Abstract
Understanding of the functional states and clonal dynamics of T cells after immune checkpoint blockade (ICB) is valuable for improving these therapeutic strategies. Here we performed Smart-seq2 single-cell RNA sequencing (scRNA-seq) analysis on 3,110 peripheral T cells of non-small cell lung cancer (NSCLC) patients before and after the initiation of programmed cell death protein 1 (PD-1) blockade. We identified individual peripheral T cell clones based on the full-length T cell receptor (TCR) sequences and monitored their dynamics during immunotherapy. We found a higher cytotoxic activity in the tumor-related CD4+ T cell clones than in the CD8+ T cell clones. Based on a large tumor-related CD4+ T cell clone, we observed a dramatically decreased abundance after progression, as well as a reduction in the percentage of PD-1+ T cells. We also detected 25 genes, such as CXCR4, DUSP2 and ZFP36, that were noticeably upregulated or downregulated following progression. In addition, the pseudotime trajectory of CD8+ T cell clones corresponded to the treatment time points, showing a decreased activity in the “cytokine and cytokine receptor interaction” pathway. These analyses provided an insight into the dynamics of peripheral T cell clones during PD-1 blockade in NSCLC.
Electronic supplementary material
The online version of this article (10.1007/s00262-020-02642-4) contains supplementary material, which is available to authorized users.
Keywords: PD-1, Cancer immunotherapy, Single cell sequencing, Non-small cell lung cancer
Introduction
The immune checkpoint blockade (ICB) that targets programmed cell death protein 1 (PD-1) has led to clinical success in treating many human tumor types [1]. Despite the unprecedented durable response rates, many patients do not benefit from the treatment (primary resistance), and some responders relapse after a period of initial response (acquired resistance). Identifying the influencing factors for response to PD-1 blockade, therefore, remains an urgent need for understanding and expanding the use of such cancer immunotherapy.
PD-1 blockade can overcome T cell dysfunction resulting from the suppressive tumor microenvironment. Multiple mechanisms of T cell biology have been linked to the different responses of individuals, and the predictive biomarkers of ICB have been investigated comprehensively. The first well-described T cell biomarker was the number of infiltrating CD8+ T cells detected before [2] or during early treatment [3]. Subsequently, several studies have revealed a connection between ICB response and T cell states, such as T cell activation, exhaustion, cytotoxicity, and interferon responses [4–6]. Other identified biomarkers include the abundance of partially exhausted CD8+ T cells in responding tumors [7] and the magnitude of T cell reinvigoration in relation to pre-treatment tumor burden in blood [8].
Genomic analyses of bulk tumors have revealed the biological pathways for tumor cells to evade immune surveillance, implicating multiple reliable response biomarkers [9–12]. While a deconvolution analysis of bulk RNA sequencing (RNA-seq) can infer the cell composition of the tumor [13], it cannot adequately illustrate the cancer-immune interplay at single-cell resolution. Single-cell RNA sequencing (scRNA-seq) is able to overcome this limitation. Recently, scRNA-seq analysis has been demonstrated for its advantage in illustrating immune landscapes of multiple human tumor types at single-cell resolution [14–16].
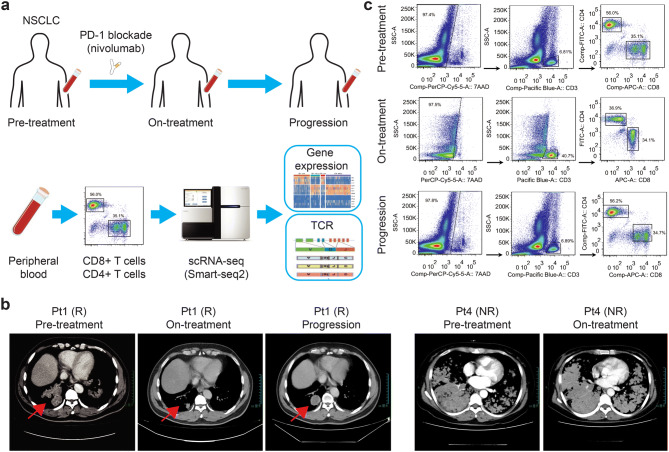
An assessment of patients’ longitudinal data can provide an insight into acquired resistance to ICB [17]. Since tumor biopsy is hard to obtain and peripheral blood is the minimally invasive biopsy from progressed non-small cell lung cancer (NSCLC) patients, patients’ blood cells are ideal for searching potential biomarkers. Although researchers have analyzed the changes of peripheral T cells during anti-PD-1 treatment in NSCLC patients through bulk RNA-seq or T cell receptor (TCR) sequencing, they have not conducted a granular-level investigation on single cells [18, 19]. Here we applied large-scale Smart-seq2 scRNA-seq on longitudinal peripheral T cells of NSCLC patients with PD-1 blockade, identified individual peripheral T cell clones based on the full-length TCR sequences and monitored their dynamics during immunotherapy (Fig. 1a).
Fig. 1.
The scheme of study design and sample collection. a Overview of the study design. Peripheral blood from NSCLC patients with anti-PD-1 treatment were collected at multiple time points during the treatment, including pre-treatment, on-treatment, and progression. Single-cell RNA sequencing was applied to individual T cells, and the output data were used for expression analysis and TCR profiling. b Computerized tomography (CT) scans of pleural in responder Pt1 at pre-treatment, on-treatment and progression. CT scans of pleural in nonresponder Pt4 at pre-treatment and on-treatment. c Gating strategy for single T cell sorting. CD8+ T cells and CD4+ T cells were enriched by sorting 7AAD−CD3+CD8+ and 7AAD−CD3+CD4+, respectively
Materials and methods
Study design
This study relies on a phase III, open-label, randomized, multiple centers study of Nivolumab compared to Docetaxel for patients with advanced or metastatic NSCLC, who had previously received conventional therapy (Check-Mate 078). The four eligible patients had histologically confirmed stage IV adenocarcinoma or squamous cell carcinoma, with Eastern Corporation Oncology Group (ECOG) performance status of zero. Tumor response for patients was defined by Response Evaluation Criteria in Solid Tumors (RECIST v1.1). Partial response: target lesion reduction more than 30%; progression disease: target lesion increase more than 20%; stable disease: target lesion size change between partial response and progressive disease. All patients received nivolumab at a dose of 3 mg/kg body weight every 3 weeks. Treatment beyond progression was permitted if protocol defined criteria were met, including investigator assessed clinical benefit, no rapid disease progression, no unacceptable toxicity and a stable performance status.
