Abstract
Dimerization of simian virus 40 T-antigen hexamers (TAgH) into double hexamers (TAgDH) on model DNA replication forks has been found to greatly stimulate T-antigen DNA helicase activity. To explore the interaction of TAgDH with DNA during unwinding, we examined the binding of TAgDH to synthetic DNA replication bubbles. Tests of replication bubble substrates containing different single-stranded DNA (ssDNA) lengths indicated that efficient formation of a TAgDH requires ≥40 nucleotides (nt) of ssDNA. DNase I probing of a substrate containing a 60-nt ssDNA bubble complexed with a TAgDH revealed that T antigen bound the substrate with twofold symmetry. The strongest protection was observed over the 5′ junction on each strand, with 5 bp of duplex DNA and ∼17 nt of adjacent ssDNA protected from nuclease cleavage. Stimulation of the T-antigen DNA helicase activity by an increase in ATP concentration caused the protection to extend in the 5′ direction into the duplex region, while resulting in no significant changes to the 3′ edge of strongest protection. Our data indicate that each TAgH encircles one ssDNA strand, with a different strand bound at each junction. The process of DNA unwinding results in each TAgH interacting with a greater length of DNA than was initially bound, suggesting the generation of a more highly processive helicase complex.
DNA helicases are a diverse group of enzymes which unwind duplex DNA at the expense of nucleoside or deoxynucleoside triphosphate hydrolysis (2, 21). DNA unwinding serves to activate the duplex DNA for biological transactions and is required for processes such as DNA replication, repair, and recombination. DNA helicases are invariably found in higher oligomeric states, generally dimers and hexamers. The simian virus 40 (SV40) large T antigen (8, 18), for example, forms hexamers (TAgH) (13, 16, 17, 20) which are seen as propeller-shaped particles that contain a central channel (14). Other members of the hexameric DNA helicase group also form ring-like structures, including the Escherichia coli DnaB protein (4, 15), the T4 gene 41 protein (6), and the bacteriophage T7 gp4 protein (7), the latter capable of encircling single-stranded DNA (ssDNA).
In an ATP-dependent reaction, T antigen recognizes DNA fork structures, binding as a TAgH and to a lesser extent as a double hexamer (TAgDH) (16, 17, 20). Previous nuclease footprinting of the TAgH bound to a DNA fork indicated that the helicase recognizes the ssDNA/duplex DNA (dsDNA) junction (16). The TAgH primarily bound the ssDNA strand with the 3′ tail (16), consistent with the 3′→5′ DNA helicase activity of T antigen (9, 22). The TAgDH form is >10-fold more active as a DNA helicase than the TAgH (17) and is competent to bridge two DNA unwinding forks (20), leading to the suggestion that the TAgDH is the entity which acts as a DNA helicase during DNA replication.
We characterized the binding and unwinding of synthetic DNA replication bubbles by the TAgDH. Our data demonstrate that the binding of a TAgDH is strongly dependent on the ssDNA length of the replication bubble. Footprinting of a TAgDH bound to a replication bubble substrate shows that each TAgH is associated with a different ssDNA/dsDNA junction and indicates that each hexamer encircles a different strand. Stimulation of the DNA helicase activity suggests that the general association of T antigen with the DNA substrate changes during DNA unwinding, resulting in T antigen binding a greater length of ssDNA.
MATERIALS AND METHODS
DNA substrates.
