Abstract
Increased proteolytic activity of cysteine cathepsins has long been known to facilitate malignant progression, and it has also been associated with tumor-promoting roles of myeloid-derived suppressor cells (MDSCs). Consequently, cysteine cathepsins have gained much attention as potential targets for cancer therapies. However, cross-talk between tumor cells and MDSCs needs to be taken into account when studying the efficacy of cathepsin inhibitors as anti-cancer agents. Here, we demonstrate the potential of the MDA-MB-231 breast cancer cell line to generate functional MDSCs from CD14+ cells of healthy human donors. During this transition to MDSCs, the overall levels of cysteine cathepsins increased, with the largest responses for cathepsins L and X. We used small-molecule inhibitors of cathepsins L and X (i.e., CLIK-148, Z9, respectively) to investigate their functional impact on tumor cells and immune cells in this co-culture system. Interactions with peripheral blood mononuclear cells reduced MDA-MB-231 cell invasion, while inhibition of cathepsin X activity by Z9 restored invasion. Inhibition of cathepsin L activity using CLIK-148 resulted in significantly increased CD8+ cytotoxicity. Of note, inhibition of cathepsins L and X in separate immune or tumor cells did not promote these functional changes. Together, our findings underlie the importance of tumor cell–immune cell interactions in the evaluation of the anti-cancer potential of cysteine cathepsin inhibitors.
Electronic supplementary material
The online version of this article (10.1007/s00262-020-02592-x) contains supplementary material, which is available to authorized users.
Keywords: Cysteine cathepsins, Cathepsin inhibition, Myeloid-derived suppressor cells, Cancer, Immunosuppression, MDA-MB-231 cells
Introduction
Myeloid-derived suppressor cells (MDSCs) are recognized as one of the most important drivers of cancer-related immune suppression [1]. Mouse models have provided valuable insights into the interactions between cancer cells and their microenvironment; however, with MDSCs, this information has been clouded by differences between the human and mouse immune systems [2]. In mice, MDSCs can be extracted from spleen, bone marrow, and tumor tissues, while in humans, peripheral blood remains their preferred source. Studies on human MDSCs have thus been limited due to practical and ethical constraints, and they have mainly focused on evaluation of the effects of cancer treatments on the numbers and immunosuppressive activities of peripheral blood MDSCs [3, 4]. Furthermore, isolated MDSCs have a relatively short half-life ex vivo [5]. Those limitations could be overcome by a reliable in vitro model that mimics MDSCs of human cancer patients.
Apart from immunosuppression, dysregulated proteolysis is another feature of tumor progression. Importantly, tumor-infiltrating myeloid cells have been shown to overexpress peptidases that have distinct roles in tumorigenesis [6, 7]. The cysteine cathepsins (cat) are a family of papain-related lysosomal peptidases that comprise 11 members, including catB, L, K, S, and X, which have been identified as potential diagnostic and prognostic biomarkers in solid tumors and blood cancers [8]. Studies in mice have shown that catB, L, and S promote tumor-associated macrophage survival [9] and that catS supports their polarization to the pro-tumoral M2 type [10]. CatB facilitates MDSC expansion through the induction of tumor necrosis factor-α secretion [11], and MDSC-mediated tumor-cell resistance to chemotherapy through activation of the Nlrp3 inflammasome [12]. Taken together, these few studies have indicated that the cat provide myeloid cells with a tumor-promoting phenotype. However, the role of cat in human MDSCs has not been studied to date.
To provide better understanding of the role of cats in human MDSCs, we generated MDSCs from MDA-MB-231 cells and CD14+ cells. With the use of specific inhibitors of catL and X, we investigated the implications of these peptidases in MDSC–tumor cell cross-talk. Importantly, we show that catL and X inhibition has significant functional consequences for reciprocal MDSC–tumor cell interactions.
Materials and methods
Cell lines
The PC-3 (CRL-1435), U-87 MG (HTB-1), MDA-MB-231 (HTB-26), HOS (CRL-1543), and Raji (CCL-86) cell lines were all from ATCC and maintained in humidified chamber at 37 °C, 5% CO2. PC-3 cells were cultured in complete 1:1 advanced DMEM and F12 (Gibco); U87 MG and HOS cells in complete advanced DMEM; and MDA-MB-231 and Raji cells in complete advanced RPMI 1640 medium (Gibco). Unless stated otherwise, the complete culture media were supplemented with 10% fetal bovine serum (Gibco), 100 U/mL penicillin, 0.1 mg/mL streptomycin, and 2 mM l-glutamine (Sigma-Aldrich). For co-culture experiments, U87 MG, HOS, and PC-3 cells were gradually adjusted to growth in complete advanced RPMI 1640 medium.
Isolation of peripheral blood mononuclear cells
Collection of the peripheral blood mononuclear cells (PBMCs) for purpose of the study was approved by The National Medical Ethics Committee of the Ministry of Health, Republic of Slovenia, under the designated number 0120-279/2017-3. The human PBMCs were isolated from the blood of healthy volunteer donors by differential density gradient centrifugation, at the Blood Transfusion Centre of Slovenia, following the institutional guidelines. The PBMCs were maintained in complete advanced RPMI 1640 medium. CD14+ monocytes and CD8+ T cells were isolated from the PBMCs using positive magnetic bead selection and MACS LS separation columns (Miltenyi Biotec), according to manufacturer protocol.
Selection of optimal tumor cell line for in vitro generation of human myeloid-derived suppressor cells
PC-3, MDA-MB-231, and U87 MG cells were seeded at 2 × 105 cells/well, and HOS cells at 1.5 × 105 cells/well, in 2 mL complete medium. They were left to adhere for 24 h prior to addition of CD14+ monocytes (2 × 106 cells/1 mL). After 1-day medium was refreshed, and following 72 h of co-culture cells were harvested. To isolate the pure MDSC populations, the collected cells were separated using CD11b+ magnetic beads and then counted and analyzed by flow cytometry (Attune NxT; Thermo Fischer Scientific). As reported, MDA-MB-231 cells do not express CD11b marker [13]. For the controls, CD14+ cells were stimulated with 1000 U/mL GM-CSF (R&D Systems) or 1000 U/mL GM-CSF plus IL-4 (Miltenyi Biotec), without and with 1 µM prostaglandin PGE2 (Tocris Bioscience).
T cell suppression assay
To evaluate the immunosuppression of the MDSCs, CD8+ T cells were labeled with 0.2 µM carboxyfluorescein succinimidyl ester (CFSE) (Invitrogen) for 15 min at 37 °C. The labeled CD8+ T cells were then resuspended in complete media supplemented with IL-2 (Bachem) and added to the purified MDSCs or tumor cells at 1:4 or 1:2 ratio. The T cells were activated by anti-CD3/CD28 antibody-coated magnetic beads (Dynabeads human T-activator; Thermo Fischer Scientific) and left to proliferate for 72 h. The T-cell proliferation was analyzed with the FlowJo software, version 7.6.5 (TreeStar), and is expressed as percentages of T-cell proliferation in the presence of the MDSCs relative to T-cell proliferation in their absence.
