Abstract
The Rex trans-regulatory protein of human T-cell leukemia virus type 1 (HTLV-1) is required for the nuclear export of incompletely spliced and unspliced viral mRNAs and is therefore essential for virus replication. Rex is a nuclear phosphoprotein that directly binds to its cis-acting Rex response element RNA target sequence and constantly shuttles between the nucleus and cytoplasm. Moreover, Rex induces nuclear accumulation of unspliced viral RNA. Three protein domains which mediate nuclear import-RNA binding, nuclear export, and Rex oligomerization have been mapped within the 189-amino-acid Rex polypeptide. Here we identified a different region in the carboxy-terminal half of Rex which is also required for biological activity. In inactive mutants with mutations that map within this region, as well as in mutants that are deficient in Rex-specific multimerization, Rex trans activation could be reconstituted by fusion to a heterologous leucine zipper dimerization interface. The intracellular trafficking capabilities of wild-type and mutant Rex proteins reveal that biologically inactive and multimerization-deficient Rex mutants are still efficiently translocated from the nucleus to the cytoplasm. This observation indicates that multimerization is essential for Rex function but is not required for nuclear export. Finally, we are able to provide an improved model of the HTLV-1 Rex domain structure.
Human T-cell leukemia virus type 1 (HTLV-1) (66) is the causative agent of adult T-cell leukemia, an aggressive malignancy of human T lymphocytes (38, 67, 96). In addition, HTLV-1 has been associated with a chronic neurodegenerative disorder termed tropical spastic paraparesis (28) or HTLV-1-associated myelopathy (61). As for every replication-competent retrovirus, the HTLV-1 genome contains the gag, pol, and env genes, which encode the viral structural proteins and enzymes. In addition to structural proteins, HTLV-1 encodes at least two trans-regulatory proteins, Tax and Rex, which are essential for virus replication (reviewed in references 18, 29, and 78). The Tax protein activates the viral long terminal repeat promoter element, resulting in enhanced transcription of all viral genes (15, 79). Furthermore, Tax also activates cellular promoters of various genes and stimulates the growth of T lymphocytes (16, 26, 44, 75). In contrast, the rex gene product acts at the posttranscriptional level, permitting the expression of the viral structural proteins (37, 45). In the absence of Rex, the unspliced and incompletely spliced viral transcripts are retained in the nucleus and either spliced to completion or subjected to degradation. When Rex is present, however, these transcripts are exported from the nucleus to the cytoplasm, where they are either translated or packaged as genomes into progeny virions (36).
Previous studies have demonstrated that Rex is a 27-kDa phosphoprotein that accumulates at steady state in the nucleoli of expressing cells (1, 39, 50, 60). Rex binds directly and specifically to its cis-acting RNA target sequence, the Rex response element (RxRE) (6, 7, 12, 30–32, 90, 91). The RxRE is a highly stable 255-nucleotide RNA stem-loop structure, which is encoded by sequences within the retroviral 3′ long terminal repeat, thereby making it an integral sequence element of all viral mRNAs (3, 36, 76, 90). In addition to its role in mediating Rex responsiveness, the RxRE also plays a role in the 3′ processing of viral primary transcripts. The viral polyadenylation signal is separated from the 3′ cleavage site by the RxRE sequence, a distance that does not allow processing of the 3′ ends of the viral primary transcripts. Formation of the correct RxRE secondary structure, however, brings the two elements in close proximity to each other, thereby permitting correct polyadenylation (2).
So far, mutational analyses have revealed three distinct functional domains in the rex gene product. A basic domain rich in arginine residues, which maps to amino acids (aa) 1 to 19 in the 189-aa Rex protein, is critically required for the sequence-specific binding of the RxRE RNA sequence (12, 30, 83) and also serves as a nuclear localization signal (13, 51, 60, 71, 77). It has been suggested that a region which maps to aa 57 to 66 is involved in Rex oligomer formation (9, 92). Finally, a protein activation or effector domain, which is required for interaction of Rex with one or multiple cellular cofactors, is located between aa 79 and 99 (93). In fact, various proteins have been reported to bind to this domain and/or to mediate Rex effector function. These include the nucleoporin-like proteins hRIP/Rab (10, 11, 25), the yeast factor Rip1p (85), and eukaryotic initiation factor 5A (48). An essential feature of the Rex activation domain is a sequence motif rich in leucine residues (40) that appears to be a target for the export factor CRM1 (24, 27, 63, 81) and acts as a nuclear export signal (10, 49, 64). As the protein contains both nuclear export and import signals, Rex is able to constantly shuttle between the nucleus and cytoplasm of the host cell (64).
Another human pathogenic retrovirus, human immunodeficiency virus type 1 (HIV-1), encodes a regulatory protein of similar activity, termed Rev (for a recent review, see reference 68). Like Rex, Rev is a shuttle protein that induces the cytoplasmic accumulation of unspliced and incompletely spliced viral mRNA species. Similarly, Rev binds to its highly structured cis-acting Rev response element (RRE) sequence, which is part of the viral unspliced and incompletely spliced mRNA species. Although HTLV-1 Rex and HIV-1 Rev lack any significant sequence homology, Rex is able to functionally substitute for Rev in HIV-1, by rescuing replication of a Rev-deficient HIV-1 provirus (72). The molecular basis for this cross-activation is the ability of HTLV-1 Rex to bind the heterologous HIV-1 RRE (3, 12, 21, 36, 46, 80, 91). This finding also suggests that both Rev and Rex access the same cellular pathway for nuclear export of their unspliced and incompletely spliced viral mRNAs. In support of this notion, it has previously been shown that HIV-1 Rev interacts with the same cellular activation domain binding proteins as Rex (8, 11, 25, 73, 85). Finally, extensive mutational analysis of Rex has allowed the identification of mutant proteins that inhibit not only the function of the homologous Rex wild-type protein on the RxRE but also the function of the heterologous Rev trans activator on the RRE, in a dominant-negative (trans-dominant) manner (14, 71). Taken together, these data led to the general notion that the biological activity and molecular mode of action of HTLV-1 Rex are identical to those of HIV-1 Rev.
This study was undertaken to characterize in more detail the regions in the HTLV-1 Rex protein which are required for biological activity. A series of Rex mutants were characterized according to their potential to stimulate expression of intron-containing mRNA and nuclear export function. Our data demonstrate that a previously unrecognized region in the carboxy-terminal half of Rex is essential for function but not for the intracellular trafficking of Rex and suggest that this region is required for protein oligomerization.
MATERIALS AND METHODS
Molecular clones.
The expression plasmid pcRex and the vectors encoding the Rex mutants RexM6, RexM7, RexM13, RexIW18, and RexΔ5 have been described in detail elsewhere (71, 72, 92, 93). The parental vector pBC12/CMV (17) and the construct pBC12/CMV/βGal (73) were used in transfection experiments to maintain constant input DNA levels and for internal control of transfection efficiencies, respectively. pDM128/CMV/RxRE is a Rex-responsive reporter construct containing the bacterial chloramphenicol acetyltransferase (CAT) gene and was constructed by ligating the 1.7-kb XbaI-BglII fragment from the Rev-specific reporter construct pDM128 (41) between the SalI and BglII sites of pgTat-RxRE (39). Plasmid pRRX is a Rex-responsive reporter construct that is derived from the 3′ half of the HTLV-1 proviral genome and gives rise to a full-length primary transcript and a spliced derivative (33).
