Abstract
Natural killer (NK) cells are critically involved in anti-tumor immunity by targeting tumor cells. In this study, we show that intratumoral NK cells from NSCLC patients expressed elevated levels of the immune checkpoint receptor PD-1 on their cell surface. In contrast to the expression of activating receptors, PD-1+ NK cells co-expressed more inhibitory receptors compared to PD-1− NK cells. Intratumoral NK cells were less functional compared to peripheral NK cells, and this dysfunction correlated with PD-1 expression. Tumor cells expressing PD-L1 inhibited the functionality of PD-1+ NK cells in ex vivo models and induced PD-1 clustering at the immunological synapse between NK cells and tumor cells. Notably, treatment with PD-1 blockade was able to reverse PD-L1-mediated inhibition of PD-1+ NK cells. Our findings highlight the therapeutic potential of PD-1+ NK cells in immune checkpoint blockade and could guide the development of NK cell-stimulating agents in combination with PD-1 blockade.
Electronic supplementary material
The online version of this article (10.1007/s00262-020-02558-z) contains supplementary material, which is available to authorized users.
Keywords: Cancer immunotherapy, Immune checkpoint inhibitor, Inhibitory receptor, Resistance, Innate immunity
Introduction
Cancer immunotherapy targeting the PD-1/PD-L1 inhibitory axis is nowadays considered to be one of the treatment pillars of non-small cell lung cancer (NSCLC) [1–8]. Despite significant improvements in survival and durability of responses, the majority of patients are still resistant to PD-1/PD-L1 blockade [9]. Several markers and mechanisms of resistance to ICB have been described, but the determinants of responses and/or failures are still poorly defined. There is increasing evidence that a lower burden of non-synonymous somatic mutations or genomic alterations in individual genes correlates with a lack of clinical benefit and immune resistance [10–12]. Recent data on patients with melanoma indicate that attenuation of MHC class I protein expression does not predict responsiveness to PD-1 blockade. This finding suggests that immune recognition beyond MHC restriction may be necessary for successful responses and other cell types than T cells might be involved in mediating anti-tumor immunity upon PD-1 blockade [13].
Natural killer cells, a subset of innate lymphoid cells, constitute a first line of defense against microbial infections and cancer development [14, 15]. They can control cancers directly by rapid killing of tumor cells through perforin and granzymes [16] and/or the ligation of death receptor-mediated pathways [17]. Moreover, NK cells secrete a wide array of cytokines and chemokines to regulate the activities of other immune cells in the tumor microenvironment [18]. While their role in cancer immunosurveillance has been demonstrated in models of primary and metastatic tumors [19], NK cell density in human tumors and their prognostic impact are largely dependent on the histological origin of the malignant cell [20–22]. In lung cancer, NK cells accumulate mainly in the tumor stroma without direct contact with tumor cells and appear not to impact the clinical outcome of chemo- or radiotherapy [23, 24]. Nevertheless, their involvement in the response to immunotherapy remains to be determined.
NK cell effector functions are tightly controlled by a balance of signals from both activating and inhibitory receptors [24]. Inhibitory killer immunoglobulin-like receptor family (KIRs), Ly49 and CD94/NKG2A receptors function to detect MHC class I self-ligands and inhibit NK cell activation to prevent the destruction of nontransformed cells [25]. In turn, activating receptors including NKG2D, the natural cytotoxicity receptors NKp46, NKp30, NKp44 as well as DNAM-1 and CD16 may induce an immune response [26]. We recently found that allelic variants of KIR3DS1/L1 correlate with resistance to PD-1/PD-L1 blockade in lung cancer, which indicates that NK cells may be implicated in the generation of anti-tumor immune responses during checkpoint inhibitor therapy [27]. There is indeed evidence that NK cells upregulate inhibitory receptors such as PD-1, T-cell immunoreceptor with Ig and ITIM domains (TIGIT), and T-cell immunoglobulin and mucin-3 (TIM-3) during cancer progression [28–30]. A comprehensive characterization of the abundance, phenotype and functional significance of tumor-infiltrating NK cells expressing those inhibitory receptors, particularly in NSCLC, is still missing.
The tumor environment can suppress NK cell functionality through different mechanisms [31]. Like T cells, intratumoral NK cells may become dysfunctional, which prevents them from displaying optimal anti-tumor responses, while their counterparts in the peripheral circulation maintain full or only slightly reduced effector functions [23, 32]. Dysfunctional NK cells in cancer are characterized by a high expression of NK cell-associated inhibitory receptors, reduced expression of activating receptors, as well as a decreased expression of the transcription factors TBET and EOMES [28, 32, 33]. The higher levels of inhibitory receptors expressed on NK cells have been associated with more aggressive and invasive tumors [32].
In this study, we show that tumor-infiltrating NK cells from NSCLC patients expressed the immune checkpoint receptor PD-1 on their surface. Moreover, we investigated the expression of other inhibitory and activating receptors on PD-1+ and PD-1− NK cell subsets and found that PD-1+ NK cells co-expressed more inhibitory receptors compared to PD-1− NK cells. We also provide evidence that the dysfunction of intratumoral NK cells correlates with increasing levels of PD-1 surface expression. Notably, treatment with PD-1 blocking antibodies was able to reverse PD-L1-mediated inhibition of NK cells, highlighting the therapeutic potential of PD-1+ NK cells in immune checkpoint blockade.
Material and methods
Cell lines
K562 cells and HEK293T cells were cultured in IMDM and DMEM, respectively, and supplemented with 10% heat-inactivated FBS (PAN Biotech), 100 ng/ml penicillin/streptomycin (Sigma), 2 mM L-glutamine (Sigma), 1 mM sodium pyruvate (Sigma) and 1% MEM non-essential amino acids (Sigma). The melanoma cell line NA8-Mel was cultured in RPMI-1640 (Sigma) supplemented as described above for IMDM and DMEM. NK92 cells were cultured in αMEM (w/o ribo/deoxyribonucleosides, with 2 mM L-glutamine and 1.5 g/l sodium bicarbonate) (Thermo Fisher) supplemented with 0.2 mM inositol, 0.1 mM 2-mercaptoethanol, 0.02 mM folic acid, 300 U/ml recombinant human IL-2 (PeproTech), 12.5% horse serum (Thermo Fisher), 12.5% heat-inactivated FBS and 100 ng/ml penicillin/streptomycin. Cells were confirmed to be negative for mycoplasma by PCR as described after every freeze–thaw cycle and then passaged every 2–3 days for a maximum of ten passages [34].
Cloning of PD-1 and PD-L1 expression vectors
The human PD-1/PD-L1 open reading frames (ORFs) were amplified by PCR from a pCMV6 human cDNA clone (RC210364, Origene) and a pCMV-XL4 human cDNA clone (SC115168, Origene), respectively. The ORFs were extended with overlapping ends (both sides) complementary to the pRRL.PPT.SFFV.EGFP.pre-expression vector (kindly provided by Drs. Baum and Schambach at Hannover Medical School, Germany [35]) together with an XbaI cut site and a Kozak sequence element. The amplified fragments were fused into the pRRL.PPT.SFFV.EGFP.pre expression vector between the sFFV promoter and the IRES-EGFP fragment using the InFusionHD kit (Clontech, Takara Bio). Both plasmids were amplified in Stbl3 E. coli (in-house) and purified using NucleoBond Xtra Midi Plus kits (Macherey Nagel). Inserts were sequenced by Sanger sequencing (Microsynth AG) to confirm correct integration. Empty IRES-GFP vectors were used as controls.