Transwell assay
To determine whether CD14+ monocytes require contact with tumor cells to become suppressive, MDA-MB-231 tumor cells (0.5 × 105 cells/well in 0.5 mL) were seeded in 24-well culture plates. The next day, 4 × 105 CD14+ cells together with 1.6 × 105 CFSE-labeled CD8+ T cells were placed either on the top of the microporous membranes of the transwell inserts, which physically separate CD14+ cells from tumor cells, or they were layered directly onto the tumor cells. Dynabeads were used to activate the T cells, and their proliferation was assessed by flow cytometry. The data were analyzed using the FlowJo software.
Flow cytometry
For flow cytometry analysis of the cell phenotypes, cells were washed with FACS buffer (2% FBS, 2 mM EDTA, in PBS, pH 7.4) and incubated with fluorochrome-conjugated primary antibodies and FcR blocking reagent (Miltenyi Biotec) for 30 min at 4 °C. The cells were then washed with PBS and analyzed by flow cytometry (Attune NxT). Four-color cell-surface staining was performed, using the following antibody panel: HLA-DR FITC (AC122), CD14 PE-Vio 770 (TÜK4), CD33 APC (REA775) (Miltenyi Biotec), and CD11b V450 (ICRF44) (BD Biosciences). For detection of STAT3 phosphorylation, cells were fixed and permeabilized with ice-cold methanol and incubated at − 20 °C for 20 min. Cells were washed with PBS and incubated with primary rabbit anti-phospho STAT3 (Tyr705) antibodies (1:100; 9145; Cell Signaling Technology) for 1 h at 4 °C. Samples and isotype controls were incubated with Alexa Fluor 488-conjugated anti-rabbit secondary antibodies (1/500; Invitrogen) in dark for 30 min at room temperature, and staining was analyzed by flow cytometry (Attune NxT).
Confocal microscopy
For the confocal microscopy, 2 × 104 cells in PBS were seeded onto 12-well microscope slides and left to adhere for 30 min at 37 °C. The cells were then fixed with 10% formalin for 20 min, permeabilized with 0.1% Triton X-100 in PBS for 5 min, and blocked with 3% bovine serum albumin in PBS for 30 min. Goat anti-catX (AF934; R&D Systems) and rabbit anti-LAMP-1 (SAB3500285; Sigma-Aldrich) antibodies were diluted 1:100 and incubated with the cells for 1 h at room temperature. Fluorochrome-labeled secondary antibodies (dilution, 1:1000; Invitrogen) were incubated for 30 min at room temperature. Slides were mounted using ProLong Gold Antifade Reagent with DAPI (Cell Signaling Technology). Images were captured with confocal microscope (LSM 710; Carl Zeiss) and exported using the ZEN 2012 SP1 black edition software, version 8.1 (Carl Zeiss).
MDA-MB-231/peripheral blood mononuclear cell co-cultures
To evaluate the role of the cat in MDSC–tumor cell interactions, the MDA-MB-231 cells were seeded at 7.5 × 105 cells/10 mL in T75 cell culture flasks and left for 24 h prior to addition of the PBMCs (20 × 106 cells/5 mL per flask). Where applicable, the catX inhibitor Z9 [14], the catL inhibitor CLIK-148 [15], or their combination was added to the cell co-cultures at the final concentrations of 10 µM and 1 µM, respectively, with DMSO (0.1%) used as the control. We also tested the impact of inhibitors and DMSO on PBMC viability and concluded they were not cytotoxic at indicated concentrations (data not shown).
Conditioned medium collection
After 72 h of MDA-MB-231/PBMC co-culture, the adherent cells were washed and left in serum-free medium for 24 h. Next, the conditioned medium was collected and concentrated tenfold using ultrafiltration tubes (cutoff, 10 kDa; Amicon) at 4000 ×g for 15 min. The concentrated conditioned medium was aliquoted and stored at − 80 °C until further use.
Cell lysate preparation
For the protein expression analysis using Western blotting, the cells were lysed in RIPA buffer [150 mM NaCl, 50 mM Tris, 1% Nonidet P-40, 0.5% Na-deoxycholate, 0.1% Na-dodecyl sulfate (SDS), 0.004% Na-azide, pH 8.0] with added protease and phosphatase inhibitor cocktail (Thermo Scientific). The cell lysates were freeze-thawed, sonicated, and centrifuged at 16,000 ×g at 4 °C for 15 min. The protein concentrations in the collected supernatants were determined using kits (DC-Protein Assay kits; Bio-Rad Laboratories), according to the manufacturer instructions.
Western blotting
Aliquots of the cell lysates containing 15–30 µg total protein were resolved by SDS-PAGE (12% Tris–glycine gels) and transferred onto nitrocellulose membranes using a dry blotting system (iBlot 2; Thermo Fisher Scientific). The membranes were blocked with 5% nonfat dried milk, in Tris-buffered saline with 0.05% Tween-20 for catX and L, and in PBS with 0.05% Tween-20 for pSTAT3, STAT3, catS, B, and legumain, for 1 h at room temperature. The antibodies (and their dilutions) were: goat anti-catX (1:1000; AF934; R&D Systems); goat anti-catS (1:1000; AF1183; R&D Systems); mouse anti-catL (1:5000; C4618; Sigma-Aldrich); sheep anti-catB (1:2500; prepared at Jozef Stefan Institute); mouse anti-legumain (1:200; sc-133234, B-8; Santa Cruz Biotechnology); mouse anti-pSTAT3 (1:500; sc-5086, B-7; Santa Cruz Biotechnology); and mouse anti-STAT3 (1:500; sc-8019, F-2; Santa Cruz Biotechnology). After washing, the membranes were incubated with the horse-radish peroxidase (HRP)-conjugated secondary antibodies: anti-mouse (1:5000; Jackson Immunoresearch), anti-sheep (1:5000; Jackson Immunoresearch), anti-rabbit (1:5000; Jackson Immunoresearch), and anti-goat (1:2500; Santa Cruz Biotechnology). SuperSignal West Dura Extended Duration Substrate (Thermo Scientific) was used for detection, and the images were acquired on G:Box system (Syngene). The band intensities were quantified using the GeneTools software. When needed, the membranes were stripped with stripping buffer (100 mM 2-mercaptoethanol, 2% SDS, and 62.5 mM Tris/HCl, pH 5.7) for 1 h at 65 °C.