PD-L1 expression detection by immunohistochemistry
The PD-L1 expression of formalin-fixed, paraffin-embedded (FFPE) specimens were measured by PD-L1 IHC 22C3 pharmDx analysis on the DAKO Autostainer Link 48 (Agilent Technologies) and VENTANA PD-L1 (SP142) assays on Ventana BenchMark (Ventana Medical System).
Extraction, target capture and sequencing of plasma DNA
The peripheral blood was centrifuged in the Streck tubes at 1600 g for 20 min at room temperature to separate the plasma. The QIAamp Circulating Nucleic Acid Kit (Qiagen) was used to extract circulating tumor DNA (ctDNA) from the plasma. The ctDNA libraries were prepared by Accel-NGS 2S Plus DNA Library Kit (SWIFT) with unique identifiers to tag individual DNA molecules. The captured libraries for plasma ctDNA were loaded into the NextSeq 500 (Illumina) to run 75 bp paired-end sequencing according to the manufacturer’s instructions.
The details of our established pipeline based on the cancer gene panel NCC-GP150 can be found elsewhere [20]. The 150 genes for panel sequencing are listed in Supplementary Table 1. The ctDNA variant-calling method was integrated with digital barcodes to tag the individual DNA molecules. All the raw variants were then filtered SNP by the population frequency > 0.015 in dbSNP, 1000Genome and ESP6500. Meanwhile, variants were seemed as germline and were removed if their allele frequencies were larger than 30%.
Neoantigen prediction
The 4-digit HLA class I alleles of each patient were determined by OptiType [21]. For each non-synonymous mutation, we predicted the binding affinity IC50 between the mutant peptides (9–11 amino acids) and HLA types using NetMHCpan (version 4.0) [22]. The binding affinity threshold for neoantigens was IC50 ≤ 500 nM.
Single-cell preparation and sequencing
The peripheral blood mononuclear cells were isolated using HISTOPAQUE-1077 (Sigma-Aldrich) solution as previously described [16]. We sorted CD8+ and CD4+ T cells by flow cytometry. Single-cell suspensions were stained with antibodies against CD3, CD4 and CD8 (anti-human CD3, UCHT1; anti-human CD4, OKT4; anti-human CD8, OKT8) for flow cytometry, performed on a BD Aria III instrument. CD8+ T cells and CD4+ T cells were enriched and separated into 96-well plates by gating 7AAD−CD3+CD8+ and 7AAD−CD3+CD4+ T cells, respectively.
Single-cell transcriptome amplifications were performed according to the Smart-seq2 protocol [23]. The amplified cDNA products were purified with 1 × Agencourt XP DNA beads (Beckman). A procedure of quality control was performed following the first round of purification, which included the detection of CD3D by quantitative PCR (forward primer, 5′-TCATTGCCACTCTGCTCC-3′; reverse primer, 5′-GTTCACTTGTTCCGAGCC-3′) and fragment analysis by analyzer AATI. For those single-cell cDNA pass quality control (cycle threshold < 30), the cDNA products were further purified with 0.5 × Agencourt XP DNA beads, and the concentration of each sample was quantified by Qubit HsDNA kits (Invitrogen). Multiplex libraries were constructed and amplified using the TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme Biotech). The libraries were then purified with Agencourt XP DNA beads and pooled for quality assessment by fragment analyzer. Finally, purified libraries were analyzed by an Illumina HiSeq 4000 sequencer with 150-bp paired-end reads.
Quality control and preprocessing of scRNA-seq data
Low-quality scRNA-seq read pairs were filtered out if at least one end of the read pair met one of the following criteria: (1) ‘N’ bases account for ≥ 10% read length; (2) bases with quality < 5 account for ≥ 50% read length; and (3) the read contains adaptor sequence. The filtered read pairs were processed using Kallisto (version 0.43.0) [24] and human genome reference GRCh38 to obtain the gene expression matrix. Low-quality cells were filtered if the library size < 100,000 or the number of expressed genes < 1000 (counts larger than zero). Furthermore, if the proportion of mitochondrial gene counts was larger than 10%, these cells were discarded. After discarding genes with average counts of fewer than one, a total of 11,535 genes and 3,042 cells were retained in the final expression table.
Unsupervised clustering analysis
The expression tables of CD8+ T cells and CD4+ T cells were fed into the unsupervised clustering pipeline separately. The top 1000 genes with the largest variance were selected for single-cell consensus clustering (SC3) [25]. Hierarchical clustering with complete agglomeration was performed on the SC3 consensus matrix and 2 clusters were inferred. The t-SNE method implemented in R package Rtsne was used for clustering visualization.
When the clustering results were obtained, one-way ANOVA implemented by R function ‘aov’ was performed to identify the signature genes among the clusters. R function TukeyHSD was used to identify which cluster pairs showed a significant difference. A gene was defined as being significantly differentially expressed based on the following criteria: (1) adjusted P value of F test < 0.05 (Benjamini–Hochberg method); (2) the absolute difference of any one significant cluster pair > 1 (P value of Tukey’s honest significant difference method).
T cell receptor analysis
TraCeR was used to determine the T cell receptor (TCR) sequences of each T cell [26]. The outputs of TraCeR include the assembled nucleotide sequences for both α- and β-chains, the coding potential of the nucleotide sequences (that is, productive or not), the translated amino acid sequence, the complementarity-determining region 3 (CDR3) sequences and the estimated TPM value of α- or β-chains. Only cells with TPM values larger than 10 for the α- and β-chains were kept. For cells with two or more α- or β-chains assembled, the αβ-pair that was productive and of the highest expression level was defined as the dominant αβ-pair in the corresponding cell. If two cells had identical α–β pairs, those were identified as clonal T cells.
