Abstract
Breast cancer (BCa) is a heterogeneous disease with different histological, prognostic and clinical aspects. Therefore, the need for identification of novel biomarkers for diagnosis, prognosis and monitoring of disease, as well as treatment outcome prediction remains at the forefront of research. The search for circulating elements, obtainable by simple peripheral blood withdrawal, which may serve as possible biomarkers, constitutes still a challenge. In the present study, we have evaluated the expression of 6 circulating miRNAs, (miR-16, miR-21, miR-23α, miR-146α, miR-155 and miR-181α), in operable BCa patients, with non-metastatic, invasive ductal carcinoma, not receiving neoadjuvant chemotherapy. These miRNAs, known to be involved in both tumor cell progression and immune pathways regulation, were analyzed in relation to circulating cytokines, tumor immune-cell infiltration and established prognostic clinicopathological characteristics. We have identified three different clusters, with overall low (C1), moderate (C2) or high (C3) expression levels of these six circulating miRNAs, which define three distinct groups of non-metastatic BCa patients characterized by different clinicopathological and immune-related characteristics, with possibly different clinical outcomes. Our data provide the proof-of-principle to support the notion that, up- or down-regulation of the same circulating miRNA may reflect different prognosis in BCa. Nonetheless, the prognostic and/or predictive potential of these three “signatures” needs to be further evaluated in larger cohorts of BCa patients with an, at least, 5-year clinical follow-up.
Electronic supplementary material
The online version of this article (10.1007/s00262-018-2252-7) contains supplementary material, which is available to authorized users.
Keywords: MiRNAs signatures, Breast cancer, Tumor infiltration, Biomarkers, Cytokines/chemokines
Introduction
Breast cancer (BCa) is a heterogeneous disease with different histological, prognostic and clinical aspects. This heterogeneity is strengthened by the dynamic interactions among the different cells composing the solid tumor mass (malignant cells, nonmalignant cells including immune cell subpopulations) [1]. Our better understanding of BCa at both the cellular and molecular levels has increased greatly during the past few years and this resulted in the establishment of new therapeutics. Nevertheless, further improvement of better stratification among patients is still required [2]. Thus, the need for identification of novel biomarkers for diagnosis, prognosis and monitoring of disease, as well as treatment outcome prediction remains at the front of research.
A biomarker represents an indicator of a time-depended biological state that can objectively measure and compare normal biological and pathogenic processes, or pharmacological responses to a therapeutic protocol [3]. The search for circulating elements, obtainable by simple peripheral blood withdrawal, which may serve as biomarker candidates, reflecting individual cancer characteristics, constitutes still a challenge [4].
Non-coding RNAs serve as promising biomarkers due to their involvement in gene expression regulation and in the development of BCa. Recent studies have focused on shedding light on the functions of long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), and few of them have investigated how lncRNAs and miRNAs are transcriptionally regulated [5].
MiRNAs are stably expressed in body fluids (e.g. serum and plasma), which are easily, non-invasively accessible, through liquid biopsies, thus allowing the evaluation and monitoring of patients [6, 7]. miRNAs are a group of small endogenous, single-stranded noncoding RNAs with approximately 19–24 nucleotides that function in the post-transcriptional regulation of gene expression by binding to the 3′ untranslated region (UTR) of the target mRNAs. The interactions between miRNAs and mRNAs, result in the destabilization of the target mRNA molecule and interfere with translation [8, 9]. Recent evidence supports that there is an interaction between miRNAs and lncRNAs, reducing their regulatory effect on mRNAs [10]. Moreover, because a single miRNA may have many potential mRNA targets, the abnormal miRNA expression is capable to influence the expression pattern of several mRNAs and proteins and, consequently, be involved in the initiation of many diseases, such as cancer [11]. A common trait of human cancers is the differential miRNA pattern of expression between cancer patients as compared with healthy individuals and a global miRNA deregulation [12–16]. Furthermore, miRNAs are also essential regulators of the differentiation, activation, proliferation and function of all the components of the immune system [17].
The role of the immune system as a key mediator in the multistep process of cancer development and progression is well established [18, 19]. As mentioned above, tumor microenvironment contains different types of immune cells, which contribute to regulate the fine balance between anti- and pro-tumor signals [20]. In this context, mechanisms of crosstalk between cancer and immune cells remain to be extensively elucidated. Moreover, there is an interplay between tumor microenvironment and peripheral blood. Therefore, circulating elements (cytokines/chemokines and miRNAs) may correlate with parameters in the tumor microenvironment. To this end, in the present study we have evaluated the expression of 6 circulating miRNAs, (miR-16, miR-21, miR-23α, miR-146α, miR-155 and miR-181α), in operable breast cancer patients, with invasive ductal carcinoma, not receiving neoadjuvant chemotherapy. The aforementioned miRNAs have been selected on the basis of their thorough investigation in cancer and particularly BCa, but also for their involvement in immune processes regulation [8, 17, 21, 22]. These miRNAs are also involved in NF-kB signaling network participating in negative or positive regulation: NF-kB is a transcriptional factor that regulates crucial genes affecting innate and adaptive immunity, cell proliferation, inflammation, and tumor development [23]. The miRNAs were analyzed in relation to circulating cytokines/chemokines, tumor immune-cell infiltration and established prognostic clinicopathological characteristics. These correlations may reflect mechanistic interrelations leading to multiparameter and functional prognostic signatures.
Materials and methods
All patients (n = 48) had non-metastatic invasive ductal carcinoma. None of these patients was treated with neoadjuvant chemotherapy or was enrolled to other research protocols, nor did have any history of other cancer or additional serious health problems. Blood samples from patients with invasive breast cancer were collected, one day prior to surgery, at the St. Savas Cancer Hospital in Athens, between February 2014 and May 2015. Peripheral blood mononuclear cells were isolated from blood using Ficoll–Hypaque gradient and stored in liquid nitrogen before use. Sera were stored at − 80 °C.
These 48 patients, also belonged to a larger cohort of BCa patients along with an additional larger group of BCa patients diagnosed between 2000 and 2010, represent a cohort which has been analyzed for the prognostic value of intratumoral immune infiltrates, as recently reported [24]. There are no clinical follow-up data available yet for any of these prospectively enrolled patients with pre-surgery obtained peripheral blood.
Peripheral blood samples (10 ml) were obtained from healthy volunteers. Aliquots of sera and PBMCs from these individuals were kept frozen and used as control groups.
Measurement of serum- and PBMCs-derived miRNAs
miRNA was extracted from serum using Qiagen miRNeasy Serum/Plasma kit (Qiagen, Hilden, Germany) and from PBMCs using Qiagen miRNeasyMini KIT according to the manufacturer’s instructions. cDNAs generated with the miScript II RT Kit (Qiagen, Hilden, Germany) and used as a template for real-time PCR with the miScript SYBR Green PCR Kit (Qiagen, Hilden, Germany). For the detection of mature miRNAs, we used miScript Primer Assays (Hs_miR-21_2; Hs_miR-181α_2; Hs_miR-146α_1; Hs_miR-23α_2; Hs_miR-16_2; Hs_miR-155_2) (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. As spike in control was used miRNeasy Serum/Plasma Spike-In Control C-39 (Qiagen, Hilden, Germany). DDCt method was used for data analysis and melting curve analysis for the specificity of miRNAs expression levels.
