Abstract
The Epstein–Barr virus (EBV) establishes lifelong infections in > 90% of the human population. Although contained as asymptomatic infection by the immune system in most individuals, EBV is associated with the pathogenesis of approximately 1.5% of all cancers in humans. Some of these EBV-associated tumors have been successfully treated by the infusion of virus-specific T-cell lines. Recent sequence analyses of a large number of viral isolates suggested that distinct EBV strains have evolved in different parts of the world. Here, we assessed the impact of such sequence variations on EBV-specific T-cell immunity. With the exceptions of EBNA2 and the EBNA3 family of proteins, an overall low protein sequence disparity of about 1% was noted between Asian viral isolates, including the newly characterized M81 strain, and the prototypic EBV type 1 and type 2 strains. However, when T-cell epitopes including their flanking regions were compared, a substantial proportion was found to be polymorphic in different EBV strains. Importantly, CD4+ and CD8+ T-cell clones specific for viral epitopes from one strain often showed diminished recognition of the corresponding epitopes in other strains. In addition, T-cell recognition of a conserved epitope was affected by amino acid exchanges within the epitope flanking region. Moreover, the CD8+ T-cell response against polymorphic epitopes varied between donors and often ignored antigen variants. These results demonstrate that viral strain heterogeneity may impair antiviral T-cell immunity and suggest that immunotherapeutic approaches against EBV should preferably target broad sets of conserved epitopes including their flanking regions.
Electronic supplementary material
The online version of this article (10.1007/s00262-018-2118-z) contains supplementary material, which is available to authorized users.
Keywords: T-cell therapy, Epstein–Barr virus, Strain variation, Epitope, Immunity
Introduction
Epstein–Barr virus (EBV) is a ubiquitous γ-herpesvirus associated with the development of various lymphoid and epithelial malignancies such as post-transplant lymphoproliferative disease (PTLD), Burkitt and Hodgkin lymphoma, T cell and NK cell lymphoma, as well as nasopharyngeal and gastric carcinoma [1, 2]. Despite the worldwide prevalence of the virus, incidence rates of some EBV-associated tumor entities vary greatly among geographical regions [3]. Besides local dietary, genetic, and environmental co-factors, EBV subtypes with enhanced pathogenicity may exist in locally restricted areas and contribute to this geographic predilection [4]. This notion is supported by the recent characterization of M81, a viral strain isolated from a Chinese nasopharyngeal carcinoma (NPC). Compared to common EBV strains, M81 shows enhanced epitheliotropism and lytic activity, and at least some of these phenotypic alterations have been ascribed to polymorphisms in viral proteins [5, 6].
Historically, EBV strains have been classified into type 1 and type 2 according to their DNA sequence divergence within the EBV nuclear antigen 2 (EBNA2) and the linked variation in EBNA3 genes [2, 7]. However, recent sequence information obtained from a large number of viral isolates from different sample types and locations worldwide identified geographic variations in EBV strains independent of the type 1/type 2 classification [5, 7, 8]. Moreover, these studies revealed variability in known immune epitopes and provided evidence for the selection of viral protein sequences by the host immune system [8].
Immunity to EBV is mediated primarily by T cells specific for epitopes derived from various latent and lytic cycle antigens of the virus [9, 10]. Hence, patients with T-cell dysfunction are at increased risk of developing life-threatening EBV-associated lymphoproliferative diseases. Moreover, reconstitution of EBV-specific immunity by the adoptive transfer of virus-specific T-cell lines has been shown to prevent and cure PTLD in transplant recipients as well as some other EBV-associated disorders in immune competent individuals [11–13]. Originally, such T-cell lines were generated by repeated stimulation of peripheral blood T cells with EBV-transformed lymphoblastoid B-cell lines (LCL) in vitro and were shown to recognize a broad set of latent and lytic cycle EBV antigens [14–16]. Recently, more rapid protocols for the production of virus-specific T-cell preparations have been developed that mostly rely on the selection of EBV-specific T cells with a limited set of antigens or peptide epitopes that are almost exclusively derived from the prototypic laboratory EBV strain B95.8 [17–19]. Given the recent findings on sequence diversity among EBV strains, it is still unknown whether immunotherapeutic and vaccine approaches that are based on B95.8 will be sufficiently cross-protective against EBV strains across the world.
Thus, the aim of our study was to (1) compare protein sequences of three distantly related EBV strains; the newly identified, NPC-derived strain M81 and the prototypic, B-cell-derived EBV type 1 and type 2 strains B95.8 and AG876, respectively, (2) inspect all published EBV T-cell epitopes for polymorphisms in these viruses, (3) investigate recognition of polymorphic epitopes by virus-specific CD8+ and CD4+ T-cell clones and (4), assess T-cell immunity against polymorphic epitopes ex vivo.
Materials and methods
Sequence analysis
DNA and protein sequences of different EBV strains were obtained from NCBI: B95.8 (GenBank Nr. V01555.2), M81 (GenBank Nr. KF373730.1), AG876 (GenBank Nr. DQ279927.1), C666-1 (GenBank Nr. AM182486.1), and HKNPC1 (GenBank Nr. JQ009376). Sequence comparisons were performed using MatLab and validated by Vector NTI Advance (Version 11.5). Flanking regions were defined as the five N- and C-terminal amino acids flanking published epitopes.
Cell culture
LCL was established by incubating peripheral blood mononuclear cells (PBMC) with virus supernatant, and LCL and the EBV-negative Burkitt lymphoma cell line DG75 cultured as described [16].
T-cell lines were generated by stimulating PBMC with autologous, irradiated LCL (80 Gy) as antigen-presenting cells (APC). EBV-specific T-cell clones were established by limiting dilution of reactive lines [16]. The CD4+ T-cell clone EBNA3C-SDD [20] was kindly provided by Dr. G. Taylor (Birmingham).
Cytokine secretion by the T cells was measured by incubating 5 × 104 target cells with 5 × 104 T cells in a final volume of 200 µl LCL medium (RPMI-1640, 10% FCS, 2 mM l-glutamine, 1% non-essential amino acids, 1 mM Na-pyruvate, 50 µg/ml gentamycin) for 18 h [21]. The supernatants were analyzed for the presence of interferon-gamma (IFN-γ) by enzyme-linked immunosorbent assay (Mabtech).
ELISpot assays were performed using the T-Track® human IFN-γ ELISpot kit following the instructions of the manufacturer (Lophius). Rested PBMC were either stimulated with 5 µM of the different EBV peptides, phytohemagglutinine (PHA) (positive control), or were left unstimulated (negative control). Plates were analyzed using the ELISpot reader ImmunoSpot® S6 and spots were counted using the CTL ImmunoSpot 5.4 Professional DC Software.
