Abstract
Transmissible gastroenteritis virus (TGEV), an enteric coronavirus of swine, is a potent inducer of alpha interferon (IFN-α) both in vivo and in vitro. Incubation of peripheral blood mononuclear cells with noninfectious viral material such as inactivated virions or fixed, infected cells leads to early and strong IFN-α synthesis. Previous studies have shown that antibodies against the virus membrane glycoprotein M blocked the IFN induction and that two viruses with a mutated protein exhibited a decreased interferogenic activity, thus arguing for a direct involvement of M protein in this phenomenon. In this study, the IFN-α-inducing activity of recombinant M protein expressed in the absence or presence of other TGEV structural proteins was examined. Fixed cells coexpressing M together with at least the minor structural protein E were found to induce IFN-α almost as efficiently as TGEV-infected cells. Pseudoparticles resembling authentic virions were released in the culture medium of cells coexpressing M and E proteins. The interferogenic activity of purified pseudoparticles was shown to be comparable to that of TGEV virions, thus establishing that neither ribonucleoprotein nor spikes are required for IFN induction. The replacement of the externally exposed, N-terminal domain of M with that of bovine coronavirus (BCV) led to the production of chimeric particles with no major change in interferogenicity, although the structures of the TGEV and BCV ectodomains markedly differ. Moreover, BCV pseudoparticles also exhibited interferogenic activity. Together these observations suggest that the ability of coronavirus particles to induce IFN-α is more likely to involve a specific, multimeric structure than a definite sequence motif.
Transmissible gastroenteritis virus (TGEV), a highly enteropathogenic virus of swine, belongs to the Coronavirus genus, a group of enveloped viruses with a large (about 30-kb), positive-stranded RNA molecule as a genome (14; reviewed in reference 29). Like those of other coronaviruses, TGEV virions are built from three major proteins: the N protein (47 kDa), with which the genomic RNA is packaged, and two envelope glycoproteins, S (220 kDa) and M (29 to 36 kDa) (29). Trimers of S protein form the prominent coronavirus spikes (12). M, the most abundant component of coronavirions, is a polytopic protein spanning the membrane bilayer three times (reviewed in reference 42). When synthesized in the absence of other viral components, M protein tends to accumulate in the Golgi complex as detergent-insoluble, polymeric structures, presumably as part of its retention mechanism (26, 27). A third, small transmembrane protein, designated E (formerly sM), has been found to be present at a low copy number in TGEV virions (19), as well as in infectious bronchitis virus (IBV) and mouse hepatitis virus (MHV) (35, 51). Coronavirus assembly has been shown to take place at membranes of the intermediate compartment, between the endoplasmic reticulum and the Golgi complex (25). Recently, substantial progress toward an understanding of the mechanism of assembly has been made, with the demonstration that coexpression of the MHV M and E proteins leads to the formation and secretion of particles closely resembling authentic virions (6, 48).
In vivo, TGEV replication takes place primarily in the differentiated enterocytes covering the villi of the small intestine (reviewed in reference 15). An ectoenzyme abundantly expressed at the brush border, aminopeptidase N, has been shown to act as a receptor for virus entry into target cells (11). In infected animals, high titers of interferon (IFN) are detected at both the local and systemic levels (28). The observed IFN activity is primarily alpha IFN (IFN-α), thus making enterocytes unlikely to represent the major IFN-producing cells. Subsequent studies have revealed that incubation of swine peripheral blood mononuclear (PBM) cells from naive animals with TGEV-infected, fixed cultures or purified virus, as well as in vivo injection of UV-inactivated virions, leads to a marked and early IFN-α synthesis (8, 38). Together, these observations support the view that such an induction pathway, involving nonreplicating viral elements, mainly accounts for the strong IFN response observed in TGEV infection.
There is now substantial evidence that IFN-α synthesis can be triggered by inert viral structures, indicating that replication and/or gene expression is not required for efficient IFN induction at least in certain categories of lymphoid cells. This kind of IFN-α-inducing activity has been reported for enveloped viruses belonging to many different groups, including paramyxo-, orthomyxo-, lenti-, rhabdo-, corona-, and herpesviruses (reviewed in reference 16). In vitro studies using virus-induced blood leukocyte preparations have allowed the definition of the IFN-α-secreting cells as a small but highly productive leukocyte subpopulation, referred to as natural interferon-producing (NIP) cells, able to synthesize IFN-α after a brief contact with noninfectious viral structures (16). Studies performed with human and pig blood leukocytes have shown that NIP cells are nonphagocytic, nonadherent cells that lack T, B, and natural killer cell lineage-specific markers but express CD4 and major histocompatibility complex class II molecules (36, 44, 47).
The stimulation of IFN synthesis is assumed to be determined by an interaction of the NIP cell surface with one or several viral components, but a limited number of studies have been aimed at understanding the nature of these viral components. Based on inhibitory effects of relevant antibodies, viral glycoproteins such as human herpes simplex virus protein D, human immunodeficiency virus (HIV) type 1 gp120, Newcastle disease virus HN, and viral hemorrhagic septicemia virus G protein have been proposed to play a role in IFN induction (17, 23, 34, 41). In the case of TGEV, two lines of evidence have pointed to a role of the membrane protein M as the major inducing component. First, within a panel of antibodies directed to the TGEV structural proteins, only two blocked IFN induction, with both of them recognizing the short N-terminal ectodomain of M (8). Second, two of the epitope mutants selected toward the above antibodies exhibited a markedly decreased interferogenic activity. The substitutions identified in both mutant M genes mapped within the ectodomain and impaired the glycosylation of the protein, without affecting the virus infectivity (32).
The use of recombinant expression is a straightforward approach in assessing any interferogenic activity of individual viral proteins. Recently, two viral proteins have been reported to induce IFN-α production by lymphohematopoietic cells independently from any other viral component. Isolated recombinant HIV gp120 protein exhibited some interferogenic activity for human PBM cells (2, 7). Cells expressing HN protein of human parainfluenza virus type 4A had an ability to induce IFN in mouse spleen cells (22). In contrast, our attempts at demonstrating interferogenic activity associated with recombinant TGEV M protein, transiently or stably expressed in mammalian cells, consistently failed (3, 4). Similar negative results were obtained when TGEV virion-derived envelope proteins reconstituted into virosomes were used (39). An interesting possibility suggested by these results was that interaction of M protein with NIP cells would not be sufficient in itself to trigger IFN synthesis, because either the putative interferogenic determinant needs to be presented in a specific context (such as the lipid bilayer) or the interferogenicity is subordinated to a polymeric structure. In order to test this hypothesis, the incidence of the expression of other viral proteins upon IFN-α induction by TGEV M was examined. It was found that coexpression of M and E proteins allowed the formation of pseudoparticles which exhibit an interferogenic activity similar to that of complete virions.
