Abstract
Tumors can utilize a diverse repertoire of immunosuppressive mechanisms to evade attack by the immune system. Despite promising success with blockade of immune checkpoints like PD-1 the majority of patients does not respond to current immunotherapies. The degradation of tryptophan into immunosuppressive kynurenine is an important immunosuppressive pathway. Recent attempts to target the key enzymes of this pathway—IDO1 and TDO2—have so far failed to show therapeutic benefit in the clinic, potentially caused by insufficient target engagement. We, therefore, sought to add an alternative, highly efficient approach to block the degradation of tryptophan by inhibiting the expression of IDO1 and TDO2 using locked nucleic acid (LNA)-modified antisense oligonucleotides (ASOs). We show that LNA-modified ASOs can profoundly inhibit the expression of IDO1 and TDO2 in cancer cells in vitro without using a transfection reagent with IC50 values in the sub-micromolar range. We furthermore measured kynurenine production by ASO-treated cancer cells in vitro and observed potently reduced kynurenine levels. Accordingly, inhibiting IDO1 expression in cancer cells in an in vitro system leads to increased proliferation of activated T cells in coculture. We furthermore show that combined treatment of cancer cells in vitro with IDO1-specific ASOs and small molecule inhibitors can reduce the production of kynurenine by cancer cells in a synergistic manner. In conclusion, we propose that a combination of LNA-modified ASOs and small molecule inhibitors should be considered as a strategy for efficient blockade of the degradation of tryptophan into kynurenine in cancer immunotherapy.
Electronic supplementary material
The online version of this article (10.1007/s00262-019-02438-1) contains supplementary material, which is available to authorized users.
Keywords: IDO1, TDO2, Immunotherapy, Antisense oligonucleotide, Locked nucleic acid
Introduction
It has been shown in the recent past that therapeutically blocking immunosuppressive mechanisms is a promising strategy in oncologic indications to help the immune system to win the fight against cancer cells. Several drugs have been approved in the last years that target the PD-1/PD-L1 axis [1, 2] and CTLA-4 [3]. However, a majority of patients does not respond to existing immunotherapies, possibly caused by the existence of a plethora of further immunosuppressive mechanisms. Degradation of tryptophan by the enzymes indoleamine 2,3-dioxygenase 1 (IDO1) [4] and tryptophan 2,3-dioxygenase (TDO2) [5] has been identified as an important pathway in the suppression of anti-tumor immune responses. IDO1 and TDO2 can exert their immunosuppressive function in two ways. On the one hand, depletion of tryptophan leads to starvation of effector immune cells that sense the reduction in tryptophan levels by a protein termed general control nonderepressible 2 kinase (GCN2) [6]. On the other hand, degradation products of tryptophan like kynurenine have been shown to bind to the aryl hydrocarbon receptor (AhR) which has an impact on a plethora of immune cells and favors tolerogenic immune responses [7]. These findings have led to the development of small molecules that are intended to block the enzymatic function of IDO1 [8, 9] and TDO2 [10]. Promising success was achieved in early clinical trials where the IDO1 inhibitor INCB24360 led to normalized kynurenine plasma levels in patients with advanced malignancies [11] and to remarkable overall response rates in combination with anti-PD-1 antibodies [12, 13]. However, those findings could not be confirmed so far in larger patient cohorts in randomized clinical trials [14] which led to a recent halt of several phase III clinical trials. Given the fact that tryptophan-degrading enzymes play an important role in the suppression of anti-tumor immune responses, we sought to add an additional modality to block the degradation of tryptophan by IDO1 and TDO2 that has a different mode of action and pharmacokinetic properties as compared to small molecules. We targeted the expression of IDO1 and TDO2 with locked nucleic acid (LNA) gapmer antisense oligonucleotides (ASOs) that have a fully phosphorothioate (PTO) backbone, which protects them from degradation by nucleases [15]. The typical gapmer design results in an increased target affinity due to the LNA-modified flanks and simultaneously the non LNA-modified “gap” allows recruitment of RNase H which leads to target RNA cleavage upon binding. In contrast to older generations of ASOs, LNA gapmers efficiently and sequence specifically suppress target gene expression without the use of transfection reagents in vitro [16, 17], a process called gymnosis. Importantly, LNA gapmers exert their effects in vivo after systemic administration in different tissues including tumors without the need for a formulation or delivery reagent [17, 18].
We here show, that treatment of cells with LNA-modified ASOs with specificity for IDO1 or TDO2 results in potent target knockdown in vitro, without the use of a transfection reagent. Accordingly, kynurenine levels in the supernatant of tumor cells treated with IDO1 or TDO2 ASOs are markedly decreased. Moreover, the proliferative capacity of T cells in coculture with IDO1-expressing tumor cells was increased, when tumor cells were treated with IDO1-specific LNA-modified ASOs. Combining an IDO1 ASO with an IDO1 small molecule inhibitor leads to a synergistic effect with regard to the suppression of kynurenine production in a cell culture model. In conclusion, we identified IDO1- and TDO2-specific LNA-modified ASOs as a highly promising additional modality for the inhibition of tryptophan degradation in the context of cancer immunotherapy.
Materials and methods
Antisense oligonucleotides
ASOs with a length of 13–17 nucleotides were selected based on the human mRNA of IDO1 (NM_002164.5) or TDO2 (NM_005651.3). LNA-modified gapmers having a fully PTO backbone were ordered at Exiqon or Eurogentec and dissolved in H2O (stock concentration: 1 mM).
Sequences of selected IDO1 and TDO2 ASOs and control oligonucleotides are shown in Supplementary Table 1.
Cell culture media
RPMI or DMEM supplemented with antimycotic-antibiotic (1%), sodium pyruvate (1%) and heat-inactivated fetal bovine serum (10%) were used for cell culture experiments (RPMIfs and DMEMfs). Cell culture reagents were obtained from Gibco. In selected experiments, tryptophan (Sigma) was added to the cell culture medium.