Plasmid construction and transfection of cells
All expression constructs are derivatives of the pCMV/cyto vector (Invitrogen). The open reading frames of EBNA1, EBNA3C, latent membrane protein 2A (LMP2A), and BZLF1 were amplified from cDNA of cell lines infected with the B95.8, M81, or AG876 viral strains by PCR. EBNA1 genes were cloned without glycine–alanine (GA) and adjacent repeats, hence consisting of amino acids 1–91 and 333–646. Gene variants were generated by PCR-based site-directed mutagenesis. All viral gene constructs carried a hexahistidine tag (His6) at the C-terminus. The restricting HLA molecules were cloned from cell lines positive for these alleles by PCR. Identity of all genes was verified by sequencing. Cells were transfected with plasmids coding for the antigen and the restricting HLA molecule by electroporation [22]. Transfection efficiency was monitored by co-transfection of a GFP expression plasmid. Peptides were designed according to the B95.8, M81, and AG876 sequences and purchased from JPT Peptide Technologies GmbH (> 90% purity).
Western blot and FACS analysis
RIPA buffer containing Protease Inhibitor Cocktail (Roche) was used to prepare whole cell extracts for Western blot analysis. Samples were separated in SDS-PAA gels, blotted onto PVDF membranes (GE-Healthcare) and incubated with the following antibodies: anti-His6 (clone 3D9, provided by Dr. E. Kremmer; Helmholtz Zentrum München), anti-tubulin (Abcam), anti-GAPDH (Millipore) and HRP-conjugated secondary antibodies against mouse IgG (GE-Healthcare). Bound antibodies were visualized by chemiluminescence (ECL plus, GE Healthcare).
Statistical analysis
Statistical significance of the data was analyzed using GraphPad Prism5. P values were calculated using unpaired t test or two-sample t test with Bonferroni correction. P values were indicated as follows: P > 0.05 n.s.; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Results
Protein sequence disparities between viral strains
Sequence information from a large number of viral isolates revealed a non-random distribution of non-synonymous polymorphisms across the viral genome and a clear separation of Asian strains from type 1 and type 2 EBV [5, 6, 8, 23]. To investigate the impact of these polymorphisms on viral immunogenicity, we compared the protein sequences between three NPC-derived virus isolates (C666-1, HKNPC1, and M81) and the B-cell-derived type 1 and type 2 reference strains B95.8 and AG876, respectively. With M81 set as reference sequence, the homologous protein sequences of the different EBV strains were aligned and the percentage of disparity calculated (Supplementary Fig. 1). After subdividing viral proteins into functional groups, an overall 6.6% disparity of the other strains to M81 was noted in latency proteins (B95.8 3.7%; AG876 17.5%; C666-1 4.0%; HKNPC1 1.0%). Consistent with the classification of M81 as type 1 strain [5], EBNA2 and EBNA3 protein sequences from M81 and AG876 were highly dissimilar (Supplementary Fig. 1a). By contrast, the overall sequence disparity in lytic cycle proteins between the four strains and M81 was only about 0.5% (Supplementary Fig. 1b–d). A larger disparity of approximately 0.8% was noted for the B-cell-derived viral strains B95.8 and AG876 than for the NPC-derived viral isolates C666-1 (0.1%) and HKNPC1 (0.2%), suggesting that polymorphisms in lytic cycle proteins rather correlate with the geographic region and/or the cell type from which the virus was isolated than with the EBV type. To investigate whether sequence variations in lytic cycle proteins were randomly distributed or overrepresented in particular gene products, lytic cycle proteins were further subdivided into functional groups (Supplementary Fig. 1b–f). Despite its enhanced epitheliotropism, M81 proteins involved in epithelial cell entry displayed an average degree of disparity from B cell-derived viral strains. However, M81 proteins with reported immunoevasive function differed from their homologues in B95.8 and AG876 by 1.2%, suggesting that these sequence variations might impact on immunogenicity (Supplementary Fig. 1e, f).
Many T-cell epitopes identified in B95.8 are polymorphic in M81 and AG876
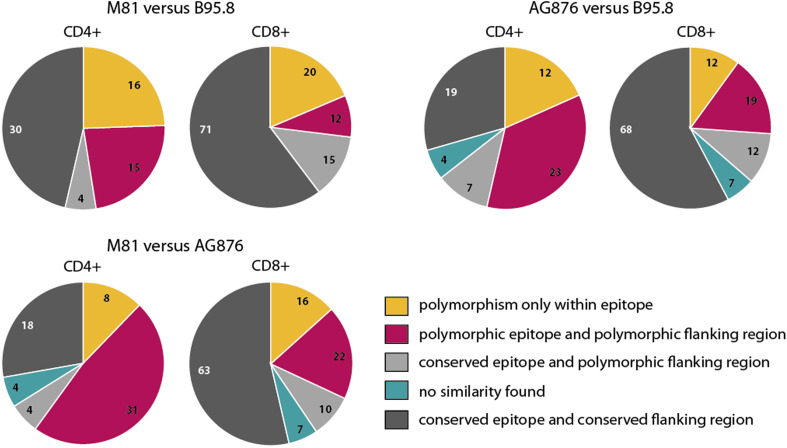
To investigate the role of the sequence disparities in immune recognition, published EBV T-cell epitopes were analyzed with respect to polymorphisms in B95.8, M81, and AG876. In total, 183 T-cell epitopes were collected from the literature, of which 65 were recognized by CD4+ T cells and 118 by CD8+ T cells. Almost all T-cell epitopes were derived from B95.8. Since amino acid sequences flanking the epitope can affect antigen processing and binding to MHC molecules [24, 25], five N-terminal and five C-terminal amino acids immediately adjacent to the published epitope were defined as “flanking region” and included in this analysis. A list of the epitopes and flanking regions that are polymorphic in at least one of the viral strains is provided in Supplementary table 1. Remarkably, about half of all CD4+ and one-third of all CD8+ T-cell epitopes and/or flanking regions proved to be polymorphic (Fig. 1). The highest disparity was noted when the two type 1 strains B95.8 and M81 were compared to the type 2 strain AG876. Nevertheless, even 48% of all CD4+ epitopes and 27% of all CD8+ epitopes differed between the two type 1 strains, indicating that sequence variations between viral strains of the same type might affect immune recognition as well.
Fig. 1.