(Part of these data have been presented at the International Symposium on Coronaviruses and Arteriviruses, Segovia, Spain, May 1997.)
MATERIALS AND METHODS
Virus and antibodies.
The Purdue-115 strain was used as a TGEV source and propagated in ST cells as previously described (31). The following monoclonal antibodies (MAbs) were used: 25.22 and 3.60, directed to the N-terminal and cytosolic regions of TGEV M, respectively (31, 32); C29, directed to the C-terminal region of TGEV E (19); 53.13 and 5.1, specific for TGEV S and N proteins, respectively (31); and 27P22 as an anti-bovine coronavirus (BCV) E antibody (supplied by J. F. Vautherot, Jouy-en-Josas, France).
PBM cells.
Nonadherent porcine PBM cells were obtained from heparinized blood by Ficoll density centrifugation on MSL (density, 1.077 g/ml; Eurobio, Paris, France) followed by adherent-cell depletion on tissue culture flasks as described previously (36). PBM cells were suspended in RPMI 1640 medium supplemented with 10% fetal calf serum and antibiotics for IFN-α induction assays.
IFN-α induction.
PBM cells were induced to produce IFN-α by overnight incubation at 37°C with TGEV in 96-well microplates. Nonadherent cells were incubated at a final concentration of 8 × 106 cells per ml in a total volume of 0.2 ml with TGEV or pseudoparticles at various dilutions, as shown in Results. In some of the experiments, PBM cells (8 × 106 cells per ml in a total volume of 0.5 ml) were incubated overnight in 24-well plates with infected or transfected cell monolayers fixed with 0.25% glutaraldehyde as described previously (8). Cell supernatants were collected and stored at −20°C before IFN-α immunoassay.
IFN-α immunoassay.
A specific enzyme-linked immunosorbent assay (ELISA) for porcine IFN-α was performed as previously described (10, 46) by using MAb K9 for coating and MAb F17 as a second antibody. In each assay, our internal standard of recombinant porcine IFN-α was included.
M antigen quantification.
M antigen associated with purified pseudoparticles and virions was quantified by an ELISA as follows. Serial threefold dilutions in 0.1 M sodium carbonate-bicarbonate buffer (pH 9.6) were incubated overnight in a 96-well plate (PROBIND); coated wells were saturated in phosphate-buffered saline–0.05% Tween 20–3% bovine serum albumin for 2h at 37°C and then incubated for 90 min at 37°C with MAb 25.22, 3.60, or 51.13 (as a negative control for pseudoparticles) ascites fluid diluted 1/1,000 in the same buffer. After washing, the samples were incubated with anti-immunoglobulin G (heavy plus light chains) rabbit antibodies conjugated with alkaline phosphatase (1 μg/ml; Biosys) for 1 h at 37°C, washed, and incubated for 15 to 30 min with para-nitrophenyl phosphate (Sigma) diluted at 1 mg/ml in 0.05 M sodium carbonate-bicarbonate–10 mM MgCl2 buffer (pH 9.8). The reaction was stopped by addition of 0.1 M NaOH. The optical density was measured at 405 nm on a Titertek Multiscan MCC/340. M and S antigens associated with fixed cell monolayers were quantified by the same procedure.
Plasmid construction.
TGEV and BCV M and E homologous and chimeric genes have been cloned into plasmid vectors providing the bacteriophage T7 transcription-regulatory sequences. All ligation junctions and sequences of inserts generated by PCR were confirmed by automated sequencing (Applied Biosystems 373A). The oligonucleotide primers used are listed in Table 1.
TABLE 1.
Amplimers used
| Primer | Sequence (5′-3′)a | Restriction site | Positionb | Gene |
|---|---|---|---|---|
| 2190 | AGCACTGCAAGCTTGAACTA | HindIII | −7 | TGEV M |
| 2282 | GTTGGCGAATTCGAAGTTTAGTTATACCATATG | EcoRI | 778 | TGEV M |
| 3247 | CTAAACACCATGGCGATTTTGTTAATATTAGCGTGTG | NcoI | 28 | TGEV M |
| 2414 | GAAGAAGGGATCCATAACTATGGCGTTTCC | BamHI | 11 | TGEV E |
| 2030 | TTGTCTAGATCAAGCAAGGAGTG | XbaI | 235 | TGEV E |
| 66830 | CCGATTCCATGGCTAGTGTAACTACACCAGCACCA | NcoI | 27 | BCV M |
| 66841 | AGGGATCCTTAAAGTTTAGATATTATTTCTC | BamHI | 678 | BVC M |
| 627 | ATGTTTATGGCTGATGC | 17 | BCV E | |
| 519 | GTAAACTGGTGCTGGTGTAGTTAC | 262 | BCV E | |
| 2373 | ATAGACCAGCTGAAGTTCCATTCCTTTAGGAA | 61 | BCV M | |
| 72916 | GGAACTTCAGCTGGTCTATAA | 181 | TGEV M | |
| 85138 | TCCAGTTCGCGAGATGCCAAATAAGATCGCC | 133 | TGEV M | |
| 85139 | TTGGCATCTCGCGAACTGGAACTTTTCTTTGGGTATTATAC | 97 | BCV M | |
| 2119 | TAATACGACTCACTATAGG | T7 promoter | ||
| 85137 | AGTTGAATGCATAATTTAAAAGTTGCCAAAAATGCC | NsiI | 98 | BCV E |
| 85136 | TTAAATTATGCATTCAACTTTGCGGTATGTG | NsiI | 145 | BCV E |
Overlapping regions for primer pairs 2373-72916 and 85138-85139 are in boldface; restriction sites are underlined.
Position of the last nucleotide of the primer relative to that of the first coding nucleotide.
(i) M gene constructs.
