ABSTRACT
There is no doubt that today’s life sciences would look very different without the availability of millions of research antibody products. Nevertheless, the use of antibody reagents that are poorly characterized has led to the publication of false or misleading results. The use of laboratory animals to produce research antibodies has also been criticized. Surprisingly, both problems can be addressed with the same technology. This review charts today’s maze of different antibody formats and the various methods for antibody production and their interconnections, ultimately concluding that sequence-defined recombinant antibodies offer a clear path to both improved quality of experimental data and reduced use of animals.
KEYWORDS: Monoclonal antibodies; polyclonal antibodies; recombinant antibodies; multiclonal antibodies; non-animal derived antibodies, phage display, 3R, immunoglobulin, library, somatic hypermutation
Introduction
The development of methods for the generation of antibodies has forever changed research, diagnostics, and therapy. Behring’s antitoxins (now called polyclonal antibodies),1 Milstein and Köhler’s hybridoma technology to generate monoclonal antibodies,2 and finally the development of antibody phage display by Breitling, McCafferty, Barbas and colleagues3–5 deeply influenced medicine, immunology, molecular biology, biochemistry, and biotechnology. Immunofluorescence literally changed the way we look at biological molecules6 by allowing a single protein to be identified through a microscope. Building on developments that started in the mid-1980s, antibodies are now used in more than 160 new medications (www.antibodysociety.org/resources/approved-antibodies/) to treat cancer, infections, autoimmune and other diseases, with many more expected.7 The reason for this widespread use is the unique ability of antibodies to recognize a specific molecule among tens of thousands of other biomolecules and bind to it with extraordinary affinity. This particular capability has contributed to the evolutionary success of longer-living animals including ourselves, as it provides a very effective weapon system against parasites, microbes and viruses that otherwise would rapidly overwhelm us. As these small organisms evolve much more rapidly than we do, our bodies routinely generate antibodies recognizing molecules that the human species may never have encountered before. This marvellous capability makes antibodies useful for research, as we can generate immunoglobulins to almost all molecules of suitable size that provide a defined structure and a minimum of interactions to assure high-affinity binding. This is illustrated by the creation of antibodies to molecules not generally considered antigens, such as Buckminsterfullerene8 or polyethylene glycol.9 This vast structural variability of millions of different molecular interaction surfaces originates from a clever loop arrangement at the tips of the immunoglobulin’s arms.10 The structural diversity is achieved by DNA fragment recombination and even new DNA synthesis in B cell development.11 Later, during antigen-dependent B cell differentiation and clonal selection, every individual antibody-producing cell undergoes clonal expansion, and can further improve the antibody it produces by somatic hypermutation.12
Today, we can imitate all essential steps of this antibody generation process in vitro,14 allowing some restrictions of our own immune system to be bypassed, such as the bias introduced by the necessity to present peptides on the major histocompatibility complex (MHC), or the limited options for heavy-chain/light-chain pairing in individual B cells. This approach also allows antibody generation to be done without any animal experiments,15 which has led companies to market them as “vegan antibodies”. Qualifying antibodies as vegan would mandate that generation, analysis, and production are achieved without the use of animals or animal-derived materials. There are currently quite a number of different ways to discover new antibodies against a desired target and produce them in sufficient quantities (Figure 1). Many cross-connections between these methods further complicate the picture, so that the use of animals or animal-derived materials is not always readily apparent. Even de novo generated antibody sequences designed by artificial intelligence (AI)16 rely on the structural guidance of nature’s antibody repertoire, as the AI systems cannot be successful without training on the sequences and structures of countless animal-derived immunoglobulins. However, this strict approach does not reveal actual animal use in a specific antibody generation project. In the analysis and discussion below, materials such as donor blood-derived human antibody DNA or the human HEK293 production cell line are not considered animal-derived because they do not originate from animals, which typically include mice, rats, hamsters, rabbits, horses, goats, sheep and guinea pigs but may also include monkeys, chickens, dogs, llamas, and sharks.
Figure 1.
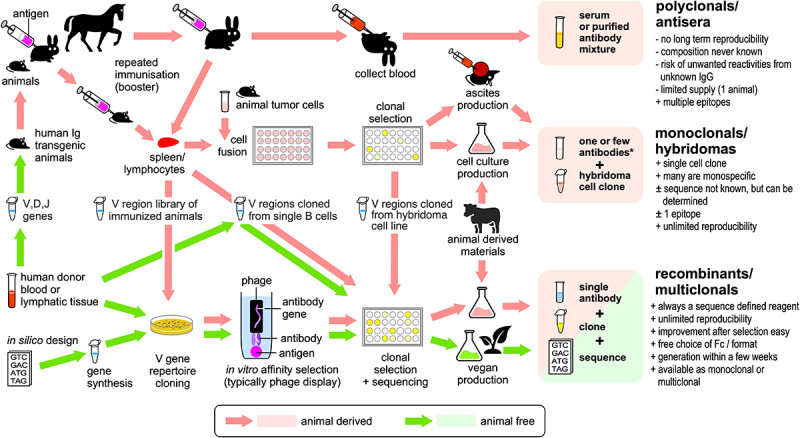
The intricate paths of antibody generation. Organisms are shaded in black. Importantly, recombinant antibodies are not always non-animal derived, as today the various advantages of the recombinant format are utilized both by animal- and non-animal-derived antibodies. Vast antibody variable region gene libraries, which can be obtained from human donor blood or by in silico design, offer animal-free access to antibody genes. However, the use of gene libraries does not always guarantee animal-free products, as they can be used in transgenic animals or obtained after animal immunization. It is also often overlooked that, after the initial animal-free generation of an antibody clone, animal materials may be used to produce these antibodies in useful quantities. Conversely, today any recombinant animal- or non-animal-derived antibody could also be produced in a vegan cell culture system. Since this map shows several paths consisting entirely of green arrows, “vegan” antibodies are indeed a reality.
*, see13
How to make antibodies
For the first 85 years of their use, antibodies were only available in the form of blood serum or polyclonal immunoglobulin fractions obtained from it. Although these crude antibody preparations are always undefined in respect of their true specificity spectrum, they saved the lives of children with diphtheria and other diseases,17 and substantially expanded our understanding of histology and cell biology.6
The Use of polyclonal antibodies, however, has drawbacks, as only 0.5–5% of the immunoglobulins in a polyclonal reagent bind to their intended target.18 Polyclonal serum is composed of an antibody mixture resulting from all immune responses of the donor animal, which may be independent from the immunization with the desired antigen. Further, this composition varies from batch to batch, because the immune response is different in every individual animal19, and even between subsequent blood samples taken at different times from the same animal.20 The additional specificities are of course unwanted, but never mapped, resulting in poorly defined reagents. While affinity chromatography on the antigen to enrich the desired specificities is possible,21 this is only offered for a small fraction of the catalog products. Using the flow-through of an affinity chromatography on antigens that must not be recognized by the final polyclonal product allows the reduction of cross-reactivities. This approach is most prominently used to remove inter-species cross-reactivity of secondary antibodies.
Despite these approaches to improve their performance, the shortcomings of polyclonal antibodies are responsible for numerous false scientific conclusions.18,22 Fifteen papers published on the dementia-related protein C9ORF72 that generated 3,500 first-layer citations and 66,000 secondary citations involve work with an antibody that does not recognize C9ORF72.23 Many antibodies underperforming in a validation study of more than 600 antibodies were used in a large number of published articles.24 Poorly characterized and ill-defined antibodies were in large part responsible for the failure to replicate the scientific results of 47 of 53 landmark preclinical studies.25 In a western blot analysis of 13,000 antibodies, only about 45% recognized their expected targets.26 While this could be explained in part by the denaturation of epitopes, Berglund et al. found that more than half of around 6,000 routinely used commercial monoclonal and polyclonal antibodies recognized more than their specified targets.27 Numerous authors have lamented the substantial amounts of money wasted per annum for antibodies that do not work or produce false results and the much higher follow-up cost for our society.28 Human lives are also at stake: In addition to research work, clinical trials and diagnostic tests for cancer were based on the use of antibodies that have since proven to not bind to the assumed target or were nonspecific.29–33 Knowing all of this, it is surprising that polyclonals still represent the largest product group for research antibodies (Figure 2). But why are research products that obviously are “littering the field with false findings”34 still so popular? Sadly, this is not driven by their performance, but mainly due to their much lower price and lack of knowledge about the alternatives.15
Figure 2.
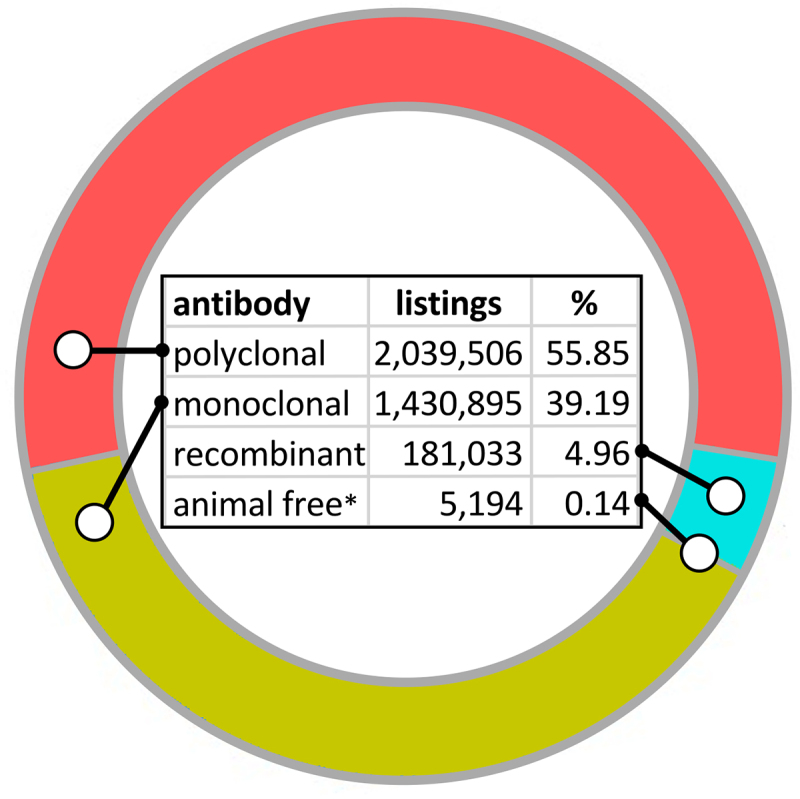
Biocompare.com search for different research antibody products, as of February 2024. The search engine could not distinguish between different products derived from the same original antibody preparation, e.g., with different labels or packaging sizes. As different labels are more commonly offered for secondary antibodies, which are mostly polyclonals, this segment may be slightly overrepresented.