A high proportion of all T-cell epitopes identified in B95.8 is polymorphic in M81 and AG876. Amino acid sequences of published CD4+ (n = 65) and CD8+ (n = 118) T-cell epitopes in B95.8 and their flanking regions were compared with the corresponding sequences in M81 (left) and AG876 (right). A comparison of epitope sequences between M81 and AG876 is shown underneath. Flanking regions were defined as five amino acids immediately before and after the published T-cell epitopes. Numbers in pie charts represent the number of epitopes that fall into each of the color-coded categories. Epitopes located in protein regions that are not present in the homologues of other viruses are denoted as “no similarity found”
Polymorphisms in viral epitopes can impair or enhance recognition by CD8+ T cells
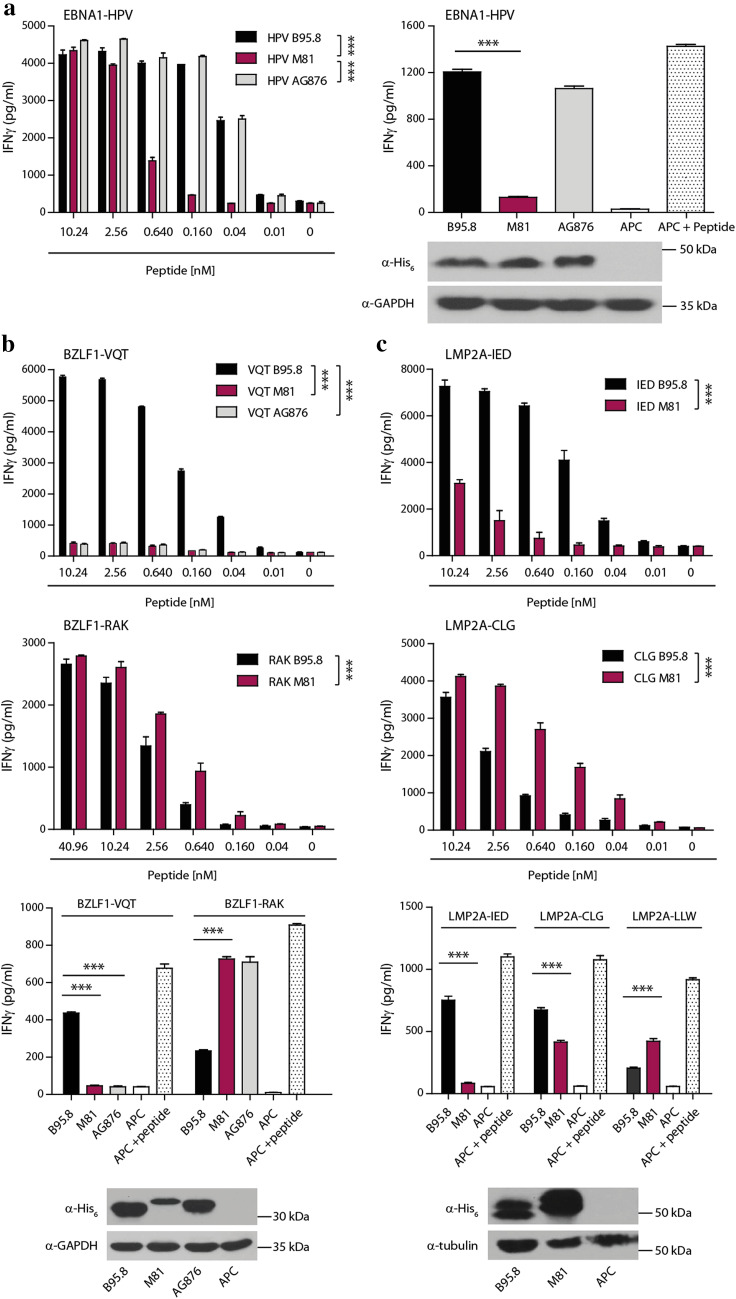
Amino acid exchanges in T-cell epitopes can affect antigen processing, binding to MHC molecules and recognition by the T-cell receptor [26, 27]. To directly assess the effect of the identified polymorphisms within epitopes on T-cell recognition, five CD8+ T-cell clones recognizing different latent and lytic cycle antigens from B95.8 were probed with polymorphic epitopes from other viral strains (Table 1). When titrated amounts of the three different EBNA1-HPV peptide epitopes were pulsed onto HLA-B*3501-positive target cells, the exchange of glutamic to aspartic acid (E>D) at position 5 of the M81 epitope led to a strong reduction in T-cell recognition. By contrast, the glycine to alanine substitution at position 4 in the AG876 sequence had no effect on the recognition by the HPV-specific T-cell clone (Fig. 2a).
Table 1.
Polymorphic published CD8+ T-cell epitopes and flanking regions analyzed in vitro
| EBNA1-HPV | BZLF1-VQT | BZLF1-RAK | LMP2A-IED | LMP2A-CLG | |
|---|---|---|---|---|---|
| HLA | HLA-B*3501 | HLA-B*1501 | HLA-B*0801 | HLA-B*4001 | HLA-A*0201 |
| Epitope coordinates (B95.8) | AA 407–417 | AA 121–129 | AA 190–197 | AA 200–208 | AA 426–434 |
| B95.8 |
RRPFF HPVGEADYFEY HQEGG |
GDNST VQTAAAVVF ACPGA |
ASRKC RAKFKQLL QHYRE |
CLTWR IEDPPFNSL LFALL |
GPVFM CLGGLLTMV AFAVW |
| M81 |
RRPFF HPVG D ADYFEY LQEGG |
GDNST VQ P AAAVV L ACPGA |
ASRKC RAKFK H LL QHYRE |
CLTWR IEDPPFNS I LFALL |
GPVFM S LGGLLTMV AGAVW |
| AG876 |
RRPFF HPV A EADYFEY HQEGG |
GDNST VQPAAAVVF ACPGA |
# | * | * |
AA amino acids
*Same as B95.8 sequence
#Same as M81 sequence
Fig. 2.
Polymorphisms in CD8+ T-cell epitopes affect T-cell recognition. Five CD8+ T-cell clones specific for different polymorphic EBV epitopes were tested for recognition of the corresponding epitopes in B95.8, M81, and AG876. a Antigen-presenting cells were loaded with titrated amounts of the EBNA1-HPV epitope peptides and then probed with the T cells (left graph). Presentation of endogenous antigen was assessed by transfecting EBV-negative DG75 cells with expression constructs for HLA-B*3501 and glycine–alanine (GA) repeat-deleted EBNA1 versions of the three viruses, and co-culturing transfected cells with HPV-specific T cells (right graph). Target cells transfected with the HLA molecule alone (APC) served as negative control, and APC pulsed with the peptide epitope from B95.8 as positive control (APC+ Peptide). Expression levels of recombinant EBNA1 was analyzed by Western blot (bottom) using an antibody directed against the His6-tag present in all transfected antigens. GAPDH (in a, b) and tubulin (in c) served as gel loading controls. Recognition of exogenously loaded peptide epitopes BZLF-VQT and -RAK (b), and LMP2A-IED and -CLG (c), and of endogenously expressed full-length protein was analyzed in the same way using T-cell clones specific for these epitopes derived from B95.8 virus. T cells specific for the non-polymorphic epitope LMP2A-LLW were used as internal control. Statistical significance was evaluated using unpaired t test. In dose–response curves, two-sample t test with Bonferroni correction was used. Data are shown as mean ± SD of n = 3 and are from single experiments representative of at least three independent experiments
To assess whether these amino acid exchanges affected T-cell recognition of endogenously expressed antigens, His6-tagged versions of the three different EBNA1 genes were cloned and expression plasmids transfected into EBV-negative DG75 cells together with the restricting HLA-B*3501 molecule. Consistent with the peptide titration experiments, cells transfected with EBNA1 from B95.8 and AG876 were readily recognized, whereas cells transfected with the M81 protein were not. All three EBNA1 proteins were expressed at similar levels, implying that the E>D amino acid exchange abrogated T-cell recognition (Fig. 2a).