Treatment of cells with oligonucleotides in vitro
EFO-21 cells (9000 per well), SKOV-3 cells (6000 per well), MDA-MB-453 cells (15,000 per well) or A-172 cells (4500 per well) were seeded in DMEMfs in 96-well plates and treated with the indicated oligonucleotide at the indicated concentration for three (EFO-21 and SKOV-3) or four (A-172) days without the use of a transfection reagent.
QuantiGene mRNA expression analysis
Target expression on mRNA level was determined using bDNA assay (QuantiGene SinglePlex Assay Kit 96-Well plate format and QuantiGene Sample Processing Kit for cultured cells, Thermo Fisher Scientific). The following probe sets were used: human IDO1 (SA-12156); human TDO2 (SA-15255); human HPRT1 (SA-10030); human GAPDH (SA-10001). All reagents were purchased from Affymetrix/Thermo Fisher Scientific.
IDO1 protein expression analysis in dendritic cells by flow cytometry
DC were generated as described previously [19]. In brief, CD14+ monocytes were differentiated into DC with GM-CSF (Peprotech, 20 ng/ml) and IL-4 (Peprotech, 100 ng/ml) and treated with the indicated oligonucleotide at 6.25 µM. 48 h later, DC were matured by adding TNF-α (Peprotech, 55 ng/ml), IL-1β (Peprotech, 2 ng/ml), IFN-γ (Peprotech, 250 ng/ml), prostaglandin E2 (PGE2) (Sigma, 250 ng/ml) and R848 (InvivoGen, 1000 ng/ml). Cells were harvested 24 h later. Cells were stained with CD83AF488 (Biolegend 305314, clone HB15e, 4 µg/ml) and CD86PerCP/Cy5.5 (Biolegend 305420, clone IT2.2, 4 µg/ml) for gating of mature DC. The following isotype control antibodies were used: IgG1 κ AF488 (Biolegend 400129, clone MOPC-21, 4 µg/ml) and IgG2b, κ (Biolegend 400338, MPC-11, 4 µg/ml). Cells were incubated at 4 °C in the dark for 20 min and washed with 150 µl FACS buffer (1x PBS + 5%FCS) (centrifugation: 500g for 5 min). Supernatants were flicked off and cells were washed with 200 µl FACS buffer (centrifugation: 500g for 5 min). Supernatants were flicked off and cells were resuspended in 100 µl IC fixation reagent (eBioscience) and incubated at 4 °C for 1 h. Cells were washed two times in 1x permeabilization buffer (eBioscience) and stained in 50 µl permeabilization buffer for 20 min at 4 °C with a hIDO1 specific antibody (eBioscience 50-9477-41, clone eyedio, 240 ng/ml) or the respective isotype control (eBioscience 50-4714-82, clone P3.6.2.8.1, 240 ng/ml). After two washing steps, cells were resuspended in FACS buffer and analyzed on an ACEA NovoCyte Flow Cytometer.
Determination of kynurenine levels by ELISA
For determination of the kynurenine concentration in cell culture supernatants a kynurenine ELISA (ImmuSmol) was performed according to the manufacturer’s instructions with slight modifications. In brief, 10 µl of standard or cell culture supernatant was pipetted into a v-bottom 96-well plate. 250 µl acylation buffer and 25 µl acylation reagent were added per well and plates were incubated at 37 °C for 90 min. 20 µl of the reaction was transferred into a kynurenine microtiter plate and 50 µl kynurenine antiserum was added per well. Plates were covered with adhesive foil and incubated for 15–20 h at 4 °C. Plates were washed 4 times and 100 µl enzyme conjugate was added per well. After incubation at room temperature, plates were washed 4 times and 100 µl substrate was added per well. 100 µl stop solution per well was added after incubation at room temperature for 20 min and absorbance was read at 450 nm on a BMG Clariostar reader.
Blood samples
PBMC were obtained from leukapheresis products. In brief, fresh leukapheresis products were diluted in PBS and carefully layered over Biocoll (Biochrom). After centrifugation at 800g for 20 min with the brake turned off, the layer containing PBMC was carefully harvested and washed twice in PBS.
Coculture of EFO-21 with human CD3+ T cells
EFO-21 cells were seeded in a flat bottom 96-well plate in 50 µl. ASO was added in 50 µl RPMIfs at the indicated concentration. CD3+ T cells were isolated on day three of the experiment out of PBMC using CD3 microbeads (Miltenyi) according to the manufacturer’s instructions and T cells were labeled with Cell Proliferation Dye eFluor 450. Therefore, cells were pelleted for 5 min at 500g after CD3 purification and resuspended in 500 µl PBS. Proliferation dye was added in 500 µl PBS (final concentration: 10 µM), the suspension was vortexed and incubated at 37 °C for 10 min. 10 ml RPMIfs were added and cells were incubated on ice for 5 min. After two washing steps (centrifugation: 500g for 5 min) cells were resuspended in RPMIfs and counted. 50,000 T cells per well were added to the pretreated EFO-21 cells together with ImmunoCult T cell activator (final concentration: 12.5 µl/ml) in 10 µl RPMIfs. Proliferation of cells was analyzed by flow cytometry 4 days later.