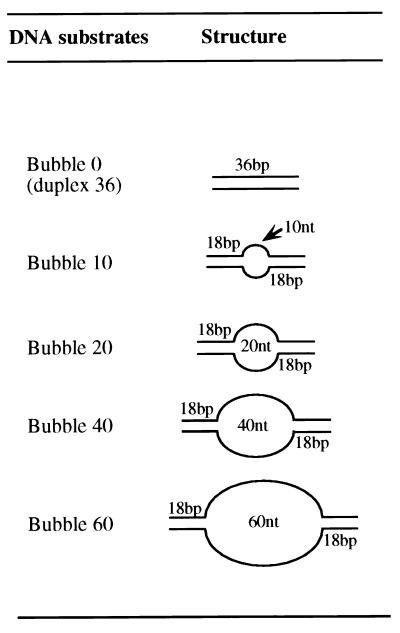
The DNA bubble substrates were assembled from two partially complementary oligonucleotides. The top and bottom strand sequences of the replication bubble 60 substrate were as follows (duplex regions underlined): top, 5′ TCT ACC TGG ACG ACC GGG (GACT)15 GGG CCA GCA GGT CCA TCA; bottom, 5′ TGA TGG ACC TGC TGG CCC (GACT)15 CCC GGT CGT CCA GGT AGA. The bubble 10, bubble 20, and bubble 40 substrates had identical sequences in the two duplex flanks and only differed in the number of d(GACT) repeats (which prevent the formation of stable secondary structures). For example, the top and bottom ssDNA region for the bubble 20 substrate contained a (GACT)5 repeat. The sequence of the bubble 0 substrate, modified to inhibit the formation of a secondary structure, was as follows: top, 5′ TTC TGT GAC TAC CTG GAC GAC CGG GTG ACT AGT TGC; bottom, 5′ GCA ACT AGT CAC CCG GTC GTC CAG GTA GTC ACA GAA. Before annealing, the top or bottom strand oligonucleotide was 5′ 32P-labeled with [γ-32P]ATP and T4 polynucleotide kinase to a specific activity of ∼1 × 106 cpm/pmol. Oligonucleotides were then annealed in reactions (50 to 100 μl) containing 5 to 10 pmol each of the top and bottom strands, 50 mM Tris-HCl (pH 8.0), and 10 mM MgCl2. Reaction mixtures were heated to 95°C and then cooled overnight. Complete annealing was verified by native gel electrophoresis and autoradiography of the reaction products.
Binding of T antigen to the replication bubble substrates.
The SV40 T antigen was expressed in Sf9 insect cells infected with recombinant baculovirus and immunoaffinity purified as previously described (1). The effect of ssDNA bubble length on the binding of TAgH and TAgDH was tested by using standard binding reaction mixtures (25 μl) containing 20 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 0.5 mM dithiothreitol, 0.1 mg bovine serum albumin per ml, 30 μM ATP, 12.5 ng of competitor DNA (Bluescript II SK digested with PvuII), and T antigen (as indicated in the figure legends). After preincubation of the reaction mixture for 10 min at 37°C, the 32P-labeled DNA substrate (0.1 pmol) was added and the reaction mixture was incubated for an additional 8 min at 37°C. The preincubation step is included because incubation of T antigen with ATP prior to the addition of the DNA substrate was found to increase the formation of TAgH and decreased nonspecific binding of T antigen to duplex DNA (data not shown). Reaction mixtures were then cross-linked by the addition of glutaraldehyde (final concentration, 0.05%). The reaction products were separated by nondenaturing electrophoresis (17) and autoradiographed. Protection was quantified by scanning gels directly with a Molecular Dynamics PhosphorImager, using ImageQuant software, and by analysis of scanned autoradiograms, using NIH image (version 1.61). Both methods of analysis yielded similar results. When the stabilities of TAgDH-DNA substrate complexes were investigated, ATP (1 mM) was added after the initial 10-min preincubation. The reaction mixtures were incubated for various times and subjected to glutaraldehyde cross-linking, as described above.
DNase I footprinting of T antigen bound to the replication bubble substrates.
Standard binding reactions (25 μl), containing 0.1 pmol of the 32P-labeled bubble 60 substrate and T antigen (as indicated in the figure legends) were prepared as described above, except that the reactions were not cross-linked. CaCl2 was then added to a final concentration of 2.5 mM, and reaction mixtures were incubated at room temperature for 5 min prior to the addition of 0.2 U of DNase I (Boehringer Mannheim). There was no effect of CaCl2 on the stability of the TAgDH-DNA substrate complexes (data not shown). After incubation for 30 s at room temperature, reactions were stopped by the addition of quench buffer (0.5% sodium dodecyl sulfate and 20 mM EDTA) and 270 μl of water. The reaction mixtures were extracted with phenol, and, following the addition of glycogen (20 μg) as carrier, the DNA was precipitated with ethanol. The dried pellets were subjected to denaturing gel electrophoresis and autoradiography. To footprint the helicase-active complexes, reaction mixtures were prepared as described above. ATP was then added to a final concentration of 1 mM, the reaction mixtures were further incubated for various times, and the DNA was digested with DNase I as described above. Protection from DNase I cleavage was quantitated as described above.