Similar experiments were performed for BZLF1 which carries two polymorphic T-cell epitopes, VQT and RAK, against which CD8+ T-cell clones were available (Table 1). In case of the BZLF1-VQT epitope, HLA-B*1501 positive target cells pulsed with the B95.8, but not the M81 and AG876 peptide epitopes, were recognized by VQT-specific CD8+ T-cell clones, indicating that threonine (T) at position 3, that is exchanged for proline (P) in M81 and AG876, is essential for CD8+ T-cell recognition. Since this T>P substitution completely abrogated T-cell recognition, it remained unknown whether the phenylalanine to leucine (F>L) exchange at position 9 of the epitope in M81 would also affect T-cell recognition. Interestingly, the exchange of glutamine for histidine (Q>H) in the RAK epitope of M81 and AG876 enhanced CD8+ T-cell recognition in peptide titration experiments (Fig. 2b). Similar results were obtained when endogenous processing and presentation of the BZLF1 epitopes were assessed in transfection experiments. While VQT-specific T cells recognized target cells transfected with BZLF1 from B95.8 only, RAK-specific T cells recognized target cells transfected with BZLF1 from M81 and AG876 more efficiently than from B95.8. Notably, the level of recognition was inverted when the RAK epitopes in BZLF1 from B95.8 and M81 were swapped (Supplementary Fig. 2), indicating that these differences in recognition were indeed due to the single amino acid exchange in the epitope and not caused by differences in immunogenicity of the full-length BZLF1 proteins.
Similar experiments were performed with two polymorphic epitopes of LMP2A, IED and CLG, against which CD8+ T-cell clones were available (Fig. 2c). Compared to B95.8, a leucine to isoleucine exchange (L>I) at position 9 of the IED epitope in M81 resulted in a diminished recognition by IED-specific T cells after exogenous peptide loading as well as after endogenous protein expression. Interestingly, the cysteine to serine (C>S) exchange at position 1 of the CLG epitope in M81 enhanced T-cell recognition in peptide titration experiments, while recognition of antigen from endogenous sources was diminished. The latter was not due to reduced M81 antigen expression as demonstrated by Western blot analyses, which revealed a much higher protein expression level for LMP2A from M81 than from B95.8. To assess whether LMP2A from M81 was less efficiently processed or had stronger immune evasive functions than LMP2A from B95.8 [28], we analyzed presentation of the conserved LLW epitope after transfection of APC with the different LMP2A expression plasmids. The enhanced recognition of the LLW epitope from LMP2A of M81 indicated that both proteins efficiently accessed endogenous presentation pathways and that the lower recognition of the CLG epitope from M81 was due to a less efficient generation of this epitope. Collectively, these findings indicated that polymorphisms within T-cell epitopes can affect both antigen processing and recognition by CD8+ T cells.
Polymorphisms within epitopes and flanking regions impact on CD4+ T-cell recognition
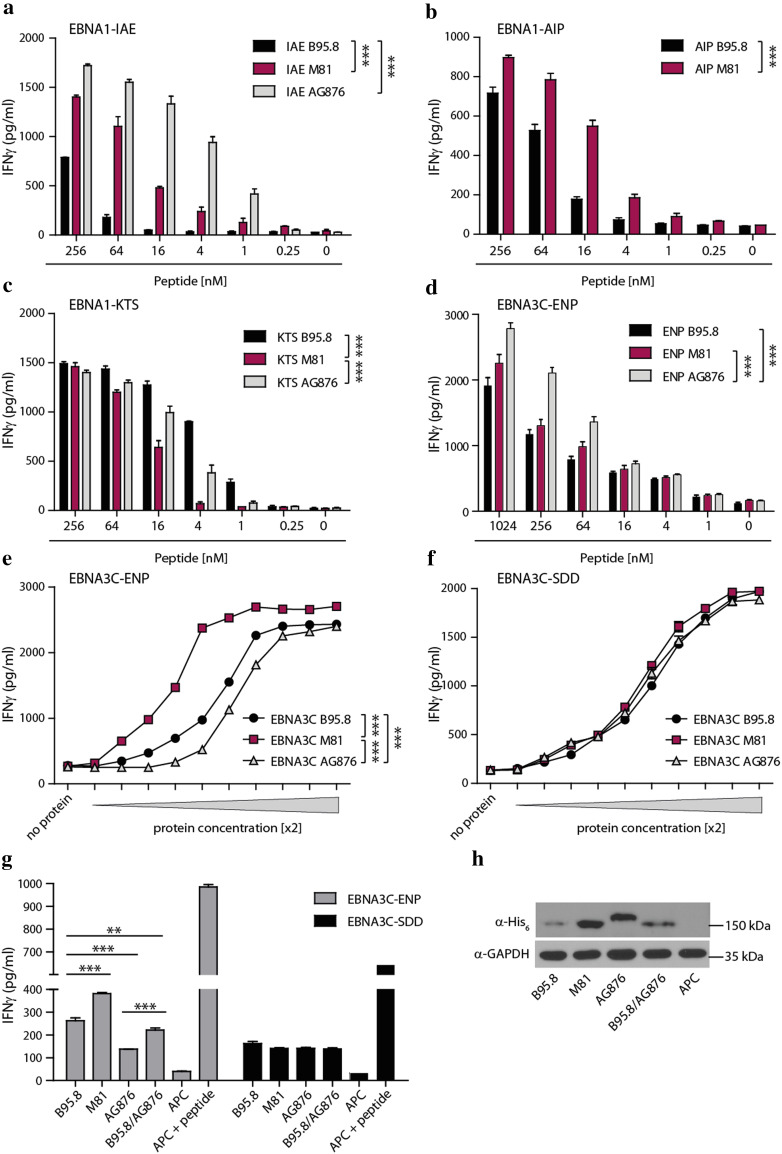
Analogous experiments were performed with CD4+ T-cell clones recognizing four different polymorphic epitopes in EBNA1 and EBNA3C (Table 2). When titrated amounts of the various polymorphic peptide epitopes were pulsed onto target cells and then probed with the T cells, recognition by CD4+ T cells was affected in all cases (Fig. 3a–d). T-cell responses against the IAE and AIP epitopes of EBNA1 as well as the EBNA3C-ENP epitope were significantly increased when peptide epitopes from M81 and AG876 were used (Fig. 3a, b, d), while T-cell reactivity against the EBNA1-KTS epitope from M81 was reduced in comparison to AG876 and B95.8 (Fig. 3c).
Table 2.
Polymorphic published CD4+ T-cell epitopes and flanking regions analyzed in vitro
| EBNA1-IAE | EBNA1-AIP | EBNA1-KTS | EBNA3C-ENP | |
|---|---|---|---|---|
| HLA | DQB1*0402 | DRB1*1301 | DRB1*1101 | DRB1*0801 |
| Epitope coordinates (B95.8) | AA 481–500 | AA 527–541 | AA 514–528 | AA 325–339 |
| B95.8 |
PKFEN IAEGLRALLARSHVE RTTDE |
RGTAL AIPQCRLTPLSRLPF GMAPG |
VYGGS KTSLYNLRRGTALAI PQCRL |
NAPPN ENPYHARRGIKEHVI QNAFR |
| M81 |
PKFEN IAEGLR V LLARSHVE RTTEE |
RGIAL A V PQCR I TPLSRLPF GMAPG |
VYGGS KTSLYNLRRG I ALA V PQCRI |
NAPPN ENPYHARRGIK D HVI QNAFR |
| AG876 |
QKFEN IAEGLR L LLAR C HVE RTTED |
* |
VYGGS KTSLYNLRRG IG LAI PQCRL |
NAPPN ENPYHARRGIKE Q VI QKAFL |
*Same as B95.8 sequence
AA amino acids
Fig. 3.