Measurement of serum cytokines
Measurement of IL-1Ra, IL-9, IL-8 IL-10 was simultaneously performed by Luminex using the human premixed multi-analyte kit (R&D systems) according to the manufacturer’s instructions. RANTES/Chemokine ligand 5 and TGF-β determinations were performed by separate Luminex-based kits (R&D systems) as described in [25].
Assessment of tumor-infiltrating leukocytes
Formalin-fixed paraffin-embedded (FFPE) tissue blocks were obtained after surgery from the Saint Savas Pathology Department and analyzed for tumor immune cell infiltration along with the cohort of retrospectively investigated patients, described in detail in our previous report [24]. Immunostaining with CD4 (4B12, 1:40; Biogenex), CD8 (SP16, 1:80; Thermo Scientific, USA) and CD163 (10D6, 1:400; Biocare) was performed for all patients using the Leica Bond III automation (Leica Biosystems, Germany) and Leica detection kit (Leica Biosystems, Newcastle, UK), as previously described. Quantification of infiltrating cells/mm2 was performed as described in [24].
Patient HLA typing
HLA genotyping was performed as recently described [25].
Statistical analysis
The initial clustering was performed after data normalization, using the ratios of the obtained miRNA relative expression values (xi) to the median of the respective patient miRNA investigated (δi) by hierarchical clustering using Ward’s Method in IBM SPSS 24. After the creation of the 3 clusters, we used QUEST classification trees in IBM SPSS modeler 18.1 to evaluate separately the prediction power of 7 predictors; the ratios of the 6 miRNAs as well as the sum of their ratios ∑(xi × δi). After determining the sum as the strongest predictor, we used a second QUEST classification tree to determine the predictor importance of the rest of the predictors. The preliminary constructed algorithm, miRNASCORE consists of the sum of the ratios (main predictor), plus the patients’ xi/δi ratio for each miRNA multiplied by the predictor importance (pi) from the second QUEST classification (secondary predictors).The ROC curves were generated using IBM SPSS 24 and the Jouden’s index was calculated using the formula J = sensitivity + specificity − 1. Contingency analysis, as well as the non-parametric Mann–Whitney t tests and Kruskal–Wallis tests were performed using GraphPad Prism 7.00.
Results
Tumor cell infiltration, circulating chemokines/cytokines and miRNAs
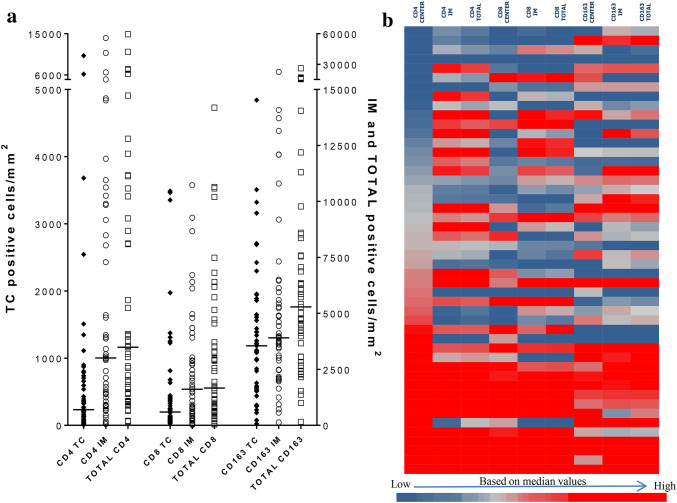
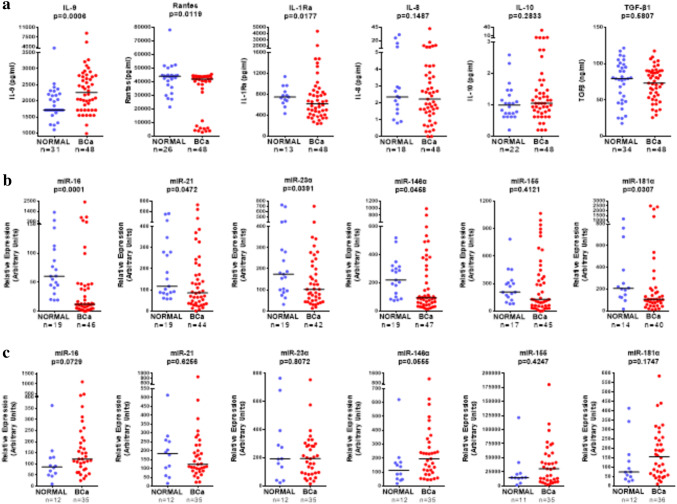
In our recent study [24] we have extensively investigated the density and spatial distribution of immune cells, namely CD8+ and CD163+, in 162 patients with invasive non-metastatic breast cancer. For 97 of them, who belonged to a retrospectively analyzed cohort of patients, clinical follow-up was available, making it possible to establish combination signatures of the infiltrating cells with clinical outcome (retrospective group), thus introducing a novel prognostic immune signature. For 48 of the 162 patients, comprising the prospective cohort of patients, serum and PBMCs, prospectively collected one day prior to surgery, were available. The clinicopathological characteristics of this prospective cohort are presented in Suppl. Table 1. In these patients, apart from CD8+ and CD163+ tumor-immune cell infiltration, we also evaluated distributions of CD4+ cells (Fig. 1a). The heatmap (Fig. 1b) was generated using the median of the CD4, CD8 and CD163 positive cells infiltrating the tumor center (TC), or the invasive margin (IM) and total (i.e. the sum of TC and IM) from all patients of, both, the retrospective and the prospective cohort of our study. Moreover, the levels of serum chemokines/cytokines (Rantes, IL-8, IL-9, IL-10, IL-1Rα, TGF-β1) (Fig. 2a) as well as of serum (Fig. 2b) and PBMC (Fig. 2c) miRNAs (miR-16, miR-21, miR-23α, miR-146α miR-155, miR-181α) were evaluated. Circulating factors were also determined in sera and PBMCs from healthy donors. IL-8, IL-10 and TGF-β1 did not significantly differ between BCa patients and healthy donors (Fig. 2a). Rantes and IL-1Ra were detected in lower levels in BCa patients (Fig. 2a). On the contrary, BCa patients had higher levels of circulating IL-9 (Fig. 2a).
Fig. 1.
Tumor cell infiltration. a Differential distribution of CD4, CD8 and CD163 cells in the tumor center (TC), invasive margin (IM) and as a sum of the two (TOTAL). The numbers represent the amount of positive cells/ mm2. IM and TOTAL cell numbers are presented on the right Y axis because of the generally lower infiltration of the TC. b Heatmap of the CD4, CD8 and CD163 positive cells infiltrating TC, IM and as a total. The heatmap colors were generated using the median of these populations from the total patients of both the retrospective and the prospective cohort of our study
Fig. 2.
