabilize HIF-1α, which promotes the transcription of target genes, including interleukin (IL-1β) [37]. These findings present an intricate pathway in macrophages whereby mtROS act as a signal that links metabolic reprogramming with transcriptional regulation during inflammation.
Redox signaling can also affect many facets of macrophage function including NF-κB signaling, pathogen recognition, and inflammasome activation, by PTM of target cysteines. S-nitrosylation, palmitoylation, glutathionylation, and oxidation of cysteine residues are examples of PTMs regulating protein function in macrophages and are discussed in detail below.
5.1. Redox regulation of NF-κB signaling in macrophages
NF-κB is a master transcription factor that plays a central role in regulation of inflammatory gene expression in macrophages [123]. As such, NF-κB activity must be carefully regulated. Under normal conditions, NF-κB subunits are sequestered in the cytosol by a family of inhibitors, so called inhibitors of NF-κB (IκB) [123]. One member of this family, IκBα, contains an N-terminal phosphorylation site that is the target of the multi-subunit IκB kinase (IKK) complex. Upon an inflammatory signal, IKK phosphorylates IκBα targeting it for ubiquitination and subsequent proteasomal degradation. This liberates NF-κB, which transfers to the nucleus and drives the transcription of a plethora of inflammatory genes [123]. NF-κB can promote ROS and RNS production by activating expression of genes encoding proteins that generate ROS and RNS such as XOR [124], NOS1 [125], COX-2 [126], NOX2 [127] and iNOS [128]. However, many studies have suggested that NF-κB can also suppress ROS production by increasing the expression of antioxidant enzymes. For example, antioxidants such as SOD1 and SOD2, glutathione peroxidase-1 (GPX1), and thioredoxin are transcriptional targets of NF-κB [[129], [130], [131], [132], [133]].
Interestingly, ROS can reciprocally regulate NF-κB activity (Fig. 2 and Table 1). Early studies suggested that ROS can directly control NF-κB by oxidizing Cys62, in the conserved Rel homology domain (RHD) of the p50 subunit to inhibit NF-κB DNA binding and transcriptional activity [[134], [135], [136]]. Cys62 of p50 can also undergo glutathionylation and S-nitrosylation, both of which impair DNA binding and recognition [137,138]. The p65 subunit of NF-κB is also regulated by RNS. In peritoneal and RAW264.7 macrophages, it was shown that iNOS-mediated S-nitrosylation of Cys38 of the p65 subunit reduces p50-p65 heterodimer formation and decreases NF-κB DNA binding [139,140]. Interestingly, this specific residue can also undergo a sulfhydration (-SSH), a PTM in which hydrogen sulfide reacts with a cysteine disulfide. Sulfhydration of Cys38 increases DNA binding and transcription activity of NF-κB in macrophages [141].
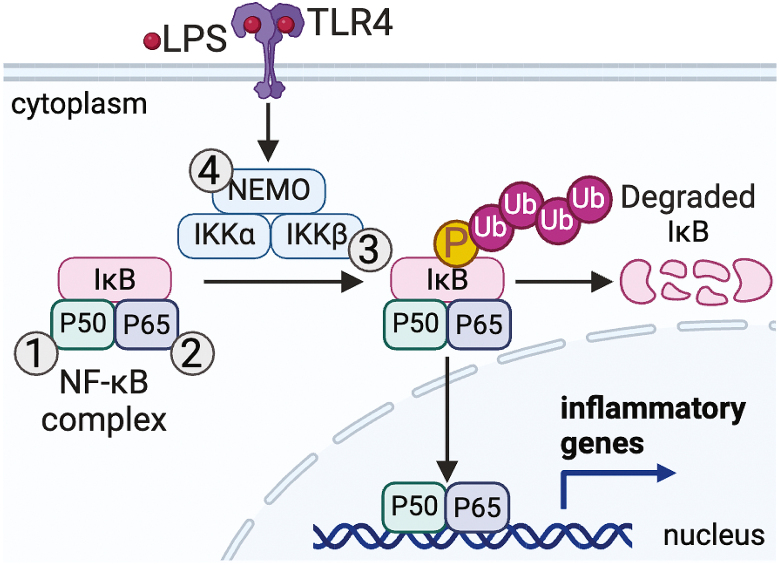
Fig. 2.
Cysteine PTM of proteins of the NF-κB pathway.
NF-κB is a master transcription factor that plays a central role in regulation of inflammatory gene expression in macrophages, therefore, its activity must be carefully regulated. Several components of the NF-κB pathway are redox-regulated. ROS can oxidize cysteine (Cys)62 of the p50 subunit to inhibit NF-κB DNA binding and transcriptional activity (1). Cys62 of p50 can also undergo glutathionylation and S-nitrosylation, both of which impair DNA binding and recognition. S-nitrosylation of Cys38 of the p65 subunit reduces p50-p65 heterodimer formation and decreases NF-κB DNA binding while sulfhydration of Cys38 increases DNA binding and transcription activity of NF-κB in macrophages (2). Glutathionylation Cys179 of inhibitor of NF-κB (IκB) kinase (IKK)β suppresses its kinase activity thereby dampening NF-κB activity (3). An intermolecular disulfide bond between Cys54 and Cys347 of NF-κB essential modulator (NEMO), which is a subunit of IKK, is critical for the stabilization of NEMO, assembly of the IKK complex, and NF-κB activation (4).
Table 1.
Cysteine PTM of proteins of the NFκB pathway.
| Protein | Cysteine(s) | PTM | Effect on NFκB activity | System | Validation | Reference (s) | |
|---|---|---|---|---|---|---|---|
| 1 | p50 | 62 | Homodimer disulfide | Increase DNA binding | Recombinant protein | Point mutation (PM) recombinant protein | [134,136] |
| p50 | 62 | Glutathionylation | Decrease DNA binding | Recombinant protein | PM in recombinant protein | [137] | |
| p50 | 62 | S-nitrosylation | Decrease DNA binding | Recombinant protein | PM in recombinant protein | [138] | |
| 2 | p65 | 38 | S-nitrosylation | Decrease DNA binding | Peritoneal macrophages & RAW264.7 | PM in HEK293T | [139,140] |
| p65 | 38 | Sulfhydration | Increase DNA binding | Peritoneal macrophages & RAW264.7 | PM in RAW264.7 | [141] | |
| 3 | IKKβ | 179 | Glutathionylation | Inhibit NFκB activity | Alveolar epithelial cells (C10) | PM in C10 cells | [142] |
| 4 | NEMO | 54 & 347 | Intramolecular disulfide bond | Promote NFκB activity | Peritoneal macrophages & BMDMs | PM in peritoneal macrophages and BMDMs | [30] |
Redox signaling can also regulate NF-κB signaling by modifying proteins controlling its activity. For example, Cys179 of IKKβ is subject to glutathionylation which suppresses its kinase activity thereby dampening NF-κB activity [142]. This PTM can be removed by glutaredoxin-1, demonstrating the complex network of redox-regulation of NF-κB that likely adapts in response to stimulation. Furthermore, bacterial infection of mouse primary macrophages (both peritoneal macrophages and BMDMs) promotes mtROS production in a TLR2-dependent manner to activate NF-κB essential modulator (NEMO), which is a subunit of IKK [30]. ROS stimulate the formation of a homodimer by promoting an intermolecular disulfide bond between Cys54 and Cys347 [143]. Disulfide bond formation is critical for the stabilization of NEMO, assembly of the IKK complex, NF-κB activation, and the subsequent secretion of NF-κB-dependent pro-inflammatory cytokines in activated macrophages [30,143] (Fig. 2 and Table 1).
Interestingly, H2O2 can also activate casein kinase II to enhance phosphorylation of tyrosine 42 in IκBα which leads to degradation of IκBα in an IKK-independent manner [144]. A later study using human KBM-5 myeloid cells showed that H2O2 induces phosphorylation and degradation of IκBα by activating spleen tyrosine kinase [145]. These observations suggest that ROS regulate the activity of additional kinases upstream of IκBα. However, the detailed mechanism for this in macrophages is still elusive.
As well as redox modification of proteins, lipids can also undergo oxidation to alter NF-κB activity and macrophage function. Lipid peroxidation occurs when polyunsaturated fatty acids (PUFAs) are oxidized by ROS including hydroxyl (HO•) and hydroperoxyl (HO•2) radicals [146]. Malondialdehyde (MDA) and 4-hydroxynonenal (4-HNE) are the two main byproducts of lipid peroxidation and are widely used as biomarkers for lipid peroxidation [147,148] and are highly prevalent in many diseases including neurodegenerative diseases [149], cancer [150], and metabolic disease [151]. In macrophages, both MDA and 4-HNE have been reported to modulate the NF-κB-mediated inflammatory response [152,153], although the mechanistic basis for this is poorly defined. The roles of lipid peroxidation in various biological processes and cell types have been extensively reviewed elsewhere (see [146] for further details).
5.2. Redox-regulation of PPRs
Macrophages can be activated by PAMPs and DAMPs that are sensed by PRRs such as TLRs, the cyclic GMP-AMP synthase-stimulator of interferon genes (cGAS-STING) pathway, and nucleotide oligomerization domain (NOD)–like receptors (NLRs). It has recently emerged that several members of the PRR family are redox-regulated.
Palmitoylation of cysteines has emerged as an important regulator of PRRs in macrophages. Palmitoylation of TLRs, the cytosolic NLRs, and cGAS-STING has been reported [33,34,[154], [155], [156]].
Palmitoylation of Cys113 of the TLR4 adaptor protein myeloid differentiation protein 88 (MyD88) upon TLR4 stimulation enhances the activity of this pathway [156]. Additional TLR adaptors such as tyrosine-protein kinase LYN (Cys3) and MyD88 adaptor-like protein (MAL; Cys91) have been reported to undergo palmitoylation and glutathionylation, respectively, in macrophages upon stimulation. These PTMs enhance TLR activity [34,157].
Upon recognition of cytosolic DNA cGAS-STING promotes the production of type I interferons (IFNs) [158]. Cys88 and Cys91 of STING undergo palmitoylation which is essential for STING complex assembly into multimeric complexes at the Golgi apparatus and subsequent recruitment of downstream signaling factors including TANK-binding kinase 1 (TBK1) [154]. Interestingly, Cys88/91 are S-nitrosylated in virally-infected macrophages, which blocks palmitoylation of these residues and subsequent STING activity [33].
NOD1/2, NLRs which detect peptidoglycan from invading microbes, are also redox regulated. Palmitoylation of several cysteine residues on NOD1 (Cys 558, 567, & 952) and NOD2 (Cys 395 & 1033) is essential for NOD1/2 membrane recruitment during bacterial challenge in macrophages [159]; (Fig. 3 and Table 2). NLR family pyrin domain containing 3 (NLRP3) is also regulated by redox signaling; this is discussed below.
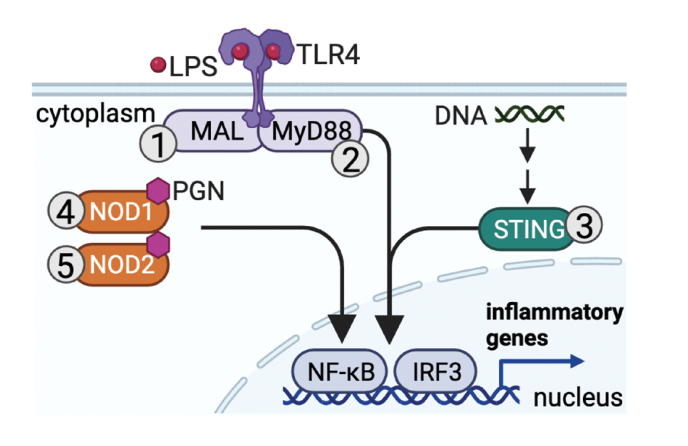
Fig. 3.
Cysteine PTMs in the regulation of pattern recognition receptors (PRRs).
PRRs such as TLRs, the cyclic GMP-AMP synthase-stimulator of interferon genes (cGAS-STING) pathway, and nucleotide oligomerization domain (NOD)–like receptors (NLRs) are redox-regulated. Glutathionylation of myeloid differentiation protein 88 (MyD88) adaptor-like protein (MAL; Cys91; 1) and palmitoylation of Cys113 of MyD88 (2) enhance TLR activity. Palmitoylation of Cys88 and Cys91 of STING (3) is essential for STING signaling. Palmitoylation of several cysteine residues on NOD1 (Cys558, 567, & 952; 4) and NOD2 (Cys 395 & 1033; 5) is required for NOD1/2 membrane recruitment during bacterial challenge in macrophages.
Table 2.
Cysteine PTMs in the regulation of PRRs.
| Protein | Cysteine(s) | PTM | Effect | System | Validation | Reference (s) | |
|---|---|---|---|---|---|---|---|
| 1 | MAL | 91 | Glutathionylation | Promote MyD88-MAL interaction and TLR4 activity | BMDMs | Point mutation (PM) in iBMDMs | [157] |
| 2 | MyD88 | 113 | Palmitoylation | Increase TLR4 activity | THP-1 & RAW264.7 | PM in HEK293T cells | [156] |
| 3 | STING | 88 & 91 | Palmitoylation | Promote TBK1 interaction and STING signaling | BMDMs | PM in HEK293T | [154] |
| STING | 88 & 91 | S-nitrosylation | Block palmitoylation and STING signaling | THP-1 & BMDMs | No PM validation | [33] | |
| 4 | NOD1 | 558, 567, & 952 | Palmitoylation | Increase NOD signaling & membrane association | BMDMs | PM in RAW264.7 | [159] |
| 5 | NOD2 | 395 & 1033 | Palmitoylation | Increase NOD signaling & membrane association | BMDMs | PM in RAW264.7 | [159] |
In response to LPS, 100 unique cysteine sites in THP-1 monocytes and peritoneal macrophages have been reported to undergo glutathionylation [160]. One example is Cys91 of MAL. Glutathionylation of Cys91 promotes the interaction between MAL and MyD88, which drives TLR signal transduction [157].