Recognition by CD4+ T cells is affected by polymorphisms within the epitope and the flanking region. Target cells were pulsed with titrated amounts of peptide epitopes from B95.8, M81, or AG876 and then probed with CD4+ T-cell clones specific for the epitopes EBNA1-AIP (a), EBNA1-IAE (b), EBNA1-KTS (c), and EBNA3C-ENP (d). Target cells were incubated with increasing amounts (twofold increments) of recombinant and purified EBNA3C protein of the indicated viruses and then probed with the EBNA3C-specific CD4+ T cells recognizing the polymorphic ENP epitope (e) or the conserved SDD epitope (f). Recognition of endogenously expressed antigen was assessed by transfecting DG75 cells with expression constructs for HLA-DRB1*0801 and -DQB1*0501, and His6-tagged EBNA3C proteins from the three viruses as well as a B95.8-derived EBNA3C mutant in which the C-terminal flanking region was exchanged for the corresponding AG876 sequence (B95.8/AG876). Transfected cells were co-cultured with ENP- and SDD-specific CD4+ T cells (g). Target cells transfected with the HLA molecules alone (APC) served as negative controls, APC pulsed with the cognate peptide epitope as positive controls. EBNA3C expression levels were analyzed by Western blot (h). GAPDH served as gel loading control. Statistical significance was evaluated using unpaired t test. In dose–response curves, two-sample t test with Bonferroni correction was used. Data are shown as mean ± SD of n = 3 and are representative of at least three independent experiments
Since the flanking regions had not been included in the tested peptide epitopes but also carried amino acid exchanges (Table 2), it remained unknown whether they affected T-cell recognition as well. Using EBNA3C as example, full-length proteins of the three viruses were recombinantly expressed, increasing amounts loaded onto target cells and T-cell recognition assessed (Fig. 3e). For adjusting protein concentrations, a second EBNA3C-specific CD4+ T-cell clone was used, which recognized a non-polymorphic epitope (SDD) embedded in conserved flanking regions in all three viral strains (Fig. 3f, g). Despite a better recognition of the EBNA3C-ENP epitope from AG876 in peptide titration experiments (Fig. 3d), significantly reduced T-cell activation levels were achieved with exogenous (Fig. 3e), or endogenously expressed (Fig. 3g) EBNA3C from AG876 than from B95.8 and especially from M81 (Fig. 3e–g). Since EBNA3C from AG876 and M81 were expressed at similar and consistently higher levels than the B95.8 homologue, variations in transfection efficiency or expression level were unlikely to account for these results (Fig. 3h). To further address the role of flanking regions in antigen presentation, the C-terminal flanking region of EBNA3C from B95.8 was exchanged for the corresponding, polymorphic sequence from AG876, and the resulting B95.8/AG876 chimera included in this analysis. After expression in DG75 cells, recognition of the chimeric antigen by ENP-specific T cells was significantly diminished in comparison to the original B95.8 version, demonstrating that in addition to polymorphisms in epitopes, sequence variations in flanking regions can also affect CD4+ T-cell recognition.
CD8+ T-cell responses in healthy virus carriers rarely cover all antigen variants
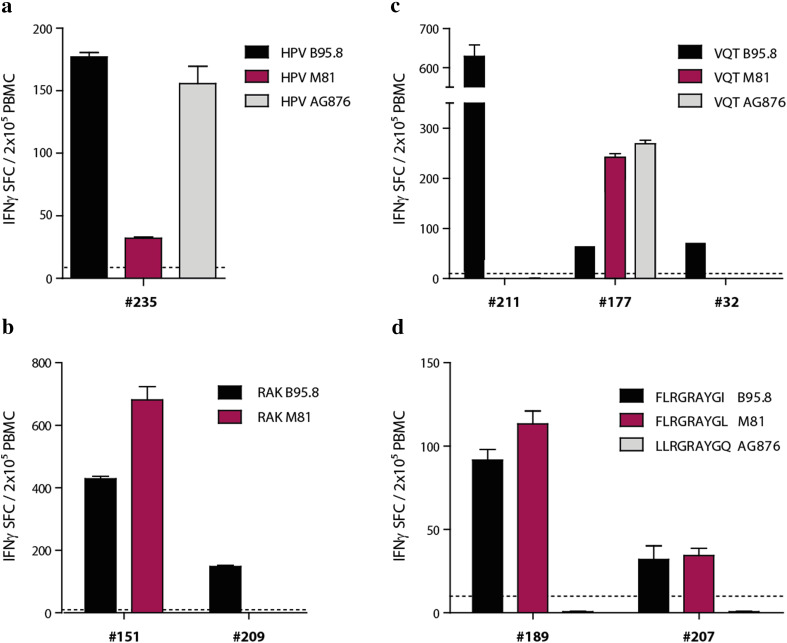
The virus-specific T-cell response is known to encompass numerous T-cell specificities that are directed against the same epitope. Thus, some of these T-cell specificities might be able to recognize antigen variants and thereby afford protection against infection with different viral strains. To address this possibility, the CD8+ T-cell response against polymorphic peptide epitopes was quantified in peripheral blood of healthy virus carriers ex vivo (Fig. 4). The magnitude of the CD8+ T-cell responses against the different HPV (Fig. 4a) and RAK peptide epitopes (Fig. 4b) in donors #235 and #151, respectively, was almost identical to the cytokine secretion patterns observed with the T-cell clones that had been isolated from these donors (Fig. 2a, b). Thus, the CD8+ T cell responses in these donors either were dominated by T cells with a similar recognition pattern as the investigated T-cell clones, or included T cells specific for all antigen variants but with different frequencies. Interestingly, donor #209 only showed responses against the RAK epitope from B95.8, suggesting that different donors can mount responses against all or individual variants. In the case of the VQT epitope, donors #211 and also #32 (from whom T-cell clones had been isolated) responded to the B95.8 variant only, whereas donor #177 had mounted an immune response against all three variants. To further substantiate these findings, the HLA-B*0801-restricted FLR epitope derived from EBNA3A, which varies in all three viral strains, was included in this analysis (Fig. 4d). Both tested donors showed responses against the epitopes from B95.8 and M81 but not from AG876. These results suggest that the CD8+ T-cell response in healthy virus carriers is mostly directed against individual polymorphic epitopes and may ignore antigen variants.
Fig. 4.
The CD8+ T-cell response against polymorphic peptide epitopes varies between donors and is often strain-specific. PBMCs from EBV-seropositive donors (identified by numbers) with appropriate HLA class I types were screened ex vivo for T-cell responses against the polymorphic EBNA1-HPV (a), BZLF1-RAK (b), BZLF1-VQT (c), and EBNA3A-FLR (d) peptide epitopes in IFNγ ELISpot assays. Results are expressed as the mean number of spot forming cells (SFC) in 2 × 105 PBMCs ± SD from biological triplicates. Dashed lines denote limit of detection. T-cell clones specific for HPV, VQT and RAK epitopes that were used in experiments are shown in Fig. 2 were derived from donors #235, #32, and #151
Discussion
Although sequence heterogeneity among EBV isolates from different parts of the world is well-established, the implications of strain variation on the efficacy of vaccine and immunotherapeutic approaches that are predominantly based on antigens derived from the laboratory EBV strain B95.8, have remained elusive. Here, we compared protein sequences in B95.8, AG876, and M81, three distantly related EBV strains, and assessed the impact of sequence disparity on T-cell immunity.