Bulk TCR β-chain sequencing
Bulk TCR β-chain sequencing was performed on pre-treatment tumor biopsies. Genomic DNA from tumor formalin-fixed, paraffin-embedded (FFPE) tissues was extracted by the DNeasy Tissue Kit (Qiagen) following the standard protocols. Generally, the CDR3 libraries were constructed by two-round PCRs. During the first round of amplification, only ten cycles were used to amplify CDR3 fragments using the primers specific against each V and J genes, and 600 ng of DNA was used as template for each sample. The target fragments of multiplex-PCR products were purified on magnetic beads (Agencourt, Beckman). In the second round, amplification was performed using universal primers. Size selection was run by agar gel electrophoresis and target fragments between 200–350 bp was retrieved and purified by QIAquick Gel Purification Kit (Qiagen). After gel purification, the PCR product was subjected to sequencing using the NextSeq500 (Illumina).
Differential gene expression analysis
Differentially expressed gene (DEGs) among samples at multiple treatment stages were detected by the linear model and the empirical Bayes method with R package limma (version 3.36.3). We used stringent significance thresholds for adjusted P < 0.01 (Benjamini–Hochberg multiple testing correction) and absolute log2 (fold change) > 2.
Trajectory analysis
To characterize the potential process of T cell state changes and determine the pseudotime trajectory of the expanded T cell clone, we applied the Monocle algorithm (version 2) [27] with the top 500 signature genes based on the rank of F statistic generated by ANOVA. Cells were ordered through the inferred pseudotime to indicate their differentiation progress. Then the trajectory was inferred after dimension reduction and cell ordering with the default parameters of R package Monocle. Gene set enrichment analysis (GSEA) was performed by R package fgsea.
Results
Patient enrollment and single T cell sequencing
Four patients (Pt1, Pt2, Pt3 and Pt4) with stage IV NSCLC were enrolled in this study and received PD-1 blockade nivolumab as the second-line treatment (Table 1). Pt1, Pt2 and Pt4 were diagnosed with adenocarcinoma, and Pt3 with squamous cell carcinoma. Pt1, Pt2 and Pt3 initially responded to the immunotherapy, but progressed later. Pt1 achieved partial response after 12 weeks (four cycles of PD-1 blockade), while Pt2 and Pt3 only achieved stable disease during the treatment. The progression-free survival of Pt1, Pt2 and Pt3 were 4.3 months, 8.5 months and 11 months, respectively. Pt4 was a nonresponder and did not benefit from the immunotherapy (Fig. 1b). Their pre-treatment biopsies showed programmed death-ligand 1 (PD-L1) expression from 5 to 40% by immunohistochemistry (Table 1). In contrast, the pre-treatment biopsy of Pt4 showed no PD-L1 expression, consistent with its non-responsiveness [28].
Table 1.
The NSCLC patients information and numbers of T cells sequenced
| Patient ID | Clinical Information | Molecular Information | Number of Collected Cells | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Response | Cancer Type | Age | Sex | Stage | Time Points | PD-L1 (%) | Neoantigens | CD8+ T cells | CD4+ T cells | Summary | |
| Pt 1 | Y | lung adenocarcinoma | 54 | M | IV | Pre/On/Pro | 20 | ERBB4R1250Q, KRASG12D | − /477/137 | − /418/120 | 1152 |
| Pt 2 | Y | lung adenocarcinoma | 51 | F | IV | Pre/On/Pro | 5 | NFE2L2G31A, KRASG12V | 68/151/168 | 76/130/147 | 740 |
| Pt 3 | Y | lung squamous cell carcinoma | 59 | M | IV | Pre/On/Pro | 40 | TP53R158L | 48/317/244 | 50/201/255 | 1115 |
| Pt 4 | N | lung adenocarcinoma | 75 | F | IV | Pre | 0 | Not detected | 51 | 52 | 103 |
Pre/On/Pro represent the time points pre-treatment, on-treatment and progression
We identified the genotypes of Human Leukocyte Antigen class I (HLA-I) genes for each patient based on the RNA-seq data, and detected somatic mutations by 150-gene panel sequencing on the pre-treatment circulating tumor DNA (ctDNA). Combining the HLA-I and mutation data, we predicted the personalized neoantigens for each patient (Table 1). At least one neoantigen was identified in the three responders, such as frequent driver mutations ERBB4R1250Q, KRASG12D, NFE2L2G31A, KRASG12V and TP53R158L. No neoantigen, however, was found in Pt4, consistent with the fact that Pt4 was a nonresponder [29].
To monitor the T cell response to the immunotherapy, we collected blood samples from each responder before treatment (pre-treatment) and every 6 weeks (two cycles of PD-1 blockade) during treatment (on-treatment), until the tumor progressed (progression) (Fig. 1c). For the nonresponder, we only collected the pre-treatment blood sample. In total, we obtained Smart-seq2 [23] scRNA-seq data of 3,110 individual CD8+ and CD4+ T cells. After quality control and filtering, we kept 3,042 (97.8%) cells for the subsequent analyses, including 1,624 CD8+ and 1,418 CD4+ T cells.