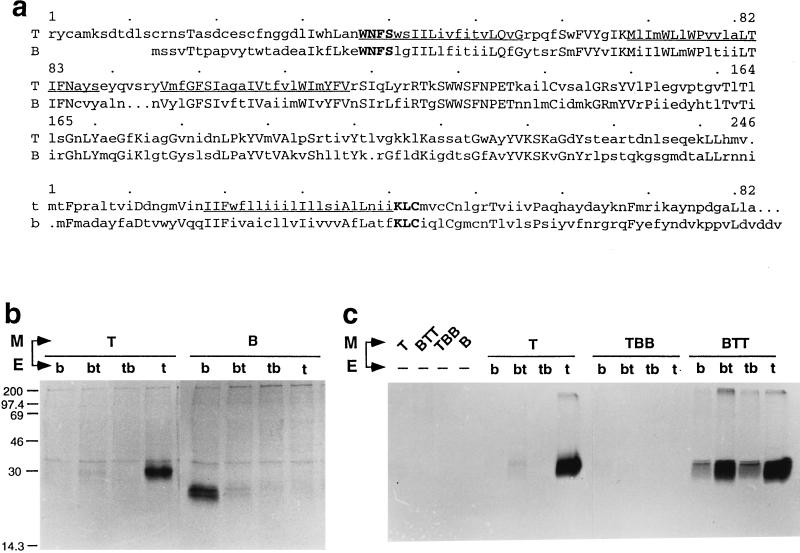
TGEV M cDNA was obtained by reverse transcription-PCR of total mRNA from TGEV-infected ST cells with primers 2190 and 2282 and cloned between the HindIII and EcoRI sites of pcDNA1 (Invitrogen Corporation) to give plasmid pTM2. The BCV M gene was PCR amplified from p174 (45) with primers 66830 and 66841 and cloned into the EcoRV site of pBluescript II SK(−) (Stratagene). The BTT chimera, which encodes TGEV N-proximal sequence fused with BCV N-distal sequence at the homologous stretch WNFS (see Fig. 5a), was constructed by insertion of PCR fragments between the NcoI and EcoRI sites of pTM1 (provided by B. Moss). The fragments were obtained by two-step amplification of p174 and pTM2 with primers 66830-2373 and 72916-2282, respectively, followed by amplification with primers 66830 and 2282, respectively. The reciprocal chimera TBB, i.e., encoding TGEV N-proximal sequence fused with BCV N-distal sequence, was obtained by cloning into the EcoRV site of pBluescript a PCR fragment obtained by the same strategy described above with primer pairs 3247-85138 and 85139-66831 followed by 3247 and 66831.
FIG. 5.
Production of pseudoparticles by coexpression of heterologous and/or chimeric M and E genes of TGEV and BCV viruses. (a) Alignments of the sequences of mature TGEV and BCV M proteins (T and B, respectively) and of TGEV and BCV E genes (t and b, respectively). The predicted transmembrane segments are underlined in the TGEV sequences. Identical amino acids are in uppercase letters. In boldface are shown two short homologous regions selected to fuse TGEV and BCV sequences in the chimeric M or E gene constructs used in these experiments. Sequence data are from references 33 (TGEV M), 30 (BCV M), 19 (TGEV E), and 1 (BCV E). (b and c) Various pairs of homologous and chimeric constructs were expressed through the vT7-3 expression system. Particle assembly and secretion were assessed by monitoring the release of soluble, radiolabeled M material in the culture medium of transfected cells. Sedimentable material was collected by centrifugation through a glycerol cushion and analyzed by SDS–12% PAGE and autoradiography. The E chimera designated bt corresponds to the BCV amino-half sequence fused with the TGEV carboxy-half sequence, and tb is the reciprocal construct. The M chimera designated TBB corresponds to the TGEV N-terminal ectodomain fused with the remaining part of BCV M sequence, and vice versa for the BTT chimera. TBB and BTT chimeric proteins were expressed at equivalent levels as judged by immunoprecipitation assays of lysates of transfected cells with MAbs 25.22 and 3.60, respectively. Numbers on the left of panel b are molecular weights in thousands.
(ii) E gene constructs.
The TGEV E gene was PCR amplified from pTG2.15 (37) with primers 2414 and 2030 and cloned between the BamHI and XbaI sites of pcDNA1 to give plasmid pcsM. In this construct, Thr at position 2 is replaced by Ala as a consequence of the optimization of the ATG context. The BCV E gene was amplified from pT7T3NS3 (50) with primers 627 and 519 and cloned between the EcoRI and BamHI sites of pcDNA3 to give plasmid p3NS3. The bt chimera, specifying a protein with TGEV N-terminal and BCV C-terminal sequences fused at the homologous stretch KLC (see Fig. 5a), and the reciprocal chimera tb were constructed by use of an NsiI site present within the KLC stretch. For bt, PCR fragments amplified from p3NS3 with primers 2119 and 85137 were cloned after BamHI-NsiI restriction into the corresponding sites of pcsM to give pc34. For tb, PCR fragments amplified from pT7T3NS3 with primers 85136 and 2119 were cloned into the EcoRV site of pBluescript. After BamHI-NsiI restriction, the insert was subcloned into the corresponding sites of pTMsM, to give pTM43. This plasmid was subjected to EcoRI-BamHI digestion and ligation to remove the T7 promoter issued from pT7T3NS3.
(iii) Other constructs.
The TGEV N gene was cloned into pcDNA1 as an EcoRV fragment from pTG2.18 (37) to give the plasmid pcN. pZG35 was used to express the full-length TGEV S protein under control of the T7 promoter (20).
Transfection, metabolic labeling, and immunoprecipitation.
Confluent monolayers of RK13 cells were infected with vaccinia virus vTF7.3 (18) at a multiplicity of infection of 5. At 1 h postinfection the cells were transfected with Lipofectamine containing the appropriate plasmids in Optimem as recommended by the manufacturer (GibcoBRL). At 3 to 5 h after DNA transfection, the cells were metabolically labeled by using methionine-depleted medium (GibcoBRL) containing 2% fetal calf serum and 100 μCi of [35S]methionine-cysteine (Tran35S-label; ICN Biomedicals) per ml. At 10 h postinfection the medium was collected. The cells were lysed with 1× immunoprecipitation buffer (50 mM Tris [pH 8], 40 mM EDTA, 0.5% Na deoxycholate, 0.5% Nonidet P-40, 0.5 ml of aprotinin per liter), and the lysate was centrifuged at 4°C for 10 min at 10,000 × g. Immunoprecipitation was performed with MAbs and protein A, and the resulting samples were analyzed by reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography, as previously described (31).
Velocity sedimentation.
Culture medium of vTF7.3-infected cells expressing M and E proteins or of TGEV-infected cells labeled from 5 to 10 h postinfection was cleared at 10,000 × g for 10 min and loaded on a linear 20 to 45% sucrose gradient. After centrifugation for 4 h at 25,000 rpm in a Beckman SW40 rotor at 4°C, 20 fractions were collected from the bottom, diluted 1:1, and centrifuged for 15 min at 300,000 × g. Pelleted material was analyzed by SDS-PAGE as described above.
Purification of pseudoparticles.