A bacteriophage M13-based oligonucleotide-directed mutagenesis system (United States Biochemicals, Cleveland, Ohio) was used to introduce in-frame NheI restriction sites into the coding region of pcRex, which permitted the subsequent construction of a series of Rex internal deletion mutants (RexID1 to RexID6). Vectors expressing leucine zipper-Rex fusion proteins (Zip-Rex) were generated by fusing synthetic oligonucleotides that encode the GCN4-derived leucine zipper element (NH2-MDPKLQRMKQLEDKVEELLSKNYHLENEVARLKKLVGG-COOH) (42) in front of the rex gene in the respective pcRex vectors. Plasmids expressing glutathione S-transferase (GST)-Rex fusion proteins (pGEX-Rex and pGEX-RexM13) were generated by cloning the respective PCR-generated (59) rex coding regions between the BamHI and EcoRI sites of the bacterial expression vector pGEX-3X (Pharmacia Biotech, Freiburg, Germany). Finally, the coding regions of all constructs generated in the course of this study were confirmed by DNA sequence analysis.
Cell culture, transfections, and assays.
The cell lines COS and HeLa were maintained and transfected with either DEAE-dextran and chloroquine or calcium phosphate, as previously described (89, 97). 293T cells were cultured in Dulbecco modified Eagle medium with 10% fetal calf serum and transfected by using the Lipofectamine reagent according to the protocol of the manufacturer (Gibco BRL, Eggenstein, Germany).
Rex protein expression was evaluated by transfection of 1.5 × 105 COS cells with 300 ng of the various Rex expression plasmid DNAs. The transfected-cell cultures were radiolabelled with [35S]cysteine at ∼48 h posttransfection, followed by immunoprecipitation with a polyclonal anti-Rex antibody (14) and electrophoresis on sodium dodecyl sulfate-polyacrylamide gels as described previously (14, 39).
Rex trans activation was investigated by cotransfection of 2.5 × 105 COS cells with 250 ng of pDM128/CMV/RxRE DNA and 250 ng of pBC12/CMV/βGal DNA, together with 250 ng of pcRex (positive control), pBC12/CMV (negative control), or mutant Rex expression plasmid. At ∼60 h posttransfection, cell lysates were prepared and the levels of β-galactosidase activity were measured as described previously (89). These values were subsequently used to determine the amount of cell extract to be assayed for CAT by an enzyme-linked immunosorbent assay (Boehringer GmbH, Mannheim, Germany).
The effect of Rex on accumulation of unspliced HTLV-1-derived mRNAs was evaluated by cotransfection of 6 × 105 293T cells with 1 μg of pRRX DNA together with 3 μg of pBC12/CMV (negative control), pcRex (positive control), or mutant Rex expression plasmid. At ∼48 h posttransfection, total cellular RNA was isolated and subjected to Northern analysis by using an intron-specific hybridization probe as previously described (33).
A total of 2.5 × 105 HeLa cells (HeLaneoRRE) (97), grown on glass coverslips, were transfected with 5 μg of expression plasmid to determine the subcellular localization of Rex proteins by indirect immunofluorescence.
Purification of GST-Rex fusion proteins.
Wild-type Rex and RexM13 were expressed as carboxy-terminal fusions to GST in Escherichia coli BL21. The fusion proteins were purified from crude lysates by affinity chromatography with glutathione-Sepharose 4B according to the specifications of the manufacturer (Pharmacia Biotech). Eluted proteins were analyzed by Rex-specific Western analysis, pooled, concentrated by ultrafiltration with a PM10 filter device (Amicon Inc., Beverly, Mass.), and stored at −70°C.
Immunofluorescence studies and microinjection.
The nucleocytoplasmic shuttling capabilities of Rex proteins were investigated with transfected HeLa cells. At ∼40 h posttransfection, cell cultures were supplemented with 50 μg of cycloheximide (Sigma, Deisenhofen, Germany) per ml to inhibit protein synthesis. After 30 min, 5 μg of actinomycin D (Sigma) per ml was added in order to inhibit gene transcription. After a further incubation for 2.5 h, cells were fixed with paraformaldehyde and subcellular localization of Rex proteins was determined by indirect immunofluorescence as described previously (54). The primary rabbit anti-Rex polyclonal antibody (14) was used at a 1:250 dilution. The second antibody, rhodamine-conjugated goat anti-rabbit immunoglobulin G (IgG), was used at a 1:200 dilution.
HeLa cells (HeLaneoRRE) constitutively expressing the HIV-1 RRE sequence (97) were comicroinjected into the nucleus with GST-Rex (1.5 mg/ml) or GST-RexM13 and rabbit IgG (1.0 mg/ml) (injection control) by using a CompiC INJECT computer-assisted injection system (Cellbiology Trading, Hamburg, Germany). Cells were fixed at 30 min postinjection with paraformaldehyde, and the injected proteins were visualized by indirect immunofluorescence analysis as described previously (8). The primary mouse anti-GST monoclonal antibody (Serotec, Oxford, United Kingdom) was used at a 1:50 dilution. The secondary antibodies, fluorescein isothiocyanate-conjugated goat anti-rabbit IgG and Texas red-conjugated goat anti-mouse IgG, were used at a 1:100 dilution.
RESULTS
Functional analysis of the HTLV-1 Rex trans-activator protein.
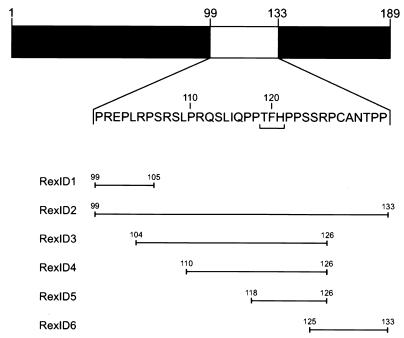
Previous studies in which various Rex regions were deleted in order to identify distinct protein domains indicated that a functionally critical region in the 189-aa Rex protein might be located somewhere between the carboxy-terminal border of the activation domain at aa 99 and aa 132 (39, 93). To test this hypothesis and to investigate the function of this protein region, we generated a series of mutants that are characterized by internal deletions between aa 99 and 133 in the carboxy-terminal half of the Rex protein (RexID1 to RexID6) (Fig. 1). In addition, various prototypic Rex control mutants which have been previously described as trans-activation negative (Table 1) were included in the study. In particular, the missense substitutions in RexM6 and RexM7 (71) and the deletion in RexΔ5 (92) have been speculated to inhibit Rex oligomer formation (9, 92). The RexIW18 protein is nonfunctional due to complete deletion of the Rex activation domain (10, 40, 49, 93). Finally, RexM13 has been described as a dominant-negative point mutant that binds the RxRE RNA with wild-type efficiency but has a negative effect on the Rex effector domain (9, 12, 71). As it has been suggested that some of these mutations impair the capacity of Rex to form multimers, we also engineered variants of all mutants in which a dimerization interface was provided by a heterologous sequence. For this, the leucine zipper domain of the yeast transcription factor GCN4, which has previously been shown to be a protein motif that forms stable dimers (20, 42, 62), was fused in front of the various rex genes, creating in-frame leucine zipper-Rex (Zip-Rex) fusion proteins. Protein expression was then confirmed for all constructs by Rex-specific immunoprecipitation analysis (Fig. 2A) with radiolabelled protein extracts of transiently transfected COS cells and a polyclonal anti-Rex antiserum directed against the Rex carboxy terminus (14). As noted previously (14, 71), even small alterations to the Rex amino acid sequence affected the electrophoretic mobility of some of the mutant proteins (e.g., RexM6 versus RexM7; Fig. 2A, lanes 13 and 14). This apparent difference in molecular weight probably reflects altered posttranslational modification (e.g., phosphorylation) (1) of these Rex mutant proteins. In general, however, all rex expression vectors appeared to produce comparable levels of protein.
FIG. 1.
Localization of HTLV-1 Rex internal deletion mutants. A series of small internal deletion mutants, designated Rex ID1 to Rex ID6, with mutations between aa 99 and 133 in the carboxy-terminal half of the 189-aa Rex protein were generated. This amino acid sequence is shown in the expanded section. Residues mutated in the previously published sequence of the biologically inactive RexM13 protein (71) are underlined. Deleted protein regions are indicated by bars.