Analysis of T cell proliferation by flow cytometry
Human T cells (labeled with Proliferation Dye eFluor 450) were harvested from cocultures with EFO-21 cells, transferred to 96-well u-bottom plates and washed with FACS buffer containing 10,000 123 count eBeads (eBioscience 01-1234-42) per well (1x PBS, 5% FCS) (centrifugation: 500g for 5 min). Supernatants were flicked off and cells were washed with 200 µl FACS buffer (centrifugation: 500g for 5 min). After flick off of supernatants, cells were resuspended in 50 µl staining solution containing the following antibodies: anti-CD45PE (Biolegend 368510, clone 2D1, 125 ng/ml), anti-CD4BV510 (Biolegend 317444, clone OKT4, 1 µg/ml), anti-CD8APC (eBioscience 17-0088-42, clone RPA-T8, 120 ng/ml) and 7-AAD for life/dead cell exclusion (Biolegend 420404, 1 µg/ml). The following isotype control antibodies were used: mouse IgG1 κ PE (Biolegend 400114, clone MPOC-21, 125 ng/ml), mouse IgG2b κ BV510 (Biolegend 400345, clone MPC-11, 1 µg/ml) and mouse IgG1 κ APC (Biolegend 400122, clone MOPC-21, 120 ng/ml). Cells were incubated at 4 °C in the dark for 20 min and washed with 150 µl FACS buffer (centrifugation: 500g for 5 min). Supernatants were flicked off and cells were washed with 200 µl FACS buffer (centrifugation: 500g for 5 min). Supernatants were flicked off and cells were resuspended in 60 µl FACS buffer and analyzed on an ACEA NovoCyte flow cytometer. Absolute cell counts were calculated by dividing the number of recorded cells in the respective gate by the number of recorded counting beads multiplied by 10,000. Proliferation index was calculated using the formula: , whereas “i” is the generation number and N is the absolute cell count in the respective generation.
Statistics
IC50 values were estimated using the four parameter nonlinear logistic regression model. Therefore, the log base 10 of the ASO concentration in mole per liter was plotted against the expression of human IDO1. Concentration of ASO in the “mock-treated cells” condition was set to log10 (10−12).
Group comparisons were done using one-way ANOVA followed by Dunnett’s multiple comparison test.
Drug interaction was investigated using the Loewe model, where the synergy score quantifies the excess over the expected response if the two drugs are the same compound.
Synergy scores, indicating the elevation over an expected response if both drugs were the same compound, and the resulting landscapes were calculated and plotted applying the functions of the SynergyFinder package (version 1.8.0) [20] in R [21] (version 3.5.1).
Results
Identification of LNA-modified ASOs with specificity for IDO1 or TDO2 that potently suppress target expression
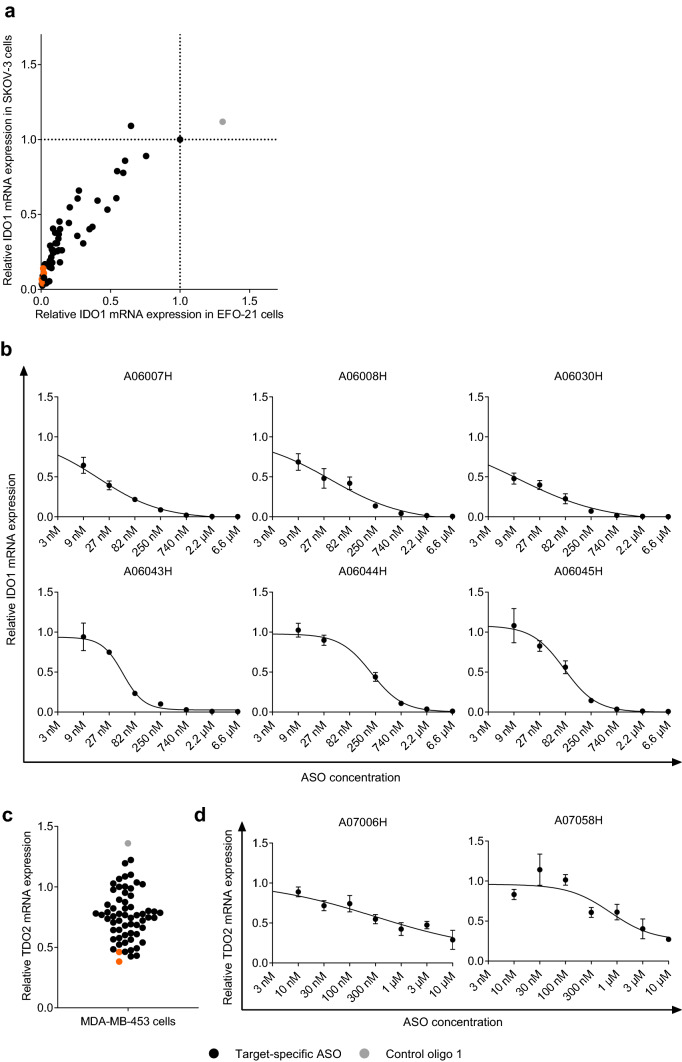
We treated the human ovarian carcinoma cell lines EFO-21 and SKOV-3 for 3 days with 56 IDO1-specific ASOs that bind to different regions of the IDO1 mRNA without the use of a transfection reagent. As shown in Fig. 1a, the majority of ASOs was able to knockdown IDO1 mRNA expression in both cell lines by > 50% (corresponding to a relative IDO1 mRNA expression of < 0.5). Of note, treatment of cells with the control oligo 1 that has no sequence complementarity to any human or mouse RNA did not affect IDO1 mRNA expression in both cell lines. We selected six ASOs with high activity in EFO-21 and SKOV-3 cells targeting unique binding sites on the IDO1 mRNA, namely A06007H, A06008H, A06030H, A06043H, A06044H and A06045H (marked in orange in Fig. 1a). We used those ASOs to determine the dose-dependency of IDO1 mRNA knockdown and the half-maximal inhibitory concentration (IC50). As shown in Fig. 1b and Table 1, all tested ASOs inhibit IDO1 mRNA expression in a dose-dependent manner with IC50 values in the low nanomolar range and a maximum knockdown efficacy of up to 99.7% at a concentration of 6.6 µM. Of note, treatment of human DC with a selected IDO1-specific ASO led to an IDO1 protein knockdown of ~ 70% (corresponding to a relative IDO1 expression of 0.3) (Supplementary Figure S2).
Fig. 1.