To determine the cat activities, the cell lysates and concentrated conditioned medium were incubated with 10 µM DCG-04 (gift from Prof. Bogyo, Stanford University), for 60 min at room temperature. The samples were then denaturated, resolved by SDS-PAGE, and blotted as described above. The DCG-04 biotinylated probe was detected with HRP-conjugated streptavidin (1:5000; Jackson Immunoresearch).
Real-time cell invasion and migration assays
The invasion and migration of the MDA-MB-231 cells were monitored using a real-time cell analyzer (xCELLigence; ACEA Biosciences) as described before [16]. Briefly, membranes of CIM-plate were coated with fibronectin (10 µg/mL; BD Biosciences) at the bottom and layered with Matrigel (1 mg/mL; Corning Life Sciences) or fibronectin at the top for invasion and migration, respectively. Subsequently, the lower compartments were loaded with complete medium, and CIM-plates were assembled. Serum-starved MDA-MB-231 cells were plated in the top chambers (40,000 cells/well) in serum-free medium. Impedance was monitored for 48 h at 15-min time interval, and the data were analyzed using the RTCA software, version 1.2.1 (ACEA Biosciences).
Cytotoxicity assay
Antigen-specific cytotoxic CD8+ cells were obtained by stimulation of the PBMCs with bacterial superantigens: Staphylococcus enterotoxins-A (Sigma-Aldrich), -B (Sigma-Aldrich), and -E (Toxin Technology) [17]. Briefly, PBMCs from MDA-MB-231 cell co-cultures were pulsed with a cocktail of Staphylococcus enterotoxins-A, -B, and -E (0.1, 0.2, 0.1 µg/mL, respectively) for 1 h and then resuspended in complete medium containing 200 U/mL IL-2, at a cell density of 2 × 106 cells/mL. The cells were left to proliferate for 5 days to 7 days, with the medium and IL-2 refreshed every second day.
In the cytotoxicity assays, the expanded CD8+ cells were used as the effector cells, and Raji cells were used as the target cells. First, Raji cells were incubated with the Staphylococcus enterotoxin cocktail for 45 min and labeled during the last 15 min with CFSE (0.2 µM). Meanwhile, different concentrations of effector cells were plated in 96-well U-bottomed plates. Next, the target cells (1 × 104) were added to the effector cells and centrifuged at 300 ×g for 1 min. After 3 h, the cells were collected, washed with cold PBS, and incubated on ice with the cell viability dye 7-aminoactinomycin D (7-AAD; Sigma-Aldrich), for 10 min. Target cell lysis was determined using the FlowJo software as the proportion of 7-AAD and CFSE-positive events relative to the total CFSE-positive cells. The data are expressed as lytic activity (LU (40%)/107 cells), calculated according to Eq. (1):
| 1 |
where T is the number of target cells and X40% is the E:T ratio required to lyse 40% of the target cells [18].
Statistics
Statistical analysis was performed using GraphPad Prism 6, and the data were evaluated using Student’s t tests or ANOVA. p < 0.05 was considered to be indicative of statistical significance.
Results
MDA-MB-231 cells promote generation of myeloid-derived suppressor cells from human monocytes in a contact-dependent manner
Several protocols to establish in vitro MDSCs from healthy human PBMCs have been introduced using either combinations of cytokines [19–21], tumor-cell-conditioned media [22], or co-culture with tumor cells [23–25]. We used the latter approach, as it is particularly useful to study reciprocal interactions between the tumor cells and the MDSCs.
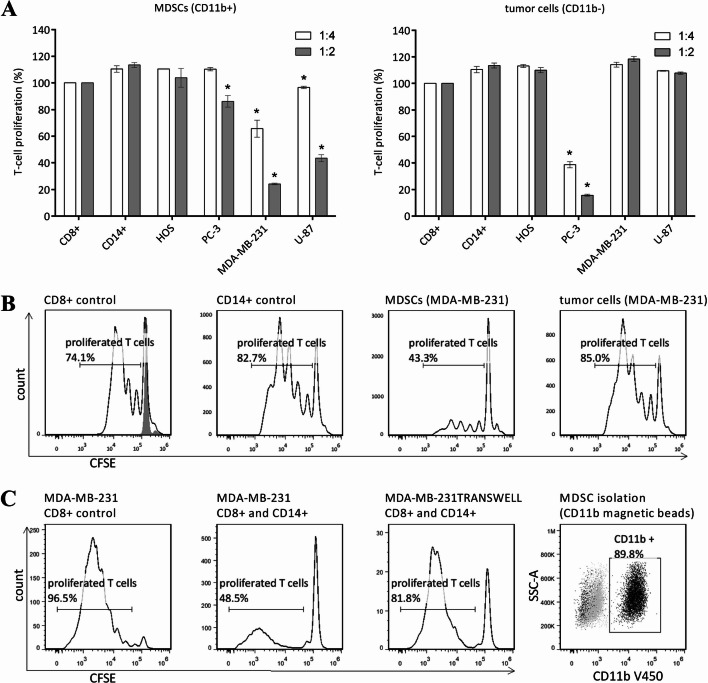
The MDA-MB-231 breast cancer and U-87 glioblastoma tumor cell lines induced the generation of immunosuppressive MDSCs from CD14+ cells. In contrast, neither the HOS osteosarcoma nor the PC-3 prostate cancer cell line evoked any immunosuppressive MDSC activity. To ensure that the suppressive effects were indeed mediated solely by the MDSCs generated, the tumor cell fraction (CD11b− cells) was also tested for T-cell proliferation. Interestingly, PC-3 tumor cells showed potent reduction of T-cell proliferation, while the other three cell lines did not (Fig. 1a). As the MDA-MB-231 cells induced the MDSCs with the most potent immunosuppressive activity (illustrated in Fig. 1b), they were adopted as the model cell line.
Fig. 1.
Immunosuppressive potential of the in vitro generated myeloid-derived suppressor cells (MDSCs). a The CD11b-positive (CD11b+) and -negative (CD11b−) cell fractions were isolated from tumor cell/CD14+ cell co-cultures and added to activated carboxyfluorescein succinimidyl ester CFSE-labeled T lymphocytes at different ratios (CD11b+/− cells:CD8+ cells, 1:4, 1:2). T-cell proliferation was assessed after 72 h by flow cytometry. Data are mean ± standard error of the means. *p < 0.05, indicating significant reduction in T-cell proliferation in comparison with fresh CD14+ cells. b Representative T-cell proliferation when the T cells were cultured alone, with fresh CD14+ cells, with MDSCs (isolated from co-cultures with MDA-MB-231 tumor cells), or with MDA-MB-231 tumor cells (ratio 1:4) (as indicated). Proliferation of activated CD8+ cells (black line) was 100% (solid gray, resting CD8+ cells). Fresh CD14+ cells were incubated with CD8+ cells as an additional control to exclude intrinsic immunosuppressive effects of CD14+ cells. c Representative T-cell proliferation as T cells alone (i) in comparison with when cultured with CD14+ cells in direct contact with (ii) or physically separated from (by transwell insert membrane) (iii) the MDA-MB-231 tumor cells. Only CD14+ cells that were in direct contact with tumor cells reduced T-cell proliferation. d Confirmation of cell purity after isolation of CD11b+ cells (MDSCs). After each separation, the purities of the isolated cell populations were around 95% and 90% for CD14+ cell and CD11b+ cells, respectively, as assessed by flow cytometry. Gray, isotype control; black, antibody-stained cells
Additionally, we asked whether the growth factors and cytokines released by MDA-MB-231 cells into the culture medium would suffice to invoke the MDSC immunosuppression. However, in the present model, the CD14+ cells that were physically separated from the tumor cells by the transwell insert membranes showed no suppressive activity, which suggested that direct contact between the tumor cells and the CD14+ cells was necessary for MDSC generation (Fig. 1c).