Circulating chemokines/cytokines and miRNAs in BCa patients. a Scatter plots representing the levels of cytokines we measured (IL-9, Rantes, IL-1Ra, IL-8, IL-10, TGF-β1), in the sera of healthy donors (blue) and BCa patients(red). The horizontal lines represent the medians. Scatter plots representing the levels of miRNAs we measured (miR-16, miR21, miR-23α, miR-146α, miR-155, miR-181α) in arbitrary units in the sera (b) and in PBMCs (c) of healthy donors (blue) and BCa patients (red). The horizontal lines represent the medians
Serum miRNAs, with the exception of miR-155, showed statistically significant lower relative expression in BCa patients compared to healthy donors (Fig. 2b). However, some high expressors could be detected among BCa patients. No statistically significant differences were observed in PBMCs’ miR-21, miR-23α and miR-155 relative expression levels between BCa patients and healthy donors, in contrast to a strong trend for increased miR-16, miR-146α and miR-181α levels in BCa patients (Fig. 2c).
Correlations of tumor cell infiltration, circulating chemokines/cytokines and miRNAs
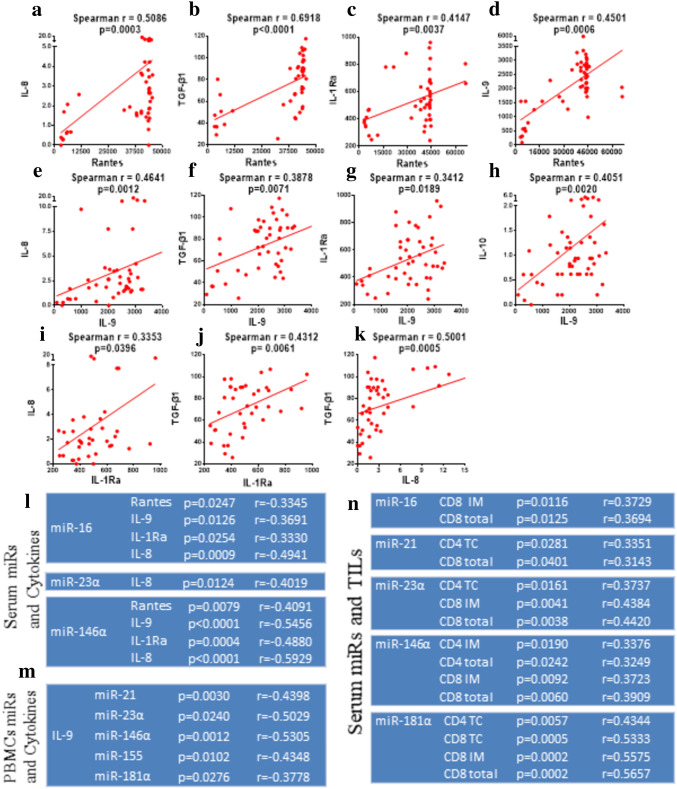
We next sought to find possible correlations among the peripheral factors examined in the previous section (i.e. chemokines and cytokines), but also between these factors and TILs. Interestingly, we could detect significant correlations among chemokines/cytokines in the group of BCa patients (Fig. 3) which could not be observed among healthy donors (suppl. Figure 1). Significant correlations were found when comparing levels of Rantes or IL-9 with each of IL-8 (Fig. 3a, e), TGF-β1(Fig. 3b, f) and IL-1Rα (Fig. 3c, g). A strong correlation was observed when comparing Rantes with IL-9 (Fig. 3d). For IL-10 and Rantes there was a strong trend (p = 0.0786; data not shown). IL-8 also significantly correlated with TGF-β1 (Fig. 3k) and with IL-1Ra (Fig. 3i). Moreover, serum miRNAs significantly correlated versus each other, a finding that was also seen in PBMC miRNAs (suppl. Table 2 and suppl. Table 3). Notwithstanding, such correlations were absent when comparing serum and PBMC miRNAs (data not shown). Serum miR-16 and miR-146α correlated with Rantes, IL-9, IL-1Ra and IL-8 (Fig. 3l). IL-8 also correlated with miR-23α. In line with recent reports [26], the strongest correlation was detected between miR-146α and IL-8, but also between miR-146α and IL-9. This latter cytokine was also found to be related with PBMC miRNAs (except miR-16) (Fig. 3m).
Fig. 3.
Correlations of Tumor cell infiltration, circulating chemokines/cytokines and miRNAs. a–kX–Y plots representing the main correlations between some of the circulating chemokines/cytokines we measured in BCa patients. The Spearman r coefficient, as well as p values are presented above their respected graphs and the regression line was plotted, representing the correlation generated for each set of cytokines/chemokines. l–n Tables representing the observed correlations between circulating miRNAs and cytokines (l), PBMC miRNAs and cytokines (m) and circulating miRNAs and TILS (n) in BCa patients. P values and Spearman coefficient (r) are presented next to each relation
With the exception of miR-155, all serum miRNAs showed correlations with tumor-infiltrating CD4+ or CD8+ cells levels, in TC, IM or both (T) (Fig. 3n). On the other hand, only IL-1Ra correlated with CD8 infiltration in IM (data not shown), while no relation was observed between PBMCs miRNAs and tumor infiltration (data not shown).
Patient stratification using serum derived miRNAs
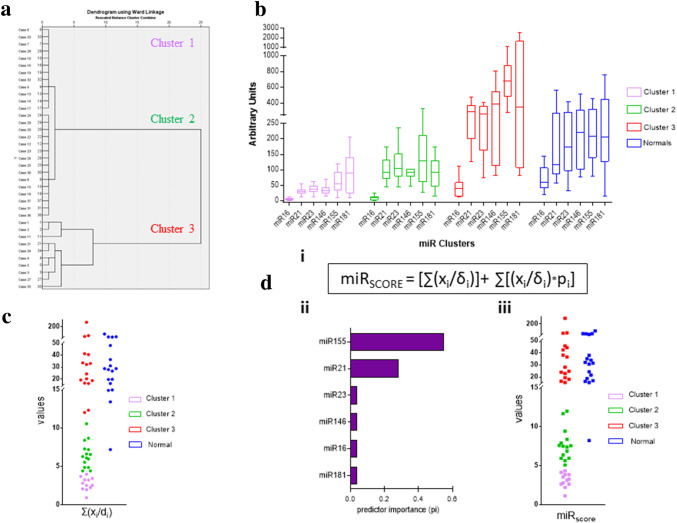
Hierarchical clustering of the normalized values [value (xi)/median (δi)], for each miRNA from 37 patients belonging to the prospective group of patients with no missing values for all of the six miRNAs (without including normal donors), resulted in three distinct clusters (Fig. 4a). Cluster one (C1) consists of patients with uniformly low levels of miRNA expression, whereas cluster two (C2), includes patients with moderate levels of miRNA expression. Finally, the third cluster (C3) generally includes patients with the highest levels of miRNA expression, but with no consistent pattern (Fig. 4b). Taking into account these clusters, we tested if the sum of the individual miRNA normalized values xi/δi, indicative of the overall miRNA expression for each patient, could classify the patients within specific clusters. Using QUEST classification to calculate the predicting potential of the sum, as well as of the individual ratios xi/δi for each of the 6 miRNAs, we confirmed that only the sum of the ratios ∑(xi/δi) (Suppl. Figure 2) could predict the classification of each patient in the respective cluster, although the upper and lower limits between the clusters were not clearly distinguishable (Fig. 4c). This problem was surmounted by creating an algorithm, named miRSCORE (Fig. 4d, i), which incorporates the predicting potential of the 6 miRNAs individually. This was accomplished after a second QUEST classification including all the six predictors, attributing to each one miRNA a predictor importance factor. Each predictor importance factor (Fig. 4d, ii) was multiplied with the corresponding xi/δi miRNA values and added to the sum of the ratios for each patient, resulting in the final miRSCORE algorithm, with an improved discriminative potential among clusters (Fig. 4d, iii). One more advantage of the miRSCORE is that patients with missing values, can be securely assigned as C3 patients if their miRSCORE is higher than the lower limit of cluster 3. After generating their respective ROC curves, each cluster appeared to have clear discrimination (C1 sensitivity = specificity = 1, C2 sensitivity = specificity = 1), with no overlapping patients (Suppl. Figure 3). The values of miRSCORE at which we observed the maximum combination of specificity–sensitivity, (miRSCORE-thresholdC1 = 4.71, miRSCORE-thresholdC2 = 13.75), will be generally considered as the distinctive thresholds, separating the 3 clusters, for patient stratification. By evaluating the normal control group based on the BCa patients clustering analyses, all, but one, of the normal donors, were found to be clustered in C3, both from hierarchical clustering and from scoring with miRSCORE. Although there were statistically significant differences in the expression levels of miR-146α (p = 0.0031), miR-155 (p = 0.0031) and miR-181α (p = 0.0083) between the groups of normal donors and patients in C3 (Suppl. Figure 4a), there was no consistent pattern for discriminating between these two groups.