Targeting the RNA expression of IDO1 and TDO2 using LNA-modified ASOs. Relative IDO1 (a, b) or TDO2 (c, d) mRNA expression normalized to HPRT1 expression as compared to mock-treated cells (set to 1) is depicted. The mean of technical triplicates is shown. a IDO1 mRNA expression was determined in SKOV-3 and EFO-21 cells that had been treated with IDO1-specific LNA-modified ASOs (black and orange circles) or the control oligo 1 (grey circle) without the use of a transfection reagent for 3 days (concentration: 10 µM). ASOs that were selected for IC50 determination are marked in orange. b IDO1 mRNA expression was determined in EFO-21 cells that had been treated with different concentrations of IDO1-specific LNA-modified ASOs without the use of a transfection reagent for 3 days. IC50 values were determined as described in Materials and Methods and are shown in Table 1. c TDO2 mRNA expression was determined in MDA-MB-453 cells that had been treated with TDO2-specific LNA-modified ASOs (black and orange circles) or the control oligo 1 (grey circle) without the use of a transfection reagent for 3 days (concentration: 5 µM). ASOs that were selected for IC50 determination are marked in orange. d TDO2 mRNA expression was determined in MDA-MB-453 cells that had been treated with different concentrations of TDO2-specific LNA-modified ASOs without the use of a transfection reagent for 3 days. IC50 values were determined as described in Materials and Methods and are shown in Table 2
Table 1.
IC50 values and knockdown efficacy at 6.6 µM of IDO1-specific ASOs
| ASO ID | IC50 (nM) | R2 | Knockdown efficacy @ 6.6 µM (%) |
|---|---|---|---|
| A06007H | 17 | 0.98 | 99.7 |
| A06008H | 36 | 0.97 | 94 |
| A06030H | 11 | 0.98 | 98 |
| A06043H | 49 | 0.97 | 95 |
| A06044H | 205 | 0.98 | 89 |
| A06045H | 78 | 0.97 | 93 |
Among 30 screened cell lines, we identified the human breast cancer cell line MDA-MB-453 and the human glioblastoma cell line A-172 as cell lines with high TDO2 mRNA expression. We treated those cells for 3 days without the use of a transfection reagent with 59 ASOs that bind to different regions of the TDO2 mRNA. As shown in Fig. 1c, a subset of those ASOs resulted in a knockdown of TDO2 mRNA expression of > 50% (corresponding to a relative TDO2 mRNA expression of < 0.5). Treatment with the control oligo 1 had no negative impact on TDO2 mRNA expression. We selected the twelve most potent candidates and, furthermore, analyzed TDO2 knockdown in A-172 cells. We selected two ASOs that showed the most potent target knockdown in MDA-MB-453 and A-172 cells, namely A07006H and A07058H (marked in orange in Fig. 1c and Supplementary Figure S3) for further investigation with regard to dose-dependency of TDO2 mRNA knockdown and IC50 in MDA-MB-453 cells. As shown in Fig. 1d and Table 2, both tested ASOs led to a dose-dependent inhibition of TDO2 mRNA expression with IC50 values in the nanomolar range and a knockdown efficacy of ~ 70% at a concentration of 10 µM.
Table 2.
IC50 values and knockdown efficacy at 10 µM of TDO2-specific ASOs
| ASO ID | IC50 (nM) | R2 | Knockdown efficacy @ 10 µM (%) |
|---|---|---|---|
| A07006H | 368 | 0.83 | 71 |
| A07058H | 842 | 0.75 | 73 |
Downstream effects of IDO1 or TDO2 knockdown by LNA-modified ASOs in cancer cells
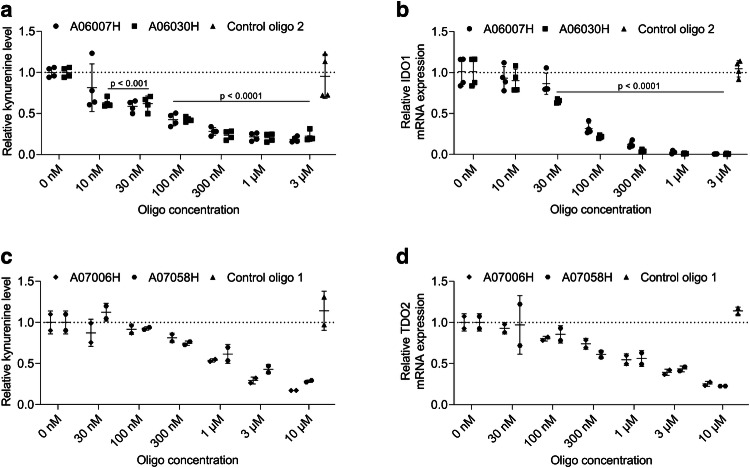
We sought to investigate the effect of IDO1 or TDO2 knockdown in cancer cell lines on their capacity to degrade tryptophan into kynurenine. Medium of ASO-treated cells was, therefore, supplemented with tryptophan and kynurenine levels were determined in the supernatants 24 h later. As shown in Fig. 2a treatment of EFO-21 cells with different concentrations of the two most potent IDO1-specific ASOs (A06007H and A06030H) indeed resulted in a significant dose-dependent reduction of kynurenine levels in the supernatant of EFO-21 cells. In contrast, kynurenine levels in the supernatant of EFO-21 cells did not change significantly as compared to mock-treated cells after treatment with control oligo 2 that has no sequence complementarity to any human or mouse RNA. As shown in Fig. 2b, the reduction of kynurenine levels correlates with IDO1 mRNA expression. Similarly, we observed a dose-dependent reduction of kynurenine levels in TDO2 ASO-treated MDA-MB-453 cells (Fig. 2c). The observed reduction in kynurenine levels correlates with TDO2 knockdown on the mRNA level (Fig. 2d).
Fig. 2.