RESULTS
It has been proposed that members of the hexameric DNA helicase family encircle one (7) or both (11) DNA strands during DNA unwinding. To understand how T antigen interacts with DNA during helicase action, we examined the binding of T antigen to a variety of synthetic replication bubble substrates that differed in the length of the central ssDNA bubble (Fig. 1). Previous footprinting studies indicated that T antigen associates primarily with one ssDNA strand of a synthetic replication fork (16). If the TAgH encircles this strand, binding of a TAgDH to the bubble substrate would require the central ssDNA region to be a certain critical length, likely a minimum of twice the length of ssDNA bound by a TAgH. Alternatively, T antigen may interact with this strand using a binding site on the outside of the TAgH. Because productive DNA unwinding can occur on substrates containing a 3′ ssDNA overhang of less than 5 nucleotides (nt) (22), TAgDH-bubble complex formation would be expected to be primarily dependent on the presence of two ssDNA/dsDNA junctions and not to require relatively large lengths of ssDNA.
FIG. 1.
Model replication bubble substrates used to investigate DNA binding by T antigen. The substrates, varying in the ssDNA length, were prepared as discussed in the Materials and Methods.
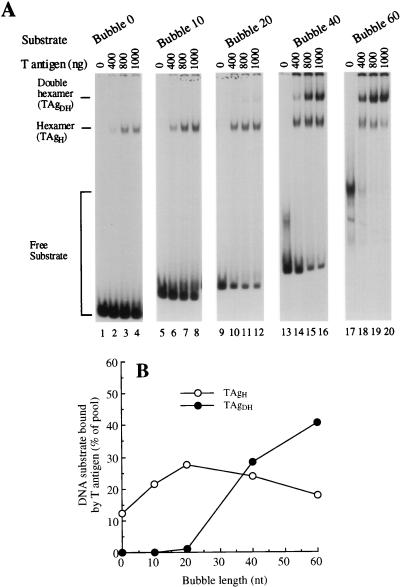
Binding was tested in the presence of a low concentration of ATP (30 μM) to support T antigen-DNA complex formation yet not induce significant unwinding of the substrate (reference 16 and data not shown). Using a gel retardation assay, titration of increasing levels of T antigen caused the bubble 0, bubble 10, and bubble 20 substrates to be bound by increasing amounts of a TAgH, while TAgDH-bubble complexes were observed only at low levels (Fig. 2A). The binding of a TAgDH increased markedly when the ssDNA bubble was lengthened to 40 nt and reached >40% of the substrate pool (at 800 ng of T antigen) using a 60 nt bubble. As the bubble length was increased to 40 and 60 nt, a decrease in the binding of the TAgH was observed, suggesting that it could transform into a TAgDH on these substrates. The amount of TAgH and TAgDH binding as a function of bubble length was quantitated and is shown for 800 ng of T antigen (Fig. 2B). We found that significant binding of a TAgDH to the replication bubble substrates requires a critical ssDNA length that was a minimum of 40 nt. Because the binding of a TAgDH requires the ssDNA length to be relatively large, these data are not consistent with a model in which each TAgH interacts with the ssDNA/dsDNA junction by using a binding site on the outer face of the TAgH. Rather, the data support a model in which each TAgH encircles one of the two strands at a replication fork.
FIG. 2.
Formation of TAgDH-bubble complexes promoted by ssDNA bubble length that is a minimum of 40 nt. (A) Increasing amounts of T antigen were preincubated at 37°C for 10 min with 30 μM ATP. To the reaction mixture was then added the appropriate 32P-labeled DNA substrate (0.1 pmol) and the mixtures were incubated for an additional 8 min. The T antigen-DNA complexes were fixed by glutaraldehyde cross-linking, separated from the free substrate by native gel electrophoresis (5% polyacrylamide), and visualized by autoradiography. The amounts of T antigen used (400, 800, and 1,000 ng) correspond to a molar excess of TAgH over substrate of 8.1, 16, and 20, respectively. The positions of the free substrate and the substrate bound by TAgH or TAgDH are indicated. (B) Quantitative analysis of the T-antigen oligomeric state bound to the replication bubble substrates, as a function of the ssDNA bubble length. From the experiment in panel A, bands corresponding to TAgH- and TAgDH-DNA substrate complexes were excised and the radioactivity was measured by scintillation counting. We show the results of using 800 ng of T antigen, indicating the fraction of DNA substrate (in percent) bound either by a TAgH or TAgDH.