5.3. Redox-regulation of the NLRP3 inflammasome
ROS also modulate the NLRP3 inflammasome, a protein complex that mediates IL-1β processing and secretion [31,161]. NLRP3 is essential for the cleavage and activation of caspase-1. Once active, caspase-1 cleaves and activates IL-1β [162] and the pore-forming protein gasdermin D (GSDMD) which is required for IL-1β secretion and the induction of an inflammatory form of cell death called pyroptosis [[163], [164], [165]]. All known activators of NLRP3 promote mtROS production and mtROS have been shown to activate NLRP3 [166,167]. mtROS promote the dissociation of thioredoxin-interacting protein (TXNIP) from thioredoxin. This supports the interaction between NLRP3 and TXNIP, and subsequent NLRP3 activation, IL-1β processing and secretion [166,168]. It should be noted that this occurs specifically in response to mtROS and is NOX-independent [169,170]. mtROS also promote the release of oxidized mitochondrial DNA that binds and activates NLRP3 [171].
Recently, NIMA-related kinase 7 (NEK7) was identified as a NLRP3-binding protein that drives NLRP3 oligomerization and activation [172]. Interestingly, ROS enhance NEK7 phosphorylation and its interaction with NLRP3 [173]. How ROS promote phosphorylation is still unclear although inhibition of certain phosphatases by ROS has been proposed as one possible mechanism. Another study demonstrated that the NLRP3-NEK7 interaction is mediated by deglutathionylation of Cys253 on NEK7 [174].
GSDMD activity is also redox-regulated. Several studies have demonstrated a direct link between redox signaling and GSDMD activity. ROS oxidize Cys192 of GSDMD to enhance its pore-forming activity [[175], [176], [177]]. Indeed, macrophages in which Cys192 of GSDMD is mutated to an alanine, show impaired pore formation and pyroptosis [177]. Intriguingly, this same cysteine residue (Cys192) is sensitive to palmitoylation. Palmitoylation of Cys192 of GSDMD leads to membrane translocation of GSDMD and the subsequent pore formation and pyroptosis [175,176]. Both pharmacological inhibition [178] and genetic mutation [177] of Cys192 of GSDMD in macrophages inhibits GSDMD activity and pore formation (Fig. 4 and Table 3).
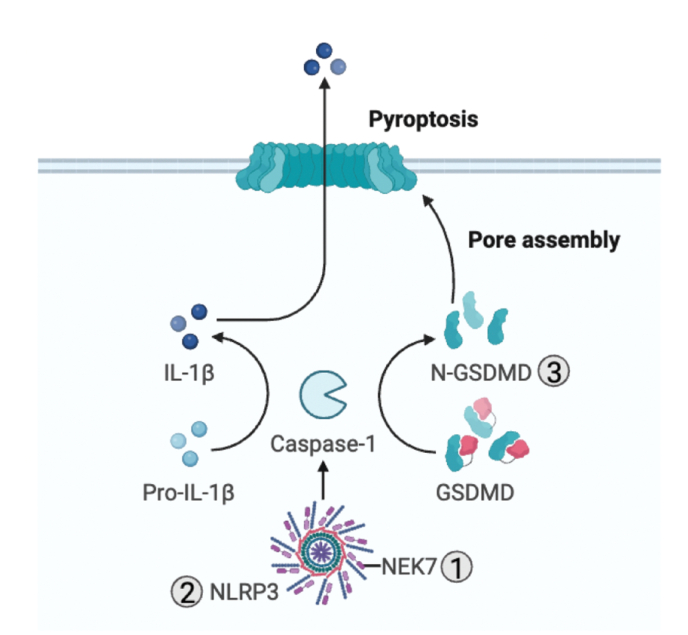
Fig. 4.
Redox regulation of NLRP3.
ROS modulate the NLRP3 inflammasome, a protein complex that mediates IL-1β processing and secretion. ROS enhance NIMA-related kinase 7 (NEK7) phosphorylation and its interaction with NLRP3 through a poorly defined mechanism. Additionally, deglutathionylation of Cys253 on NEK7 is important for its interaction with NLRP3 (1). Itaconate derivatives targets Cys548 of the NLRP3 inflammasome to prevent its interaction with NEK7 (2). ROS oxidize Cys192 of GSDMD to enhance its pore-forming activity (3) while palmitoylation of Cys192 of GSDMD leads to membrane translocation of GSDMD and the subsequent pore-formation and pyroptosis. Furthermore, ROS promote cleavage of GSDMD, mobilization to the plasma membrane, oligomerization and eventually pore formation. GSDMD is also regulated by succination and alkylation by itaconate.
Table 3.
Cysteine PTMs in the regulation of NLRP3.
| Protein | Cysteine(s) | PTM | Effect | System | Validation | Reference (s) | |
|---|---|---|---|---|---|---|---|
| 1 | NEK7 | 253 | Glutathionylation | Inhibit NLRP3 activity | BMDMs | Point mutation (PM) in HEK293T cells | [174] |
| 2 | NLRP3 | 548 | Alkylation by itaconate | Inhibit NLRP3 activity and interaction with NEK7 | BMDMs | No PM validation | [193] |
| 3 | GSDMD | 77 | Alkylation by itaconate | Activate pyroptosis | BMDMs | No PM validation | [192] |
| GSDMD | 192 | Palmitoylation | Promote GSDMD signaling | THP-1, BMDMs, & iBMDMs | PM in iBMDMs and HEK293T cells | [[175], [176], [177]] | |
| GSDMD | 192 | Succination by fumarate | Inhibit GSDMD signaling and interaction with Caspase-1 | THP-1 & BMDMs | No PM validation | [204] |
Furthermore, ROS promote caspases 1, 4, and 5-mediated cleavage of GSDMD, resulting in macrophage pyroptosis [163,179,180], while XOR-derived ROS activate a MAP3K signaling cascade in macrophages to promote mobilization of GSDMD to the plasma membrane [181]. Additionally, mtROS production via the Ragulator-Rag complex acts downstream of GSDMD cleavage to promote GSDMD oligomerization in the plasma membrane, which eventually leads to pore formation [182].
5.4. The role of RNS in redox signaling in macrophages
RNS can also modulate macrophage function through modification of cysteine residues and metabolic reprogramming. As discussed above, S-nitrosylation of Cys38 of the p65 subunit of NF-κB decreases NF-κB activity [139,140] while S-nitrosylation of Cys88/91 of STING impairs STING activity [33]. Additionally, NO has been reported to regulate NLRP3 activation as virally-infected, IFNγ-stimulated BMDMs from iNOS-deficient mice produce more mature IL-1β than BMDMs from wildtype mice [183]. Total levels of S-nitrosylated NLRP3 protein were decreased in iNOS-deficient BMDMs [183]; however, the specific cysteine residue(s) on NLRP3 subjected to this PTM are unknown. Furthermore, NO can alter the metabolic state of pro-inflammatory macrophages by inhibiting several metabolic enzymes including complex I [184], isocitrate dehydrogenase (IDH) [184], pyruvate dehydrogenase (PDH) [89], and aconitase 2 by disrupting Fe–S clusters in these enzymes [89]. These studies also demonstrated that NO modifies cysteine residues on IDH (Cys 113, 133, 154, & 418) [184] and PDH (Cys unidentified) [89] that may contribute to the regulatory function of NO.
5.5. The role of metabolites in redox signaling in macrophages
In the past decade it has emerged that metabolites are important signaling molecules that regulate macrophage function [37,41,109,185]. The mechanisms by which metabolites signal to regulate macrophage function are diverse, but modulation of ROS production and redox modification of cysteine residues in target protein represent key mechanisms. Three examples of this are succinate, fumarate and itaconate.
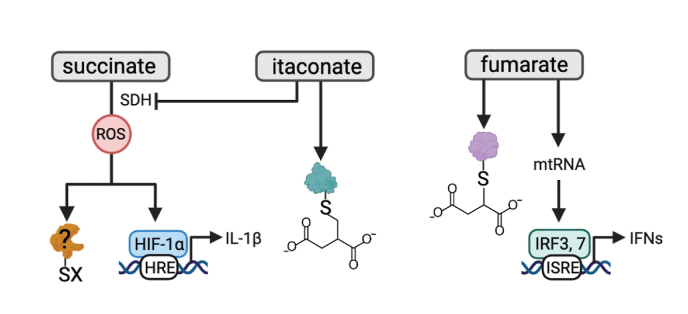
As mentioned above, succinate is a pro-inflammatory molecule that, upon oxidation, promotes complex I-derived mtROS, and inflammation in a HIF-1α-dependent manner [37,122]. Whether succinate-induced mtROS promotes oxidation of specific cysteine residues in target proteins to modulate inflammatory function remains to be determined but it is unlikely that HIF-1α is the sole target of mtROS in activated macrophages (Fig. 5).
Fig. 5.
Redox regulation of macrophages by metabolites.
Metabolites also contribute to redox signaling in macrophages. Oxidation of succinate by succinate dehydrogenase (SDH), promotes complex I-derived mtROS production, and inflammation in a hypoxia inducible factor (HIF)-1α-dependent manner. It remains to be determined if mtROS driven by succinate oxidation promotes oxidation of specific cysteine residues in target proteins to modulate inflammatory function. Itaconate has potent immunomodulatory effects in macrophages. Two major mechanisms underpinning the immunomodulatory activity of itaconate are SDH inhibition and 2,3-dicarboxypropylation or alkylation of cysteine residues. Several alkylation targets of itaconate have emerged. These include cysteines on KEAP1, the NLRP3 inflammasome, gasdermin D (GSDMD), residues on several metabolic enzymes, including glyceraldehyde 3-phosphate dehydrogenase (GAPDH), aldolase A (ALDOA), and lactate dehydrogenase A (LDHA). Fumarate reacts with thiols in cysteine residues in a PTM termed succination. Many of the protein succination targets of fumarate are shared with the alkylation targets of itaconate including KEAP1, GSDMD, and GAPDH. Fumarate can also promote the release of mtRNA, and retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5) activity to promote interferon signaling.
Itaconate is another key immunomodulatory metabolite in macrophages. Itaconate is a metabolite first discovered 150 years ago as a byproduct of fermentation in fungi and was thought to be a potential Krebs cycle intermediate by Hans Krebs [186]. However, it was not until the 2010s that the role of itaconate in macrophage biology was appreciated. Itaconate is produced from aconitate as a divergence from the TCA cycle. This reaction is catalyzed by aconitate decarboxylase (ACOD1), an enzyme encoded by immunoresponsive gene 1 (Irg1), which is uniquely expressed in myeloid cells, and most particularly in macrophages [187]. Irg1 expression and itaconate production are extremely sensitive to inflammatory stimuli such as LPS, other TLR agonists, and inflammatory cytokines [[187], [188], [189]]. Itaconate has potent immunomodulatory effects in macrophages. These include limiting mtROS production [190,191], inhibition of inflammasome activation [190,192,193], control of pro-inflammatory cytokines [41,194], and rewiring of macrophage metabolism [190,195,196].
Two major mechanisms underpinning the immunomodulatory activity of itaconate are succinate dehydrogenase (SDH) inhibition and cysteine alkylation. Itaconate is a competitive inhibitor of SDH due to its structural similarity to the SDH substrate, succinate [190,191]. This limits succinate oxidation and subsequent mtROS production and HIF-1α-mediated IL-1β transcription [190,191]. More recently, it was discovered that itaconate modifies cysteine residues in target proteins, a PTM defined as 2,3-dicarboxypropylation, or alkylation [41], to exert an anti-inflammatory effect. The first evidence of this PTM came from the finding that, an itaconate derivative alkylates multiple cysteine sites on Kelch like ECH associated protein 1 (KEAP1), promoting Nuclear factor erythroid 2-related factor 2 (NRF2) activity and the transcription of downstream anti-inflammatory genes [41]. This effect was corroborated in ACOD1-knockout macrophages, which fail to produce itaconate [194]. Since this finding, several cysteine targets of itaconate have been identified. An itaconate derivative targets Cys548 of the NLRP3 inflammasome to prevent its interaction with NEK7, subsequently inhibiting NLRP3 activation and IL-1β processing [193]. Itaconate has also been shown to alkylate Cys77 of GSDMD although the functional relevance of this remains to be determined [192]. In another study, itaconate was found to alkylate Cys212 on transcription factor EB (TFEB), a master regulator of lysosomal biogenesis, to promote nuclear translocation of TFEB in LPS-treated macrophages [197]. Interestingly, mutation of this cysteine residue to a serine in macrophage cell lines and in mice inhibits lysosomal biogenesis and antibacterial defense [197]. Additionally, a cell-permeable derivative of itaconate was shown to inhibit Janus kinase 1 (JAK1) activity by alkylating several cysteine residues on JAK1 (Cys 817, 716, & 944), leading to impaired alternative activation of macrophages [198]. Itaconate has also been shown to inhibit glycolysis by modifying specific cysteine residues on several metabolic enzymes, including glyceraldehyde 3-phosphate dehydrogenase (GAPDH) [196], aldolase A (ALDOA) [195], and lactate dehydrogenase A (LDHA) [195]. As glycolysis is a metabolic signature of pro-inflammatory macrophages, this demonstrates the important role of itaconate as a redox modifier acting on multiple immune and metabolic pathways to regulate macrophage inflammatory response (Fig. 5 and Table 4). This is not an exhaustive list of the functional effects of itaconate on macrophage function. Several detailed reviews of itaconate and its cysteine targets have been published [199,200].
Table 4.