Consistent with previous reports [5, 23], the proteomes of M81 and other NPC-derived viral strains differed from that of B95.8 by approximately 1.2%. With 3.2%, the overall sequence disparity between M81 and the type 2 strain AG876 was markedly higher due to the well-documented divergence in EBNA2 and EBNA3 family of proteins.
When published T-cell epitopes were compared between B95.8 and M81, more than 50% of all CD4+ T-cell epitopes and almost 40% of all CD8+ T-cell epitopes were found to be polymorphic either in the published epitope sequence itself and/or in the epitope flanking regions. The percentage of polymorphic epitopes was even higher when the type 1 strains B95.8 and M81 were compared to the type 2 strain AG876; more than 70% of all CD4+ and more than 40% of all CD8+ T-cell epitopes and their flanking regions were found to be polymorphic. However, one has to keep in mind that many of the published CD4+ T-cell epitopes were defined using overlapping peptide libraries of 15 or 20 amino acids in length (Supplementary table 1). Yet, the core sequence of CD4+ T-cell epitopes usually encompasses only 9 amino acids [29]. Consequently, some of the identified polymorphisms may reside outside the core epitope or its flanking regions and the total number of polymorphisms in CD4+ T-cell core epitopes and/or flanking regions may have been over-estimated.
Amino acid exchanges within or in close proximity to T-cell epitopes can affect T-cell recognition by altering antigen processing, presentation, and binding to MHC and the T-cell receptor (TCR) [30–34]. To account for this multitude of possible effects, peptide titration experiments were complemented with studies on full-length proteins either exogenously loaded onto, or endogenously expressed in APCs. These experiments demonstrated that polymorphisms within or next to EBV epitopes can affect T-cell recognition in different and unpredictable ways. Some amino acid exchanges enhanced (e.g., BZLF1-RAK, B95.8 versus M81), retained (e.g., EBNA1-HPV, B95.8 versus AG876), impaired (EBNA1-HPV, B95.8 versus M81), or abrogated T-cell recognition (BZLF1-VQT, B95.8 versus M81 and AG876). In many cases, more than one order of magnitude higher concentrations of the variant peptides were required for T-cell activation, which may compromise or even abolish recognition of target cells infected with the variant viral strain.
Amino acid exchanges occurred in regions known to interact with the TCR as well as in anchor residues [32]. In most cases, the impact of these amino acid exchanges on T-cell activation cannot be predicted and needs to be verified immunologically. Furthermore, the contrasting results obtained in T-cell assays with “CLG” peptide-pulsed versus LMP2A-transfected target cells implied that single amino acid exchanges can impact on antigen processing and presentation that is not recapitulated by exogenous loading of peptide epitopes.
In the case of CD4+ T cells, divergent results were obtained when EBNA3C “ENP” epitope-specific CD4+ T cells were probed with target cells that had been pulsed with epitope peptides versus incubated with exogenous full-length EBNA3C protein or probed with target cells endogenously expressing EBNA3C, suggesting that amino acid exchanges can also affect antigen processing and presentation on MHC II. Here, this effect was mediated by epitope flanking regions, either by altering protein processing, e.g., by destroying or creating protease cleavage sites, or by modulating TCR binding when located at the C-terminus [31, 34].
Unexpectedly, the EBNA1-IAE epitope variants from AG876 and M81 were much better recognized than the B95.8-derived peptide. This T-cell clone had been established from PBMC of a healthy European virus carrier by repeated stimulation with recombinant EBNA1 protein from B95.8, which is thought to predominate in carriers of the Western world [22]. In light of the newly described EBV strain heterogeneity and the demonstrated effects of polymorphisms on T-cell recognition, we sought to characterize the viral strain carried by this donor, established spontaneous LCL from peripheral blood and sequenced the EBNA1 gene. Surprisingly, the “IAE” epitope sequence in this strain (QKFEN-IAEGLRTLLARCHVE-RTTDE) differed from all three strains examined, and target cells pulsed with this IAE peptide were recognized by the autologous T cells with highest affinity (data not shown). Whether such strain differences also accounted for the increased T-cell recognition of some epitopes derived from M81 or AG876 (e.g., BZLF1-RAK) and if so, whether T cells isolated from a donor infected with a B95.8-homologous strain would still show better recognition of the corresponding epitopes from M81 and AG876, remains to be established.
Furthermore, the CD8+ T-cell response against polymorphic epitopes appears to vary between donors and to be focused on strain-specific epitope variants. Whether these donor-specific differences in the natural CD8+ T-cell response against EBV were due to the infection with different viral strains and/or genetic differences between donors (e.g., HLA type), is currently not known. Given the large number of polymorphic T-cell epitopes and different viral strains around the world [8], and the potential implications of this antigen variety on immunity against infection with strain variants, this issue warrants further investigation. Additional studies are also needed to assess whether polymorphisms in T-cell epitopes are immune-driven escape mutants and whether antigenicity and pathogenicity of a viral strain correlate.
These findings have implications for immune monitoring, vaccine design, as well as immunotherapy of EBV-associated disorders. In immunosuppressed transplant recipients and cancer patients, virus-specific T-cell responses are often monitored using overlapping peptide libraries covering mostly latent cycle proteins e.g. EBNA3 [35, 36]. These peptide libraries are based on B95.8 sequences and may incompletely measure virus-specific T-cell responses if the patient is infected with a different viral strain. Likewise, current approaches for the design of therapeutic EBV vaccines, which aim at increasing and sustaining the number of virus-specific T cells in persistently infected individuals, are mostly based on latency proteins from B95.8 virus [37–39]. Such vaccines may fail to boost relevant T-cell responses in vaccinees infected with strain variants. Furthermore, EBV-specific T-cell preparations for the treatment of PTLD in hematopoietic stem cell transplant recipients are generally derived from the graft donor or, when virus-naïve, from partially HLA-matched third party donors [11, 38, 40]. In most cases, B95-8 sequence-derived peptide mixes are used as source of antigen for the generation of virus-specific T-cell lines [17, 18, 40–43]. However, T-cell donor and recipient may be infected with different viral strains, which may result in diminished clinical efficacy of the infused T cells. Consequently, peptide-selected T-cell preparations for clinical use should preferably focus on conserved T-cell epitopes or target a broad set of viral antigens. The future incorporation of sequence information from all published virus isolates into our epitope registry is expected to identify viral epitopes that are conserved in all strains or at least in subgroups, such as those prevalent in certain geographical regions. Alternatively, diagnostic methods for rapid virus typing in B cells from T-cell donor and recipient and/or in tumor tissue may be developed to facilitate the identification of customized sets of antigens for individualized T-cell therapy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The excellent technical support by Grit Müller-Neumann and Dorothea Seubert is greatly appreciated.