Myeloid-derived suppressor cells generated by MDA-MB-231 cells are distinct from cytokine-induced myeloid-derived suppressor cells, macrophages, and dendritic cells
As MDSCs share their immunosuppression with other regulatory myeloid cell types, specifically dendritic cells and macrophages, confirmation of their distinct phenotypic features was required. Expression of the consensus cell-surface markers was evaluated for MDA-MB-231-derived MDSCs in comparison with cytokine-derived MDSCs, dendritic cells, and macrophages.
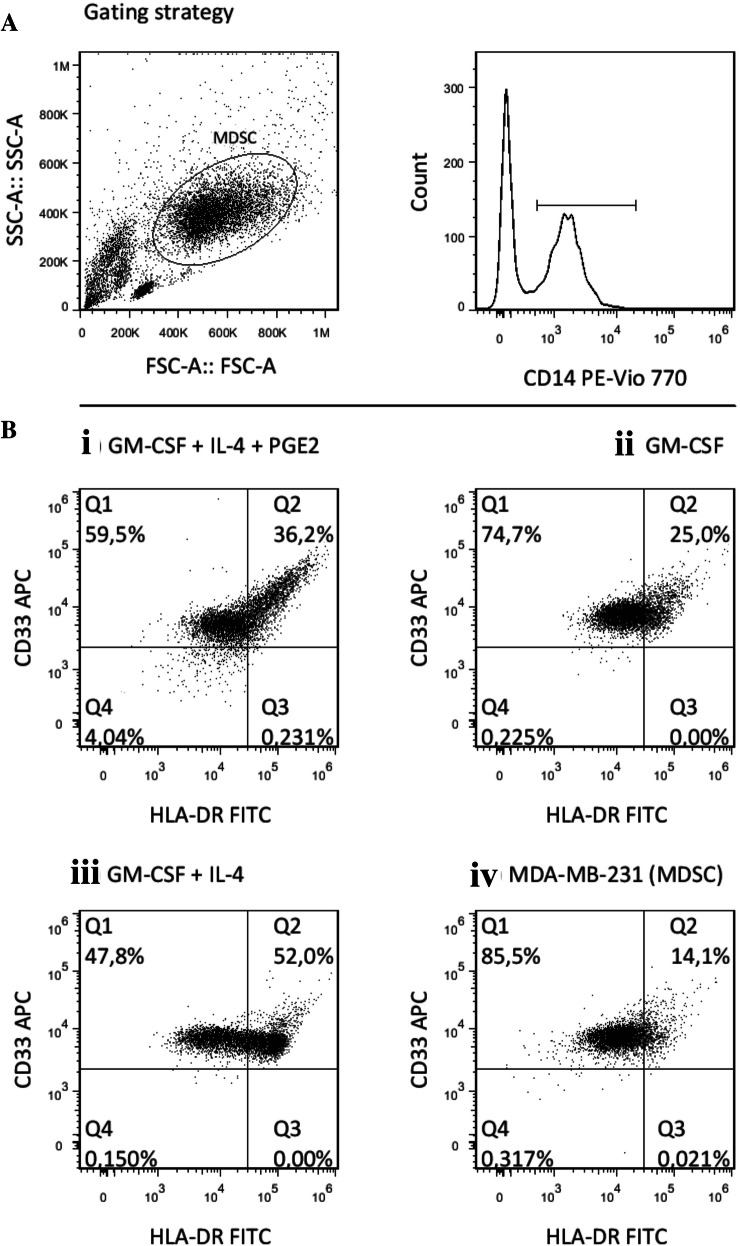
For each of these cell types, the CD14+ CD11b+ cells were analyzed for HLA-DR versus CD33 expression. The high proportions of cells with the CD14+ CD11b+, HLA-DR low, and CD33+ phenotype indicated effective generation of MDSCs from the MDA-MB-231 tumor cell co-cultures (Fig. 2).
Fig. 2.
CD14+ cells from the co-cultures with the MDA-MB-231 tumor cells show typical myeloid-derived suppressor cell (MDSC) phenotype, and are distinct from other myeloid populations. Representative dot plots for the phenotype comparisons. a MDA-MB-231-generated MDSCs according to gating for CD14+ CD11b+ expression. b HLA-DR and CD33 expression for the cytokine-induced MDSCs (i), macrophages (ii), dendritic cells (iii) and MDA-MB-231-generated MDSCs (iv). Cells in quadrant Q1 correspond to the MDSC phenotype
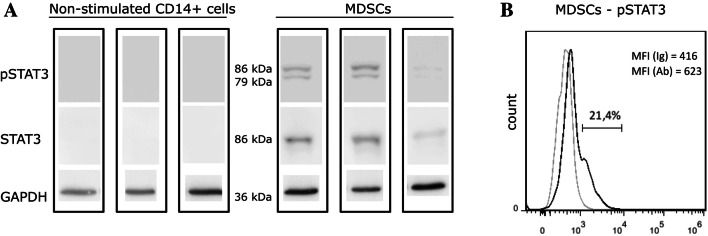
Activation of the transcriptional factor STAT3 was then investigated, as another hallmark that indicates bona-fide MDSC generation (Fig. 3). The STAT3 protein was below the limit of detection in fresh CD14+ cells; however, its expression was significantly increased after co-culture with the MDA-MB-231 cells. Of note, phosphorylation of STAT3 in the MDA-MB-231 cells also increased after co-culture with PBMC (Supplementary Figure 1).
Fig. 3.