TABLE 1.
Summary of Rex mutant proteinsa
| Mutant | Positions (aa)b | Mutations | Domain affectedc | Reference |
|---|---|---|---|---|
| RexM6 | 5, 60 | Y,I→D,L | M | 71 |
| RexM7 | 64, 65 | Y,W→D,L | M | 71 |
| RexΔ5 | 58–70 | Deleted | M | 92 |
| RexIW18 | 86–104 | Deleted | AD | 93 |
| RexM13 | 119–121 | T,F,H→D,L | M | 71 |
| RexID1 | 99–105 | Deleted | M | This study |
| RexID2 | 99–133 | Deleted | M | This study |
| RexID3 | 104–126 | Deleted | M | This study |
| RexID4 | 110–126 | Deleted | M | This study |
| RexID5 | 118–126 | Deleted | M | This study |
| RexID6 | 125–133 | Deleted | M | This study |
Variants expressing leucine zipper-Rex (Zip-Rex) fusion proteins were constructed by fusing sequences encoding the GCN4 leucine zipper element (42) in frame in front of the respective rex gene.
Positions mutated within the 189-aa HTLV-1 Rex protein.
M, multimerization domain; AD, activation-effector domain.
FIG. 2.
Functional characterization of HTLV-1 Rex mutant proteins. (A) Radioimmunoprecipitation of wild-type (WT) and mutant Rex proteins. COS cell monolayers were transiently transfected with either a negative control plasmid (neg.) (lane 1), pcRex (lane 2), or the mutant rex expression vectors indicated (see also Table 1). At ∼48 h posttransfection, cultures were radiolabelled with [35S]cysteine and subjected to immunoprecipitation with a polyclonal anti-Rex antibody (14). Precipitated proteins were resolved on sodium dodecyl sulfate–14% polyacrylamide gels and visualized by autoradiography. Molecular mass standards (in kilodaltons) are at the left. (B) Rex trans-activation capacity was determined by cotransfection of COS cell monolayers with the Rex-responsive reporter plasmid pDM128/CMV/RxRE, the mutant Rex expression plasmids indicated, and the constitutive internal control vector pBC12/CMV/βGal. Data are expressed as percentages of wild-type Rex activity (set to 100%), and the error bars represent the standard deviations from three independent experiments. All CAT values were adjusted for transfection efficiency by determining the level of β-galactosidase in each culture and were corrected for background (mock) activity.
Next we tested the biological activities of these Rex mutant proteins in trans-activation assays. For this, we employed the Rex-responsive reporter construct pDM128/CMV/RxRE. This plasmid contains the CAT indicator gene and the RxRE target sequence of Rex. These elements are positioned between HIV-1 splice sites and are under control of the cytomegalovirus immediate-early promoter (41, 56). As shown in previous studies (10, 32, 40, 49), RNA produced from this type of Rex reporter construct contains a single intron containing the CAT gene and RxRE sequence, which is removed when the RNA is spliced. However, in the presence of a functional Rex protein, unspliced message is exported to the cytoplasm, resulting in high levels of CAT protein and thereby providing an assay system for Rex functionality. Figure 2B summarizes the activities of the various Rex mutants in this reporter system. These experiments were internally controlled for varying transfection efficiencies by inclusion of the constitutive control vector pBC12/CMV/βGal (73). All CAT values are expressed as a percentage of wild-type Rex activity (set arbitrarily to 100%; Fig. 2B, bar 2) and have been corrected for background (mock) activity. The controls RexM6, RexM7, RexΔ5, RexIW18, and RexM13 (Fig. 2B, bars 12 to 16) were, as reported previously (71, 92, 93), inactive in this assay. The trans-activation phenotypes of our newly generated mutants revealed wild-type activities for RexID1 and RexID6 (Fig. 2B, bars 17 and 22). In contrast, RexID2 to RexID5 were characterized by complete nonfunctionality (Fig. 2B, bars 18 to 21). Importantly, in-frame fusion of the GCN4 leucine zipper dimerization domain to RexID2 to RexID5 restored Rex function to a maximal level of ∼80% of wild-type activity (Fig. 2B, bars 8 to 11). The reconstitution of biological activity was also seen in the case of RexM13 (Fig. 2B, bar 7), which contains a point mutation that maps to the protein region that is covered by the overlapping internal deletions in RexID2 to RexID5 (Table 1). Furthermore, the leucine zipper element also rescued Rex activity to a similar extent in the mutants RexM6, RexM7, and RexΔ5 (Fig. 2B, bars 3 to 5), which have previously been shown to be important for Rex oligomerization (9, 92). In contrast, Rex activity could not be reconstituted in the activation domain mutant RexIW18 (Fig. 2B, bar 6).
In a previous study we were able to show that HTLV-1 Rex activity not only affects the nucleocytoplasmic transport of viral RNA, as demonstrated in the pDM128/CMV/RxRE-based trans-activation assay, but also interferes with intron excision, thereby inducing nuclear accumulation of unspliced viral RNAs (33). As a second indicator of Rex function, we also tested selected prototypic Rex deletion mutants for their capacity to stimulate the intracellular accumulation of intron-containing HTLV-1-derived mRNAs. For this, 293T cells were cotransfected with the HTLV-1-derived Rex-responsive reporter construct pRRX (33) and various Rex expression plasmids. Total cellular RNAs were isolated at 48 h posttransfection, separated by gel electrophoresis, and subjected to Northern analysis. The pRRX plasmid contains sequences derived from the 3′ half of the proviral genome, including genuine HTLV-1 splice donor and splice acceptor sites and the full-length RxRE sequence. Since the pRRX plasmid alone does not express Rex, cotransfection with a Rex expression vector allowed the effect of Rex on the accumulation of unspliced RNAs to be monitored by using an intron-specific hybridization probe. As shown in Fig. 3, coexpression of an unrelated control vector (lane 1) or the trans-activation-negative mutant RexID2, RexID3, or RexID4 (lanes 3, 5 and 7, respectively) failed to induce significant levels of unspliced HTLV-1-specific mRNAs in the transfected cell cultures. In agreement with previous data (33), coexpression of the Rex wild-type protein resulted in a marked increase in unspliced RNA (Fig. 3, lane 2). As in the CAT indicator assay, the leucine zipper-Rex fusion proteins Zip-RexID2, Zip-RexID3, and Zip-RexID4 displayed activities which were comparable to that of the wild-type Rex protein (Fig. 3, lanes 4, 6, and 8).
FIG. 3.
Effect of HTLV-1 Rex on viral mRNA splicing. The mutant rex genes were cotransfected together with pRRX into 293T cells. The plasmid pRRX is derived from the 3′ half of the HTLV-1 provirus and produces spliced and unspliced HTLV-1 RNAs, whose amounts can be regulated by Rex. Total cellular RNA from transfected cells was separated on a 1% formaldehyde agarose gel, blotted onto a nylon membrane, and hybridized to an intron-specific radioactive probe. Lane 1, pBC12/CMV (unrelated control vector), lane 2, pcRex, lanes 3 to 8, plasmids expressing the indicated Rex mutants. WT, wild type; neg., negative control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Taken together, these data suggest that a region in HTLV-1 Rex, located between aa 106 and 124, is essential for Rex-specific trans activation. Furthermore, the consistent reconstitution of Rex biological activity by a heterologous dimerization domain in otherwise inactive and multimerization-deficient mutants (e.g., RexM6, RexM7, and RexM13) (9) suggests that this region of Rex is involved in the formation of Rex oligomeric complexes.
Intracellular trafficking of HTLV-1 Rex mutants.