Dose-dependent reduction of kynurenine production in IDO1 or TDO2 LNA-modified ASO-treated cancer cells. Cancer cells (a, b EFO-21, c, d MDA-MB-453) were treated with different concentrations of IDO1-specific (a, b), TDO2-specific (c, d) ASOs or the control oligo 2 (a, b concentration: 3 µM) or the control oligo 1 (c, d concentration: 10 µM) for 3 days without the use of a transfection reagent. Afterwards, medium was replaced and supplemented with tryptophan (concentration: 100 µM). Supernatants were harvested 24 h after medium replacement and kynurenine levels were determined by ELISA. Relative kynurenine levels as compared to mock-treated cells (set to 1) are depicted. IDO1 mRNA (b) or TDO2 mRNA expression (d) was analyzed in cells 24 h after medium replacement and relative mRNA expression is depicted compared to mock-treated cells (set to 1). a, b The mean ± SD of two independent experiments run in duplicates is shown. c, d The mean of duplicates ± SD from an experiment is shown. a, b Significant differences compared to mock-treated cells were determined using one-way ANOVA followed by Dunnett’s multiple comparison test
Investigation of the effect of ASO-mediated IDO1 knockdown in cancer cells on the proliferation of T cell in coculture
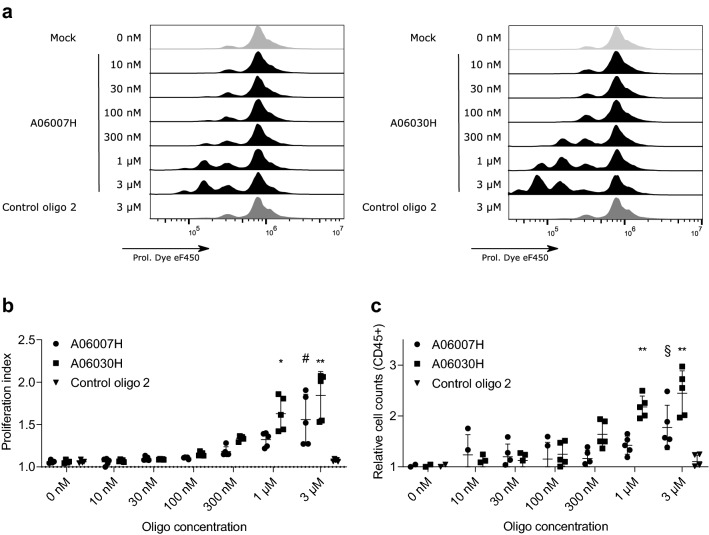
We investigated the effect of IDO1 knockdown on the proliferation of human T cells in coculture. Therefore, CD3+ T cells were added to EFO-21 cells that had been pretreated with IDO1-specific ASOs at different concentrations for 3 days. T cells were polyclonally stimulated and their proliferative capacity was determined 4 days later. Supplementary Figure 1 shows a detailed gating strategy of flow cytometry data. As shown in Fig. 3, we observed a significantly increased proliferation index (a and b) and cell count (c) when EFO-21 cells had been pretreated with 3 µM and 1 µM of an IDO1-specific ASO. Of note, pretreatment with the control oligo 2 at a concentration of 3 µM had no effect on T cell proliferation.
Fig. 3.
Pretreatment of EFO-21 cells with IDO1-specific ASOs results in increased proliferation of T cells in coculture. EFO-21 cells were treated with different concentrations of IDO1-specific ASOs or the control oligo 2 (concentration: 3 µM) for 3 days without the use of a transfection reagent. Afterwards, CD3-enriched human T cells were labeled with a proliferation dye, added to EFO-21 cells and activated using CD3/28/2 tetrameric antibody complexes. On day four after start of the coculture proliferation of T cells (a) shown as proliferation index (b) and T cell counts (c) were determined by flow cytometry. Relative values are depicted as compared to mock-treated cells (0 nM). The mean ± SD of two independent experiments run in duplicates and triplicates, respectively, is depicted. Significant differences compared to mock-treated cells were determined using one-way ANOVA followed by Dunnett’s multiple comparison test. **p < 0.005, *p < 0.05, #p = 0.0687, §p = 0.1245
Comparison of the effect of an IDO1-specific ASO and an IDO1-specific small-molecule inhibitor on kynurenine production in human cancer cells
We sought to compare the effect of an IDO1-specific ASO and the IDO1-specific small molecule inhibitor Epacadostat on kynurenine production in human cancer cells. Therefore, cells were treated with the most potent IDO1-specific ASO (A06030H) for 3 days before replacing the medium with tryptophan-supplemented medium. In the case of Epacadostat, cells were treated with Epacadostat during the incubation period with tryptophan supplemented medium. Kynurenine levels were measured in the supernatant 24 h after addition of tryptophan supplemented medium. As shown in Fig. 4, treatment of cells with both compounds resulted in a reduction of kynurenine levels as compared to mock-treated cells.
Fig. 4.