Myeloid-derived suppressor cells (MDSCs) generated from the MDA-MB-231 tumor cell/CD14+ cell co-cultures express pSTAT3. a Representative Western blotting of phosphorylated STAT3 (pSTAT3) and total STAT3 (STAT3) expression in nonstimulated CD14+ cells (left) and MDSCs (right) from three different donors. GAPDH expression was used as the loading control. b Flow cytometry analysis of pSTAT3 in MDSCs. Gray line, isotype control; black line, pSTAT3 antibody-stained cells. MFI: median fluorescence intensity
Cysteine cathepsins are up-regulated during myeloid-derived suppressor cell generation from monocytes
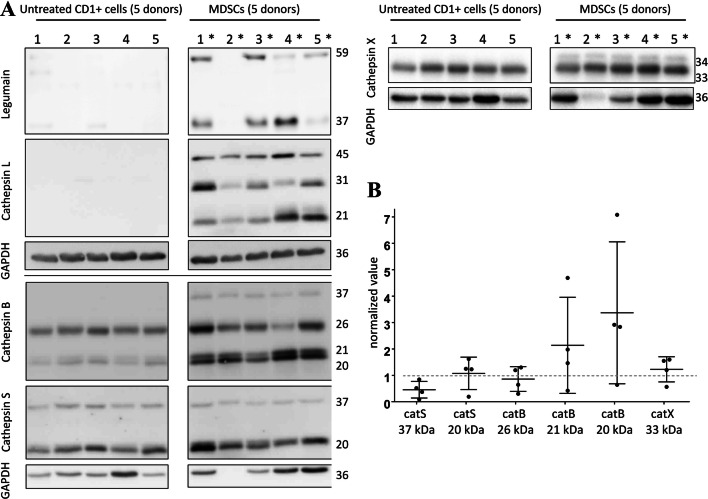
Having successfully established in vitro MDSC model, we turned our attention to the role of the cat in MDSC function. The protein expression levels were evaluated for MDSCs that were isolated from five different PBMC donors, using Western blotting (Fig. 4a, samples 1*–5*), and compared to the fresh CD14+ cells from the same donors (Fig. 4a, samples 1–5). CatB and S were both up-regulated, and the stronger bands in low molecular weight range indicate their increased processing (Fig. 4). The most striking differences, however, were seen for catL protein expression, which was not detected in the fresh monocytes, but it was abundant in the isolated MDSCs.
Fig. 4.
Expression of selected cysteine cathepsins in the MDA-MB-231-generated myeloid-derived suppressor cells (MDSCs) compared to the untreated fresh monocytes from the same donors. a Representative Western blotting for expression of legumain and cathepsins L (top left), B, S (bottom left), and X (top right) in MDSCs shows increased protein levels and processing, of cathepsin B and S. Similar to cathepsin B, the pro-form and active form of cathepsin X was increased. Lanes 1-5, cell lysates from fresh untreated CD14+ cells; lanes 1*–5*, MDSC lysates from matching donors. GAPDH was used as the loading control. b Semi-quantitative analysis of the levels of cathepsins S (catS), B (catB), and X (catX) expression relative to the untreated fresh monocytes (MDSC samples normalized for GAPDH loading control and their matched untreated CD14+ control samples). Only the samples with bands detected for both fresh monocytes and MDSCs were compared
Previous studies have shown that aspartic endopeptidase legumain acts as a pro-catL convertase [26]. Accordingly, here legumain was shown to coincide with de-novo catL expression. Accumulation of the catX pro-form was also seen for these MDSCs (Fig. 4a), and its active form was also increased (Fig. 4b). For catX we also performed immunofluorescence staining to indicate its distribution across different types of myeloid cells (Supplementary Figure 2). These data suggest that catL may play important role in regulation of key MDSC functions. In addition to protein expression analysis, we evaluated catL and X activity in MDSCs samples generated without and with catL inhibitor CLIK-148, using activity-based probe DCG-04 (Supplementary Figure 3).
With the knowledge that catL acts as a pro-catX convertase [27], we focused on catL and X to determine how inhibition of their individual proteolytic activities, or of both, influenced the tumor cell–MDSC interactions.
Differential impact of cathepsin L and cathepsin X inhibition on tumor cell invasion in the presence of peripheral blood mononuclear cells
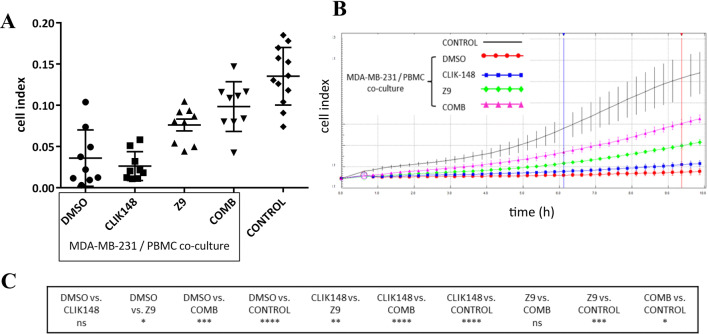
Next, we exploited the generation of MDSCs from MDA-MB-231 cells to study the immune cell–tumor cell interactions in a MDA-MB-231/PBMC co-culture. The effects of PBMCs and catL and/or catX inhibitors on the MDA-MB-231 cell invasion were examined in real-time assays (Fig. 5). Importantly, neither the PBMCs nor the cat inhibitors were present during the invasion assays. Contrary to expectations, the co-culture of the MDA-MB-231 cells with PBMCs markedly reduced the MDA-MB-231 cell invasion. However, this invasion was not affected further with the addition of the catL inhibitor CLIK-148. Notably, the catX inhibitor Z9 increased the intrinsic invasion of the MDA-MB-231 cells, and the combination of both of these inhibitors (i.e., CLIK-148 plus Z9) showed an even greater increase. Additional analyses were performed where the MDA-MB-231 cells were treated with the catL and/or catX inhibitors without PBMCs. In this case, no significant differences were seen for the invasion of the MDA-MB-231 cells between different inhibitor treatments (Supplementary Figure 4a).
Fig. 5.
Intrinsic invasion of the MDA-MB-231 cells is modulated in the presence of peripheral blood mononuclear cells (PBMCs) and the cathepsin L/X inhibitors. MDA-MB-231 cells were co-cultured with PBMCs and treated with the cathepsin L inhibitor CLIK-148 or the cathepsin X inhibitor Z9, or the combination of both, for 72 h. DMSO-treated co-cultures and MDA-MB-231 cells without PBMCs and inhibitors added served as controls. a PBMCs suppressed MDA-MB-231 cell invasion. Treatment with the cathepsin X inhibitor (Z9) partially, and in combination with cathepsin L inhibitor even more so (COMB), restored the invasion of the MDA-MB-231 cells, while treatment with the cathepsin L inhibitor alone had not effects (CLIK148). Data are mean ± standard error of the mean for three independent experiments, each performed in triplicate (n = 9). b Representative curves of MDA-MB-231 cell invasion through Matrigel, driven by the serum gradient between the upper and lower compartments. The two vertical lines show the time interval used for determination of cell index. c Statistical significance of the differences between the different treatments. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
Several cat can degrade the extracellular matrix and might contribute to the observed differences in invasion. We examined the activities of the secreted cat in the culture medium using a pan-cat activity-based probe, DCG-04 [28]. No differences in the profiles of the secreted active cat were observed between the DMSO control of the MDA-MB-231 cell/PBMC co-cultures and those treated with the catL and/or catX inhibitors (Supplementary Figure 4b). Furthermore, the migration assays gave similar results to the invasion assays, suggesting that it was unlikely that the differences in intracellular degradation of Matrigel were responsible for changes in the invasion profile (Supplementary Figure 4c).