Fig. 4.
Hierarchical clustering and miRSCORE algorithm. a Dendrogram generated using the xi/δi values in hierarchical clustering with Ward’s Method. The three distinct clusters are presented (from top to bottom: C1, C2, C3). b Tukey plots of the levels of circulating miRNAs levels, in the BCa patients classified by hierarchical clustering, measured in arbitrary units, between C1 (pink), C2 (green), C3 (red) patients and healthy donors (Blue). c Scatter diagram depicting the sum of the ratios (xi/δi) in BCa patients (left) and healthy donors (right). BCa patients are distinguished further between their clusters (C1 pink, C2 green, C3 red). d, i Our preliminary algorithm, miRSCORE, contains 2 variables: the sum of the ratios xi/δi (as presented in C) and the sum of each patient’s ratio (xi/δi) after being multiplied first with the respective miRNAs predictor importance (pi). ii The predictor importance (pi) values for each of our 6 miRNAs in descending order as it was measured using QUEST classification trees. iii Scatter diagram depicting the miRSCORE values for BCa patients (left) and Healthy Donors (right). BCa patients are distinguished further between their clusters [C1 pink (n = 12), C2 green (n = 15), C3 red (n = 16)]
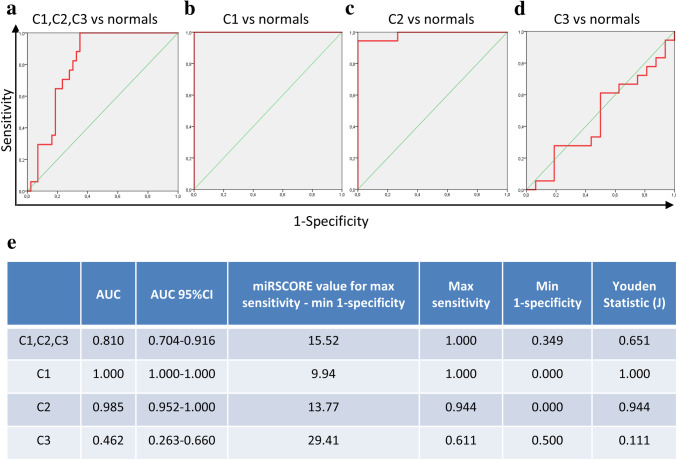
To investigate any possible diagnostic value of miRSCORE, we generated ROC curves to determine our algorithm’s ability to distinguish between BCa patients and healthy donors (Fig. 5). By examining our total patient population, we were presented a ROC curve with diagnostic potential (AUC = 0.810; 95%CI 0.704–0.916, SensitivityMAX = 1.000, SpecificityMAX = 0.651, J = 0.651), reaching a maximum combination of sensitivity–specificity at the threshold of miRSCORE-threshold = 15.52. After observing the individual discriminatory capabilities within each cluster, our previous remark was confirmed. The ability of our algorithm in diagnosis was reduced, due to the high overlap between healthy donors and C3 patients (AUC = 0.462; 95%CI 0.263–0.660). The algorithm values within C1 (AUC = 1.000) and C2 (AUC = 0.985; 95%CI 0.952–1.000) showed a clear distinction between healthy donors and BCa patients. Moreover, there were differences among the clusters and normal donors in terms of serum cytokine levels (Suppl. Figure 4b and 4c), with more prominent those for IL-9 (p = 0.0030), IL-1Ra (p = 0.0153) and IL-8 (p = 0.0058). However, we were not able to establish any characteristic profile that could improve the classification power of the miRSCORE.
Fig. 5.