* Only 27 antibodies made by animal-free methods were listed in this database; this label was obviously not yet provided for all recombinant antibodies made by these methods. The recombinant antibody website https://afa.ehstaging.net/listed 5194 different animal-free antibody products by the same date, which would still represent only 0.14%.
The polyclonal nature of serum-derived antibodies also has a positive aspect: immune responses of the immunized animals typically lead to the expansion and differentiation of several antigen-specific B cells that in most cases cover different epitopes on the antigen of interest. The resulting serum therefore offers better chances to recognize the antigen in different experimental settings where parts of the epitopes may have changed due to denaturation (i.e., in western blots or fixed tissue sections), and the recognition of several epitopes on the same molecule can increase the detection signal in an assay.35 These advantages can be exploited, as the presence of the unknown fraction of unwanted IgGs in polyclonal preparations is excluded. Multiclonal antibodies,36 composed of an exactly defined mixture of sequence-defined recombinant antibody clones, provide the polyclonal advantage, resulting in elimination of false positive signals, and they provide lower background because they do not contain any potentially misleading off-target binders.37
The development of hybridoma technologies in 1975 was a substantial advance in antibody generation methods.38 Hybridoma generation relies on the fusion of two cells: a B cell encoding the desired antibody and a tumor cell that provides the immortality necessary for unlimited future cultivation to produce the monoclonal. Monoclonal antibodies originating from a single immortalized B cell revolutionized many areas of life sciences. For example, the knowledge explosion in molecular immunology was initiated and driven by a rapidly growing collection of monoclonal antibodies to lymphocyte surface markers. Monoclonal antibody clones literally defined cluster of differentiation (CD) antigens, allowing researchers for the first time to distinguish between the many different cell types of the adaptive immune system, which until then had all looked the same under the microscope.39 They also enabled the development of the first therapeutic monoclonal antibodies from the 1980s onwards.40
For decades, monoclonals provided the gold standard for specificity, as they represented a huge improvement in specificity over polyclonal serum products. Recent results, however, explain why some monoclonals are not monospecific.27 First, two or more VHDJH or VLJL recombination patterns of immunoglobulin heavy or light chains have been found in many single B cells.41 Second, due to the cell fusion that leads to hybridomas, these are initially tetraploid and carry four different antibody alleles. Hybridoma cells grow like cancers and are not regulated by a surrounding immune system. As a result, the number of chromosomes found in hybridomas varies widely; some hybridomas were shown to have developed up to 50 additional chromosomes.42–45 A multicentric study that sequenced expressed antibody genes of 185 hybridomas, including a large number of viable research catalog products, showed that only 68.1% of the analyzed hybridomas expressed only one of their four alleles, while the rest produced additional antibody chains (mainly light chains), which resulted in the secretion of more than one antibody by many truly monoclonal cell lines.13 This led to false positive reactivities and lower sensitivity.
This problem can be solved by cloning the variable regions of the antibodies from the hybridomas to generate recombinant versions. Many methods are available to do this, most prominently by PCR46,47 or next-generation sequencing.48,49 Once the correct pair of VH and VL is identified, these can be re-inserted into the genes encoding the chains for a full IgG and efficiently produced in mammalian cell culture.50 Nowadays, both transient production methods with HEK293 cells51,52 and stable cell lines, typically CHO,53 are well established for recombinant IgG and often provide yields of over 5 g/l. All recombinant immunoglobulins resulting from such a clone have the same structure and specificity. Importantly, this recombinant approach can also be used if the hybridoma is contaminated with bacteria or fungi, produces antibodies with undesirable glycosylations, does not adapt to the cultivation medium or is no longer sufficiently productive. These problems54,55 were usually solved by a passage of the hybridoma line through the abdomen of a living mouse,56 where they generate an ascites tumor that is very painful for the laboratory animals.57
Due to the knowledge of their sequence, recombinant antibodies are therefore always structurally defined reagents, in contrast to the vast majority of monoclonal and polyclonal research antibodies available from commercial suppliers. As a result, recombinant antibodies improve the reproducibility of research because they can be provided as long as the sequence information is known – a remarkable logistic advantage compared to the efforts necessary to keep hybridomas available in liquid nitrogen tanks. A substantial fraction of experiments published with monoclonals cannot be repeated because the hybridoma clones are no longer available. In contrast, databases collecting antibody sequences have been established to support unlimited reproducibility of experiments with recombinant antibodies.58
Beyond IgG: the choice of different antibody formats
Once available, antibody variable region DNA can be cloned into vectors encoding various other formats or other Ig types. Most methods using E.coli as expression host and most in vitro selection methods use just antigen-binding portions of the antibodies, like single-chain variable fragments (scFvs),59,60 antigen-binding fragments (Fabs)61 or scFab.62 Regardless of the format in which the antibodies were originally selected, the final application format is usually different today, as format conversions are uncomplicated in recombinant production systems. Various antibody formats are currently used for research (Figure 3).
Figure 3.
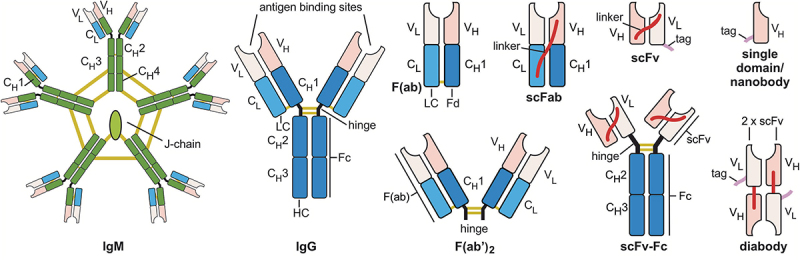
Antibody formats most frequently used in research assays: IgM, Immunoglobulin M; IgG, Immunoglobulin G; F(ab’)2, divalent fragment antigen binding; F(ab), fragment antigen binding, now usually named Fab; scFab, single chain fragment antigen binding; scFv-Fc, single chain fragment variable::Fc fusion protein; scFv, single chain fragment variable; diabody, dimerized scFv; and single domain antibodies/nanobody. Constant regions are colored dark blue (γ chains), green (µ chains), or light blue (κ or λ chains); variable (antigen binding) regions in pink. Yellow lines indicate disulfide bonds, red lines peptide linker, purple lines peptide tags. C, constant region, V, variable region, HC, heavy chain; LC light chain. Elements are not drawn to scale.
IgG is the most commonly used antibody format in research. IgG molecules offer two antigen-binding sites and thus improved apparent affinity in all assays where antigen density and localization support avidity effects. Their Fc region is easily recognized by secondary antibodies for detection, and protein A enables efficient purification of all molecules containing an Fc moiety in one step.63 Polyclonal antibodies are often supplied as IgG-enriched fractions, and most hybridoma-derived antibodies are supplied as purified IgG fractions. However, some hybridomas only produce IgM,64 which provides higher avidity due to their 10-12 antigen-binding sites, but must be carefully checked for cross-reactivity, as IgM represent an earlier stage of antibody maturation in the B cell and typically have lower monovalent affinities per antigen-binding site.
Antibody fragments can be either generated on the protein level by chemical/proteolytic cleavage of animal-derived IgG preparations, or by respective design of the antibody genes for recombinant production.65 F(ab’)2 are generated by enzymatic cleavage of immunoglobulins from animals or humans.66 They have two antigen-binding sites of an IgG but can be used in assays that would be interfered with by the presence of Fc parts, e.g., when using material with a high content of Fc receptors. Due to their smaller size, they penetrate tissue more efficiently. They allow double staining when combined with an IgG, as they can be detected by anti-Fab-specific secondary antibodies that do not interfere with anti-Fc-mediated antibody detection.
Fragments antigen-binding, F(ab), also known as Fab, can be produced either by chemical reduction of the disulfide bonds in the hinge of F(ab’)2,67 or recombinantly in E. coli or eukaryotes.68 Fabs provide monovalent antigen binding, which in some assays can result in much weaker signals compared to the IgG from which they are derived due to the lack of avidity. Recombinant Fab are expressed from two separate open reading frames (encoding the light chain and the corresponding Fd fragment). They are commonly used in phage display and mammalian cell display, and are the most efficient format for the expression of functional antibodies by yeast surface display.69 Single chain fragments antigen binding (scFab) are a variant of Fabs in which the light chain and the corresponding Fd fragment (VH and CH1) are joined by a peptide linker to be expressed from a single open reading frame.62 They can only be produced recombinantly. Their main advantage is that only one gene fragment needs to be transferred when subcloning the Fab fragment into other formats.70 However, scFab:Fc fusions (single chain immunoglobulin G, scIgG) tend to form higher-order aggregates.71 ScFab have also been used to generate mammalian cell surface expression libraries for in vitro selection using transposon technology.72
Single chain fragment variable:Fc fusion proteins (scFv-Fc) offer the advantage of full compatibility with common secondary anti-Fc fragment antibodies for their detection and generic protein A affinity chromatography for their purification, while requiring cloning of only one gene fragment encoding both the VH and VL regions that form the complete antigen-binding site,73 for example, after in vitro selection from an scFv gene library. ScFv-Fc makes up a large part of the non-animal derived catalog antibodies, as their subcloning and production are less costly compared to conversion to full IgG, while they can be used just like IgG.
Single chain fragment variable (scFv) is the smallest stable constructs to contain the full antigen-binding site.59,60 As they are produced with high yield in E. coli, they are often used in antibody phage display libraries. ScFv provides monovalent antigen binding, but because they lack structural stabilization by the constant regions, some of them can form bivalent homodimers (diabodies), which may have a higher apparent affinity due to avidity. This can lead to a different performance of the antibodies in assays where avidity plays a role compared to assays that only detect monovalent binding. Diabody formation can be forced by shortening the linker between the V regions.74 This provides a simple way to produce bivalent antibodies in E. coli. However, it should be noted that in diabodies the flexibility in the spatial orientation of the two antigen-binding sites (paratopes) is severely restricted due to their rather rigid conformation, whereas the antigen-binding sites of naïve IgG have much greater freedom of movement to contact their antigens due to the flexibility of the hinge region.75 As a result, the gain in apparent affinity by avidity may differ between a diabody and an IgG with identical paratopes.