As noted above, nuclear export of HTLV-1 Rex protein is mediated by the protein activation domain. The mutation of critical amino acid residues in the activation domain results in Rex mutant proteins that are nuclear export deficient and also inactive when tested in trans-activation assays (10, 49, 64). These data indicate that the nuclear export of Rex is essential for its biological activity. Therefore, in this study, we also evaluated the nucleocytoplasmic trafficking capabilities of wild-type and mutant Rex proteins in HeLa cells. Although Rex is a protein that constantly shuttles between the nucleus and cytoplasm, previous studies have shown that Rex accumulates at steady state in the nuclei and nucleoli of transfected cells (1, 39, 50, 60). However, inhibition of RNA synthesis by actinomycin D induces the relocation of Rex from the nucleus to the cytoplasm (19, 64, 82). The molecular basis for this effect appears to be that nuclear import of shuttle proteins depends on continuous transcription. This was shown originally for the hnRNP A1 protein (65), a factor which appears to be involved in the transport of mRNA, and subsequently for the HIV-1 Rev trans activator (47, 57, 70, 82, 95).
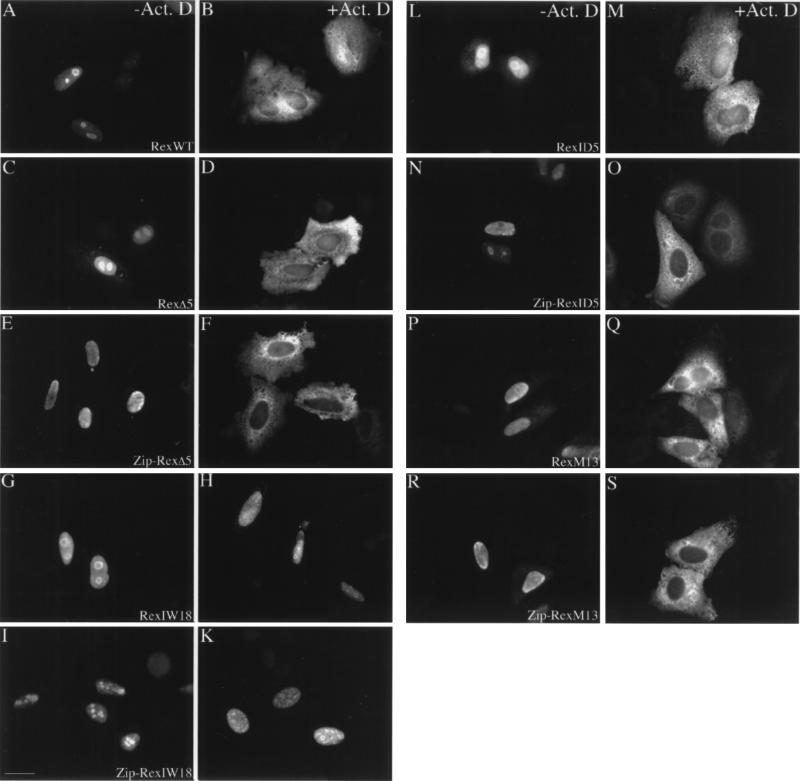
Figure 4 shows the subcellular distribution of the various Rex proteins in transfected HeLa cells, as determined by indirect immunofluorescence. Clearly, wild-type Rex displayed its predominantly nuclear-nucleolar localization in untreated cells (Fig. 4A). In contrast, and as expected, significant amounts of the protein were found in the cytoplasm in cells exposed to 5 μg of actinomycin D per ml and 50 μg of cycloheximide per ml, which was added in order to inhibit de novo protein synthesis. This actinomycin D-dependent cytoplasmic accumulation of Rex was not observed in the activation-deletion mutants RexIW18 (Fig. 4G and H) and Zip-RexIW18 (Fig. 4I and K), which served as negative controls in this assay. However, the mutants RexΔ5, RexID5, and RexM13 relocated to the cytoplasm in the presence of actinomycin D (Fig. 4C, L, and P versus Fig. 4D, M, and Q). This was also seen in experiments using the respective leucine zipper-Rex variants (Fig. 4E, N, and R versus Fig. 4F, O, and S) and was comparable to the actinomycin D-induced cytoplasmic accumulation of the wild-type Rex protein (Fig. 4B). These data suggest that trans-activation-negative mutants, such as RexΔ5, RexID5, and RexM13, are still capable of translocating from the nucleus to the cytoplasm.
FIG. 4.
Subcellular localization of Rex mutant proteins in transfected HeLa cells. Cells transfected with the indicated Rex mutants were either incubated in medium alone (−Act.D) (A, C, E, G, I, L, N, P, and R) or exposed to 50 μg of cycloheximide per ml plus 5 μg of the transcription inhibitor actinomycin D per ml (+Act. D) (B, D, F, H, K, M, O, Q, and S). After fixation, the subcellular locations of the various Rex proteins were determined by indirect immunofluorescence with a rabbit polyclonal anti-Rex antibody (14) and rhodamine-conjugated secondary antibody. WT, wild type. Bar, 20 μm.
We next wanted to investigate Rex nuclear export more directly. Experimentally this can be carried out independent of nuclear import, by microinjection of GST fusion proteins into the nuclei of human somatic cells. As demonstrated previously for GST-Rev proteins, active export occurs within minutes after microinjection and can be easily visualized by indirect immunofluorescence (8, 74). This is possible because nuclear export of Rev has been shown to be significantly more efficient than its reimport into the nucleus (84).
As shown clearly in Fig. 5B, microinjection of GST-Rex together with rabbit IgG, which was included to establish the site of injection, into the nuclei of HeLa cells resulted in nucleocytoplasmic translocation of GST-Rex. In contrast, the coinjected rabbit IgG remained in the cell nucleus (Fig. 5A). Next we investigated the export capacity of a biologically inactive Rex mutant, which is representative of the type of mutants described in this study. GST-RexM13 protein was injected together with rabbit IgG into HeLa cell nuclei as before. Indirect immunofluorescence analysis clearly demonstrated translocation of the GST-RexM13 mutant protein from the nucleus to the cytoplasm (Fig. 5C and D). These experiments confirm that nonfunctional Rex molecules can be exported from the nucleus.
FIG. 5.
Nuclear export of HTLV-1 Rex in human somatic cells. The nuclei of HeLa cells were microinjected with wild-type GST-Rex (A and B) or GST-RexM13 (C and D) fusion proteins together with rabbit IgG. At 30 min after microinjection, the cells were fixed and subjected to double-label indirect immunofluorescence analysis. The GST-Rex proteins were visualized by using a mouse monoclonal anti-GST antibody followed by Texas red-conjugated goat anti-mouse IgG (B and D). The microinjected rabbit IgG, which served to control the site of injection, was detected with a fluorescein isothiocyanate-conjugated goat anti-rabbit antibody (A and C). WT, wild type. Bar, 20 μm.
DISCUSSION
By constructing small internal deletions, in this study we have identified a region of 19 amino acid residues (aa 106 to 124) (Fig. 6) in HTLV-1 Rex that is required for biological activity. Interestingly, we were able to rescue at least partial Rex activity by fusing our inactive mutants in frame to the heterologous GCN4 leucine zipper element. This effect was detectable in a standard Rex trans-activation assay, which measures the cytoplasmic expression of RxRE-containing mRNAs (Fig. 2B), as well as in an independent assay that measures the Rex-dependent increase of unspliced RNA (Fig. 3). Furthermore, the finding that the leucine zipper element also reconstituted the function of mutants that were previously reported to be defective in their ability to form Rex homomultimers (RexM6, RexM7, and RexM13) (9) suggests that the region between aa 106 and 124 in Rex functions as a protein oligomerization domain. However, whether a homodimer is sufficient to create a fully functional Rex complex cannot be concluded from our experiments using the leucine zipper element. It is indeed conceivable that multiple Rex dimers are able to interact with the RxRE sequence, thereby creating a higher-order oligomeric complex. Unfortunately, it was not possible to investigate the multimerization status of our Rex deletion mutants in vitro by RNA gel retardation analysis, because these proteins repeatedly proved to be unstable when expressed as recombinant proteins in E. coli.