Differential impact of cathepsin L and cathepsin X inhibition on CD8+ T-cell cytotoxicity in the presence of tumor cells
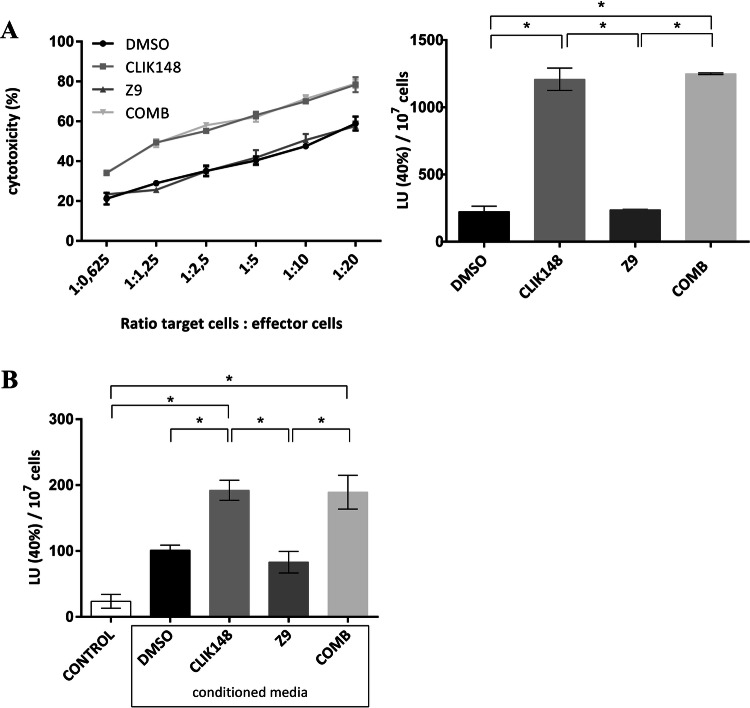
Following investigation of the functional impact of the immune cells on the tumor cells, we asked whether the properties of the immune cells were also differentially modulated in the presence of the tumor cells and the cat inhibitors. Therefore, CD8+ cells from PBMC/MDA-MB-231 cell co-cultures that were treated with DMSO or the cat inhibitors were assayed for cytotoxicity. Interestingly, the catL inhibitor CLIK-148 potently augmented the CD8+ cell cytotoxicity, while the cytotoxicity was not changed for the Z9-treated cells, compared to the DMSO control (Fig. 6a). The combination of both of these cat inhibitors showed the same CD8+ cell cytotoxicity as for CLIK-148 alone (Fig. 6a). Again, the cat inhibitors were not present during the course of experiment, which meant that the intrinsic changes to the cytotoxic cells during co-culture period accounted for the different behaviors of the immune cells.
Fig. 6.
Cytotoxicity of CD8+ cells from peripheral blood mononuclear cells (PBMC)/tumor cell co-cultures is modulated by cathepsin L inhibition. a PBMCs from co-cultures with MDA-MB-231 (without and with cathepsin L, cathepsin X inhibitors) were stimulated with Staphylococcus enterotoxins, and the CD8+ cells were left to expand. To test their cytotoxicity, they were plated with target Raji cells at different ratios (CD8+ cells:Raji, 1:0.625, 1:1.25, 1:2.5, 1:5, 1:10, 1:20; as indicated). After 2.5 h, the target-cell viability was determined using flow cytometry. Left: Killing curves. Data are means ± standard errors from one of the two independent experiments performed in duplicate. Right: Data expressed as lytic activities. *p < 0.05. bStaphylococcus enterotoxin-reactive CD8+ cells were expanded from an allogenic donor and incubated overnight with conditioned medium from cathepsin inhibitor or DMSO-treated tumor cell/PBMC co-cultures. Cytotoxicity was assayed the day after (as above). The additional control was CD8+ cells incubated in fresh cell culture medium (n = 4)
In addition, CD8+ cells from an allogenic donor were used, with an overnight incubation in conditioned medium harvested from inhibitor-treated co-cultures of PBMCs and MDA-MB-231 cells. The next day, the cytotoxicities were measured (Fig. 6b). These matched the data from the previous experimental setup.
Discussion
Here, we have developed an in vitro model of human MDSCs from co-cultures of tumor cell line MDA-MB-231 with human donor CD14+ cells, to investigate the role of cysteine peptidases cats L and X in MDSC–tumor cell interactions.
To the best of our knowledge, the MDA-MB-231 cell line has not yet been shown to generate functional human monocytic MDSCs. Jiang et al. used MDA-MB-231 cells in co-culture with CD33+ myeloid progenitors from peripheral blood; however, their focus was on immature MDSCs (i.e., CD45+ CD13+ CD33+ CD14− CD15−) [29]. Interestingly, in a canonical study by Lechner et al., MDSCs were generated from cells of every tumor type, except for breast cancer [23]. This discrepancy with the present study might well be explained by differences in the experimental setup. Generation of MDSCs by the glioblastoma U-87 cell line was not surprising, as this had already been shown by Rodrigues et al. [25]. However, Rodrigues et al. concluded that the U-87 cell line generated poorly immunosuppressive MDSCs, which might be attributed to the assay they used for immunosuppressive activity (i.e., apoptosis induction, instead of T-cell proliferation). In line with other studies [25, 30], we established the need for the close proximity of the CD14+ cells and the tumor cells in order to generate MDSCs.
We demonstrated that MDA-MB-231-generated MDSCs express the typical MDSC surface phenotype. In comparison with fresh CD14+ cells, CD14 surface expression was down-regulated in MDA-MB-231-generated MDSC (data not shown). This observation is in agreement with Rodrigues et al. [25], Lechner et al. [31], and Casacuberta-Serra et al. [19], but contrary to the findings of Mao et al. [30]. In conclusion, the differences between in vitro MDSC models appear to be not only due to variations in experimental protocols and characterization strategies, but also to the inherent MDSC heterogeneity and plasticity [32, 33]. Indeed, in vitro generated MDSCs can be phenotypically and functionally similar, yet not identical [31].
MDSCs are usually associated with increased invasive behavior of tumor cells [34–37]. Therefore, our finding that the presence of PBMCs reduced tumor cell invasion appeared counterintuitive. However, this finding is not definite as MDA-MB-231 control lacked vehicle in comparison to MDA-MB-231/PBMC control. Nevertheless, we also observed that PBMC increased the levels of phosphorylated STAT3 in the MDA-MB-231 cells. As reported by Banerjee et al., STAT3 in MDA-MB-231 cells might act in a tumor-suppressive manner [38].