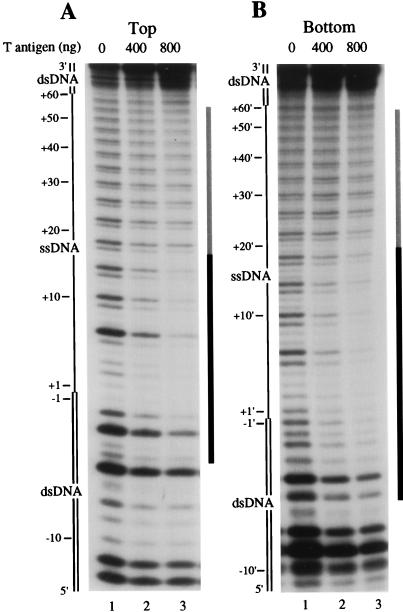
The binding of T antigen to the bubble 60 substrate, 5′-32P-labeled on the top (Fig. 3A) or bottom (Fig. 3B) strand, was examined by DNase I footprinting. We found that 800 ng of T antigen led to strong protection (>85%) on the 5′ half of the top-labeled substrate, encompassing 5 bp of the duplex DNA at the 5′ ssDNA/dsDNA junction and 16 nt of adjacent ssDNA. Moderate protection (>35% and <55%) was detected in the remaining 3′ ssDNA and 5′ dsDNA regions. A very similar pattern of protection was observed by using a substrate that was 5′-32P-labeled on the bottom strand (Fig. 3B; 18 nt of ssDNA protected) and on the bubble 60 substrate labeled on the 3′ end (data not shown). We believe that the protection on the extreme 5′ end of the duplex DNA is relatively nonspecific (i.e., not dependent on the ssDNA/dsDNA junction) because control experiments with completely duplex fragments also showed a similar moderate protection by T antigen (data not shown). Nonspecific protection of duplex DNA ends was also observed for binding of the RuvB helicase to Holliday junction intermediates (11).
FIG. 3.
DNase I footprinting analysis of static complexes formed on the synthetic bubble 60 substrate by T antigen. The bubble 60 substrate, 5′-32P-labeled on the top (A) or bottom (B) strand, was incubated with 400 or 800 ng of T antigen. Reaction mixtures were then treated with 0.2 U of DNase I, and the reaction products were separated by denaturing polyacrylamide gel electrophoresis. The ssDNA region of the substrate is designated as a single line, while the duplex DNA is designated as a double line. Positions on the substrate are indicated with respect to the 5′ ssDNA/dsDNA junction, with duplex DNA or ssDNA positions indicated as negative or positive numbers, respectively. Regions of strong or moderate protection are indicated by a black or gray line, respectively, on the right side of each panel. Note that we can not examine the 5 nt of DNA on the 5′ terminus because of the inability of DNase I to digest this DNA.
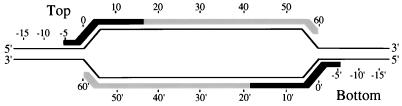
The summarized protection pattern revealed that a TAgDH bound with twofold symmetry to the replication bubble substrate (Fig. 4). As we previously observed that a TAgH protected only one of the two ssDNA strands on a synthetic replication fork (16), our data indicate that the primary interactions of each TAgH are with different ssDNA strands at each junction, with each hexamer bound to the 5′ portion of the single-stranded region. Combined with the clear dependence of TAgDH binding on the ssDNA bubble length, our data are consistent with a model in which a TAgH encircles a different ssDNA strand at each ssDNA/dsDNA junction.
FIG. 4.
Summary of nuclease footprinting analysis. The dried gels or autoradiographs were analyzed as described in Materials and Methods. Strong protection of >85% compared to the absence of T antigen is indicated by a black line above or below the protected strand, while moderate protection (>35% and <55%) is indicated by a gray line above or below the protected strand.