Production of lentivirus
Low-passage HEK293T cells were cultured in DMEM medium and transfected with the packaging plasmid pCMV-delta8.9, the envelope plasmid VSV-G and either PD-1-GFP, PD-L1-GFP or empty GFP sFFV transfer plasmids, respectively, in combination with PEI 25 K (Polysciences Inc.) at a ratio of 1:3. After two days, supernatants were collected and filtered through a 0.45-μm PES filter. Supernatants containing lentiviral PD-L1 particles were stored at − 80 °C until usage. Supernatants containing lentiviral PD-1 particles were concentrated by ultracentrifugation at 40,000 × g for 2 h at 4 °C, resuspended in complete αMEM medium and immediately used for transduction.
Generation of PD-1+GFP+-NK92 cell line
One day prior to transduction, NK92 cells were expanded in fresh complete αMEM medium containing 300 U/ml IL-2. The following day, cells were re-stimulated with 100 U/ml IL-2 and 100 ng/ml IL-12 (PeproTech) for 2 h without medium exchange. Cells were then collected by centrifugation and resuspended in freshly concentrated supernatant containing lentiviral PD-1 particles or the vector control particles. Cells were seeded in 96-well flat-bottom plates coated with 5 μg/cm2 Retronectin (Takara Bio). Afterward, cells were expanded in complete αMEM medium containing 300 U/ml IL-2 for 2 weeks. GFP+/PD-1+GFP+NK92 cells were then sorted by flow cytometry, and the pool of sorted cells was used in all subsequent experiments.
Generation of PD-L1+GFP+K562 and PD-L1+GFP+NA8-Mel cell lines
K562 cells were collected by centrifugation and resuspended in supernatant containing either lentiviral particles coding for sFFV-PD-L1-IRES-GFP or sFFV-IRES-GFP control as well as 8 μg/ml polybrene. Nine days after transduction, several clones of GFP+/PD-L1+GFP+ K562 cells were sorted by flow cytometry. Cell lines from these single cell clones were validated for expression of GFP and GFP + PD-L1. NA8-Mel cells were transduced by replacing media with lentiviral supernatants and polybrene as described above followed by medium replacement one day after transduction. Seven days after transduction, pools of GFP+/PD-L1+GFP+ NA8-Mel cells were sorted and cultured for 1 week. The expression of PD-L1 and GFP was validated by flow cytometry.
Isolation of healthy human blood cells and tumor-infiltrating lymphocytes
Human peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Histopaque-1077 (Sigma) from buffy coats obtained from 23 healthy blood donors (HD) (Blood Bank, University Hospital Basel) and from 19 NSCLC patients. Fresh tumor tissues were collected from 16 patients with NSCLC undergoing surgery at the University Hospital Basel, Switzerland. Detailed HD/patient characteristics are provided in Table 1. Staging was based upon the seventh edition of the AJCC/UICC tumor–node–metastasis (TNM) staging system. Tumor lesions were mechanically dissociated and digested using accutase (PAA), collagenase IV (Worthington), type V hyaluronidase from bovine testes (Sigma) and DNAse type IV (Sigma), directly after excision. Single-cell suspensions were prepared. All samples were stored in 10% DMSO/90% FCS in liquid nitrogen until further usage.
Table 1.
Cohort of healthy donors and NSCLC patients
| Healthy donors PBMCs | NSCLC PBMCs | NSCLC TILs | |
|---|---|---|---|
| Characteristics | N = 23 | N = 19 | N = 16 |
| Age, median years (range) | 48 (19–72) | 67.6 (42–80) | 68.2 (54–83) |
| Sex | |||
| Male (%) | 12 (52.2%) | 11 (47.8%) | 9 (56%) |
| Female (%) | 11 (58%) | 8 (42%) | 7 (44%) |
| Stage | |||
| I (%) | NA | 8 (42.1%) | 4 (25%) |
| II (%) | 6 (31.6%) | 7 (43.8%) | |
| III (%) | 4 (21%) | 4 (25%) | |
| IV (%) | 1 (5.3%) | 1 (6.3%) | |
Antibodies and flow cytometry
Cryopreserved tumor digests or PBMCs were thawed, washed, resuspended in PBS and blocked with human Fc/receptor/inhibitor (eBioscience). Dead cells were stained with fixable viability dyes (Biolegend or eBioscience). For surface staining, cells were washed, resuspended in FACS buffer (PBS supplemented with 2 mMEDTA, 0.1% Na-Azide, 2% FCS) and stained with appropriate antibodies for 30 min at 4 °C. All antibodies used in this study are listed in Supplementary Table S1. For intracellular (cytoplasmic) staining, cells were fixed and permeabilized using IC Fixation Buffer and Permeabilization Buffer (eBioscience). For staining of the nuclear proteins, the Fixation/Permeabilization kit (eBioscience) was used. After staining, cells were analyzed on a BD LSR Fortessa Cell analyzer (BD Bioscience). Data were collected using the BD FACS Diva Software version 7 and further analyzed with FlowJo v10.1.6 (Tree Star Inc.) and GraphPad Prism v7.0a (GraphPad Software Inc.). An example gating strategy can be found in Supplementary Fig. S1a. All results show integrated fluorescence area on a biexponential scale.
Baseline characterization of NK cells
Patient samples (PBMCs and TILs) and PBMCs from HDs were comprehensively characterized by multicolor flow cytometry. NK cells were identified by gating for live CD45+, CD3−, CD56+ and then divided into subpopulations based on the expression of the described markers.
FACS-based cell sorting
For functional assays, NK cells from PBMCs and TILS were isolated by flow cytometry-based sorting. In brief, cells were thawed, washed and stained with appropriate antibodies for 30 min at 4 °C. Following incubation, cells were washed, resuspended in FACS buffer and filtered. Sorting of cells was performed using a FACSAria III (BD), and the purity of sorted populations was routinely tested.