FIG. 6.
Domain structure of the HTLV-1 Rex trans-activator protein. The four distinct functional regions located within the 189-aa Rex protein are indicated by boxes (see text for details). Interaction of Rex with its RxRE target sequence and nuclear-nucleolar localization of the protein are mediated by the Rex amino terminus (aa 1 to 19; hatched box). The protein activation domain, which contains a leucine-rich peptide core motif that serves as a nuclear export signal, is located in the center of the protein (aa 79 to 99; cross-hatched box). Regions involved in the multimerization of Rex are localized in both the amino-terminal and carboxy-terminal halves of the protein (aa 57 to 66 and aa 106 to 124; filled boxes). The amino acid sequences of these multimerization domains are shown as expanded sections. The Rex sequences indicated by grey boxes are dispensable for in vivo Rex function (39) and appear to serve a structural rather than a directly functional role in Rex. NLS, nuclear localization signal; NES, nuclear export signal.
Nevertheless, we are now able to suggest an improved model of the Rex domain structure (Fig. 6). The Rex amino terminus (aa 1 to 19) is required for both the nuclear accumulation of Rex (13, 60, 71, 77) and its direct binding to the RxRE RNA (12, 30, 83). Therefore, this region can be considered the Rex RNA binding and nuclear localization domain. The activation domain, required for the interaction of Rex with cellular cofactors, is located in the center of the protein (aa 79 to 99) (93) and is characterized by hydrophobic residues (commonly leucine) (40) that constitute a nuclear export signal (10, 49, 64). So far, two regions have been identified that appear to be involved in Rex multimer formation and are therefore operationally referred to as multimerization domains. The more amino-terminal region stretches from aa 57 to 66. The notion that this region constitutes a multimerization domain originated from the finding that this sequence can be functionally replaced by a region of HIV-1 Rev that has been implicated in the formation of Rev homomultimers (53, 55, 92). Point mutants with mutations that map within this Rex region (e.g., RexM6 and RexM7) (Table 1) failed to form homomultimeric complexes in a mammalian cell-based two-hybrid assay (9). In contrast, the second multimerization region, identified in this study, localizes to the carboxy-terminal half of Rex and maps to aa 106 to 124. The only inactive point mutant described so far with a mutation that maps within this region, namely, RexM13 (Table 1) (71), has also been reported to lack the intrinsic capacity of wild-type Rex for protein-protein interaction (9). Obviously, our findings with the leucine zipper dimerization interface further support the notion that both regions, which are separated by the protein activation domain, participate in Rex oligomerization. It should be noted that a similar domain arrangement also occurs in the HIV-1 Rev trans-activator protein. Two multimerization domains are separated by the RNA binding-nuclear localization domain in the amino-terminal half of Rev (35, 53, 55, 86, 88), which forms a helix-loop-helix motif (4), thereby permitting the creation of a single exposed hydrophobic oligomerization interface (87, 88). It remains to be seen whether biophysical measurements are able to confirm a similar structural organization of the multimerization interface in HTLV-1 Rex.
Random mutational analysis of the rex gene allowed the identification of dominant-negative inhibitors for both HTLV-1 Rex and HIV-1 Rev function (14, 71). By comparing the positions of the mutations introduced within the Rex domain structure (Fig. 6), it is evident that all of the mutations that gave rise to trans-dominant Rex repressors targeted residues within either the activation domain or the multimerization domains (14, 71). These data suggest that both the interaction of Rex with cellular cofactors via its activation domain and the formation of oligomeric Rex complexes via the multimerization domains are equally required for full Rex biological activity.
Inspection of the amino acid sequence of the Rex multimerization domains reveals no known or apparent motifs that indicate the structural basis of this protein-protein interaction. The high proline content of the multimerization domains does not contrast with the overall composition of Rex and suggests a mechanically rather rigid protein chain that does not allow formation of any of the known protein contact structural motifs. The multimerization domains may be close together in the functional fold of the protein, thereby forming an interaction surface. It is most likely that the hydrophobic residues (Y59, I60, Y64, W65, L109, L114, and F120) (Fig. 6) play a role in contact formation. A histidine residue at position 121 could also be involved in specific hydrogen bonds between the interacting proteins. The reconstitution of function by chimeric constructs with amino-terminal leucine zipper elements is remarkable, as it demonstrates the modular architecture of Rex, whereby multimerization functionality can be swapped to other regions of the protein. It is conceivable that the function of such multimerization mutants can usually be restored only when the structure has been destabilized rather than irreversibly disrupted. In this case the zipper element can support the protein-protein interactions that result in the formation of multimers and that would otherwise be too weak.
As noted before, Rex mutants that are deficient in nuclear export due to disruption of the protein activation domain also lack trans-activation capacity (10, 49, 64). As this correlated with data generated for the HIV-1 Rev protein (22, 54, 56, 58, 94), and as Rev-mediated viral mRNA transport is known to occur independent of any pre-mRNA splicing events (23), it seemed likely that Rex, like Rev, acts primarily at the level of nuclear export. However, evaluation of the nuclear export capacity of biologically inactive Rex mutants in this study (Fig. 4 and 5) suggested that, although it is required, the nuclear export activity is not sufficient for Rex-mediated trans activation. For example, the RexM13 protein is multimerization deficient (9) and biologically inactive in trans-activation assays (Fig. 2) (71). Despite this, RexM13 still binds to its RxRE RNA target sequence (12) and is also exported from the nucleus (Fig. 4Q and Fig. 5D) with wild-type efficiency. Thus, the recruitment of multiple Rex monomers appears to be required for an activity other than the one seen in nuclear export. This notion is directly supported by a recent study in which conditional Rex-human estrogen receptor (ER) fusion molecules were investigated (69): in the absence of hormone, Rex-ER protein remained in the cytoplasm but was relocated into the nucleus, and particularly to the nuclear pore complex (NPC), in a hormone-dependent manner; this localization also correlated with Rex trans activation. Importantly, the biologically inactive RexM7-ER chimera exhibited NPC colocalization comparable to that of the wild-type protein in these experiments, providing evidence that intranuclear translocation of Rex to the NPC is independent of the oligomeric status of Rex.
Although our study does not directly address the question of which functions other than nuclear export are executed by the HTLV-1 Rex protein, it is likely that these activities take place at the level of viral pre-mRNA splicing for a number of reasons. For example, it has been shown that unspliced HTLV-1 RNA accumulates in the nuclei and cytoplasm of transiently transfected cells in the presence of Rex (43). Furthermore, we have been able to provide evidence that expression of Rex increases the levels of unspliced viral RNA by reducing the rates of intron excision and degradation in the nucleus (33). The most direct confirmation of this idea, however, comes from a recent study in which it was demonstrated that the Rex protein of HTLV-2 acts as a potent inhibitor of in vitro splicing reactions by interfering with an early step in spliceosome assembly (5). It is important that HTLV-1 Rex has previously been reported to bind a protein that is associated with the splicing factor ASF/SF2 (52). Finally, indirect evidence that Rex has functions other than the one linked to its activation domain (e.g., nuclear export) originates from a study in which the biological activities of HIV-1 Rev and HTLV-1 Rex in Jurkat T cells were compared (34). It was shown that although the activation domain function was intact, Rex appeared to be biologically inactive in these cells.