Receiver operating characteristic (ROC) curve analysis of our clusters. ROC curves showing the distinction of C1, C2, C3 patients (a), C1 (b), C2 (c) and C3 (d) BCa patient algorithm values, with those of normal donors. Table (e) presents some of the important ROC curve metrics such as the AUC (with the 95%CI), the algorithm value at which the best combination of sensitivity–specificity is reached along with the coordinates of this curve point (max sensitivity, min 1-specificity) as well as the Youden statistic (J)
Clinicopathological characteristics and tumor immune signatures of patients in the different miRNA clusters
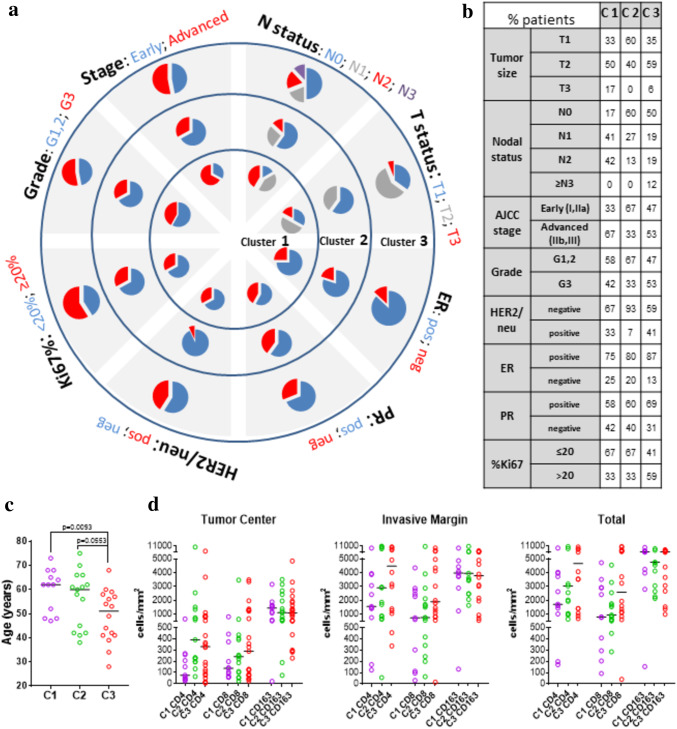
The above-defined clusters, each representing one distinct serum miRNA profile, either immune-related or cancer-related, or both, were checked if they comprise patients with distinct clinicopathological characteristics (Fig. 6a). At a first glance, the clusters seem to differ regarding age, with C3 including mostly patients diagnosed with breast cancer at a younger age than C1 and C2 (Fig. 6c). It is interesting that the majority (> 60%) of patients with the C2 miRNA signature exhibit a more favorable prognostic profile according to TNM staging, tumor grade and low Ki67 (Fig. 6a, b). Furthermore, the majority (93%) of C2 patients are HER2/neu negative. No substantial differences were observed in all 3 clusters regarding hormone receptors, ER and/or PR. C3 cluster, as opposed to C1, comprised more patients with aggressive disease, as evidenced, by younger age, and increased frequencies of patients with less differentiated (grade 3) and highly proliferating (Ki67 > 20%) tumors (Fig. 6b). C1 and C2 are composed of patients with different nodal status (p = 0.0497). Moreover, there are important tendencies when combining clinicopathological characteristics, especially nodal status and HER2/neu expression, p = 0.0502, nodal status & Stage, p = 0.0537. There is also, a very strong trend p = 0.0501, when we collectively examine nodal status, HER2/neu status and Stage between patients of the two clusters. Furthermore, there are significant differences between the C2 and C3 patients regarding HER2/neu status (p = 0.0184). In addition, we observed trends by examining HER2/neu status combined with Ki67% and HER2/neu status combined with Stage between C2 and C3, (p = 0.0657 and p = 0.0919, respectively). The combination of these three parameters (i.e. HER2/neu, Stage and Ki67%) slightly differentiated the two clusters leaning towards significance (p = 0.1516). Similarly, C1 and C3 patients exhibited a trend for the nodal status combined with Ki67% (p = 0.1795). It is worth mentioning that we consider these trends significant, since the comparison of the percentages resulted in strong statistical significances (p < 0.0001), indicating that not reaching statistical significance might be attributed to the small number of patients in some groups.
Fig. 6.
Clinicopathological characteristics and tumor infiltration of patients in the different miRNA clusters. a Complex pie-chart representing the differential distribution of the main clinicopathological characteristics in each vector (T), nodal status (N), AJCC staging, histological Grade, Ki67%, HER2/neu and Hormone Receptor Status. Each circle represents a different cluster, with C1 in the center, C2 in the middle and C3 on the outside of the circle. The colors ranging from blue, grey, red, purple, represent clinicopathological characteristics with ascending worse prognosis. b The age distribution of the BCa patients within our clusters. C1 patients are presented with pink, C2 with green and C3 with red. c Table with the distribution of clinicopathological characteristics between the clusters in percentages, as presented in a. d Differential distribution of CD4, CD8 and CD163 positive cells infiltrating the TC, IM and total. Values are colored in purple for C1, in green for C2 and in red for C3. The horizontal lines represent each group’s median
There were also correlations between the clusters and tumor immune infiltration. We could detect differences in CD4 and CD8 T cell infiltration, with a statistically significant difference in TC CD4 between C1 and C2 (p = 0.0054), and an almost significant difference between C1 and C3 (p = 0.0547). CD163 tumor infiltration did not differ among clusters (Fig. 6d). Based on the combination of tumor CD8 and CD163 cell infiltration in the TC and the IM, we have previously defined a favorable signature (FCIS, with high (H) CD8 cell infiltration in the TC and low (L) in the IM (i.e. CD8 HL) and/or CD163 LH), an unfavorable (UCIS, CD8 LH or CD163 HL and none of the good profiles, CD8 HL/CD163 LH) and a remaining group of all other combinations (REST). These combined signatures were related to the clinical outcome of a retrospectively evaluated cohort of 97 patients. When comparing the percent distributions of each infiltration signature within each cluster (suppl. Figure 5a), there were differences among clusters (p = 0.0021). C1 and C3 were very similar, whereas there was an increased frequency of patients with the FCIS signature and lower UCIS patients in the C2 cluster.
HLA and miRNA clusters
We have previously presented data suggesting that some HLA alleles are related to better prognosis and/or better prediction of clinical response after immunization [27–29]: HLA-A*24, HLA-A3 superfamily alleles (A*03, A*11, A*31, A*33, A*68), and HLA-DRB1*11 expressing patients have a prognostic advantage compared to HLA-A*02 expressors. Based on these preliminary observations, we compared the distribution of patients expressing each allele among the different miRNA clusters. Cluster 3 comprised a significantly increased number of HLA-A*02+ and the lowest number of HLA-A*24+ and HLA-DRB1*11+ patients, whereas all C1 patients expressed alleles of the A3 superfamily. The lowest number of HLA-A*02+ patients was observed in C2 (suppl. Figure 5b).
Discussion
To our knowledge, this is the first report to show that the evaluation of 6 serum miRNAs (miR-16, miR-21, miR-23α, miR-146α, miR-155 and miR-181α) defines three different groups/clusters of BCa patients with different clinicopathological characteristics, tumor infiltration status, circulating cytokines/chemokines levels and HLA allele expression frequencies.
In a cohort of non-metastatic and non-neoadjuvant treated BCa patients with invasive ductal carcinoma, the overall pre-surgery serum miRNAs expression levels were found to be lower than those quantified in normal individuals. However, by carefully observing the diverse expression of these miRNAs among individual patients, it was apparent that each miRNA could be either overexpressed/upregulated or downregulated (or even remain within normal range), thus probably reflecting different disease situations. Circulating miR-21 and miR-155 have been extensively investigated in BCa and in most cases they were reported to be upregulated [30] and thus suggested to harbor diagnostic potential. However, there are also studies documenting decreased levels of these two miRNAs in patients with invasive BCa compared to controls [31], which is in agreement with our findings. Similarly, serum miR-16 and miR-181α levels in BCa have been found either up- or down-regulated by different groups and correlated with high risk, and tumor development and metastasis, respectively [15, 32]. Circulating levels of miR-23α and miR-146α have not so far been investigated in BCa, although their upregulation has been documented in other cancer types [21, 22]. Signatures incorporating some of the above miRNAs have been suggested by several groups to possess an important diagnostic impact [8].