FACS-based NK cell killing assay
One day prior to the experiment, 20,000 sorted NK cells from TILs or PBMCs were seeded into 96-well U-bottom plates and cultured in complete RPMI-1640 in the presence or absence of 20 ng/ml IL-15 (PeproTech). The next day, K562 cells were stained with Vybrant DiO Dye (Molecular Probes) according to the manufacturer’s instructions for 25 min at 37 °C and then washed in complete medium. NK cells from HDs/patients and K562 cells were then co-cultured at an E/T ratio of 2:1 for 6 h at 37° C (duplicates). NK cells cultured without target cells were used as spontaneous cell death control. Following incubation, cells were washed and resuspended in FACS buffer. Shortly before sample acquisition, propidium iodide (PI; BD) was added to each sample to stain for dead cells and samples were acquired using a Beckmann Cytoflex flow cytometer. Cells were gated as shown in Supplementary Fig. S4a. Specific NK cell killing was determined by the following equation:
Calcein release-based NK cell killing assay
GFP+/PD-1+GFP+ NK92 cells were cultured as described above. Cells were stimulated with 20 ng/ml IL-15 overnight, and the next day, they were washed in serum-free RPMI-1640 and stained with Calcein-AM Viability Dye (eBioscience) for 30 min 37 °C. Stained cells were resuspended in assay medium (RPMI-1640 with 10% FBS and 5 mM HEPES) and seeded into a 96-well V-bottom plate. Next, 10 μg/ml nivolumab (Opdivo, Bristol-Meyer-Squib) or IgG4 isotype control (Ultra-LEAF purified, Biolegend) was added to the wells and incubated for 15 min at 37 °C. Pre-stimulated GFP+/PD-1+GFP+ NK92 cells were washed, resuspended in assay medium and added to the wells at an E/T ratio of 2:1. Maximum lysis was induced with assay medium containing 2% Triton-X100 (Sigma). Cells were co-cultured for 4 h. Following incubation, calcein was measured from supernatants using a Biotek Plate Reader at 490 nm ex / 520 nm em. Specific NK cell killing was determined by the following equation:
NK cell degranulation
Sorted NK cells from TILs or PBMCs were co-cultured with K562 cells for 6 h at 37 °C as described in the FACS-based NK cell killing assay. In addition, 20 ng/ml anti-CD107a antibody was added. After 1 h of incubation, degranulation was blocked using 1X Monensin (Biolegend) and 1X Brefeldin A (eBioscience). Following incubation, the accumulation of IFNγ and TNFα within NK cells was investigated by flow cytometry. Cells were gated as shown in Supplementary Fig. S4b.
Cytokine-mediated PD-1 upregulation on peripheral NK cells
Frozen PBMCs from HDs were thawed, washed and resuspended in complete RPMI-1640 medium. Cells were seeded into 96-well round-bottom plates and rested for 2 h at 37 °C. Afterward, various combinations of recombinant human IL-15 (25 ng/μl), IL-18 (20 ng/μl) and IL-12 (25 ng/μl) were added to the cells and incubated for 48 h at 37 °C. PD-1 upregulation was investigated by flow cytometry (Cytoflex, Beckmann Coulter).
Immune synapse analysis
Two days prior to the experiment, GFP+/PD-1+GFP+NK92 cells were cultured at 0.5*10^6 cells/ml in complete αMEM medium containing 300 U/ml IL-2. One day prior, GFP+/PD-L1+GFP+ K562 cells were stained with 1 μM Cell Trace Violet (Thermo Fisher Scientific) according to the manufacturers’ instructions for 15 min at 37 °C. Stained cells were resuspended in serum-free RPMI-1640 and incubated with 20 μg/ml of atezolizumab (Tecentriq, Roche) for 15 min at 37 °C. GFP+/PD-1+GFP+ NK92 cells were washed, resuspended in serum-free RPMI-1640 and co-cultured with K562 at an E/T of 2:1 at 37 °C. After 15 min, cells were immediately fixed for 20 min at RT using 4% PFA. Cell clusters were washed with permeabilization buffer (eBioscience) and subsequently stained with anti-PD-1-AlexaFluor647 antibody (clone EH12.1, BD Phosphoflow) for 30 min at RT. Cells were analyzed using an MKII Imagestream Analyzer (EMD Millipore). One-to-one cell ratio clusters were gated as shown in Supplementary Fig. S5a using the IDEAS software (EMD Millipore). An interface mask between Cell Trace Violet+/PD-1+ cells (synapse between PD-1+GFP+NK92 cells and PD-L1+GFP+K562 target cells) of width 6 was created. Next, an intensity concentration ratio feature for the PD-1 signal was calculated for this interface and compared to the total PD-1 mask (immune synapse versus total cell). The geometric mean of the intensity concentration ratio distribution of one-to-one cell clusters was used for further analysis.
Statistical analysis
The statistical analysis and graph preparation were performed using the software package Prism version 8.0 a (GraphPad Software, La Jolla, CA). Functional data are representative of at least three experiments. Data were considered statistically significant with p values < 0.05. Normality tests were used to choose parametric or nonparametric tests. Data are shown as mean ± standard deviation with symbols representing individual patients or donors where applicable. The individually applied statistical tests are described in the figure legends of each figure. For Fig. 2, Boolean gating was performed to identify differences in the expression of activating and inhibitory receptors as well as transcription factors on PD-1+ and PD-1−intratumoral NK cells from NSCLC TILs. The obtained differences were statistically analyzed using a two-way ANOVA with Sidak’s multiple comparison test. Patients were paired (“Subjects”) and the variance was analyzed for two-factor (1) “Co-Expression” (in categories 0, 1, 2, 3 and 4 other receptors) and (2) cell type (”PD-1+ NK” and “PD-1− NK”).
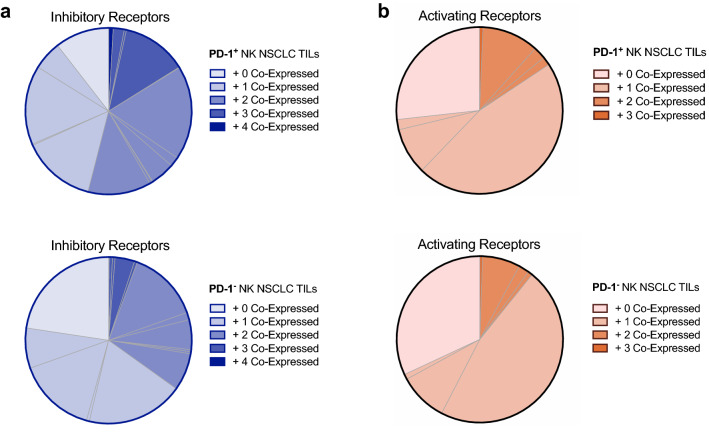
Fig. 2.