Single domain antibodies76 and nanobodies77 consist of only half of the antigen-binding site of an IgG (which comes from the heavy chain), as they have no light chain. They can be produced efficiently in E. coli. Some variants of them can efficiently fold in the cytoplasm,78 making them in particular suited to be used as intrabodies.79 Nanobodies derived from heavy-chain only IgG antibodies found in the Camelidae family have an enlarged complementary-determining region 1 (CDR1) and an elongated CDR3 loop that facilitates the binding of antigens with a groove and allow binding to a different structural spectrum of antigens than IgG.80
ScFvs, diabodies and nanobodies can be used for research purposes, but usually require an additional incubation step for detection, as they do not have constant regions/Fc fragments that can be recognized by conventional secondary antibodies. Although the conserved framework sequences within V regions of some scFvs and nanobodies may be recognized by some preparations of anti-IgG polyclonals, this is not guaranteed for every single fragment due to their high variability. Therefore, peptide tags (usually between 4-15 amino acids residues in length) are added to the scFv or nanobodies to enable a generic detection and/or purification procedure. The c-myc tag is often used for this purpose. It can be detected either with the animal-derived monoclonal antibody Myc1-9E1081 or the affinity/stability-enhanced “vegan” Hypermyc antibody.82 His-tags provide simple generic purification,83,84 the FLAG-tag provides both purification and detection,85 Strep-tag86 or Avi-tag87 allow to use avidin and streptavidin-based reagents, and many other tags are available. To avoid the use of tags, scFv can be converted into Fab or IgG by subcloning. However, this can affect stability and antibody function,88 and some scFvs loose binding affinity when converted to full IgG format. Therefore, in vitro selections using surface display systems should be performed in formats as close as possible to the final application.89
To be useful in the laboratory, antibodies must not only bind to an antigen with high specificity and affinity but also have additional properties such as long-term stability, low aggregation in solution, high expression yield for low-cost production and more. These “developability” factors90 must be taken into account when producing antibodies for the low-margin research markets. While the “natural” IgG format is considered the most inherently stable variant, not all in vitro selection systems are capable of producing complete IgG molecules, which usually require mammalian cells for efficient production and correct folding. Gene libraries with a semi-synthetic design, in which natural complementarity-determining regions (CDRs) of antibodies are grafted into antibody frameworks of known “well-functioning” antibodies,91 or fully synthetic designs based on very stable scaffolds92 offer a way to generate good research reagents with minimal effort.
Since some scFvs and the widely used scFv-Fc fusions derived from in vitro selections have a higher tendency to aggregate, this parameter must be checked early in the antibody selection process to exclude molecules with developability liabilities. It is important to note that these are properties of individual scFv molecules, as the scFv format is by no means always less stable. Very robust scFv reagents can be obtained directly from libraries93 or by in vitro optimization.94 This is also illustrated by many clinically used fusion proteins that contain scFvs, like the chimeric antigen receptors (CAR) where the largest group of extracellular-binding regions are scFvs,95 or bispecific antibodies96 and immunotoxins.97 For research applications, however, conversion to a full IgG is often the chosen solution to provide full compatibility to the assays established for animal-derived antibodies. This conversion can also be used to add functionalities that are not easily available from animal-derived antibodies. For example, by cloning the variable regions of different antibodies into vectors encoding Fc parts of different species, antibodies originating from the same library can be converted to appear as IgG from different species, and detected by differently labeled detection reagents. Combined in one experiment, their use in combination with Fc-specific anti-rabbit, anti-rat, anti-human, or anti-mouse secondary antibodies for detection allows for multicolor analyzes not possible in the same way with a set of mouse monoclonals or rabbit polyclonals.98 Conversion to other formats like IgA, IgE or IgM are also possible in case the respective functionality is needed. Even non-canonical amino acids can be inserted at defined positions during cell culture production to provide functionalities not achievable with animal-produced antibodies.99,100 Last but not least, the availability of antibody variable (V) region DNA has allowed construction of a vast and growing zoo of non-IgG-like fusion proteins. The resulting bispecific antibodies,101 immunotoxins,102 or CAR-T cells103 have already provided many new and successful therapeutic approaches.
Making antibodies without using animals
All the advantages of recombinant immunoglobulins also apply to antibodies developed entirely in vitro because these methods are based on antibody genes that are recombinantly expressed in cultivated cells, so that their genetic blueprint is always available. The first successful and still prevalent method to generate antibodies completely without immunization and independent from a living immune system is antibody phage display.3–5 It employs highly diverse gene libraries of rearranged immunoglobulin genes (1010-1011 different clones), obtained either from B cells (often human), chemical synthesis or by a mix of both. These libraries encode a collection of variable (V) regions of antibodies that structurally determine the paratope (antigen-binding surface) of the antibody, and thus its specificity and affinity. “Universal” (also named “naïve”) antibody gene libraries can be made without the use of animals.104 These libraries are cloned into a phagemid vector4 which, when present together with a helper phage genome in the same E. coli cell, leads to the production of antibody phage that have a physical linkage of the antibody protein to the DNA encoding it. Mixtures of antibody-phage, containing billions of different functional antibody fragments, each linked to their encoding genes, are then incubated with immobilized antigen. While the vast majority of antibody-carrying phage does not interact with the antigen, a few antibody-phage are bound to the antigen and can be eluted, after washing away the unbound ones. As they are phage, after elution they are able to very efficiently re-infect E. coli. The antibody-phage also carry a resistance marker that allows only the antibody-gene carrying bacteria to multiply. DNA encoding antigen-specific antibody genes can easily be isolated from the resulting E. coli colonies. This process, called “panning”, relies on individual single molecule interactions to identify the sequences of recombinant monoclonal antibodies.
The antibody DNA can be ligated into many different vectors providing a final antibody format of choice. Typically, it is cloned into suitable IgG expression vectors to produce the full monospecific immunoglobulin.105 These immunoglobulins carry all functions of IgG from animal sources, and consequently are applicable to any assay that has been developed for animal-derived antibodies,15 as they are structurally identical to them. More recently developed in vitro antibody selection methods, like yeast display106 or mammalian cell display,107,108 are based on the same principle: clonal selection from vast DNA repertoires using the antigen-binding function of antibodies that are physically attached to the cell carrying the encoding gene. These methods offer the advantage of glycosylation and the display of larger antibody formats including full IgG, which is extremely inefficiently expressed in E. coli. Although the diversity of the repertoire of antibody genes that can be utilized with eukaryotic cell display methods still is several orders of magnitude lower than the diversity of 1010-1011 different antibody genes typically used in phage display,109 complex selections can be made in cell display systems that would be much more difficult to perform with phage display. For example, with appropriate labeling in the selection step of flow cytometry, both antibody expression and antigen binding can be quantified simultaneously for each individual antibody-expressing cell. With the right gating in flow cytometry, significantly higher enrichment factors can be achieved than with phage panning, reducing the number of selection rounds required. A further advantage of these approaches is that they enable integrated selection for antibody developability. Here, the mammalian display can select not only for antigen binding but also simultaneously for well-expressing and stable antibody candidates. This is best achieved by displaying the “final” full IgG format. In addition, both display methods can be combined, using a phage display library with a diversity that cannot be achieved with cellular display in the first round of selection to obtain more initial binders, followed by batch subcloning into a cellular display system for refinement 110.
Phage display and the other surface display methods allow the generation of specific immunoglobulins against everything that can bind to antibodies with reasonable affinity, as they are not restricted by the bottleneck of peptide presentation on MHC (Major Histocompatibility Complex). This allows to obtain antibodies against antigens that cannot be handled by a living immune system, like toxins or non-peptidic materials. In addition, most in vitro display libraries use random pairing of light and heavy chains, which means that the chance of achieving an optimal VH/VL match is much greater than in any B cell. This is a significant advantage compared to antibody selection in animals, where B cells can only produce two rearranged versions of the light chain per heavy chain.111 This limitation is one reason why repeated immunizations (boosts) are necessary in animals to increase affinities by somatic hypermutation. In contrast, by using in vitro display libraries with random VH/VL pairing, similar affinity increases (>1000 ×) can be achieved without introducing point mutations, just by the identification of a better fitting light chain from repertoires of > 106 VL region genes.112 This process, named “chain shuffling”, exchanges the light chain of a given antibody with the full light-chain gene repertoire before performing a new phage, yeast or mammalian display selection.113 Since both chains of the resulting antibodies have sequences with a much lower number of mutations compared to the human germline, the probability of developing adverse immune reactions in patients treated with these antibodies should be lower than with somatically hypermutated IgG. Combined with careful adaptation of the selection conditions, even the kinetic properties of antigen binding can deliberately be changed while keeping specificity and affinity identical.114 Of course, somatic hypermutation can also be reproduced in vitro by adding point mutations prior to in vitro selection.94
The complete control over the biochemical milieu at the very moment of antibody-phage selection is another advantage of in vitro selections.114,115 Antibodies with pre-adapted properties can be obtained in this way. For example, the buffer conditions of the final assay the antibody has to work in can be used during its selection, yielding only antibodies that survive these buffer conditions. Even the kinetic properties of an antibody can be adjusted: simple prolongation of the washing steps during the panning selects for antibodies with increased stability and slower dissociation.94 Unwanted cross-reactivities can be counter-selected against by adding soluble competitors during the panning step. The other way round, sequential affinity selections on different homologous proteins can identify a common structural motive.116 This can be used to select for antibodies that cross-react with homolog proteins from different species – a feature that is important for preclinical development that relies on animal models to demonstrate efficacy and test for target-related toxicity.
In the future, combining sequencing of a person’s entire B-cell receptor immune repertoire (BCR sequencing) with deep-learning algorithms to identify disease related antibody sequences117 or de-novo prediction of antigen binding site (paratope) structures16 may offer additional ways to identify specific antibodies.
Why do different origins of antibodies matter?