Taken together, these various lines of evidence suggest that HTLV-1 Rex trans activation is the sum of multiple Rex activities, including those at the level of nuclear export and, presumably, splicing of viral RNAs. While HTLV-1 Rex nuclear export, and therefore cofactor interaction via its activation domain, apparently takes place in the absence of Rex multimerization, the formation of Rex oligomers appears to be required for full biological activity. It is hoped that mapping of the regions in HTLV-1 Rex that mediate these homomultimeric protein-protein interactions will provide the basis to allow elucidation of the Rex protein structure in greater detail.
ACKNOWLEDGMENTS
We thank Warner C. Greene for the RexM6, RexM7, and RexM13 constructs and Nicole Hirschmann and Lotte Hofer for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB466) and Johannes and Frieda Marohn-Stiftung.
REFERENCES
- 1.Adachi Y, Copeland T D, Takahashi C, Nosaka T, Ahmed A, Oroszlan S, Hatanaka M. Phosphorylation of the Rex protein of human T-cell leukemia virus type I. J Biol Chem. 1992;267:21977–21981. [PubMed] [Google Scholar]
- 2.Ahmed Y F, Gilmartin G M, Hanly S M, Nevins J R, Greene W C. The HTLV-1 Rex response element mediates a novel form of mRNA polyadenylation. Cell. 1991;64:727–737. doi: 10.1016/0092-8674(91)90502-p. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed Y F, Hanly S M, Malim M H, Cullen B R, Greene W C. Structure-function analyses of the HTLV-1 Rex and HIV-1 Rev RNA response elements: insights into the mechanism of Rex and Rev action. Genes Dev. 1990;4:1014–1022. doi: 10.1101/gad.4.6.1014. [DOI] [PubMed] [Google Scholar]
- 4.Auer M, Gremlich H-U, Seifert J-M, Daly T J, Parslow T G, Casari G, Gstach H. Helix-loop-helix motif in HIV-1 Rev. Biochem. 1994;33:2988–2996. doi: 10.1021/bi00176a031. [DOI] [PubMed] [Google Scholar]
- 5.Bakker A, Li X, Ruland C T, Stephens D W, Black A C, Rosenblatt J D. Human T-cell leukemia virus type 2 Rex inhibits pre-mRNA splicing in vitro at an early stage of spliceosome formation. J Virol. 1996;70:5511–5518. doi: 10.1128/jvi.70.8.5511-5518.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballaun C, Farrington G K, Dobrovnik M, Rusche J, Hauber J, Böhnlein E. Functional analysis of human T-cell leukemia virus type I rex-response element: direct RNA binding of Rex protein correlates with in vivo activity. J Virol. 1991;65:4408–4413. doi: 10.1128/jvi.65.8.4408-4413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baskerville S, Zapp M, Ellington A D. High-resolution mapping of the human T-cell leukemia virus type 1 Rex-binding element by in vitro selection. J Virol. 1995;69:7559–7569. doi: 10.1128/jvi.69.12.7559-7569.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bevec D, Jaksche H, Oft M, Wöhl T, Himmelspach M, Pacher A, Schebesta M, Koettnitz K, Dobrovnik M, Csonga R, Lottspeich F, Hauber J. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science (Washington, DC) 1996;271:1858–1860. doi: 10.1126/science.271.5257.1858. [DOI] [PubMed] [Google Scholar]
- 9.Bogerd H, Greene W C. Dominant negative mutants of human T-cell leukemia virus type I Rex and human immunodeficiency virus type 1 Rev fail to multimerize in vivo. J Virol. 1993;67:2496–2502. doi: 10.1128/jvi.67.5.2496-2502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogerd H P, Fridell R A, Benson R E, Hua J, Cullen B R. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogerd H P, Fridell R A, Madore S, Cullen B R. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 12.Bogerd H P, Huckaby G L, Ahmed Y S, Hanly S M, Greene W C. The type I human T-cell leukemia virus (HTLV-1) Rex trans-activator binds directly to the HTLV-1 Rex and the type 1 human immunodeficiency virus Rev RNA response elements. Proc Natl Acad Sci USA. 1991;88:5704–5708. doi: 10.1073/pnas.88.13.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Böhnlein E, Berger J, Hauber J. Functional mapping of the human immunodeficiency virus type 1 Rev RNA binding domain: new insights into the domain structure of Rev and Rex. J Virol. 1991;65:7051–7055. doi: 10.1128/jvi.65.12.7051-7055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Böhnlein S, Pirker F P, Hofer L, Zimmermann K, Bachmayer H, Böhnlein E, Hauber J. Transdominant repressors for human T-cell leukemia virus type I Rex and human immunodeficiency virus type 1 Rev function. J Virol. 1991;65:81–88. doi: 10.1128/jvi.65.1.81-88.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cann A J, Rosenblatt J D, Wachsman W, Shah N P, Chen I S. Identification of the gene responsible for human T-cell leukemia virus transcriptional regulation. Nature (London) 1985;318:571–574. doi: 10.1038/318571a0. [DOI] [PubMed] [Google Scholar]
- 16.Cross S L, Feinberg M B, Wolf J B, Holbrook N J, Wong Staal F, Leonard W J. Regulation of the human interleukin-2 receptor alpha chain promoter: activation of a nonfunctional promoter by the transactivator gene of HTLV-1. Cell. 1987;49:47–56. doi: 10.1016/0092-8674(87)90754-9. [DOI] [PubMed] [Google Scholar]
- 17.Cullen B R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 18.Cullen B R. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol Rev. 1992;56:375–394. doi: 10.1128/mr.56.3.375-394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Agostino D M, Ciminale V, Zotti L, Rosato A, Chieco Bianchi L. The human T-cell lymphotropic virus type 1 Tof protein contains a bipartite nuclear localization signal that is able to functionally replace the amino-terminal domain of Rex. J Virol. 1997;71:75–83. doi: 10.1128/jvi.71.1.75-83.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellenberger T E, Brandl C J, Struhl K, Harrison S C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 21.Felber B K, Derse D, Athanassopoulos A, Campbell M, Pavlakis G N. Cross-activation of the Rex proteins of HTLV-1 and BLV and of the Rev protein of HIV-1 and nonreciprocal interactions with their RNA responsive elements. New Biol. 1989;1:318–330. [PubMed] [Google Scholar]
- 22.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 23.Fischer U, Meyer S, Teufel M, Heckel C, Lührmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 25.Fritz C C, Zapp M L, Green M R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature (London) 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 26.Fujii M, Sassone Corsi P, Verma I M. c-fos promoter trans-activation by the tax1 protein of human T-cell leukemia virus type I. Proc Natl Acad Sci USA. 1988;85:8526–8530. doi: 10.1073/pnas.85.22.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 28.Gessain A, Barin F, Vernant J C, Gout O, Maurs L, Calender A, de The G. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet. 1985;ii:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 29.Gitlin S D, Dittmer J, Reid R L, Brady J N. The molecular biology of human T-cell leukemia viruses. In: Cullen B R, editor. Human retroviruses. Oxford, United Kingdom: Oxford University Press; 1993. pp. 159–192. [Google Scholar]
- 30.Grassmann R, Berchtold S, Aepinus C, Ballaun C, Boehnlein E, Fleckenstein B. In vitro binding of human T-cell leukemia virus rex proteins to the rex-response element of viral transcripts. J Virol. 1991;65:3721–3727. doi: 10.1128/jvi.65.7.3721-3727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green P L, Yip M T, Xie Y, Chen I S. Phosphorylation regulates RNA binding by the human T-cell leukemia virus Rex protein. J Virol. 1992;66:4325–4330. doi: 10.1128/jvi.66.7.4325-4330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gröne M, Hoffmann E, Berchtold S, Cullen B R, Grassmann R. A single stem-loop structure within the HTLV-1 Rex response element is sufficient to mediate Rex activity in vivo. Virology. 1994;204:144–152. doi: 10.1006/viro.1994.1518. [DOI] [PubMed] [Google Scholar]