Co-expression of inhibitory and activating receptors in PD-1+/PD-1− NK cell subpopulations in NSCLC patients. Co-expression of inhibitory (a) and activating (b) receptors in PD-1+/PD-1− NK cell subpopulations in NSCLC TILs. The number of receptors co-expressed on either population is indicated by color opacity within the pie chart. The individual subpopulations described in Supplementary Tables S3 and S4 are delimited by grey lines. Statistics are described in Supplementary Table S2
Results
NK cells of NSCLC patients express the immune checkpoint PD-1
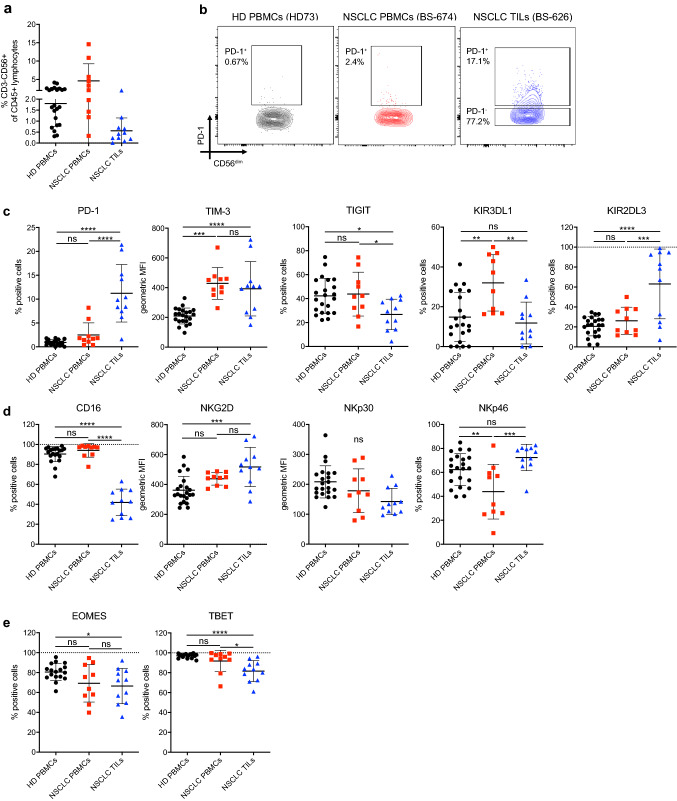
We previously showed that a variant of a killer immunoglobulin receptor expressed on NK cells may mediate resistance to PD-1 blockade in NSCLC [27]. Here, we aimed to obtain a broader overview of the expression of activating and inhibitory receptors including immune checkpoints PD-1, TIM-3, TIGIT on NK cells from NSCLC patients. We characterized NK cells from PBMCs and tumors of 16 patients by flow cytometry for the expression of PD-1, TIM-3, TIGIT, as well as NK cell inhibitory/activating receptors KIR3DL1, KIR2DL3, CD16, NKG2D, NKp30, NKp46 and transcription factors TBET and EOMES (Supplementary Fig. S1). PBMCs from healthy donors (HD) served as controls (Table 1). Detection of these proteins was not affected by cryopreservation (Supplementary Fig. S2a-b). There was a minor reduction in staining intensities by the enzymatic digestion used for tumor samples. However, this did not interfere with identification of positive and negative cells (Supplementary Fig. S2a). NK cells comprised 0.56 ± 0.58% (mean ± standard deviation, range 0.31 – 4.16%) of all CD45 + lymphocytes in the tumor (Fig. 1a). Cell surface PD-1 expression was observed on 11.2 ± 6% (1.59 – 21.4%) of intratumoral NK cells. In contrast, only 2.5 ± 2.5% (0.39 – 7.8%) of NK cells in PBMCs from NSCLC patients expressed PD-1, while PD-1 was absent on NK cells from HDs (Fig. 1b and c). PD-1 expression on NK cells also did not correlate with PD-1 expression on CD8 T cells (Supplementary Fig. S3a). The expression of TIM-3 was similar on NK cells in NSCLC TILs compared to NSCLC PBMCs (Fig. 1c), while fewer intratumoral NK cells expressed TIGIT compared to peripheral blood NK cells of NSCLC patients. NK cells from patients’ PBMCs expressed higher levels of KIR3DL1 compared to both intratumoral NK cells and healthy donor NK cells, while higher levels of KIR2DL3 were observed on intratumoral NK cells only (Fig. 1c). We detected significantly lower expression of the activating receptor CD16 on intratumoral NK cells (Fig. 1d), while the expression of NKG2D and NKp30 was similar on intratumoral NK cells compared to peripheral NK cells from NSCLC patients. The expression of NKp46 was lower in peripheral NK cells from patients compared to both intratumoral and healthy donor NK cells. The transcription factor TBET was significantly reduced in intratumoral NK cells compared to peripheral NK cells of NSCLC patients, whereas EOMES was unchanged (Fig. 1e). It should be mentioned that healthy donors were significantly younger than patients in the NSCLC PBMCs and TIL cohorts (Supplementary Fig. S3b). Nevertheless, we observed no correlation between the expression of PD-1 on NK cells with age in healthy donors or NSCLC TILs. Only a weak negative correlation with age in NSCLC PBMCs was observed (Supplementary Fig. S3c), while there was no correlation between age and any of the other measured proteins (Supplementary Fig. S3c–m).
Fig. 1.
Characterization of NK cell infiltration and immune checkpoint expression in NSCLC patients. a Frequency of NK cells in HD PBMCs, NSCLC PBMCs and NSCLC TILs relative to CD45+ cells. b Contour plot for PD-1 flow cytometry staining from one representative HD PBMC, NSCLC PBMC and NSCLC TIL sample. c Expression of inhibitory receptors, d expression of activating receptors and e expression of transcription factors by NK cells from HD PBMCs, NSCLC PBMCs and NSCLC TILs measured by flow cytometry. One-way ANOVA with Tukey’s multiple comparison test was performed on all data. p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
PD-1+ NK cells co-express more inhibitory receptors than PD-1− NK cells in NSCLC patients
PD-1 expressed on NK cells could be a direct target of therapeutic PD-1 blocking antibodies, which supports a potential role for NK cells in immunotherapy. We hypothesized that the expression of PD-1 on NK cells may correlate with the expression pattern of other receptors and therefore influence functionality as observed with tumor-infiltrating T cells in NCSLC [36]. Therefore, we investigated the co-expression of TIM-3, TIGIT, KIR3DL1, KIR2DL3, the activating receptors CD16, NKG2D and NKp30 as well as the transcription factors TBET and EOMES on intratumoral PD-1+ or PD-1− NK cell subsets. As seen in Fig. 2a, PD-1+ NK cells more frequently co-expressed other inhibitory receptors (TIM-3, TIGIT, KIR2DL3 and KIR3DL1) compared to PD-1− NK cells (p = 0.018 for interaction between cell type and co-expression, Supplementary Table S2). NK cells that co-expressed none of the four inhibitory receptors (“ + 0 Co-Expression,” light shading) show a trend toward being more frequent in the PD-1− NK cell subset (p = 0.1, Supplementary Tables S2 and S3). Notably, the co-expression of activating receptors (Fig. 2b and Supplementary Tables S2 and S4) and of the two transcription factors (Supplementary Fig. S3n) did not differ significantly between PD-1+ and PD-1− NK cells.
Tumor-infiltrating NK cells display impaired functionality compared to NK cells from healthy donors
We next explored how the difference in the receptor repertoire between tumor-infiltrating and peripheral NK cells impacts NK cell functionality. We noted a trend for lower cytotoxic activity of intratumoral NK cells (p = 0.12) against the MHC-I-deficient tumor cell line K562 compared to peripheral patient NK cells (Fig. 3a). In addition, degranulation measured by CD107a accumulation on the cell surface upon co-culture with K562 cells was decreased on intratumoral NK cells from NSCLC patients compared to peripheral NK cells from healthy donors (Fig. 3b). Notably, also the degranulation capacity did not correlate with the age of the patients and donors (Supplementary Fig. S4c). Similarly, intratumoral NK cells produced less IFN-γ (Fig. 3b, Supplementary Fig. S4d). We detected no significant difference in TNF-α production (Fig. 3b). Notably, polyfunctionality, defined as the combined capacity for cytotoxicity, IFN-γ and TNF-α release, was reduced in intratumoral NK cells (Fig. 3c, Supplementary Fig. S4e) and correlated with PD-1 expression (Fig. 3d). In the absence of PD-L1 on K562 target cells, no functional difference between intratumoral PD-1+ and PD-1− NK cells could be detected (Supplementary Fig. S4f).
Fig. 3.