Clustering T cells based on gene expression profiles
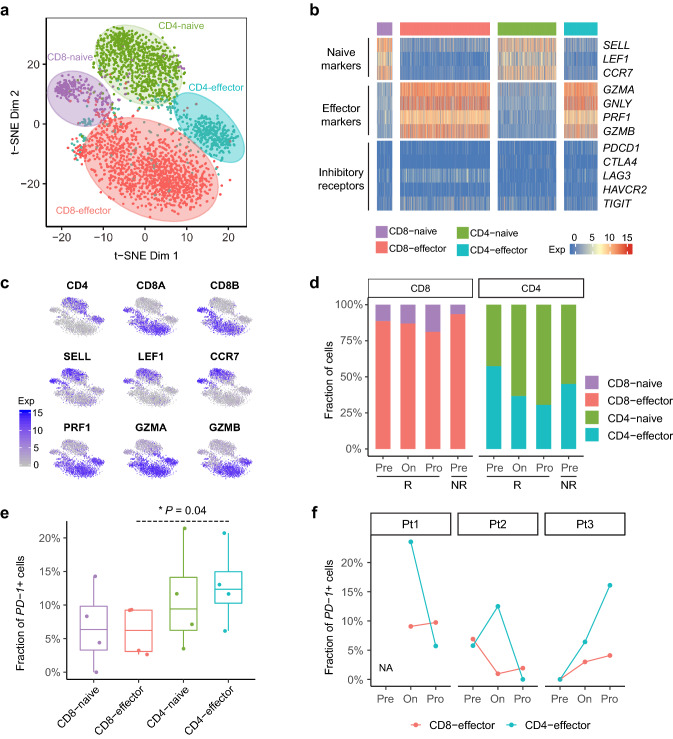
To reveal the functional subtypes of the T cell population, we performed unsupervised clustering analysis on CD8+ and CD4+ T cells separately. Four stable clusters were obtained, including CD8-naïve, CD8-effector, CD4-naïve and CD4-effector, which contained 237, 1,387, 902 and 516 cells, respectively (Fig. 2a). The cells in the CD8-naïve and CD4-naïve clusters specifically expressed “naïve” marker genes, such as SELL, LEF1 and CCR7 (Fig. 2b, c); while the CD8-effector and CD4-effector clusters were characterized by the high expression of GZMA, GNLY, PRF1 and GZMB, which are known to be associated with cytotoxic function [30]. Notably, the percentage of the CD8-effector and CD4-effector T cells decreased in the responders along the anti-PD-1 treatment course (Fig. 2d).
Fig. 2.
T cell subtype analysis based on single-cell gene expression. a The t-SNE projection of 3,042 single T cells from four patients, showing the formation of 4 main clusters illustrated in different colors. Cluster C8-naive: naive CD8+ T cells; C8-effector: effector CD8+ T cells; CD4-naive: naive CD4+ T cells; CD4-effector: effector CD4+ T cells. b Heatmap of four T cell clusters with signatures genes, which included naive, cytotoxic and exhaustion markers. c Expression levels of the selected genes across 3,042 single T cells illustrated in t-SNE plots. Selected genes included CD4, CD8A, CD8B, SELL, LEF1, CCR7, PRF1, GZMA, GZMB. d The percentages of cells in different clusters at pre-treatment, on-treatment and progression. The Pre/On/Pro represent the time points of pre-treatment, on-treatment and progression. The R and NR represent the responder and the nonresponder, respectively. e The percentage of PD-1+ T cells in each T cell cluster. f The percentage of PD-1+ T cells at each time point in three responders
In our dataset, 6.0% (97/1,624) of CD8+ and 10.5% (149 / 1,418) of CD4+ T cells were PD-1 positive, which was consistent with the previous report [31]. The CD4-effector cluster enriched PD-1+ T cells compared with the CD8-effector cluster (P = 0.04, two-sided Student’s t test, Fig. 2e). In all of the three responders, we detected an increase of PD-1+ T cells in the CD8-effector cluster following the tumor progression (Fig. 2f). After the anti-PD-1 treatment, the percentage of PD-1+ T cells in the CD4-effector cluster increased in Pt2 and Pt3. After the progression, the percentages of PD-1+ T cells in the CD4-effector cluster decreased in Pt1 and Pt2, while it increased in Pt3.
As an essential proliferation marker, Ki-67 positivity has been reported to decrease in T cells of ICB-treated NSCLC patients after the tumor progression [32]. In our dataset, however, only 1.8% (29/1,624) of CD8+ and 2.6% (36/1,418) of CD4+ T cells expressed Ki-67, and we did not observe a decreasing pattern.
Identifying T cell clones based on TCR sequences
TCRs can be used to define T cell lineages. We used TraCeR [26] to assemble the full-length sequences of TCR α- and β-chains, and determined the TCR sequences for 98.2% (2,988/3,042) of the peripheral T cells. The T cells with identical paired αβ-chains were regarded as an identical clone from the same ancestry. In total, 78.0% (2,373/3,042) of the T cells had paired TCR αβ-chains, belonging to 1,379 clones.
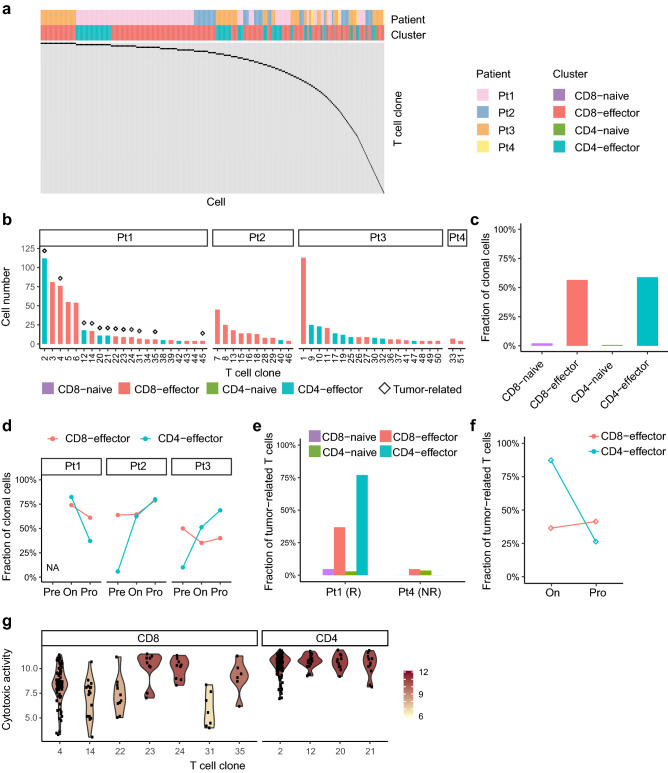
Overall, we detected 113 clonally-expanded T cell clones, containing at least two cells (Fig. 3a). Large expanded clones, with at least 6 cells, were all comprised the effector T cells (Fig. 3b). Different T cell clusters exhibited distinct fractions of clonal T cells. The CD8-effector and CD4-effector clusters had 56.5% (784/1,387) and 58.9% (304/516) of clonal T cells, respectively; while the CD8-naive and CD4-naive clusters had only 2.1% (5/237) and 0.7% (6/902) of clonal T cells, respectively (Fig. 3c). After the anti-PD-1 treatment, the percentage of clonal T cells in the CD4-effector cluster increased from 5.8% (3/52) to 62.5% (5/8) in Pt2 and from 10.0% (1/10) to 51.3% (40/78) in Pt3. Following the tumor progression, we observed decreased fractions of clonal T cells in the CD8-effector and CD4-effector clusters in Pt1, but increased fractions in Pt2 and Pt3 (Fig. 3d).