Cysteine alkylation targets of itaconate.
| Protein | Cysteine(s) | PTM | Effect | System | Validation | Reference (s) |
|---|---|---|---|---|---|---|
| KEAP1 | 151, 257, 288, 273, & 297 | Alkylation by itaconate derivative | Activate NRF2 | BMDMs | Point mutation (PM) in COS-1 cells (Cys151) | [41] |
| RIPK3 | 360 | Alkylation by itaconate | Activate RIPK3 | RAW264.7 | PM in RAW264.7 | [316] |
| TFEB | 212 (270 in mice) | Alkylation by itaconate | Promote TFEB nuclear translocation | THP-1, iBMDMs, & BMDMs | PM in THP-1, iBMDMs, & mice | [197] |
| JAK1 | 817, 716, & 944 | Alkylation by itaconate derivative | Inhibit JAK1 | THP-1 | No PM validation | [198] |
| LDHA | 84 | Alkylation by itaconate | Inhibit lactate production | BMDMs & RAW264.7 | PM in RAW264.7 | [195] |
| GAPDH | 22 | Alkylation by itaconate | Inhibit glycolysis | BMDMs & RAW264.7 | PM in RAW264.7 | [196] |
| ALDOA | 73, 339 | Alkylation by itaconate | Inhibit glycolysis | BMDMs & RAW264.7 | PM in RAW264.7 | [195] |
| NLRP3 | 548 | Alkylation by itaconate derivative | Inhibit NLRP3 activity and interaction with NEK7 | BMDMs | No PM validation | [193] |
| GSDMD | 77 | Alkylation by itaconate | Activate pyroptosis | BMDMs | No PM validation | [192] |
Fumarate is another metabolite linked to inflammatory macrophage function and ROS production [201,202]. Fumarate, like itaconate, is mildly electrophilic and thereby reacts with thiols in cysteine residues in a PTM termed succination [40]. Many of the protein succination targets of fumarate are shared with the alkylation targets of itaconate including KEAP1 [203], GSDMD [204], and GAPDH [205]. For example, GSDMD Cys192 has been reported to be succinated, albeit these experiments were carried out with monomethyl fumarate as opposed to endogenous fumarate [204] (Fig. 5).
A new role for fumarate in the regulation of cytokine production in macrophages has recently emerged. In response to LPS, the expression of fumarate hydratase (FH), the enzyme converting fumarate to malate, decreases, and flux through the argininosuccinate shunt increases leading to fumarate accumulation [202]. This accumulated fumarate was found to decrease Il10 expression in LPS-activated macrophages, potentially via succination of the transcription factor cFos, and consequently to boost Tnf expression [202]. A second study exploring the effect of FH in kidney cancer identified a potent upregulation of the type I IFN response upon FH loss. Mechanistically, fumarate accumulation in the kidney was found to promote the release of mtDNA in mitochondria-derived vesicles and activation of the cGAS-STING DNA-sensing pathway [201]. Interestingly, in macrophages, FH loss, but not direct fumarate treatment, was shown to promote mtRNA release, retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5) activity to enhance IFN signaling [202]. Precisely how fumarate stimulates mtDNA release remains to be elucidated.
Citrate can also modulate redox signaling in macrophages. LPS-activated macrophages accumulate citrate in the cytoplasm due to inhibition of isocitrate dehydrogenase and an increase in mitochondrial citrate carrier activity [35,206]. Citrate accumulation enhances NO, ROS, and prostaglandin E2 (PGE2) production in macrophages [206,207]. Mechanistically, the conversion of cytosolic citrate to acetyl-CoA generates NADPH, which is a substrate for NOS and NOX enzymes [207,208]. Additionally, acetyl-CoA is an essential metabolite in the arachidonic acid pathway, which is the likely link between citrate and increased production of PGE2 [206].
6. ROS signaling in alternatively-activated macrophages
While ROS are typically associated with an inflammatory response in macrophages, ROS signaling also supports the function of alternatively-activated macrophages. Alternatively-activated macrophages play a critical role in tissue repair and resolution of inflammation but can also exert pro-oncogenic effects (see review [209] for further details). Alternative-activation of macrophages by the anti-inflammatory cytokine IL-4 leads to a decrease in phagosomal activity, NOX2 expression, and decreased ROS production [210]. However, it has also been shown that macrophage polarization to an alternatively- activated state requires NOX-mediated ROS [39,211]. Furthermore, mtROS promote polarization of alveolar macrophages towards an alternatively-activated phenotype in a STAT6-dependent manner during lung fibrosis [212]. A similar phenotype was observed in intestinal macrophages in a pre-clinical model of colitis [213]. However, the mechanistic basis for this remains to be elucidated. ROS also support an alternatively activated, pro-tumorigenic profile in macrophages in cancer. This is discussed below.
7. Regulation of redox signaling by the antioxidant response in macrophages
Phenotypic plasticity is a hallmark of macrophages and enables them to switch between pro- and anti-inflammatory states in response to environmental cues. Many redox modifications in macrophages can be reversed by the antioxidant response. Several studies have shown that both SOD1 and 2 are highly expressed upon TLR stimulation in macrophages [214,215]. Additionally, loss-of-function mutations in SOD1 and 2 exacerbate oxidative stress/ROS production in macrophages while gain-of-function mutations reduce ROS production [[216], [217], [218]]. Similarly, GSH and its associated glutathione peroxidases (GPxs), which use GSH to reduce H2O2 to H2O, regulate intracellular ROS levels and limit the inflammatory response in macrophages [[219], [220], [221]]. Interestingly, both SODs and GPxs are downstream targets of NF-κB, which is regulated by ROS [222]. Taken together, these data suggest that NF-κB and ROS are linked in a feedback loop to prevent a hyperinflammatory response.
NRF2 is an antioxidant transcription factor that regulates macrophage activation by modulating ROS production. Under normal condition, NRF2 is trapped in the cytoplasm by KEAP1, an adaptor protein for the Cullin 3 E3 ubiquitin ligase complex and targeted for proteasomal degradation. Upon oxidative stress, oxidation of several cysteine residues in KEAP1 (including Cys151, Cys273, & Cys288) promotes its dissociation from NRF2. NRF2 subsequently translocates to the nucleus to transcribe target antioxidant genes [223]. Early evidence in a pre-clinical model of sepsis showed that NRF2 deficiency leads to hyperactive macrophages and greater mortality. This is linked with the ability of NRF2 to suppress the NOX pathway, thereby limiting cytosolic ROS production [224,225]. Interestingly, it was recently shown that ROS can also act as an upstream signal to further stabilize NRF2 via oxygen sensors mammalian sterile 20-like kinases 1 and 2 (MST1/2) [226]. This is another example of a feedback loop in place to maintain redox homeostasis. NO is another important regulator of NRF2. Macrophages with impaired NO production have decreased NRF2-dependent gene expression in response to LPS + IFNγ-stimulation [227].
8. The role of ROS and macrophages in disease
8.1. Chronic granulomatous disease
Dysregulation of redox signaling in macrophages has been linked to several pathologies and one of the most best examples is chronic granulomatous disease (CGD). This is a rare immunodeficiency where the anti-microbial response in innate immune cells is compromised. Macrophages from CGD patients are unable to activate phagocytosis, which can result in life-threatening consequences such as bacterial/fungal abscesses in the lung, liver, and spleen [228]. Over 400 mutations linked with CGD have been identified, primarily occurring in the NADPH oxidase complex, including subunits gp91phox, p22phox, p67phox, p47phox, and Rac 2 [229]. These mutations attenuate the NOX pathway, which is critical for phagosomal ROS production, and suppress downstream pro-inflammatory cytokine production in macrophages [[230], [231], [232]]. Furthermore, studies have shown that peripheral blood mononuclear cells (PBMCs) from CGD patients exhibit higher caspase-1 activity and IL-1β secretion in the presence of NOX inhibitors, suggesting that additional factors, independent of ROS, contribute to the pathology of CGD [233,234]. Increased NF-κB activity [235] and mtROS production [236] have also been reported in PBMCs isolated from CGD patients. Although the full scope of the hyperinflammatory phenotype that leads to CGD pathophysiology is still unclear, it has been suggested that a combination of chronic, unresolved infections and intrinsic dysregulation of inflammation are contributing factors [237].
8.2. Inflammatory bowel disease
Infiltration of inflammatory macrophages to the intestine is one of the hallmarks of chronic gastrointestinal diseases such as colitis and Crohn's disease, which are generally known as inflammatory bowel disease (IBD) [238]. Bacterial or viral infections that hyperactivate macrophages and cause oxidative stress/damage to the gastrointestinal tract and can promote IBD [[239], [240], [241]]. Elevated levels of cellular and mitochondrial ROS in PBMCs have been reported in IBD patients [170]. Furthermore, in young children, variants in components of the NOX2 complex, that cause a partial loss of function, have been linked with disease susceptibility [242]. SNP variants of neutrophil cytosolic factor 4, a gene encoding for the p40phox subunit of the NOX complex, have also been associated with an increased risk of IBD [243,244].
8.3. Rheumatoid arthritis
Redox dysfunction in macrophages is one of many hallmarks of sterile inflammatory diseases. For example, rheumatoid arthritis (RA), an autoimmune disease that affects the synovial joints, is characterized by a state of chronic inflammation which has been linked with macrophages with aberrant metabolic and immune function (see review [245] for further details). Tissue-resident macrophages from the synovial fluid of RA patients show a pro-inflammatory signature with high abundance of inflammatory cytokines such as IL-1β and TNF [[246], [247], [248]]. Elevated ROS/RNS production and oxidative stress are commonly present in the synovial tissues of RA patients, suggesting a link between disease pathogenesis and redox dysfunction in macrophages [249,250]. However, direct measurements of ROS/RNS in arthritic macrophages have not been performed. Interestingly, in mouse models of RA, HIF1-α was implicated as an important driver of the inflammatory signature of macrophages [251]. This could be due to the hypoxic environment of the RA synovial joints [252].
8.4. Neuroinflammatory diseases
Neuroinflammatory diseases such as Alzheimer's disease, Parkinson's disease, and multiple sclerosis are characterized by chronic inflammation of the central nervous system, which can promote neuronal degeneration leading to cognitive impairment and motor dysfunction [253]. Oxidative stress has been widely considered as a hallmark of neuroinflammatory diseases (see review [254] for further details). Aberrant activation of the NLRP3 inflammasome has also been reported in Alzheimer's disease patients [255]. Moreover, depletion or pharmacological inhibition of NLRP3 in microglial cells isolated from a mouse model of Alzheimer's has been shown to be protective against disease pathogenesis, suggesting that NLRP3 is one of the drivers of the disease [[255], [256], [257]]. Although a direct link has not been made in patients, analysis in a mouse model of Parkinson's disease has shown a correlation between increased ROS production and NLRP3 activity in microglia upon neurotoxin-induced damage [258]. GSDMD activation has been linked with the hyperinflammatory phenotype observed in peripheral myeloid and microglial cells, and disease progression in the experimental autoimmune encephalomyelitis mouse model of multiple sclerosis [259,260]. With recent findings highlighting the importance of Cys192 of GSDMD in protein function, several inhibitors, including necrosulfonamide, dimethyl fumarate, and disulfiram, have been utilized to inhibit GSDMD activity by targeting Cys192 [178,204,261]. These drugs are being tested in several disease models, including neuroinflammatory diseases (this is described in detail in Ref. [262]).
8.5. Obesity/metabolic disease
Dysfunctional macrophages are also a characteristic of obesity-associated metabolic disease. Obesity is a global epidemic with widespread health consequences including type 2 diabetes (T2D), cancer, non-alcoholic fatty liver disease, and cardiovascular disease. Large numbers of macrophages infiltrate the adipose tissue during obesity [263]. These cells are chronically activated by inflammatory insults such as high levels of free fatty acids [264] and have been extensively linked with obesity-associated inflammation, cytokine production, and progression of T2D [[263], [264], [265], [266], [267], [268]]. The earliest studies to link inflammation and obesity-associated insulin resistance demonstrated that TNF decreases the activity of the insulin receptor [23,269]. Additionally, activation of the NLRP3 inflammasome, which is triggered by ROS, has also been found to contribute to insulin resistance [[270], [271], [272]]. Obesity also promotes Nos2 expression in adipose tissue macrophages [264]. Whether this is merely a marker of inflammatory macrophages or results in increased NO production is unclear. Additionally, myeloid-specific deletion of NOX2 was shown to decrease adiposity, macrophage infiltration to adipose tissue, inflammation, and to improve glucose homeostasis in a model of diet-induced obesity [273]. These studies suggest a link between the pathogenesis of obesity and oxidative stress driven by macrophages. However, it is important to note that the authors did not assess ROS production, nor did they examine the mechanistic basis for these observations. These data all point towards a relationship between dysfunctional redox signaling in macrophages, inflammation, and the pathology of T2D and metabolic disease; however, the precise mechanistic details of this interplay remain to be elucidated.
8.6. Cancer
In the tumor microenvironment, macrophages are reprogrammed to become pro-oncogenic and support cancer cell growth and metastasis, and mediate immunosuppressive effects on adaptive immune cells [274,275]. ROS have been suggested to support this by driving the release of cytokines such as transforming growth factor-β, IL-6, and IL-13 from cancer cells that polarize macrophages to this phenotype [276]. In cancer models of leukemia, lung, breast, liver, and ovarian cancers, either pharmacological inhibition or genetic perturbation (such as NOX knockout) of ROS limits tumorigenesis and the pro-tumor activity of macrophages [39,[277], [278], [279], [280], [281]]. In ovarian and triple negative breast cancers, ROS induction in macrophages has been shown to upregulate programmed death-ligand 1 (PD-L1), which impairs cytotoxic T cell activity, promotes an immunosuppressive phenotype, and increases immune evasion by tumor cells [278,282]. Furthermore, in lung cancer, ROS have been found to be crucial for the differentiation of macrophages to a pro-tumor phenotype [39]. Limiting ROS production decreased the abundance of pro-tumor macrophages and suppressed tumorigenesis in pre-clinical models of lung cancer [39]. In colon cancer, increased ROS production in macrophages leads to DNA damage to intestinal epithelial cells and enhanced tumor-cell adhesion, both of which promote cancer growth [283,284]. Furthermore, RNS promote fibrosis in pancreatic stellate cells, leading to increased collagen deposition to support the progression of pancreatic cancer [285]. In a metastatic breast cancer model, NO can play a pro-tumorigenic role, promoting tumor growth and metastasis in mice [286]. These studies demonstrate a link between redox homeostasis and tumorigenesis, but further data are required to understand the cell and anatomical specificity of this relationship.
9. New approaches and methodology to detect cysteine PTMs
Due to the crucial role of cysteine redox PTMs in diverse cellular functions, many methods have been developed to investigate, characterize, and discover functional cysteines on a proteome-wide scale. Early methods have been developed to computationally predict redox-sensitive cysteines [287] or identify cysteine sites undergoing specific modifications via chemical labeling and mass spectrometry (MS) [[288], [289], [290]]. However, many of these approaches lack the power to determine cysteine modification stoichiometrically.