The starting material was produced from five 162-cm2 flasks (Costar) of RK13 cells coexpressing the TGEV M and E genes. Each monolayer was infected with vTF7.3 at a multiplicity of infection of 3 and then transfected with 10 ml of a 50-ml mixture containing 40 and 120 μg of pTMM2 and pcsM plasmid DNAs, respectively, and 1.2 ml of Lipofectin in Optimem. After 5 h, the monolayers were rinsed and maintained in Eagle minimal essential medium plus 10% fetal calf serum. The culture media were collected at 24 h postinfection and processed as described for TGEV virions (31). The resulting concentrated material was loaded onto two 20 to 45% sucrose gradients and centrifuged for 3.30 h in an SW27 rotor. Pseudoparticle material was visible as a discrete, opalescent band which was collected directly.
Electron microscopy.
Formvar-carbon-coated grids were put in contact with a purified pseudoparticle suspension for 2 min, rinsed with phosphate-buffered saline plus 2% bovine serum albumin, and incubated for 30 min with anti-TGEV M antibody 25.22 and then with protein G coupled to 10-nm-diameter gold particles (British Biocell International). Grids were viewed on a Philips EM12 electron microscope after staining with 2% phosphotungstic acid (pH 6.8).
RESULTS
IFN induction by cells coexpressing TGEV M and E proteins.
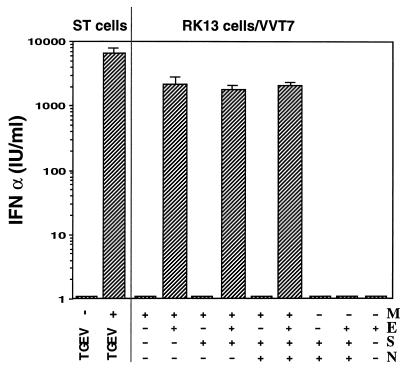
The vaccinia virus-T7-driven system was used to express the M protein alone or together with one or several of the other structural proteins, including the minor structural protein E. After fixation, the cell monolayers were incubated with swine PBM cells, and the IFN activity present in the culture medium was quantified (Fig. 1). As a striking result, cotransfection of M and E genes was found to be an absolute requirement for IFN induction. The level of IFN production was higher than 103 IU/ml, a titer which is not far from that observed when TGEV-infected, fixed cell cultures are used as an inducer in the same assay. The IFN induction was parallel to expression of M antigen at the surface of the fixed cells, as determined by an ELISA test (data not shown). This is consistent with the notion that accessibility of M to PBM cells is a prerequisite for IFN induction. In addition, IFN induction was found not to be enhanced by the additional expression of the virion proteins S and/or N. Both proteins were found to be readily detectable in cultures cotransfected with the four structural genes, based on ELISA and indirect immunofluorescence tests. From these results, it was concluded that the coexpression of M and E was both necessary and sufficient to induce IFN synthesis. This phenomenon appears to be independent of the cell context, as the same conclusion was reached when Cos7 or ST cells were used for coexpression (data not shown) instead of RK13 cells as done for Fig. 1.
FIG. 1.
Induction of IFN-α by cell cultures expressing TGEV structural proteins. Vaccinia virus vT7-3-infected RK13 cells were transfected with Lipofectamine containing one or a combination of plasmids, each containing one of the TGEV genes, as indicated. The monolayers were fixed with 0.25% glutaraldehyde at 10 h postinfection and then incubated overnight with PBM cells (107/ml) for IFN induction. TGEV- or mock-infected ST cell monolayers were processed in the same way for comparison. The interferon titer in the maintenance medium was determined by an ELISA test specific for IFN-α.
TGEV pseudoparticles are released from cells coexpressing M and E.
During the course of these studies, it was reported that the coexpression of MHV M and E proteins leads to the assembly and secretion of particles closely resembling authentic coronavirions (6, 48). Accordingly, a possible explanation of the above-mentioned observation was that the formation of similar viral structures was a crucial parameter for TGEV M-mediated induction of IFN. The following experiments were performed in order to formally demonstrate that formation and release of pseudoparticles also occurred in TGEV M- and E-expressing cells.
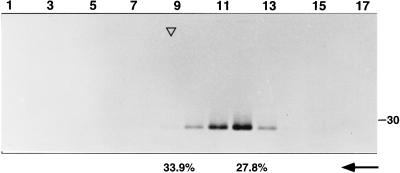
First, the material present in the medium from radioisotopically labeled, cotransfected or infected cultures was analyzed by ultracentrifugation in sucrose gradients. As shown in Fig. 2, M protein material sedimented as a discrete peak, with a velocity close to but lower than that of TGEV virions. No E protein could be detected in this experiment, presumably because it was incorporated at a low ratio in the pseudoparticles, similar to that found for TGEV and MHV virions (19, 51).
FIG. 2.
Velocity centrifugation analysis of M protein material released from cells coexpressing TGEV M and E genes. The culture medium from vT7-3-infected, transfected, and labeled RK13 cells was centrifuged on a 20 to 45% linear sucrose gradient. The material present in each fraction was pelleted and analyzed directly by SDS–12% PAGE and autoradiography. The position (top of the peak) of labeled TGEV virions sedimented in a parallel gradient is shown by an open triangle. The sucrose concentrations in the fractions corresponding to the sedimentation peaks of pseudoparticles and virions, respectively, are shown (arrow).
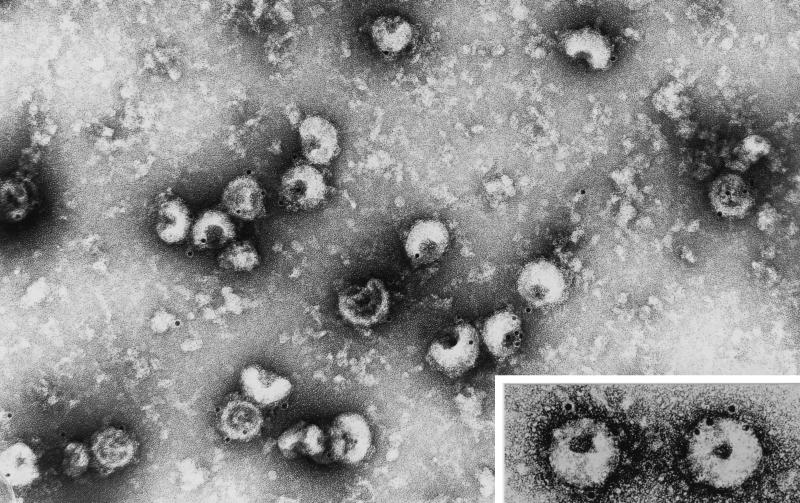
Second, in an attempt to visualize the recombinant pseudoparticles, extracellular material derived from coexpressing cultures was purified by sucrose gradient centrifugation and analyzed by electron microscopy after immunogold labeling (Fig. 3). Negative staining revealed a population of relatively homogeneous, spherical particles, the diameter of which was estimated to be 110 ± 10 nm (n = 30), very similar to that determined for TGEV virions (spikes excluded). Thus, the data overall confirmed those obtained for MHV pseudovirions (48).