Fig. 3.
T cell clone analysis based on the TCR sequences. a TCR distribution of clonally-expanded T cells from four patients. Each column represents an individual cell, and each row represents a distinct T cell clone. b The cell numbers of expanded T cell clones. Only clones with ≥ 4 cells were showed. The diamond above the bar represents the tumor-related clone. c The percentage of clonal T cells in each T cell cluster. d The percentage of clonal T cells at each time point in three responders. e The percentage of tumor-related T cells in each cluster in responder Pt1 and nonresponder Pt4. f The percentages of tumor-related T cell in responder Pt1 at on-treatment and progression. g Violin plot showing the cytotoxic activity of tumor-related CD8+ and CD4+ T cell clones of Pt1. The cytotoxic activity was defined as the log-average (geometric mean) of expression of four cytotoxic genes, GZMA, GNLY, PRF1 and GZMB
There exists clonal overlap between peripheral T cells and tumor-infiltrating T cells, and it has been reported that certain T cells in both peripheral blood and tumor tissue could recognize some specific tumor antigens [33]. Therefore, we performed TCR β-chain sequencing on pre-treatment tumor biopsies of Pt1 and Pt4. The T cells in peripheral blood were defined as tumor-related, if their TCR β-sequences could be detected in the tumor tissue of a given patient. In responder Pt1, 32.4% (196/605) of CD8+ and 34.0% (180/530) of CD4+ T cells in blood were tumor-related; while in nonresponder Pt4, only 4.4% (2/46) of CD8+ and 2.0% (1/51) of CD4+ T cells in blood were tumor-related (Fig. 3e). These results indicated that blood T cells from the responder were more likely to be mobilized to the tumor tissue than those from the nonresponder.
Pt1 was a responder whose on-treatment and progression samples were all collected, so we examined this patient in particular. We found that the effector T cells in the peripheral blood of Pt1, compared with naïve T cells, were associated with tumor-infiltrating T cells to a greater extent (Fig. 3e). Among the top ten clones in the peripheral blood of Pt1, three of the six CD8+ and all of the four CD4+ T cell clones were tumor-related (Fig. 3b). After the tumor progression in Pt1, we observed a dramatic decrease in the fraction of CD4-effector T cells that were tumor-related (Fig. 3f). However, we only observed a slight increase in the fraction of tumor-related CD8-effector T cells. We then defined the cytotoxic activity in terms of the log-average (geometric mean) of the expression of four cytotoxic genes, GZMA, GNLY, PRF1 and GZMB. Notably, among the large tumor-related T cell clones with at least six cells, we found a higher cytotoxic activity in the CD4+ T cell clones than that in the CD8+ T cell clones (P < 0.001, two-sided Student’s t test, Fig. 3g). These results suggested that tumor-related CD4+ rather than CD8+ T cell clones played a more important role in killing the tumor.
Dynamics of individual T cell clones during anti-PD-1 treatment
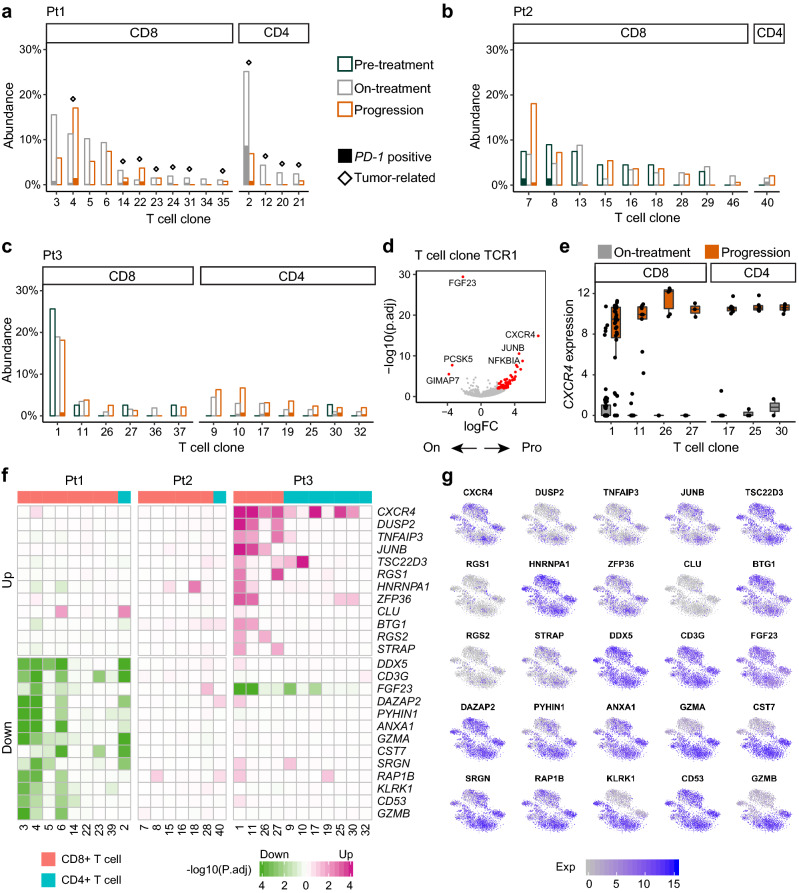
We subsequently investigated the changes in individual T cell clones in peripheral blood during the anti-PD-1 immunotherapy. For the expanded T cell clones occupying more than 1% of the entire cell population, we characterized their abundances along the treatment. In Pt1, all such CD4+ T cell clones could be detected in the tumor tissue (Fig. 4a). The largest CD4+ T cell clone TCR2 showed a dramatic decrease of the abundance from 25.1% (104/414) at on-treatment to 6.9% (8/116) at progression. Of note, the clone TCR2 had 33.0% (37/112) of PD-1+ T cells, which was the highest percentage among all clones. The percentage of PD-1+ T cells in the clone TCR2 decreased from 34.6% (36/104) at on-treatment to 12.5% (1/8) at progression. Because the mechanism of anti-PD-1 therapy requires PD-1 as the binding target, we suggested that the CD4+ T cell clone TCR2 might directly respond to the PD-1 blockade and its decreased fraction at progression hampered the efficacy of the treatment. In Pt2, most of CD8+ T cell clones were present in the pre-treatment blood (Fig. 4b). The percentage of the CD4-effector cells was reduced sharply at on-treatment and progression in Pt2, and clonal CD4+ T cells also decreased. In Pt3, the CD8+ T cell clone TCR1 was the largest clone with 113 cells. Although the fraction of the clone TCR1 decreased slightly after progression, it remained the dominant clone in peripheral blood (Fig. 4c). We also observed six newly-emerging CD4+ T cell clones after the treatment in Pt3.