The development of chemical proteomic tools to quantitatively measure cysteine reactivity has been pioneered by the Cravatt group. The principle of these methods focuses on activity-based protein profiling (ABPP), which is a strategy using specific chemical probes to determine protein functional state in the native proteome based on the protein reactivity towards the probes. Early work by this group determined that some moderately reactive electrophiles can serve as selective ABPP probes for cysteine thiols [291]. This proof-of-concept experiment quickly evolved into a proteome-scale method to quantify cysteine reactivity in native biological systems called isotopic tandem orthogonal proteolysis ABPP (isoTOP-ABPP). This tool combines a cleavable biorthogonal thiol probe and MS to isotopically label, and detect, cysteines on a proteome-wide level in intact cells [292]. The introduction of isoTOP-ABPP has marked the first time that redox-sensitive cysteines can be intrinsically characterized in a native biological system. Many derivatives of isoTOP-ABPP have been utilized to profile reactive cysteines in various redox contexts including cysteine targets of hydrogen peroxide [293], lipid peroxidation [294], sulfenylation [295], S-nitrosylation [296], and phosphorylation-dependent cysteine redox PTMs [297]. Drug discovery is perhaps the most powerful application of isoTOP-ABPP. In particular, this method has been applied to identify cysteine targets of small-molecule covalent drugs [[298], [299], [300]] or to discover targetable cysteine sites in “undruggable” proteins [301]. Many efforts have been made to improve the cysteine coverage and scalability of the original method. As of now, isoTOP-ABPP can be multiplexed to cover up to 17,000 cysteines [302] and streamlined for screening of large ligand libraries (∼300 small-molecule drugs) [[302], [303], [304]].
A caveat of these ABPP probes is that they are designed to only label free cysteines that are reactive and, therefore, do not represent the complete cysteine redox landscape. To overcome this limitation, many methods have been developed to measure PTMs on redox-sensitive cysteines. Various proteomics approaches have been developed to stoichiometrically quantify cysteine PTMs in cells, including OxMRM [305], OxiTMT [306], RacTMT [307], and OxICAT [308,309]. Such stoichiometric assessment provides important information about the functional significance of a PTM on any given cysteine. Notably, OxICAT can also be combined with a proximity labeling technique called TurboID to explore the cysteine proteome at a subcellular level. Two recent independent studies have used this approach to study redox-sensitive cysteines in various cellular compartments in macrophages [310,311]. One study identified ∼300 highly oxidized cysteines out of 599 unique cytosolic cysteine-containing peptides detected in LPS-stimulated Raw 264.7 macrophages [310]. The other study identified 32 highly oxidized cysteines out of 559 unique mitochondrial cysteine peptides detected in LPS + IFNγ-treated immortalized BMDMs (iBMDMs) [311]. These data suggest that the redox proteome of inflammatory macrophages is regulated at the subcellular level. However, a significant drawback of these methods is the relatively low proteome coverage, with ∼4300 cysteine-containing peptides quantified [308]. A recent method by the Chouchani group introduced a novel molecular label called the cysteine-reactive phosphate tag (CPT). CPT contains an iodoacetamide moiety to label cysteines and a phosphate group that allows enrichment of labelled peptide [312]. This enrichment process takes advantage of an established pipeline from the phosphoproteomics field that uses immobilized metal affinity chromatography (IMAC) to separate phosphopeptides from the proteome [313,314]. The CPT method can quantify the % reversible modification of ∼34,000 unique cysteine sites, an exponential coverage boost compared with other tools [312]. CPT can be extended to facilitate covalent drug discovery. For example, CPT has been applied in a drug screening study to discover a selective covalent inhibitor that targets an active site cysteine in creatine kinases [315].
9.1. Methods to measure PTM by itaconate
As an important cysteine PTM in macrophages, methods to examine cysteine alkylation mediated by itaconate have been developed. One of these methods is adopted from isoTOP-ABPP to measure cysteine reactivity and tailored specifically to detect itaconate alkylation by using a customized thiol-reactive itaconate-competitive probe (1-OH-Az) [195]. Using this probe, Qin and colleagues. detected 412 cysteines that were sensitive to itaconate treatment out of 766 quantified cysteines. They identified Cys73 and 339 on ALDOA and Cys84 on LDHA as targets of itaconate. Itaconate-mediated alkylation at these sites was found to inhibit enzymatic activity of ALDOA and LDHA [195]. This approach provides a unique platform to deconvolute functional cysteine targets specific to itaconate. However, like the parental isoTOP-ABPP approach, it has a limited coverage of the cysteine proteome.
Another method utilizes an alkyne analogue of the cell-permeable itaconate derivative 4-octyl itaconate (4OI) termed ITalk [316]. ITalk alkylates cysteine targets of itaconate while the alkyne group of the probe allows for enrichment of labelled sites. Using this approach, 1926 proteins were detected that contained residues modified by the itaconate derivative. From this dataset, the authors identified, among other targets, that itaconate alkylates Cys360 on receptor-interacting serine/threonine-protein kinase 3 (RIPK3). RIPK3 activation is essential for the induction of a form of programmed cell death called necroptosis. Modification of Cys360 was shown to be required for RIPK3 phosphorylation and activation by itaconate [316]. Precisely how alkylation of Cys360 by itaconate promotes its activity remains to be determined. Although ITalk can significantly increase cysteine coverage, it is important to note that the cysteine reactivity of the probe is likely to over-represent that of endogenous itaconate.
10. Conclusions/Future directions
The essential and versatile functions of redox signaling in many cellular processes including the regulation of inflammatory macrophages are indisputable. Moreover, ROS and RNS have been evidenced by numerous studies to be much more than non-discriminatory damaging molecules, but rather to form an intricate signaling network that modulates protein function through redox signaling. Cysteine, one of the least abundant amino acids, is the hub of redox signaling as the primary target for PTM by ROS, RNS, and metabolites.
Macrophages have an intimate link with redox signaling, with a plethora of evidence demonstrating how cysteine PTMs alter the inflammatory function of these cells. Despite the extensive developments made in the field of redox signaling in the last 30 years, there are still several challenges and questions that remain. For instance, it is still unclear what dictates the form of PTM (oxidation, S-nitrosylation, glutathionylation, alkylation, etc.) to which a cysteine is subjected. This could simply be a substrate concentration-mediated effect or something more intricate. An example of this is GSDMD Cys192 which is subject to oxidation, succination, and palmitoylation [[175], [176], [177],192,204]. These distinct PTMs are likely to alter protein function, activity, and interactions in diverse ways. Indeed, oxidation and palmitoylation of Cys192 enhance GSDMD activity while succination represses GSDMD activity. The relative contributions and temporal regulation of these PTMs remains to be established. It should be noted however, that the approaches used to identify modification of GSDMD Cys192 vary widely, and some allow for stoichiometric determination of cysteine engagement while others do not. As such, it is difficult to determine unequivocally the relative role for each PTM in the regulation of GSDMD activity.
Furthermore, it is poorly understood what defines cysteine thiol redox sensitivity. Using the Oximouse dataset, a correlation between the local electrostatic environment and highly modified cysteines was identified [312]. More specifically, there was a selection against proximal acidic amino acids and a selection for the basic amino acid arginine for a cysteine to be highly oxidized. It is thought that the positively-charged side chain of arginine can stabilize the negatively- charged thiolate which is known to be more reactive than its corresponding thiol. The opposite is true for negatively-charged amino acids. Intriguingly, the same is true for proximal phosphogroups. As such, a local phosphorylated amino acid antagonizes cysteine oxidation.
The potential reactivity of a cysteine may also be dictated by the intrinsic electrostatic state of the cysteinyl residue where the thiolate (-S-) form is a powerful nucleophile and highly susceptible to oxidation [94]. As the formation of thiol vs thiolate depends on pH, it is probable that the local pH in cells can influence cysteine reactivity. This could be particularly relevant for inflammatory macrophages which are highly glycolytic and therefore are likely to have a decrease in intracellular pH upon activation. The probability for a cysteine to be modified could also depend on the proximity of the target protein to the site of ROS or metabolite production. On the other hand, target proteins could be recruited to the site of ROS/metabolite production. These questions warrant further structural and biochemical investigation to fully understand the regulatory landscape of redox signaling in macrophages.
Finally, functional validation of potential redox-sensitive cysteines in a native cell system should become a gold standard to determine the biological relevance of a newly identified redox signal/target. There is a persistent lack of these essential validation experiments in many studies, especially in macrophages. Functional validation of a specific cysteine residue involves endogenous mutation of the target cysteine to a different amino acid (most often alanine or serine). This is now attainable, both in vivo and in vitro, via genome editing technology such as CRISPR-Cas9 and prime editing [317,318]. However, this is still extremely challenging in macrophages for several reasons: 1) low transfection efficiency, 2) low cell survival due to incompatible delivery systems, and 3) low editing efficiency due to impaired DNA double-strand break repair [319]. These approaches, in combination with the global stoichiometric assessment of the redox proteome using new techniques described above, are likely to significantly advance our knowledge of redox signaling in macrophages in years to come.
CRediT authorship contribution statement
Nhien Tran: Writing – review & editing, Writing – original draft. Evanna L. Mills: Writing – review & editing, Writing – original draft.
Declaration of competing interest
None.
Acknowledgements
Images were created with Biorender.com. This work was supported by NIH-NIDKK (4R00 DK123321-04), The Mathers Foundation (MF-2204-02617), Claudia Adams Barr Program in Innovative Basic Cancer Research, American Cancer Society (DBG-23-983219-01-TBE), Parker Institute for Cancer Immunotherapy, DF/HCC SPORE in Breast Cancer (1P50CA168504-08), DFCI Innovations Research Fund, the Anna Fuller Fund, and the Chan Zuckerberg Institute.
Data availability
No data was used for the research described in the article.
References
- 1.Forman H.J., Ursini F., Maiorino M. An overview of mechanisms of redox signaling. J. Mol. Cell. Cardiol. 2014;73:2–9. doi: 10.1016/j.yjmcc.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCord J.M., Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J. Biol. Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 3.Bae Y.S., et al. Epidermal growth factor (EGF)-induced generation of hydrogen peroxide. Role in EGF receptor-mediated tyrosine phosphorylation. J. Biol. Chem. 1997;272:217–221. doi: 10.1074/jbc.272.1.217. [DOI] [PubMed] [Google Scholar]
- 4.Beltran B., Orsi A., Clementi E., Moncada S. Oxidative stress and S-nitrosylation of proteins in cells. Br. J. Pharmacol. 2000;129:953–960. doi: 10.1038/sj.bjp.0703147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sundaresan M., Yu Z.X., Ferrans V.J., Irani K., Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 6.Zheng M., Aslund F., Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 7.Woo H.A., et al. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science. 2003;300:653–656. doi: 10.1126/science.1080273. [DOI] [PubMed] [Google Scholar]
- 8.Chandel N.S., et al. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. U. S. A. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandel N.S., Vander Heiden M.G., Thompson C.B., Schumacker P.T. Redox regulation of p53 during hypoxia. Oncogene. 2000;19:3840–3848. doi: 10.1038/sj.onc.1203727. [DOI] [PubMed] [Google Scholar]
- 10.Chandel N.S., Trzyna W.C., McClintock D.S., Schumacker P.T. Role of oxidants in NF-kappa B activation and TNF-alpha gene transcription induced by hypoxia and endotoxin. J. Immunol. 2000;165:1013–1021. doi: 10.4049/jimmunol.165.2.1013. [DOI] [PubMed] [Google Scholar]
- 11.Nemoto S., Takeda K., Yu Z.X., Ferrans V.J., Finkel T. Role for mitochondrial oxidants as regulators of cellular metabolism. Mol. Cell Biol. 2000;20:7311–7318. doi: 10.1128/MCB.20.19.7311-7318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J.W., Helmann J.D. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 13.Lou Y.W., et al. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. FEBS J. 2008;275:69–88. doi: 10.1111/j.1742-4658.2007.06173.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y.Y., et al. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J. Biol. Chem. 2008;283:35265–35272. doi: 10.1074/jbc.M805287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canet-Aviles R.M., et al. The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackinton J., et al. Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1. J. Biol. Chem. 2009;284:6476–6485. doi: 10.1074/jbc.M806599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park M.D., Silvin A., Ginhoux F., Merad M. Macrophages in health and disease. Cell. 2022;185:4259–4279. doi: 10.1016/j.cell.2022.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Odegaard J.I., et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paolicelli R.C., et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 20.Klein I., et al. Kupffer cell heterogeneity: functional properties of bone marrow derived and sessile hepatic macrophages. Blood. 2007;110:4077–4085. doi: 10.1182/blood-2007-02-073841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carey B., Trapnell B.C. The molecular basis of pulmonary alveolar proteinosis. Clin. Immunol. 2010;135:223–235. doi: 10.1016/j.clim.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohyama M., et al. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 2009;457:318–321. doi: 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hotamisligil G.S., Shargill N.S., Spiegelman B.M. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 24.Guillot-Sestier M.V., et al. Microglial metabolism is a pivotal factor in sexual dimorphism in Alzheimer's disease. Commun. Biol. 2021;4:711. doi: 10.1038/s42003-021-02259-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills E.L., et al. UCP1 governs liver extracellular succinate and inflammatory pathogenesis. Nat. Metab. 2021;3:604–617. doi: 10.1038/s42255-021-00389-5. Epub 2021 May 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ioannou M., et al. Microbe capture by splenic macrophages triggers sepsis via T cell-death-dependent neutrophil lifespan shortening. Nat. Commun. 2022;13:4658. doi: 10.1038/s41467-022-32320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matzinger P. Tolerance, danger, and the extended family. Annu. Rev. Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 28.Alvarez-Dominguez C., Carrasco-Marin E., Lopez-Mato P., Leyva-Cobian F. The contribution of both oxygen and nitrogen intermediates to the intracellular killing mechanisms of C1q-opsonized Listeria monocytogenes by the macrophage-like IC-21 cell line. Immunology. 2000;101:83–89. doi: 10.1046/j.1365-2567.2000.00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utermöhlen O., Karow U., Löhler J.r., Krönke M. Severe impairment in early host defense against Listeria monocytogenes in mice deficient in acid sphingomyelinase 1. J. Immunol. 2003;170:2621–2628. doi: 10.4049/jimmunol.170.5.2621. [DOI] [PubMed] [Google Scholar]