FIG. 3.
Electron microscopy of TGEV pseudoparticles. Particles secreted from RK13 cells coexpressing the M and E genes were purified by a procedure similar to that used for TGEV virions (see Materials and Methods). Particles were viewed after immunogold labeling with an anti-M ectodomain antibody and negative staining. The insert shows two pseudoparticles at a twofold-higher magnification.
TGEV pseudoparticles exhibit interferogenic activity.
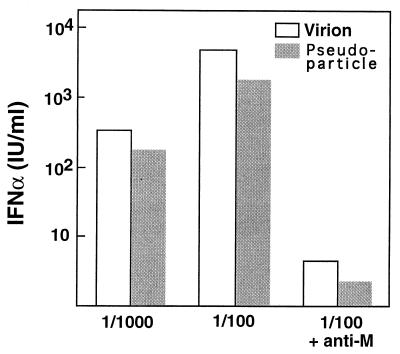
Experiments were next performed to learn whether material released in the medium from cultures cosynthesizing M and E through vaccinia virus-T7 expression exhibited interferogenic activity. A substantial, dose-dependent IFN-inducing activity was found to be associated with sedimentable extracellular material collected after pelleting through a glycerol cushion (data not shown). Importantly, no interferogenic material was released from cultures expressing M protein alone. A key issue was whether the specific interferogenic activity of the recombinant particles compared to that of authentic TGEV virions. To address this, the contents of M antigen in sucrose gradient-purified preparations of recombinant pseudoparticles and virions were determined by ELISA, and then dilutions of either material giving approximately the same signal were allowed to incubate with PBM cells. These experiments showed that the interferogenic activity (i) persisted after purification of the pseudoparticles; (ii) was comparable for two batches produced and purified independently; (iii) was close to that of virions, i.e., about fivefold less according to the data shown in Fig. 4, which were obtained with the purified batch having the highest M antigen concentration; and (iv) was inhibited by an antibody directed to the N-terminal ectodomain of the M protein to the same extent as for TGEV virions.
FIG. 4.
IFN-α-inducing activity of recombinant pseudoparticles and TGEV virions. Purified recombinant particles and virions were obtained as described in Materials and Methods. Based on the results of an ELISA, the suspensions were adjusted to have approximately the same M antigen content and then incubated with PBM cells at the indicated dilutions. The blocking effect of anti-M MAb 25.22 on IFN induction is shown on the right. The ratio of virions to PBM cells was estimated to be 100 ng per 2 × 105 cells at the 1/100 dilution.
IFN induction by chimeric TGEV-BCV pseudoparticles.
The possibility of generating fully interferogenic particles by transient expression of two cloned viral genes made it tempting to use a genetic approach in an attempt to identify the molecular determinant(s) implicated in this phenomenon. To this end, it was decided to generate chimeric particles through the use of the BCV E and M genes, which were previously cloned in the laboratory (45, 50). A first series of experiments were designed with the aim of answering more specifically two questions: (i) whether apart from its apparent morphogenic role, the minor component E protein plays any role in the interferogenicity and (ii) whether the latter activity would be altered by a change in the M ectodomain, assuming that this is the main region of the protein interacting with the IFN-producing cells.
The structures of the relevant constructs and the particle release results obtained are presented in Fig. 5. Extracellular M material was readily detected in the culture medium of cells coexpressing homologous (TGEV or BCV) M and E proteins, whereas very little or no release was observed when either M-E heterologous constructs or M–half-chimeric E constructs (tb and bt) were coexpressed (Fig. 5b). In contrast, the M chimera called BTT, in which the TGEV M ectodomain was replaced by that of BCV, was found to allow efficient particle formation (Fig. 5c). Furthermore, this BTT chimera appeared to lead to pseudoparticle formation with each of the four E gene constructs. This finding was unexpected in view of our unsuccessful attempts to obtain particle formation through the coexpression of eight different TGEV-BCV chimeric M proteins (3), as shown here for the TBB chimera.
Interestingly, when the BTT chimera was analyzed for IFN induction, it was found to promote IFN synthesis as efficiently as the parental TGEV M protein when coexpressed with TGEV E protein (Fig. 6a). The BCV M-E pair also induced IFN efficiently. When BTT was coexpressed with BCV E (BTT plus b), a significant but lower IFN-inducing activity was observed, consistent with the fact that this pair allowed less efficient particle formation than the BTT-t pair or homologous pairs (Fig. 5c). Strikingly, no IFN-inducing activity could be detected in any of the cultures which expressed a pair of constructs unable to promote particle release (Fig. 6a and data not shown). As shown above with purified pseudoparticles (Fig. 4), a marked inhibition of IFN induction was observed when an antibody specific for the TGEV M ectodomain was added during incubation with PBM cells (Fig. 6b). However, as expected, no inhibition occurred in the case of chimeric pseudoparticles produced with the BTT construct, which carries the BCV M ectodomain.
FIG. 6.
Induction of IFN-α by cell cultures producing homologous or chimeric pseudoparticles of TGEV and BCV. The experiments were performed as described in the legend to Fig. 1. The coexpressed genes are indicated as uppercase letters on the x axis for M constructs and by lowercase letters for E constructs. For panel b, an antibody specific for the ectodomain of TGEV M protein was added in the culture medium during the incubation with PBM cells.
DISCUSSION
As a main achievement, this study demonstrated that recombinant TGEV pseudoparticles, lacking spikes and ribonucleoprotein, can substitute for authentic virions to trigger efficient IFN-α synthesis by swine leukocytes. It also revealed that BCV pseudoparticles exhibit basically this same property, thus shedding new insights into the possible nature of the viral structure responsible for this intriguing biological feature. Finally, based on attempts at generating chimeric TGEV-BCV pseudoparticles, it was indicated that the formation of coronavirus particles involves highly specific, possibly multiple interactions.
The pseudoparticles were generated through the coexpression of the major virion component, i.e., the membrane M glycoprotein, and the minor, small membrane protein E (formerly called sM) and were found to be the same size as TGEV virions. This observation confirms that the notion of nucleocapsid-independent assembly, first described for the murine coronavirus MHV (6, 24, 48), extends to TGEV, which is representative of a distinct genetic subset. The E protein-mediated release of M material has also been observed with BCV (this study) and feline infectious peritonitis virus (49), two viruses closely related to MHV and TGEV, respectively. It thus can be postulated that such a mechanism, in which the M and E proteins act as necessary and sufficient partners for budding, is a general feature of coronaviruses, although this remains to be formally established for IBV, which belongs to a third genetic group.