The properties of polyclonal, monoclonal, and recombinant antibodies, in particular their specificity, false positive reactivities, recognized epitopes and reproducibility, differ significantly (Figure 1 right). More rigorous validation certainly would be beneficial for all types of research antibodies. A systematic study analyzing 614 commercially available antibodies targeting 65 protein targets24 underlined the paramount importance of validation of every individual antibody for each application. Polyclonal, hybridoma-derived, and recombinant antibodies were validated for their performance in western blot, immunofluorescence, and immunoprecipitation. Within this set of antibodies, recombinant antibodies performed better when directly compared to hybridoma-derived monoclonals and polyclonals directed at the same protein. Also, some of the assumed advantages of polyclonals were not evident, as these did not outperform the monoclonal antibodies in immunoprecipitation and were universally worse across the three applications tested. In another study, recombinant IgG cloned from hybridoma lines performed better than the IgG purified directly from supernatants from that same monoclonal hybridoma line.13 In conclusion, we should at least minimize the risks of false positive reactivities that originate in the undefined nature of polyclonals118 by using monoclonal, or even better, recombinant antibodies whenever possible. Here, we should be aware that additional reactivities can be present in some hybridoma-derived antibodies despite the origin from a single clone.13 However, the latter risk is far lower than that originating from the use of polyclonal antibodies. Luckily, the availability of suitable recombinant technologies enables the conversion of both reagent types to completely defined reagents with known polypeptide sequence.
As long as the animal that produced a polyclonal serum is still available, single B cell cloning119 or V region libraries generated from lymphatic tissue or blood combined with phage or yeast display120 can be used to save the information on the immune response against the antigen used for immunization, by generating one or several individual recombinant antibody clones, which should provide similar specificities completely without the contamination by a large fraction of unknown IgG. Most importantly, this conversion allows unlimited production of these antibodies in the future, in contrast to the limited amount of serum-based polyclonals, which is defined by the blood volume that a single animal can provide over its lifespan. Even the multi-epitope recognition advantage of polyclonal antibodies can be restored in recombinant form by using multiclonals37 that avoid the disadvantages of serum products.
The advantages of the recombinant formats and vast gene libraries inspired the creation of many approaches that use a mix of animal-based and in vitro methods. As a consequence, antibody phage display is often not animal free. Antibody gene repertoires isolated from immunized animals are frequently used to combine the advantages of the in vitro selection process with the results of immunizing a natural immune system. For example, nanobodies can be identified from phage display libraries generated from the blood cells of immunized llamas or camels.121
Another approach is offered by transgenic animals, which were originally created to avoid adverse side effects induced by a patient’s immune response against murine therapeutic antibodies.122 While genetic engineering allows for many different approaches to convert a mouse monoclonal to a more “human-like” sequence (humanization),123,124 a more direct approach was made possible by cloning the entire antibody gene repertoire of humans into mice, rats or rabbits which had their own IgG genes knocked out.125–127 Immunization of these “human IgG-transgenic” animals leads to the production of antibodies using the human sequence repertoire – a major source of the currently used therapeutic antibodies.
By amplification of the antibody genes from single B cells,119 recombinant versions of antibodies can be generated very quickly, as recently demonstrated during the SARS-CoV-2 pandemic.128 The race to produce therapeutic antibodies against the coronavirus also highlighted the potential of the phage display antibody to generate antibodies against antigens previously unknown to the human immune system. Before the first COVID patient materials were available in Europe, universal phage display libraries provided SARS-CoV-2 neutralizing antibodies within a few weeks, as the only material that was needed was the sequence information of the antigen.129 So, antibody phage display demonstrated the capability to identify binders against newly emerging pathogens more rapidly than any animal immunization-based approach can,130 and today’s naïve/universal antibody libraries for in vitro surface display now match immune sources for the affinities of antibodies that can be obtained.91,131–133
With all of these advantages, one might wonder why recombinant antibodies are not used much more frequently. In fact, while they represent less than 5% of the research reagents (Figure 2), all monoclonal antibody products currently used in human therapy are recombinant. The explanation for this dramatic difference in the acceptance of an obviously superior technology is complicated but is clearly based on their intended use. A report by the European Union Reference Laboratory for Alternatives to Animal Testing (EURL ECVAM)15 found that a combination of a lack of knowledge about recombinant formats and scientific misconceptions, including preconceptions about new methods stemming from issues encountered during the early days of the technology, contributed to the causes. The most important factors, however, are the limited availability and the usually higher price. Even antibodies derived from hybridomas, which have been available for almost half a century and have much better properties, have not been able to replace polyclonals as research reagents for the same reason (Figure 2), as they are around 5–10 times more expensive than polyclonals. Generation of new recombinant antibodies currently costs at least the same or even more than hybridoma generation, mainly due to their still small market share. Wider adoption of recombinant research reagents that meet the optimal standards available today would therefore require significant investments to broaden the industrial base for their supply, which should also lead to a significant reduction in price. Notably, the current research catalog prices of animal-free recombinants are not higher when compared to animal-derived antibodies.
Vegan antibodies?
The technology currently available can provide DNA blueprints for highly specific antibodies with high affinity without the need for animal experiments. In particular, antibody phage display has yielded many research antibodies.134,135 Non-animal-derived antibodies are used in diagnostics without being identified as such, and a growing number of antibodies obtained by animal-free methods are used as therapeutics.136 Consequently, the ECVAM report concluded that methods that do not involve animals can now yield research antibodies as good as animal-derived antibodies,15 and thousands are already available in catalogs today (Figure 2; see also https://afa.ehstaging.net/).
Nevertheless, it is not always easy for researchers who want to avoid animal use to understand which antibodies fulfill this criterion. Importantly, in vitro antibody production methods are not a priori animal-free. To identify those approaches that would qualify as “vegan” according to the practical criteria for animal use set forth in the introduction, we first have to look at the origins of the V region DNA. Antibody V genes can be collected from the genomes or transcriptomes of non-immunized or immunized animal or human donors, or they can be synthesized chemically, typically in the form of “synthetic” libraries.137–139 The latter are based on one or a few antibody V region genes (frameworks) taken from databases. Only the loops directly contacting the antigen (CDRs) are randomized during DNA synthesis to provide the structural diversity within the antigen binding site.140 Even in vitro cell-based methods to replicate both primary and secondary antibody diversification mechanisms were conceived.141
While human antibody libraries usually qualify as non-animal derived, not all antibodies labeled as “human antibodies” do. They could have been produced in human IgG transgenic animals after immunization,125–127 or originate from humanizing a hybridoma.123,124 A pragmatic approach in respect of minimizing animal use would be to check whether the antibody diversity originates from animal immunization or directly from an animal used for this purpose, or whether it just uses antibody structures originating from databases or from human blood donations.
Another route to recombinant antibodies uses V genes identified by sequencing hybridomas. This is a valuable path to further utilize the huge trove of historically created monoclonal antibodies. It would be detrimental to the progress of science to forego their use, since the animal experiments for their generation are finished, and if done right, no new animals are needed for their ongoing production. Instead of using the ascites method56 to save deteriorating or contaminated hybridoma lines, recombinant production could be used. Moreover, an “Antibody Heritage Program”142 to generate recombinant versions of all available hybridomas would not cause more animal use, but assure the long-term survival of valuable monoclonals and upgrade them with the advantages of the recombinant format. This would even allow their fully vegan production – of course, not qualifying them as vegan antibodies, but it would be a step to reduce animal suffering.
Although they are still rarely found in research reagent catalogs, a large number of alternative molecular scaffolds has been developed over the past decades that could replace the antibody molecule in many usual assays.143 However, many of them need different detection systems, for example by using anti-tag antibodies, which takes away some of their original advantages and still limits their routine use in research today. All of these are initially identified by in vitro selection methods analogous to those used for antibody generation, and that do not require animals.
While antibody discovery from gene libraries can be done with or without animal experiments, the same is also true for their recombinant production (Figure 1). Here, sequences obtained by gene synthesis from databases or from clones established decades ago that encode the constant regions of different animals are always required to reconstruct the V regions into full IgG. Only this format is fully interchangeable for animal-derived antibodies in typical research applications.15 While human Fc parts that meet the “non-animal-derived” criterion could be used in many applications, they are less suitable when analyzing human samples and tissues that naturally contain immunoglobulins. Consequently, recombinant IgG versions with the constant parts from animals such as mice, rats, rabbits will be needed for research. Of course, new animal experiments will never be necessary to use this constant region DNA, as all the necessary gene fragments have long been made available in commercially sourced vectors.
The actual use of animal materials in the protein production step of non-animal-derived in vitro generated antibodies is often much more difficult to identify. While the inhumane ascites method to produce monoclonals from hybridoma tumors in mice56 has been used to generate monoclonals, it is not used for the production of recombinant antibodies. Nevertheless, in vitro production in cell culture often relied on animal materials, such as fetal calf serum in the growth medium. However, defined media that completely avoid the use of animal-derived reagents are available for both hybridomas144 and recombinant antibody-expressing cells.51,52 In addition, various systems for the plant-based production of antibodies have been developed,145 offering an undisputable vegan solution. Along the same lines, antibodies can be made in yeasts, which have proven to be particularly useful to provide defined glycosylation of the antibodies. For example, glycoengineered Komagataella (formerly named Pichia) pastoris strains were used for the production of humanized mAbs with glycosylation patterns optimized for therapeutic use.146–148
Various materials derived from animals may also be used in the assays that are an integral part of the discovery and production processes. Bovine serum albumin or skimmed milk powder is typically used as blocking reagents in the ELISA screening required to identify the correct clones or test the correct function of production batches. These animal-derived reagents can be replaced by chemically synthesized or plant-derived materials (Abcalis, personal communication). While we cannot completely avoid animal use in the development of therapeutic antibodies,149 we now have a choice to go the “vegan” way of antibody making for many research applications.
The author would like to reiterate his opinion that legal restrictions on the use of animals for the production of antibodies would considerably hinder research and the development of drugs.149 Rather, efforts should be made to improve general access to non-animal-derived antibodies, allowing more researchers to recognize the intrinsic advantages of in vitro-derived antibodies. Wider use of animal-free methods for the generation of antibodies should be encouraged, as the resulting use of defined recombinant reagents would be beneficial not only for the animals but also for the quality and reproducibility of our experimental results.