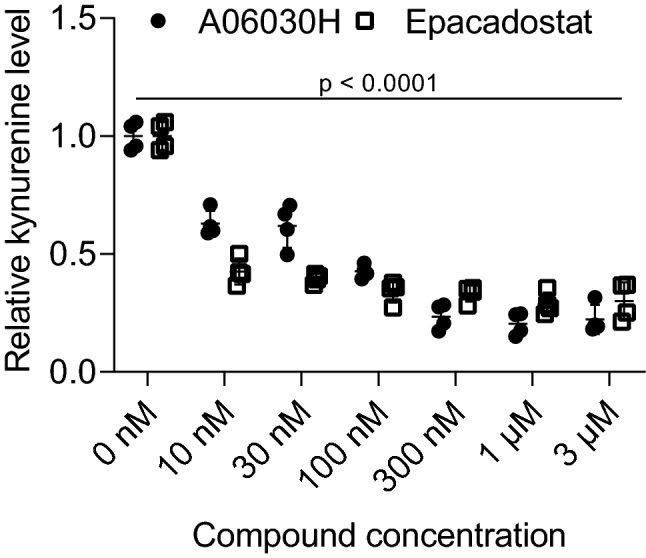
Comparison of the effect of an IDO1-specific ASO and an IDO1-specific small molecule inhibitor on kynurenine production in human cancer cells. EFO-21 cells were treated with the IDO1-specific ASO A06030H and Epacadostat at different concentrations. In the case of ASO, cells were treated for 3 days before adding tryptophan supplemented medium for 24 h. In the case of Epacadostat, cells were treated with the small molecule during incubation in tryptophan supplemented medium for 24 h. Kynurenine levels were measured by ELISA 24 h after the start of incubation in tryptophan supplemented medium. The mean of four replicates from two independent experiments is shown. Significant differences compared to mock-treated cells were determined using one-way ANOVA followed by Dunnett’s multiple comparison test
Effect of the combination of an IDO1 small molecule inhibitor with an IDO1-specific ASO on kynurenine production in human cancer cells
As small molecule inhibitors have a shorter half-life and different mode of action compared to ASOs, combining both modalities to address one target could be advantageous. It has been shown by us and others that ASOs can lead to a target knockdown for several days even in the absence of ASO [18]. IDO1 ASOs could, therefore, lead to persistent downregulation of IDO1 mRNA expression and IDO1-specific small molecule inhibitors would block activity of residual IDO1 molecules not suppressed by ASO. Combination of both approaches could, therefore, result in an increased overall suppression of kynurenine accumulation. We investigated in a cell culture system the effect of a combination of the IDO1 small molecule inhibitor Epacadostat and the potent IDO1-specific ASO A06030H on kynurenine production. Therefore, cells were pretreated with three different concentrations of A06030H (3 µM, 200 nM, 30 nM) or left untreated for 3 days, followed by treatment with different concentrations (10 nM–3 µM) of Epacadostat for 24 h. Thereafter, kynurenine levels were measured in the supernatant. As shown in Fig. 5a, pretreatment of EFO-21 cells with different concentrations of the IDO1-specific ASO followed by treatment with different concentrations of Epacadostat led to a higher reduction in kynurenine levels at all tested combinations as compared to Epacadostat alone. We used the Loewe additivity model [22] to test if the observed effect of the combination of IDO1 ASO with the small molecule inhibitor Epacadostat was synergistic. The model tests, if the concentration of two drugs given in combination that is needed to achieve a therapeutic effect is lower as the concentration that is needed of each compound in a monotherapy setting to achieve the same therapeutic effect. As shown in Fig. 5b and c, the observed effect was indeed synergistic according to Loewe over a variety of combinations (average Loewe score: 6.135) with the most pronounced effect observed at the combination of 30 nM ASO and 30 nM Epacadostat (Loewe score: 18).
Fig. 5.
Combination of an IDO1 small molecule inhibitor with an IDO1-specific ASO results in a synergistic kynurenine reduction in a human cell culture system. EFO-21 cells were treated with the IDO1-specific ASO A06030H at 3 different concentrations or left untreated for 3 days. Thereafter, Epacadostat was added at different concentrations and medium was supplemented with tryptophan (final concentration: 100 µM). Supernatant was harvested 24 h later and kynurenine levels were measured by ELISA. Relative kynurenine levels as compared to mock-treated cells are depicted (a). The mean of four replicates from two independent experiments is shown. 2D (b) and 3D (c) synergy maps of the combination responses, were calculated based on the Loewe model using the SynergyFinder R package [20]. Red areas in the graph indicate synergistic effects, which thus seem to be most pronounced for low ASO concentrations. Pooled data from two independent experiments were analyzed
Discussion
Despite promising approaches to unleash endogenous anti-tumor immune responses, a majority of patients does not benefit from currently available therapies [23–26]. The reason for this could be the existence of a variety of suppressive mechanisms that hinder the immune system from attacking tumor cells. One of those mechanisms is the degradation of tryptophan by enzymes like IDO1 and TDO2. In the present study, we show that inhibiting IDO1 and TDO2 with LNA-modified ASOs is feasible and a highly potent approach to block the degradation of tryptophan in cancer immunotherapy. We demonstrate that knockdown of IDO1 and TDO2 in cancer cell lines leads to reduced kynurenine levels in cell culture supernatants. Accordingly, proliferation of T cells is increased in coculture with IDO1 ASO-treated tumor cells. We furthermore show that combinatorial treatment of human cancer cells with the IDO1 small molecule inhibitor Epacadostat and an IDO1-specific ASOs leads to a synergistic reduction of kynurenine production. Due to the lack of a suitable cell culture model, the IDO1 small molecule inhibitor could not be combined with a TDO2-specific ASO.
Strategies to inhibit tryptophan degradation to kynurenine currently rely solely on small molecule inhibitors [8, 9, 27], which have so far failed to show therapeutic benefit in phase III clinical trials and there is only one report so far of a clinical trial with TDO2 small molecule inhibitors [28]. However, there is compelling evidence that TDO2 and especially IDO1 remain important targets in cancer therapy. IDO1 is overexpressed in a variety of tumors [4, 29] and has been associated with poor prognosis for example in patients with glioblastoma multiforme [30] and esophageal cancer [31]. Furthermore, the relevance of IDO1 as therapeutic target has been shown in different mouse models of cancer, especially in combination with chemotherapy [32, 33] and checkpoint inhibitors [34]. Suboptimal blockade of the enzymatic function of IDO1 could be a reason for limited efficacy of small molecule inhibitors and a multimodal approach targeting IDO1 could potentially lead to sufficient inhibition of tryptophan degradation by IDO1. Indeed, due to the short half-life of small molecules (plasma half-life of 2.9 h for Epacadostat), treatment of patients with Epacadostat was done twice daily [14]. It has been shown in whole blood stimulation assays from patients treated with Epacadostat that the maximal inhibitory capacity of Epacadostat peaks 2 h after application and then starts to decline over time [11]. Modified ASOs in contrast have a distinguished pharmacokinetics that is in large part determined by backbone chemistry [35].
After intravenous (i.v.) or subcutaneous (s.c.) administration of PTO-modified oligonucleotides, there is a fast decline from peak plasma concentration within few hours with a synchronous increase in tissue levels. The first rapid elimination phase is followed by a slower terminal elimination phase with half-lives of up to several weeks that are paralleled by a slow decay in tissues. Intracellular ASOs have a half-live- of 2–4 weeks. Prolonged target suppression in cells has been reported that lasts substantially even after removal of the ASO from cell culture supernatants [18]. Taken together, much longer target exposure times and sustained target engagement can be achieved by modified ASOs compared with small molecule inhibitors such as Epacadostat.