The induction of leukocytic IFN-α by recombinant TGEV pseudoparticles essentially recapitulates the characteristics previously established for authentic virions. (i) The specific interferogenic activity of purified pseudoparticles, estimated on the basis of the M protein content, closely approached that of TGEV virions. The small difference observed (Fig. 4) may be irrelevant, since pseudoparticles and virions were produced in two different cell systems. (ii) Antibodies directed to the M ectodomain were found to prevent the IFN induction, as shown to be the case for TGEV virions. (iii) Coexpression of the S protein had no discernible effect on IFN stimulation by pseudoparticle-producing cultures. This finding is in agreement with our earlier observations that antibodies directed to either the S protein or its cellular receptor APN had no effect on IFN induction by TGEV-infected cells or virions (8, 39). In the future, it might be of interest to see whether recombinant pseudoparticles made of unglycosylated M exhibit a reduced interferogenic activity, as previously demonstrated for TGEV mutant viruses encoding M protein with an altered glycosylation site (32). It would also be worth examining the interferogenic activity of TGEV pseudoparticles toward PBM cells from different animal species, in order to compare the resulting cell spectrum with that of virions, as it has been previously shown that IFN-α induction by TGEV was not restricted to PBM cells from the swine species (8).
An important finding of this study was that the ability to induce strong IFN-α synthesis by PBM cells is not a trait unique to TGEV pseudoparticles, as BCV recombinant particles were found to exhibit basically the same property. This observation raises the question of whether the two viruses have a common interferogenic determinant. Stimulation of IFN synthesis must imply a physical interaction between pseudoparticle determinants and a subpopulation of PBM cells, the so-called NIP cells (see the introduction). The N-terminal ectodomain of the M molecule is stoichiometrically the most abundant viral constituent exposed at the outer face of the pseudoparticles, and this region was considered in our earlier studies as a likely candidate for mediating such an interaction (32). It should be pointed out, however, that the N-terminal ectodomains of TGEV and BCV differ notably in size and amino acid composition (Fig. 5a). The M ectodomain of TGEV potentially has a more complex structure than that of BCV and, of possible relevance, is associated with a functional signal peptide, in contrast to the case for most coronaviruses (33). Moreover, the carbohydrate moiety of M, previously shown to play a role in IFN induction by TGEV (9, 32), differs between the two viruses: N and O glycosylation for TGEV and BCV, respectively (13, 30). In addition, it was established during this study that a TGEV recombinant particle, in which the ectodomain of M protein was replaced by that of BCV, was as interferogenic as the homologous pseudoparticles. Together, these findings raise the possibility that the N ectodomain itself may not be the sole or the major determinant of the interferogenic activity. It cannot be excluded, however, that TGEV and BCV particles may interact with different subpopulations of IFN-producing cells. Addressing this issue relies on the progress of ongoing studies on the identification and isolation of the NIP cells.
As an extension of the present study, we have examined whether the ability to induce IFN-α is a general, so-far-unrecognized feature of coronaviral particles. As reported elsewhere, all of the eight viruses tested so far, including IBV, proved to be potent inducers for swine PBM cells, irrespective of their natural host specificity (5). Inspection of the reported M polypeptide sequences failed to reveal any conserved, contiguous sequence motif which could account for this function. Based on the accepted topology of the M protein in the lipid membrane (42, 43), three regions might contain a determinant accessible to the effector cells. Both the N-terminal ectodomain and the short junction between the transmembrane segments 2 and 3 are widely divergent among the three coronavirus groups. The third region includes a C-terminal domain which would become externally exposed, assuming that some M virion-associated molecules comprise a fourth transmembrane segment, as was recently proposed for TGEV (40). Within this region, a 5-amino-acid stretch (YVKSK) is overall well conserved, except in IBV. In this respect, an antibody directed to the TGEV M endodomain failed to prevent IFN induction (8, 32). Finally, an alignment of the E protein sequences (19) did not reveal any obvious candidate consensus motif either. Accordingly, it seems more plausible that the signal for IFN induction relies on a common structural feature than that it relies on a specific amino acid sequence motif.
In this study, we attempted to generate chimeric TGEV-BCV pseudoparticles as a possible approach to delineate an interferogenic determinant. Together, the results led us to conclude that a homologous, direct or indirect, interaction between the M and E proteins is required for efficient assembly, at least in the absence of other viral components. No particle formation could be observed through the expression of TGEV M along with BCV E and vice versa. Restoration of particle formation through the coexpression of various combinations of M and E chimeric constructs was subsequently attempted. These experiments, the details of which will be reported in a separate paper, proved to be generally unsuccessful (3). At least two levels of blockade can be envisioned: M-M interaction, which is known to occur (27) and is an obvious prerequisite for particle formation, and M-E interaction. The observed incompatibility could reflect a complex process, in which several domains of the M and E polypeptide chains would be implicated in stereospecific interactions. It should be pointed out that no direct protein-protein interaction between M and E has been seen yet. Comparative studies performed with four coronaviruses failed to reveal noticeable differences in their budding sites in infected cells (25). However, subtle differences in the subcellular localization of the TGEV and BCV proteins could account, at least in part, for the lack of assembly of chimeric particles. In this respect, the production of particles by use of the chimeric construct BTT, encoding a TGEV M protein in which the ectodomain was replaced by its BCV counterpart, is intriguing. Indeed, coexpression of BTT not only with TGEV E but also with either of the half-chimeric E proteins (tb or bt) and even BCV E protein allowed particle formation, as if the specificity of M-E interaction was, at least in part, abolished. Clearly, additional studies are needed to address these important issues.
The mechanism by which inert viral particles, either free or associated with the cell surface, commit leukocytes to synthesize IFN-α remains elusive. To date, most of the viral proteins recognized as playing a role in the induction are actually glycoproteins. On this basis, an interaction via a lectin-like activity has been proposed as a common and central event for triggering IFN production (21). In the case of TGEV, there is indeed substantial evidence that the carbohydrate moiety attached to the M protein does contribute to the interferogenic activity. Removal of complex, Golgi-modified oligosaccharides but not of mannose-rich oligosaccharides by treatment of purified virions with appropriate glycosidases was shown to markedly reduce, but not abolish, the IFN induction (9). Also, the notable decrease in IFN-inducing ability of two TGEV mutants was correlated with amino acid changes in the ectodomain of the M protein, which either suppressed or impaired the N-linked glycosylation (32).
For several reasons, however, an alternative scenario should be considered, in which the carbohydrate chain is only one component of the stimulus necessary for signal transduction. First, there is both direct and indirect evidence against a role of the spike S protein, a heavily glycosylated protein (34% carbohydrate) (13), in IFN induction (see above). Second, envelope protein-liposome mixtures prepared from purified virions repeatedly failed to induce IFN, despite the fact that these structures expressed both M and S representative epitopes; furthermore, such virosomes appeared to compete with TGEV for IFN induction (39). Third, all of our attempts to stimulate IFN production by using nonlysed cells, intracellular membranes, or detergent-solubilized material from cultures singly expressing TGEV M (reference 4 and this study) were unsuccessful. Conversely, IFN induction was observed each time that particle formation was observed.