Cytotoxicity, degranulation and cytokine production by NK cells in response to K562 cells. a Cytotoxicity analyzed using a flow cytometry-based killing assay with sorted NK cells from HD PBMCs, NSCLC PBMCs and NSCLC TILs co-cultured with K562 wild-type cells for 4 h at an E/T ratio of 1:2. b Sorted NK cells from HD PBMCs, NSCLC PBMCs and NSCLC TILs co-cultured with K562 wild-type cells for 6 h at an E/T ratio of 2:1. NK cell degranulation (CD107a exposure), secretion of IFN-γ and TNF-α was measured by flow cytometry. c Polyfunctionality (CD107a + IFN-γ + and TNF-α + NK cells) from the same data as in B. d Polyfunctionality of NK cells correlated with cell-surface expression of PD-1 on CD56 + NK cells. Linear correlation was performed on logarithmically transformed data. A normality test (Anderson–Darling test) was performed on the data in graphs (a) and (b). For data that passed the test (cytotoxicity, CD107a and TNF-α), an unpaired 1-way ANOVA with Tukey’s multiple comparison test was performed. When the normality test failed (p < 0.05), a nonparametric Kruskal–Wallis test was performed with Dunn’s multiple comparison correction (IFN-γ + , polyfunctionality). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
PD-1 blockade can rescue PD-L1-mediated inhibition of PD-1+ NK cells
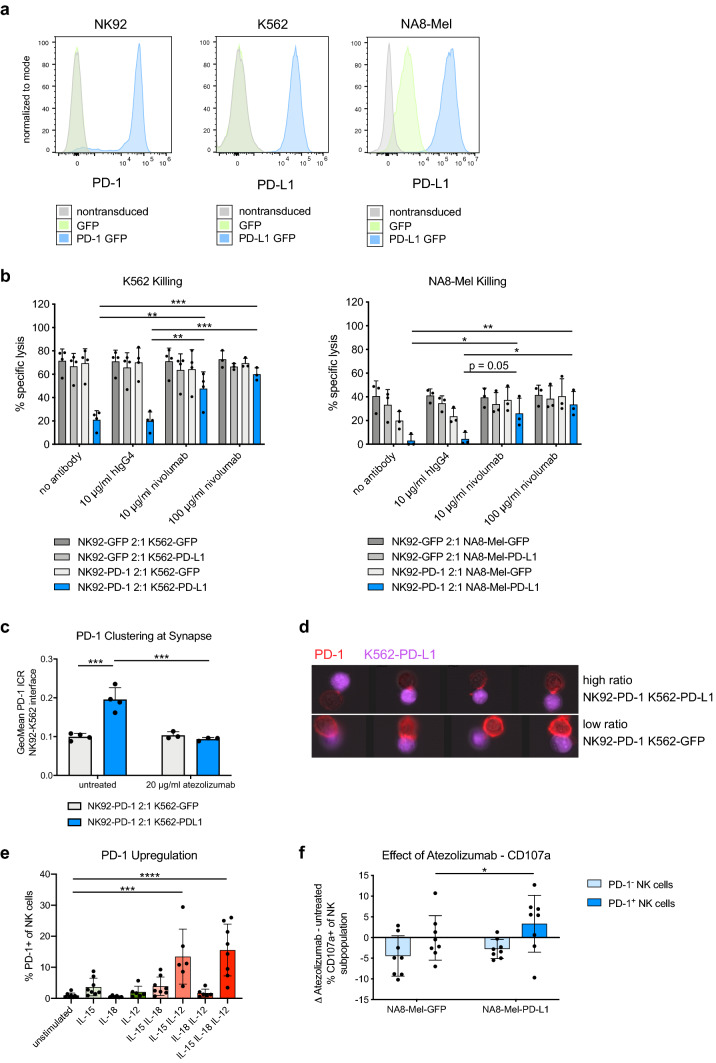
To assess whether PD-1+ NK cells can be inhibited by PD-L1 on tumor cells, we established an NK92 cell line model with overexpression of PD-1. In addition, we generated PD-L1-IRES-GFP- or GFP-only-overexpressing tumor cell lines from MHC-I-deficient K562 cells and MHC-I-proficient NA8-Mel cells (Fig. 4a, hereafter called K562-PD-L1/GFP cells or NA8-Mel-PD-L1/GFP cells, respectively). When NK92-PD-1 cells were exposed to PD-L1-overexpressing K562 and NA8-Mel, the cytotoxic capacity was profoundly decreased (Fig. 4b). This effect could be relieved with increasing concentrations of nivolumab (Fig. 4b). We observed no difference in cytotoxicity between NK92-PD-1 and NK92-GFP cells against K562-GFP cells, while cytotoxicity was slightly decreased against NA8-Mel-GFP cells, which was antagonized by nivolumab treatment. This is likely explained by the endogenous expression of PD-L1 on NA8-Mel cells (Fig. 4a, b).
Fig. 4.
PD-1 blockade restores NK cell functionality. a Histogram of PD-1 and PD-L1 measured by antibody-based staining and flow cytometry. Left panel: PD-1 on NK92 cells transduced with a lentiviral vector encoding for PD-1 GFP or GFP alone. Middle and right panel: PD-L1 expression of the different K562 and NA8-Mel cell lines transduced with PD-L1 GFP or GFP lentiviral vectors. b Effects of nivolumab on specific target cell lysis measured by calcein-AM release after 2 h of coculture of NK92-PD-1/GFP effector cells with K562 (left) or NA8-Mel (right) target cells expressing either PD-L1 GFP or GFP only. Paired two-way ANOVA with Tukey’s multiple comparison test. c PD-1 clustering measured by ImageStream at the cell–cell synapse between NK92-PD-1 cells and K562-PD-L1/GFP cells. Quantified geometric mean of the intensity concentration ratio between the cell–cell interface and the total cell area (see methods for details). Two-way ANOVA with Tukey’s multiple comparison test. d Representative images from the ImageStream analysis quantified in c showing NK92-K562 interactions with a high intensity–concentration ratio (top panel, PD-1 clustered at synapse) or low ratio (bottom, no clustering). e Upregulation of PD-1 on peripheral NK cells from healthy donors after in vitro stimulation with the indicated combinations of recombinant human IL-15 (25 ng/μl), IL-18 (20 ng/μl) and IL-12 (25 ng/μl). Unpaired one-way ANOVAs with Tukey’s multiple comparison test. f Change in degranulation between atezolizumab (20 μg/ml) and untreated PD-1+ and PD-1− NK cells pre-cultured for 2 days with recombinant human IL-15 (25 ng/μl) and IL-12 (25 ng/μl), which were then co-cultured with NA8-Mel-PD-L1/GFP. Paired two-way ANOVA with Tukey’s multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
To study how PD-1 on NK cells interacts with PD-L1 on tumor cells, we next analyzed the clustering of PD-1 at the NK92 cell/tumor cell immunological synapse. Doublets of single NK92 and K562 cells were readily identified by imaging flow cytometry (Supplementary Fig. S5). The clustering of PD-1 at the cell–cell interface increased when NK92-PD-1 cells were co-cultured with K562-PD-L1 cells compared to K562-GFP cells. Moreover, the clustering of PD-1 was abrogated when the PD-L1 blocking antibody atezolizumab was present during the co-culture (Fig. 4c, d).