- 30.Herb M., et al. Mitochondrial reactive oxygen species enable proinflammatory signaling through disulfide linkage of NEMO. Sci. Signal. 2019;12:eaar5926. doi: 10.1126/scisignal.aar5926. [DOI] [PubMed] [Google Scholar]
- 31.Dostert C., et al. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allan E.R., et al. NADPH oxidase modifies patterns of MHC class II-restricted epitopic repertoires through redox control of antigen processing. J. Immunol. 2014;192:4989–5001. doi: 10.4049/jimmunol.1302896. [DOI] [PubMed] [Google Scholar]
- 33.Hansen A.L., et al. Nitro-fatty acids are formed in response to virus infection and are potent inhibitors of STING palmitoylation and signaling. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E7768–E7775. doi: 10.1073/pnas.1806239115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borzecka-Solarz K., et al. Association of Lyn kinase with membrane rafts determines its negative influence on LPS-induced signaling. Mol. Biol. Cell. 2017;28:1147–1159. doi: 10.1091/mbc.E16-09-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jha A.K., et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Garaude J., et al. Mitochondrial respiratory-chain adaptations in macrophages contribute to antibacterial host defense. Nat. Immunol. 2016;17:1037–1045. doi: 10.1038/ni.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mills E.L., et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016;167:457–470 e413. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li F., et al. Regulation of HIF-1alpha stability through S-nitrosylation. Mol. Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., et al. ROS play a critical role in the differentiation of alternatively activated macrophages and the occurrence of tumor-associated macrophages. Cell Res. 2013;23:898–914. doi: 10.1038/cr.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adam J., et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20:524–537. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mills E.L., et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holmstrom K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 43.Bedard K., Krause K.H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 44.Babior B.M., Curnutte J.T., Kipnes B.S. Pyridine nucleotide-dependent superoxide production by a cell-free system from human granulocytes. J. Clin. Invest. 1975;56:1035–1042. doi: 10.1172/JCI108150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reczek C.R., Chandel N.S. ROS-dependent signal transduction. Curr. Opin. Cell Biol. 2015;33:8–13. doi: 10.1016/j.ceb.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sies H., Jones D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020;21:363–383. doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- 48.Panday A., Sahoo M.K., Osorio D., Batra S. NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell. Mol. Immunol. 2015;12:5–23. doi: 10.1038/cmi.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee C.F., Qiao M., Schroder K., Zhao Q., Asmis R. Nox4 is a novel inducible source of reactive oxygen species in monocytes and macrophages and mediates oxidized low density lipoprotein-induced macrophage death. Circ. Res. 2010;106:1489–1497. doi: 10.1161/CIRCRESAHA.109.215392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franchini A.M., Hunt D., Melendez J.A., Drake J.R. FcgammaR-driven release of IL-6 by macrophages requires NOX2-dependent production of reactive oxygen species. J. Biol. Chem. 2013;288:25098–25108. doi: 10.1074/jbc.M113.474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bylund J., Campsall P.A., Ma R.C., Conway B.A., Speert D.P. Burkholderia cenocepacia induces neutrophil necrosis in chronic granulomatous disease. J. Immunol. 2005;174:3562–3569. doi: 10.4049/jimmunol.174.6.3562. [DOI] [PubMed] [Google Scholar]
- 52.Sim S., et al. NADPH oxidase-derived reactive oxygen species-mediated activation of ERK1/2 is required for apoptosis of human neutrophils induced by Entamoeba histolytica. J. Immunol. 2005;174:4279–4288. doi: 10.4049/jimmunol.174.7.4279. [DOI] [PubMed] [Google Scholar]
- 53.Han D., Antunes F., Canali R., Rettori D., Cadenas E. Voltage-dependent anion channels control the release of the superoxide anion from mitochondria to cytosol. J. Biol. Chem. 2003;278:5557–5563. doi: 10.1074/jbc.M210269200. [DOI] [PubMed] [Google Scholar]
- 54.Turrens J.F., Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem. J. 1980;191:421–427. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quinlan C.L., et al. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J. Biol. Chem. 2012;287:27255–27264. doi: 10.1074/jbc.M112.374629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muller F.L., Liu Y., Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J. Biol. Chem. 2004;279:49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- 57.Bleier L., et al. Generator-specific targets of mitochondrial reactive oxygen species. Free Radic. Biol. Med. 2015;78:1–10. doi: 10.1016/j.freeradbiomed.2014.10.511. [DOI] [PubMed] [Google Scholar]
- 58.Vane J.R., et al. Inducible isoforms of cyclooxygenase and nitric-oxide synthase in inflammation. Proc. Natl. Acad. Sci. U. S. A. 1994;91:2046–2050. doi: 10.1073/pnas.91.6.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McNally J.S., Saxena A., Cai H., Dikalov S., Harrison D.G. Regulation of xanthine oxidoreductase protein expression by hydrogen peroxide and calcium. Arterioscler. Thromb. Vasc. Biol. 2005;25:1623–1628. doi: 10.1161/01.ATV.0000170827.16296.6e. [DOI] [PubMed] [Google Scholar]
- 60.Veith A., Moorthy B. Role of cytochrome P450s in the generation and metabolism of reactive oxygen species. Curr Opin Toxicol. 2018;7:44–51. doi: 10.1016/j.cotox.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Byrne R.S., Hansch R., Mendel R.R., Hille R. Oxidative half-reaction of arabidopsis thaliana sulfite oxidase: generation of superoxide by a peroxisomal enzyme. J. Biol. Chem. 2009;284:35479–35484. doi: 10.1074/jbc.M109.067355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.del Rio L.A., et al. Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J. Exp. Bot. 2002;53:1255–1272. doi: 10.1093/jexbot/53.372.1255. [DOI] [PubMed] [Google Scholar]
- 63.Harding H.P., et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 64.Fang F.C. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 65.Palmieri E.M., McGinity C., Wink D.A., McVicar D.W. Nitric oxide in macrophage immunometabolism: hiding in plain sight. Metabolites. 2020;10 doi: 10.3390/metabo10110429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burke S.J., et al. Regulation of iNOS gene transcription by IL-1β and IFN-γ requires a coactivator exchange mechanism. Mol. Endocrinol. 2013;27:1724–1742. doi: 10.1210/me.2013-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kamijo R., et al. Requirement for transcription factor IRF-1 in NO synthase induction in macrophages. Science. 1994;263:1612–1615. doi: 10.1126/science.7510419. [DOI] [PubMed] [Google Scholar]
- 68.Xie Q.W., Kashiwabara Y., Nathan C. Role of transcription factor NF-kappa B/Rel in induction of nitric oxide synthase. J. Biol. Chem. 1994;269:4705–4708. doi: 10.1016/S0021-9258(17)37600-7. [DOI] [PubMed] [Google Scholar]
- 69.Vazquez-Torres A., Jones-Carson J., Mastroeni P., Ischiropoulos H., Fang F.C. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J. Exp. Med. 2000;192:227–236. doi: 10.1084/jem.192.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iovine N.M., et al. Reactive nitrogen species contribute to innate host defense against Campylobacter jejuni. Infect. Immun. 2008;76:986–993. doi: 10.1128/IAI.01063-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vijayan V., et al. Human and murine macrophages exhibit differential metabolic responses to lipopolysaccharide - a divergent role for glycolysis. Redox Biol. 2019;22 doi: 10.1016/j.redox.2019.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weinberg J.B., et al. Human mononuclear phagocyte inducible nitric oxide synthase (iNOS): analysis of iNOS mRNA, iNOS protein, biopterin, and nitric oxide production by blood monocytes and peritoneal macrophages. Blood. 1995;86:1184–1195. doi: 10.1182/blood.V86.3.1184.1184. [DOI] [PubMed] [Google Scholar]
- 73.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang B.H., Rue E., Wang G.L., Roe R., Semenza G.L. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 1996;271:17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- 75.Chandel N.S., et al. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J. Biol. Chem. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 76.Guzy R.D., et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metabol. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 77.Pan Y., et al. Multiple factors affecting cellular redox status and energy metabolism modulate hypoxia-inducible factor prolyl hydroxylase activity in vivo and in vitro. Mol. Cell Biol. 2007;27:912–925. doi: 10.1128/MCB.01223-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qutub A.A., Popel A.S. Reactive oxygen species regulate hypoxia-inducible factor 1alpha differentially in cancer and ischemia. Mol. Cell Biol. 2008;28:5106–5119. doi: 10.1128/MCB.00060-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang J., et al. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat. Cell Biol. 2013;15:1186–1196. doi: 10.1038/ncb2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kang Y., Liu R., Wu J.-X., Chen L. Structural insights into the mechanism of human soluble guanylate cyclase. Nature. 2019;574:206–210. doi: 10.1038/s41586-019-1584-6. [DOI] [PubMed] [Google Scholar]
- 81.Wink D.A., et al. Inhibition of cytochromes P450 by nitric oxide and a nitric oxide-releasing agent. Arch. Biochem. Biophys. 1993;300:115–123. doi: 10.1006/abbi.1993.1016. [DOI] [PubMed] [Google Scholar]
- 82.Cleeter M.W., Cooper J.M., Darley-Usmar V.M., Moncada S., Schapira A.H. Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide. Implications for neurodegenerative diseases. FEBS Lett. 1994;345:50–54. doi: 10.1016/0014-5793(94)00424-2. [DOI] [PubMed] [Google Scholar]
- 83.Brown G.C., Cooper C.E. Nanomolar concentrations of nitric oxide reversibly inhibit synaptosomal respiration by competing with oxygen at cytochrome oxidase. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1994;356:295–298. doi: 10.1016/0014-5793(94)01290-3. [DOI] [PubMed] [Google Scholar]
- 84.Abu-Soud H.M., Ichimori K., Nakazawa H., Stuehr D.J. Regulation of inducible nitric oxide synthase by self-generated NO. Biochemistry. 2001;40:6876–6881. doi: 10.1021/bi010066m. [DOI] [PubMed] [Google Scholar]
- 85.Abu-Soud H.M., et al. Neuronal nitric oxide synthase self-inactivates by forming a ferrous-nitrosyl complex during aerobic catalysis. J. Biol. Chem. 1995;270:22997–23006. doi: 10.1074/jbc.270.39.22997. [DOI] [PubMed] [Google Scholar]
- 86.Poulos T.L. Heme enzyme structure and function. Chem. Rev. 2014;114:3919–3962. doi: 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chouchani E.T., et al. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat. Med. 2013;19:753–759. doi: 10.1038/nm.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Welter R., Yu L., Yu C.-A. The effects of nitric oxide on electron transport complexes. Arch. Biochem. Biophys. 1996;331:9–14. doi: 10.1006/abbi.1996.0276. [DOI] [PubMed] [Google Scholar]
- 89.Palmieri E.M., et al. Nitric oxide orchestrates metabolic rewiring in M1 macrophages by targeting aconitase 2 and pyruvate dehydrogenase. Nat. Commun. 2020;11:698. doi: 10.1038/s41467-020-14433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Read A.D., Bentley R.E., Archer S.L., Dunham-Snary K.J. Mitochondrial iron-sulfur clusters: structure, function, and an emerging role in vascular biology. Redox Biol. 2021;47 doi: 10.1016/j.redox.2021.102164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Drapier J.C., Hibbs J.B., Jr. Differentiation of murine macrophages to express nonspecific cytotoxicity for tumor cells results in L-arginine-dependent inhibition of mitochondrial iron-sulfur enzymes in the macrophage effector cells. J. Immunol. 1988;140:2829–2838. [PubMed] [Google Scholar]
- 92.Drapier J.C., Hibbs J.B., Jr. Murine cytotoxic activated macrophages inhibit aconitase in tumor cells. Inhibition involves the iron-sulfur prosthetic group and is reversible. J. Clin. Invest. 1986;78:790–797. doi: 10.1172/jci112642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lundberg J.O., Weitzberg E. Nitric oxide signaling in health and disease. Cell. 2022;185:2853–2878. doi: 10.1016/j.cell.2022.06.010. [DOI] [PubMed] [Google Scholar]
- 94.Poole L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015;80:148–157. doi: 10.1016/j.freeradbiomed.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Anastasiou D., et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gravina S.A., Mieyal J.J. Thioltransferase is a specific glutathionyl mixed disulfide oxidoreductase. Biochemistry. 1993;32:3368–3376. doi: 10.1021/bi00064a021. [DOI] [PubMed] [Google Scholar]
- 97.Prinarakis E., Chantzoura E., Thanos D., Spyrou G. S-glutathionylation of IRF3 regulates IRF3-CBP interaction and activation of the IFN beta pathway. EMBO J. 2008;27:865–875. doi: 10.1038/emboj.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mitchell D.A., Vasudevan A., Linder M.E., Deschenes R.J. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 2006;47:1118–1127. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- 99.Tomatis V.M., Trenchi A., Gomez G.A., Daniotti J.L. Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duncan J.A., Gilman A.G. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS) J. Biol. Chem. 1998;273:15830–15837. doi: 10.1074/jbc.273.25.15830. [DOI] [PubMed] [Google Scholar]
- 101.Camp L.A., Hofmann S.L. Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J. Biol. Chem. 1993;268:22566–22574. [PubMed] [Google Scholar]
- 102.Soyombo A.A., Hofmann S.L. Molecular cloning and expression of palmitoyl-protein thioesterase 2 (PPT2), a homolog of lysosomal palmitoyl-protein thioesterase with a distinct substrate specificity. J. Biol. Chem. 1997;272:27456–27463. doi: 10.1074/jbc.272.43.27456. [DOI] [PubMed] [Google Scholar]