- 33.Gröne M, Koch C, Grassmann R. The HTLV-1 Rex protein induces nuclear accumulation of unspliced viral RNA by avoiding intron excision and degradation. Virology. 1996;218:316–325. doi: 10.1006/viro.1996.0200. [DOI] [PubMed] [Google Scholar]
- 34.Hamaia S, Casse H, Gazzolo L, Duc Dodon M. The human T-cell leukemia virus type 1 Rex regulatory protein exhibits an impaired functionality in human lymphoblastoid Jurkat T cells. J Virol. 1997;71:8514–8521. doi: 10.1128/jvi.71.11.8514-8521.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammerschmid M, Palmeri D, Ruhl M, Jaksche H, Weichselbraun I, Böhnlein E, Malim M H, Hauber J. Scanning mutagenesis of the arginine-rich region of the human immunodeficiency virus type 1 Rev trans activator. J Virol. 1994;68:7329–7335. doi: 10.1128/jvi.68.11.7329-7335.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanly S M, Rimsky L T, Malim M H, Kim J H, Hauber J, Duc Dodon M, Le S-Y, Maizel J V, Cullen B R, Greene W C. Comparative analysis of the HTLV-1 Rex and HIV-1 Rev trans-regulatory proteins and their RNA response elements. Genes Dev. 1989;3:1534–1544. doi: 10.1101/gad.3.10.1534. [DOI] [PubMed] [Google Scholar]
- 37.Hidaka M, Inoue J, Yoshida M, Seiki M. Post-transcriptional regulator (rex) of HTLV-1 initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J. 1988;7:519–523. doi: 10.1002/j.1460-2075.1988.tb02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinuma Y, Nagata K, Hanaoka M, Nakai M, Matsumoto T, Kinoshita K I, Shirakawa S, Miyoshi I. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc Natl Acad Sci USA. 1981;78:6476–6480. doi: 10.1073/pnas.78.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofer L, Weichselbraun I, Quick S, Farrington G K, Böhnlein E, Hauber J. Mutational analysis of the human T-cell leukemia virus type I trans-acting rex gene product. J Virol. 1991;65:3379–3383. doi: 10.1128/jvi.65.6.3379-3383.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hope T J, Bond B L, McDonald D, Klein N P, Parslow T G. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J Virol. 1991;65:6001–6007. doi: 10.1128/jvi.65.11.6001-6007.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hope T J, Huang X, McDonald D, Parslow T G. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J C, O’Shea E K, Kim P S, Sauer R T. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science (Washington, DC) 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 43.Inoue J, Itoh M, Akizawa T, Toyoshima H, Yoshida M. HTLV-1 Rex protein accumulates unspliced RNA in the nucleus as well as in cytoplasm. Oncogene. 1991;6:1753–1757. [PubMed] [Google Scholar]
- 44.Inoue J, Seiki M, Taniguchi T, Tsuru S, Yoshida M. Induction of interleukin 2 receptor gene expression by p40x encoded by human T-cell leukemia virus type 1. EMBO J. 1986;5:2883–2888. doi: 10.1002/j.1460-2075.1986.tb04583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inoue J, Yoshida M, Seiki M. Transcriptional (p40x) and post-transcriptional (p27x-III) regulators are required for the expression and replication of human T-cell leukemia virus type I genes. Proc Natl Acad Sci USA. 1987;84:3653–3657. doi: 10.1073/pnas.84.11.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Itoh M, Inoue J, Toyoshima H, Akizawa T, Higashi M, Yoshida M. HTLV-1 rex and HIV-1 rev act through similar mechanisms to relieve suppression of unspliced RNA expression. Oncogene. 1989;4:1275–1279. [PubMed] [Google Scholar]
- 47.Kalland K-H, Szilvay A M, Brokstad K A, Sætrevik W, Haukenes G. The human immunodeficiency virus type 1 Rev protein shuttles between the cytoplasm and nuclear compartments. Mol Cell Biol. 1994;14:7436–7444. doi: 10.1128/mcb.14.11.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katahira J, Ishizaki T, Sakai H, Adachi A, Yamamoto K, Shida H. Effects of translation initiation factor eIF-5A on the functioning of human T-cell leukemia virus type I Rex and human immunodeficiency virus Rev inhibited trans dominantly by a Rex mutant deficient in RNA binding. J Virol. 1995;69:3125–3133. doi: 10.1128/jvi.69.5.3125-3133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim F J, Beeche A A, Hunter J J, Chin D J, Hope T J. Characterization of the nuclear export signal of human T-cell lymphotropic virus type 1 Rex reveals that nuclear export is mediated by position-variable hydrophobic interactions. Mol Cell Biol. 1996;16:5147–5155. doi: 10.1128/mcb.16.9.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiyokawa T, Seiki M, Iwashita S, Imagawa K, Shimizu F, Yoshida M. p27x-III and p21x-III, proteins encoded by the pX sequence of human T-cell leukemia virus type I. Proc Natl Acad Sci USA. 1985;82:8359–8363. doi: 10.1073/pnas.82.24.8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kubota S, Nosaka T, Cullen B R, Maki M, Hatanaka M. Effects of chimeric mutants of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex on nucleolar targeting signals. J Virol. 1991;65:2452–2456. doi: 10.1128/jvi.65.5.2452-2456.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo Y, Yu H, Peterlin B M. Cellular protein modulates effects of human immunodeficiency virus type 1 Rev. J Virol. 1994;68:3850–3856. doi: 10.1128/jvi.68.6.3850-3856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madore S J, Tiley L S, Malim M H, Cullen B R. Sequence requirements for Rev multimerization in vivo. Virology. 1994;202:186–194. doi: 10.1006/viro.1994.1334. [DOI] [PubMed] [Google Scholar]
- 54.Malim M H, Böhnlein S, Hauber J, Cullen B R. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 55.Malim M H, Cullen B R. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 56.Malim M H, McCarn D F, Tiley L S, Cullen B R. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meyer B E, Malim M H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 58.Meyer B E, Meinkoth J L, Malim M H. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals. J Virol. 1996;70:2350–2359. doi: 10.1128/jvi.70.4.2350-2359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 60.Nosaka T, Siomi H, Adachi Y, Ishibashi M, Kubota S, Maki M, Hatanaka M. Nucleolar targeting signal of human T-cell leukemia virus type I rex-encoded protein is essential for cytoplasmic accumulation of unspliced viral mRNA. Proc Natl Acad Sci USA. 1989;86:9798–9802. doi: 10.1073/pnas.86.24.9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Osame M, Usuku K, Izumo S, Ijichi N, Amitani H, Igata A, Matsumoto M, Tara M. HTLV-1 associated myelopathy, a new clinical entity. Lancet. 1986;i:1031–1032. doi: 10.1016/s0140-6736(86)91298-5. [DOI] [PubMed] [Google Scholar]
- 62.O’Shea E K, Klemm J D, Kim P S, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science (Washington, DC) 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 63.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science (Washington, DC) 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 64.Palmeri D, Malim M H. The human T-cell leukemia virus type 1 posttranscriptional trans-activator Rex contains a nuclear export signal. J Virol. 1996;70:6442–6445. doi: 10.1128/jvi.70.9.6442-6445.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pinol Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature (London) 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 66.Poiesz B J, Ruscetti F W, Gazdar A F, Bunn P A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;85:7124–7128. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poiesz B J, Ruscetti F W, Reitz M S, Kalyanaraman V S, Gallo R C. Isolation of a new type C retrovirus (HTLV) in primary uncultured cells of a patient with Sezary T-cell leukaemia. Nature (London) 1981;294:268–271. doi: 10.1038/294268a0. [DOI] [PubMed] [Google Scholar]