The cat are regarded as promising targets for cancer treatments [14]. However, before their inhibitors can be translated into clinical practice, a more detailed understanding is needed of the role of the cat in the complex tumor microenvironment. Remarkably, few studies have focused on the effects of cat inhibition on cell types other than tumor cells. Recently, concerns were raised regarding the use of catB, L, S, and X inhibitors as anti-tumor agents, as Edgington-Mitchell et al. [7] reported increased osteoclast formation in the presence of cat inhibitors, which might exacerbate MDSC-mediated cancer to bone metastasis.
Importantly, catX inhibition reverted the suppressive effects of the PBMCs on MDA-MB-231 cell invasion. Furthermore, combined inhibition of catX and L here led to even greater increases in MDA-MB-231 cell invasion. An explanation for this additive effect might be that under catL inhibition, conversion of pro-catX to active catX is abrogated, and thus contributes to an even lower overall catX activity.
While inhibition of catX activity changed the intrinsic invasion of the MDA-MB-231 cells, down-regulation of catL activity resulted in enhanced cytotoxicity of the CD8+ T cells. On the other hand, catL is a known activator of the pore-forming protein perforin in T cells, and its inhibition has been shown to decrease their cytotoxicity in vitro [39]. The dual effects of cat inhibitor have been reported before; in a study by Yan et al. [40] treatment of regulatory T cells with a catS inhibitor alone was shown to enhance their immunosuppressive activity, but under the same conditions, addition of tumor-cell-conditioned medium reverted this effect and contributed to enhanced CD8+ T-cell immunity.
Taken together, in vitro models for validation of potential targets in cancers often lack the complexity of the immune-cell component. Herein, we show that catL and X are important for distinct functional changes to the tumor cells and immune cells, respectively, which might only be revealed through the tumor cell–immune cell co-culture system. All in all, our data underline the need for careful examination of cysteine peptidase inhibitors in the context of tumor cell–immune cell interactions, to better predict their impact on tumor progression in vivo.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to acknowledge Matthew Bogyo (Stanford University) for the DCG-04 probe, and Christopher Berrie for critical review of the manuscript before submission.
Abbreviations
- Cat
Cathepsin(s)
- CFSE
Carboxyfluorescein succinimidyl ester
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- IL
Interleukin
- MDSCs
Myeloid-derived suppressor cells
- PBMCs
Peripheral blood mononuclear cells
- PBS
Phosphate-buffered saline
- PG
Prostaglandin
Author contributions
TJ performed the experiments and cooperated with UŠ on the work with the PBMCs. TJ analyzed the data and prepared the figures. TJ, AP, UPF, and JK designed the framework for the study. AP and UPF offered technical support. TJ drafted the paper, and the other co-authors (AP, UPF, UŠ, JK) revised and complemented its content.
Funding
This study was supported by the Research Agency of Republic of Slovenia (Grants P4-0127, J4-1776, J4-8227 and J4-6811, to JK).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Ethic approval for PBMCs No. 0120-279/2017-3.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Groth C, Hu X, Weber R, et al. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br J Cancer. 2019;120:16–25. doi: 10.1038/s41416-018-0333-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassetta L, Baekkevold ES, Brandau S, et al. Deciphering myeloid-derived suppressor cells: isolation and markers in humans, mice and non-human primates. Cancer Immunol Immunother. 2019;68:687–697. doi: 10.1007/s00262-019-02302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruger AM, Dorhoi A, Esendagli G, et al. How to measure the immunosuppressive activity of MDSC: assays, problems and potential solutions. Cancer Immunol Immunother. 2019;68:631–644. doi: 10.1007/s00262-018-2170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kochan G. Human MDSCs. In: Escors D, Talmadge JE, Breckpot K, Van Ginderachter JA, Kochan G, editors. Myeloid-derived suppressor cells and cancer. Springer briefs in immunology. Cham: Springer; 2016. pp. 39–48. [Google Scholar]
- 5.Ostrand-Rosenberg S, Fenselau C. Myeloid-derived suppressor cells: immune-suppressive cells that impair antitumor immunity and are sculpted by their environment. J Immunol. 2018;200:422–431. doi: 10.4049/jimmunol.1701019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akkari L, Gocheva V, Kester JC, et al. Distinct functions of macrophage-derived and cancer cell-derived cathepsin Z combine to promote tumor malignancy via interactions with the extracellular matrix. Genes Dev. 2014;28:2134–2150. doi: 10.1101/gad.249599.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgington-Mitchell LE, Rautela J, Duivenvoorden HM, et al. Cysteine cathepsin activity suppresses osteoclastogenesis of myeloid-derived suppressor cells in breast cancer. Oncotarget. 2015;6:8–10. doi: 10.18632/oncotarget.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fonović M, Turk B. Cysteine cathepsins and their potential in clinical therapy and biomarker discovery. PROTEOMICS - Clin Appl. 2014;8:416–426. doi: 10.1002/prca.201300085. [DOI] [PubMed] [Google Scholar]
- 9.Salpeter SJ, Pozniak Y, Merquiol E, et al. A novel cysteine cathepsin inhibitor yields macrophage cell death and mammary tumor regression. Oncogene. 2015;34:6066–6078. doi: 10.1038/onc.2015.51. [DOI] [PubMed] [Google Scholar]
- 10.Yang M, Liu J, Shao J, et al. Cathepsin S-mediated autophagic flux in tumor-associated macrophages accelerate tumor development by promoting M2 polarization. Mol Cancer. 2014;13:43. doi: 10.1186/1476-4598-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gounaris E, Tung CH, Restaino C, et al. Live imaging of cysteine-cathepsin activity reveals dynamics of focal inflammation, angiogenesis, and polyp growth. PLoS ONE. 2008;3:e2916. doi: 10.1371/journal.pone.0002916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruchard M, Mignot G, Derangère V, et al. Chemotherapy-triggered cathepsin B release in myeloid-derived suppressor cells activates the Nlrp3 inflammasome and promotes tumor growth. Nat Med. 2013;19:57–64. doi: 10.1038/nm.2999. [DOI] [PubMed] [Google Scholar]
- 13.Wu QD, Wang JH, Condron C, et al. Human neutrophils facilitate tumor cell transendothelial migration. Am J Physiol Cell Physiol. 2001;280:C814-22. doi: 10.1152/ajpcell.2001.280.4.C814. [DOI] [PubMed] [Google Scholar]