We have recently shown that systemic application of LNA-modified ASOs with specificity for CD39 can lead to potent suppression of target expression in different tumor stromal cells like regulatory T cells and myeloid cell subsets such as tumor-associated macrophages (TAMs) and MDSC [18]. As the enzymatic activity of IDO1 is an important mediator of the suppressive activity of myeloid cells in the tumor microenvironment [36, 37], efficient and persistent suppression of IDO1 expression in those cells could contribute to increased tryptophan and reduced kynurenine levels in the tumor microenvironment. Furthermore, there is also evidence for efficient knockdown of gene expression in tumor cells after systemic treatment with LNA-modified ASOs (data not shown).
In conclusion, we demonstrate in this study, that expression of IDO1 and TDO2 can efficiently be inhibited by LNA-modified ASOs which leads to decreased degradation of tryptophan as measured by kynurenine production.
We are aware that our study is confined to an in vitro experimental setting which investigates target down-regulation and reduction in kynurenine production using ASO technology. The therapeutic efficacy of inhibiting IDO1 and/or TDO2 expression by LNA-modified ASOs needs to be investigated in appropriate animal tumor models in the future. This will be instrumental in assessing in vivo activity and potential combination therapies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- AhR
Aryl hydrocarbon receptor
- ASO
Antisense oligonucleotide
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GCN2
General control nonderepressible 2 kinase
- HPRT1
Hypoxanthine phosphoribosyltransferase 1
- IDO1
Indoleamine 2,3-dioxygenase 1
- LNA
Locked nucleic acid
- PGE2
Prostaglandin E2
- PTO
Phosphorothioate
- TAM
Tumor associated macrophage
- TDO2
Tryptophan 2,3-dioxygenase
Author contributions
RK planned the study and experiments, analyzed data, performed statistical analysis and wrote the manuscript. SM performed bioinformatics analysis, performed statistical analysis of the data and wrote the manuscript. MS and LH performed experiments. AZ provided conceptual advice, interpreted data and wrote the manuscript. FJ planned the study, interpreted data and wrote the manuscript.
Funding
This work has been supported by a Grant from the Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung, BMBF) (Grant number: 031B0459) and a research Grant provided to AZ from Secarna.
Compliance with ethical standards
Conflict of interest
Richard Klar, Sven Michel, Monika Schell, Lisa Hinterwimmer and Frank Jaschinski are employed by Secarna. Richard Klar and Frank Jaschinski are holding patents for IDO1 and TDO2 ASOs, Sven Michel is holding a patent for TDO2 ASOs. Alfred Zippelius received research funding from Secarna Munich.
Ethical approval
Leukapheresis from healthy individuals was collected after informed consent following requirements of the local ethical board and principles of the Helsinki Declaration. The study was approved by the ethics commission of the Technical University of Munich (ethics commission reference: 329/16 S, study approval date: 9-19-2016).
Informed consent
Healthy donors consented in written form that their personal information was anonymized after leukapheresis and their PBMC were used to investigate the effect of antisense oligonucleotide-mediated knockdown of immunosuppressive factors in vitro.
Animal source
Not applicable.
Cell line authentication
EFO-21 (purchased from DSMZ), SKOV-3 (purchased from ATCC), MDA-MB-453 (purchased from ATCC) and A-172 (purchased from ATCC) cells were used in this study. Authentication of cell lines was not required as cells were purchased from professional vendors and master cell banks were generated shortly after taking the original vials into culture. Cells were kept in culture for a maximum time of 3 weeks to minimize the risk of contaminations.
Note on previous publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Richard Klar, Email: richard.klar@secarna.com.
Frank Jaschinski, Email: frank.jaschinski@secarna.com.
References
- 1.Herbst RS, Baas P, Kim D-W, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet Lond Engl. 2016;387:1540–1550. doi: 10.1016/S0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
- 2.Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 3.Hodi FS, O’Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uyttenhove C, Pilotte L, Théate I, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 5.Opitz CA, Litzenburger UM, Sahm F, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 6.Lee GK, Park HJ, Macleod M, et al. Tryptophan deprivation sensitizes activated T cells to apoptosis prior to cell division. Immunology. 2002;107:452–460. doi: 10.1046/j.1365-2567.2002.01526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apetoh L, Quintana FJ, Pot C, et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol. 2010;11:854–861. doi: 10.1038/ni.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Shin N, Koblish HK, et al. Selective inhibition of IDO1 effectively regulates mediators of antitumor immunity. Blood. 2010;115:3520–3530. doi: 10.1182/blood-2009-09-246124. [DOI] [PubMed] [Google Scholar]
- 9.Crosignani S, Bingham P, Bottemanne P, et al. Discovery of a novel and selective indoleamine 2,3-dioxygenase (IDO-1) inhibitor 3-(5-fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione (EOS200271/PF-06840003) and its characterization as a potential clinical candidate. J Med Chem. 2017;60:9617–9629. doi: 10.1021/acs.jmedchem.7b00974. [DOI] [PubMed] [Google Scholar]