Abbreviations
- EBNA
Epstein–Barr virus nuclear antigen
- EBV
Epstein–Barr virus
- GA
Glycine–alanine
- His6
Hexahistidine tag
- IFN-γ
Interferon-gamma
- LCL
Lymphoblastoid B-cell line
- LMP
Latent membrane protein
- NPC
Nasopharyngeal carcinoma
- PTLD
Post-transplant lymphoproliferative disease
Author contributions
U. Behrends and J. Mautner conceived and designed the study. A. Cirac, S. Stützle, M. Dieckmeyer, and D. Adhikary contributed to the acquisition, analysis, and interpretation of data. A. Cirac and M. Dieckmeyer performed the statistical analysis. A. Moosmann, N. Körber, T. Bauer, K. Witter, and H-J. Delecluse provided essential reagents and technical and material support. A. Cirac, U. Behrends and J. Mautner wrote the paper and all authors made substantial contributions to data analysis and interpretation, manuscript editing, review and approval.
Funding
This study was supported by the German Center for Infection Research - DZIF (TTU 07.804). A. Cirac was supported by the Technische Universität München (TUM) Graduate School.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from all individual participants included in the study. All procedures involving human participants were in accordance with the ethical standards of the institutional research committee of the Technische Universität München and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Footnotes
Josef Mautner and Uta Behrends are joint senior authors.
References
- 1.Longnecker RM, Kieff E, Cohen JI. Epstein–Barr virus. In: Knipe DM, Howley PM, editors. Fields virology. 6. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2013. pp. 1898–1959. [Google Scholar]
- 2.Rickinson AB, Kieff E. Epstein–Barr Virus. In: Knipe DM, Howley PM, editors. Fields virology. 5. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2575–2627. [Google Scholar]
- 3.Young LS, Rickinson AB. Epstein–Barr virus: 40 years on. Nat Rev Cancer. 2004;4(10):757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 4.Chang CM, Yu KJ, Mbulaiteye SM, Hildesheim A, Bhatia K. The extent of genetic diversity of Epstein–Barr virus and its geographic and disease patterns: a need for reappraisal. Virus Res. 2009;143(2):209–221. doi: 10.1016/j.virusres.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai MH, Raykova A, Klinke O, Bernhardt K, Gartner K, Leung CS, Geletneky K, Sertel S, Munz C, Feederle R, Delecluse HJ. Spontaneous lytic replication and epitheliotropism define an Epstein–Barr virus strain found in carcinomas. Cell Rep. 2013;5(2):458–470. doi: 10.1016/j.celrep.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Feederle R, Klinke O, Kutikhin A, Poirey R, Tsai MH, Delecluse HJ. Epstein–Barr virus: from the detection of sequence polymorphisms to the recognition of viral types. Curr Top Microbiol Immunol. 2015;390(Pt 1):119–148. doi: 10.1007/978-3-319-22822-8_7. [DOI] [PubMed] [Google Scholar]
- 7.Farrell PJ. Epstein–Barr virus strain variation. Curr Top Microbiol Immunol. 2015;390(Pt 1):45–69. doi: 10.1007/978-3-319-22822-8_4. [DOI] [PubMed] [Google Scholar]
- 8.Palser AL, Grayson NE, White RE, Corton C, Correia S, Ba Abdullah MM, Watson SJ, Cotten M, Arrand JR, Murray PG, Allday MJ, Rickinson AB, Young LS, Farrell PJ, Kellam P. Genome diversity of Epstein–Barr virus from multiple tumor types and normal infection. J Virol. 2015;89(10):5222–5237. doi: 10.1128/JVI.03614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor GS, Long HM, Brooks JM, Rickinson AB, Hislop AD. The immunology of Epstein–Barr virus-induced disease. Annu Rev Immunol. 2015;33:787–821. doi: 10.1146/annurev-immunol-032414-112326. [DOI] [PubMed] [Google Scholar]
- 10.Long HM, Taylor GS, Rickinson AB. Immune defence against EBV and EBV-associated disease. Curr Opin Immunol. 2011;23(2):258–264. doi: 10.1016/j.coi.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 11.Gottschalk S, Rooney CM. Adoptive T-cell immunotherapy. Curr Top Microbiol Immunol. 2015;391:427–454. doi: 10.1007/978-3-319-22834-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merlo A, Turrini R, Dolcetti R, Zanovello P, Rosato A. Immunotherapy for EBV-associated malignancies. Int J Hematol. 2011;93(3):281–293. doi: 10.1007/s12185-011-0782-2. [DOI] [PubMed] [Google Scholar]
- 13.Bollard CM, Gottschalk S, Torrano V, Diouf O, Ku S, Hazrat Y, Carrum G, Ramos C, Fayad L, Shpall EJ, Pro B, Liu H, Wu MF, Lee D, Sheehan AM, Zu Y, Gee AP, Brenner MK, Heslop HE, Rooney CM. Sustained complete responses in patients with lymphoma receiving autologous cytotoxic T lymphocytes targeting Epstein–Barr virus latent membrane proteins. J Clin Oncol. 2014;32(8):798–808. doi: 10.1200/JCO.2013.51.5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rooney CM, Smith CA, Ng CY, Loftin SK, Sixbey JW, Gan Y, Srivastava DK, Bowman LC, Krance RA, Brenner MK, Heslop HE. Infusion of cytotoxic T cells for the prevention and treatment of Epstein–Barr virus-induced lymphoma in allogeneic transplant recipients. Blood. 1998;92(5):1549–1555. [PubMed] [Google Scholar]
- 15.McAulay KA, Haque T, Urquhart G, Bellamy C, Guiretti D, Crawford DH. Epitope specificity and clonality of EBV-specific CTLs used to treat posttransplant lymphoproliferative disease. J Immunol. 2009;182(6):3892–3901. doi: 10.4049/jimmunol.0803572. [DOI] [PubMed] [Google Scholar]
- 16.Adhikary D, Behrends U, Boerschmann H, Pfunder A, Burdach S, Moosmann A, Witter K, Bornkamm GW, Mautner J. Immunodominance of lytic cycle antigens in Epstein–Barr virus-specific CD4+ T cell preparations for therapy. PLoS One. 2007;2:e583. doi: 10.1371/journal.pone.0000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moosmann A, Bigalke I, Tischer J, Schirrmann L, Kasten J, Tippmer S, Leeping M, Prevalsek D, Jaeger G, Ledderose G, Mautner J, Hammerschmidt W, Schendel DJ, Kolb HJ. Effective and long-term control of EBV PTLD after transfer of peptide-selected T cells. Blood. 2010;115(14):2960–2970. doi: 10.1182/blood-2009-08-236356. [DOI] [PubMed] [Google Scholar]
- 18.Icheva V, Kayser S, Wolff D, Tuve S, Kyzirakos C, Bethge W, Greil J, Albert MH, Schwinger W, Nathrath M, Schumm M, Stevanovic S, Handgretinger R, Lang P, Feuchtinger T. Adoptive transfer of Epstein–Barr virus (EBV) nuclear antigen 1-specific t cells as treatment for EBV reactivation and lymphoproliferative disorders after allogeneic stem-cell transplantation. J Clin Oncol. 2013;31(1):39–48. doi: 10.1200/JCO.2011.39.8495. [DOI] [PubMed] [Google Scholar]