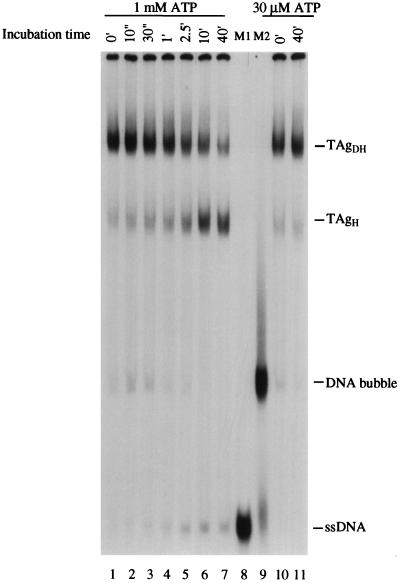
We examined the interaction of the T-antigen DNA helicase with the replication bubble substrate under conditions that support DNA unwinding. The binding of T antigen to the bubble 60 substrate was examined by a gel retardation assay. T antigen (800 ng) was first incubated with the substrate, using 30 μM ATP to allow complex formation, and the DNA helicase activity was then stimulated by the addition of ATP to 1 mM (see reference 17). After various incubation times, the T antigen-bubble 60 complexes were subjected to gel retardation analysis (Fig. 5). In the presence of 1 mM ATP, a gradual dissociation of TAgDH to TAgH was observed. After incubation for 40 min, the amount of the TAgDH-DNA bubble complex decreased from 53 to 33% of the substrate pool, while the amount of TAgH showed a corresponding increase from 20 to 45%. Control incubation of the T antigen-bubble complex for 40 min in the presence of 30 μM ATP showed no significant dissociation of TAgDH to TAgH (Fig. 5, lane 11). Over this 40-min time course, the amount of observable ssDNA increased from 0 to 12% of total substrate, an underestimate because a significant fraction of the generated ssDNA remains bound to T antigen after cross-linking (e.g., see reference 17). DNA helicase assays indicate that 45% of the substrate is denatured during a 40-min incubation (data not shown). These data indicate that the oligomeric state of T antigen bound to the DNA decreases during active DNA unwinding. We postulate that, upon complete unwinding of the substrate, the two TAgH molecules separate and segregate to individual ssDNA strands, resulting in each strand being bound by one TAgH.
FIG. 5.
Activation of the T-antigen DNA helicase on the bubble 60 substrate results in a conversion of TAgDH to TAgH. After preliminary incubation of 800 ng of T antigen with 30 μM ATP, the 32P-labeled replication bubble 60 was added to allow T antigen-DNA complex formation. The ATP concentration was then raised to 1 mM, and the reaction mixtures were further incubated for the indicated times (lanes 1 to 7). The reaction mixtures were subjected to glutaraldehyde cross-linking, and the different T antigen-DNA complexes were separated by native gel electrophoresis and visualized by autoradiography. Control reactions contained T antigen in the presence of 30 μM ATP for 0 (lane 10) or 40 (lane 11) min. Lanes 8 and 9 contain as markers the top 32P-labeled ssDNA strand (M1) and the bubble 60 substrate (M2), respectively.
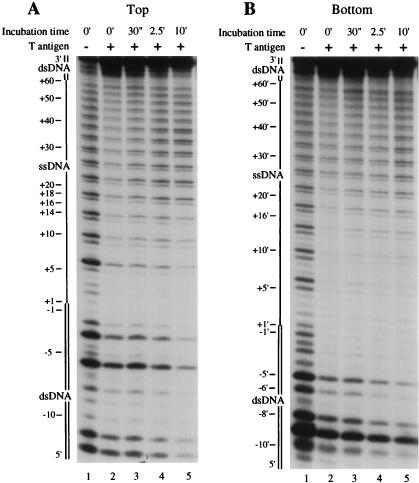
We examined the interaction of the activated T antigen DNA helicase with the bubble 60 substrate by DNase I footprinting (Fig. 6). T antigen was bound to the substrate in the presence of 30 μM ATP, and the DNA helicase activity was then activated by increasing the ATP concentration to 1 mM. After various times, the DNA labeled on the top (Fig. 6A) or bottom (Fig. 6B) strand was subjected to DNase I digestion. A control digestion performed in the absence of increased ATP levels showed that T antigen most strongly protected 16 nt of the 5′ portion of the ssDNA bubble on the top strand (from nt +1 to +16; Fig. 6A), as seen above. Upon activation of the helicase activity for 30 s, the protection of the 3′ ssDNA (between nt +17 to +60) was reduced, and the footprint on the 5′ ssDNA was more clearly delineated between nt +1 and +16. Protection of the duplex DNA (nt −5 to −1) changed only slightly. As the incubation time with 1 mM ATP increased, the protection of the 3′ ssDNA (nt +17 to +60) lessened further, while the boundary of protection at nt +16 remained unchanged (i.e., note that the difference in intensity of bands +14 and +18 is maintained at 30 s and 10 min). In contrast, incubation with 1 mM ATP caused T antigen to strongly protect the 5′ end of the DNA strand. After 10 min, the 5′ DNA that was initially duplex became resistant to nuclease digestion (>90% protection).