- 10.Pei Z, Mendonca R, Gazzard L, et al. Aminoisoxazoles as potent inhibitors of tryptophan 2,3-dioxygenase 2 (TDO2) ACS Med Chem Lett. 2018;9:417–421. doi: 10.1021/acsmedchemlett.7b00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beatty GL, O’Dwyer PJ, Clark J, et al. First-in-human phase 1 study of the oral inhibitor of indoleamine 2,3-dioxygenase-1 Epacadostat (INCB024360) in patients with advanced solid malignancies. Clin Cancer Res Off J Am Assoc Cancer Res. 2017;23:3269–3276. doi: 10.1158/1078-0432.CCR-16-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell TC, Hamid O, Smith DC, et al. Epacadostat plus pembrolizumab in patients with advanced solid tumors: phase I results from a multicenter, open-label phase I/II trial (ECHO-202/KEYNOTE-037) J Clin Oncol Off J Am Soc Clin Oncol. 2018 doi: 10.1200/JCO.2018.78.9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez RP, Riese MJ, Lewis KD, et al. Epacadostat plus nivolumab in patients with advanced solid tumors: preliminary phase I/II results of ECHO-204. J Clin Oncol. 2017;35:3003. doi: 10.1200/JCO.2017.35.15_suppl.3003. [DOI] [Google Scholar]
- 14.Long GV, Dummer R, Hamid O, et al. Epacadostat (E) plus pembrolizumab (P) versus pembrolizumab alone in patients (pts) with unresectable or metastatic melanoma: results of the phase 3 ECHO-301/KEYNOTE-252 study. J Clin Oncol. 2018;36:108. doi: 10.1200/JCO.2018.36.15_suppl.108. [DOI] [Google Scholar]
- 15.Eckstein F. Nucleoside phosphorothioates. Annu Rev Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- 16.Stein CA, Hansen JB, Lai J, et al. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2010;38:e3–e3. doi: 10.1093/nar/gkp841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaschinski F, Korhonen H, Janicot M. Design and selection of antisense oligonucleotides targeting transforming growth factor beta (TGF-β) isoform mRNAs for the treatment of solid tumors. In: Walther W, Stein U, editors. Gene therapy of solid cancers: methods and protocols. New York: Springer; 2015. pp. 137–151. [DOI] [PubMed] [Google Scholar]
- 18.Kashyap AS, Thelemann T, Klar R, et al. Antisense oligonucleotide targeting CD39 improves anti-tumor T cell immunity. J Immunother Cancer. 2019 doi: 10.1186/s40425-019-0545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beck B, Dörfel D, Lichtenegger FS, et al. Effects of TLR agonists on maturation and function of 3-day dendritic cells from AML patients in complete remission. J Transl Med. 2011;9:151. doi: 10.1186/1479-5876-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ianevski A, He L, Aittokallio T, Tang J. SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinform Oxf Engl. 2017;33:2413–2415. doi: 10.1093/bioinformatics/btx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R Core Team . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 22.Loewe S. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung. 1953;3:285–290. [PubMed] [Google Scholar]
- 23.McDermott D, Lebbé C, Hodi FS, et al. Durable benefit and the potential for long-term survival with immunotherapy in advanced melanoma. Cancer Treat Rev. 2014;40:1056–1064. doi: 10.1016/j.ctrv.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Robert C, Thomas L, Bondarenko I, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364:2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 25.Weber JS, D’Angelo SP, Minor D, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16:375–384. doi: 10.1016/S1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- 26.Robert C, Long GV, Brady B, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 27.Cady SG, Sono M. 1-Methyl-dl-tryptophan, beta-(3-benzofuranyl)-dl-alanine (the oxygen analog of tryptophan), and beta-[3-benzo(b)thienyl]-dl-alanine (the sulfur analog of tryptophan) are competitive inhibitors for indoleamine 2,3-dioxygenase. Arch Biochem Biophys. 1991;291:326–333. doi: 10.1016/0003-9861(91)90142-6. [DOI] [PubMed] [Google Scholar]
- 28.Papadopoulos K, Eder P, Piha-Paul SA et al (2019) First-in-human phase I study of M4112, the first dual inhibitor of indoleamine 2,3-dioxygenase-1 and tryptophan 2,3-dioxygenase 2, in patients with advanced solid malignancies. In: AACR. Atlanta, p abstract no. CT011/6
- 29.Théate I, van Baren N, Pilotte L, et al. Extensive profiling of the expression of the indoleamine 2,3-dioxygenase 1 protein in normal and tumoral human tissues. Cancer Immunol Res. 2015;3:161–172. doi: 10.1158/2326-6066.CIR-14-0137. [DOI] [PubMed] [Google Scholar]
- 30.Zhai L, Ladomersky E, Lauing KL, et al. Infiltrating T cells increase IDO1 expression in glioblastoma and contribute to decreased patient survival. Clin Cancer Res. 2017;23:6650–6660. doi: 10.1158/1078-0432.CCR-17-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiyozumi Y, Baba Y, Okadome K, et al. IDO1 expression is associated with immune tolerance and poor prognosis in patients with surgically resected esophageal cancer. Ann Surg. 2018 doi: 10.1097/SLA.0000000000002754. [DOI] [PubMed] [Google Scholar]
- 32.Muller AJ, DuHadaway JB, Donover PS, et al. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat Med. 2005;11:312–319. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- 33.Hou D-Y, Muller AJ, Sharma MD, et al. Inhibition of indoleamine 2,3-dioxygenase in dendritic cells by stereoisomers of 1-methyl-tryptophan correlates with antitumor responses. Cancer Res. 2007;67:792–801. doi: 10.1158/0008-5472.CAN-06-2925. [DOI] [PubMed] [Google Scholar]
- 34.Spranger S, Koblish HK, Horton B, et al. Mechanism of tumor rejection with doublets of CTLA-4, PD-1/PD-L1, or IDO blockade involves restored IL-2 production and proliferation of CD8(+) T cells directly within the tumor microenvironment. J Immunother Cancer. 2014;2:3. doi: 10.1186/2051-1426-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geary RS, Norris D, Yu R, Bennett CF. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Yu J, Du W, Yan F, et al. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO Expression and correlate with lymph node metastasis in patients with breast cancer. J Immunol. 2013;190:3783–3797. doi: 10.4049/jimmunol.1201449. [DOI] [PubMed] [Google Scholar]
- 37.Muller AJ, Sharma MD, Chandler PR, et al. Chronic inflammation that facilitates tumor progression creates local immune suppression by inducing indoleamine 2,3 dioxygenase. Proc Natl Acad Sci. 2008;105:17073–17078. doi: 10.1073/pnas.0806173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.