- 14.Fonović UP, Mitrović A, Knez D, et al. Identification and characterization of the novel reversible and selective cathepsin X inhibitors. Sci Rep. 2017;7:1–11. doi: 10.1038/s41598-017-11935-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katunuma N, Murata E, Kakegawa H, et al. Structure based development of novel specific inhibitors for cathepsin L and cathepsin S in vitro and in vivo. FEBS Lett. 1999;458:6–10. doi: 10.1016/S0014-5793(99)01107-2. [DOI] [PubMed] [Google Scholar]
- 16.Mirkovic B, Markelc B, Butinar M, et al. Nitroxoline impairs tumor progression in vitro and in vivo by regulating cathepsin B activity. Oncotarget. 2015;6:19027–19042. doi: 10.18632/oncotarget.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webb SR, Gascoigne NRJ. T-cell activation by superantigens. Curr Opin Immunol. 1994;6:467–475. doi: 10.1016/0952-7915(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 18.Valiathan R, Lewis JE, Melillo AB, et al. Evaluation of a flow cytometry-based assay for natural killer cell activity in clinical settings. Scand J Immunol. 2012;75:455–462. doi: 10.1111/j.1365-3083.2011.02667.x. [DOI] [PubMed] [Google Scholar]
- 19.Casacuberta-Serra S, Parés M, Golbano A, et al. Myeloid-derived suppressor cells can be efficiently generated from human hematopoietic progenitors and peripheral blood monocytes. Immunol Cell Biol. 2017;95:538–548. doi: 10.1038/icb.2017.4. [DOI] [PubMed] [Google Scholar]
- 20.Obermajer N, Kalinski P. Generation of myeloid-derived suppressor cells using prostaglandin E2. Transplant Res. 2012;1:15. doi: 10.1186/2047-1440-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heine A, Held SAE, Schulte-Schrepping J, et al. Generation and functional characterization of MDSC-like cells. Oncoimmunology. 2017;6:e1295203. doi: 10.1080/2162402X.2017.1295203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okada SL, Simmons RM, Franke-Welch S, et al. Conditioned media from the renal cell carcinoma cell line 786.O drives human blood monocytes to a monocytic myeloid-derived suppressor cell phenotype. Cell Immunol. 2018;323:49–58. doi: 10.1016/j.cellimm.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Lechner MG, Megiel C, Russell SM, et al. Functional characterization of human Cd33+ And Cd11b+ myeloid-derived suppressor cell subsets induced from peripheral blood mononuclear cells co-cultured with a diverse set of human tumor cell lines. J Transl Med. 2011;9:90. doi: 10.1186/1479-5876-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panni RZ, Sanford DE, Belt BA, et al. Tumor-induced STAT3 activation in monocytic myeloid-derived suppressor cells enhances stemness and mesenchymal properties in human pancreatic cancer. Cancer Immunol Immunother. 2014;63:513–528. doi: 10.1007/s00262-014-1527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrigues JC, Gonzalez GC, Zhang L, et al. Normal human monocytes exposed to glioma cells acquire myeloid-derived suppressor cell-like properties. Neuro Oncol. 2010;12:351–365. doi: 10.1093/neuonc/nop023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou L, Cooley J, Swanson R, et al. The protease cathepsin L regulates Th17 cell differentiation. J Autoimmun. 2015;65:56–63. doi: 10.1016/j.jaut.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pečar Fonović U. Efficient removal of cathepsin L from active cathepsin X using immunoprecipitation technique. Acta Chim Slov. 2009;56:985–988. [Google Scholar]
- 28.Greenbaum DC, Arnold WD, Lu F, et al. Small molecule affinity fingerprinting. A tool for enzyme family subclassification, target identification, and inhibitor design. Chem Biol. 2002;9:1085–1094. doi: 10.1016/s1074-5521(02)00238-7. [DOI] [PubMed] [Google Scholar]
- 29.Jiang M, Chen J, Zhang W, et al. Interleukin-6 trans-signaling pathway promotes immunosuppressive myeloid-derived suppressor cells via suppression of suppressor of cytokine signaling 3 in breast cancer. Front Immunol. 2017;8:1840. doi: 10.3389/fimmu.2017.01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao Y, Poschke I, Wennerberg E, et al. Melanoma-educated CD14+ cells acquire a myeloid-derived suppressor cell phenotype through COX-2-dependent mechanisms. Cancer Res. 2013;73:3877–3887. doi: 10.1158/0008-5472.CAN-12-4115. [DOI] [PubMed] [Google Scholar]
- 31.Lechner MG, Liebertz DJ, Epstein AL. Characterization of cytokine-induced myeloid-derived suppressor cells from normal human peripheral blood mononuclear cells. J Immunol. 2010;185:2273–2284. doi: 10.4049/jimmunol.1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elliott LA, Doherty GA, Sheahan K, Ryan EJ. Human tumor-infiltrating myeloid cells: phenotypic and functional diversity. Front Immunol. 2017 doi: 10.3389/fimmu.2017.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Movahedi K, Guilliams M, Van den Bossche J, et al. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood. 2008;111:4233–4244. doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- 34.Idorn M, Kollgaard T, Kongsted P, et al. Correlation between frequencies of blood monocytic myeloid-derived suppressor cells, regulatory T cells and negative prognostic markers in patients with castration-resistant metastatic prostate cancer. Cancer Immunol Immunother. 2014;63:1177–1187. doi: 10.1007/s00262-014-1591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.OuYang L-Y, Wu X-J, Ye S-B, et al. Tumor-induced myeloid-derived suppressor cells promote tumor progression through oxidative metabolism in human colorectal cancer. J Transl Med. 2015;13:47. doi: 10.1186/s12967-015-0410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen M-F, Kuan F-C, Yen T-C, et al. IL-6-stimulated CD11b+ CD14+ HLA-DR-myeloid-derived suppressor cells, are associated with progression and poor prognosis in squamous cell carcinoma of the esophagus. Oncotarget. 2014;5:8716–8728. doi: 10.18632/oncotarget.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Zhang L, Wang H, et al. Tumor-induced CD14+HLA-DR−/low myeloid-derived suppressor cells correlate with tumor progression and outcome of therapy in multiple myeloma patients. Cancer Immunol Immunother. 2015;64:389–399. doi: 10.1007/s00262-014-1646-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banerjee K, Pru C, Pru JK, Resat H. STAT3 knockdown induces tumor formation by MDA-MB-231 cells. Clin Oncol Res. 2018 doi: 10.31487/j.COR.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konjar Š, Sutton VR, Hoves S, et al. Human and mouse perforin are processed in part through cleavage by the lysosomal cysteine proteinase cathepsin L. Immunology. 2010;131:257–267. doi: 10.1111/j.1365-2567.2010.03299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan X, Wu C, Chen T, et al. Cathepsin S inhibition changes regulatory T-cell activity in regulating bladder cancer and immune cell proliferation and apoptosis. Mol Immunol. 2017;82:66–74. doi: 10.1016/j.molimm.2016.12.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.