- 103.Abrami L., et al. Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains. Nat. Chem. Biol. 2021;17:438–447. doi: 10.1038/s41589-021-00753-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hirai A., et al. Geranylgeranylated rho small GTPase(s) are essential for the degradation of p27Kip1 and facilitate the progression from G1 to S phase in growth-stimulated rat FRTL-5 cells. J. Biol. Chem. 1997;272:13–16. doi: 10.1074/jbc.272.1.13. [DOI] [PubMed] [Google Scholar]
- 105.Molnar G., Dagher M.C., Geiszt M., Settleman J., Ligeti E. Role of prenylation in the interaction of Rho-family small GTPases with GTPase activating proteins. Biochemistry. 2001;40:10542–10549. doi: 10.1021/bi011158e. [DOI] [PubMed] [Google Scholar]
- 106.Roberts P.J., et al. Rho Family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J. Biol. Chem. 2008;283:25150–25163. doi: 10.1074/jbc.M800882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Farnsworth C.C., Seabra M.C., Ericsson L.H., Gelb M.H., Glomset J.A. Rab geranylgeranyl transferase catalyzes the geranylgeranylation of adjacent cysteines in the small GTPases Rab1A, Rab3A, and Rab5A. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11963–11967. doi: 10.1073/pnas.91.25.11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.West A.P., Shadel G.S., Ghosh S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011;11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mills E.L., Kelly B., O'Neill L.A.J. Mitochondria are the powerhouses of immunity. Nat. Immunol. 2017;18:488–498. doi: 10.1038/ni.3704. [DOI] [PubMed] [Google Scholar]
- 110.Nauseef W.M. The phagocyte NOX2 NADPH oxidase in microbial killing and cell signaling. Curr. Opin. Immunol. 2019;60:130–140. doi: 10.1016/j.coi.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bae Y.S., et al. Macrophages generate reactive oxygen species in response to minimally oxidized low-density lipoprotein: toll-like receptor 4- and spleen tyrosine kinase-dependent activation of NADPH oxidase 2. Circ. Res. 2009;104:210–218. doi: 10.1161/CIRCRESAHA.108.181040. 221pp. following 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim S.Y., et al. Pro-inflammatory hepatic macrophages generate ROS through NADPH oxidase 2 via endocytosis of monomeric TLR4-MD2 complex. Nat. Commun. 2017;8:2247. doi: 10.1038/s41467-017-02325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sanjuan M.A., et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 114.Huang J., et al. Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6226–6231. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gluschko A., et al. The beta(2) integrin mac-1 induces protective LC3-associated phagocytosis of Listeria monocytogenes. Cell Host Microbe. 2018;23:324–337 e325. doi: 10.1016/j.chom.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 116.Vouldoukis I., et al. The killing of Leishmania major by human macrophages is mediated by nitric oxide induced after ligation of the Fc epsilon RII/CD23 surface antigen. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7804–7808. doi: 10.1073/pnas.92.17.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mastroeni P., et al. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J. Exp. Med. 2000;192:237–248. doi: 10.1084/jem.192.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Muñoz-Fernández M.A., Fernández M.A., Fresno M. Activation of human macrophages for the killing of intracellular Trypanosoma cruzi by TNF-alpha and IFN-gamma through a nitric oxide-dependent mechanism. Immunol. Lett. 1992;33:35–40. doi: 10.1016/0165-2478(92)90090-b. [DOI] [PubMed] [Google Scholar]
- 119.Geng J., et al. Kinases Mst1 and Mst2 positively regulate phagocytic induction of reactive oxygen species and bactericidal activity. Nat. Immunol. 2015;16:1142–1152. doi: 10.1038/ni.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pearce E.L., Pearce E.J. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rodriguez-Prados J.C., et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J. Immunol. 2010;185:605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- 122.Tannahill G.M., et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Taniguchi K., Karin M. NF-kappaB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol. 2018;18:309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 124.Xu P., Huecksteadt T.P., Hoidal J.R. Molecular cloning and characterization of the human xanthine dehydrogenase gene (XDH) Genomics. 1996;34:173–180. doi: 10.1006/geno.1996.0262. [DOI] [PubMed] [Google Scholar]
- 125.Li Y., et al. Regulation of neuronal nitric oxide synthase exon 1f gene expression by nuclear factor-kappaB acetylation in human neuroblastoma cells. J. Neurochem. 2007;101:1194–1204. doi: 10.1111/j.1471-4159.2006.04407.x. [DOI] [PubMed] [Google Scholar]
- 126.Deng W.G., Zhu Y., Wu K.K. Up-regulation of p300 binding and p50 acetylation in tumor necrosis factor-alpha-induced cyclooxygenase-2 promoter activation. J. Biol. Chem. 2003;278:4770–4777. doi: 10.1074/jbc.M209286200. [DOI] [PubMed] [Google Scholar]
- 127.Anrather J., Racchumi G., Iadecola C. NF-kappaB regulates phagocytic NADPH oxidase by inducing the expression of gp91phox. J. Biol. Chem. 2006;281:5657–5667. doi: 10.1074/jbc.M506172200. [DOI] [PubMed] [Google Scholar]
- 128.Xie Q.W., Kashiwabara Y., Nathan C. Role of transcription factor NF-kappa B/Rel in induction of nitric oxide synthase. J. Biol. Chem. 1994;269:4705–4708. [PubMed] [Google Scholar]
- 129.Jones P.L., Ping D., Boss J.M. Tumor necrosis factor alpha and interleukin-1beta regulate the murine manganese superoxide dismutase gene through a complex intronic enhancer involving C/EBP-beta and NF-kappaB. Mol. Cell Biol. 1997;17:6970–6981. doi: 10.1128/MCB.17.12.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rojo A.I., Salinas M., Martin D., Perona R., Cuadrado A. Regulation of Cu/Zn-superoxide dismutase expression via the phosphatidylinositol 3 kinase/Akt pathway and nuclear factor-kappaB. J. Neurosci. 2004;24:7324–7334. doi: 10.1523/JNEUROSCI.2111-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schreiber J., et al. Coordinated binding of NF-kappaB family members in the response of human cells to lipopolysaccharide. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5899–5904. doi: 10.1073/pnas.0510996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Djavaheri-Mergny M., Javelaud D., Wietzerbin J., Besancon F. NF-kappaB activation prevents apoptotic oxidative stress via an increase of both thioredoxin and MnSOD levels in TNFalpha-treated Ewing sarcoma cells. FEBS Lett. 2004;578:111–115. doi: 10.1016/j.febslet.2004.10.082. [DOI] [PubMed] [Google Scholar]
- 133.Kairisalo M., Korhonen L., Blomgren K., Lindholm D. X-linked inhibitor of apoptosis protein increases mitochondrial antioxidants through NF-kappaB activation. Biochem. Biophys. Res. Commun. 2007;364:138–144. doi: 10.1016/j.bbrc.2007.09.115. [DOI] [PubMed] [Google Scholar]
- 134.Toledano M.B., Leonard W.J. Modulation of transcription factor NF-kappa B binding activity by oxidation-reduction in vitro. Proc. Natl. Acad. Sci. U. S. A. 1991;88:4328–4332. doi: 10.1073/pnas.88.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Matthews J.R., Kaszubska W., Turcatti G., Wells T.N., Hay R.T. Role of cysteine 62 in DNA recognition by the P50 subunit of NF-kappa B. Nucleic Acids Res. 1993;21:1727–1734. doi: 10.1093/nar/21.8.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Matthews J.R., Wakasugi N., Virelizier J.L., Yodoi J., Hay R.T. Thioredoxin regulates the DNA binding activity of NF-kappa B by reduction of a disulphide bond involving cysteine 62. Nucleic Acids Res. 1992;20:3821–3830. doi: 10.1093/nar/20.15.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pineda-Molina E., et al. Glutathionylation of the p50 subunit of NF-kappaB: a mechanism for redox-induced inhibition of DNA binding. Biochemistry. 2001;40:14134–14142. doi: 10.1021/bi011459o. [DOI] [PubMed] [Google Scholar]
- 138.Matthews J.R., Botting C.H., Panico M., Morris H.R., Hay R.T. Inhibition of NF-kappaB DNA binding by nitric oxide. Nucleic Acids Res. 1996;24:2236–2242. doi: 10.1093/nar/24.12.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kelleher Z.T., Matsumoto A., Stamler J.S., Marshall H.E. NOS2 regulation of NF-kappaB by S-nitrosylation of p65. J. Biol. Chem. 2007;282:30667–30672. doi: 10.1074/jbc.M705929200. [DOI] [PubMed] [Google Scholar]
- 140.Marshall H.E., Stamler J.S. Inhibition of NF-kappa B by S-nitrosylation. Biochemistry. 2001;40:1688–1693. doi: 10.1021/bi002239y. [DOI] [PubMed] [Google Scholar]
- 141.Sen N., et al. Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol. Cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reynaert N.L., et al. Dynamic redox control of NF-kappaB through glutaredoxin-regulated S-glutathionylation of inhibitory kappaB kinase beta. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13086–13091. doi: 10.1073/pnas.0603290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Herscovitch M., et al. Intermolecular disulfide bond formation in the NEMO dimer requires Cys54 and Cys 347. Biochem. Biophys. Res. Commun. 2008;367:103–108. doi: 10.1016/j.bbrc.2007.12.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schoonbroodt S., et al. Crucial role of the amino-terminal tyrosine residue 42 and the carboxyl-terminal PEST domain of I kappa B alpha in NF-kappa B activation by an oxidative stress. J. Immunol. 2000;164:4292–4300. doi: 10.4049/jimmunol.164.8.4292. [DOI] [PubMed] [Google Scholar]
- 145.Takada Y., et al. Hydrogen peroxide activates NF-kappa B through tyrosine phosphorylation of I kappa B alpha and serine phosphorylation of p65: evidence for the involvement of I kappa B alpha kinase and Syk protein-tyrosine kinase. J. Biol. Chem. 2003;278:24233–24241. doi: 10.1074/jbc.M212389200. [DOI] [PubMed] [Google Scholar]
- 146.Ayala A., Munoz M.F., Arguelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014;2014 doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Esterbauer H., Cheeseman K.H. Determination of aldehydic lipid peroxidation products: malonaldehyde and 4-hydroxynonenal. Methods Enzymol. 1990;186:407–421. doi: 10.1016/0076-6879(90)86134-h. [DOI] [PubMed] [Google Scholar]
- 148.Benedetti A., Comporti M., Esterbauer H. Identification of 4-hydroxynonenal as a cytotoxic product originating from the peroxidation of liver microsomal lipids. Biochim. Biophys. Acta. 1980;620:281–296. doi: 10.1016/0005-2760(80)90209-x. [DOI] [PubMed] [Google Scholar]
- 149.Sanyal J., et al. Plasma levels of lipid peroxides in patients with Parkinson's disease. Eur. Rev. Med. Pharmacol. Sci. 2009;13:129–132. [PubMed] [Google Scholar]
- 150.VanderVeen L.A., Hashim M.F., Shyr Y., Marnett L.J. Induction of frameshift and base pair substitution mutations by the major DNA adduct of the endogenous carcinogen malondialdehyde. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14247–14252. doi: 10.1073/pnas.2332176100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cohen G., et al. Role of lipid peroxidation and PPAR-delta in amplifying glucose-stimulated insulin secretion. Diabetes. 2011;60:2830–2842. doi: 10.2337/db11-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee S.J., Kim C.E., Seo K.W., Kim C.D. HNE-induced 5-LO expression is regulated by NF-kappaB/ERK and Sp1/p38 MAPK pathways via EGF receptor in murine macrophages. Cardiovasc. Res. 2010;88:352–359. doi: 10.1093/cvr/cvq194. [DOI] [PubMed] [Google Scholar]
- 153.Shanmugam N., et al. Proinflammatory effects of advanced lipoxidation end products in monocytes. Diabetes. 2008;57:879–888. doi: 10.2337/db07-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Haag S.M., et al. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559:269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- 155.Chesarino N.M., et al. Chemoproteomics reveals Toll-like receptor fatty acylation. BMC Biol. 2014;12:91. doi: 10.1186/s12915-014-0091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kim Y.C., et al. Toll-like receptor mediated inflammation requires FASN-dependent MYD88 palmitoylation. Nat. Chem. Biol. 2019;15:907–916. doi: 10.1038/s41589-019-0344-0. [DOI] [PubMed] [Google Scholar]
- 157.Hughes M.M., et al. Solution structure of the TLR adaptor MAL/TIRAP reveals an intact BB loop and supports MAL Cys91 glutathionylation for signaling. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E6480–E6489. doi: 10.1073/pnas.1701868114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lu Y., et al. Palmitoylation of NOD1 and NOD2 is required for bacterial sensing. Science. 2019;366:460–467. doi: 10.1126/science.aau6391. [DOI] [PubMed] [Google Scholar]
- 160.Ullevig S.L., et al. Protein S-glutathionylation mediates macrophage responses to metabolic cues from the extracellular environment. Antioxidants Redox Signal. 2016;25:836–851. doi: 10.1089/ars.2015.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tschopp J., Schroder K. NLRP3 inflammasome activation: the convergence of multiple signalling pathways on ROS production? Nat. Rev. Immunol. 2010;10:210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 162.Thornberry N.A., et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 163.He W.T., et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res. 2015;25:1285–1298. doi: 10.1038/cr.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Liu X., et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shi J., et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 166.Zhou R., Yazdi A.S., Menu P., Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 167.Groß C.J., et al. K(+) efflux-independent NLRP3 inflammasome activation by small molecules targeting mitochondria. Immunity. 2016;45:761–773. doi: 10.1016/j.immuni.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 168.Zhou R., Tardivel A., Thorens B., Choi I., Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 169.van Bruggen R., et al. Human NLRP3 inflammasome activation is Nox 1-4 independent. Blood. 2010;115:5398–5400. doi: 10.1182/blood-2009-10-250803. [DOI] [PubMed] [Google Scholar]
- 170.Dashdorj A., et al. Mitochondria-targeted antioxidant MitoQ ameliorates experimental mouse colitis by suppressing NLRP3 inflammasome-mediated inflammatory cytokines. BMC Med. 2013;11:178. doi: 10.1186/1741-7015-11-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shimada K., et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.He Y., Zeng M.Y., Yang D., Motro B., Nunez G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 2016;530:354–357. doi: 10.1038/nature16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shi H., et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 2016;17:250–258. doi: 10.1038/ni.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hughes M.M., et al. Glutathione transferase omega-1 regulates NLRP3 inflammasome activation through NEK7 deglutathionylation. Cell Rep. 2019;29:151–161 e155. doi: 10.1016/j.celrep.2019.08.072. [DOI] [PubMed] [Google Scholar]
- 175.Balasubramanian A., et al. 2023. Palmitoylation of Gasdermin D Directs its Membrane Translocation and Pore Formation in Pyroptosis. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Du G., et al. bioRxiv; 2023. ROS-Dependent Palmitoylation Is an Obligate Licensing Modification for GSDMD Pore Formation. [DOI] [Google Scholar]