In conclusion, our data lend support to the view that the IFN-α-inducing activity of coronavirus M protein could rely upon its incorporation into a polymeric, specifically organized structure. Moreover, we suggest that such a notion might be relevant for viruses other than coronaviruses. Taking HIV as an example, it is striking that the ability of virions to trigger IFN-α induction in PBM cells is dramatically higher than that of soluble gp120 protein (17). Densely arranged and ordered repetitive structures are a hallmark of infectious agents, including viruses, and it has been proposed that B cells can discriminate between harmless and harmful nonself antigens according to pattern or antigen organization (52). Along the same lines, it can be speculated that the spatial organization is one of the factors determining the ability of NIP cells to differentiate between endogenous glycoproteins and nonself, virus-associated glycoproteins.
ACKNOWLEDGMENTS
We thank Emmanuel Kut for technical support, Patrice Vende for DNA sequence confirmation, Stephan Chilmonczyk for assistance with electron microscope experiments, Pascal Boireau for the plasmids containing the BCV M and E genes, Jean-François Vautherot for anti-BCV E protein MAbs, and Roger Morris for editing of the English.
REFERENCES
- 1.Abraham S, Kienzle T E, Lapps W E, Brian D A. Sequence and expression analysis of potential non-structural proteins of 4.9, 4.8, 12.7 and 9.5 kDa encoded between the spike and membrane protein genes of bovine coronavirus. Virology. 1990;177:488–495. doi: 10.1016/0042-6822(90)90513-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ankel H, Capobianchi M R, Castiletti C, Dianzani F. Interferon induction by HIV glycoprotein 120: role of the V3 loop. Virology. 1994;205:34–43. doi: 10.1006/viro.1994.1617. [DOI] [PubMed] [Google Scholar]
- 3.Baudoux P. Ph. D. thesis. Paris-Grignon, France: Institut National Agronomique; 1996. [Google Scholar]
- 4.Baudoux P, Charley B, Laude H. Recombinant expression of the TGEV membrane protein. Adv Exp Med Biol. 1995;380:305–310. doi: 10.1007/978-1-4615-1899-0_49. [DOI] [PubMed] [Google Scholar]
- 5.Baudoux, P., L. Besnardeau, C. Carrat, P. Rottier, B. Charley, and H. Laude. Interferon alpha inducing property of coronavirus particles and pseudoparticles. Adv. Exp. Med. Biol., in press. [DOI] [PubMed]
- 6.Boss E C W, Luytjes W, van der Meulen H, Koerten H K, Spaan W J M. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capobianchi M R, Ankel H, Ameglio F, Paganelli R, Pizzoli P M, Dianzani F. Recombinant glycoprotein of human immunodeficiency virus is a potent interferon inducer. AIDS Res Hum Retroviruses. 1992;8:575–579. doi: 10.1089/aid.1992.8.575. [DOI] [PubMed] [Google Scholar]
- 8.Charley B, Laude H. Induction of interferon alpha by transmissible gastroenteritis coronavirus: role of transmembrane glycoprotein E1. J Virol. 1988;62:8–11. doi: 10.1128/jvi.62.1.8-11.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charley B, Lavenant L, Delmas B. Glycosylation is required for coronavirus TGEV to induce an efficient production of IFNα by blood mononuclear cells. Scand J Immunol. 1991;33:435–440. doi: 10.1111/j.1365-3083.1991.tb01792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Arce H D, Artursson K, L’Haridon R, Perers A, La Bonnardière C, Alm G V. A sensitive immunoassay for porcine interferon-α. Vet Immunol Immunopathol. 1992;30:319–327. doi: 10.1016/0165-2427(92)90102-v. [DOI] [PubMed] [Google Scholar]
- 11.Delmas B, Gelfi J, L’Haridon R, Vogel L K, Sjöström H, Norén O, Laude H. Aminopeptidase is a major receptor for the enteropathogenic coronavirus TGEV. Nature (London) 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delmas B, Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J Virol. 1990;64:5367–5375. doi: 10.1128/jvi.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delmas B, Laude H. Carbohydrate-induced conformational changes strongly modulate the antigenicity of coronavirus TGEV glycoproteins S and M. Virus Res. 1991;20:107–120. doi: 10.1016/0168-1702(91)90103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eleouët J F, Rasschaert D, Lambert P, Levy L, Vende P, Laude H. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology. 1995;206:817–822. doi: 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enjuanes L, Van der Zeijst B A M. Molecular basis of transmissible gastroenteritis epidemiology. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 337–376. [Google Scholar]
- 16.Fitzgerald-Bocarsly P. Human natural interferon-α producing cells. Pharmacol Ther. 1993;60:39–62. doi: 10.1016/0163-7258(93)90021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francis M L, Meltzer M S. Induction of IFN-α by HIV-1 in monocyte-enriched PBMC requires gp120-CD4 interaction but not viral replication. J Immunol. 1993;151:2208. [PubMed] [Google Scholar]
- 18.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godet M, L’Haridon R, Vautherot J F, Laude H. TGEV coronavirus ORF4 encodes a membrane protein that is incorporated into virions. Virology. 1992;188:666–675. doi: 10.1016/0042-6822(92)90521-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godet M, Rasschaert D, Laude H. Processing and antigenicity of entire and anchor-free spike glycoprotein S of TGEV coronavirus TGEV expressed by recombinant baculovirus. Virology. 1991;184:732–740. doi: 10.1016/0042-6822(91)90544-L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito Y. Induction of interferon by virus glycoprotein(s) in lymphoid cells through interaction with the cellular receptors via lectin-like action: an alternative interferon induction mechanism. Arch Virol. 1994;138:187–198. doi: 10.1007/BF01379125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito Y, Bando H, Komada H, Tsurudome M, Nishio M, Kawano M, Matsumura H, Kusagawa S, Yuasa T, Ohta H, Ikemura M, Watanabe N. HN proteins of human parainfluenza type 4A virus expressed in cell lines transfected with a cloned cDNA have an ability to induce interferon in mouse spleen cells. J Gen Virol. 1994;75:567–572. doi: 10.1099/0022-1317-75-3-567. [DOI] [PubMed] [Google Scholar]
- 23.Jestin A, Cherbonnel M. Interferon induction in mouse spleen cells by the Newcastle disease virus (NDV) HN protein. Ann Rech Vet. 1991;22:365–372. [PubMed] [Google Scholar]