Acknowledgment
We thank Janice M. Reichert for her valuable comments to improve the manuscript.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure statement
The author is associated with companies that employ animal-free antibody generation, antibody engineering and clinical development of antibodies (mAb-factory GmbH, Yumab GmbH, Abcalis GmbH, Mabswitch Inc., Corat Therapeutics GmbH).
References
- 1.von Behring E, Kitasato S.. Über das Zustandekommen der Diphtherie-Immunität und der Tetanus-Immunität bei Thieren. Deutsche Medizinische Wochenzeitschrift. 1890;16:1113–13. [Google Scholar]
- 2.Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975. Aug 7;256(5517):495–97. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 3.Barbas CF, Kang AS, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA. 1991. Sep 15;88(18):7978–82. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitling F, Dübel S, Seehaus T, Klewinghaus I, Little M. A surface expression vector for antibody screening. Gene. 1991. Aug 15;104(2):147–53. doi: 10.1016/0378-1119(91)90244-6. [DOI] [PubMed] [Google Scholar]
- 5.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990. Dec 6;348(6301):552–54. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 6.Coons AH. Immunofluorescence. Public Health Rep. 1960. Oct;75(10):937–43. [PMC free article] [PubMed] [Google Scholar]
- 7.Crescioli S, Kaplon H, Chenoweth A, Wang L, Visweswaraiah J, Reichert JM. Antibodies to watch in 2024. Mabs-austin. 2024;16(1):2297450. doi: 10.1080/19420862.2023.2297450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen BX, Wilson SR, Das M, Coughlin DJ, Erlanger BF. Antigenicity of fullerenes: antibodies specific for fullerenes and their characteristics. Proc Natl Acad Sci U S A. 1998. Sep 1;95(18):10809–13. doi: 10.1073/pnas.95.18.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee CC, Su YC, Ko TP, Lin LL, Yang CY, Chang SSC, Roffler SR, Wang AHJ. Structural basis of polyethylene glycol recognition by antibody. J Biomed Sci. 2020. Jan 7;27(1):12. doi: 10.1186/s12929-019-0589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu TT, Kabat EA. An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J Exp Med. 1970. Aug 1;132(2):211–50. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Early P, Huang H, Davis M, Calame K, Hood L. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell. 1980. Apr;19(4):981–92. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- 12.Chi X, Li Y, Qiu X. V(D)J recombination, somatic hypermutation and class switch recombination of immunoglobulins: mechanism and regulation. Immunology. 2020. Jul;160(3):233–47. doi: 10.1111/imm.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradbury ARM, Trinklein ND, Thie H, Wilkinson IC, Tandon AK, Anderson S, Bladen CL, Jones B, Aldred SF, Bestagno M. et al. When monoclonal antibodies are not monospecific: Hybridomas frequently express additional functional variable regions. Mabs-austin. 2018. Jun;10(4):539–46. doi: 10.1080/19420862.2018.1445456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs P, Dübel S, Breitling F, Braunagel M, Klewinghaus I, Little M. Recombinant human monoclonal antibodies. Basic principles of the immune system transferred to E. coli. Cell Biophys. 1992;21(1–3):81–91. doi: 10.1007/BF02789480. [DOI] [PubMed] [Google Scholar]
- 15.Barroso J, Halder M, Whelan M. EURL ECVAM recommendation on non-animal-derived antibodies. Luxembourg: Publications Office of the European Union; 2020. [Google Scholar]
- 16.Syrlybaeva R, Strauch EM, Valencia A. Deep learning of protein sequence design of protein–protein interactions. Bioinformatics. 2023. Jan 1;39(1):btac733. doi: 10.1093/bioinformatics/btac733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hifumi T, Yamamoto A, Ato M, Sawabe K, Morokuma K, Morine N, Kondo Y, Noda E, Sakai A, Takahashi J. et al. Clinical serum therapy: benefits, cautions, and potential applications. Keio J Med. 2017. Dec 25;66(4):57–64. doi: 10.2302/kjm.2016-0017-IR. [DOI] [PubMed] [Google Scholar]
- 18.Bradbury A, Plückthun A. Reproducibility: Standardize antibodies used in research. Nature. 2015. Feb;518(7537):27–29. doi: 10.1038/518027a. [DOI] [PubMed] [Google Scholar]
- 19.Hjelm B, Forsström B, Löfblom J, Rockberg J, Uhlén M and Lu S. Parallel Immunizations of Rabbits Using the Same Antigen Yield Antibodies with Similar, but Not Identical, Epitopes. PLoS ONE. 2012;7(12):e45817. doi: 10.1371/journal.pone.0045817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Kong Y, Yu X, Yu W, Wen K, Shen J, Wang Z. Characteristics of rabbit hapten-specific and germline-based BCR repertoires following repeated immunization. One Health Adv. 2023 Jun 29;1(1):17. doi: 10.1186/s44280-023-00013-z. [DOI] [Google Scholar]
- 21.Englebienne P, Doyen G. An improved method for isolation of specific antibodies by affinity chromatography; application to an antiserum to testosterone. J Immunol Methods. 1983. Aug 26;62(2):197–204. doi: 10.1016/0022-1759(83)90247-8. [DOI] [PubMed] [Google Scholar]
- 22.Bradbury AM, Plückthun A. Antibodies: validate recombinants once. Nature. 2015. Apr;520(7547):295–295. doi: 10.1038/520295b. [DOI] [PubMed] [Google Scholar]
- 23.Laflamme C, McKeever PM, Kumar R, Schwartz J, Kolahdouzan M, Chen CX, You Z, Benaliouad F, Gileadi O, McBride HM. et al. Implementation of an antibody characterization procedure and application to the major ALS/FTD disease gene C9ORF72. Elife. 2019. Oct 15;8:e48363. doi: 10.7554/eLife.48363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayoubi R, Ryan J, Biddle MS, Alshafie W, Fotouhi M, Bolivar SG, Ruiz Moleon V, Eckmann P, Worrall D, McDowell I. et al. Scaling of an antibody validation procedure enables quantification of antibody performance in major research applications. eLife. 2023;12:RP91645. doi: 10.7554/eLife.91645.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012. Mar 28;483(7391):531–33. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 26.Algenäs C, Agaton C, Fagerberg L, Asplund A, Björling L, Björling E, Kampf C, Lundberg E, Nilsson P, Persson A. et al. Antibody performance in western blot applications is context-dependent. Biotechnol J. 2014. Mar;9(3):435–45. doi: 10.1002/biot.201300341. [DOI] [PubMed] [Google Scholar]
- 27.Berglund L, Björling E, Oksvold P, Fagerberg L, Asplund A, Szigyarto CAK, Persson A, Ottosson J, Wernérus H, Nilsson P. et al. A genecentric human protein atlas for expression profiles based on antibodies. Mol Cell Proteomics. 2008. Oct;7(10):2019–27. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Voskuil JLA, Bandrowski A, Begley CG, Bradbury ARM, Chalmers AD, Gomes AV, Hardcastle T, Lund-Johansen F, Plückthun A, Roncador G. et al. The antibody society’s antibody validation webinar series. Mabs-austin. 2020. Jan 1;12(1):1794421. doi: 10.1080/19420862.2020.1794421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elliott S, Busse L, Bass MB, Lu H, Sarosi I, Sinclair AM, Spahr C, Um M, Van G, Begley CG. et al. Anti-Epo receptor antibodies do not predict Epo receptor expression. Blood. 2006. Mar 1;107(5):1892–95. doi: 10.1182/blood-2005-10-4066. [DOI] [PubMed] [Google Scholar]
- 30.Andersson S, Sundberg M, Pristovsek N, Ibrahim A, Jonsson P, Katona B, Clausson C-M, Zieba A, Ramström M, Söderberg O. et al. Insufficient antibody validation challenges oestrogen receptor beta research. Nat Commun. 2017. Jun 15;8(1):15840. doi: 10.1038/ncomms15840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schrohl AS, Pedersen HC, Jensen SS, Nielsen SL, Brünner N. Human epidermal growth factor receptor 2 (HER2) immunoreactivity: specificity of three pharmacodiagnostic antibodies. Histopathology. 2011. Nov;59(5):975–83. doi: 10.1111/j.1365-2559.2011.04034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaezi AE, Bepler G, Bhagwat NR, Malysa A, Rubatt JM, Chen W, Hood BL, Conrads TP, Wang L, Kemp CE. et al. Choline phosphate cytidylyltransferase-α is a novel antigen detected by the anti-ERCC1 antibody 8F1 with biomarker value in patients with lung and head and neck squamous cell carcinomas. Cancer. 2014. Jun 15;120(12):1898–907. doi: 10.1002/cncr.28643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukinavičius G, Lavogina D, Gönczy P, Johnsson K. [Letter to the editor]: Commercial Cdk1 antibodies recognize the centrosomal protein Cep152. Biotechniques. 2013. Sep;55(3):111–14. doi: 10.2144/000114074. [DOI] [PubMed] [Google Scholar]
- 34.Baker M. Reproducibility crisis: Blame it on the antibodies. Nature. 2015. May 21;521(7552):274–76. doi: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- 35.Miyanishi S. General properties and kinds of antibodies for enzyme immunoassay–characteristics and advantages of polyclonal and monoclonal antibodies. Nihon Rinsho. 1995. Sep;53(9):2149–53. [PubMed] [Google Scholar]
- 36.Haurum J, Bregenholt S. Recombinant polyclonal antibodies: therapeutic antibody technologies come full circle. IDrugs. 2005. May;8(5):404–09. [PubMed] [Google Scholar]
- 37.Wenzel EV, Russo G, Dübel S. Multiklonale Antikörper als Ersatz für Zweitantikörper aus Seren. Biospektrum. 2020. Jun;26(4):416–17. doi: 10.1007/s12268-020-1401-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Köhler G, Milstein C. Continuous culture of fused cells secreting antibody of predefined specificity. Nature. 1975;256(5517):495–97. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 39.Zola H, Swart B. The human leucocyte differentiation antigens (HLDA) workshops: the evolving role of antibodies in research, diagnosis and therapy. Cell Res. 2005. Sep;15(9):691–94. doi: 10.1038/sj.cr.7290338. [DOI] [PubMed] [Google Scholar]