To evaluate if differential expression levels of all six miRNAs could distinguish between different groups of BCa patients at diagnosis, we applied hierarchical clustering which revealed three distinct groups of patients, namely C1 with low overall miRNA expression, C2 with intermediate overall expression and C3 with high overall expression. Using an algorithm to classify individuals within each cluster, no normal donor was found exhibiting C1 miRNA levels profile, and only one out of 18 (5.5%) was classified within the C2 patient group. Thus, C1 and C2 miRNA signatures have a potential diagnostic role, which may not be the case for C3 because more than one-third of the BCa patients (37%), belonging to this cluster had a miRNA profile which was very similar to that defined in the majority of the normal population. This was further confirmed by ROC analyses using as cutoff the C2 cutoff value, where sensitivity was very high (94.4%), but specificity fairly exceeded a 60%, rendering the overall diagnostic potential of the signature quite poor.
Interestingly, we found that patients classified in the three different groups possess different clinicopathological characteristics with established prognostic relevance, thus offering possible prognostic miRNA signatures. Those belonging to C2 seem to have the best overall profile, as more than two-third of them were at earlier stages (I or IIa), with differentiated (low/intermediate grade) tumor cells, exhibiting low proliferation rate (< 20% Ki67 expression), bearing hormone receptors and lacking HER2/neu oncoprotein overexpression. The latter could be considered nowadays as a disadvantage, since more than 90% of the C2 patients are considered HER2/neu negative and thus could not benefit from the very effective therapeutic option of Herceptin treatment [33].On the other hand, C1 and C3 patients do not substantially differ in most clinicopathological parameters, apart from age and Ki67 expression, with patients in C3 being significantly younger at diagnosis and comprising the highest percentage of > 20% Ki67 expressors. These two characteristics, along with other minor differences in nodal status and tumor size, may imply that patients in C3 exhibit a worse prognostic profile, according to classical clinicopathological prognostic characteristics.
Additionally, the three clusters of patients displayed different immune-related characteristics. Although the tumors among the three groups were similarly infiltrated by CD163+ macrophages, they differed in their T cell content. C1-classified patients had the lowest numbers of CD4 and CD8 T cells infiltrating their tumors, both in the TC and the IM, in contrast to patients belonging to the C3 cluster who exhibited the highest T cell infiltration. C2 BCa patients displayed rather high numbers of CD4 TILs both in the TC and the IM, but the median levels of CD8 TILs in the TC were high and in the IM were low. Consequently, C2 patients comprised the highest frequency of patients with the favorable infiltration profile, i.e. FCIS [24], while more UCIS patients were found within the C3 cluster. These findings further support the notion that patients with the C1 miRNA signature and, more profoundly, with the C3 signature might have a worse prognosis than those classified within the C2 group. Since serum miRNA levels positively correlated with CD4 and CD8 tumor T cell infiltration, and no correlation was observed with PBMC miRNAs, it is reasonable to assume that circulating miRNAs reflect the tumor microenvironment immune situation. Since the expression of miR-16, miR-21 and miR-146 [34] have been reported to positively correlate with memory T cells, and increased miR-23α and miR-155 levels with the presence of CD8 T cells in tumors [35, 36], their combined circulating levels support the notion that patients with the C3 miRNA signature may have more immunogenic tumors represented by higher adaptive immune cell infiltration than patients belonging to the C1 cluster. However, the high levels of miR-23α may hamper their functional potential, since high miR-23 expression was related to impaired cytotoxic T lymphocyte function [35], while miR-21, miR-146α and miR-155 positively control T regulatory cell functional competence [37–39],supporting an underlying immune-related tumor escape mechanism in C3 patients. It can be hypothesized that patients with the C3 miRNA profile could represent good candidates for immunotherapeutic interventions, aiming at reactivating their preexisting endogenous immune responses [40]. On the other hand, the low overall expression of the six miRNAs of the C1 signature correlated with low T cell tumor infiltration, implying that patients within this group bear poorly immunogenic tumors. Thus, strategies aiming at increasing tumor recognition and infiltration by immune cells might improve the clinical outcome of these patients.
Although PBMC-miRNA levels did not correlate with serum miRNAs, nor with circulating cytokine/chemokine levels (apart from IL-9), cluster signatures were compatible with circulating cytokine/chemokine profiles of the distinct groups: high levels of miR-16, miR-21, miR-146 and miR-155 have been previously reported to be negatively correlated with cytokines/chemokines production, including IL-8, IL-10, Rantes [41, 42], mostly through their positive or negative role in the NF-kB signaling network [23].Whether circulating miRNAs are implicated in PBMC functionality, independently of their subpopulations endogenous miRNA levels, cannot be excluded in the context of cell-to-cell interaction [43]. It remains to be elucidated whether the deregulation of miRNAs (along with the NF-kB pathway) is fundamental for tumor development and progression.
The MHC is crucial in anticancer T cell immunity and some HLA alleles have been correlated with some cancer types. For instance, the HLA-A2 allele has been correlated with worse prognosis in ovarian, prostate, lung and breast cancer, while HLA-A24 has been associated with better clinical outcome in prostate and breast cancer [27–29, 44, 45]. Furthermore, some miRNAs have been reported to act as regulators of different HLA loci [18]. The significantly increased frequency of HLA-A2 negative patients, along with an increased frequency of HLA-A24 expressing patients detected in C2, are also indicative for a better prognostic profile as opposed to the C3 group which exhibits these HLA genotypes inversely.
An important limitation of the present study is the low numbers of BCa patients analyzed among the 3 clusters, as well as that of normal individuals. Nevertheless, our data constitute a clear proof-of-principle supporting the notion that, the up- or down-regulation of the same circulating miRNAs may be associated with plausible different prognosis in BCa. This may have an important impact given that most of the published reports so far in the field refer to the diagnostic (and much less to the prognostic/predictive) relevance of either upregulation (or overexpression) or downregulation of each specified miRNA, even if those are included in signatures with other miRNAs [8, 14]. Discrepancies in circulating miRNA expression levels among different research groups are attributed mainly to methodological differences [46]. However, in the present study, sera were collected, handled and molecularly analyzed under identical conditions, thus avoiding any methodological interference for our data.
In conclusion, we present strong evidence that three different signatures of six circulating miRNAs (miR-16, miR-21, miR-23α, miR-146α, miR-155 and miR-181α) define three distinct groups of non-metastatic BCa patients with invasive ductal carcinoma, characterized by different clinicopathological and immune-related characteristics, with possibly different clinical outcome. Nonetheless, the prognostic and/or predictive potential of these signatures needs to be further evaluated in larger cohorts of BCa patients with an, at least, 5-year clinical follow-up.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- AJCC
American Joint Committee on Cancer
- BCa
Breast cancer
- DAB
Diaminobenzidine
- ER
Estrogen receptor
- FCIS
Favorable combined immune signatures
- FFPE
Formalin-fixed, paraffin-embedded
- HE
Hematoxylin Eosin
- HER2
Human epidermal growth factor receptor 2
- HLA
Human leukocyte antigen
- IL
Interleukin
- IL-1Rα
Interleukin 1 receptor antagonist
- IM
Invasive margin
- LncRNA
Long noncoding RNA
- MHC
Major histocompatibility complex
- miRNA
MicroRNA
- mRNA
Messenger RNA
- NF-kB
Nuclear factor kappa beta
- PR
Progesterone receptor
- ROC
Receiver operating characteristic
- T
Tumor size
- TC
Tumor center
- TGF-β
Transforming growth factor beta
- Treg
T regulatory (cell)
- UCIS
Unfavorable combined immune signatures
- UTR
Untranslated region
Author contributions
Sotirios P. Fortis designed and performed research, analyzed data, and wrote the manuscript; Christoforos K. Vaxevanis performed research, analyzed data and wrote the manuscript; Louisa G. Mahaira, Michael Sofopoulos, Nectaria N. Sotiriadou, Amalia Dinou, Niki Arnogiannaki, Catherine Stavropoulos-Giokas, contributed to experimental design, and performed research; Dimitris Thanos contributed to experimental design and data analysis; Constantin N. Baxevanis contributed to experimental design, data analysis, and wrote the manuscript; Sonia A. Perez supervised the study, contributed to experimental design, data analysis, and wrote the manuscript. All authors read and approved the manuscript.