FIG. 6.
DNase I footprinting analysis of T antigen-bubble 60 substrate complexes under conditions that support DNA unwinding. T antigen (800 ng) was incubated in the presence of 30 μM ATP with the replication bubble 60 substrate that was 5′-32P-labeled on the top (A) or the bottom (B) strand. The level of ATP was then increased to 1 mM, and, at various times, the reaction mixtures were subjected to digestion with DNase I. Digestion products were analyzed by denaturing polyacrylamide gel electrophoresis and autoradiography. Positions on the substrate are as indicated in the legend to Fig. 3.
A similar observation was noted for the replication bubble labeled on the 5′ end of the bottom strand (Fig. 6B). During the initial 30-s incubation with 1 mM ATP, the protection by T antigen of the 3′ two-thirds of the bubble (from nt +17′ to +60′) was reduced, while the footprint over the 5′ ssDNA (nt +1′ to +16′) was unaffected. As the time after DNA helicase activation increased to 10 min, no further change in the 3′ footprint boundary at nt +16′ was observed. The protection of the 5′ end of the substrate was seen to greatly increase between 30 s and 10 min (note the decrease in the intensity of bands corresponding to nt −5′, −6′, −8′, and −10′ in lane 5 compared to lane 3). On both strands, therefore, activation of the T-antigen helicase caused the overall amount of DNA that was strongly protected by T antigen to increase in the 3′→5′ direction, consistent with the polarity of the T-antigen DNA helicase activity. Note that although the length of DNA that was strongly protected by T antigen increased, we observed a reduction in the oligomeric state of T antigen bound to the substrate (Fig. 5). The increased protection on the 5′ end of the DNA substrate is therefore not a result of the binding of additional molecules of T antigen but rather is a consequence of the T-antigen helicase activity.
DISCUSSION
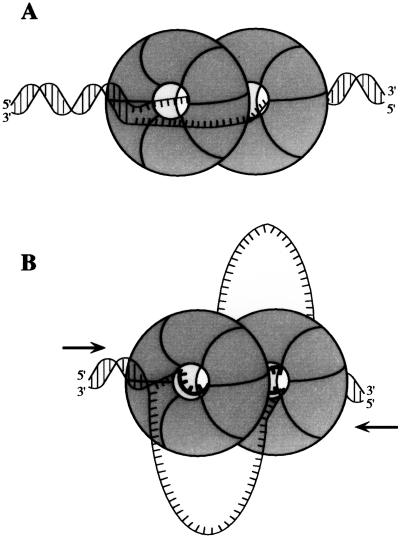
Our combined footprinting and binding data suggest that two TAgH molecules initially bind symmetrically to the bubble substrate (Fig. 7A). At each junction, one TAgH encircles the ssDNA strand whose 5′ end attaches to the junction. The ssDNA strand that passes through the center of one TAgH is thus located on the outside of the adjacent TAgH molecule. Upon an increase in the ATP concentration, the duplex DNA on each flank becomes unwound and one of the two strands moves through the central channel (Fig. 7B). DNA unwinding leads to the T antigen interacting with a greater length of ssDNA and causes the generation of two ssDNA loops.
FIG. 7.
Model of the TAgDH binding to and unwinding of the synthetic replication bubble. See text for details.
The ability of T antigen to encircle one of the two ssDNA strands is supported by our observation that the binding of a TAgDH to the replication bubble requires the central ssDNA region to be a minimum of 40 nt. Our footprinting analysis showed that each TAgH strongly protects 16 to 18 nt of ssDNA from DNase I cleavage on only one of the two strands at the junction. Thus, the binding of two hexamers would be expected to require a ssDNA bubble that was a minimum of ∼34 nt, assuming that the hexamers are arranged side by side. An alternative model in which T antigen interacts with the ssDNA/dsDNA junction using an outside face of the TAgH is argued against because T antigen can efficiently unwind DNA molecules containing a 3′ ssDNA overhang of less than 5 nt (22). That is, TAgDH binding to the bubble substrate using DNA recognition sites on the outside face of the TAgH would predict a greatly reduced dependence for ssDNA length and would not require a 40-nt ssDNA bubble.