- 19.Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: lessons from Epstein–Barr virus. Annu Rev Immunol. 2007;25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- 20.Taylor GS, Long HM, Haigh TA, Larsen M, Brooks J, Rickinson AB. A role for intercellular antigen transfer in the recognition of EBV-transformed B cell lines by EBV nuclear antigen-specific CD4+ T cells. J Immunol. 2006;177(6):3746–3756. doi: 10.4049/jimmunol.177.6.3746. [DOI] [PubMed] [Google Scholar]
- 21.Adhikary D, Behrends U, Moosmann A, Witter K, Bornkamm GW, Mautner J. Control of Epstein–Barr virus infection in vitro by T helper cells specific for virion glycoproteins. J Exp Med. 2006;203(4):995–1006. doi: 10.1084/jem.20051287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mautner J, Pich D, Nimmerjahn F, Milosevic S, Adhikary D, Christoph H, Witter K, Bornkamm GW, Hammerschmidt W, Behrends U. Epstein–Barr virus nuclear antigen 1 evades direct immune recognition by CD4+ T helper cells. Eur J Immunol. 2004;34(9):2500–2509. doi: 10.1002/eji.200324794. [DOI] [PubMed] [Google Scholar]
- 23.Kwok H, Wu CW, Palser AL, Kellam P, Sham PC, Kwong DL, Chiang AK. Genomic diversity of Epstein–Barr virus genomes isolated from primary nasopharyngeal carcinoma biopsy samples. J Virol. 2014;88(18):10662–10672. doi: 10.1128/JVI.01665-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker A, Skibbe K, Steinmann E, Pfaender S, Kuntzen T, Megger DA, Groten S, Sitek B, Lauer GM, Kim AY, Pietschmann T, Allen TM, Timm J. Distinct escape pathway by hepatitis C virus genotype 1a from a dominant CD8+ T cell response by selection of altered epitope processing. J Virol. 2015;90(1):33–42. doi: 10.1128/JVI.01993-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steers NJ, Currier JR, Jobe O, Tovanabutra S, Ratto-Kim S, Marovich MA, Kim JH, Michael NL, Alving CR, Rao M. Designing the epitope flanking regions for optimal generation of CTL epitopes. Vaccine. 2014;32(28):3509–3516. doi: 10.1016/j.vaccine.2014.04.039. [DOI] [PubMed] [Google Scholar]
- 26.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11(12):823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 27.Desai DV, Kulkarni-Kale U. T-cell epitope prediction methods: an overview. Methods Mol Biol. 2014;1184:333–364. doi: 10.1007/978-1-4939-1115-8_19. [DOI] [PubMed] [Google Scholar]
- 28.Rancan C, Schirrmann L, Huls C, Zeidler R, Moosmann A. Latent membrane protein LMP2A impairs recognition of EBV-infected cells by CD8+ T Cells. PLoS Pathog. 2015;11(6):e1004906. doi: 10.1371/journal.ppat.1004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holland CJ, Cole DK, Godkin A. Re-directing CD4(+) T cell responses with the flanking residues of MHC class II-bound peptides: the core is not enough. Front Immunol. 2013;4:172. doi: 10.3389/fimmu.2013.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ossendorp F, Eggers M, Neisig A, Ruppert T, Groettrup M, Sijts A, Mengede E, Kloetzel PM, Neefjes J, Koszinowski U, Melief C. A single residue exchange within a viral CTL epitope alters proteasome-mediated degradation resulting in lack of antigen presentation. Immunity. 1996;5(2):115–124. doi: 10.1016/S1074-7613(00)80488-4. [DOI] [PubMed] [Google Scholar]
- 31.Hastings KT. GILT: shaping the MHC class II-restricted peptidome and CD4(+) T cell-mediated immunity. Front Immunol. 2013;4:429. doi: 10.3389/fimmu.2013.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- 33.Neefjes J, Ovaa H. A peptide’s perspective on antigen presentation to the immune system. Nat Chem Biol. 2013;9(12):769–775. doi: 10.1038/nchembio.1391. [DOI] [PubMed] [Google Scholar]
- 34.Cole DK, Gallagher K, Lemercier B, Holland CJ, Junaid S, Hindley JP, Wynn KK, Gostick E, Sewell AK, Gallimore AM, Ladell K, Price DA, Gougeon ML, Godkin A. Modification of the carboxy-terminal flanking region of a universal influenza epitope alters CD4(+) T-cell repertoire selection. Nat Commun. 2012;3:665. doi: 10.1038/ncomms1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tischer S, Dieks D, Sukdolak C, Bunse C, Figueiredo C, Immenschuh S, Borchers S, Stripecke R, Maecker-Kolhoff B, Blasczyk R, Eiz-Vesper B. Evaluation of suitable target antigens and immunoassays for high-accuracy immune monitoring of cytomegalovirus and Epstein–Barr virus-specific T cells as targets of interest in immunotherapeutic approaches. J Immunol Methods. 2014;408:101–113. doi: 10.1016/j.jim.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 36.Korber N, Behrends U, Hapfelmeier A, Protzer U, Bauer T. Validation of an IFNgamma/IL2 FluoroSpot assay for clinical trial monitoring. J Transl Med. 2016;14(1):175. doi: 10.1186/s12967-016-0932-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor GS, Steven NM. Therapeutic vaccination strategies to treat nasopharyngeal carcinoma. Chin Clin Oncol. 2016;5(2):23. doi: 10.21037/cco.2016.03.20. [DOI] [PubMed] [Google Scholar]
- 38.Smith C, Khanna R. The Development of prophylactic and therapeutic EBV vaccines. Curr Top Microbiol Immunol. 2015;391:455–473. doi: 10.1007/978-3-319-22834-1_16. [DOI] [PubMed] [Google Scholar]
- 39.Cohen JI. Epstein–barr virus vaccines. Clin Transl Immunol. 2015;4(1):e32. doi: 10.1038/cti.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bollard CM, Heslop HE. T cells for viral infections after allogeneic hematopoietic stem cell transplant. Blood. 2016;127(26):3331–3340. doi: 10.1182/blood-2016-01-628982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haque T, Wilkie GM, Jones MM, Higgins CD, Urquhart G, Wingate P, Burns D, McAulay K, Turner M, Bellamy C, Amlot PL, Kelly D, MacGilchrist A, Gandhi MK, Swerdlow AJ, Crawford DH. Allogeneic cytotoxic T-cell therapy for EBV-positive posttransplantation lymphoproliferative disease: results of a phase 2 multicenter clinical trial. Blood. 2007;110(4):1123–1131. doi: 10.1182/blood-2006-12-063008. [DOI] [PubMed] [Google Scholar]
- 42.O’Reilly RJ, Prockop S, Hasan AN, Koehne G, Doubrovina E. Virus-specific T-cell banks for ‘off the shelf’ adoptive therapy of refractory infections. Bone Marrow Transpl. 2016;51(9):1163–1172. doi: 10.1038/bmt.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bollard CM. Improving T-cell therapy for Epstein–Barr virus lymphoproliferative disorders. J Clin Oncol. 2013;31(1):5–7. doi: 10.1200/JCO.2012.43.5784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.