- 24.Kim K H, Narayan K, Makino S. Assembled coronavirus from complementation of two defective interfering RNAs. J Virol. 1997;71:3922–3931. doi: 10.1128/jvi.71.5.3922-3931.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klumperman J, Krijnse Locker J, Meijer A, Horzinek M C, Greuze H J, Rottier P J M. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krijnse Locker J, Griffith G, Horzinek M C, Rottier P J M. O-glycosylation of the coronavirus M protein: differential localization of sialyltransferase in N- and O-glycosylation. J Biol Chem. 1992;267:14094–14101. doi: 10.1016/S0021-9258(19)49683-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krijnse Locker J, Opestelten D J E, Ericsson M, Horzinek M C, Rottier P J M. Oligomerization of a trans-Golgi/trans-Golgi network retained protein occurs in the Golgi complex and may be part of its retention. J Biol Chem. 1995;270:8815–8821. doi: 10.1074/jbc.270.15.8815. [DOI] [PubMed] [Google Scholar]
- 28.La Bonnardière C, Laude H. High interferon titer in newborn pig intestine during experimentally induced viral enteritis. Infect Immun. 1981;32:28–31. doi: 10.1128/iai.32.1.28-31.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Research. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lapps W, Hogue B G, Brian D. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology. 1987;157:47–57. doi: 10.1016/0042-6822(87)90312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laude H, Chapsal J M, Gelfi J, Labiau S, Grosclaude J. Antigenic structure of transmissible gastroenteritis coronavirus. I. Properties of monoclonal antibodies directed against virion proteins. J Gen Virol. 1986;67:119–130. doi: 10.1099/0022-1317-67-1-119. [DOI] [PubMed] [Google Scholar]
- 32.Laude H, Gelfi J, Lavenant L, Charley B. Single amino acid changes in the viral glycoprotein M affect induction of alpha interferon by the coronavirus transmissible gastroenteritis virus. J Virol. 1992;66:743–749. doi: 10.1128/jvi.66.2.743-749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laude H, Rasschaert D, Huet J C. Sequence and N-terminal processing of the transmembrane protein EI of the coronavirus transmissible gastroenteritis virus. J Gen Virol. 1987;68:1687–1693. doi: 10.1099/0022-1317-68-6-1687. [DOI] [PubMed] [Google Scholar]
- 34.Lebon P, Commoy-Chevalier M J, Robert-Galliot B, Chany C. Different mechanisms for α and β interferon induction. Virology. 1982;119:504–507. doi: 10.1016/0042-6822(82)90109-x. [DOI] [PubMed] [Google Scholar]
- 35.Liu D X, Inglis S C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowacki W, Charley B. Enrichment of coronavirus-induced interferon-producing blood leukocytes increases the interferon yield per cell: a study with pig leukocytes. Res Immunol. 1993;144:111–120. doi: 10.1016/0923-2494(93)80066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasschaert D, Gelfi J, Laude H. Enteric coronavirus TGEV: partial sequence of the genomic RNA, its organization and expression. Biochimie. 1987;69:591–600. doi: 10.1016/0300-9084(87)90178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riffault S, Carrat C, Besnardeau L, La Bonnardière C, Charley B. In vivo induction of interferon-α in pig by non-infectious coronavirus: tissue localization and in situ phenotypic characterization of interferon-α-producing cells. J Gen Virol. 1997;78:2483–2487. doi: 10.1099/0022-1317-78-10-2483. [DOI] [PubMed] [Google Scholar]
- 39.Riffault S, Grosclaude J, Vayssier M, Laude H, Charley B. Reconstituted coronavirus TGEV virosomes lose the virus ability to induce porcine interferon-alpha production. Vet Res. 1997;28:105–114. [PubMed] [Google Scholar]
- 40.Risco C, Anton I M, Sune C, Pedregosa A M, Martin-Alonso J M, Parra F, Carrascosa J L, Enjuanes L. Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion. J Virol. 1995;69:5269–5277. doi: 10.1128/jvi.69.9.5269-5277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogel-Gaillard C, Chilmonczyk S, de Kinkelin P. In vitro induction of interferon-like activity from rainbow trout leucocytes stimulated by Egtved virus. Fish Shellfish Immunol. 1993;3:383–394. [Google Scholar]
- 42.Rottier P J M. The coronavirus membrane glycoprotein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 115–139. [Google Scholar]
- 43.Rottier P J M, Welling G W, Welling-Wester S, Niesters H G, Lenstra J A, Van der Zeijst B A M. Predicted membrane topology of the coronavirus protein E1. Biochemistry. 1986;25:1335–1339. doi: 10.1021/bi00354a022. [DOI] [PubMed] [Google Scholar]
- 44.Sandberg K, Eloranta M L, Johannisson A, Alm G V. Flow cytometric analysis of natural interferon-α producing cells. Scand J Immunol. 1991;34:565–576. doi: 10.1111/j.1365-3083.1991.tb01580.x. [DOI] [PubMed] [Google Scholar]
- 45.Savoysky E, Boireau P, Finance C, Laporte J. Sequence and analysis of BECV F15 matrix protein. Res Virol. 1990;141:411–425. doi: 10.1016/0923-2516(90)90042-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Splichal I, Bonneau M, Charley B. Ontogeny of interferon alpha secreting cells in the porcine fetal hematopoietic organs. Immunol Lett. 1994;43:203–208. doi: 10.1016/0165-2478(94)90224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svensson H, Johanisson A, Nikkilä T, Alm G V, Cederblad B. The cell surface phenotype of human natural interferon-α producing cells as determined by flow cytometry. Scand J Immunol. 1996;44:164–172. doi: 10.1046/j.1365-3083.1996.d01-289.x. [DOI] [PubMed] [Google Scholar]
- 48.Vennema H, Godeke G-J, Rossen J W A, Vorhout W F, Horzinek M C, Opstelten D J, Rottier P J M. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vennema, H., and P. J. M. Rottier. Unpublished data.
- 50.Woloszyn N, Boireau P, Laporte J. Nucleotide sequence of the bovine enteric coronavirus F15 mRNA 5 and mRNA 6 unique regions. Nucleic Acids Res. 1990;18:1303. doi: 10.1093/nar/18.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu X, Bi W, Weiss S R, Leibowitz J L. Mouse hepatitis gene 5b protein is a new virion envelope protein. Virology. 1994;202:1018–1023. doi: 10.1006/viro.1994.1430. [DOI] [PubMed] [Google Scholar]
- 52.Zinkernagel R M. Immunology taught by viruses. Science. 1996;271:173–178. doi: 10.1126/science.271.5246.173. [DOI] [PubMed] [Google Scholar]