Our previous analysis of T antigen-replication fork complexes showed that a TAgH protected 4 bp of top strand dsDNA and 10 nt of contiguous ssDNA at the ssDNA/dsDNA junction, with little protection observed on the other strand (16). Although we find a similar region of duplex protection using the replication bubble substrate, the ssDNA that was strongly protected by T antigen was longer on the bubble substrate (16 and 18 nt on the top and bottom strands, respectively) compared to the fork (10 nt). This discrepancy may be explained by the difference between TAgH and TAgDH binding. For example, because we found previously that the TAgDH was much more active as a DNA helicase than the TAgH (17), the TAgH may change its conformation upon dimerization, leading to an altered protection pattern. Alternatively, the binding of a TAgDH may cause greater steric hindrance that prevents nuclease access to the ssDNA, compared to TAgH binding to a fork. We note that the moderate protection of the 3′ portion of the ssDNA was not observed on the comparable bottom strand of a synthetic replication fork (16). Our data therefore lead to the suggestion that the steric constraints of a TAgDH bound to the replication bubble cause greater protection of this region, for example, by close contact of the 3′ portion of the ssDNA strand with the outside of each TAgH.
Footprinting of the activated T antigen helicase revealed that, during unwinding of the substrate, the 3′ boundary of the most strongly protected region (position +16 on each strand) did not change appreciably, even though the 5′ DNA that was initially duplex became protected. This observation was surprising because we expected to observe a ∼20 nt footprint migrate in the 3′→5′ direction as T antigen unwound the substrate. We found that the 30 nt of DNA that was bound by T antigen after unwinding was protected to a similar extent (>80%; data not shown) over the entire region. Thus, these results can not be explained by the presence of two populations of T antigen during DNA unwinding, an active pool that moved along the DNA and an inactive pool that remained bound at the initial binding site. Our data therefore indicate that, upon DNA unwinding, T antigen maintains contacts with the ssDNA at the 3′ edge of initial strong protection. DNA unwinding causes each TAgH to bind additional ssDNA as it is formed, perhaps by facilitating an increase in the fraction of TAgH subunits binding DNA. At the completion of DNA unwinding, each TAgH protects a greater length of DNA than was initially bound. Since the initiation of DNA unwinding causes T antigen to bind more DNA, our data also suggest that a more highly processive DNA helicase would result. In other words, the association of T antigen with a greater length of DNA could allow formation of more protein-DNA contacts, thereby yielding a higher-affinity complex.
Interestingly, previous data from our laboratory showed that ethylation of phosphate residues at the ssDNA/dsDNA junction, but not at phosphate residues downstream of the junction, inhibited DNA fork denaturation, leading us to suggest that T antigen formed a more stable complex with the DNA fork upon the initiation of DNA unwinding (16). We also note that the hexameric bacteriophage T7 gp4 protein has been proposed to bind the contacted DNA strand with each monomer during DNA unwinding (10). Our data predict that further increases in the duplex DNA length ahead of the fork will, at some critical DNA length, lead to deprotection of the original junction during DNA unwinding.
While we observe continued association of T antigen with the initial ssDNA/dsDNA junction during DNA unwinding, it should be emphasized that our studies were performed using purified T antigen. Because other replication factors, such as human replication protein A or DNA polymerase α/DNA primase (12, 19), are known to physically interact with T antigen, it is possible that these other factors modulate the interaction of T antigen with the ssDNA as it is generated at the DNA replication fork.
During the initiation of SV40 DNA replication, T antigen binds to the viral origin as a double hexamer (13). Previous binding studies have indicated that each TAgH assembles around the duplex DNA (5). The amount of DNA melting (8 bp in the early palindrome) (3) appears insufficient to allow one ssDNA strand to pass through the center of the TAgH on each origin half. These data therefore suggest that, during denaturation of the SV40 origin, the T-antigen–DNA complex undergoes a topological change. One of the two encircled strands must pass through the hexameric T-antigen ring to lead to the final helicase-active entity.