To investigate the reinvigoration of freshly obtained PD-1+ NK cells after PD-(L)1 blockade, we exposed NK cells from healthy donors to cytokines previously associated with PD-1 upregulation [37, 38]. Of note, the combination of IL-15 and IL-12 led to a robust PD-1 upregulation on NK cells (Fig. 4e). Whereas IL-15 only led to a minimal change in PD-1 expression, treatment with IL-18 had no effect on PD-1 expression. Upon co-incubation with PD-L1+tumor cells, atezolizumab specifically enhanced the degranulation of ex vivo generated PD-1+ NK cells (Fig. 4f). These data further indicate that PD-1/PD-L1-targeting immunotherapies can target and impact the functionality of primary human PD-1+ NK cells.
Discussion
NK cells are key mediators of anti-tumor immunity and express a variety of activating and inhibitory receptors, which are critical for their effector functions [39]. Although NK cell infiltration in NSCLC has been investigated [23], little is known about the expression of immune checkpoint receptors on intratumoral NK cells. In this study, we found that a significant fraction of NK cells infiltrated in NSCLC expressed PD-1 and co-expressed other inhibitory receptors such as TIM-3 or TIGIT. In addition, we provide evidence that PD-L1-mediated inhibition of PD-1+ NK cells can be reversed by clinically used PD-1/PD-L1 blocking antibodies.
PD-1+ NK cells were previously identified in patients with multiple myeloma [40], lymphoproliferative disorders [41], renal cell carcinoma [42], Kaposi sarcoma [43], digestive cancers [37], and in pleural effusions of ovarian cancer patients [29]. In contrast to a recent work that identifies a rare subset of peripheral PD-1-expressing NK cells in healthy individuals [29], we were only able to detect some expression of PD-1 in circulating NK cells from NSCLC patients. In contrast, though with substantial inter-patient variability, up to 21% of intratumoral NK cells of NSCLC patients expressed PD-1, which is comparable to findings in other cancer types [29, 37, 42]. Interestingly, PD-1 expression on NK cells and CD8 T cells appears to be independently regulated in the tumor as the expression of PD-1 on both subsets did not correlate. Furthermore, we observed increased expression of TIM-3 in both peripheral and intratumoral NK cells of NSCLC patients. TIM-3+ NK cells were previously identified in the peripheral blood of advanced melanoma patients, and TIM-3 blockade was reported to rescue dysfunctional NK cells [28].
NK cells are known to stochastically vary the expression of activating and inhibitory receptors. The integration of negative and positive stimuli together with cell-intrinsic fine-tuning determines whether NK cells are activated and mediate their effector functions [39]. The observation that PD-1+ NK cells more frequently expressed other inhibitor receptors may have implications for their response when exposed to tumor cells expressing inhibitory ligands. The increase of certain KIRs such as KIR2DL3 on intratumoral NK cells is particularly interesting as KIR-targeting agents such as lirilumab (KIR2DL1/2/3-targeting mAb) are tested in multiple clinical trials for different cancer types, often together with PD-(L)1 inhibitors [44]. In agreement with observations in patients with Kaposi sarcoma [43], we found substantial expression of the investigated activating receptors CD16, NKG2D and NKp30 on PD-1+ and PD-1− NK cell subsets. Therefore, we believe that these findings further support the clinical development of novel NK cell engagers which have demonstrated activation of NK cells when co-targeting activating NK cell receptors [45].
The mechanisms which drive the upregulation of PD-1 on NK cells in cancer patients are unclear. PD-1 expression correlated with human cytomegalovirus seropositivity, which suggests a link between PD-1 upregulation and NK cell-mediated anti-viral activity [29]. In agreement with previous work [37, 38], combined treatment with IL-15 and IL-12 caused a significant upregulation of PD-1 on human NK cells. The synergistic effect of these cytokines is intriguing because IL-12 is typically produced by dendritic cells and was recently found to be essential for a successful response to PD-1 blocking agents [46]. We speculate that PD-1+ NK cells may be involved in interactions between NK cells and stimulatory DCs in the tumor microenvironment as recently observed for human cancer, particularly for patients responding to immunotherapy [47].
NK cell hyporesponsiveness has previously been described for a fraction of NK cells that did not express ligands for MHC-I molecules or in humans that lack MHC-I presentation [48, 49]. We found that NK cells from NSCLC patients were mostly limited in their degranulation capacity and polyfunctionality, whereas cytokine secretion and cytotoxicity were less affected. Moreover, our findings indicate that the expression of PD-1 on NK cells might correlate with NK hyporesponsiveness. However, in the absence of PD-L1 expression on target cells, we detected no functional differences between PD-1+ or PD-1− NK cells in response to MHC-I-deficient K562 cells. Interestingly, in a recent study, PD-1+ NK cells in murine tumor models were even more functional than PD-1− NK cells in the absence of PD-L1 [50]. Our experiments showed that PD-L1-expressing tumor cells substantially inhibited cytotoxic killing by PD-1+ NK cells. This inhibition could be reversed by a PD-L1 blocking antibody. We additionally observed PD-1 clustering at the NK-tumor cell–cell interface, which indicates that modulation of the activating and inhibitory signals at the immunological synapse is likely involved in PD-1 signaling in NK cells.
Our findings highlight the importance of PD-1 expression on NK cells and show that PD-1 blockade in patients may directly result in NK cell re-activation. In agreement with our data, Hsu and colleagues recently showed that PD-1+ NK cells in murine tumor models are necessary to mediate the full therapeutic benefit of PD-1 blocking antibodies [50]. Intriguingly, we recently reported on the KIR3DS1 gene variant, which is expressed mainly in NK cells and correlates with resistance to PD-1 blockade in NSCLC patients [27]. Laugney et al. revealed that lung cancers of different lineages display variable susceptibility toward NK cell-mediated immune surveillance [51]. Our findings highlight the need to better understand the contribution of PD-1+ NK cells to cancer immunotherapy responses. A limitation of our study is that the functional assays were performed either in a model system or with healthy donor human NK cells due to the low abundance of NK cells in patient tumors. Further investigations should aim at analyzing intratumoral NK cells from patients undergoing immunotherapy and increasing the resolution to a single-cell level. These investigations will shed further light on the exact role of PD-1+ NK cells upon immune checkpoint blockade with PD-1/PD-L1-targeting agents.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgement
We thank the FACS Core Facility of the DBM of the University of Basel for sorting cells used in this study. Moreover, we thank Prof. Dr. Baum and Prof. Dr. Axel Schambach (Medizinische Hochschule Hannover, Germany) for providing the pRRL.PPT.SFFV.EGFP.pre expression vector. We thank Dr. Ana Luisa Pinto Correia and Priska Auf der Maur for critical input on the manuscript. We also thank all the patients that allowed the use of their material and made this work possible.