- 40.Dübel S, Reichert JM. Handbook of therapeutic antibodies. 2nd. Weinheim: Wiley-VCH; 2014. [Google Scholar]
- 41.Shi Z, Zhang Q, Yan H, Yang Y, Wang P, Zhang Y, Deng Z, Yu M, Zhou W, Wang Q. et al. More than one antibody of individual B cells revealed by single-cell immune profiling. Cell Discov. 2019;5(1):64. doi: 10.1038/s41421-019-0137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kontsek P, Novák M, Kontseková E. Karyotype analysis of hybridomas producing monoclonal antibodies against different antigens. Folia Biol (Praha). 1988;34:99–104. [PubMed] [Google Scholar]
- 43.Rodova MA, LA T, Kushch AA, Novokhatskiĭ AS. Karyological analysis of hybridoma lines producing monoclonal antibodies to viral antigens. Tsitol Genet. 1985;19:425–28. [PubMed] [Google Scholar]
- 44.Wollweber L, Münster H, Hoffmann S, Siller K, Greulich KO. Early phase karyotype analysis of chromosome segregation after formation of mouse-mouse hybridomas with chromosome painting probes. Chromosome Res. 2000;8(1):37–44. doi: 10.1023/A:1009223102068. [DOI] [PubMed] [Google Scholar]
- 45.Zhil’tsova MA, Trofimova MN, Novikov VV.. Karyological analysis of hybridoma cells after prolonged cultivation. Zh Mikrobiol Epidemiol Immunobiol. 1989. Jun;6:99–102. [PubMed] [Google Scholar]
- 46.Breitling, F., and Dübel, S. (1997). Cloning and Expression of Single Chain Fragments [scFv] from Mouse and Rat Hybridomas. In 'Molecular Diagnosis of Infectious Diseases' in the series 'Methods in Molecular Medicine, Vol 13', U. Reischl, ed. (Humana Press Inc//Totowa, NY), pp. 581-591. [DOI] [PubMed] [Google Scholar]
- 47.Larrick JW, Danielsson L, Brenner CA, Abrahamson M, Fry KE, Borrebaeck CA. Rapid cloning of rearranged immunoglobulin genes from human hybridoma cells using mixed primers and the polymerase chain reaction. Biochem Biophys Res Commun. 1989. May 15;160(3):1250–56. doi: 10.1016/S0006-291X(89)80138-X. [DOI] [PubMed] [Google Scholar]
- 48.Mitchell KG, Gong B, Hunter SS, Burkart-Waco D, Gavira-O’Neill CE, Templeton KM, Goethel ME, Bzymek M, MacNiven LM, Murray KD. et al. High-volume hybridoma sequencing on the NeuroMabSeq platform enables efficient generation of recombinant monoclonal antibodies and scFvs for neuroscience research. Sci Rep. 2023. Sep 27;13(1):16200. doi: 10.1038/s41598-023-43233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subas Satish HP, Zeglinski K, Uren RT, Nutt SL, Ritchie ME, Gouil Q, Kluck, RM. Nab-seq: an accurate, rapid, and cost-effective method for antibody long-read sequencing in hybridoma cell lines and single B cells. Mabs-austin. 2022;14(1):2106621. doi: 10.1080/19420862.2022.2106621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frenzel A, Hust M, Schirrmann T. Expression of recombinant Antibodies. Front Immunol. 2013. Jul 29;4:217. doi: 10.3389/fimmu.2013.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Backliwal G, Hildinger M, Chenuet S, Wulhfard S, De Jesus M, Wurm FM. Rational vector design and multi-pathway modulation of HEK 293E cells yield recombinant antibody titers exceeding 1 g/l by transient transfection under serum-free conditions. Nucleic Acids Res. 2008. Sep;36(15):e96. doi: 10.1093/nar/gkn423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jäger V, Büssow K, Wagner A, Weber S, Hust M, Frenzel A, Schirrmann T. High level transient production of recombinant antibodies and antibody fusion proteins in HEK293 cells. BMC Biotechnol. 2013. Jun 26;13(1):52. doi: 10.1186/1472-6750-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reinhart D, Damjanovic L, Kaisermayer C, Sommeregger W, Gili A, Gasselhuber B, Castan A, Mayrhofer P, Grünwald-Gruber C, Kunert R. et al. Bioprocessing of recombinant CHO-K1, CHO-DG44, and CHO-S: CHO expression hosts favor either mAb production or biomass synthesis. Biotechnol J. 2019. Mar;14(3):e1700686. doi: 10.1002/biot.201700686. [DOI] [PubMed] [Google Scholar]
- 54.Leenaars M, Hendriksen CFM. Critical steps in the production of polyclonal and monoclonal antibodies: evaluation and recommendations. ILAR J. 2005;46(3):269–79. doi: 10.1093/ilar.46.3.269. [DOI] [PubMed] [Google Scholar]
- 55.Council NR. Monoclonal antibody production [Internet]. Washington, DC: The National Academies Press; 1999. https://nap.nationalacademies.org/catalog/9450/monoclonal-antibody-production [PubMed] [Google Scholar]
- 56.Yokoyama WM. Production of monoclonal antibody supernatant and ascites fluid. Curr Protoc Mol Biol. Jul 2008;83(1):Chapter 11:Unit 11.10. doi: 10.1002/0471142727.mb1110s83. [DOI] [PubMed] [Google Scholar]
- 57.Jackson LR, Trudel LJ, Fox JG, Lipman NS. Monoclonal antibody production in murine ascites. I. Clinical and pathologic features. Lab Anim Sci. 1999. Feb;49(1):70–80. [PubMed] [Google Scholar]
- 58.Lima WC, Gasteiger E, Marcatili P, Duek P, Bairoch A, Cosson P. The ABCD database: a repository for chemically defined antibodies. Nucleic Acids Res. 2020. Jan 8;48(D1):D261–4. doi: 10.1093/nar/gkz714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bird RE, Hardman KD, Jacobson JW, Johnson S, Kaufman BM, Lee SM, Lee T, Pope SH, Riordan GS, Whitlow M. et al. Single-chain antigen-binding proteins. Science. 1988. Oct 21;242(4877):423–26. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- 60.Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotný J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R. et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci USA. 1988. Aug;85(16):5879–83. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nisonoff A, Wissler FC, Lipman LN, Woernley DL. Separation of univalent fragments from the bivalent rabbit antibody molecule by reduction of disulfide bonds. Arch Biochem Biophys. 1960. Aug;89(2):230–44. doi: 10.1016/0003-9861(60)90049-7. [DOI] [PubMed] [Google Scholar]
- 62.Hust M, Jostock T, Menzel C, Voedisch B, Mohr A, Brenneis M, Kirsch MI, Meier D, Dübel S. Single chain Fab (scFab) fragment. BMC Biotechnol. 2007;7(1):14. doi: 10.1186/1472-6750-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goding JW. Use of staphylococcal protein a as an immunological reagent. J Immunol Methods. 1978;20:241–53. doi: 10.1016/0022-1759(78)90259-4. [DOI] [PubMed] [Google Scholar]
- 64.Köhler G, Hengartner H, Shulman MJ. Immunoglobulin production by lymphocyte hybridomas. Eur J Immunol. 1978. Feb;8(2):82–88. doi: 10.1002/eji.1830080203. [DOI] [PubMed] [Google Scholar]
- 65.Nelson AL. Antibody fragments: hope and hype. Mabs-austin. 2010;2(1):77–83. doi: 10.4161/mabs.2.1.10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Medgyesi GA, Gergely J. Structural studies of immunoglobulins. 3. Susceptibility of human gamma G globulins to peptic hydrolysis and papain sensitivity of F(ab’)2 fragments. Immunochemistry. 1969. May;6(3):473–79. doi: 10.1016/0019-2791(69)90304-8. [DOI] [PubMed] [Google Scholar]
- 67.Dorrington KJ, Carneiro L, Munro DS. The proteolysis of immunoglobulin G with long-acting thyroid-stimulating activity. Biochem J. 1966. Mar;98(3):858–61. doi: 10.1042/bj0980858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Plückthun A, Glockshuber R, Skerra A, Stadmüller J. Properties of FV and Fab fragments of the antibody McPC603 expressed in E. coli. Behring Inst Mitt. 1990. Dec;87:48–55. [PubMed] [Google Scholar]
- 69.Sivelle C, Sierocki R, Ferreira-Pinto K, Simon S, Maillere B, Nozach H. Fab is the most efficient format to express functional antibodies by yeast surface display. Mabs-austin. 2018. Jul;10(5):720–29. doi: 10.1080/19420862.2018.1468952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanna R, Cardarelli L, Patel N, Blazer LL, Adams JJ, Sidhu SS. A phage-displayed single-chain Fab library optimized for rapid production of single-chain IgGs. Protein Sci. 2020. Oct;29(10):2075–84. doi: 10.1002/pro.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schirrmann T, Menzel C, Hust M, Prilop J, Jostock T, Dübel S. Oligomeric forms of single chain immunoglobulin (scIgG). Mabs-austin. 2010. Jan;2(1):73–76. doi: 10.4161/mabs.2.1.10784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang J, Rader C, Peng H. A mammalian cell display platform based on scFab transposition. Antib Ther. 2023. Jul;6(3):157–69. doi: 10.1093/abt/tbad009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li SL, Liang SJ, Guo N, Wu AM, Fujita-Yamaguchi Y. Single-chain antibodies against human insulin-like growth factor I receptor: expression, purification, and effect on tumor growth. Cancer Immunol Immunother. 2000. Jul;49(4–5):243–52. doi: 10.1007/s002620000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmiedl A, Zimmermann J, Scherberich JE, Fischer P, Dübel S. Recombinant variants of antibody 138H11 against human gamma-glutamyltransferase for targeting renal cell carcinoma. Hum Antibodies. 2006;15(3):81–94. doi: 10.3233/HAB-2006-15303. [DOI] [PubMed] [Google Scholar]