- 68.Pollard V W, Malim M H. The HIV-1 Rev protein. 1998. Annu. Rev. Microbiol., in press. [DOI] [PubMed] [Google Scholar]
- 69.Rehberger S, Gounari F, DucDodon M, Chlichlia K, Gazzolo L, Schirrmacher V, Khazaie K. The activation domain of a hormone inducible HTLV-1 Rex protein determines colocalization with the nuclear pore. Exp Cell Res. 1997;233:363–371. doi: 10.1006/excr.1997.3562. [DOI] [PubMed] [Google Scholar]
- 70.Richard N, Iacampo S, Cochrane A. HIV-1 Rev is capable of shuttling between the nucleus and cytoplasm. Virology. 1994;204:123–131. doi: 10.1006/viro.1994.1516. [DOI] [PubMed] [Google Scholar]
- 71.Rimsky L, Duc Dodon M, Dixon E P, Greene W C. Trans-dominant inactivation of HTLV-1 and HIV-1 gene expression by mutation of the HTLV-1 Rex transactivator. Nature (London) 1989;341:453–456. doi: 10.1038/341453a0. [DOI] [PubMed] [Google Scholar]
- 72.Rimsky L, Hauber J, Dukovich M, Malim M H, Langlois A, Cullen B R, Greene W C. Functional replacement of the HIV-1 rev protein by the HTLV-1 rex protein. Nature (London) 1988;335:738–740. doi: 10.1038/335738a0. [DOI] [PubMed] [Google Scholar]
- 73.Ruhl M, Himmelspach M, Bahr G M, Hammerschmid F, Jaksche H, Wolff B, Aschauer H, Farrington G K, Probst H, Bevec D, Hauber J. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans-activation. J Cell Biol. 1993;123:1309–1320. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schatz O, Oft M, Dascher C, Schebesta M, Rosorius O, Jaksche H, Dobrovnik M, Bevec D, Hauber J. Interaction of the HIV-1 Rev cofactor eukaryotic initiation factor 5A with ribosomal protein L5. Proc Natl Acad Sci USA. 1998;95:1607–1612. doi: 10.1073/pnas.95.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmitt I, Rosin O, Rohwer P, Gossen M, Grassmann R. Stimulation of cyclin-dependent kinase activity and G1- to S-phase transition in human lymphocytes by the human T-cell leukemia/lymphotropic virus type 1 Tax protein. J Virol. 1998;72:633–640. doi: 10.1128/jvi.72.1.633-640.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seiki M, Inoue J-I, Hidaka M, Yoshida M. Two cis-acting elements responsible for posttranscriptional trans-regulation of gene expression of human T-cell leukemia virus type I. Proc Natl Acad Sci USA. 1988;85:7124–7128. doi: 10.1073/pnas.85.19.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Siomi H, Shida H, Nam S H, Nosaka T, Maki M, Hatanaka M. Sequence requirements for nucleolar localization of human T cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell. 1988;55:197–209. doi: 10.1016/0092-8674(88)90043-8. [DOI] [PubMed] [Google Scholar]
- 78.Smith M R, Greene W C. Molecular biology of the type I human T-cell leukemia virus (HTLV-1) and adult T-cell leukemia. J Clin Invest. 1991;87:761–766. doi: 10.1172/JCI115078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sodroski J, Rosen C, Goh W C, Haseltine W. A transcriptional activator protein encoded by the x-lor region of the human T-cell leukemia virus. Science (Washington, DC) 1985;228:1430–1434. doi: 10.1126/science.2990028. [DOI] [PubMed] [Google Scholar]
- 80.Solomin L, Felber B K, Pavlakis G N. Different sites of interaction for Rev, Tev, and Rex proteins within the Rev-responsive element of human immunodeficiency virus type 1. J Virol. 1990;64:6010–6017. doi: 10.1128/jvi.64.12.6010-6017.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 82.Stauber R, Gaitanaris G A, Pavlakis G N. Analysis of trafficking of Rev and transdominant Rev proteins in living cells using green fluorescent protein fusions: transdominant Rev blocks the export of Rev from the nucleus to the cytoplasm. Virology. 1995;213:439–449. doi: 10.1006/viro.1995.0016. [DOI] [PubMed] [Google Scholar]
- 83.Stong R C, Korsmeyer S J, Parkin J L, Arthur D C, Kersey J H. Human acute leukemia cell line with the t(4;11) chromosomal rearrangement exhibits B lineage and monocytic characteristics. Blood. 1985;65:21–31. [PubMed] [Google Scholar]
- 84.Stutz F, Izaurralde E, Mattaj I W, Rosbash M. A role for nucleoporin FG repeat domains in export of human immunodeficiency virus type 1 Rev protein and RNA from the nucleus. Mol Cell Biol. 1996;16:7144–7150. doi: 10.1128/mcb.16.12.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 86.Tan R, Chen L, Buettner J A, Hudson D, Frankel A D. RNA recognition by an isolated α helix. Cell. 1993;73:1031–1040. doi: 10.1016/0092-8674(93)90280-4. [DOI] [PubMed] [Google Scholar]
- 87.Thomas S L, Hauber J, Casari G. Probing the structure of the HIV-1 Rev trans-activator protein by functional analysis. Protein Eng. 1997;10:103–107. doi: 10.1093/protein/10.2.103. [DOI] [PubMed] [Google Scholar]
- 88.Thomas S L, Oft M, Jaksche H, Casari G, Heger P, Dobrovnik M, Bevec D, Hauber J. Functional analysis of the human immunodeficiency virus type 1 Rev protein oligomerization interface. J Virol. 1998;72:2935–2944. doi: 10.1128/jvi.72.4.2935-2944.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tiley L S, Madore S J, Malim M H, Cullen B R. The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev. 1992;6:2077–2087. doi: 10.1101/gad.6.11.2077. [DOI] [PubMed] [Google Scholar]
- 90.Toyoshima H, Itoh M, Inoue J, Seiki M, Takaku F, Yoshida M. Secondary structure of the human T-cell leukemia virus type 1 Rex-responsive element is essential for Rex regulation of RNA processing and transport of unspliced RNAs. J Virol. 1990;64:2825–2832. doi: 10.1128/jvi.64.6.2825-2832.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Unge T, Solomin L, Mellini M, Derse D, Felber B K, Pavlakis G N. The Rex regulatory protein of human T-cell lymphotropic virus type I binds specifically to its target site within the viral RNA. Proc Natl Acad Sci USA. 1991;88:7145–7149. doi: 10.1073/pnas.88.16.7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weichselbraun I, Berger J, Dobrovnik M, Bogerd H, Grassmann R, Greene W C, Hauber J, Böhnlein E. Dominant-negative mutants are clustered in a domain of the human T-cell leukemia virus type I Rex protein: implications for trans dominance. J Virol. 1992;66:4540–4545. doi: 10.1128/jvi.66.7.4540-4545.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weichselbraun I, Farrington G K, Rusche J R, Böhnlein E, Hauber J. Definition of the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex protein activation domain by functional exchange. J Virol. 1992;66:2583–2587. doi: 10.1128/jvi.66.4.2583-2587.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 95.Wolff B, Cohen G, Hauber J, Meshcheryakova D, Rabeck C. Nucleocytoplasmic transport of the Rev protein of human immunodeficiency virus type 1 is dependent on the activation domain of the protein. Exp Cell Res. 1995;217:31–41. doi: 10.1006/excr.1995.1060. [DOI] [PubMed] [Google Scholar]
- 96.Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc Natl Acad Sci USA. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zimmermann K, Weber S, Dobrovnik M, Hauber J, Böhnlein E. Expression of chimeric Neo-Rev response element sequences interferes with Rev-dependent HIV-1 Gag expression. Hum Gene Ther. 1992;3:155–161. doi: 10.1089/hum.1992.3.2-155. [DOI] [PubMed] [Google Scholar]