Funding
This study was supported by grant GER_1968 (acronym ISPEBREAST) to Constantin Baxevanis from a bilateral research and innovation cooperation, funded by the General Secretariat for Research and Technology (GSRT) of the Ministry of Education, Research and Religious Affairs of the Hellenic Republic and the German Federal Ministry for Education and Research (BMBF), and a donation to Sonia Perez from the Haegeman-Goossens family, Netherlands.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by the Institutional Review Board of St. Savas Cancer Hospital (IRB-ID 6079/448/10-6-13).
Ethical standards
All prospectively enrolled patients signed a written informed consent, approved by the Review Board at St. Savas Cancer Hospital. All data of other retrospectively analyzed patients were obtained from an anonymized database constructed for the purposes of a previous study [24]. Healthy volunteers presented as blood donors at the Blood Collection and Transfusion Department of Saint Savas Hospital. They fulfilled all requirements for blood donation. Volunteers consented verbally, and no personal information was recorded. All procedures were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- 1.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochimica et Biophysica Acta. 2010;1805(1):105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mannello F, Ligi D. Resolving breast cancer heterogeneity by searching reliable protein cancer biomarkers in the breast fluid secretome. BMC Cancer. 2013;13:344. doi: 10.1186/1471-2407-13-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai JP, Bell R, Buckman S, Burckart GJ, Eichler HG, Fang KC, Goodsaid FM, Jusko WJ, Lesko LL, Meibohm B, Patterson SD, Puig O, Smerage JB, Snider BJ, Wagner JA, Wang J, Walton MK, Weiner R. Translational biomarkers: from preclinical to clinical a report of 2009 AAPS/ACCP Biomarker Workshop. AAPS J. 2011;13(2):274–283. doi: 10.1208/s12248-011-9265-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravelli A, Reuben JM, Lanza F, Anfossi S, Cappelletti MR, Zanotti L, Gobbi A, Senti C, Brambilla P, Milani M, Spada D, Pedrazzoli P, Martino M, Bottini A, Generali D, Marrow Transplantation S. Solid tumor working party of European B. Breast cancer circulating biomarkers: advantages, drawbacks, and new insights. Tumour Biol J Int Soc Oncodev Biol Med. 2015;36(9):6653–6665. doi: 10.1007/s13277-015-3944-7. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Li Y, Wang Q, Zhang X, Wang D, Tang HC, Meng X, Ding X. Identification of an lncRNA-miRNA-mRNA interaction mechanism in breast cancer based on bioinformatic analysis. Mol Med Rep. 2017;16(4):5113–5120. doi: 10.3892/mmr.2017.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amorim M, Salta S, Henrique R, Jeronimo C. Decoding the usefulness of non-coding RNAs as breast cancer markers. J Transl Med. 2016;14:265. doi: 10.1186/s12967-016-1025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Zhang Y, Li Q, Li J, Ma X, Xing J, Rong S, Wu Z, Tian Y, Li J, Jia L. MiRNAs predict the prognosis of patients with triple negative breast cancer: a meta-analysis. PloS One. 2017;12(1):e0170088. doi: 10.1371/journal.pone.0170088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahrami A, Aledavood A, Anvari K, Hassanian SM, Maftouh M, Yaghobzade A, Salarzaee O, ShahidSales S, Avan A. The prognostic and therapeutic application of microRNAs in breast cancer: tissue and circulating microRNAs. J Cell Physiol. 2018;233(2):774–786. doi: 10.1002/jcp.25813. [DOI] [PubMed] [Google Scholar]
- 9.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA–LncRNA interactions. Methods Mol Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 11.Chin LJ, Slack FJ. A truth serum for cancer—microRNAs have major potential as cancer biomarkers. Cell Res. 2008;18(10):983–984. doi: 10.1038/cr.2008.290. [DOI] [PubMed] [Google Scholar]
- 12.van Schooneveld E, Wildiers H, Vergote I, Vermeulen PB, Dirix LY, Van Laere SJ. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res: BCR. 2015;17:21. doi: 10.1186/s13058-015-0526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farina NH, Ramsey JE, Cuke ME, Ahern TP, Shirley DJ, Stein JL, Stein GS, Lian JB, Wood ME. Development of a predictive miRNA signature for breast cancer risk among high-risk women. Oncotarget. 2017;8(68):112170–112183. doi: 10.18632/oncotarget.22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamam R, Hamam D, Alsaleh KA, Kassem M, Zaher W, Alfayez M, Aldahmash A, Alajez NM. Circulating microRNAs in breast cancer: novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017;8(9):e3045. doi: 10.1038/cddis.2017.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5(10):1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Backes C, Meese E, Keller A. Specific miRNA disease biomarkers in blood, serum and plasma: challenges and prospects. Mol Diagn Ther. 2016;20(6):509–518. doi: 10.1007/s40291-016-0221-4. [DOI] [PubMed] [Google Scholar]
- 17.Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol. 2016;16(5):279–294. doi: 10.1038/nri.2016.40. [DOI] [PubMed] [Google Scholar]
- 18.Eichmuller SB, Osen W, Mandelboim O, Seliger B (2017) Immune modulatory microRNAs involved in tumor attack and tumor immune escape. J Natl Cancer Inst 109 (10). 10.1093/jnci/djx034 [DOI] [PubMed]
- 19.Paladini L, Fabris L, Bottai G, Raschioni C, Calin GA, Santarpia L. Targeting microRNAs as key modulators of tumor immune response. J Exp Clin Cancer Res. 2016;35:103. doi: 10.1186/s13046-016-0375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taube JM, Galon J, Sholl LM, Rodig SJ, Cottrell TR, Giraldo NA, Baras AS, Patel SS, Anders RA, Rimm DL, Cimino-Mathews A. Implications of the tumor immune microenvironment for staging and therapeutics. Mod Pathol. 2018;31(2):214–234. doi: 10.1038/modpathol.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell research. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 22.Labbaye C, Testa U. The emerging role of MIR-146A in the control of hematopoiesis, immune function and cancer. J Hematol Oncol. 2012;5:13. doi: 10.1186/1756-8722-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-kappaB signaling. J Mol Cell Biol. 2011;3(3):159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fortis SP, Sofopoulos M, Sotiriadou NN, Haritos C, Vaxevanis CK, Anastasopoulou EA, Janssen N, Arnogiannaki N, Ardavanis A, Pawelec G, Perez SA, Baxevanis CN. Differential intratumoral distributions of CD8 and CD163 immune cells as prognostic biomarkers in breast cancer. J Immunother Cancer. 2017;5:39. doi: 10.1186/s40425-017-0240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen N, Fortis SP, Speigl L, Haritos C, Sotiriadou NN, Sofopoulos M, Arnogiannaki N, Stavropoulos-Giokas C, Dinou A, Perez S, Pawelec G, Baxevanis CN, Shipp C. Peripheral T cell responses to tumour antigens are associated with molecular, immunogenetic and cellular features of breast cancer patients. Breast Cancer Res Treat. 2017;161(1):51–62. doi: 10.1007/s10549-016-4037-z. [DOI] [PubMed] [Google Scholar]