- 75.Zhang X, Zhang L, Tong H, Peng B, Rames MJ, Zhang S, Ren G. 3D structural fluctuation of IgG1 antibody revealed by individual particle electron tomography. Sci Rep. 2015;5(1):9803. doi: 10.1038/srep09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Drabek D, Janssens R, de Boer E, Rademaker R, Kloess J, Skehel J, Grosveld F. Expression cloning and production of human heavy-chain-only antibodies from murine transgenic plasma cells. Front Immunol. 2016;7:619. doi: 10.3389/fimmu.2016.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82(1):775–97. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 78.Wagner TR, Rothbauer U. Nanobodies right in the middle: intrabodies as toolbox to visualize and modulate antigens in the living cell. Biomolecules. 2020 Dec 21;10(12):1701. doi: 10.3390/biom10121701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang C, Ötjengerdes RM, Roewe J, Mejias R, Marschall ALJ. Applying antibodies inside cells: principles and recent advances in neurobiology, virology and oncology. BioDrugs. 2020. Aug;34(4):435–62. doi: 10.1007/s40259-020-00419-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jin BK, Odongo S, Radwanska M, Magez S. Nanobodies: a review of generation, diagnostics and therapeutics. Int J Mol Sci. 2023 Mar 22;24(6):5994. doi: 10.3390/ijms24065994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell biol. 1985. Dec;5(12):3610–16. doi: 10.1128/MCB.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Russo G, Unkauf T, Meier D, Wenzel EV, Langreder N, Schneider KT, K-T Schneider, Wiesner R, Bischoff R, Stadler V, Dübel, S.. In vitro evolution of myc- tag antibodies: in-depth specificity and affinity analysis of Myc1-9E10 and Hyper-Myc. Biol Chem. 2022;403(5–6):479–494. doi: 10.1515/hsz-2021-0405. [DOI] [PubMed] [Google Scholar]
- 83.Sulkowski E. Immobilised metal affinity chromatography. Trends Biotechnol. 1985;3(1):1–7. doi: 10.1016/0167-7799(85)90068-X. [DOI] [Google Scholar]
- 84.Dübel S, Breitling F, Klewinghaus I, Little M. Regulated secretion and purification of recombinant antibodies in E. coli. Cell Biophys. 1992;21(1–3):69–79. doi: 10.1007/BF02789479. [DOI] [PubMed] [Google Scholar]
- 85.Booth RJ, Grandison PM, Prestidge RL, Watson JD. The use of a ‘universal’ yeast expression vector to produce an antigenic protein of Mycobacterium leprae.” Immunol Lett 19, no. 1 (1988. 19): 65–69. doi: 10.1016/0165-2478(88)90121-6. [DOI] [PubMed] [Google Scholar]
- 86.Schmidt TG, Koepke J, Frank R, Skerra A. Molecular interaction between the Strep-tag affinity peptide and its cognate target, streptavidin. J Mol Biol. 1996;255(5):753–66. doi: 10.1006/jmbi.1996.0061. [DOI] [PubMed] [Google Scholar]
- 87.Li Y, Sousa R. Expression and purification of E. coli BirA biotin ligase for in vitro biotinylation. Protein Expr Purif. 2012. Mar;82(1):162–67. doi: 10.1016/j.pep.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quintero-Hernández V, Juárez-González VR, Ortíz-León M, Sánchez R, Possani LD, Becerril B. The change of the scFv into the Fab format improves the stability and in vivo toxin neutralization capacity of recombinant antibodies. Mol Immunol. 2007. Feb;44(6):1307–15. doi: 10.1016/j.molimm.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 89.Steinwand M, Droste P, Frenzel A, Hust M, Dübel S, Schirrmann T. The influence of antibody fragment format on phage display based affinity maturation of IgG. Mabs-austin. 2014. Feb;6(1):204–18. doi: 10.4161/mabs.27227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mieczkowski C, Zhang X, Lee D, Nguyen K, Lv W, Wang Y, Zhang Y, Way J, Gries J-M. Blueprint for antibody biologics developability. Mabs-austin. 2023;15(1):2185924. doi: 10.1080/19420862.2023.2185924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Azevedo Reis Teixeira A, Erasmus MF, D’Angelo S, Naranjo L, Ferrara F, Leal-Lopes C, Durrant O, Galmiche C, Morelli A, Scott-Tucker A. et al. Drug-like antibodies with high affinity, diversity and developability directly from next-generation antibody libraries. Mabs-austin. 2021;13(1):1980942. doi: 10.1080/19420862.2021.1980942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fellouse FA, Esaki K, Birtalan S, Raptis D, Cancasci VJ, Koide A, Jhurani P, Vasser M, Wiesmann C, Kossiakoff AA. et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol. 2007 Nov 2;373(4):924–40. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 93.Al-Halabi L, Balck A, Michalzik M, Fröde D, Büttgenbach S, Hust M, Schirrmann T, Dübel S. Recombinant antibody fragments allow repeated measurements of C-reactive protein with a quartz crystal microbalance immunosensor. Mabs-austin. 2013. Feb;5(1):140–49. doi: 10.4161/mabs.22374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thie H, Toleikis L, Li J, von Wasielewski R, Bastert G, Schirrmann T, Esteves IT, Behrens CK, Fournes B, Fournier N. et al. Rise and Fall of an Anti-MUC1 Specific Antibody. PLOS ONE. 2011 Jan 14;6(1):e15921. doi: 10.1371/journal.pone.0015921. Chu HW, editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gacerez AT, Arellano B, Sentman CL. How chimeric antigen receptor design affects adoptive T cell therapy. J Cell Physiol. 2016. Dec;231(12):2590–98. doi: 10.1002/jcp.25419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huehls AM, Coupet TA, Sentman CL. Bispecific T-cell engagers for cancer immunotherapy. Immunol Cell Biol. 2015. Mar;93(3):290–96. doi: 10.1038/icb.2014.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pastan IH, Pai LH, Brinkmann U, Fitzgerald DJ. Recombinant toxins: new therapeutic agents for cancer. Ann NY Acad Sci. 1995;758(1):345–54. doi: 10.1111/j.1749-6632.1995.tb24840.x. [DOI] [PubMed] [Google Scholar]
- 98.Moutel S, El Marjou A, Vielemeyer O, Nizak C, Benaroch P, Dübel S, Perez F. A multi-Fc-species system for recombinant antibody production. BMC Biotechnol. 2009;9(1):14. doi: 10.1186/1472-6750-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hanaee-Ahvaz H, Cserjan-Puschmann M, Mayer F, Tauer C, Albrecht B, Furtmüller PG, Wiltschi B, Hahn R, Striedner G. Antibody fragments functionalized with non-canonical amino acids preserving structure and functionality - a door opener for new biological and therapeutic applications. Heliyon. 2023. Dec;9(12):e22463. doi: 10.1016/j.heliyon.2023.e22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rakotoarinoro N, Dyck YFK, Krebs SK, Assi MK, Parr MK, Stech M. A disruptive clickable antibody design for the generation of antibody-drug conjugates. Antibody Ther. 2023. Oct;6(4):298–310. doi: 10.1093/abt/tbad023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brinkmann U, Kontermann RE. The making of bispecific antibodies. Mabs-austin. 2017;9(2):182–212. doi: 10.1080/19420862.2016.1268307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alewine C, Hassan R, Pastan I. Advances in Anticancer Immunotoxin Therapy. Oncologist. 2015. Feb;20(2):176–85. doi: 10.1634/theoncologist.2014-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sun D, Shi X, Li S, Wang X, Yang X, Wan M. CAR‑T cell therapy: A breakthrough in traditional cancer treatment strategies (Review). Mol Med Rep. 2024. Mar;29(3):47. doi: 10.3892/mmr.2024.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Y. Evolution of phage display libraries for therapeutic antibody discovery. Mabs-austin. 2023;15(1):2213793. doi: 10.1080/19420862.2023.2213793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xiao X, Douthwaite JA, Chen Y, Kemp B, Kidd S, Percival-Alwyn J, Smith A, Goode K, Swerdlow B, Lowe D. et al. A high-throughput platform for population reformatting and mammalian expression of phage display libraries to enable functional screening as full-length IgG. Mabs-austin. 2017 Aug 18;9(6):996–1006. doi: 10.1080/19420862.2017.1337617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997. Jun;15(6):553–57. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 107.Akamatsu Y, Pakabunto K, Xu Z, Zhang Y, Tsurushita N. Whole IgG surface display on mammalian cells: Application to isolation of neutralizing chicken monoclonal anti-IL-12 antibodies. J Immunol Methods. 2007 Oct 31;327(1–2):40–52. doi: 10.1016/j.jim.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 108.Ho M, Nagata S, Pastan I. Isolation of anti-CD22 Fv with high affinity by Fv display on human cells. Proc Natl Acad Sci U S A. 2006 Jun 20;103(25):9637–42. doi: 10.1073/pnas.0603653103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Valldorf B, Hinz SC, Russo G, Pekar L, Mohr L, Klemm J, Doerner A, Krah S, Hust M, Zielonka S. et al. Antibody display technologies: selecting the cream of the crop. Biol Chem. 2022 Apr 26;403(5–6):455–77. doi: 10.1515/hsz-2020-0377. [DOI] [PubMed] [Google Scholar]
- 110.Ferrara F, Naranjo LA, Kumar S, Gaiotto T, Mukundan H, Swanson B, Bradbury ARM. Using phage and yeast display to select hundreds of monoclonal antibodies: application to antigen 85, a tuberculosis biomarker. PLOS ONE. 2012;7(11):e49535. doi: 10.1371/journal.pone.0049535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Feddersen RM, Van Ness BG. Corrective recombination of mouse immunoglobulin kappa alleles in Abelson murine leukemia virus-transformed pre-B cells. Mol Cell biol. 1990. Feb;10(2):569–76. doi: 10.1128/MCB.10.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marks JD. Antibody affinity maturation by chain shuffling. Methods Mol Biol. 2004;248:327–43. [DOI] [PubMed] [Google Scholar]