ACKNOWLEDGMENTS
We thank Xiang-Peng Kong, David Frendewey, Ruben Abagyan, and Ken Marians for helpful discussions and advice. We thank Cristina Iftode, Jennifer Garner, Yaron Daniely, and Mehboob Shivji for critical reading of the manuscript.
This research was supported by NIH grant AI29963 and by Kaplan Cancer Center Developmental Funding and Kaplan Cancer Center Support Core Grant (NCI P30CA16087).
REFERENCES
- 1.Borowiec J. Inhibition of structural changes in the simian virus 40 core origin of replication by mutation of essential origin sequences. J Virol. 1992;66:5248–5255. doi: 10.1128/jvi.66.9.5248-5255.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borowiec J A. DNA helicases. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 545–574. [Google Scholar]
- 3.Borowiec J A, Hurwitz J. Localized melting and structural changes in the SV40 origin of DNA replication induced by T antigen. EMBO J. 1988;7:3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bujalowski W, Klonowska M M, Jezewska M J. Oligomeric structure of Escherichia coli primary replicative helicase DnaB protein. J Biol Chem. 1994;269:31350–31358. [PubMed] [Google Scholar]
- 5.Dean F B, Borowiec J A, Eki T, Hurwitz J. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J Biol Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
- 6.Dong F, Gogol E P, von Hippel P H. The phage T4-coded DNA replication helicase (gp41) forms a hexamer upon activation by nucleoside triphosphate. J Mol Biol. 1995;270:7462–7473. doi: 10.1074/jbc.270.13.7462. [DOI] [PubMed] [Google Scholar]
- 7.Egelman E H, Yu X, Wild R, Hingorani M M, Patel S S. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc Natl Acad Sci USA. 1995;92:3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 9.Goetz G S, Dean F B, Hurwitz J, Matson S W. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J Biol Chem. 1988;263:383–392. [PubMed] [Google Scholar]
- 10.Hacker K J, Johnson K A. A hexameric helicase encircles one DNA strand and excludes the other during DNA unwinding. Biochemistry. 1997;36:14080–14087. doi: 10.1021/bi971644v. [DOI] [PubMed] [Google Scholar]
- 11.Hiom K, West S C. Branch migration during homologous recombination: assembly of a RuvAB-Holliday junction complex in vitro. Cell. 1995;80:787–793. doi: 10.1016/0092-8674(95)90357-7. [DOI] [PubMed] [Google Scholar]
- 12.Hurwitz J, Dean F B, Kwong A D, Lee S-H. The in vitro replication of DNA containing the SV40 origin. J Biol Chem. 1990;265:18043–18046. [PubMed] [Google Scholar]
- 13.Mastrangelo I A, Hough P V C, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 14.San Martín M C, Gruss C, Carazo J M. Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J Mol Biol. 1997;268:15–20. doi: 10.1006/jmbi.1997.0952. [DOI] [PubMed] [Google Scholar]
- 15.San Martín M C, Stamford N P J, Dammerova N, Dixon N E, Carazo J M. A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J Mol Biol. 1995;114:167–176. doi: 10.1006/jsbi.1995.1016. [DOI] [PubMed] [Google Scholar]
- 16.SenGupta D J, Borowiec J A. Strand-specific recognition of a synthetic DNA replication fork by the SV40 large tumor antigen. Science. 1992;256:1656–1661. doi: 10.1126/science.256.5064.1656. [DOI] [PubMed] [Google Scholar]
- 17.Smelkova N V, Borowiec J A. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J Virol. 1997;71:8766–8773. doi: 10.1128/jvi.71.11.8766-8773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stahl H, Dröge P, Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986;5:1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stillman B. Smart machines at the DNA replication fork. Cell. 1994;78:725–728. doi: 10.1016/s0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 20.Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West S C. DNA helicases: new breeds of translocating motors and molecular pumps. Cell. 1996;86:177–180. doi: 10.1016/s0092-8674(00)80088-4. [DOI] [PubMed] [Google Scholar]
- 22.Wiekowski M, Schwarz M W, Stahl H. Simian virus 40 large T antigen DNA helicase. J Biol Chem. 1988;263:436–442. [PubMed] [Google Scholar]