Abbreviations
- ATCC
American type culture collection
- DBM
Department of biomedicine
- HD
Healthy donor
- ICB
Immune checkpoint blockade
- KIR
Killer immunoglobulin-like receptor
- LAG-3
Lymphocyte activation gene-3
- NSCLC
Non-small cell lung cancer
- NK
Natural killer cell
- ORF
Open reading frame
- PD-1
Programmed cell death protein 1
- PD-L1
Programmed cell death 1 ligand 1
- TIGIT
T-cell immunoreceptor with Ig and ITIM domains
- TIL
Tumor-infiltrating lymphocyte
- TIM-3
T-cell immunoglobulin and mucin 3
Author contributions
FU, AZ and MPT conceived the idea for the study. FU, MPT and AZ interpreted the data, made the figures and wrote the manuscript. MPT, MK, MAS, FU and AZ planned the experiments. MPT, MK, MS and PH performed and analyzed the experiments. DL, MW and SS provided samples. HL and AZ collected the clinical data and ethical board approvals.
Funding
This work was supported by grants from the Swiss National Science Foundation (320030_162575 to Alfred Zippelius) and a Research Fund of the University of Basel (to Franziska Uhlenbrock).
Compliance with ethical standards
Conflict of interest
Heinz Läubli and Alfred Zippelius received research funding from Bristol-Myers Squibb. Alfred Zippelius received consulting/advisor fees from Bristol-Myers Squibb, Merck Sharp& Dohme, Hoffmann–La Roche, NBE Therapeutics, Secarna, ACM Pharma and Hookipa and maintains further non-commercial research agreements with Secarna, Hookipa, Roche and Beyondsprings. The authors declare that there are no other conflicts of interest.
Ethical approval and ethical standards
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee (Ethikkommission Nordwestschweiz, Study Approval Number EK321/10) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Written informed consent was obtained from all individual participants included in the study for the use of their specimens in research and publication. This article does not contain any studies with animals performed by any of the authors.
Cell line authentication
K562 cells (ATCC CCL-243) and HEK293T cells (ATCC CRL-3216) were purchased from ATCC, which provided detailed cell line authentication documentation. NA8-Mel was kindly provided by Dr. Romero (University of Lausanne) and their authenticity was confirmed by HLA-A2 positivity and cell morphology. NK92 cells (ATCC CRL-2407) were kindly provided by Dr. Bentires-Alj (University of Basel, Switzerland) and authenticity was certified by ATCC.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Franziska Uhlenbrock and Alfred Zippelius these authors jointly directed this work.
Contributor Information
Marcel P. Trefny, Email: marcel.trefny@unibas.ch
Alfred Zippelius, Email: alfred.zippelius@usb.ch.
References
- 1.Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WEE, Poddubskaya E, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non–Small-Cell Lung Cancer. N Engl J Med. 2015;373(2):123–35. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373(17):1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csöszi T, Fülöp A, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 4.Hellmann MD, Ciuleanu T-E, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, et al. Nivolumab plus Ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378(22):2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378(22):2078–2092. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 6.Socinski MA, Jotte RM, Cappuzzo F, Orlandi F, Stroyakovskiy D, Nogami N, et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N Engl J Med. 2018;378(24):2288–2301. doi: 10.1056/NEJMoa1716948. [DOI] [PubMed] [Google Scholar]
- 7.Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gümüş M, Mazières J, et al. Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N Engl J Med. 2018;379(21):2040–2051. doi: 10.1056/NEJMoa1810865. [DOI] [PubMed] [Google Scholar]
- 8.Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, et al. Overall survival with durvalumab after chemoradiotherapy in Stage III NSCLC. N Engl J Med. 2018;379(24):2342–2350. doi: 10.1056/NEJMoa1809697. [DOI] [PubMed] [Google Scholar]
- 9.Garon EB, Hellmann MD, Rizvi NA, Carcereny E, Leighl NB, Ahn M-J, et al. Five-year overall survival for patients with advanced non-small-cell lung cancer treated with pembrolizumab: results from the Phase I KEYNOTE-001 Study. J Clin Oncol. 2019 doi: 10.1200/jco.19.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odorizzi PM, Pauken KE, Paley MA, Sharpe A, Wherry EJ. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8 + T cells. J Exp Med. 2015;212(7):1125–1137. doi: 10.1084/jem.20142237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in Melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant Lung Adenocarcinoma. Cancer Discov. 2018;8(7):822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodig SJ, Gusenleitner D, Jackson DG, Gjini E, Giobbie-Hurder A, Jin C, et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci Transl Med. 2018;10(450):3342. doi: 10.1126/scitranslmed.aar3342. [DOI] [PubMed] [Google Scholar]
- 14.López-Soto A, Gonzalez S, Smyth MJ, Galluzzi L. Control of metastasis by NK cells. Cancer Cell. 2017;32(2):135–154. doi: 10.1016/j.ccell.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 2016;16(1):7–19. doi: 10.1038/nrc.2015.5. [DOI] [PubMed] [Google Scholar]
- 16.Voskoboinik I, Smyth MJ, Trapani JA. Perforin-mediated target-cell death and immune homeostasis. Nat Rev Immunol. 2006;6(12):940–952. doi: 10.1038/nri1983. [DOI] [PubMed] [Google Scholar]
- 17.Screpanti V, Wallin RPA, Grandien A, Ljunggren H-G. Impact of FASL-induced apoptosis in the elimination of tumor cells by NK cells. Mol Immunol. 2005;42(4):495–499. doi: 10.1016/j.molimm.2004.07.033. [DOI] [PubMed] [Google Scholar]
- 18.Fauriat C, Long EO, Ljunggren H-G, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115(11):2167–2176. doi: 10.1182/blood-2009-08-238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guillerey C, Smyth MJ. NK cells and cancer immunoediting. Curr Top Microbiol Immunol. 2016;395:115–145. doi: 10.1007/82_2015_446. [DOI] [PubMed] [Google Scholar]
- 20.Remark R, Alifano M, Cremer I, Lupo A, Dieu-Nosjean M-C, Riquet M, et al. Characteristics and clinical impacts of the immune environments in colorectal and renal cell carcinoma lung metastases: influence of tumor origin. Clin Cancer Res. 2013;19(15):4079–4091. doi: 10.1158/1078-0432.CCR-12-3847. [DOI] [PubMed] [Google Scholar]
- 21.Sconocchia G, Arriga R, Tornillo L, Terracciano L, Ferrone S, Spagnoli GC. Melanoma cells inhibit NK cell functions-letter. Cancer Res. 2012;72(20):5428–5429. doi: 10.1158/0008-5472.CAN-12-1181. [DOI] [PubMed] [Google Scholar]
- 22.Delahaye NF, Rusakiewicz S, Martins I, Ménard C, Roux S, Lyonnet L, et al. Alternatively spliced NKp30 isoforms affect the prognosis of gastrointestinal stromal tumors. Nat Med. 2011;17(6):700–707. doi: 10.1038/nm.2366. [DOI] [PubMed] [Google Scholar]
- 23.Platonova S, Cherfils-Vicini J, Damotte D, Crozet L, Vieillard V, Validire P, et al. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. 2011;71(16):5412–5422. doi: 10.1158/0008-5472.CAN-10-4179. [DOI] [PubMed] [Google Scholar]