Fig. 4.
Dynamics of individual T cell clones during the treatment. a, b, c The abundances of individual T cell clones at each time point in Pt1 (a), Pt2 (b) and Pt3 (c). The diamond above the bar represents the tumor-related clone. The height of solid bar represents the percentage of PD-1+ T cells. d Volcano plot showing DEGs between on-treatment and progression time points in the T cell clone TCR1. e Boxplot showing gene expression of CXCR4 in T cell clones from Pt3. f Heatmap showing the frequent DEGs between on-treatment and progression time points in individual T cell clones. g Expression levels of the frequent DEGs illustrated in t-SNE plots
We next compared gene expression profiles between on-treatment and progression samples to explore the dynamic changes in each peripheral T cell clone (Fig. 4d, e). We conducted differential gene expression analyses on 27 T cell clones from the three responders, including 18 CD8+ and 9 CD4+ T cell clones. In total, we detected 524 differentially expressed genes (DEGs), including 178 upregulated and 346 downregulated genes (Fig. 4f, g). The heatmap of changes for the 25 most frequently upregulated and downregulated genes showed distinct patterns between T cell clones from different patients. Several well-known T cell functional genes, such as CXCR4, DUSP2 and ZFP36, were upregulated after progression in multiple T cell clones (Fig. 4e, f). Meanwhile, we observed decreased GZMA and GZMB expression in T cell clones of Pt1 (Fig. 4f).
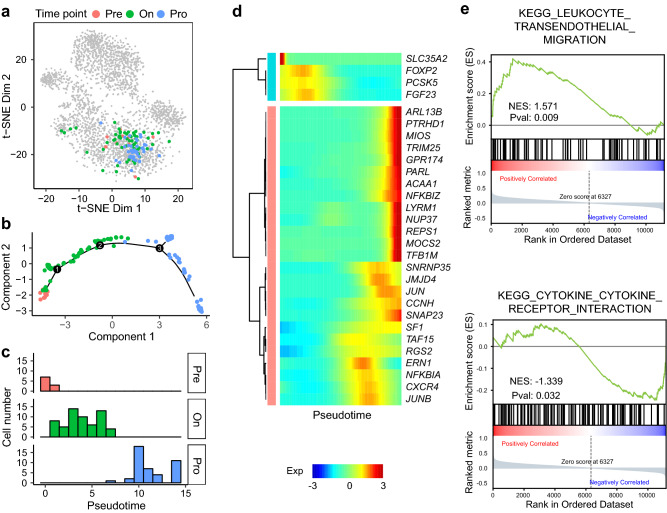
The abundance of the T cell clone TCR1, which was the dominant clone in the peripheral blood of Pt3 (Fig. 5a), decreased from 25.6% (10/39) at pre-treatment to 18.9% (60/317) at on-treatment to 18.1% (43/237) at progression. We subsequently evaluated how the gene expression landscape was altered in this clone during the anti-PD-1 immunotherapy. We used Monocle [27] to order these cells along a pseudotime trajectory on the basis of similarities in their expression patterns (Fig. 5b). The inferred developmental trajectory suggested a continuous structure, with progression cells positioned at the opposite direction of pre-treatment cells. The pseudotime trajectory corresponded to the real time points, from pre-treatment to on-treatment to progression (Fig. 5c). We next identified pseudotime-dependent genes, which were the differentially expressed genes on the different paths along the pseudotime trajectory (Fig. 5d). Based on the pseudo-temporal expression profiles of these pseudotime-dependent genes, we conducted gene set enrichment analysis (GSEA). The genes involved in the “leukocyte transendothelial migration” pathway were upregulated and the genes involved in the “cytokine and cytokine receptor interaction” pathway were downregulated along the pseudotime (Fig. 5e). These results suggested that downregulation of cytokine in the clone TCR1 might lead to dysfunction after progression, although the abundance of this clone remained relatively high in peripheral blood.
Fig. 5.
Trajectory analysis and related genes of CD8+ T cell clone TCR1. a Cells of the T cell clone TCR1 illustrated in t-SNE plot. b Pseudotime trajectory of the T cell clone TCR1 by Monocle. c Distribution of cells from different time points in pseudotime inferred by trajectory analysis. d Expression level of pseudotime-dependent genes. e Gene set enrichment analysis showing that cells of the T cell clone TCR1 exhibited higher activity in the “leukocyte transendothelial migration” pathway and lower activity in the “cytokine and cytokine receptor interaction” pathway along the pseudotime
Discussion
In summary, we applied Smart-seq2 scRNA-seq to study longitudinal peripheral T cells throughout the course of anti-PD-1 treatment. T cells, as the direct target of PD-1 blockade, are highly heterogeneous, and only a subset of T cell clones are responsive to the tumor-related antigens [33]. Recently, Wu et al. observed that expanded clonotypes in the tumor can also typically be detected in peripheral blood, suggesting that the identification of expanded clones in blood may characterize the TCR composition of clinically relevant intratumoral T cells [34]. Fairfax et al. also found that post-ICB peripheral CD8+ clonality can provide information regarding long-term treatment response in metastatic melanoma patients [35]. Consequently, it is necessary to divide T cells into different clones based on their TCR sequences. Traditional TCR sequencing uses only TCR β-chains to define the clones. By taking advantage of Smart-seq2 scRNA-seq, we assembled the full-length TCR sequences with paired αβ-chains and tracked the individual T cell clones. Moreover, we defined tumor-related T cells in peripheral blood by comparing TCR β-sequences of peripheral T cells and tumor-infiltrating T cells. In this study, we observed distinct patterns in T cell clone dynamics among the three responders. The fraction of tumor-related CD4+ T cell clones was reduced after progression in Pt1, while the fraction of expanded CD4+ T cell clones increased slightly in Pt2 and Pt3. T cell clones from the individual patient had similar DEGs between on-treatment and progression samples, while T cell clones from the different patients had distinct gene expression profiling. Based on the observations from Pt1, who exhibited partial response to the initial immunotherapy, we hypothesized that the dominant PD-1+CD4+ T cell clone in peripheral blood indicated a better response to anti-PD-1 treatment. Meanwhile, a decreased abundance of the dominant PD-1+CD4+ T cell clone in peripheral blood might be a potential biomarker of acquired resistance to PD-1 blockade. In addition, we developed an interactive website (https://118.190.148.166:3838/lcpd) for analyzing and visualizing dynamic changes in each T cell clone.