- 177.Devant P., et al. Gasdermin D pore-forming activity is redox-sensitive. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.112008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hu J.J., et al. FDA-approved disulfiram inhibits pyroptosis by blocking gasdermin D pore formation. Nat. Immunol. 2020;21:736–745. doi: 10.1038/s41590-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Semino C., Carta S., Gattorno M., Sitia R., Rubartelli A. Progressive waves of IL-1beta release by primary human monocytes via sequential activation of vesicular and gasdermin D-mediated secretory pathways. Cell Death Dis. 2018;9:1088. doi: 10.1038/s41419-018-1121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang Y., et al. Mitochondrial ROS promote macrophage pyroptosis by inducing GSDMD oxidation. J. Mol. Cell Biol. 2019;11:1069–1082. doi: 10.1093/jmcb/mjz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bradfield C.J., et al. Biphasic JNK signaling reveals distinct MAP3K complexes licensing inflammasome formation and pyroptosis. Cell Death Differ. 2023;30:589–604. doi: 10.1038/s41418-022-01106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Evavold C.L., et al. Control of gasdermin D oligomerization and pyroptosis by the Ragulator-Rag-mTORC1 pathway. Cell. 2021;184:4495–4511 e4419. doi: 10.1016/j.cell.2021.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mishra B.B., et al. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome–dependent processing of IL-1β. Nat. Immunol. 2013;14:52–60. doi: 10.1038/ni.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bailey J.D., et al. Nitric oxide modulates metabolic remodeling in inflammatory macrophages through TCA cycle regulation and itaconate accumulation. Cell Rep. 2019;28:218–230.e217. doi: 10.1016/j.celrep.2019.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.O'Neill L.A., Pearce E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Holmes F.L. 1993. Oxford University Press. [Google Scholar]
- 187.Michelucci A., et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Strelko C.L., et al. Itaconic acid is a mammalian metabolite induced during macrophage activation. J. Am. Chem. Soc. 2011;133:16386–16389. doi: 10.1021/ja2070889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Degrandi D., Hoffmann R., Beuter-Gunia C., Pfeffer K. The proinflammatory cytokine-induced IRG1 protein associates with mitochondria. J. Interferon Cytokine Res. 2009;29:55–67. doi: 10.1089/jir.2008.0013. [DOI] [PubMed] [Google Scholar]
- 190.Lampropoulou V., et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metabol. 2016;24:158–166. doi: 10.1016/j.cmet.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Cordes T., et al. Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J. Biol. Chem. 2016;291:14274–14284. doi: 10.1074/jbc.M115.685792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bambouskova M., et al. Itaconate confers tolerance to late NLRP3 inflammasome activation. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hooftman A., et al. The immunomodulatory metabolite itaconate modifies NLRP3 and inhibits inflammasome activation. Cell Metabol. 2020;32:468–478 e467. doi: 10.1016/j.cmet.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bambouskova M., et al. Electrophilic properties of itaconate and derivatives regulate the IkappaBzeta-ATF3 inflammatory axis. Nature. 2018;556:501–504. doi: 10.1038/s41586-018-0052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Qin W., et al. S-glycosylation-based cysteine profiling reveals regulation of glycolysis by itaconate. Nat. Chem. Biol. 2019;15:983–991. doi: 10.1038/s41589-019-0323-5. [DOI] [PubMed] [Google Scholar]
- 196.Liao S.T., et al. 4-Octyl itaconate inhibits aerobic glycolysis by targeting GAPDH to exert anti-inflammatory effects. Nat. Commun. 2019;10:5091. doi: 10.1038/s41467-019-13078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhang Z., et al. Itaconate is a lysosomal inducer that promotes antibacterial innate immunity. Mol. Cell. 2022;82:2844–2857 e2810. doi: 10.1016/j.molcel.2022.05.009. [DOI] [PubMed] [Google Scholar]
- 198.Runtsch M.C., et al. Itaconate and itaconate derivatives target JAK1 to suppress alternative activation of macrophages. Cell Metabol. 2022;34:487–501 e488. doi: 10.1016/j.cmet.2022.02.002. [DOI] [PubMed] [Google Scholar]
- 199.Day E.A., O'Neill L.A.J. Protein targeting by the itaconate family in immunity and inflammation. Biochem. J. 2022;479:2499–2510. doi: 10.1042/BCJ20220364. [DOI] [PubMed] [Google Scholar]
- 200.Peace C.G., O'Neill L.A. The role of itaconate in host defense and inflammation. J. Clin. Invest. 2022;132 doi: 10.1172/JCI148548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zecchini V., et al. Fumarate induces vesicular release of mtDNA to drive innate immunity. Nature. 2023;615:499–506. doi: 10.1038/s41586-023-05770-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hooftman A., et al. Macrophage fumarate hydratase restrains mtRNA-mediated interferon production. Nature. 2023;615:490–498. doi: 10.1038/s41586-023-05720-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Linker R.A., et al. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain. 2011;134:678–692. doi: 10.1093/brain/awq386. [DOI] [PubMed] [Google Scholar]
- 204.Humphries F., et al. Succination inactivates gasdermin D and blocks pyroptosis. Science. 2020;369:1633–1637. doi: 10.1126/science.abb9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kornberg M.D., et al. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science. 2018;360:449–453. doi: 10.1126/science.aan4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Infantino V., Iacobazzi V., Menga A., Avantaggiati M.L., Palmieri F. A key role of the mitochondrial citrate carrier (SLC25A1) in TNFalpha- and IFNgamma-triggered inflammation. Biochim. Biophys. Acta. 2014;1839:1217–1225. doi: 10.1016/j.bbagrm.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Infantino V., Iacobazzi V., Palmieri F., Menga A. ATP-citrate lyase is essential for macrophage inflammatory response. Biochem. Biophys. Res. Commun. 2013;440:105–111. doi: 10.1016/j.bbrc.2013.09.037. [DOI] [PubMed] [Google Scholar]
- 208.Palmieri E.M., et al. Acetylation of human mitochondrial citrate carrier modulates mitochondrial citrate/malate exchange activity to sustain NADPH production during macrophage activation. Biochim. Biophys. Acta. 2015;1847:729–738. doi: 10.1016/j.bbabio.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 209.Mantovani A., Allavena P., Marchesi F., Garlanda C. Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov. 2022;21:799–820. doi: 10.1038/s41573-022-00520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Balce D.R., et al. Alternative activation of macrophages by IL-4 enhances the proteolytic capacity of their phagosomes through synergistic mechanisms. Blood. 2011;118:4199–4208. doi: 10.1182/blood-2011-01-328906. [DOI] [PubMed] [Google Scholar]
- 211.Xu Q., et al. NADPH oxidases are essential for macrophage differentiation. J. Biol. Chem. 2016;291:20030–20041. doi: 10.1074/jbc.M116.731216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.He C., Ryan A.J., Murthy S., Carter A.B. Accelerated development of pulmonary fibrosis via Cu,Zn-superoxide dismutase-induced alternative activation of macrophages. J. Biol. Chem. 2013;288:20745–20757. doi: 10.1074/jbc.M112.410720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Formentini L., et al. Mitochondrial ROS production protects the intestine from inflammation through functional M2 macrophage polarization. Cell Rep. 2017;19:1202–1213. doi: 10.1016/j.celrep.2017.04.036. [DOI] [PubMed] [Google Scholar]
- 214.Li P., et al. Comparative proteomic analysis of polarized human THP-1 and mouse RAW264.7 macrophages. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.700009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Rakkola R., Matikainen S., Nyman T.A. Proteome analysis of human macrophages reveals the upregulation of manganese-containing superoxide dismutase after toll-like receptor activation. Proteomics. 2007;7:378–384. doi: 10.1002/pmic.200600582. [DOI] [PubMed] [Google Scholar]
- 216.Hwang J., et al. SOD1 suppresses pro-inflammatory immune responses by protecting against oxidative stress in colitis. Redox Biol. 2020;37 doi: 10.1016/j.redox.2020.101760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Israelson A., et al. Macrophage migration inhibitory factor as a chaperone inhibiting accumulation of misfolded SOD1. Neuron. 2015;86:218–232. doi: 10.1016/j.neuron.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Dimayuga F.O., et al. SOD1 overexpression alters ROS production and reduces neurotoxic inflammatory signaling in microglial cells. J. Neuroimmunol. 2007;182:89–99. doi: 10.1016/j.jneuroim.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Cheng F., et al. Impact of glutathione peroxidase-1 deficiency on macrophage foam cell formation and proliferation: implications for atherogenesis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0072063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yang X., et al. Inhibition of glutathione production induces macrophage CD36 expression and enhances cellular-oxidized low density lipoprotein (oxLDL) uptake. J. Biol. Chem. 2015;290:21788–21799. doi: 10.1074/jbc.M115.654582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Salzano S., et al. Linkage of inflammation and oxidative stress via release of glutathionylated peroxiredoxin-2, which acts as a danger signal. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12157–12162. doi: 10.1073/pnas.1401712111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Morgan M.J., Liu Z.G. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011;21:103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Yamamoto T., et al. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kong X., Thimmulappa R., Kombairaju P., Biswal S. NADPH oxidase-dependent reactive oxygen species mediate amplified TLR4 signaling and sepsis-induced mortality in Nrf2-deficient mice. J. Immunol. 2010;185:569–577. doi: 10.4049/jimmunol.0902315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Thimmulappa R.K., et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J. Clin. Invest. 2006;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wang P., et al. Macrophage achieves self-protection against oxidative stress-induced ageing through the Mst-Nrf2 axis. Nat. Commun. 2019;10:755. doi: 10.1038/s41467-019-08680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.McNeill E., et al. Regulation of iNOS function and cellular redox state by macrophage Gch1 reveals specific requirements for tetrahydrobiopterin in NRF2 activation. Free Radic. Biol. Med. 2015;79:206–216. doi: 10.1016/j.freeradbiomed.2014.10.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Anjani G., et al. Recent advances in chronic granulomatous disease. Genes Dis. 2020;7:84–92. doi: 10.1016/j.gendis.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mittal M., Siddiqui M.R., Tran K., Reddy S.P., Malik A.B. Reactive oxygen species in inflammation and tissue injury. Antioxidants Redox Signal. 2014;20:1126–1167. doi: 10.1089/ars.2012.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Fernandez-Boyanapalli R.F., et al. Impaired apoptotic cell clearance in CGD due to altered macrophage programming is reversed by phosphatidylserine-dependent production of IL-4. Blood. 2009;113:2047–2055. doi: 10.1182/blood-2008-05-160564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rahman F.Z., Hayee B., Chee R., Segal A.W., Smith A.M. Impaired macrophage function following bacterial stimulation in chronic granulomatous disease. Immunology. 2009;128:253–259. doi: 10.1111/j.1365-2567.2009.03112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Pizzolla A., et al. Reactive oxygen species produced by the NADPH oxidase 2 complex in monocytes protect mice from bacterial infections. J. Immunol. 2012;188:5003–5011. doi: 10.4049/jimmunol.1103430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.van de Veerdonk F.L., et al. Reactive oxygen species–independent activation of the IL-1β inflammasome in cells from patients with chronic granulomatous disease. Proc. Natl. Acad. Sci. USA. 2010;107:3030–3033. doi: 10.1073/pnas.0914795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Meissner F., et al. Inflammasome activation in NADPH oxidase defective mononuclear phagocytes from patients with chronic granulomatous disease. Blood. 2010;116:1570–1573. doi: 10.1182/blood-2010-01-264218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Bylund J., et al. Enhanced inflammatory responses of chronic granulomatous disease leukocytes involve ROS-independent activation of NF-κB. Eur. J. Immunol. 2007;37:1087–1096. doi: 10.1002/eji.200636651. [DOI] [PubMed] [Google Scholar]
- 236.Sundqvist M., et al. Elevated mitochondrial reactive oxygen species and cellular redox imbalance in human NADPH-oxidase-deficient phagocytes. Front. Immunol. 2017;8:1828. doi: 10.3389/fimmu.2017.01828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Segal B.H., Grimm M.J., Khan A.N., Han W., Blackwell T.S. Regulation of innate immunity by NADPH oxidase. Free Radic. Biol. Med. 2012;53:72–80. doi: 10.1016/j.freeradbiomed.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Na Y.R., Stakenborg M., Seok S.H., Matteoli G. Macrophages in intestinal inflammation and resolution: a potential therapeutic target in IBD. Nat. Rev. Gastroenterol. Hepatol. 2019;16:531–543. doi: 10.1038/s41575-019-0172-4. [DOI] [PubMed] [Google Scholar]
- 239.Muthupalani S., et al. Systemic macrophage depletion inhibits Helicobacter bilis-induced proinflammatory cytokine-mediated typhlocolitis and impairs bacterial colonization dynamics in a BALB/c Rag 2-/- mouse model of inflammatory bowel disease. Infect. Immun. 2012;80:4388–4397. doi: 10.1128/IAI.00530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Knutson C.G., et al. Chemical and cytokine features of innate immunity characterize serum and tissue profiles in inflammatory bowel disease. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E2332–E2341. doi: 10.1073/pnas.1222669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Erdman S.E., et al. Nitric oxide and TNF-alpha trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag 2-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 2009;106:1027–1032. doi: 10.1073/pnas.0812347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Dhillon S.S., et al. Variants in nicotinamide adenine dinucleotide phosphate oxidase complex components determine susceptibility to very early onset inflammatory bowel disease. Gastroenterology. 2014;147:680–689 e682. doi: 10.1053/j.gastro.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 243.Roberts R.L., et al. Confirmation of association of IRGM and NCF4 with ileal Crohn's disease in a population-based cohort. Gene Immun. 2008;9:561–565. doi: 10.1038/gene.2008.49. [DOI] [PubMed] [Google Scholar]
- 244.Rioux J.D., et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kurowska-Stolarska M., Alivernini S. Synovial tissue macrophages in joint homeostasis, rheumatoid arthritis and disease remission. Nat. Rev. Rheumatol. 2022;18:384–397. doi: 10.1038/s41584-022-00790-8. [DOI] [PubMed] [Google Scholar]
- 246.Zhang F., et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 2019;20:928–942. doi: 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.You S., et al. Identification of key regulators for the migration and invasion of rheumatoid synoviocytes through a systems approach. Proc. Natl. Acad. Sci. U. S. A. 2014;111:550–555. doi: 10.1073/pnas.1311239111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Soler Palacios B., et al. Macrophages from the synovium of active rheumatoid arthritis exhibit an activin A-dependent pro-inflammatory profile. J. Pathol. 2015;235:515–526. doi: 10.1002/path.4466. [DOI] [PubMed] [Google Scholar]