- 113.Kang AS, Jones TM, Burton DR. Antibody redesign by chain shuffling from random combinatorial immunoglobulin libraries. Proc Natl Acad Sci USA. 1991 Dec 15;88(24):11120–23. doi: 10.1073/pnas.88.24.11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frenzel A, Kügler J, Helmsing S, Meier D, Schirrmann T, Hust M, Dübel S. Designing Human Antibodies by Phage Display. Transfusion Medicine and Hemotherapy: Offizielles Organ Der Deutschen Gesellschaft Fur Transfusionsmedizin Und Immunhamatologie. 2017. Sep;44(5):312–18. doi: 10.1159/000479633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bradbury ARM, Sidhu S, Dübel S, McCafferty J. Beyond natural antibodies: the power of in vitro display technologies. Nat Biotechnol. 2011. Mar;29(3):245–54. doi: 10.1038/nbt.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang MY, Shu Y, Rudolph D, Prabakaran P, Labrijn AF, Zwick MB, Lal RB, Dimitrov DS. Improved breadth and potency of an HIV-1-neutralizing human single-chain antibody by random mutagenesis and sequential antigen panning. J Mol Biol. 2004 Jan 2;335(1):209–19. doi: 10.1016/j.jmb.2003.09.055. [DOI] [PubMed] [Google Scholar]
- 117.Gallo E. Current advancements in B-cell receptor sequencing fast-track the development of synthetic antibodies. Mol Biol Rep. 2024 Jan 18;51(1):134. doi: 10.1007/s11033-023-08941-0. [DOI] [PubMed] [Google Scholar]
- 118.Russo G, Theisen U, Fahr W, Helmsing S, Hust M, Köster RW, Dübel S. Sequence defined antibodies improve the detection of cadherin 2 (N-cadherin) during zebrafish development. N Biotechnol. 2018 Oct 25;45:98–112. doi: 10.1016/j.nbt.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 119.Pedrioli A, Oxenius A. Single B cell technologies for monoclonal antibody discovery. Trends Immunol. 2021. Dec;42(12):1143–58. doi: 10.1016/j.it.2021.10.008. [DOI] [PubMed] [Google Scholar]
- 120.Ridder R, Schmitz R, Legay F, Gram H. Generation of rabbit monoclonal antibody fragments from a combinatorial phage display library and their production in the yeast pichia pastoris. Bio/Technology. 1995;13(3):255–60. doi: 10.1038/nbt0395-255. [DOI] [PubMed] [Google Scholar]
- 121.Romao E, Morales-Yanez F, Hu Y, Crauwels M, De Pauw P, Hassanzadeh GG, Devoogdt N, Ackaert C, Vincke C, Muyldermans S. et al. Identification of Useful nanobodies by phage display of immune single domain libraries derived from camelid heavy chain antibodies. Curr Pharm Des. 2017;22(43):6500–18. doi: 10.2174/1381612822666160923114417. [DOI] [PubMed] [Google Scholar]
- 122.Courtenay-Luck NS, Epenetos AA, Moore R, Larche M, Pectasides D, Dhokia B, Ritter MA. Development of primary and secondary immune responses to mouse monoclonal antibodies used in the diagnosis and therapy of malignant neoplasms. Cancer Res. 1986. Dec;46(12 Pt 1):6489–93. [PubMed] [Google Scholar]
- 123.Gorman SD, Clark MR. Humanisation of monoclonal antibodies for therapy. Semin Immunol. 1990. Nov;2(6):457–66. [PubMed] [Google Scholar]
- 124.Hwang WYK, Foote J. Immunogenicity of engineered antibodies. Methods. 2005. May;36(1):3–10. doi: 10.1016/j.ymeth.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 125.Fishwild DM, O’Donnell SL, Bengoechea T, Hudson DV, Harding F, Bernhard SL, Jones D, Kay RM, Higgins KM, Schramm SR. et al. High-avidity human IgGκ monoclonal antibodies from a novel strain of minilocus transgenic mice. Nat Biotechnol. 1996. Jul;14(7):845–51. doi: 10.1038/nbt0796-845. [DOI] [PubMed] [Google Scholar]
- 126.Jakobovits A. Production of fully human antibodies by transgenic mice. Curr Opin Biotechnol. 1995. Oct;6(5):561–66. doi: 10.1016/0958-1669(95)80093-X. [DOI] [PubMed] [Google Scholar]
- 127.Osborn MJ, Ma B, Avis S, Binnie A, Dilley J, Yang X, Lindquist K, Ménoret S, Iscache A-L, Ouisse L-H. et al. High-affinity IgG antibodies develop naturally in Ig-knockout rats carrying germline human IgH/Igκ/Igλ loci bearing the rat CH region. J Immunol. 2013 Feb 15;190(4):1481–90. doi: 10.4049/jimmunol.1203041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S. et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020. Aug;584(7819):115–19. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 129.Bertoglio F, Meier D, Langreder N, Steinke S, Rand U, Simonelli L, Heine P A, Ballmann R, K-T Schneider, Roth, K D R. et al. SARS-CoV-2 neutralizing human recombinant antibodies selected from pre-pandemic healthy donors binding at RBD-ACE2 interface. Nat Commun. 2021;12:1577. doi: 10.1038/s41467-021-21609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Roth KDR, Wenzel EV, Ruschig M, Steinke S, Langreder N, Heine PA, Schneider K-T, Ballmann R, Fühner V, Kuhn P. et al. Developing recombinant antibodies by phage display against infectious diseases and toxins for diagnostics and therapy. Front Cell Infect Microbiol. 2021 Jul 7;11:697876. doi: 10.3389/fcimb.2021.697876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kellmann SJ, Hentrich C, Putyrski M, Hanuschka H, Cavada M, Knappik A, Ylera F. SpyDisplay: a versatile phage display selection system using SpyTag/SpyCatcher technology. Mabs-austin. 2023;15(1):2177978. doi: 10.1080/19420862.2023.2177978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhai W, Glanville J, Fuhrmann M, Mei L, Ni I, Sundar PD, Van Blarcom T, Abdiche Y, Lindquist K, Strohner R. et al. Synthetic antibodies designed on natural sequence landscapes. J Mol Biol. 2011. Sep 9;412(1):55–71. doi: 10.1016/j.jmb.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 133.Hoet RM, Cohen EH, Kent RB, Rookey K, Schoonbroodt S, Hogan S, Rem L, Frans N, Daukandt M, Pieters H. et al. Generation of high-affinity human antibodies by combining donor-derived and synthetic complementarity-determining-region diversity. Nat Biotechnol. 2005. Mar;23(3):344–48. doi: 10.1038/nbt1067. [DOI] [PubMed] [Google Scholar]
- 134.Colwill K, Gräslund S. Renewable protein binder working group, Gräslund S. A roadmap to generate renewable protein binders to the human proteome. Nat Methods. 2011. Jul;8(7):551–58. doi: 10.1038/nmeth.1607. [DOI] [PubMed] [Google Scholar]
- 135.Schofield DJ, Pope AR, Clementel V, Buckell J, Chapple SD, Clarke KF, Conquer JS, Crofts AM, Crowther SR, Dyson MR. et al. Application of phage display to high throughput antibody generation and characterization. Genome Biol. 2007;8(11):R254. doi: 10.1186/gb-2007-8-11-r254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nagano K, Tsutsumi Y. Phage display technology as a powerful platform for antibody drug discovery. Viruses. 2021 Jan 25;13(2):178. doi: 10.3390/v13020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bradbury ARM, Marks JD. Antibodies from phage antibody libraries. J Immunol Methods. 2004. Jul;290(1–2):29–49. doi: 10.1016/j.jim.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 138.Zhou H, Zhang YL, Lu G, Ji H, Rodi CP. Recombinant antibody libraries and selection technologies. N Biotechnol. 2011. Sep;28(5):448–52. doi: 10.1016/j.nbt.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 139.Little M, Breitling F, Dübel S, Fuchs P, Braunagel M, Seehaus T, Klewinghaus I. Universal antibody libraries on phage and bacteria. Year Immunol. 1993;7:50–55. [PubMed] [Google Scholar]
- 140.Harel Inbar N, Benhar I. Selection of antibodies from synthetic antibody libraries. Arch Biochem Biophys. 2012. Oct 15;526(2):87–98. doi: 10.1016/j.abb.2011.12.028. [DOI] [PubMed] [Google Scholar]
- 141.de Brito PM, Saruga A, Cardoso M, Goncalves J, de Brito PM. Methods and cell-based strategies to produce antibody libraries: current state. Appl Microbiol Biotechnol. 2021. Oct;105(19):7215–24. doi: 10.1007/s00253-021-11570-x. [DOI] [PubMed] [Google Scholar]
- 142.Weller MG. Quality issues of research antibodies. Anal Chem Insights. 2016;11:21. doi: 10.4137/ACI.S31614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Skerra A. Alternative non-antibody scaffolds for molecular recognition. Curr Opin Biotechnol. 2007. Aug;18(4):295–304. doi: 10.1016/j.copbio.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 144.Kovár J, Franĕk F. Serum-free medium for hybridoma and parental myeloma cell cultivation: a novel composition of growth-supporting substances. Immunol Lett. 1984;7(6):339–45. doi: 10.1016/0165-2478(84)90092-0. [DOI] [PubMed] [Google Scholar]
- 145.Donini M, Marusic C. Current state-of-the-art in plant-based antibody production systems. Biotechnol Lett. 2019. Mar;41(3):335–46. doi: 10.1007/s10529-019-02651-z. [DOI] [PubMed] [Google Scholar]
- 146.Bobrowicz P, Davidson RC, Li H, Potgieter TI, Nett JH, Hamilton SR. et al. Engineering of an artificial glycosylation pathway blocked in core oligosaccharide assembly in the yeast Pichia pastoris: production of complex humanized glycoproteins with terminal galactose. Glycobiology. 2004. Sep;14(9):757–66. doi: 10.1093/glycob/cwh104. [DOI] [PubMed] [Google Scholar]
- 147.Li H, Sethuraman N, Stadheim TA, Zha D, Prinz B, Ballew N, Bobrowicz P, Choi B-K, Cook WJ, Cukan M. et al. Optimization of humanized IgGs in glycoengineered Pichia pastoris. Nat Biotechnol. 2006. Feb;24(2):210–15. doi: 10.1038/nbt1178. [DOI] [PubMed] [Google Scholar]
- 148.Potgieter TI, Cukan M, Drummond JE, Houston-Cummings NR, Jiang Y, Li F, Lynaugh H, Mallem M, McKelvey TW, Mitchell T. et al. Production of monoclonal antibodies by glycoengineered Pichia pastoris. J Biotechnol. 2009. Feb 23;139(4):318–25. doi: 10.1016/j.jbiotec.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 149.Bradbury ARM, Dübel S, Knappik A, Plückthun A. Animal- versus in vitro -derived antibodies: avoiding the extremes. Mabs-austin. 2021. Jan 1;13(1):1950265. doi: 10.1080/19420862.2021.1950265. [DOI] [PMC free article] [PubMed] [Google Scholar]