Through scRNA-seq, several T cell functional genes were identified, showing differential expression between on-treatment and progression samples. CXCR4 is a key receptor in the crosstalk between tumor cells and their microenvironment [36]. It has been previously reported that tumor-infiltrating T cells expressed an abundance of CXCR4, and CXCL12-CXCR4 chemotaxis has been implicated in the migration of T cells in the tumor microenvironment [37]. In mice, CXCR4 blockade greatly increased T cell-mediated anti-tumor immune responses, conferring a significant survival advantage to CXCR4 antagonist-treated mice [38]. Previous evidence has shown the potential of CXCR4 blockade as the combination therapy to overcome resistance to anti-PD-1 immunotherapy [39], thus the combination therapy targeting both CXCR4 and PD-1 should be carefully evaluated in the future. DUSP2, also called PAC-1, is a member of the dual specificity protein phosphatase subfamily and has been demonstrated to be closely associated with the T cell function [40]. Previous genome-wide mRNA expression profiling has shown that DUSP2 was upregulated in the tolerant T cells but was downregulated in the rescued T cells [41]. ZFP36, also known as tristetraprolin, is an AU-rich elements (ARE)-binding factor and is expressed in hematopoietic cell lineages [42, 43]. Previous research has revealed that ZFP36 played a critical role in restraining T cell expansion and effector functions [44]. These DEGs upon progression might serve as biomarkers to monitor the progression status in NSCLC patients with anti-PD-1 treatment, and such upregulated genes could be potential targets for combination therapy to overcome the acquired resistance to anti-PD-1 treatment.
Our study also has several limitations. The high cost of Smart-seq2 method prevents us from obtaining a large sample of cases. Although the new 10× Genomics approach significantly reduces the cost of single cell sequencing, its gene detection sensitivity is much lower [45]. Therefore, we feel that we should still focus on using the most sensitive method to take the most advantage of the valuable sample of patients. Although the sample size was limited in this study, we provided a comprehensive dataset concerning HLA types, personalized neoantigens, cell clusters, TCR repertoires and T cell expression profiles for all patients. Future study with large sample size is likely to uncover the regulatory association between these molecular factors in relation to ICB response. Another potential weakness is that we did not observe consistent patterns of peripheral T cell dynamics across the three responders, which highlight the patient heterogeneity. Large cohort studies are thus needed to replicate our research and validate the findings on the biomarkers regarding anti-PD-1 treatment. With a large NSCLC cohort, a consistent dynamic pattern of peripheral T cell clones is likely to emerge, and there is a high chance of discovering more reliable biomarkers in peripheral blood. Although this study cannot definitely and comprehensively define the T cell dynamic patterns post treatment, we nonetheless feel that our study paved the road for further such studies at much larger scale, which are expected to emerge in the coming years.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Xuefang Zhang and Fei Wang for assistance with flow cytometry. We thank the Computing Platform of the CLS (Peking University).
Abbreviations
- DEG
Differentially expressed gene
- ICB
Immune checkpoint blockade
- NSCLC
Non-small cell lung cancer
- PD-1
Programmed cell death protein 1
- PD-L1
Programmed death-ligand 1
- scRNA-seq
Single-cell RNA sequencing
- TCR
T cell receptor
Author contributions
XH and JW designed experiments. RG and XH performed the experiments. FZ analyzed sequencing data. HB, KF and JD collected clinical samples. FZ and XH wrote the initial draft of the manuscript. HB, KF, ZZ and JW contributed to analysis and interpretation of data, and critically reviewed the manuscript. All authors approved the final version of the manuscript.
Funding
This project was supported by Beijing Advanced Innovation Center for Genomics at Peking University, National Natural Science Foundation of China (31530036, 91742203, 91942307, 81988101, 81630071), National Key Research and Development Project (2019YFC1315700) and CAMS Innovation Fund for Medical Sciences (CIFMS 2016-I2M-3-008).
Data availability
Sequencing raw data is available at GSA (Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences). The accession number is HRA000104. Gene expression profiling of individual T cell clones at different time points can be explored on https://118.190.148.166:3838/lcpd.
Compliance with ethical standards
Conflict of interest
The authors report no financial interests or potential conflicts of interests.
Ethical approval
This study was approved by the Research and Ethical Committee of Cancer Hospital, Chinese Academy of Medical Sciences, China and complied with relevant ethical regulations.
Informed consent
Written informed consents were provided by all patients.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fan Zhang and Hua Bai authors contributed equally.
Contributor Information
Jie Wang, Email: zlhuxi@163.com.
Xueda Hu, Email: hu_xue_da@pku.edu.cn.