- 24.Carrega P, Morandi B, Costa R, Frumento G, Forte G, Altavilla G, et al. Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56 bright CD16 - cells and display an impaired capability to kill tumor cells. Cancer. 2008;112(4):863–75. doi: 10.1002/cncr.23239. [DOI] [PubMed] [Google Scholar]
- 25.Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev. 2002;190(1):40–52. doi: 10.1034/j.1600-065X.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- 26.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 27.Trefny MP, Rothschild SI, Uhlenbrock F, Rieder D, Kasenda B, Stanczak MA, et al. A variant of a killer cell immunoglobulin-like receptor is associated with resistance to PD-1 blockade in lung cancer. Clin Cancer Res. 2019;25(10):3026–3034. doi: 10.1158/1078-0432.CCR-18-3041. [DOI] [PubMed] [Google Scholar]
- 28.da Silva IP, Gallois A, Jimenez-Baranda S, Khan S, Anderson AC, Kuchroo VK, et al. Reversal of NK-cell exhaustion in advanced melanoma by Tim-3 blockade. Cancer Immunol Res. 2014;2(5):410–422. doi: 10.1158/2326-6066.CIR-13-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pesce S, Greppi M, Tabellini G, Rampinelli F, Parolini S, Olive D, et al. Identification of a subset of human natural killer cells expressing high levels of programmed death 1: A phenotypic and functional characterization. J Allergy Clin Immunol. 2017;139(1):335–346. doi: 10.1016/j.jaci.2016.04.025. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Hou M, Liu Y. TIGIT, a novel therapeutic target for tumor immunotherapy. Immunol Invest. 2017;46(2):172–182. doi: 10.1080/08820139.2016.1237524. [DOI] [PubMed] [Google Scholar]
- 31.Vitale M, Cantoni C, Pietra G, Mingari MC, Moretta L. Effect of tumor cells and tumor microenvironment on NK-cell function. Eur J Immunol. 2014;44(6):1582–1592. doi: 10.1002/eji.201344272. [DOI] [PubMed] [Google Scholar]
- 32.Mamessier E, Sylvain A, Thibult ML, Houvenaeghel G, Jacquemier J, Castellano R, et al. Human breast cancer cells enhance self tolerance by promoting evasion from NK cell antitumor immunity. J Clin Invest. 2011;121(9):3609–22. doi: 10.1172/jci45816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gill S, Vasey AE, De Souza A, Baker J, Smith AT, Kohrt HE, et al. Rapid development of exhaustion and down-regulation of eomesodermin limit the antitumor activity of adoptively transferred murine natural killer cells. Blood. 2012;119(24):5758–68. doi: 10.1182/blood-2012-03-415364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choppa PC, Vojdani A, Tagle C, Andrin R, Magtoto L. Multiplex PCR for the detection of Mycoplasma fermentans, M. hominis and M. penetrans in cell cultures and blood samples of patients with chronic fatigue syndrome. Mol Cell Probes. 1998;12(5):301–8. doi: 10.1006/mcpr.1998.0186. [DOI] [PubMed] [Google Scholar]
- 35.Zychlinski D, Schambach A, Modlich U, Maetzig T, Meyer J, Grassman E, et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther. 2008;16(4):718–725. doi: 10.1038/MT.2008.5. [DOI] [PubMed] [Google Scholar]
- 36.Thommen DS, Schreiner J, Muller P, Herzig P, Roller A, Belousov A, et al. Progression of lung cancer is associated with increased dysfunction of T cells defined by coexpression of multiple inhibitory receptors. Cancer Immunol Res. 2015;3(12):1344–1355. doi: 10.1158/2326-6066.CIR-15-0097. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Cheng Y, Xu Y, Wang Z, Du X, Li C, et al. Increased expression of programmed cell death protein 1 on NK cells inhibits NK-cell-mediated anti-tumor function and indicates poor prognosis in digestive cancers. Oncogene. 2017;36(44):6143–6153. doi: 10.1038/onc.2017.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quatrini L, Wieduwild E, Escaliere B, Filtjens J, Chasson L, Laprie C, et al. Endogenous glucocorticoids control host resistance to viral infection through the tissue-specific regulation of PD-1 expression on NK cells. Nat Immunol. 2018;19(9):954–962. doi: 10.1038/s41590-018-0185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol. 2016;17(9):1025–1036. doi: 10.1038/ni.3518. [DOI] [PubMed] [Google Scholar]
- 40.Benson DM, Bakan CE, Mishra A, Hofmeister CC, Efebera Y, Becknell B, et al. The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT-011, a novel monoclonal anti-PD-1 antibody. Blood. 2010;116(13):2286–2294. doi: 10.1182/blood-2010-02-271874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiesmayr S, Webber SA, Macedo C, Popescu I, Smith L, Luce J, et al. Decreased NKp46 and NKG2D and elevated PD-1 are associated with altered NK-cell function in pediatric transplant patients with PTLD. Eur J Immunol. 2012;42(2):541–550. doi: 10.1002/eji.201141832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacFarlane AW, Jillab M, Plimack ER, Hudes GR, Uzzo RG, Litwin S, et al. PD-1 expression on peripheral blood cells increases with stage in renal cell carcinoma patients and is rapidly reduced after surgical tumor resection. Cancer Immunol Res. 2014;2(4):320–331. doi: 10.3892/etm.2018.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beldi-Ferchiou A, Lambert M, Dogniaux S, Vély F, Vivier E, Olive D, et al. PD-1 mediates functional exhaustion of activated NK cells in patients with Kaposi sarcoma. Oncotarget. 2016 doi: 10.18632/oncotarget.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiossone L, Vienne M, Kerdiles YM, Vivier E. Natural killer cell immunotherapies against cancer: checkpoint inhibitors and more. Semin Immunol. 2017;31:55–63. doi: 10.1016/j.smim.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Gauthier L, Morel A, Anceriz N, Rossi B, Blanchard-Alvarez A, Grondin G, et al. Multifunctional natural killer cell engagers targeting NKp46 trigger protective tumor immunity. Cell. 2019;177(7):1701–1713.e16. doi: 10.1016/J.CELL.2019.04.041. [DOI] [PubMed] [Google Scholar]
- 46.Garris CS, Arlauckas SP, Kohler RH, Trefny MP, Garren S, Piot C, et al. Successful anti-PD-1 cancer immunotherapy requires T cell-dendritic cell crosstalk involving the cytokines IFN-γ and IL-12. Immunity. 2018;49(6):1148–1161.e7. doi: 10.1016/j.immuni.2018.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, et al. A natural killer–dendritic cell axis defines checkpoint therapy–responsive tumor microenvironments. Nat Med. 2018;24(8):1178–1191. doi: 10.1038/s41591-018-0085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zimmer J, Donato L, Hanau D, Cazenave J, Tongio M, Moretta A, et al. Activity and phenotype of natural killer cells in peptide transporter (TAP)-deficient patients (Type I bare lymphocyte syndrome) J Exp Med. 1998;187(1):117–122. doi: 10.1084/JEM.187.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anfossi N, André P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25(2):331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 50.Hsu J, Hodgins JJ, Marathe M, Nicolai CJ, Bourgeois-Daigneault M-C, Trevino TN, et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J Clin Invest. 2018 doi: 10.1172/JCI99317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laughney AM, Hu J, Campbell NR, Bakhoum SF, Setty M, Lavallée V-P, et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat Med. 2020;26(2):1–11. doi: 10.1038/s41591-019-0750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.