- 26.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27(42):5643–5647. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anastasopoulou EA, Voutsas IF, Keramitsoglou T, Gouttefangeas C, Kalbacher H, Thanos A, Papamichail M, Perez SA, Baxevanis CN. A pilot study in prostate cancer patients treated with the AE37 Ii-key-HER-2/neu polypeptide vaccine suggests that HLA-A*24 and HLA-DRB1*11 alleles may be prognostic and predictive biomarkers for clinical benefit. Cancer Immunol Immunother. 2015;64(9):1123–1136. doi: 10.1007/s00262-015-1717-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stokidis S, Dinou A, Fortis SP, Vaxevanis CK, Konstantellou M, Stavropoulos-Giokas C, Perez SA, Baxevanis CN (2017) The impact of HLA-A*02 and HLA-A*24 allele expression in prostate cancer prognosis In: 3rd Symposium on Advances in Cancer Immunology and Immunotherapy, Athens, Greece, November 2–4. 10.13140/RG.2.2.34617.47202
- 29.Vaxevanis C, Anastasopoulou E, Tzonis P, Ardavanis A, Baxevanis CN, Peoples GE, Perez SA (2017) An IFN-γ response-based algorithm with predictive potential in AE37-vaccinated breast cancer patients In: 3rd Symposium on Advances in Cancer Immunology and Immunotherapy, Athens, Greece, November 2–4 2017. 10.13140/RG.2.2.24551.14248
- 30.Han JG, Jiang YD, Zhang CH, Yang YM, Pang D, Song YN, Zhang GQ. A novel panel of serum miR-21/miR-155/miR-365 as a potential diagnostic biomarker for breast cancer. Ann Surg Treat Res. 2017;92(2):55–66. doi: 10.4174/astr.2017.92.2.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurkovicova D, Smolkova B, Magyerkova M, Sestakova Z, Kajabova VH, Kulcsar L, Zmetakova I, Kalinkova L, Krivulcik T, Karaba M, Benca J, Sedlackova T, Minarik G, Cierna Z, Danihel L, Mego M, Chovanec M, Fridrichova I. Down-regulation of traditional oncomiRs in plasma of breast cancer patients. Oncotarget. 2017;8(44):77369–77384. doi: 10.18632/oncotarget.20484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang C, Tabatabaei SN, Ruan X, Hardy P. The dual regulatory role of MiR-181a in breast cancer. Cell Physiol Biochem. 2017;44(3):843–856. doi: 10.1159/000485351. [DOI] [PubMed] [Google Scholar]
- 33.Perez EA, Romond EH, Suman VJ, Jeong JH, Sledge G, Geyer CE, Jr, Martino S, Rastogi P, Gralow J, Swain SM, Winer EP, Colon-Otero G, Davidson NE, Mamounas E, Zujewski JA, Wolmark N. Trastuzumab plus adjuvant chemotherapy for human epidermal growth factor receptor 2-positive breast cancer: planned joint analysis of overall survival from NSABP B-31 and NCCTG N9831. J Clin Oncol. 2014;32(33):3744–3752. doi: 10.1200/JCO.2014.55.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naive, effector and memory CD8 T cells. PloS One. 2007;2(10):e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin R, Chen L, Chen G, Hu C, Jiang S, Sevilla J, Wan Y, Sampson JH, Zhu B, Li QJ. Targeting miR-23a in CD8 + cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J Clin Invest. 2014;124(12):5352–5367. doi: 10.1172/JCI76561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huffaker TB, Hu R, Runtsch MC, Bake E, Chen X, Zhao J, Round JL, Baltimore D, O’Connell RM. Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep. 2012;2(6):1697–1709. doi: 10.1016/j.celrep.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hippen KL, Loschi M, Nicholls J, MacDonald KPA, Blazar BR. Effects of MicroRNA on regulatory T cells and implications for adoptive cellular therapy to ameliorate Graft-versus-host disease. Front Immunol. 2018;9:57. doi: 10.3389/fimmu.2018.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.So AY, Zhao JL, Baltimore D. The Yin and Yang of microRNAs: leukemia and immunity. Immunol Rev. 2013;253(1):129–145. doi: 10.1111/imr.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, Rudensky AY. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30(1):80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baxevanis CN, Perez SA. Cancer dormancy: a regulatory role for endogenous immunity in establishing and maintaining the tumor dormant state. Vaccines (Basel) 2015;3(3):597–619. doi: 10.3390/vaccines3030597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perry MM, Williams AE, Tsitsiou E, Larner-Svensson HM, Lindsay MA. Divergent intracellular pathways regulate interleukin-1beta-induced miR-146a and miR-146b expression and chemokine release in human alveolar epithelial cells. FEBS Lett. 2009;583(20):3349–3355. doi: 10.1016/j.febslet.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 42.Testa U, Pelosi E, Castelli G, Labbaye C. miR-146 and miR-155: Two Key Modulators of Immune Response and Tumor Development. Non-Coding RNA. 2017;3(3):1–22. doi: 10.3390/ncrna3030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sohel MH. Extracellular/circulating MicroRNAs: release mechanisms, functions and challenges. Achiev Life Sci. 2016;10(2):175–186. doi: 10.1016/j.als.2016.11.007. [DOI] [Google Scholar]
- 44.Nagata Y, Hanagiri T, Mizukami M, Kuroda K, Shigematsu Y, Baba T, Ichiki Y, Yasuda M, So T, Takenoyama M, Sugio K, Nagashima A, Yasumoto K. Clinical significance of HLA class I alleles on postoperative prognosis of lung cancer patients in Japan. Lung Cancer. 2009;65(1):91–97. doi: 10.1016/j.lungcan.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 45.De Petris L, Bergfeldt K, Hising C, Lundqvist A, Tholander B, Pisa P, van der Zanden HG, Masucci G. Correlation between HLA-A2 gene frequency, latitude, ovarian and prostate cancer mortality rates. Med Oncol. 2004;21(1):49–52. doi: 10.1385/MO:21:1:49. [DOI] [PubMed] [Google Scholar]
- 46.Jarry J, Schadendorf D, Greenwood C, Spatz A, van Kempen LC. The validity of circulating microRNAs in oncology: five years of challenges and contradictions. Mol Oncol. 2014;8(4):819–829. doi: 10.1016/j.molonc.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.