References
- 1.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen P-L, Roh W, Reuben A, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016;6:827–837. doi: 10.1158/2159-8290.CD-15-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prat A, Navarro A, Paré L, et al. Immune-related gene expression profiling after pd-1 blockade in non-small cell lung carcinoma, head and neck squamous cell carcinoma, and melanoma. Cancer Res. 2017;77:3540–3550. doi: 10.1158/0008-5472.CAN-16-3556. [DOI] [PubMed] [Google Scholar]
- 6.Riaz N, Havel JJ, Makarov V, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daud AI, Wolchok JD, Robert C, et al. Programmed death-ligand 1 expression and response to the anti-programmed death 1 antibody pembrolizumab in melanoma. J Clin Oncol. 2016;34:4102–4109. doi: 10.1200/JCO.2016.67.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang AC, Postow MA, Orlowski RJ, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545:60–65. doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jerby-Arnon L, Shah P, Cuoco MS, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. 2018;175:984–997.e24. doi: 10.1016/j.cell.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulson KG, Voillet V, McAfee MS, et al. Acquired cancer resistance to combination immunotherapy from transcriptional loss of class I HLA. Nat Commun. 2018;9:3868. doi: 10.1038/s41467-018-06300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-Mutant lung adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finotello F, Trajanoski Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol Immunother. 2018;67:1031–1040. doi: 10.1007/s00262-018-2150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo X, Zhang Y, Zheng L, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Yu X, Zheng L, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 16.Zheng C, Zheng L, Yoo J-K, et al. Landscape of Infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 17.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anagnostou V, Forde PM, White JR, et al. Dynamics of tumor and immune responses during immune checkpoint blockade in non-small cell lung cancer. Cancer Res. 2019;79:1214–1225. doi: 10.1158/0008-5472.CAN-18-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osa A, Uenami T, Koyama S, et al. Clinical implications of monitoring nivolumab immunokinetics in non-small cell lung cancer patients. JCI Insight. 2018;3:e59125. doi: 10.1172/jci.insight.59125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Duan J, Cai S, et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol. 2019;5:696–702. doi: 10.1001/jamaoncol.2018.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szolek A, Schubert B, Mohr C, et al. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30:3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurtz V, Paul S, Andreatta M, et al. NetMHCpan-4.0: Improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Picelli S, Faridani OR, Björklund ÅK, et al. Full-length RNA-seq from single cells using smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 24.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 25.Kiselev VY, Kirschner K, Schaub MT, et al. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods. 2017;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stubbington MJT, Lönnberg T, Proserpio V, et al. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods. 2016;13:329–332. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu X, Hill A, Packer J, et al. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter KA, Socinski MA, Villaruz LC. PD-L1 testing in guiding patient selection for PD-1/PD-L1 inhibitor therapy in lung cancer. Mol Diagn Ther. 2018;22:1–10. doi: 10.1007/s40291-017-0308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hidalgo LG, Einecke G, Allanach K, Halloran PF. The Transcriptome of human cytotoxic T cells: similarities and disparities among allostimulated CD4+ CTL, CD8+ CTL and NK cells. Am J Transplant. 2008;8:627–636. doi: 10.1111/j.1600-6143.2007.02128.x. [DOI] [PubMed] [Google Scholar]
- 31.Thommen DS, Koelzer VH, Herzig P, et al. A transcriptionally and functionally distinct PD-1 + CD8 + T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018;24:994–1004. doi: 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osa A, Uenami T, Koyama S, et al. Clinical implications of monitoring nivolumab immunokinetics in non–small cell lung cancer patients. JCI. 2018;3:12. doi: 10.1172/jci.insight.59125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gros A, Parkhurst MR, Tran E, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med. 2016;22:433–438. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu TD, Madireddi S, de Almeida PE, et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature. 2020;579:274–278. doi: 10.1038/s41586-020-2056-8. [DOI] [PubMed] [Google Scholar]
- 35.Fairfax BP, Taylor CA, Watson RA, et al. Peripheral CD8 + T cell characteristics associated with durable responses to immune checkpoint blockade in patients with metastatic melanoma. Nat Med. 2020;26:193–199. doi: 10.1038/s41591-019-0734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burger JA, Kipps TJ. CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107:1761–1767. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- 37.Peng L, Zhuang Y, Shi Y, et al. Increased tumor-infiltrating CD8+Foxp3+ T lymphocytes are associated with tumor progression in human gastric cancer. Cancer Immunol Immunother. 2012;61:2183–2192. doi: 10.1007/s00262-012-1277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Righi E, Kashiwagi S, Yuan J, et al. CXCL12/CXCR4 blockade induces multimodal anti-tumor effects that prolong survival in an immunocompetent mouse model of ovarian cancer. Cancer Res. 2011;71:5522–5534. doi: 10.1158/0008-5472.CAN-10-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scala S. Molecular pathways: targeting the CXCR4–CXCL12 axis—untapped potential in the tumor microenvironment. Clin Cancer Res. 2015;21:4278–4285. doi: 10.1158/1078-0432.CCR-14-0914. [DOI] [PubMed] [Google Scholar]
- 40.Rohan PJ, Davis P, Moskaluk CA, et al. PAC-1: a mitogen-induced nuclear protein tyrosine phosphatase. Science. 1993;259:1763–1766. doi: 10.1126/science.7681221. [DOI] [PubMed] [Google Scholar]
- 41.Schietinger A, Delrow JJ, Basom RS, et al. Rescued tolerant cd8 t cells are preprogrammed to reestablish the Tolerant State. Science. 2012;335:723–727. doi: 10.1126/science.1214277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-α production by Tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 43.Raghavan A, Robison RL, McNabb J, et al. HuA and tristetraprolin are induced following T cell activation and display distinct but overlapping rna binding specificities. J Biol Chem. 2001;276:47958–47965. doi: 10.1074/jbc.M109511200. [DOI] [PubMed] [Google Scholar]
- 44.Moore MJ, Blachere NE, Fak JJ, et al. ZFP36 RNA-binding proteins restrain T cell activation and anti-viral immunity. ELife. 2018;7:3–57. doi: 10.7554/eLife.33057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, He Y, Zhang Q, et al. Direct comparative analysis of 10x genomics chromium and smart-seq2. BioRxiv. 2019;61:5–13. doi: 10.1016/j.gpb.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing raw data is available at GSA (Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences). The accession number is HRA000104. Gene expression profiling of individual T cell clones at different time points can be explored on https://118.190.148.166:3838/lcpd.