- 249.Khojah H.M., Ahmed S., Abdel-Rahman M.S., Hamza A.B. Reactive oxygen and nitrogen species in patients with rheumatoid arthritis as potential biomarkers for disease activity and the role of antioxidants. Free Radic. Biol. Med. 2016;97:285–291. doi: 10.1016/j.freeradbiomed.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 250.Mateen S., Moin S., Khan A.Q., Zafar A., Fatima N. Increased reactive oxygen species formation and oxidative stress in rheumatoid arthritis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0152925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Cramer T., et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Quinonez-Flores C.M., Gonzalez-Chavez S.A., Pacheco-Tena C. Hypoxia and its implications in rheumatoid arthritis. J. Biomed. Sci. 2016;23:62. doi: 10.1186/s12929-016-0281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Voet S., Srinivasan S., Lamkanfi M., van Loo G. Inflammasomes in neuroinflammatory and neurodegenerative diseases. EMBO Mol. Med. 2019;11 doi: 10.15252/emmm.201810248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Butterfield D.A., Halliwell B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat. Rev. Neurosci. 2019;20:148–160. doi: 10.1038/s41583-019-0132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Heneka M.T., et al. NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature. 2013;493:674–678. doi: 10.1038/nature11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Yin J., et al. NLRP3 inflammasome inhibitor ameliorates amyloid pathology in a mouse model of Alzheimer's disease. Mol. Neurobiol. 2018;55:1977–1987. doi: 10.1007/s12035-017-0467-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Dempsey C., et al. Inhibiting the NLRP3 inflammasome with MCC950 promotes non-phlogistic clearance of amyloid-β and cognitive function in APP/PS1 mice. Brain Behav. Immun. 2017;61:306–316. doi: 10.1016/j.bbi.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 258.Sarkar S., et al. Mitochondrial impairment in microglia amplifies NLRP3 inflammasome proinflammatory signaling in cell culture and animal models of Parkinson's disease. npj Parkinson's Disease. 2017;3:30. doi: 10.1038/s41531-017-0032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Li S., et al. Gasdermin D in peripheral myeloid cells drives neuroinflammation in experimental autoimmune encephalomyelitis. J. Exp. Med. 2019;216:2562–2581. doi: 10.1084/jem.20190377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.McKenzie B.A., et al. Caspase-1 inhibition prevents glial inflammasome activation and pyroptosis in models of multiple sclerosis. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E6065–e6074. doi: 10.1073/pnas.1722041115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Rathkey J.K., et al. Chemical disruption of the pyroptotic pore-forming protein gasdermin D inhibits inflammatory cell death and sepsis. Sci Immunol. 2018;3 doi: 10.1126/sciimmunol.aat2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Coll R.C., Schroder K., Pelegrín P. NLRP3 and pyroptosis blockers for treating inflammatory diseases. Trends Pharmacol. Sci. 2022;43:653–668. doi: 10.1016/j.tips.2022.04.003. [DOI] [PubMed] [Google Scholar]
- 263.Weisberg S.P., et al. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Lumeng C.N., Bodzin J.L., Saltiel A.R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 2007;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Spranger J., et al. Inflammatory cytokines and the risk to develop type 2 diabetes: results of the prospective population-based European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam Study. Diabetes. 2003;52:812–817. doi: 10.2337/diabetes.52.3.812. [DOI] [PubMed] [Google Scholar]
- 266.Eldor R., et al. Conditional and specific NF-kappaB blockade protects pancreatic beta cells from diabetogenic agents. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5072–5077. doi: 10.1073/pnas.0508166103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Xu H., et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Lumeng C.N., Deyoung S.M., Saltiel A.R. Macrophages block insulin action in adipocytes by altering expression of signaling and glucose transport proteins. Am. J. Physiol. Endocrinol. Metab. 2007;292:E166–E174. doi: 10.1152/ajpendo.00284.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Hotamisligil G.S., et al. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science. 1996;271:665–668. doi: 10.1126/science.271.5249.665. [DOI] [PubMed] [Google Scholar]
- 270.Stienstra R., et al. The inflammasome-mediated caspase-1 activation controls adipocyte differentiation and insulin sensitivity. Cell Metabol. 2010;12:593–605. doi: 10.1016/j.cmet.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Vandanmagsar B., et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat. Med. 2011;17:179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Wen H., et al. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat. Immunol. 2011;12:408–415. doi: 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Pepping J.K., et al. Myeloid-specific deletion of NOX2 prevents the metabolic and neurologic consequences of high fat diet. PLoS One. 2017;12 doi: 10.1371/journal.pone.0181500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Qian B.Z., Pollard J.W. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Cassetta L., Pollard J.W. A timeline of tumour-associated macrophage biology. Nat. Rev. Cancer. 2023;23:238–257. doi: 10.1038/s41568-022-00547-1. [DOI] [PubMed] [Google Scholar]
- 276.Kuo C.L., et al. Mitochondrial oxidative stress by Lon-PYCR1 maintains an immunosuppressive tumor microenvironment that promotes cancer progression and metastasis. Cancer Lett. 2020;474:138–150. doi: 10.1016/j.canlet.2020.01.019. [DOI] [PubMed] [Google Scholar]
- 277.Salpeter S.J., et al. A novel cysteine cathepsin inhibitor yields macrophage cell death and mammary tumor regression. Oncogene. 2015;34:6066–6078. doi: 10.1038/onc.2015.51. [DOI] [PubMed] [Google Scholar]
- 278.Roux C., et al. Reactive oxygen species modulate macrophage immunosuppressive phenotype through the up-regulation of PD-L1. Proc. Natl. Acad. Sci. U. S. A. 2019;116:4326–4335. doi: 10.1073/pnas.1819473116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Stixova L., Prochazkova J., Soucek K., Hofmanova J., Kozubik A. 5-Lipoxygenase inhibitors potentiate 1alpha,25-dihydroxyvitamin D3-induced monocytic differentiation by activating p38 MAPK pathway. Mol. Cell. Biochem. 2009;330:229–238. doi: 10.1007/s11010-009-0138-x. [DOI] [PubMed] [Google Scholar]
- 280.Kennel K.B., Greten F.R. Immune cell - produced ROS and their impact on tumor growth and metastasis. Redox Biol. 2021;42 doi: 10.1016/j.redox.2021.101891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Xia H., et al. Autophagic adaptation to oxidative stress alters peritoneal residential macrophage survival and ovarian cancer metastasis. JCI Insight. 2020;5 doi: 10.1172/jci.insight.141115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Li X., et al. Reactive oxygen species reprogram macrophages to suppress antitumor immune response through the exosomal miR-155-5p/PD-L1 pathway. J. Exp. Clin. Cancer Res. 2022;41:41. doi: 10.1186/s13046-022-02244-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Canli O., et al. Myeloid cell-derived reactive oxygen species induce epithelial mutagenesis. Cancer Cell. 2017;32:869–883 e865. doi: 10.1016/j.ccell.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 284.Gul N., et al. Macrophages mediate colon carcinoma cell adhesion in the rat liver after exposure to lipopolysaccharide. OncoImmunology. 2012;1:1517–1526. doi: 10.4161/onci.22303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.LaRue M.M., et al. Metabolic reprogramming of tumor-associated macrophages by collagen turnover promotes fibrosis in pancreatic cancer. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2119168119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Somasundaram V., et al. Inducible nitric oxide synthase-derived extracellular nitric oxide flux regulates proinflammatory responses at the single cell level. Redox Biol. 2020;28 doi: 10.1016/j.redox.2019.101354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Fomenko D.E., Xing W., Adair B.M., Thomas D.J., Gladyshev V.N. High-throughput identification of catalytic redox-active cysteine residues. Science. 2007;315:387–389. doi: 10.1126/science.1133114. [DOI] [PubMed] [Google Scholar]
- 288.Sethuraman M., et al. Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J. Proteome Res. 2004;3:1228–1233. doi: 10.1021/pr049887e. [DOI] [PubMed] [Google Scholar]
- 289.Baty J.W., Hampton M.B., Winterbourn C.C. Proteomic detection of hydrogen peroxide-sensitive thiol proteins in Jurkat cells. Biochem. J. 2005;389:785–795. doi: 10.1042/BJ20050337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Kim J.R., Yoon H.W., Kwon K.S., Lee S.R., Rhee S.G. Identification of proteins containing cysteine residues that are sensitive to oxidation by hydrogen peroxide at neutral pH. Anal. Biochem. 2000;283:214–221. doi: 10.1006/abio.2000.4623. [DOI] [PubMed] [Google Scholar]
- 291.Weerapana E., Simon G.M., Cravatt B.F. Disparate proteome reactivity profiles of carbon electrophiles. Nat. Chem. Biol. 2008;4:405–407. doi: 10.1038/nchembio.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Weerapana E., et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Deng X., et al. Proteome-wide quantification and characterization of oxidation-sensitive cysteines in pathogenic bacteria. Cell Host Microbe. 2013;13:358–370. doi: 10.1016/j.chom.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Wang C., Weerapana E., Blewett M.M., Cravatt B.F. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods. 2014;11:79–85. doi: 10.1038/nmeth.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Yang J., et al. Global, in situ, site-specific analysis of protein S-sulfenylation. Nat. Protoc. 2015;10:1022–1037. doi: 10.1038/nprot.2015.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Mnatsakanyan R., et al. Proteome-wide detection of S-nitrosylation targets and motifs using bioorthogonal cleavable-linker-based enrichment and switch technique. Nat. Commun. 2019;10:2195. doi: 10.1038/s41467-019-10182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Kemper E.K., Zhang Y., Dix M.M., Cravatt B.F. Global profiling of phosphorylation-dependent changes in cysteine reactivity. Nat. Methods. 2022;19:341–352. doi: 10.1038/s41592-022-01398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Backus K.M., et al. Proteome-wide covalent ligand discovery in native biological systems. Nature. 2016;534:570–574. doi: 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Blewett M.M., et al. Chemical proteomic map of dimethyl fumarate-sensitive cysteines in primary human T cells. Sci. Signal. 2016;9:rs10. doi: 10.1126/scisignal.aaf7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Tian C., et al. Multiplexed thiol reactivity profiling for target discovery of electrophilic natural products. Cell Chem. Biol. 2017;24:1416–1427 e1415. doi: 10.1016/j.chembiol.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 301.Bar-Peled L., et al. Chemical proteomics identifies druggable vulnerabilities in a genetically defined cancer. Cell. 2017;171:696–709 e623. doi: 10.1016/j.cell.2017.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Kuljanin M., et al. Reimagining high-throughput profiling of reactive cysteines for cell-based screening of large electrophile libraries. Nat. Biotechnol. 2021;39:630–641. doi: 10.1038/s41587-020-00778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Fu L., et al. A quantitative thiol reactivity profiling platform to analyze redox and electrophile reactive cysteine proteomes. Nat. Protoc. 2020;15:2891–2919. doi: 10.1038/s41596-020-0352-2. [DOI] [PubMed] [Google Scholar]
- 304.Vinogradova E.V., et al. An activity-guided map of electrophile-cysteine interactions in primary human T cells. Cell. 2020;182:1009–1026 e1029. doi: 10.1016/j.cell.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Held J.M., et al. Targeted quantitation of site-specific cysteine oxidation in endogenous proteins using a differential alkylation and multiple reaction monitoring mass spectrometry approach. Mol. Cell. Proteomics. 2010;9:1400–1410. doi: 10.1074/mcp.M900643-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Shakir S., Vinh J., Chiappetta G. Quantitative analysis of the cysteine redoxome by iodoacetyl tandem mass tags. Anal. Bioanal. Chem. 2017;409:3821–3830. doi: 10.1007/s00216-017-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Gaffrey M.J., Day N.J., Li X., Qian W.J. Resin-assisted capture coupled with isobaric tandem mass tag labeling for multiplexed quantification of protein thiol oxidation. J. Vis. Exp. 2021 doi: 10.3791/62671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Topf U., et al. Quantitative proteomics identifies redox switches for global translation modulation by mitochondrially produced reactive oxygen species. Nat. Commun. 2018;9:324. doi: 10.1038/s41467-017-02694-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Leichert L.I., et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Kisty E.A., Falco J.A., Weerapana E. Redox proteomics combined with proximity labeling enables monitoring of localized cysteine oxidation in cells. Cell Chem. Biol. 2023;30:321–336 e326. doi: 10.1016/j.chembiol.2023.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Yan T., et al. Proximity-labeling chemoproteomics defines the subcellular cysteinome and inflammation-responsive mitochondrial redoxome. Cell Chem. Biol. 2023;30:811–827 e817. doi: 10.1016/j.chembiol.2023.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Xiao H., et al. A quantitative tissue-specific landscape of protein redox regulation during aging. Cell. 2020;180:968–983 e924. doi: 10.1016/j.cell.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Huttlin E.L., et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Bekker-Jensen D.B., et al. Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries. Nat. Commun. 2020;11:787. doi: 10.1038/s41467-020-14609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Darabedian N., et al. Depletion of creatine phosphagen energetics with a covalent creatine kinase inhibitor. Nat. Chem. Biol. 2023;19:815–824. doi: 10.1038/s41589-023-01273-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Qin W., et al. Chemoproteomic profiling of itaconation by bioorthogonal probes in inflammatory macrophages. J. Am. Chem. Soc. 2020;142:10894–10898. doi: 10.1021/jacs.9b11962. [DOI] [PubMed] [Google Scholar]
- 317.Ran F.A., et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Miura H., Quadros R.M., Gurumurthy C.B., Ohtsuka M. Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat. Protoc. 2018;13:195–215. doi: 10.1038/nprot.2017.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Bauer M., et al. Human monocytes are severely impaired in base and DNA double-strand break repair that renders them vulnerable to oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 2011;108:21105–21110. doi: 10.1073/pnas.1111919109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.